Abstract
The origins of the influenza A (H1N1) pandemic of 2009 in swine are unknown, highlighting gaps in our understanding of influenza A virus ecology and evolution. Here we review how recently strengthened influenza virus surveillance in pigs has revealed that influenza virus transmission from humans to swine is far more frequent than swine-to-human zoonosis, and is central in seeding swine globally with new viral diversity. The scale of global human-to-swine transmission represents the largest ‘reverse zoonosis’ of a pathogen documented to date. Overcoming the bias towards perceiving swine as sources of human viruses, rather than recipients, is key to understanding how the bidirectional nature of the human-animal interface produces influenza threats to both hosts.
Keywords: influenza A virus, swine, pandemic, evolution, human-animal interface
Swine as reservoirs for influenza A virus diversity and pandemic threats
Influenza A viruses (IAVs) are considered to be one of the greatest threats for the next global pandemic due to the abundance of permanent animal reservoirs harboring viruses that occasionally spill over into humans [1]. For decades, swine have been thought to serve as intermediate ‘mixing vessel’ hosts (see Glossary) for the evolution of pandemic viruses because of their capacity to be infected with both avian and human influenza viruses, which can exchange genome segments via a process termed ‘reassortment’ to generate new pandemic viruses [2–4]. This threat was borne out in 2009, when the first influenza pandemic of the 21st century was caused by a novel reassortant swine H1N1 virus with genetic segments from both avian-like Eurasian swine viruses and North American swine viruses [5]. Since 2009, the genetic diversity of IAVs in swine populations (swIAVs) has expanded globally, generating additional pandemic threats such as the novel variant H3N2v swIAV that infected more than 330 humans in the United States during 2011–2013 [6]. Determining the evolutionary mechanisms that increase IAV diversity in swine is therefore important for human and animal health.
The evolutionary dynamics of IAVs are determined by their complex multi-host ecology, viral structure, and segmented genome. IAVs are single-stranded, negative-sense RNA viruses of the family Orthomyxoviridae that infect a wide range of avian and mammalian species. The genome of IAV (total length ~13 kb) is composed of eight single-stranded RNA (ssRNA) segments that encode for at least 12 viral proteins. Two surface glycoproteins, the hemagglutinin (HA) and neuraminidase (NA), are the main viral antigens and are used to describe the diversity of IAV subtypes (e.g., H3N2). Sixteen antigenically distinct HA and nine NA subtypes exist in wild aquatic birds, which are considered to be the reservoir hosts for IAV diversity [7]. Avian influenza viruses occasionally spill over into humans (e.g., H5N1 and H7N9), usually with little onward transmission. On rare occasions, major influenza pandemics occur when an avian influenza virus evolves the capacity to transmit human-to-human, as occurred in 1918, 1957, and 1968 [8,9]. Reassortment was key in generating the novel avian-human reassortant H2N2 and H3N2 viruses associated with the pandemics of 1957 and 1968 [9], respectively, although the proposed role of swine as intermediary mixing vessel hosts remains unclear. Avian influenza viruses also have evolved to be transmissible in non-human mammalian hosts: avian-origin H3N8 viruses have transmitted in horses since the 1960s [10]; avian-origin H3N2 has circulated in dogs in Asia since the mid 2000s [11]; and avian-origin H1N1 has circulated in Eurasian swine since the 1970s [12]. IAVs also can jump between mammalian species, as evidenced by the adaptation of equine H3N8 viruses to canines in the early 2000s [13] and swine H1N1 viruses to humans in 2009 [5]. One determinant of host specificity is the distribution of sialic acid receptors that bind the viral HA glycoproteins on host cells to facilitate viral entry [14]. Understanding what molecular evolutionary changes allow certain IAVs to jump between host species, and the potential role of swine as intermediary hosts, remains a central question in influenza virus research (Box 1).
Box 1. Outstanding questions.
Where did the Eurasian-North American reassortant influenza virus associated with the 2009 H1N1 human influenza pandemic evolve in swine and circulate undetected for approximately one decade?
Were swine ‘mixing vessel’ hosts that facilitated the reassortment events between avian and human viruses that generated the pandemic viruses of 1957 and 1968?
Did humans introduce the 1918 H1N1 ‘Spanish flu’ pandemic virus into pigs, or did the virus circulate and evolve in swine prior to entering the human population?
What factors make swine so susceptible to human IAVs? What is the relative importance of low immunity in swine, high density of animals facilitating onward transmission, and frequency of exposure to human viruses?
What is the full extent of human-origin IAV diversity circulating in swine globally, including in under-sampled pig populations in Latin America, Africa, Oceania, and South Asia?
What are the genetic constraints on the transmission of human viruses to swine? For example, there is no evidence that human H2N2 viruses or influenza B viruses have ever successfully transmitted in swine.
What are the roles of airborne versus contact in human-to-swine transmission, and do any biosecurity practices on farms reduce transmission between humans and swine?
Why is reassortment more frequent in swine than in humans?
How do differences in the intensity of IAV surveillance across different hosts bias our understanding of IAV ecology and inter-species transmission?
Can we further assess the pandemic threat of human-origin influenza viruses circulating in swine by determining the extent of existing cross-immunity in humans of different age groups?
In the aftermath of the 2009 H1N1 pandemic, there has been great interest in understanding the threat of swIAVs for humans, including (i) estimates of the frequency of human infection with swIAVs [15,16], (ii) assessments of the pandemic potential of various swIAVs in animal models [17,18], and (iii) increased surveillance of global swIAV diversity, including regions and continents that rarely or never previously reported swIAVs, including Africa [19], Latin America [20–23], South Asia [24,25], and Australia [26,27]. Paradoxically, the swIAV sequence data collected to assess the pandemic risk of swine for humans has demonstrated that humans transmit far more IAV diversity to swine than pigs have ever transmitted to humans, at least in terms of viruses that transmit onward in the new host (as opposed to dead-end infections). In this review we describe the central role of human-to-swine transmission events in the evolution of IAV diversity in swine. In particular, since 2009 the continual introduction of pandemic H1N1 (pH1N1) viruses from humans has expanded the genetic diversity of swIAVs globally, introducing new challenges for the control of influenza in swine, pandemic threats for humans, and important research questions related to the human-swine interface.
Humans as major sources of influenza virus diversity in swine
While there is historical evidence for influenza outbreaks in humans, poultry, horses, and even canines dating back several centuries [28], there is evidence of only one possible localized influenza virus outbreak in swine in England prior to the 1918 H1N1 ‘Spanish flu’ pandemic [29]. During early stages of the 1918 pandemic, outbreaks of the virus were identified in both humans and swine in the United States [30], and it remains impossible to determine whether humans first transmitted the virus to pigs, or vice versa, owing to the lack of data from that time [31]. From 1918 to 1998, this ‘classical’ swine H1N1 influenza virus, isolated in swine in 1930 [32], exhibited relatively limited antigenic evolution and caused low concern for swine producers. Beginning in the 1970s, the global evolution of swIAVs became much more complex with the introduction of an avian H1N1 virus and a human seasonal H3N2 ‘Port Chalmers-like’ virus into European swine herds, and subsequent reassortment events between them (a detailed historical characterization of global influenza virus diversity in swine has been provided in several recent reviews [33–35]).
Central to the global evolution of genetic diversity of swIAVs is the repeated introduction of IAVs of human seasonal virus origin in swine [21,36–41]. In contrast to major pandemic influenza events that last 1–3 years and occur sporadically, seasonal influenza viruses are associated with annual epidemics in humans that occur in winter in temperate regions. It remains difficult to estimate the full extent of human-to-swine transmission of seasonal viruses historically, owing to large gaps in swIAV surveillance geographically and in past decades (Figure 1). However, a conservative global estimate of 20 discrete human-to-swine transmission events was recently inferred by phylogenetic analysis of whole genome viral sequence data [42] (Figure 2). No clear patterns of geographical distribution or any ‘hot zone’ of human-to-swine transmission of IAVs is apparent once sampling bias is removed. Instead, these events have occurred multiple times on all four continents that conduct at least minimal swine surveillance: North America, South America, Asia, and Europe. It is highly likely that additional human-to-swine transmission events have gone undetected in countries where surveillance has been low or absent or in cases where the virus transmits in swine for less than five years. Notably, independent human-to-swine introductions are observed in countries where surveillance is not conducted regularly, including Argentina, Thailand, Vietnam, and Japan. In several countries long phylogenetic branch lengths separate the swine viruses from the most closely related seasonal human viruses, signifying circulation in swine for many years prior to recent detection (Figure 3). Notably, the HA and NA segments of human origin have persisted in swine at a higher rate compared to the internal gene segments [42], raising the possibility that reassortment with swine virus internal genes is associated with the adaptation of seasonal human IAVs to new swine hosts.
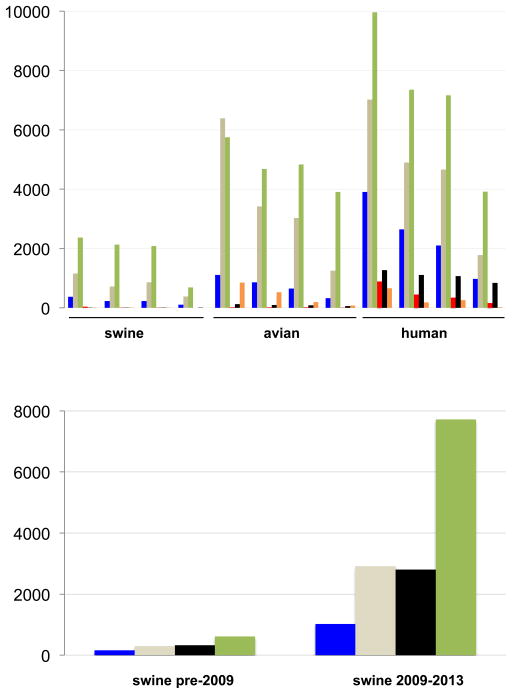
Figure 1. IAV sequence data available for swine, birds, and humans.
(a) Number of IAV sequences available at GenBank’s Influenza Virus Resource [120] for swine, birds, and humans for the full-length HA segment, full-length NA segment, whole genome, and main antigenic region of the HA (HA1), by region. The bars representing the number of viral sequences from Europe are shaded blue, Asia = light brown, North America = green, South America = red, Pacific region = black, and Africa = orange. (b) Number of swine influenza virus sequences available at GenBank for the full-length HA segment, full-length NA segment, whole genome, and partial HA gene, for the time periods 1931–2008 and 2009–2013.
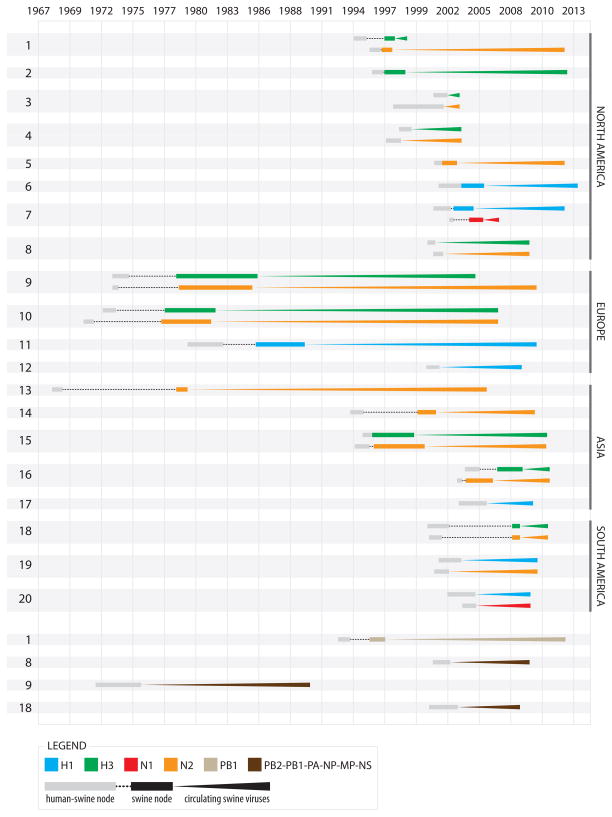
Figure 2. Introductions of human seasonal influenza viruses into swine, 1965–2013.
A summary of the 20 introductions of seasonal human IAVs into swine resulting in sustained transmission in swine (for at least one year) by segment and region. Introductions involving the HA and NA segments are depicted in the upper portion of the figure, and the subset involving internal gene segments are presented in the lower portion. Each colored line represents a human-to-swine transmission event of a segment (H1 = blue, H3 = green, N1 = red, N2 = orange, PB1 = light brown, and the full constellation of all six internal gene segments = dark brown). Each introduction is numbered 1–20. Human internal gene segments have persisted in swine for four introductions (introductions 1, 8, 9, and 18), as shown at the bottom of the figure. The timing of each human-to-swine transmission event is estimated from the times to the Most Recent Common Ancestor (tMRCAs) inferred from the maximum clade credibility (MCC) trees, with grey boxes indicating the 95% highest posterior density (HPD) interval between the swine clade and most closely related human viruses, and the black box indicating the 95% HPD interval for the swine clade only. Each line extends forward in time up to the most recently sampled swine virus of that lineage. Copyright © American Society for Microbiology, [Journal of Virology, 88, 2014, 10110–10119, doi: 10.1128/JVI.01080-14] [42] reprinted with permission.
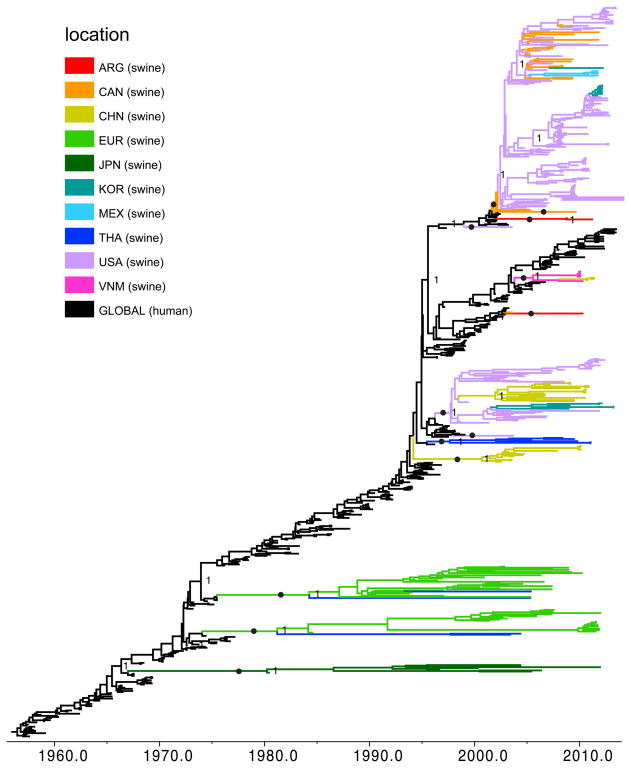
Figure 3. Phylogenetic relationships between human and swine N2 segments.
Time-scaled Bayesian MCC tree inferred for the NA (N2) sequences of 350 swine influenza viruses identified as of human seasonal virus origin and 325 human H3N2, H2N2, and H1N2 seasonal influenza viruses, collected 1957–2013. Branches of human seasonal H3N2 influenza virus origin are shaded black, while branches associated with viruses from swine are shaded by country of origin: Argentina (ARG) = red; Canada (CAN) = orange; China (CHN, including Hong Kong SAR and Taiwan) = yellow; Europe (EUR) = light green; South Korea (KOR) = teal; JPN = dark green; Mexico (MEX) = light blue; Thailand (THA) = dark blue; USA = purple; Vietnam (VNM) = pink. Posterior probabilities > 0.9 are included for key nodes, and the 13 discrete introductions of the human N2 segment into swine that are supported by high posterior probabilities and long branch lengths are identified with solid black circles. Copyright © American Society for Microbiology, [Journal of Virology, 88, 2014, 10110–10119, doi: 10.1128/JVI.01080-14] [42] reprinted with permission.
Large-scale reverse zoonosis of pH1N1 viruses
On May 2, 2009, approximately one month after the pH1N1 virus was detected in humans, pH1N1 was isolated from the first pig on a farm in Alberta, Canada in what was eventually determined to be the first documented case of human-to-swine transmission of pH1N1 [43,44]. Since then, the pH1N1 virus has transmitted repeatedly from humans to swine spanning six continents (Table 1, Figure 4), in what is arguably the largest global occurrence of reverse zoonosis of any infectious disease that has been documented to date [45]. Although the molecular clock suggests that the pH1N1 virus originated in swine approximately one decade prior to its emergence in humans [46], the virus was not detected in the large and relatively routinely sampled swine populations in North America, Europe, or Southern China prior to its emergence in humans, and where the virus originated in swine remains a key outstanding question. The most parsimonious explanation is that pH1N1 evolved in swine in Mexico, where pH1N1 first emerged in humans, or possibly a neighboring country in Central or South America where surveillance in swine is limited (Figure 1b). Still, there is no evidence that Eurasian and North American swine viruses co-circulate in any part of Latin America, and Asia is the only region to date where viruses have been detected that contain segments from both lineages that resemble pH1N1 [47]. Adding to the mystery of pH1N1’s origin, it has been demonstrated that the specific reassortment event required to generate the pH1N1 virus does not occur readily in experimentally infected pigs or cell culture [48].
Table 1.
Reports of human-origin pH1N1 viruses in swine during 2009–2014
| Location | Year | Pandemic viruses identified | Ref or GenBankaccession |
|---|---|---|---|
| North America | |||
| Canada | 2009 | pH1N1 | [121] |
| Canada | 2009 | pH1N1 | [43] |
| Mexico | 2009 | pH1N1 | [122] |
| United States | 2009–2010 | pH1N1, pH1N1 reassortantsa | [112] |
| 2010 | pH1N1 reassortants | [110] | |
| Europe | |||
| France | 2010 | pH1N1 | CCB78518 |
| Finland | 2009–2010 | pH1N1 | [123] |
| Germany | 2009–2010 | pH1N1 reassortants | [116] |
| Germany | 2009–2010 | pH1N1 reassortants | [114] |
| Hungary | 2010–2011 | pH1N1 reassortants | [124] |
| Italy | 2009 | pH1N1 | [125] |
| Italy | 2013 | pH1N1 reassortants | [117] |
| Norway | 2009 | pH1N1 | [126] |
| Norway | 2009–2010 | pH1N1 | [127] |
| Poland | 2012 | pH1N1 | AHJ57400 |
| Russia | 2009–2011 | pH1N1 | AFV31481 |
| Spain | 2010–2011 | pH1N1 | [128] |
| United Kingdom | 2010 | pH1N1 reassortants | [115] |
| United Kingdom | 2013 | pH1N1 | [129] |
| Asia | |||
| China | 2009–2010 | pH1N1 reassortants | [118] |
| China | 2010–2011 | pH1N1 reassortants | [113] |
| Japan | 2009–2010 | pH1N1 | [130] |
| India | 2009 | pH1N1 | [24] |
| Singapore | 2009 | pH1N1 | ACY82404 |
| South Korea | 2009 | pH1N1 | [131] |
| South Korea | 2009 | pH1N1 | [132] |
| Sri Lanka | 2009–2012 | pH1N1 | [25] |
| Thailand | 2009–2010 | pH1N1 | [133] |
| Thailand | 2010–2012 | pH1N1, pH1N1 reassortants | [111] |
| Vietnam | 2010–2011 | pH1N1 | [130] |
| Vietnam | [134] | ||
| South/Central America and Caribbean | |||
| Argentina | 2009 | pH1N1 | [20] |
| Brazil | 2009–2010 | pH1N1 | [22] |
| Colombia | 2009–2011 | pH1N1 | [23] |
| Costa Rica | 2010 | pH1N1 | ADW93781 |
| Cuba | 2010 | pH1N1 | [135] |
| Africa | |||
| Cameroon | 2010 | pH1N1 | [19] |
| Nigeria | 2011 | pH1N1 | [49] |
| Togo | 2013 | pH1N1 | AIO11674 |
| Pacific | |||
| Australia | 2009 | pH1N1 | [27] |
| Australia | 2009 | pH1N1 | [26] |
pH1N1 reassortants refers to influenza viruses that were generated by reassortment between pH1N1 and other co-circulating swine viruses, and therefore have some, but not all, segments of pH1N1 origin
Figure 4. Countries where pH1N1 has been identified in swine.
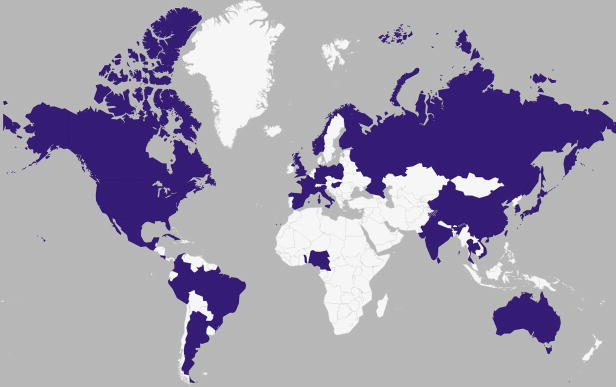
See also Table 1. Countries shaded in purple represent those where pH1N1 viruses of human origin have been detected in swine since 2009.
Since 2009, increases in the intensity and geographic range of surveillance for swIAVs (Figure 1b) have revealed at least 49 introductions of pH1N1 from humans into swine [45]. This estimate is conservative due to reliance on high bootstrap values, low availability of IAV sequence data from swine in many regions, and higher rates of sequencing of the HA and NA segments, which have not transmitted onward in swine as extensively as the internal pH1N1 gene segments due to reassortment. Detection of human-origin pH1N1 in swine herds was reported in many countries where influenza had not previously been documented as an important swine health problem, including Australia [26,27], India [24], Sri Lanka [25], Colombia [23], and Cameroon [19] (Table 1, Figure 4). The introductions of pH1N1 into the large commercial swine herds in Brazil [22] and Nigeria [49] have caused outbreaks that are of particular concern, as these possibly endemic viruses could complicate swine health and production in regions with high hog densities but lacking experience in diagnosing and controlling influenza in swine.
Refining models of influenza A virus ecology
The relatively lower barrier of cross-species transmission of pH1N1 from humans back to swine, compared to other human viruses, may be explained in part by the fact that pH1N1 was originally a swine virus. Still, the human-adapted pH1N1 shows little evidence of transmitting from swine back to humans, at least in terms of stable introductions with onward human-to-human transmission [45]. Direct comparisons of the frequency of human-to-swine versus swine-to-human IAV transmission remains complicated by substantial differences in the intensity of surveillance in humans and swine (Figure 1, Box 1). In general, the lower availability of sequence data from swine means that many human-to-swine transmission events that do not result in substantial onward transmission in swine are likely to go undetected, whereas transient spillovers of swine viruses in humans will be identified due to higher sampling in humans (e.g., the detection of >300 H3N2v spillovers from swine to humans [6]). However, additional swine data is needed to improve our understanding of inter-species transmission dynamics in both directions.
Even excluding the high number of human-to-swine transmission events of the recent pH1N1 virus, a greater number of human influenza viruses have crossed species boundaries to successfully transmit in swine (n = 20) [42] than the number of all other IAV cross-species events with sustained transmission combined. Host jumps of IAV that result in stable transmission in the new host are rare events, at least in recent decades when genetic sequence data is available. Based on genetic data, there have been three avian-to-human transmission events resulting in sustained onward transmission. These avian-to-human transmissions are associated with the 1918, 1957, and 1968 pandemics, although it remains unclear whether a proposed intermediary mammalian host facilitated the adaptation of avian viruses to humans in any of these cases [9]. Two avian-to-swine transmission events involving sustained onward transmission have been documented (triple reassortant viruses and Eurasian viruses) [12,50], three avian-to-equine transmissions (H7N7 and two H3N8 transmissions) [10,51,52], one avian-to-canine transmission (H3N2) [11], one equine-to-canine transmission (H3N8) [13], and one swine-to-human* (2009 H1N1 pandemic) [5] (*the highly localized outbreak of H1N1 swine influenza in Fort Dix, NJ in 1976 was not considered sustained transmission by our definition[53]) (Figure 5a). For simplicity we adhere to a strict definition of sustained transmission for at least one year in the secondary host, and exclude transmission events between different species of birds, which exhibit extremely low levels of host restriction [54]. Although major outbreaks of IAVs of avian, human, and swine origin also have been observed in other mammalian hosts, including domestic turkeys [55–57] and seals [58–60], at this time there is insufficient evidence of sustained onward transmission.
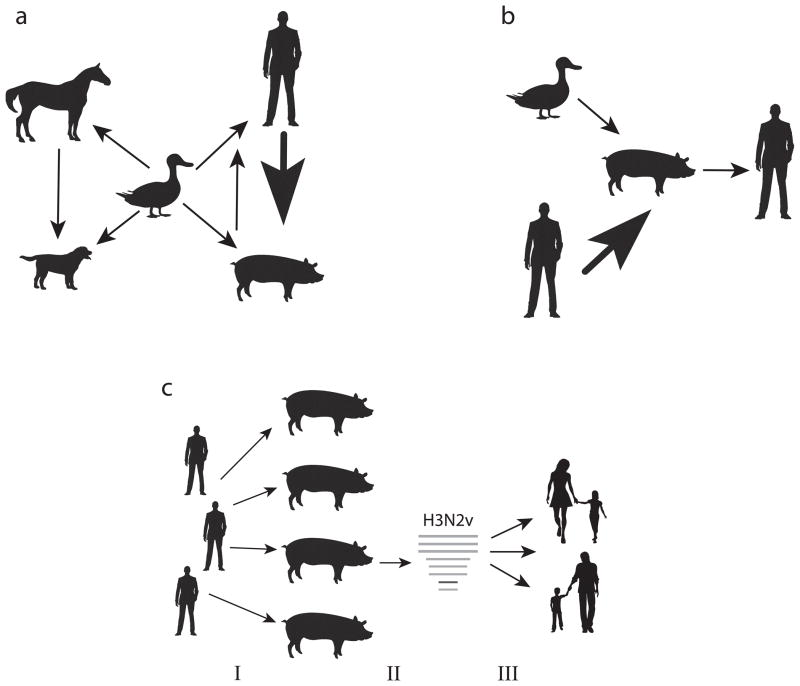
Figure 5. A model for the ecology of influenza A viruses.
(a) Cross-species transmission between avian species (thought to be the main reservoir for IAV diversity) and humans (H1N1, H2N2, and H3N2), equines [H3N8 (2x) and H7N7], canines (H3N2), and swine (H1N1 and triple reassortant PB2/PA), as well as transmission between mammalian species: equine-to-canine (H3N8), swine-to-human (H1N1), and human-to-swine (H1N1, H1N2, and H3N2, at least 20 times). Width of arrows is proportional to the number of IAV transmission events between species that result in endemic circulation of a virus in a new host (transient spillovers not included). (b) Model for the role of swine as ‘mixing vessels’ for the evolution of pandemic viruses (width of arrows also are proportional to the frequency of transmission). (c) Model for the evolution of H3N2v viruses in swine. Large-scale transmission of pH1N1 viruses from humans to swine seeds pH1N1 in swine populations globally (I); reassortment between pH1N1 viruses and co-circulating triple reassortant H3N2 viruses in the United States, generating novel reassortant H3N2v variants with seven triple reassortant H3N2 virus segments and the MP segment of pH1N1 origin (II); and transmission of > 300 H3N2v viruses from swine to humans, resulting in one adult fatality, in the United States during 2011–2013 (III).
Notably, the frequency of avian-to-swine transmission is lower than the frequency of human-to-swine transmission, by our definition of viral introductions with onward transmission (Figure 5b). To date, there is evidence of only two full or partial avian influenza viruses successfully adapting to transmit stably in swine: the avian-origin Eurasian H1N1 swine viruses [12] and the avian-origin PB2 and PA segments associated with the triple reassortant viruses that emerged in North American swine in the 1990s [50]. A wide range of avian influenza virus subtypes have transiently infected swine in Asia (H1N1 [61], H3N2 [62], H3N8 [63], H4N1 [64], H4N8 [65], H5N1 [66–68], H5N2 [69], H6N6 [70], H7N2 [71], H9N2 [72–75], H10N5 [76], and H11N6), North America (H1N1 [77], H2N3 [78], H3N3 [77], and H4N6 [79]), and Europe (H1N7 [80]). The detection of avian-swine reassortant H2N3 viruses in swine in 2006 in Missouri, USA raised a particular concern for humans, but also did not sustain viral transmission in swine, and there was no evidence of infection of humans [81]. The higher rate of observed transmission of human viruses to swine could potentially be related to surveillance bias. However, the matrix protein-based primers currently used for IAV diagnostics in swine are able to detect a broad diversity of avian and human viral subtypes. Furthermore, although the relatively low levels of surveillance in swine compared to humans and avian species (Figure 1a) will not identify all transient avian influenza virus spillovers, avian influenza viruses with sustained long-term transmission in swine are far more likely to be detected, suggesting that sustained circulation of avian influenza viruses in swine is indeed a rare event. It is possible that the biological barrier to transmission of avian influenza viruses to swine may be similarly high as for other mammals, such as humans, dogs, and horses. Recent experimental studies indicate that the barriers to avian influenza virus replication and transmission are high in both swine and humans [82,83], and the swine trachea contains a preponderance of α-2,6-linked receptors, similar to humans [84,85]. However, further understanding of the distribution of sialic acid receptors in swine and human hosts and their comparative level of binding with influenza viruses of different host origins is greatly needed.
Questions about the ecology of human-to-swine transmission
Compared to the extensive study of the zoonotic transmission of H3N2v viruses from swine to humans [86,87], including the capacity of swine with asymptomatic influenza infections to efficiently transmit viruses to humans, mainly children, within the agricultural fair setting [88], relatively little is known about the ecological and immunological circumstances under which human IAVs transmit to swine. This lack of understanding undermines efforts to evaluate potential biosecurity measures, such as vaccination of swine workers. Presumably, human-to-swine transmission is associated with peaks in human influenza activity (pandemic waves or seasonal winter epidemics in temperate regions), but data from swine is not sufficiently resolved at this time to capture such patterns. The higher rate of H3N2 transmission from human to swine compared to seasonal H1N1 viruses since 1968 is consistent with the dominance of the H3N2 subtype in humans. However, at least three introductions of reassortant human H1N2 viruses into swine have been observed [42], despite only circulating in humans from 2001–2003.
Differences in immunity between humans and swine could explain, at least in part, why viruses transmit more frequently from humans to swine than in the reverse direction. Young pigs typically live only six months before going to market. In contrast, adult swine workers are likely to build up immunity over time to swIAVs through exposure, and older humans in general may have at least partial cross-immunity to human-origin swine influenza viruses as a result of previous exposure to the human precursors of the swine viruses. The high density of pigs in commercial settings also may provide an opportunity for human-origin viruses that are imperfectly adapted to swine to continue to transmit between highly connected animals while evolving higher fitness. The co-circulation of multiple swIAVs within herds also may facilitate host jumps by providing more opportunities for reassortment. As our broad understanding of how IAVs successfully jump hosts has been limited to date by the relatively small number of host-switch events observed in nature (Figure 5a), the less constrained human-to-swine interface may provide a useful model system for investigating the interactions between immunity, ecology, and evolution during host switches.
Human-origin viral diversity reduces efficacy of swine influenza vaccines
The repeated introduction of human-origin viral diversity, particularly in the form of multiple antigenically distinct co-circulating HA and NA proteins, has greatly complicated the development of effective vaccines for the control of influenza in swine. In the United States vaccines are used primarily in sows to provide passive antibody to piglets and, to a lesser extent, during the grow/finish phase of production to decrease disease, lung lesions, and transmission [89,90]. One of the commercially available US influenza vaccines is a quadravalent whole inactivated virus (WIV) vaccine that contains four strains: classical H1 (gamma), which has circulated in swine since the 1918 pandemic, and three strains of more recent human origin: H1 delta-1, H1 delta-2, and H3 (cluster 4). Following the introduction of pH1N1 viruses from humans into US swine herds, a monovalent pH1N1 vaccine also was developed. To further complicate the matter, a phenomenon called ‘vaccine-associated enhanced respiratory disease’ (VAERD) [91] has been observed experimentally in pigs when there is substantial antigenic distance between the vaccine and challenge strains of the same H1 subtype (e.g., any of the classical, delta, and pandemic H1 viruses). Live attenuated influenza virus (LAIV) vaccines administered by the mucosal route mimic natural infection and have the potential to elicit broader cross-protective immunity [92–95] without inducing VAERD [96], and may present the best strategy for vaccinating against the substantial antigenic variation of human-origin viruses that circulate in US swine.
The future of swIAV surveillance and research
Despite the evidence, it remains counterintuitive to view humans as major sources of disease for swine. This bias is perpetuated by the imbalance in surveillance in humans versus swine (Figure 1), which obscures an accurate understanding of viral ecology in which humans and swine exchange pathogens in both directions. Sample bias continues to be a major impediment to understanding swIAV evolution and diversity, including capturing the full scale of human-to-swine transmission. Major gaps in surveillance of influenza in swine were recently highlighted by the identification of an entirely novel serotype in swine and cattle that are phylogenetically distinct from the diversity of influenza A, B, and C viruses [97]. Characterizing the global diversity of human-origin IAVs in swine will require expanding surveillance, particularly in countries with large swine populations but relatively low surveillance currently (e.g., Brazil, Vietnam, Russia). Substantial temporal and spatial biases in sampling of swine viruses (Figure 1) also make it difficult to infer the geographical origins of human-to-swine transmission events, especially when the virus is first sampled in pigs many years following initial introduction [42]. Globally, swIAVs are rarely sampled in a population-based manner, undermining our understanding of the population dynamics of co-circulating viruses over time.
Recognition of the central role of humans in the evolution of swIAV diversity has important implications for future research and surveillance, including the need for strengthened collaboration between the animal and human health sides. Relations between the human and swine influenza sides have improved considerably since the 2009 ‘swine flu’ pandemic. The regrettable naming of ‘swine flu’ perpetuated a global misperception that pigs presented an immediate disease risk for humans, which resulted in inhumane conditions and widespread pig culling in several countries [44,98] and cost the US pork industry billions of dollars from unnecessary export bans and reduced pork consumption. Although American and European swine producers regularly collect respiratory samples from sick pigs for diagnostic investigation, many of these samples are sent to private diagnostic companies to inform the design of autogenous influenza vaccines. As a result, only a relatively small number of sequences and/or isolates are made available for public research.
Our knowledge of reverse zoonosis is even lower in other species and wild animal populations that present additional challenges for surveillance [99]. During the 2009 pandemic, pH1N1 viruses of presumed human origin were isolated from a range of species, including felines [100], canines [101], and seals [59]. Human influenza B viruses (IBVs) have a history of causing major outbreaks in seals [102,103], raising the possibility that phocines sustain permanent reservoirs of human-origin IBV diversity. A number of other pathogens of human origin also have been observed in wild and domestic animals [99], although onward transmission of these pathogens is rarely substantiated. Our greater understanding of the human-swine interface for influenza invites further investigation of additional pathogens, for example rotaviruses, that are known to transmit from swine and other livestock to humans but for which the human-animal interface remains poorly understood, particularly from a bi-directional perspective.
Improving surveillance of swIAVs is also important for understanding how evolutionary processes amplify and disseminate viral diversity in pig populations. Several recent studies have revealed the importance of viral migration via swine movements [104,105] and reassortment with endemic swine viruses [106,107]. Increased whole-genome sequencing of swIAVs in recent years has been particularly important in revealing the frequency of reassortment in swine hosts and the diversity of novel IAV genotypes that are generated, including those such as H3N2v that present threats to humans [106–109] (Figure 5c). The frequency of reassortment between IAVs in swine greatly exceeds the reassortment rate observed in humans. As a case in point, pH1N1 viruses have reassorted extensively with many diverse IAVs in swine hosts [108,110–118] but show no evidence of reassorting with co-circulating seasonal H3N2 viruses in humans. The difference in reassortment rate between humans and swine is still not fully explained, but may relate to higher rates of co-infection in swine due to weaker cross-immunity and differences in fitness. Insights into population dynamics, including patterns of reassortment, require unbiased data that either have been selected randomly from passive surveillance or systematically sampled in swine through active surveillance [119], rather than sequencing ad hoc or selectively for specific genotypes of interest.
Concluding remarks
Recognizing the important role of human-to-swine transmission in the evolution of swIAV diversity requires that we refine our concept of the ‘mixing vessel’ (Figure 5b), which has perpetuated an over-simplified understanding of IAV ecology. Swine can be considered mixing vessels in the sense that pigs are more capable than humans of harboring a large number of co-circulating IAV lineages that can generate diverse combinations of segments via reassortment. But at this time there is insufficient evidence that swine are more suitable hosts than humans for stable transmission of avian viruses. In theory, humans could be regarded as ‘mixing vessel’ intermediaries for pigs during the 1968 pandemic, as there is evidence that humans transmitted these reassortant avian-origin H3N2 viruses to swine but no evidence from the field that these viruses circulated in swine prior to emergence in humans. However, as little sequence data from swIAVs are available from the time of the 1968 pandemic, it is possible that future advancements in surveillance and archival sequencing will further revise our understanding of the human-swine interface. At this time, however, it is critical that we employ our improved knowledge of IAV ecology to enhance biosecurity and vaccination strategies in swine workers, learn more about the processes of adaptive evolution involved in host jumps between humans and swine, and further expand swIAV surveillance globally.
Highlights.
Humans transmit far more influenza viruses to swine than swine transmit to humans.
Human-to-swine transmission is key to the evolution of influenza diversity in swine.
In effect, humans sow the seeds of future pandemics by infecting pigs.
A balanced view of the bi-directional nature of the human-animal interface is needed.
Acknowledgments
We would like to thank NIH intern Adaeze Ezeofor for her contributions to the organization and identification of the many journal references cited in this manuscript. We are also grateful to Drs Cecile Viboud, NIH, and Edward C. Holmes, University of Sydney, for their insightful comments on earlier drafts of this manuscript. This research was conducted within the context of the Multinational Influenza Seasonal Mortality Study (MISMS) (http://www.origem.info/misms/), an on-going international collaborative effort to understand influenza epidemiology and evolution, led by the Fogarty International Center, National Institutes of Health (NIH), with funding from the Office of Global Affairs at the US Department of Health and Human Services (DHHS) (MIN). AV is funded by US Department of Agriculture-ARS. USDA is an equal opportunity provider and employer. Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the USDA.
Glossary
- Antigenic drift
the continual evasion of host immunity by the gradual accumulation of mutations in the hemagglutinin and neuraminidase surface glycoproteins that change the antigenic structure of the influenza A virus
- Bayesian phylogeography
the inference of viral migration patterns using time-scaled phylogenies inferred in a Bayesian framework in which prior knowledge assesses the probability of model parameters in the presence of new data
- Hemagglutinin (HA)
an influenza virus surface glycoprotein, which binds to sialic acid receptors to assist viral entry into host epithelial cells. Eighteen HA serotypes are present in animal species
- Mixing vessel
a host species with the capacity to be co-infected with multiple genetically distinct viruses of different animal origins that are able to reassort to generate genetically novel viruses. Swine are considered to be ‘mixing vessels’ for reassortment between human and avian influenza viruses
- Molecular clock
the observation that the rate of molecular evolution is relatively constant for a given lineage, which provides a useful tool for estimating the timing of events in evolutionary history
- Pandemic
an influenza outbreak associated with a new viral subtype that occurs over a large geographic area (often globally)
- Reassortment
a form of genetic recombination in which two influenza viruses co-infect a single cell and exchange entire RNA segments to form genetically novel viruses
- Reverse zoonosis
an infectious disease that can be transmitted from a human to a non-human host (sometimes by a vector)
- Zoonosis
an infectious disease that can be transmitted from a non-human host to a human (sometimes by a vector)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Morens DM, et al. H7N9 avian influenza A virus and the perpetual challenge of potential human pandemicity. MBio. 2013;4:e00445–13. doi: 10.1128/mBio.00445-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scholtissek C. Pigs as the “mixing vessel” for the creation of new pandemic influenza A viruses. Med Princip Pr. 1990;2:65–71. [Google Scholar]
- 3.Kida H, et al. Potential for transmission of avian influenza viruses to pigs. J Gen Virol. 1994;75:2183–2188. doi: 10.1099/0022-1317-75-9-2183. [DOI] [PubMed] [Google Scholar]
- 4.Ma W, et al. The role of swine in the generation of novel influenza viruses. Zoo Pub Heal. 2009;56:326–337. doi: 10.1111/j.1863-2378.2008.01217.x. [DOI] [PubMed] [Google Scholar]
- 5.Garten RJ, et al. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science. 2009;325:197–201. doi: 10.1126/science.1176225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Epperson S, et al. Human infections with influenza A(H3N2) variant virus in the United States, 2011–2012. Clin Infect Dis. 2013;57:S4–S11. doi: 10.1093/cid/cit272. [DOI] [PubMed] [Google Scholar]
- 7.Webster RG, et al. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Webster RG, Laver WG. The origin of pandemic influenza. Bull World Heal Organ. 1972;47:449–452. [PMC free article] [PubMed] [Google Scholar]
- 9.Kawaoka Y, et al. Avian-to-human transmission of the PB1 gene of influenza A viruses in the 1957 and 1968 pandemics. J Virol. 1989;63:4603–4608. doi: 10.1128/jvi.63.11.4603-4608.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scholtens RG, et al. U.S. Epizootic of equine influenza, 1963. Public Heal Rep. 1964;79:393–402. [PMC free article] [PubMed] [Google Scholar]
- 11.Li S, et al. Avian-origin H3N2 canine influenza A viruses in Southern China. Infect Genet Evol. 2010;10:1286–1288. doi: 10.1016/j.meegid.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schultz U, et al. Evolution of pig influenza viruses. Virology. 1991;183:61–73. doi: 10.1016/0042-6822(91)90118-u. [DOI] [PubMed] [Google Scholar]
- 13.Crawford PC, et al. Transmission of equine influenza virus to dogs. Science. 2005;310:482–485. doi: 10.1126/science.1117950. [DOI] [PubMed] [Google Scholar]
- 14.Imai M, Kawaoka Y. The role of receptor binding specificity in interspecies transmission of influenza viruses. Curr Opin Virol. 2012;2:160–167. doi: 10.1016/j.coviro.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shinde V, et al. Triple-reassortant swine influenza A (H1) in humans in the United States, 2005–2009. N Engl J Med. 2009;360:2616–2625. doi: 10.1056/NEJMoa0903812. [DOI] [PubMed] [Google Scholar]
- 16.Shu B, et al. Genetic analysis and antigenic characterization of swine origin influenza viruses isolated from humans in the United States, 1990–2010. Virology. 2012;422:151–160. doi: 10.1016/j.virol.2011.10.016. [DOI] [PubMed] [Google Scholar]
- 17.Pascua PNQ, et al. Virulence and transmissibility of H1N2 influenza virus in ferrets imply the continuing threat of triple-reassortant swine viruses. Proc Natl Acad Sci USA. 2012;109:15900–15905. doi: 10.1073/pnas.1205576109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barman S, et al. Pathogenicity and transmissibility of North American triple reassortant swine influenza A viruses in ferrets. PLoS Path. 2012;8:e1002791. doi: 10.1371/journal.ppat.1002791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Njabo KY, et al. Pandemic A/H1N1/2009 influenza virus in swine, Cameroon, 2010. Vet Microbiol. 2012;156:189–192. doi: 10.1016/j.vetmic.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pereda A, et al. Pandemic (H1N1) 2009 outbreak on pig farm, Argentina. Emerg Infect Dis. 2010;16:304–307. doi: 10.3201/eid1602.091230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cappuccio Ja, et al. Outbreak of swine influenza in Argentina reveals a non-contemporary human H3N2 virus highly transmissible among pigs. J Gen Virol. 2011;92:2871–2878. doi: 10.1099/vir.0.036590-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rajão DS, et al. Genetic characterization of influenza virus circulating in Brazilian pigs during 2009 and 2010 reveals a high prevalence of the pandemic H1N1 subtype. Influ Other Respi Viruses. 2012;7:783–790. doi: 10.1111/irv.12072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramirez-Nieto GC, et al. First isolation and identification of H1N1 swine influenza viruses in Colombian pig farms. Health (Irvine Calif) 2012;04:983–990. [Google Scholar]
- 24.Nagarajan K, et al. Influenza A H1N1 virus in Indian pigs & its genetic relatedness with pandemic human influenza A 2009 H1N1. Indian J Med Res. 2010;132:160–167. [PubMed] [Google Scholar]
- 25.Perera HKK, et al. Swine influenza in Sri Lanka. Emerg Infect Dis. 2013;19:481–484. doi: 10.3201/eid1903.120945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holyoake PK, et al. The first identified case of pandemic H1N1 influenza in pigs in Australia. Aust Vet J. 2011;89:427–431. doi: 10.1111/j.1751-0813.2011.00844.x. [DOI] [PubMed] [Google Scholar]
- 27.Deng Y, et al. Transmission of influenza A (H1N1) 2009 pandemic viruses in Australian swine. Influ Other Respi Viruses. 2012;6:42–47. doi: 10.1111/j.1750-2659.2012.00337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morens DM, Taubenberger JK. Historical thoughts on influenza viral ecosystems, or behold a pale horse, dead dogs, failing fowl, and sick swine. Influ Other Respi Viruses. 2010;4:327–37. doi: 10.1111/j.1750-2659.2010.00148.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morens DM, Taubenberger JK. A possible outbreak of swine influenza, 1892. Lancet Infect Dis. 2014;14:169–72. doi: 10.1016/S1473-3099(13)70227-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.JSK A practical method for field diagnosis of swine diseases. Am J Vet Med. 1919;14:468–470. [Google Scholar]
- 31.Worobey M, et al. A synchronized global sweep of the internal genes of modern avian influenza virus. Nature. 2014;508:254–257. doi: 10.1038/nature13016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shope RE. The etiology of swine influenza. Science. 1931;73:214–5. doi: 10.1126/science.73.1886.214. [DOI] [PubMed] [Google Scholar]
- 33.Zhu H, et al. History of Swine influenza viruses in Asia. Curr Top Microbiol Immunol. 2013;370:57–68. doi: 10.1007/82_2011_179. [DOI] [PubMed] [Google Scholar]
- 34.Brown IH. History and epidemiology of Swine influenza in Europe. Curr Top Microbiol Immunol. 2013;370:133–146. doi: 10.1007/82_2011_194. [DOI] [PubMed] [Google Scholar]
- 35.Lorusso A, et al. Contemporary epidemiology of North American lineage triple reassortant influenza A viruses in pigs. Curr Top Microbiol Immunol. 2013;370:113–132. doi: 10.1007/82_2011_196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ottis K, et al. Human influenza A viruses in pigs: isolation of a H3N2 strain antigenically related to A/England/42/72 and evidence for continuous circulation of human viruses in the pig population. Arch Virol. 1982;73:103–108. doi: 10.1007/BF01314719. [DOI] [PubMed] [Google Scholar]
- 37.Castrucci M, et al. Genetic Reassortment between Avian and Human Influenza A Viruses in Italian Pigs. Virology. 1993;193:503–506. doi: 10.1006/viro.1993.1155. [DOI] [PubMed] [Google Scholar]
- 38.Olsen C, et al. Virologic and serologic surveillance for human, swine and avian influenza virus infections among pigs in the north-central United States. Arch Virol. 2000;145:1399–1419. doi: 10.1007/s007050070098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karasin A, et al. Identification of human H1N2 and human-swine reassortant H1N2 and H1N1 influenza A viruses among pigs in Ontario, Canada (2003 to 2005) J Clin Microbiol. 2006;44:1123–1126. doi: 10.1128/JCM.44.3.1123-1126.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu H, et al. Further evidence for infection of pigs with human-like H1N1 influenza viruses in China. Virus Res. 2009;140:85–90. doi: 10.1016/j.virusres.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 41.Karasin AI, et al. Genetic characterization of H3N2 influenza viruses isolated from pigs in North America, 1977–1999: evidence for wholly human and reassortant virus genotypes. Virus Res. 2000;68:71–85. doi: 10.1016/s0168-1702(00)00154-4. [DOI] [PubMed] [Google Scholar]
- 42.Nelson MI, Wentworth DE, Culhane MR, Vincent AL, Viboud C, LaPointe M, Holmes ECDS. Introductions and evolution of human-origin seasonal influenza A viruses in multinational swine populations. J Virol. 88:10110–10119. doi: 10.1128/JVI.01080-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Howden K, Brockhoff E. An investigation into human pandemic influenza virus (H1N1) 2009 on an Alberta swine farm. Can Vet J. 2009;50:1153–1161. [PMC free article] [PubMed] [Google Scholar]
- 44.Forgie S, Keenliside J. Swine outbreak of pandemic influenza A virus on a Canadian research farm supports human-to-swine transmission. Clin Infect Dis. 2011;52:10–18. doi: 10.1093/cid/ciq030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nelson MI, et al. Global transmission of influenza viruses from humans to swine. J Gen Virol. 2012;93:2195–2203. doi: 10.1099/vir.0.044974-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith GJD, et al. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature. 2009;459:1122–1125. doi: 10.1038/nature08182. [DOI] [PubMed] [Google Scholar]
- 47.Lam TTY, et al. Reassortment events among swine influenza A viruses in China: implications for the origin of the 2009 influenza pandemic. J Virol. 2011;85:10279–10285. doi: 10.1128/JVI.05262-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma W, et al. North American triple reassortant and Eurasian H1N1 swine influenza viruses do not readily reassort to generate a 2009 pandemic H1N1-like virus. MBio. 2014;5:e00919–13. doi: 10.1128/mBio.00919-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meseko CA, et al. Detection and isolation of 2009 pandemic influenza A/H1N1 virus in commercial piggery, Lagos Nigeria. Vet Microbiol. 2014;168:197–201. doi: 10.1016/j.vetmic.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 50.Zhou NN, et al. Genetic reassortment of avian, swine, and human influenza A viruses in American pigs. J Virol. 1999;73:8851–8856. doi: 10.1128/jvi.73.10.8851-8856.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guo Y, et al. Characterization of a new avian-like influenza A virus from horses in China. Virology. 1992;188:245–255. doi: 10.1016/0042-6822(92)90754-d. [DOI] [PubMed] [Google Scholar]
- 52.Sovinova O, et al. Isolation of a virus causing respiratory disease in horses. Acta Virol. 1958;2:52–61. [PubMed] [Google Scholar]
- 53.Kendal AP, et al. Identification and preliminary antigenic analysis of swine influenza-like viruses isolated during an influenza outbreak at Fort Dix, New Jersey. J Infect Dis. 1977;136:S381–S385. doi: 10.1093/infdis/136.supplement_3.s381. [DOI] [PubMed] [Google Scholar]
- 54.Dugan VG, et al. The evolutionary genetics and emergence of avian influenza viruses in wild birds. PLoS Path. 2008;4:e1000076. doi: 10.1371/journal.ppat.1000076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Berhane Y, et al. Molecular and antigenic characterization of reassortant H3N2 viruses from turkeys with a unique constellation of pandemic H1N1 internal genes. PLoS One. 2012;7:e32858. doi: 10.1371/journal.pone.0032858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wood GW, et al. The nucleotide sequence of the HA1 of the haemagglutinin of an HI avian influenza virus isolate from turkeys in Germany provides additional evidence suggesting recent transmission from pigs. Avian Path. 1997;26:347–55. doi: 10.1080/03079459708419217. [DOI] [PubMed] [Google Scholar]
- 57.Olsen CW, et al. Triple reassortant H3N2 influenza A viruses, Canada, 2005. Emerg Infect Dis. 2006;12:1132–5. doi: 10.3201/eid1207.060268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Callan RJ, et al. The appearance of H3 influenza viruses in seals. J Gen Virol. 1995;76:199–203. doi: 10.1099/0022-1317-76-1-199. [DOI] [PubMed] [Google Scholar]
- 59.Goldstein T, et al. Pandemic H1N1 influenza isolated from free-ranging northern elephant seals in 2010 off the central California coast. PLoS One. 2013;8:e62259. doi: 10.1371/journal.pone.0062259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Anthony SJ, et al. Emergence of fatal avian influenza in New England harbor seals. MBio. 2012;3:e00166–12. doi: 10.1128/mBio.00166-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guan Y, et al. Emergence of avian H1N1 influenza viruses in pigs in China. J Virol. 1996;70:8041–6. doi: 10.1128/jvi.70.11.8041-8046.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Su S, et al. Complete genome sequence of a novel avian-like H3N2 swine influenza virus discovered in Southern China. J Virol. 2012;86:9533. doi: 10.1128/JVI.01315-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tu J, et al. Isolation and molecular characterization of equine H3N8 influenza viruses from pigs in China. Arch Virol. 2009;154:887–90. doi: 10.1007/s00705-009-0381-1. [DOI] [PubMed] [Google Scholar]
- 64.Hu Y, et al. Complete genome sequence of a novel H4N1 influenza virus isolated from a pig in central China. J Virol. 2012;86:13879. doi: 10.1128/JVI.02726-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Su S, et al. Complete genome sequence of an avian-like H4N8 swine influenza virus discovered in southern China. J Virol. 2012;86:9542. doi: 10.1128/JVI.01475-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nidom Ca, et al. Influenza A (H5N1) viruses from pigs, Indonesia. Emerg Infect Dis. 2010;16:1515–1523. doi: 10.3201/eid1610.100508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li HY, Yu KZ, Yang HL, Xin XG, Chen JY, Zhao P, Bi YZ, Chen HL. Isolation and characterization of H5N1 and H9N2 influenza viruses from pigs in China. Chin J Prev Vet Med. 2004;26:1–6. [Google Scholar]
- 68.He L, et al. Isolation and characterization of two H5N1 influenza viruses from swine in Jiangsu Province of China. Arch Virol. 2013;158:2531–41. doi: 10.1007/s00705-013-1771-y. [DOI] [PubMed] [Google Scholar]
- 69.Lee JH, et al. Isolation and genetic characterization of H5N2 influenza viruses from pigs in Korea. J Virol. 2009;83:4205–4215. doi: 10.1128/JVI.02403-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang G, et al. Identification of an H6N6 swine influenza virus in southern China. Infect Genet Evol. 2011;11:1174–1177. doi: 10.1016/j.meegid.2011.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kwon TY, et al. Genetic characterization of H7N2 influenza virus isolated from pigs. Vet Microbiol. 2011;153:393–397. doi: 10.1016/j.vetmic.2011.06.011. [DOI] [PubMed] [Google Scholar]
- 72.Rui-Hua Z, et al. Molecular characterization and pathogenicity of swine influenza H9N2 subtype virus A/swine/HeBei/012/2008/(H9N2) Acta Virol. 2011;55:219–226. doi: 10.4149/av_2011_03_219. [DOI] [PubMed] [Google Scholar]
- 73.Peiris J, et al. Cocirculation of avian H9N2 and contemporary “human” H3N2 influenza A viruses in pigs in southeastern China: potential for genetic reassortment? J Virol. 2001;75:9679–9686. doi: 10.1128/JVI.75.20.9679-9686.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cong YL, et al. Swine infection with H9N2 influenza viruses in China in 2004. Virus Genes. 2008;36:461–469. doi: 10.1007/s11262-008-0227-z. [DOI] [PubMed] [Google Scholar]
- 75.Yu H, et al. Genetic diversity of H9N2 influenza viruses from pigs in China: a potential threat to human health? Vet Microbiol. 2011;149:254–261. doi: 10.1016/j.vetmic.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 76.Wang N, et al. Complete genome sequence of an H10N5 avian influenza virus isolated from pigs in central China. J Virol. 2012;86:13865–13866. doi: 10.1128/JVI.02687-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Karasin AI, et al. Characterization of avian H3N3 and H1N1 influenza A viruses isolated from pigs in Canada. J Clin Microbiol. 2004;42:4349–4354. doi: 10.1128/JCM.42.9.4349-4354.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ma W, et al. Identification of H2N3 influenza A viruses from swine in the United States. Proc Natl Acad Sci USA. 2007;104:20949–20954. doi: 10.1073/pnas.0710286104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Karasin aI, et al. Isolation and characterization of H4N6 avian influenza viruses from pigs with pneumonia in Canada. J Virol. 2000;74:9322–9327. doi: 10.1128/jvi.74.19.9322-9327.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Brown IH, et al. Genetic characterisation of an influenza A virus of unusual subtype (H1N7) isolated from pigs in England. Arch Virol. 1997;142:1045–50. doi: 10.1007/s007050050140. [DOI] [PubMed] [Google Scholar]
- 81.Beaudoin A, et al. Serologic survey of swine workers for exposure to H2N3 swine influenza A. Influ Other Respi Viruses. 2010;4:163–170. doi: 10.1111/j.1750-2659.2009.00127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Löndt BZ, et al. Failure to infect pigs co-housed with ducks or chickens infected experimentally with A/turkey/Turkey/1/2005 (H5N1) highly pathogenic avian influenza virus. Vet Microbiol. 2013;162L:944–8RI. doi: 10.1016/j.vetmic.2012.11.040. [DOI] [PubMed] [Google Scholar]
- 83.Moncorgé O, et al. Investigation of influenza virus polymerase activity in pig cells. J Virol. 2013;87:384–394. doi: 10.1128/JVI.01633-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Trebbien R, et al. Distribution of sialic acid receptors and influenza A virus of avian and swine origin in experimentally infected pigs. Virol J. 2011;8:434. doi: 10.1186/1743-422X-8-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Van Poucke SGM, et al. Replication of avian, human and swine influenza viruses in porcine respiratory explants and association with sialic acid distribution. Virol J. 2010;7:38. doi: 10.1186/1743-422X-7-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wong KK, et al. Transmissibility of variant influenza from swine to humans: a modeling approach. Clin Infect Dis. 2013;57(Suppl 1):S16–22. doi: 10.1093/cid/cit303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cauchemez S, et al. Using routine surveillance data to estimate the epidemic potential of emerging zoonoses: application to the emergence of US swine origin influenza A H3N2v virus. PLoS Med. 2013;10:e1001399. doi: 10.1371/journal.pmed.1001399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bowman AS, et al. Subclinical influenza virus A infections in pigs exhibited at agricultural fairs, Ohio, USA, 2009–2011. Emerg Infect Dis. 2012;18:1945–1950. doi: 10.3201/eid1812.121116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chen Q, et al. Vaccine development for protecting swine against influenza virus. Anim Heal Res Rev. 2012;13:181–195. doi: 10.1017/S1466252312000175. [DOI] [PubMed] [Google Scholar]
- 90.Van Reeth K, Ma W. Swine influenza virus vaccines: to change or not to change-that’s the question. Curr Top Microbiol Immunol. 2013;370:173–200. doi: 10.1007/82_2012_266. [DOI] [PubMed] [Google Scholar]
- 91.Gauger PC, et al. Kinetics of lung lesion development and pro-inflammatory cytokine response in pigs with vaccine-associated enhanced respiratory disease induced by challenge with pandemic (2009) A/H1N1 influenza virus. Vet Path. 2012;49:900–12. doi: 10.1177/0300985812439724. [DOI] [PubMed] [Google Scholar]
- 92.Richt JA, et al. Vaccination of pigs against swine influenza viruses by using an NS1-truncated modified live-virus vaccine. J Virol. 2006;80:11009–11018. doi: 10.1128/JVI.00787-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Masic A, et al. An eight-segment swine influenza virus harbouring H1 and H3 hemagglutinins is attenuated and protective against H1N1 and H3N2 subtypes in pigs. J Virol. 2013 doi: 10.1128/JVI.01348-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pena L, et al. Modifications in the polymerase genes of a swine-like triple-reassortant influenza virus to generate live attenuated vaccines against 2009 pandemic H1N1 viruses. J Virol. 2011;85:456–469. doi: 10.1128/JVI.01503-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Loving CL, et al. Efficacy of inactivated and live-attenuated influenza virus vaccines in pigs against infection and transmission of emerging H3N2 similar to the 2011–2012 H3N2v. J Virol. 2013;87:9895–9903. doi: 10.1128/JVI.01038-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vincent AL, et al. Live attenuated influenza vaccine provides superior protection from heterologous infection in pigs with maternal antibodies without inducing vaccine-associated enhanced respiratory disease. J Virol. 2012;86:10597–10605. doi: 10.1128/JVI.01439-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hause BM, et al. Characterization of a novel influenza virus in cattle and Swine: proposal for a new genus in the Orthomyxoviridae family. MBio. 2014;5:e00031–14. doi: 10.1128/mBio.00031-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Seef S, Jeppsson A. Is it a policy crisis or it is a health crisis? The Egyptian context--analysis of the Egyptian health policy for the H1N1 flu pandemic control. Pan Afr Med J. 2013;14:59. doi: 10.11604/pamj.2013.14.59.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Messenger AM, et al. Reverse zoonotic disease transmission (zooanthroponosis): a systematic review of seldom-documented human biological threats to animals. PLoS One. 2014;9:e89055. doi: 10.1371/journal.pone.0089055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ali A, et al. Pandemic and seasonal human influenza virus infections in domestic cats: prevalence, association with respiratory disease, and seasonality patterns. J Clin Microbiol. 2011;49:4101–5. doi: 10.1128/JCM.05415-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Song D, et al. A novel reassortant canine H3N1 influenza virus between pandemic H1N1 and canine H3N2 influenza viruses in Korea. J Gen Virol. 2012;93:551–554. doi: 10.1099/vir.0.037739-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bodewes R, et al. Recurring influenza B virus infections in seals. Emerg Infect Dis. 2013;19:511–512. doi: 10.3201/eid1903.120965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Osterhaus AD, et al. Influenza B virus in seals. Science. 2000;288:1051–3. doi: 10.1126/science.288.5468.1051. [DOI] [PubMed] [Google Scholar]
- 104.Nelson MI, et al. Spatial dynamics of human-origin H1 influenza A virus in North American swine. PLoS Path. 2011;7:e1002077. doi: 10.1371/journal.ppat.1002077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Vijaykrishna D, et al. Long-term evolution and transmission dynamics of swine influenza A virus. Nature. 2011;473:519–522. doi: 10.1038/nature10004. [DOI] [PubMed] [Google Scholar]
- 106.Lycett SJ, et al. Estimating reassortment rates in co-circulating Eurasian swine influenza viruses. J Gen Virol. 2012;93:2326–2336. doi: 10.1099/vir.0.044503-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nelson MI, et al. Genomic reassortment of influenza A virus in North American swine, 1998–2011. J Gen Virol. 2012;93:2584–2589. doi: 10.1099/vir.0.045930-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kitikoon P, et al. Genotype patterns of contemporary reassorted H3N2 virus in US swine. J Gen Virol. 2013;94:1236–1241. doi: 10.1099/vir.0.51839-0. [DOI] [PubMed] [Google Scholar]
- 109.Nelson MI, et al. Evolution of novel reassortant A/H3N2 influenza viruses in North American swine and humans, 2009–2011. J Virol. 2012;86:8872–8878. doi: 10.1128/JVI.00259-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ali A, et al. Identification of swine H1N2/pandemic H1N1 reassortant influenza virus in pigs, United States. Vet Microbiol. 2012;158:60–68. doi: 10.1016/j.vetmic.2012.02.014. [DOI] [PubMed] [Google Scholar]
- 111.Charoenvisal N, et al. Genetic characterization of Thai swine influenza viruses after the introduction of pandemic H1N1 2009. Virus Genes. 2013;47:75–85. doi: 10.1007/s11262-013-0927-x. [DOI] [PubMed] [Google Scholar]
- 112.Ducatez MF, et al. Multiple reassortment between pandemic (H1N1) 2009 and endemic influenza viruses in pigs, United States. Emerg Infect Dis. 2011;17:1624–9. doi: 10.3201/1709.110338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Fan X, et al. Emergence and dissemination of a swine H3N2 reassortant influenza virus with 2009 pandemic H1N1 genes in pigs in China. J Virol. 2012;86:2375–8. doi: 10.1128/JVI.06824-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Harder TC, et al. Expanded co-circulation of stable subtypes, emerging lineages and new sporadic reassortants of porcine influenza viruses in swine populations in Northwest Germany. J Virol. 2013;87:10460–10476. doi: 10.1128/JVI.00381-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Howard W, Essen S. Reassortant Pandemic (H1N1) 2009 Virus in Pigs, United Kingdom. Emerg Infect Dis. 2011;17:1049–1052. doi: 10.3201/eid1706.101886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Starick E, et al. Reassorted pandemic (H1N1) 2009 influenza A virus discovered from pigs in Germany. J Gen Virol. 2011;92:1184–1188. doi: 10.1099/vir.0.028662-0. [DOI] [PubMed] [Google Scholar]
- 117.Moreno A, et al. Novel H1N2 swine influenza reassortant strain in pigs derived from the pandemic H1N1/2009 virus. Vet Microbiol. 2011;149:472–477. doi: 10.1016/j.vetmic.2010.12.011. [DOI] [PubMed] [Google Scholar]
- 118.Vijaykrishna D, et al. Reassortment of pandemic H1N1/2009 influenza A virus in swine. Science (80-) 2010;328:1529. doi: 10.1126/science.1189132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Corzo CA, et al. Active Surveillance for Influenza A Virus among Swine, Midwestern United States, 2009–2011. Emerg Infect Dis. 2013;19:954–960. doi: 10.3201/eid1906.121637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bao Y, et al. The influenza virus resource at the National Center for Biotechnology Information. J Virol. 2008;82:596–601. doi: 10.1128/JVI.02005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Pasma T, Joseph T. Pandemic (H1N1) 2009 Infection in Swine Herds, Manitoba, Canada. Emerg Infect Dis. 2010;16:706–708. doi: 10.3201/eid1604.091636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Escalera-Zamudio M, et al. Characterization of an influenza A virus in Mexican swine that is related to the A/H1N1/2009 pandemic clade. Virology. 2012;433:176–182. doi: 10.1016/j.virol.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 123.Nokireki T, et al. The first detection of influenza in the Finnish pig population: a retrospective study. Acta Vet Scand. 2013;55:69. doi: 10.1186/1751-0147-55-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bányai K, et al. Genome sequence of a monoreassortant H1N1 swine influenza virus isolated from a pig in Hungary. J Virol. 2012;86:13133. doi: 10.1128/JVI.02503-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Guercio A, et al. Pandemic influenza A/H1N1 virus in a swine farm house in Sicily, Italy. J Environ Biol. 2012;33:155–157. [PubMed] [Google Scholar]
- 126.Hofshagen M, et al. Pandemic Influenza A(H1N1)v: Human to pig transmission in Norway. Euro Surveill. 2009;14:1–3. doi: 10.2807/ese.14.45.19406-en. [DOI] [PubMed] [Google Scholar]
- 127.Grøntvedt CA, et al. Influenza A(H1N1)pdm09 virus infection in Norwegian swine herds 2009/10: the risk of human to swine transmission. Prev Vet Med. 2013;110:429–434. doi: 10.1016/j.prevetmed.2013.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Martín-Valls GE, et al. Phylogeny of Spanish swine influenza viruses isolated from respiratory disease outbreaks and evolution of swine influenza virus within an endemically infected farm. Vet Microbiol. 2014;170:266–77. doi: 10.1016/j.vetmic.2014.02.031. [DOI] [PubMed] [Google Scholar]
- 129.Welsh MD, et al. Initial incursion of pandemic (H1N1) 2009 influenza A virus into European pigs. Vet Rec. 2010;166:642–645. doi: 10.1136/vr.4851. [DOI] [PubMed] [Google Scholar]
- 130.Takemae N, et al. Antigenic variation of H1N1, H1N2 and H3N2 swine influenza viruses in Japan and Vietnam. Arch Virol. 2013;158:859–876. doi: 10.1007/s00705-013-1616-8. [DOI] [PubMed] [Google Scholar]
- 131.Kim SH, et al. Outbreak of pandemic influenza (H1N1) 2009 in pigs in Korea. Vet Rec. 2011;169:155. doi: 10.1136/vr.c7464. [DOI] [PubMed] [Google Scholar]
- 132.Song MS, et al. Evidence of human-to-swine transmission of the pandemic (H1N1) 2009 influenza virus in South Korea. J Clin Microbiol. 2010;48:3204–11. doi: 10.1128/JCM.00053-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sreta D, et al. Pandemic (H1N1) 2009 virus on commercial swine farm, Thailand. Emerg Infect Dis. 2010;16:1587–1590. doi: 10.3201/eid1610.100665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Trevennec K, et al. Transmission of pandemic influenza H1N1 (2009) in Vietnamese swine in 2009–2010. Influenza Other Respi Viruses. 2012;6:348–57. doi: 10.1111/j.1750-2659.2011.00324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Pérez LJ, et al. Isolation and complete genomic characterization of pandemic H1N1/2009 influenza viruses from Cuban swine herds. Res Vet Sci. 2013;94:781–788. doi: 10.1016/j.rvsc.2012.11.018. [DOI] [PubMed] [Google Scholar]