Summary
Background
Autophagy is a conserved process mediating vacuolar degradation and recycling. Autophagy is highly upregulated upon various stresses and is essential for cell survival in deleterious conditions. Autophagy defects are associated with severe pathologies, whereas unchecked autophagy activity causes cell death. Therefore, to support proper cellular homeostasis, the induction and amplitude of autophagy activity have to be finely regulated. Transcriptional control is a critical, yet largely unexplored, aspect of autophagy regulation. In particular little is known about the signaling pathways modulating the expression of autophagy-related genes, and only a few transcriptional regulators have been identified as contributing in the control of this process.
Results
We identified Rph1 as a negative regulator of the transcription of several ATG genes and a repressor of autophagy induction. Rph1 is a histone demethylase protein, but it regulates autophagy independently of its demethylase activity. Rim15 mediates the phosphorylation of Rph1 upon nitrogen starvation, which causes an inhibition of its function. Preventing Rph1 phosphorylation or overexpressing the protein causes a severe block in autophagy induction. A similar function of Rph1/KDM4 is seen in mammalian cells, indicating that this process is highly conserved.
Conclusion
Rph1 maintains autophagy at a low level in nutrient-rich conditions; upon nutrient limitation, the inhibition of its activity is a prerequisite to the induction of ATG gene transcription and autophagy.
Keywords: autophagy, lysosome, vacuole, stress, yeast
Introduction
Macroautophagy (hereafter referred to as autophagy) is a highly conserved pathway during which portions of the cytoplasm, superfluous or damaged organelles, or invasive pathogens are delivered to the vacuole (in yeast and plants) or the lysosome (in mammals) for degradation and subsequent recycling. Morphologically, autophagy starts with the nucleation of the phagophore, the initial membrane structure, at a peri-vacuolar site called the phagophore assembly site, or PAS (Figure S1A, [1]). Through the acquisition of lipids, the phagophore then expands, surrounding its cargo, and ultimately seals to generate a double-membrane vesicle called the autophagosome. After autophagosome fusion with the vacuole, the inner membrane of the vesicle as well as its cargo are degraded and recycled into the cytosol. The macromolecules resulting from this pathway are then used by the cell to maintain cellular homeostasis during deleterious conditions [2].
Autophagy is highly upregulated upon multiple stress conditions and notably nutrient limitation where it plays critical roles in cell adaptation and survival [3]. The autophagy pathway plays a role in cellular physiology, and mammalian development as well as the immune response [4–6], and autophagy impairments are associated with various human pathologies such as cancer or metabolic diseases [7–8]. Autophagy also occurs at a constitutive basal level in physiological conditions. In yeast, the cytoplasm-to-vacuole targeting (Cvt) pathway is essential for the biosynthetic delivery of vacuolar enzymes during growth [9]. In mammals, basal autophagy functions in the quality control machinery involved in the degradation of damaged organelles and protein aggregates which, when accumulated, lead to various neurodegenerative disorders such as Huntington, Alzheimer and Parkinson diseases [10].
If a sub-optimal autophagy activity can be detrimental, a supra-optimal level can also be deleterious for the cells; uncontrolled autophagy leads to cell death [11–12], certain microbes rely on autophagy to provide nutrients [13], and cancer cells can also use autophagy to help survive in an unfavorable environment [14]. Therefore, to support proper cellular functions, rates of autophagy have to be finely regulated. Over the past 20 years, extensive molecular studies of autophagy have led to the identification of the core components of the autophagy machinery, encoded by the autophagy-related (ATG) genes (for review see ref. [15]); yet the regulatory pathways governing their functions remain largely unknown. In particular the transcriptional regulation of the ATG genes is still mostly unexplored (for review see ref. [14]).
The expression of most of the ATG genes, and their corresponding proteins, highly increases upon autophagy induction after nitrogen starvation [16]. Given the energy cost for such an induction, it has been suggested that this upregulation is critical for the normal modulation of autophagy and for the pathway to reach full amplitude when most needed. Furthermore, the reported abnormal expression of several ATG genes in various human diseases supports the proposed physiological importance of their transcriptional regulation [17–20]. Recent studies in yeast identified Ume6 as a transcriptional repressor of ATG8 [21] and showed that the level of Atg8 controls the size of the autophagosome [22]. Conversely, the level of Atg9, regulated by Pho23, controls the frequency of formation (hence the number) of autophagosomes [16]. Nevertheless, very few transcription factors have been identified as regulating the expression of ATG genes in either yeast or mammals and the contribution of the induction of specific ATG genes in autophagy activity is still poorly understood.
At the starting point of this study a search for new transcriptional regulators of autophagy was initiated by the screen of a collection of over 150 mutants lacking a single transcription factor or DNA-binding protein. Analysis of the expression of a set of ATG genes in this library led to the identification of Rph1 as a master transcriptional repressor of autophagy.
Results
Rph1 Is a Transcriptional Repressor of ATG Genes
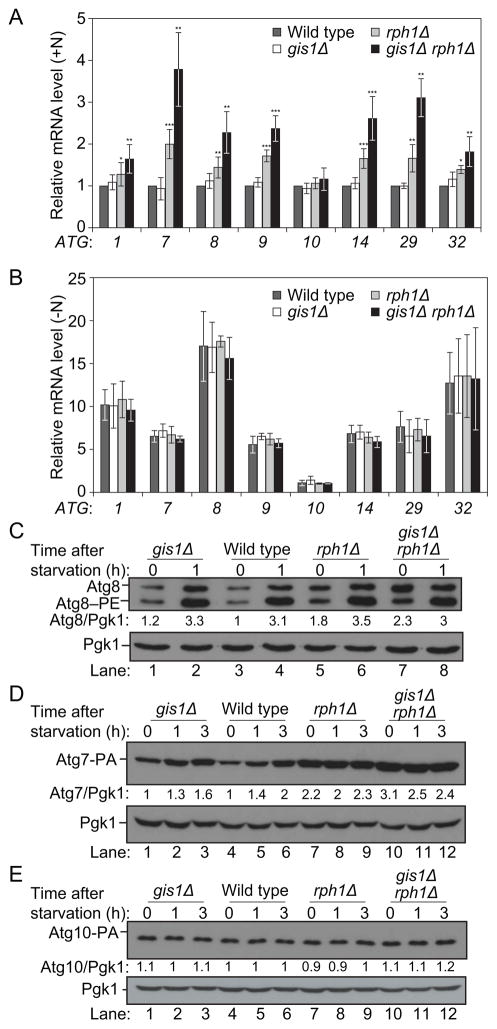
In order to identify transcriptional regulators of autophagy we compared the expression of a subset of ATG genes in wild-type cells to that in a library of DNA binding protein deletion mutants. ATG1, ATG7, ATG8, ATG9, ATG14 and ATG29 were selected as target genes because they encode proteins involved in different steps of the autophagy pathway (Figure S1A) and because they show a strong induction after nitrogen starvation, indicating that they are under transcriptional control in these conditions (Figure 1A,B; [16]). The DNA binding protein mutants displayed a range of phenotypes (data not shown), and rph1Δ in particular showed a significant upregulation of ATG7, ATG8, ATG9, ATG14 and ATG29 accompanied with a modest induction of ATG1 in growing conditions (Figure 1A, Table S1, Table S2). Similarly, the deletion of RPH1 resulted in an increased expression of ATG32 (Figure 1A), a marker of mitophagy (the selective autophagic degradation of mitochondria). In contrast, deleting RPH1 had essentially no effect on the transcription of ATG10, an autophagy-related gene that displays no change in expression in response to nitrogen deprivation (Figure 1A,B). Additionally, there was no significant difference in the mRNA levels of ATG genes between the rph1Δ strain and wild-type cells after starvation (Figure 1B). This difference between growing and starvation conditions suggests that Rph1 represses the expression of nitrogen-sensitive ATG genes specifically in nutrient-replete conditions.
Figure 1. Rph1 represses the expression of nitrogen-sensitive ATG genes in nutrient-replete conditions.
(A–B) Rph1 represses the expression of ATG1, ATG7, ATG8, ATG9, ATG14 and ATG29 in nutrient-replete conditions. Wild-type (YTS158), rph1Δ (YAB300), gis1Δ (YAB301) and gis1Δ rph1Δ (YAB302) cells were grown in YPD (+N) until mid-log phase (A) and then starved for nitrogen (−N) for 1 h (B). mRNA levels were quantified by RT-qPCR. Error bars indicate the standard deviation of the average of at least 3 independent experiments.
(C–E) Rph1 represses the expression of the Atg7 and Atg8 proteins. (C) For the analysis of Atg8, wild-type (YTS158, BY4742), rph1Δ (YAB300), gis1Δ (YAB301) and gis1Δ rph1Δ (YAB302) cells were grown in YPD until mid-log phase and then starved for nitrogen for the indicated times. Protein extracts were analyzed by western blot with anti-Atg8 and anti-Pgk1 (loading control) antisera.
(D) To analyze Atg7 abundance, a protein A tag was integrated at the chromosomal locus. Wild-type (YAB312, BY4742), rph1Δ (YAB313), gis1Δ (YAB314) and gis1Δ rph1Δ (YAB315) cells were grown in the same conditions as in (C). Protein extracts were analyzed as in (C) with an antibody that detects PA or anti-Pgk1 antiserum.
(E) To analyze Atg10 abundance, a protein A tag was integrated at the chromosomal locus. Wild-type (YAB350, BY4742), rph1Δ (YAB351), gis1Δ (YAB352) and gis1Δ rph1Δ (YAB353) cells were grown in the same conditions as in (C). Protein extracts were analyzed as in (D).
Rph1 is a DNA-binding protein, which contains two C2H2 zinc-finger motifs as well as a Jumonji C histone demethylase catalytic domain. Rph1 has a paralog, Gis1, and previous studies report that Rph1 and Gis1 have distinct as well as overlapping targets [23–26]. Whereas the deletion of GIS1 had a very minor effect on the expression of some of the ATG genes that we examined, the gis1Δ rph1Δ double mutant showed an enhanced induction of the mRNA level of these genes compared to the single RPH1 deletion in growing conditions (Figure 1A,B). This finding indicates that Rph1 plays a predominant role in the regulation of ATG genes, but that its absence can be partially compensated by Gis1.
Consistent with the changes observed at the mRNA levels, rph1Δ cells showed a strong enrichment of the proteins Atg7, Atg8, Atg9, Atg14 and Atg29 and a modest increase in Atg1 compared to the wild type; in contrast the level of Atg10 was unaffected (Figure 1C–E, Figure S1B,C). This phenotype was exacerbated in gis1Δ rph1Δ cells as suggested by the analysis of Atg7 and Atg8 (Figure 1C,D). It is worth noting that although the abundance of Atg1, Atg8, Atg9 and Atg14 was higher in rph1Δ cells in rich conditions compared to the wild type (Figure 1C, compare lanes 3 and 5, Figure S1B,C), the protein levels were still lower than those seen after starvation (Figure 1C, compare lanes 5 and 6, Figure S1B,C), indicating that other pathways are contributing to the regulation of the corresponding genes. In contrast, deleting RPH1 alone was sufficient to increase the Atg7 protein abundance to a higher level than that seen after 3 h of nitrogen starvation in the wild-type cells (Figure 1D, compare lanes 6 and 7, Figure S1B,C), suggesting that Rph1 has a preponderant role in regulating the expression of Atg7. Together these results show that Rph1 is a transcriptional repressor of ATG genes.
Rph1 Is a Negative Regulator of Autophagy
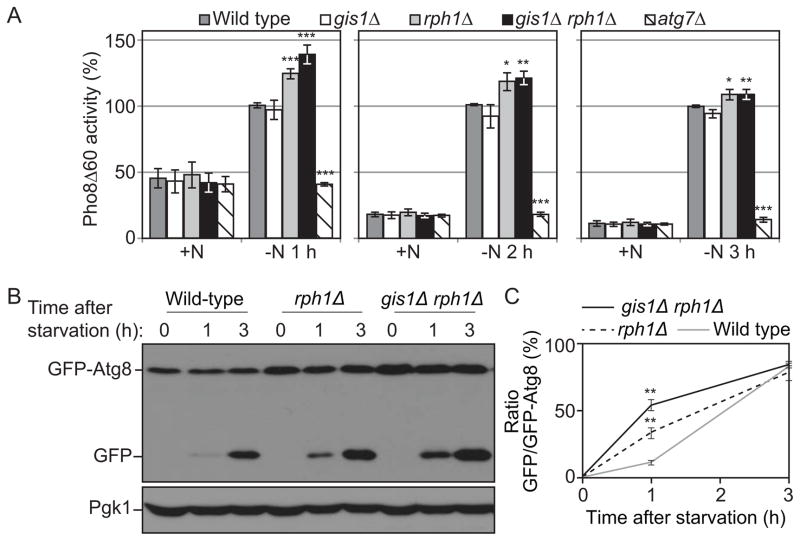
Finding that Rph1 represses the expression of ATG genes in growing conditions suggests that it might be a negative regulator of autophagy. To test this hypothesis autophagy activity was measured using a Pho8Δ60 assay in which alkaline phosphatase activity reflects autophagy amplitude [27]. After 1 h of nitrogen starvation, the wild-type cells displayed a substantial induction of alkaline phosphatase activity (50% of the activity seen at 3 h; note that the wild-type activity is set to 100% at each time point in Figure 2A), which was blocked by the deletion of ATG7 (Figure 2A). Compared to the wild type, autophagy was induced to a higher level in the rph1Δ cells and gis1Δ rph1Δ cells; in contrast the deletion of GIS1 alone had no significant effect. We extended this analysis by using a GFP-Atg8 processing assay [28]. After one hour of starvation, autophagy was induced more rapidly and to a higher extent in the rph1Δ and gis1Δ rph1Δ cells, as indicated by the level of free GFP compared to the wild-type (Figure 2B,C). Note that the enhanced autophagy activity in rph1Δ cells upon short-term nitrogen starvation was attenuated after a prolonged time of induction. This observation is consistent with the fact that deleting RPH1 increases the transcription of ATG genes only in nutrient-rich conditions; a higher abundance of Atg proteins can therefore support a jump-start in autophagy activity upon its induction by nitrogen starvation. After prolonged starvation, Atg protein levels in wild-type cells reach that of the mutant strains, abolishing the difference in autophagy activity relative to the rph1Δ and gis1Δ rph1Δ mutants.
Figure 2. Rph1 negatively regulates autophagy.
(A) Autophagy as measured by the Pho8Δ60 assay is increased in rph1Δ and gis1Δ rph1Δ cells. Wild-type (YTS158, BY4742), rph1Δ (YAB300), gis1Δ (YAB301), gis1Δ rph1Δ (YAB302) and atg7Δ (YAB292) cells were grown in YPD (+N) and then starved for nitrogen (−N) for the indicated times. The Pho8Δ60 activity was measured and normalized to the activity of wild-type cells after starvation, which was set to 100% at each time point. The data represent the average of 3 independent experiments; error bars indicate standard deviation.
(B) Autophagy as measured by the GFP-Atg8 processing is increased in rph1Δ and gis1Δ rph1Δ cells. Wild-type (YTS158, BY4742), rph1Δ (YAB300) and gis1Δ rph1Δ (YAB302) cells transformed with a CEN plasmid carrying a GFP-Atg8 construct were grown in rich selective medium and then starved for 1 and 3 h. Cells were collected and protein extracts were analyzed by western blot with anti-YFP antibody and anti-Pgk1 (loading control) antiserum.
(C) Quantification of (B). The ratio free GFP:GFP-Atg8 was measured and normalized to that of wild-type cells after 3 h starvation, which was set to 100%. Average values ± standard deviations of 2 independent experiments are indicated.
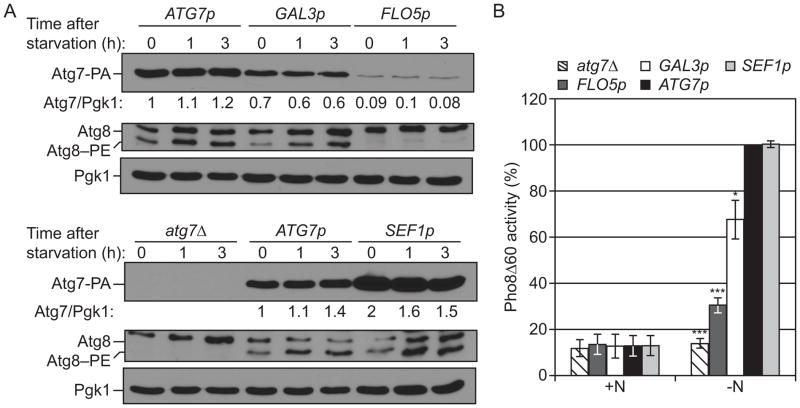
Our results indicated that Rph1 has a major role in the transcriptional regulation of ATG7 (Figure 1, Figure S1B,C). Atg7 is an E1 activating enzyme, which notably mediates the conjugation of Atg8 to PE and is critical for autophagy (Figure 2A). Nevertheless, very little is known concerning the effect of ATG7 transcriptional regulation on autophagy. To address this, an atg7Δ strain was transformed with plasmids in which protein A (PA)-tagged Atg7 was placed under the control of promoters (FLO5, GAL3, ATG7 and SEF1 in increasing order of strength) with different transcriptional activities (Figure 3A). Note that in these conditions we did not observe a strong increase in the abundance of Atg7-PA in wild-type cells after starvation (compared to Figure 1D and Figure S1B,C), which may reflect expression from a plasmid as opposed to the complete ATG7 promoter. Lowering Atg7 abundance correlated with a reduction of Atg8–PE conjugates (Figure 3A) as well as a reduction in autophagy activity (Figure 3B). The finding that Atg7 protein amount directly affects autophagy activity supports a model in which Rph1 regulates autophagy in part by controlling the expression of Atg7. Nevertheless, an increase in Atg7 protein alone was not sufficient to induce autophagy (Figure 3B, +N) suggesting that the rph1Δ phenotype results from the combined upregulation of multiple ATG genes.
Figure 3. The level of Atg7 regulates autophagy.
(A) Lowering the level of Atg7 results in a decrease in Atg8–PE conjugation and (B) a decrease in autophagy activity. atg7Δ cells (YAB292) were transformed with the ATG7p-ATG7-PA, GAL3p-ATG7-PA, FLO5p-ATG7-PA, or SEF1p-ATG7-PA plasmid or the corresponding empty plasmid (pRS416-PA). Cells were grown in rich selective medium (SMD-Ura) until mid log phase and then starved for nitrogen for the indicated times.
(A) Cells were collected and protein extracts were analyzed by western blot with either an antibody that recognizes PA, or anti-Atg8 and anti-Pgk1 (loading control) antisera.
(B) Cells were starved for 3 h. The Pho8Δ60 activity was measured and normalized to the activity of ATG7p-ATG7-PA cells, which was set to 100%. Data represent the average of 3 independent experiments.
The overexpression of Rph1 inhibits autophagy and decreases cell survival in nitrogen starvation conditions
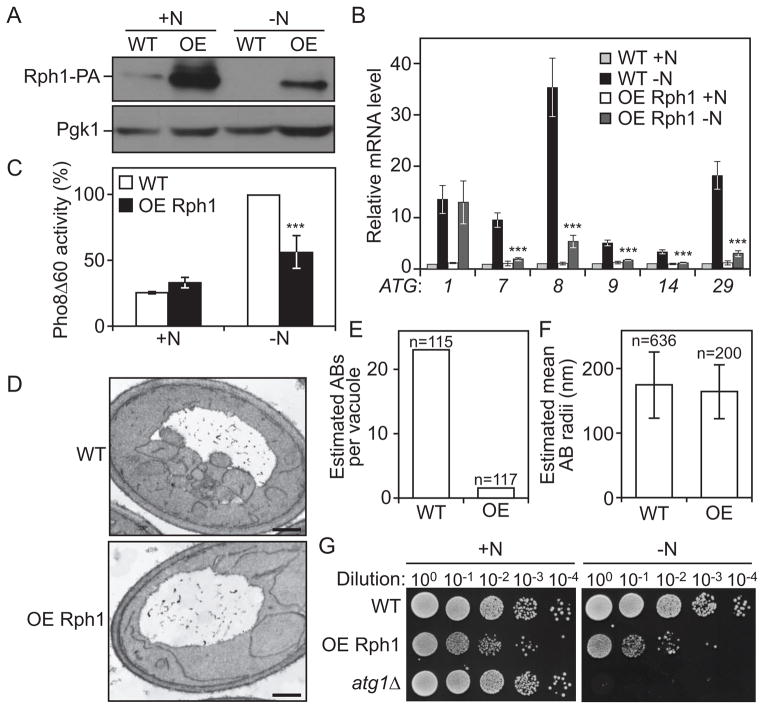
To further test the role of Rph1 in autophagy, RPH1 was overexpressed under the control of the ZEO1 promoter (Figure 4A). Overexpression of Rph1 caused a severe block in the induction of several ATG genes after starvation (Figure 4B) but had no effect on the expression of ATG1. Furthermore, the reduction in ATG transcripts correlated with a decrease in autophagy activity (Figure 4C). A decrease in autophagy flux could either result from a reduction in the number of autophagosomes or a reduction in their size. To distinguish between the two hypotheses, cells overexpressing Rph1 were further analyzed using transmission electron microscopy (TEM). After 2 h of nitrogen starvation, cells overexpressing Rph1 displayed a strong reduction in the average number of autophagic bodies accumulated in the vacuole compared to wild-type cells (Figure 4D,E, Figure S2, Table S3); in contrast there was no significant effect on the size of the autophagic bodies (Figure 4D,F, Figure S2, Table S3).
Figure 4. The overexpression of Rph1 inhibits autophagy and decreases cell survival in nitrogen starvation conditions.
(A–C) Overexpressed Rph1 (ZEO1p-RPH1-PA) results in a block in ATG genes expression and autophagy flux. Rph1-PA cells (WT, YAB323, SEY6210) and cells overexpressing Rph1-PA (OE Rph1, YAB329) were grown in YPD (+N) until mid-log phase and then starved for nitrogen (−N).
(A) Cells were starved for 2 h, and protein extracts were analyzed by western blot with either an antibody that recognizes PA or anti-Pgk1 (loading control) antiserum.
(B) Total RNA of cells in mid-log phase (+N) as well as after 1 h of nitrogen starvation (−N) was extracted and the mRNA levels were quantified by RT-qPCR. The mRNA level of individual ATG genes was normalized to the mRNA level of the corresponding gene in Rph1-PA cells (WT), which was set to 1. Data represent the average of 3 independent experiments ± standard deviation.
(C) The Pho8Δ60 activity was measured and normalized to the activity of Rph1-PA cells (WT) after 2 h of nitrogen starvation (−N), which was set to 100%. Error bars indicate standard deviation of 3 independent experiments.
(D–E) Rph1 overexpression blocks the biogenesis of autophagic bodies. Wild-type cells (WT, FRY143) and cells overexpressing Rph1 (OE Rph1, YAB346) were imaged using transmission electron microscopy after 2 h of nitrogen starvation.
(D) Representative TEM images showing a reduced accumulation of autophagic bodies in the vacuole of cells overexpressing Rph1 compared to wild type. Scale bar, 500 nm.
(E) Estimated average number of autophagic bodies (AB) per vacuole. Estimation was based on the number of autophagic body cross-sections observed by TEM [50]. Over 100 unique cells per strain were captured and analyzed.
(F) The estimated mean radii (in nm) of the original autophagic bodies (AB) observed by TEM in wild-type and OE Rph1 cells was analyzed as in (E).
(G) Rph1 overexpression reduces cell survival after prolonged nitrogen starvation. Rph1-PA cells (WT, YAB323, SEY6210), cells overexpressing Rph1-PA (OE-Rph1, YAB329) and atg1Δ cells (WLY192) were grown in YPD (+N) until mid-log phase and then starved for nitrogen for 15 days (−N). Dilutions as indicated were grown on YPD plates for 2 days, then imaged.
Next, the physiological importance of the Rph1-dependent regulation of autophagy was tested by monitoring the survival phenotype of cells overexpressing Rph1 after nitrogen starvation. Wild-type yeast cells showed no significant cell death during the time course of the experiment, whereas a complete block of autophagy in atg1Δ cells resulted in a complete loss of viability (Figure 4G). In comparison, cells overexpressing Rph1 exhibited a slight growth defect in nitrogen-replete conditions (Figure 4G, left panel) and showed a reduced survival rate after prolonged starvation (Figure 4G, right panel). Together these results suggest that overexpressed-Rph1 promotes cell death by repressing autophagosome biogenesis and thereby inhibiting autophagy.
Rph1 DNA Binding Ability but Not Histone Demethylase Activity Is Required for its Function in Autophagy
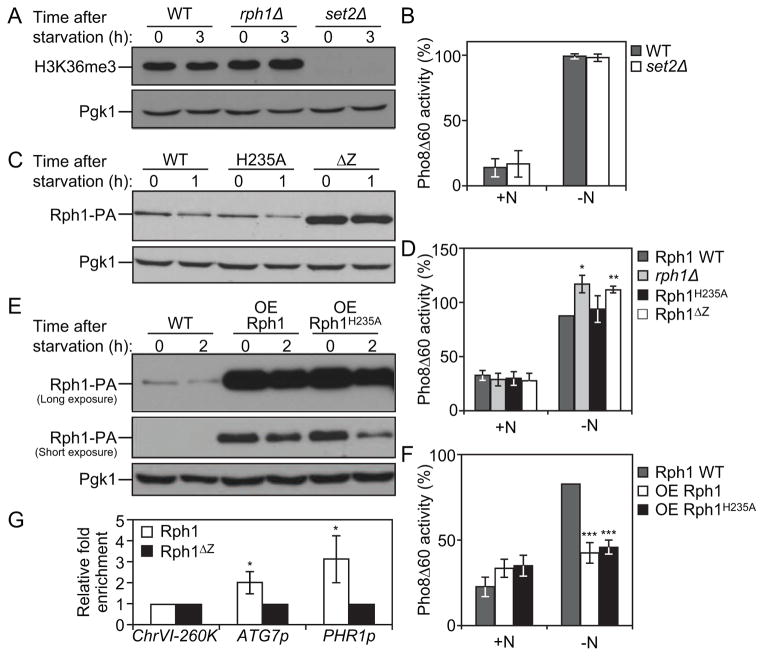
Rph1 is a Jumonji C (JmjC)-containing histone demethylase protein, which is the only demethylase targeting H3K36 tri-methylation in Saccharomyces cerevisiae [29]. Previous studies showed that the transcriptional repression of some Rph1 targets is dependent on its histone demethylase activity [30]. We therefore proposed that Rph1 may repress ATG genes transcription, and thereby autophagy, in a similar manner. This idea was first tested using a set2Δ deletion strain; Set2 is the histone methyltransferase mediating H3K36 methylation in yeast cells [31] and, in this regard, has an opposite function to that of Rph1. Nitrogen starvation did not significantly affect H3K36 tri-methylation (H3K36me3) and rph1Δ cells did not show any difference in the H3K36me3 status compared to wild-type cells (Figure 5A). In contrast, the deletion of SET2 led to a complete block in H3K36me3 marks. Nevertheless, we did not observe an autophagy defect in set2Δ cells compared to the wild type (Figure 5B), which indicates that the level of H3K36me3 does not regulate autophagy.
Figure 5. Rph1 DNA binding ability but not histone demethylase activity is required for its function in autophagy.
(A) Wild-type (YTS158), rph1Δ (YAB300) and set2Δ (YAB318) cells were grown in YPD (+N) and then starved for nitrogen (−N) for 3 h. Protein extracts were analyzed by western blot with anti-H3K36me3 antibody and anti-Pgk1 (loading control) antiserum.
(B) Wild-type (YTS158, BY4742) and set2Δ (YAB318) cells were grown in YPD (+N) and then starved for nitrogen (−N) for 3 h. The Pho8Δ60 activity was measured and normalized to the activity of wild-type cells, which was set to 100%. For panels B, D, F and G, the data represent the average of 3 independent experiments ± standard deviation.
(C–D) rph1Δ cells were transformed with the RPH1p-RPH1-PA (Rph1, Wild-type), RPH1p-RPH1H235A-PA (H235A), or RPH1p-RPH1ΔZ-PA (ΔZ) plasmids or the corresponding empty plasmid (pRS461-PA, rph1Δ). Cells were grown in rich selective medium (SMD-Ura) until mid-log phase and then starved for nitrogen for 1 h.
(C) The stability of Rph1 mutants was analyzed by western blot with either an antibody that recognizes PA or anti-Pgk1 (loading control) antiserum.
(D) The Pho8Δ60 activity was measured and normalized to the activity of wild-type Rph1 cells after 1 h of nitrogen starvation (−N), which was set to 100%.
(E–F) Rph1-PA cells (WT, YAB366) and cells overexpressing Rph1-PA (OE Rph1, YAB363) or Rph1H235A-PA (OE Rph1H235A, YAB364) were grown in YPD (+N) until mid-log phase and then starved for nitrogen for 2 h (−N).
(E) Protein extracts were analyzed by western blot with either an antibody that recognizes PA or anti-Pgk1 (loading control) antiserum.
(F) The Pho8Δ60 activity was measured and normalized to the activity of wild-type cells after 2 h of nitrogen starvation (−N), which was set to 100%.
(G) Rph1-PA binds the ATG7 promoter. rph1Δ cells transformed with the RPH1p-RPH1-PA (Rph1) or RPH1p-RPH1ΔZ-PA (Rph1ΔZ) plasmids were analyzed by ChIP. ChIP was conducted on the ATG7 promoter (ATG7p), a large non-coding region located at 260 kb on chromosome VI (ChrVI-260K) which was used as a negative control, and on the PHR1 promoter (PHR1p) which was used as a positive control. Results were normalized to the input DNA and calibrated to the ChrVI-260K PCR product; results are presented as fold-enrichment of Rph1 binding compared to Rph1ΔZ, which was set to 1.
Next, we analyzed the previously described Rph1H235A mutant in which the replacement of histidine 235 with alanine inhibits the protein’s histone demethylase activity (Figure S3A,B; [30]). Protein analysis verified the stability of Rph1H235A compared to wild-type Rph1 (Figure 5C). The rph1Δ phenotype was restored to the same extent by either wild-type Rph1 or Rph1H235A (Figure 5D, Figure S3C). In addition, cells overexpressing Rph1 or the Rph1H235A mutant showed a similar reduction in autophagy activity compared to wild-type cells (Figure 5E,F). Together these results show that the function of Rph1 in autophagy is, at least mostly, independent of its histone demethylase activity.
Previous results from a large-scale ChIP-chip (chromatin immunoprecipitation-on-chip) analysis identified several ATG genes as potential direct targets of Rph1 [32]. Consistent with this idea, a DNA motifs search in the promoter region of ATG genes identified one or multiple Rph1-binding consensus sites in 23 out of 32 ATG genes (Table S4, Table S5). In particular, our analysis revealed the presence of the environmentally regulated Rph1 binding site (TWAGGG; [26]) in the promoter of highly regulated genes such as ATG7, ATG9 and ATG14 and of a related site (AGGGG) in ATG8 and ATG29 but not in those of a modestly affected gene, ATG1, or a non-affected gene, ATG10. Therefore we tested the importance of the DNA-binding domain of the protein by constructing an Rph1ΔZ mutant, in which the deletion of both zinc-finger motifs at the C-terminal part of the protein prevents DNA binding ([31]; Figure S3A). A ChIP approach showed a 2-fold enrichment of Rph1-PA at the ATG7 promoter compared to Rph1ΔZ-PA (Figure 5G). A similar enrichment was found at the PHR1 promoter, which was used as a positive control [30], but not at the non-coding region located at the tip of chromosome VI (ChrVI-260K), which was used as a negative control. This indicates that Rph1 directly binds the promoter region of ATG7 and that this binding is significantly reduced upon deletion of the zing-finger domains of the protein. Furthermore, even though Rph1ΔZ-PA was found over-accumulated compared to Rph1-PA (Figure 5C), this mutant form of the protein was unable to rescue the ATG gene overexpression phenotype or the increased autophagy activity induction of the rph1Δ cells (Figure 5D, Figure S3C), thus showing that the DNA-binding domain of Rph1 is strictly required for its function in autophagy. Together these results show that the histone demethylase activity of Rph1 is not required for its function in autophagy and suggest that, instead, Rph1 represses the expression of ATG genes by restricting the access of the transcriptional machinery or potential activators at these loci as has been proposed for other Rph1-regulated genes [23, 33].
Rph1 Phosphorylation upon Nitrogen Starvation Releases its Repression of Autophagy
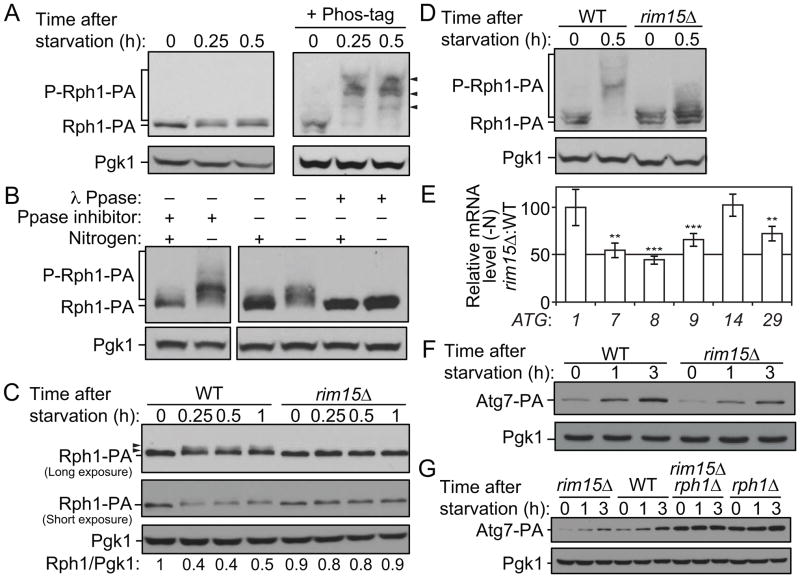
As mentioned above, the deletion of RPH1 induced the expression of ATG genes in rich conditions but had no effect on mRNA levels after nitrogen starvation (Figure 1). These results suggest a model in which Rph1 represses the transcription of these targets when cells grow in nutrient-replete medium and that upon nitrogen starvation this repression is released thereby allowing the induction of ATG genes and autophagy. Previous studies describe Rph1 as a phosphoprotein and show that Rph1 phosphorylation inhibits its activity [23]. Similarly, we observed a shift in the molecular weight of Rph1-PA as well as the appearance of a faint upper band in conditions of nitrogen depletion, suggesting that Rph1 undergoes phosphorylation at multiple sites (Figure 6A, left panel, and Figure 6C). This shift was accompanied by a modest decrease in protein level shortly after nitrogen starvation (Figure 6A,C). In order to get a better resolution of the phosphorylation status of Rph1 we used Phos-tag, a molecule that binds specifically to phosphorylated ions, thereby increasing the molecular weight of phospho-isoforms. With addition of Phos-tag, Rph1-PA was observed as several higher molecular weight bands in nitrogen starvation conditions compared to rich conditions, strongly supporting the hypothesis that Rph1 is phosphorylated upon starvation (Figure 6A, right panel). Note that this analysis also revealed Rph1-PA as a doublet in rich conditions (Figure 6A, right panel) indicating that the protein undergoes some phosphorylation even in the presence of nitrogen. Furthermore, phosphatase treatment of the protein samples prevented the changes in Rph1 mobility demonstrating that the change in migration represents genuine phosphorylation of the protein (Figure 6B).
Figure 6. Rim15-dependent phosphorylation of Rph1 upon nitrogen starvation releases its repression on ATG gene expression and autophagy.
(A–B) Rph1 is phosphorylated upon nitrogen starvation. Rph1 was chromosomally tagged with PA tag and Rph1-PA cells (YAB308) were grown in YPD until mid-log phase and then starved for nitrogen for the indicated times.
(A) Protein extracts were analyzed by western blot with or without 50 μM Phos-tag as indicated and with either an antibody that recognizes PA or anti-Pgk1 (loading control) antiserum. Arrowheads indicate shifts in molecular weight of Rph1-PA suggesting phosphorylation.
(B) Rph1-PA band shifts represent phospho-isoforms of the protein. Cells were lysed with or without phosphatase (Ppase) inhibitors. An aliquot of cell lysates was incubated at 30°C for 90 mintes in λ-phosphatase buffer with or without λ-phosphatase (λ Ppase). The reaction was stopped and proteins were precipitated by addition of 10% trichloroacetic acid. Protein extracts were analyzed by western blot as in (A) from gels containing Phos-tag.
(C–D) Rph1 phosphorylation upon nitrogen starvation is blocked in rim15Δ cells. RPH1 was chromosomally tagged with PA in wild-type (WT) and rim15Δ cells. Cells were grown in YPD until mid-log phase and then starved for nitrogen for the indicated times. WT (YAB308) and rim15Δ cells (YAB341) were collected and protein extracts were analyzed by western blot with either an antibody that recognizes PA or anti-Pgk1 (loading control) antiserum. (D) Protein extracts were analyzed by western blot as in (B).
(E) The deletion of RIM15 reduces the induction of ATG gene expression after nitrogen starvation. WT (YAB308) and rim15Δ (YAB341) cells were grown in YPD until mid-log phase and then starved for 1 h (−N). mRNA levels were quantified by RT-qPCR. The mRNA levels of individual ATG genes were normalized to the mRNA level of the corresponding gene in WT cells in nitrogen starvation condition (−N), which was set to 100. Data represent the average of at least 3 independent experiments ± standard deviation.
(F) ATG7 was chromosomally tagged with PA in WT (YAB312) and rim15Δ (YAB342) cells. Cells were grown in YPD until mid-log phase and then starved for nitrogen for the indicated times. Protein extracts were analyzed by western blot with either an antibody that recognizes PA or anti-Pgk1 (loading control) antiserum.
(G) ATG7 was chromosomally tagged with PA in WT (YAB312), rim15Δ (YAB342), rph1Δ (YAB313) and rim15Δ rph1Δ (YAB347) cells. Proteins extracts were analyzed as in (F).
(See also Figure S4.)
To test the importance of Rph1 phosphorylation for autophagy induction, we next aimed to identify the phosphorylated residues upon nitrogen starvation. In order to identify such sites we used previously published data from large-scale phosphoproteomic analyses. In particular Huber et al. [34] identified 5 sites (group II, Figure S4) at which phosphorylation was suggested to be upregulated upon treatment with rapamycin, a drug that induces autophagy; in addition the work by Bodenmiller et al. [35], proposed that phosphorylation at T411 and S412 (group I, Figure S4) is upregulated when the gene encoding Tpk2, a subunit of the autophagy repressor PKA, is deleted. Based on these data we generated Rph1 mutants where the phosphoresidues in group I or group I and II were mutated to alanine. In addition serine 429 which is also predicted to be phosphorylated was mutated to alanine because of its proximity to the residues in group I. The mutants showed only a partial reduction in phosphorylation based on gel migration, and a concomitant accumulation of the non-phosphorylated form of the protein after starvation (Figure S4B). Accordingly, autophagy activity was only reduced to a minor, yet significant, level in these mutants (Figure S4C), suggesting that additional residues that were not identified by mass spectrometry may also be involved in phospho-regulation of Rph1. Together these results show that residues in group I and II participate in the phosphorylation of Rph1 after starvation and suggest that this modification of the protein inhibits its activity upon autophagy induction.
Rim15 Mediates the Phosphorylation of Rph1 upon Nitrogen Starvation thereby inducing Autophagy
Next, we attempted to identify the kinase responsible for Rph1 phosphorylation prior to autophagy activation; Rim15 appeared to be a good candidate as it is phosphorylated and translocated into the nucleus after nitrogen starvation at which time it activates autophagy [36–38]. Analysis of Rph1-PA showed a strong block in Rph1 molecular mass shift when RIM15 was deleted (Figure 6C,D) showing that this kinase is, at least partly, responsible for Rph1 phosphorylation upon nitrogen starvation. Moreover, the inhibition of Rph1 phosphorylation in the rim15Δ cells was accompanied by a stabilization of the Rph1 protein compared to wild-type cells where the protein level decreased after nitrogen starvation. In addition, the RIM15 deletion caused a partial block in the induction of some of the ATG genes and Atg proteins that we analyzed, concomitant with the reduction of autophagy activity seen in the same conditions (Figure 6E,F and Figure S4D–F). Furthermore, deletion of RPH1 in the rim15Δ cells rescued the defect in Atg7-PA level in response to nitrogen starvation (Figure 6G), showing that Rim15 acts upstream of Rph1 to regulate ATG7 expression during autophagy.
These findings strongly support the idea that the Rim15-dependent phosphorylation of Rph1 is required for proper induction of autophagy upon nitrogen starvation. Our previous work showed that Rim15 also phosphorylates Ume6 leading to an inhibition of Ume6 activity and an upregulation of ATG8 after nitrogen starvation [21]. As previously described, the ume6Δ cells showed an increase in the expression of ATG8 in nutrient-rich conditions compared to the wild type ([21], Figure S4G); this was accompanied by an upregulation of ATG1 and ATG9. In contrast the expression of ATG7, ATG14 and ATG29 was unaffected by the deletion of UME6. Because rph1Δ cells showed higher expression of ATG7 and ATG29 we suggest that the block of, at least, ATG7 and ATG29 induction in the rim15Δ cells is specifically due to the loss of Rph1 phosphorylation and propose that this contributes to the reduction of autophagy activity in this mutant background.
The Regulation of Autophagy by Rph1/KDM4 Is Conserved from Yeast to Mammals
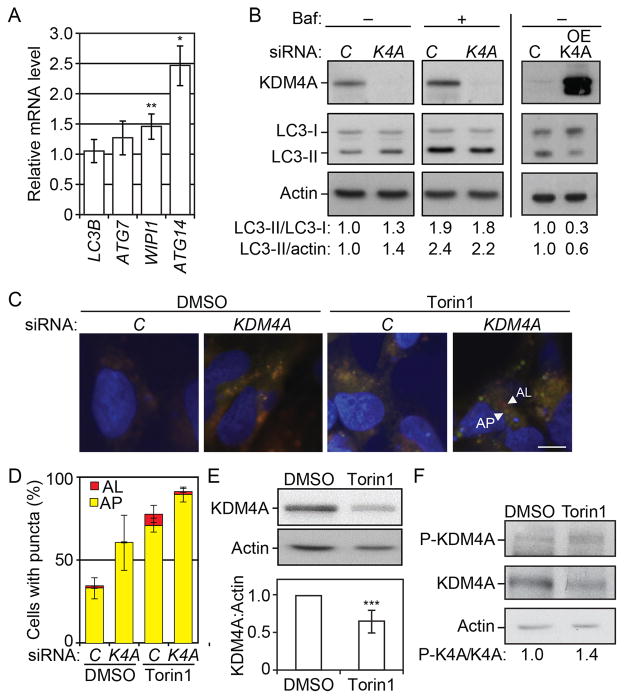
To test if the mechanism of autophagy regulation that we uncovered here was conserved in higher eukaryotes we studied the role of one mammalian homolog of RPH1, KDM4A, in the autophagy pathway. A reduction of KDM4A level was associated with an increase in the mRNA level of several ATG genes including ATG7, WIPI1 and ATG14, whereas there was essentially no effect on MAP1LC3B (Figure 7A). HeLa cells transformed with siKDM4A showed an accumulation of LC3-II compared to control cells pointing to an increase in basal autophagy in this condition (Figure 7B, left panel). To verify that the increase in LC3-II reflected an upregulation of autophagy rather than a block in flux, we treated the cells with the autophagy inhibitor bafilomycin A1. We detected a further increase in the level of LC3-II upon the addition of bafilomycin A1, suggesting that the knockdown of KDM4A did not interfere with, but instead enhanced, autophagic flux. Conversely, the overexpression of KDM4A led to a reduction in the level of LC3-II compared to control cells (Figure 7B, right panel) further supporting the conclusion that KDM4A is a repressor of autophagy.
Figure 7. KDM4 regulates autophagy in mammalian cells.
(A) Transfected cells were grown for 48 h post transfection. Total RNA was extracted and the mRNA levels were quantified by RT-qPCR. The mRNA level of individual ATG genes was normalized to the mRNA level of the corresponding gene in the control cells (transfected with non-targeting siRNAs), which was set to 1. Error bars indicate the standard deviation of at least 3 independent experiments.
(B) Left panel: HeLa cells transfected with siRNA against KDM4A (K4A) show a reduction in the expression of KDM4A and a concomitant increase in the LC3-II/LC3-I and LC3-II/actin ratio compared to the cells transfected with non-targeting siRNAs (control, C). In the presence of bafilomycin A1 (Baf), control and siKDM4A cells show a similar accumulation of LC3-II. Right panel: HeLa cells transfected with a plasmid overexpressing KDM4A (OE-KDM4A) show a decrease in the LC3-II/LC3-I and LC3-II/actin ratio compared to control cells (C). Total cell lysates were harvested 48 h after transfection and protein extracts were analyzed by western blot using antibodies to the indicated proteins.
(C–D) Cells were transfected with the mRFP-GFP-LC3 plasmid. On the subsequent day, cells were transfected with siKDM4A (KDM4) or non-targeting siRNAs (control, C). Cells were treated for 24 h with DMSO or 250 nM Torin1 as indicated.
(C) Cells were analyzed by fluorescence microscopy. Scale bar, 5 μm.
(D) Autophagy was determined by quantification of the number of cells with LC3-positive organelles. The histogram represents the mean of 3 independent experiments with SEM. AL, autolysosomes; AP, autophagosomes; K4A, KDM4A.
(E) KDM4A is degraded upon autophagy induction by Torin1. Cells were treated with 250 nM Torin1 for 24 h. Protein extracts were analyzed by western blot using antibodies to the indicated proteins. The average and standard deviation of 7 independent experiments are provided.
(F) Phosphorylation of KDM4A at Tyr547 is upregulated by treatment with Torin1. Cells were treated with Torin1 and protein extracts analyzed by western blot as in (E). K4A, KDM4A; P-KDM4A/P-K4A, phosphorylated KDM4A.
To extend this analysis, we monitored the autophagic flux using a tandem reporter construct, mRFP-GFP-LC3, a probe that allows distinction between autophagosomes (AP, GFP+ RFP+, yellow puncta, Figure 7C) and autolysosomes (AL, GFP− RFP+, red puncta, Figure 7C). Compared to the control, cells knocked down for KDM4A showed an increase in yellow puncta representing autophagosomes, in both basal autophagy (DMSO) conditions or after autophagy induction by Torin1 (Figure 7C,D). Following Torin1 treatment there was an increase in autolysosomes in both the control and siKDM4A cells, again indicating that autophagic flux was not inhibited by KDM4A depletion.
We found that the level of KDM4A was reduced in response to Torin1 treatment (Figure 7E) indicating that KDM4A is degraded in this condition. A previous phosphoproteomic analysis indicated that KDM4A is phosphorylated at residue Y547 [39]; analysis of KDM4A with a phosphospecific antibody directed towards Y547 showed that phosphorylation at this residue is upregulated by treatment with Torin1. Together, these results suggest that, similar to Rph1, the phosphorylation of KDM4A acts as a switch to promote autophagy induction in mammalian cells.
Discussion
In this study, a screen for transcription factors modulating ATG gene expression identified Rph1 as a transcriptional repressor of autophagy. We show that Rph1 is a negative regulator of the expression of, at least, ATG7, ATG8, ATG9, ATG14 and ATG29. Consistent with our results, a recent microarray analysis reported the induction of ATG7, and ATG14, as well as ATG16, ATG17, ATG23 and ATG24 in rph1Δ cells in physiological conditions [23] suggesting that Rph1 might control a large set of ATG genes.
Our results show that the deletion of RPH1 can be partially complemented by Gis1, its paralog, indicating that Gis1 plays a minor role in the regulation of ATG genes. Rph1 and Gis1 are JmjC-domain containing proteins; while Rph1 histone demethylase activity is well established [29], controversies exist concerning Gis1 [29, 40]. Whether or not Gis1 is catalytically active, its JmjC domain is not required for its transcriptional activity [24,41]. Similarly we show here that the histone demethylase activity of Rph1 is not required for its control of autophagy, but that its DNA binding domain is critical. Rph1 and Gis1 share a high homology in their C2H2 zinc finger DNA-binding domain suggesting that they could bind similar motifs, which might explain the partial redundancy between the two proteins. Yet, the deletion of RPH1 has a strong effect on the expression of ATG genes and affects autophagy activity, whereas deleting GIS1 alone did not affect autophagy suggesting that Rph1 has higher affinity for the promoters of ATG genes and plays a major role in the repression of autophagy in nitrogen-replete conditions.
The deletion of RPH1 has no effect on the expression of ATG genes after nitrogen depletion, and the phenotype of the RPH1 null strain is attenuated after prolonged starvation suggesting that the activity of the protein is repressed in this condition. Indeed, we found that Rph1 is phosphorylated in a Rim15-dependent manner upon nitrogen starvation and that this phosphorylation is accompanied with a partial degradation of the protein. Inhibition of Rph1 phosphorylation or overexpression of the protein leads to a strong block of ATG gene induction as well as autophagy activity, showing that the phosphorylation of Rph1 represses its activity and is a prerequisite for autophagy induction. Altogether, these data propose a model in which, by repressing the expression of ATG genes, Rph1 maintains autophagy at a low level in nutrient-replete conditions. Upon nitrogen starvation, Rim15 mediates Rph1 phosphorylation, which causes a partial autophagy-independent degradation of the protein and an inhibition of its activity (Figure 6, Figure S4 and data not shown). This leads to a release of Rph1 repression of ATG genes, an induction of their expression and an overall induction of autophagy activity.
Rim15 controls many aspects of the nutrient-regulatory pathways, including autophagy, by integrating signals from TORC1 and PKA [38, 42]; yet little is known about the downstream effectors in these signaling pathway. We show that Rim15 phosphorylates Rph1, thereby releasing its repression of the expression of several ATG genes including ATG7 and ATG29, upon nutrient limitation. Conversely, although Rph1 controls the expression of ATG14, its expression was not affected by the deletion of RIM15. Rim15 is under the control of both positive and negative regulators of autophagy [38], and it is therefore possible that the result of competitive effectors controlling the expression of ATG14 account for this observation. Uncovering the Rim15-dependent Rph1-mediated transcriptional control of autophagy here brings further comprehension to the overall signaling pathways leading to autophagy induction upon starvation.
How does the level of ATG gene transcription regulate autophagy activity? Several ATG genes show higher expression after nitrogen starvation ([16]; Figure 1), suggesting that this increase is required to support optimal autophagy activity; yet the physiological relevance of the induction of particular ATG genes is still largely unknown. Our previous work showed that the amount of Atg9 correlates with the number of autophagosomes [16], whereas the induction of ATG8 promotes the formation of larger autophagosomes [22] thereby leading to an increase in the amount of cargo delivered to the autophagy pathway. Here we show that Rph1 plays a major role in the control of the expression of ATG7. Upon autophagy induction, Atg7 functions in the formation of Atg8–PE conjugates, which is a prerequisite to the membrane recruitment of Atg8 and the elongation of the phagophore [45]. Our results show that negatively modulating the expression of ATG7 causes defects in autophagy activity (Figure 3), suggesting that the amount of Atg7 is rate limiting in the lipidation of the large pool of newly synthesized Atg8 after starvation. It is therefore tempting to speculate that, upon autophagy induction, a concomitant upregulation of both ATG7 and ATG8 is strictly required to support efficient autophagosome formation and thereby an increase in the magnitude of autophagy activity.
Besides its role in cell survival upon starvation, autophagy activity is also critical for cellular clearance of protein aggregates and damaged organelles, which, when accumulated, result in severe pathologies, such as neurodegenerative diseases [10]. Interestingly, a recent genomic study of sporadic Parkinson disease patients identified mutations in the ATG7 promoter causing decreased ATG7 expression [17]. In this context, our finding that the transcriptional control of ATG7 is critical for autophagy activity points to the physiological importance of the Rph1-mediated control of autophagy that we identified here. In particular, we show that the function of Rph1 is conserved from yeast to mammals. Four isoforms of KDM4, the homolog of Rph1, are present in mammalian cells. Here we show that KDM4A is a negative regulator of autophagy; whether other isoforms are also involved in this process remains to be investigated. Knocking down KDM4A increased autophagy activity, especially at base line, suggesting that modulating this novel regulatory pathway could be a good target for the development of disease therapies.
Experimental Procedures
Yeast Strains, Media and Culture
Gene disruptions were performed using a standard method [44]. Yeast cells were grown in YPD or SMD [16] and autophagy was induced in SD-N [16]. The yeast strains used in this study are listed in Table S1.
HeLa cells were purchased from Sigma and grown in MEM supplemented with penicillin/streptomycin, L-glutamine, non-essential amino acids and sodium pyruvate.
RNA and RT-qPCR
RNA was extracted using the RNeasy mini kit (Qiagen), and RNA was reverse-transcribed using the High-capacity cDNA Reverse Transcription kit (Applied Biosystems). See the supplement for details.
Chromatin Immunoprecipitation
ChIP was performed as previously described [45] with minor modifications as described in the supplement.
Transmission electron microscopy
TEM and sample analysis were performed as previously described [16].
Mammalian Cell Transfection
HeLa cells were transfected with siRNA using Lipofectamine 2000 reagent (Invitrogen) as described in the supplement.
Supplementary Material
Acknowledgments
This work was supported by NIH grant GM053396 to DJK, and by grants from the Swedish Cancer Society, the Swedish Childhood Cancer Foundation and the Swedish Research Council to BJ.
Abbreviations
- Atg
autophagy-related
- GFP
green fluorescent protein
- PA
protein A
- TEM
transmission electron microscopy
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, four figures and five tables and can be found with this article online at XXXX.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lenstra TL, Benschop JJ, Kim T, Schulze JM, Brabers NA, Margaritis T, van de Pasch LA, van Heesch SA, Brok MO, Groot Koerkamp MJ, et al. The specificity and topology of chromatin interaction pathways in yeast. Mol Cell. 2011;42:536–549. doi: 10.1016/j.molcel.2011.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 3.Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- 4.Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 5.Deretic V, Levine B. Autophagy, immunity, and microbial adaptations. Cell Host Microbe. 2009;5:527–549. doi: 10.1016/j.chom.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi CS, Shenderov K, Huang NN, Kabat J, Abu-Asab M, Fitzgerald KA, Sher A, Kehrl JH. Activation of autophagy by inflammatory signals limits IL-1beta production by targeting ubiquitinated inflammasomes for destruction. Nat Immunol. 2012;13:255–263. doi: 10.1038/ni.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang J, Klionsky DJ. Autophagy and human disease. Cell Cycle. 2007;6:1837–1849. doi: 10.4161/cc.6.15.4511. [DOI] [PubMed] [Google Scholar]
- 8.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lynch-Day MA, Klionsky DJ. The Cvt pathway as a model for selective autophagy. FEBS Lett. 2010;584:1359–1366. doi: 10.1016/j.febslet.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nixon RA. The role of autophagy in neurodegenerative disease. Nat Med. 2013;19:983–997. doi: 10.1038/nm.3232. [DOI] [PubMed] [Google Scholar]
- 11.Chen Y, Azad MB, Gibson SB. Superoxide is the major reactive oxygen species regulating autophagy. Cell Death Differ. 2009;16:1040–1052. doi: 10.1038/cdd.2009.49. [DOI] [PubMed] [Google Scholar]
- 12.Levine B, Yuan J. Autophagy in cell death: an innocent convict? J Clin Invest. 2005;115:2679–2688. doi: 10.1172/JCI26390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ravikumar B, Sarkar S, Davies JE, Futter M, Garcia-Arencibia M, Green-Thompson ZW, Jimenez-Sanchez M, Korolchuk VI, Lichtenberg M, Luo S, Massey DC, Menzies FM, Moreau K, Narayanan U, Renna M, Siddiqi FH, Underwood BR, Winslow AR, Rubinsztein DC. Regulation of mammalian autophagy in physiology and pathophysiology. Physiol Rev. 2010;90:1383–1435. doi: 10.1152/physrev.00030.2009. [DOI] [PubMed] [Google Scholar]
- 14.Jin M, Klionsky DJ. Regulation of autophagy: Modulation of the size and number of autophagosomes. FEBS Lett. 2014;588:2457–2463. doi: 10.1016/j.febslet.2014.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parzych KR, Klionsky DJ. An overview of autophagy: morphology, mechanism, and regulation. Antioxid Redox Signal. 2014;20:460–473. doi: 10.1089/ars.2013.5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin M, He D, Backues SK, Freeberg MA, Liu X, Kim JK, Klionsky DJ. Transcriptional regulation by pho23 modulates the frequency of autophagosome formation. Curr Biol. 2014;24:1314–1322. doi: 10.1016/j.cub.2014.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen D, Pang S, Feng X, Huang W, Hawley RG, Yan B. Genetic analysis of the ATG7 gene promoter in sporadic Parkinson’s disease. Neurosci Lett. 2013;534:193–198. doi: 10.1016/j.neulet.2012.12.039. [DOI] [PubMed] [Google Scholar]
- 18.Liu H, He Z, von Rutte T, Yousefi S, Hunger RE, Simon HU. Down-regulation of autophagy-related protein 5 (ATG5) contributes to the pathogenesis of early-stage cutaneous melanoma. Sci Transl Med. 2013;5:202ra123. doi: 10.1126/scitranslmed.3005864. [DOI] [PubMed] [Google Scholar]
- 19.Wang J, Pan XL, Ding LJ, Liu DY, Da-Peng L, Jin T. Aberrant expression of Beclin-1 and LC3 correlates with poor prognosis of human hypopharyngeal squamous cell carcinoma. PLoS ONE. 2013;8:e69038. doi: 10.1371/journal.pone.0069038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jo YK, Kim SC, Park IJ, Park SJ, Jin DH, Hong SW, Cho DH, Kim JC. Increased expression of ATG10 in colorectal cancer is associated with lymphovascular invasion and lymph node metastasis. PLoS ONE. 2012;7:e52705. doi: 10.1371/journal.pone.0052705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bartholomew CR, Suzuki T, Du Z, Backues SK, Jin M, Lynch-Day MA, Umekawa M, Kamath A, Zhao M, Xie Z, et al. Ume6 transcription factor is part of a signaling cascade that regulates autophagy. Proc Natl Acad Sci U S A. 2012;109:11206–11210. doi: 10.1073/pnas.1200313109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xie Z, Nair U, Klionsky DJ. Atg8 controls phagophore expansion during autophagosome formation. Mol Biol Cell. 2008;19:3290–3298. doi: 10.1091/mbc.E07-12-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liang CY, Wang LC, Lo WS. Dissociation of the H3K36 demethylase Rph1 from chromatin mediates derepression of environmental stress-response genes under genotoxic stress in Saccharomyces cerevisiae. Mol Biol Cell. 2013;24:3251–62. doi: 10.1091/mbc.E12-11-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jang YK, Wang L, Sancar GB. RPH1 and GIS1 are damage-responsive repressors of PHR1. Mol Cell Biol. 1999;19:7630–7638. doi: 10.1128/mcb.19.11.7630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang N, Oliver SG. The transcription activity of Gis1 is negatively modulated by proteasome-mediated limited proteolysis. J Biol Chem. 2010;285:6465–6476. doi: 10.1074/jbc.M109.073288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Orzechowski Westholm J, Tronnersjo S, Nordberg N, Olsson I, Komorowski J, Ronne H. Gis1 and Rph1 regulate glycerol and acetate metabolism in glucose depleted yeast cells. PLoS One. 2012;7:e31577. doi: 10.1371/journal.pone.0031577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Noda T, Klionsky DJ. The quantitative Pho8Delta60 assay of nonspecific autophagy. Methods Enzymol. 2008;451:33–42. doi: 10.1016/S0076-6879(08)03203-5. [DOI] [PubMed] [Google Scholar]
- 28.Shintani T, Klionsky DJ. Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J Biol Chem. 2004;279:29889–29894. doi: 10.1074/jbc.M404399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tu S, Bulloch EM, Yang L, Ren C, Huang WC, Hsu PH, Chen CH, Liao CL, Yu HM, Lo WS, et al. Identification of histone demethylases in Saccharomyces cerevisiae. J Biol Chem. 2007;282:14262–14271. doi: 10.1074/jbc.M609900200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang CY, Hsu PH, Chou DF, Pan CY, Wang LC, Huang WC, Tsai MD, Lo WS. The histone H3K36 demethylase Rph1/KDM4 regulates the expression of the photoreactivation gene PHR1. Nucleic Acids Res. 2011;39:4151–4165. doi: 10.1093/nar/gkr040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strahl BD, Grant PA, Briggs SD, Sun ZW, Bone JR, Caldwell JA, Mollah S, Cook RG, Shabanowitz J, Hunt DF, et al. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol Cell Biol. 2002;22:1298–1306. doi: 10.1128/mcb.22.5.1298-1306.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Venters BJ, Wachi S, Mavrich TN, Andersen BE, Jena P, Sinnamon AJ, Jain P, Rolleri NS, Jiang C, Hemeryck-Walsh C, et al. A comprehensive genomic binding map of gene and chromatin regulatory proteins in Saccharomyces. Mol Cell. 2011;41:480–492. doi: 10.1016/j.molcel.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nordberg N, Olsson I, Carlsson M, Hu GZ, Orzechowski Westholm J, Ronne H. The histone demethylase activity of Rph1 is not essential for its role in the transcriptional response to nutrient signaling. PLoS one. 2014;9:e95078. doi: 10.1371/journal.pone.0095078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huber A, Bodenmiller B, Uotila A, Stahl M, Wanka S, Gerrits B, Aebersold R, Loewith R. Characterization of the rapamycin-sensitive phosphoproteome reveals that Sch9 is a central coordinator of protein synthesis. Genes Dev. 2009;23:1929–1943. doi: 10.1101/gad.532109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bodenmiller B, Wanka S, Kraft C, Urban J, Campbell D, Pedrioli PG, Gerrits B, Picotti P, Lam H, Vitek O, Brusniak MY, Roschitzki B, Zhang C, Shokat KM, Schlapbach R, Colman-Lerner A, Nolan GP, Nesvizhskii AI, Peter M, Loewith R, von Mering C, Aebersold R. Phosphoproteomic analysis reveals interconnected system-wide responses to perturbations of kinases and phosphatases in yeast. Sci Signal. 2010;3:rs4. doi: 10.1126/scisignal.2001182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pedruzzi I, Dubouloz F, Cameroni E, Wanke V, Roosen J, Winderickx J, De Virgilio C. TOR and PKA signaling pathways converge on the protein kinase Rim15 to control entry into G0. Mol Cell. 2003;12:1607–1613. doi: 10.1016/s1097-2765(03)00485-4. [DOI] [PubMed] [Google Scholar]
- 37.Wanke V, Pedruzzi I, Cameroni E, Dubouloz F, De Virgilio C. Regulation of G0 entry by the Pho80–Pho85 cyclin–CDK complex. EMBO J. 2005;24:4271–4278. doi: 10.1038/sj.emboj.7600889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang Z, Geng J, Yen WL, Wang K, Klionsky DJ. Positive or negative roles of different cyclin-dependent kinase Pho85-cyclin complexes orchestrate induction of autophagy in Saccharomyces cerevisiae. Mol Cell. 2010;23:250–264. doi: 10.1016/j.molcel.2010.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tao WA, Wollscheid B, O’Brien R, Li XJ, Bodenmiller B, Watts JD, Hood L, Aebersold R. Quantitative phosphoproteome analysis using a dendrimer conjugation chemistry and tandem mass spectrometry. Nat Methods. 2005;2:591–598. doi: 10.1038/nmeth776. [DOI] [PubMed] [Google Scholar]
- 40.Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet. 2006;7:715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- 41.Yu Y, Neiman AM, Sternglanz R. The JmjC domain of Gis1 is dispensable for transcriptional activation. FEMS Yeast Res. 2010;10:793–801. doi: 10.1111/j.1567-1364.2010.00680.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Swinnen E, Wanke V, Roosen J, Smets B, Dubouloz F, Pedruzzi I, Cameroni E, De Virgilio C, Winderickx J. Rim15 and the crossroads of nutrient signalling pathways in Saccharomyces cerevisiae. Cell Div. 2006;1:3. doi: 10.1186/1747-1028-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Geng J, Nair U, Yasumura-Yorimitsu K, Klionsky DJ. Post-Golgi Sec proteins are required for autophagy in Saccharomyces cerevisiae. Mol Biol Cell. 2010;21:2257–2269. doi: 10.1091/mbc.E09-11-0969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Longtine MS, McKenzie A, III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 45.Govin J, Dorsey J, Gaucher J, Rousseaux S, Khochbin S, Berger SL. Systematic screen reveals new functional dynamics of histones H3 and H4 during gametogenesis. Genes Dev. 2010;24:1772–1786. doi: 10.1101/gad.1954910. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.