Abstract
Kinase inhibitors such as imatinib have dramatically improved outcomes for GIST patients, but many patients develop resistance to these treatments. While in some patients this event corresponds with mutations in the GIST driver oncogenic kinase KIT, other patients development resistance without KIT mutations. In this study, we address this patient subset in reporting a functional dependence of GIST on the FGF receptor FGFR3 and its crosstalk with KIT in GIST cells. Addition of the FGFR3 ligand FGF2 to GIST cells restored KIT phosphorylation during imatinib treatment, allowing sensitive cells to proliferate in the presence of the drug. FGF2 expression was increased in imatinib-resistant GIST cells, the growth of which was blocked by RNAi-mediated silencing of FGFR3. Moreover, combining KIT and FGFR3 inhibitors synergized to block the growth of imatinib-resistant cells. Signaling crosstalk between KIT and FGFR3 activated the MAPK pathway to promote resistance to imatinib. Clinically, an immunohistochemical analysis of tumor specimens from imatinib-resistant GIST patients revealed a relative increase in FGF2 levels, with a trend towards increased expression in imatinib-naïve samples consistent with possible involvement in drug resistance. Our findings provide a mechanistic rationale to evaluate existing FGFR inhibitors and multi-kinase inhibitors that target FGFR3 as promising strategies to improve treatment of GIST patients with de novo or acquired resistance to imatinib.
Introduction
Gastrointestinal stromal tumors (GISTs) are the most common mesenchymal neoplasms of the gastrointestinal tract with 5,000 to 6,000 new cases in the United States each year(1). The receptor tyrosine kinase (RTK) KIT is highly expressed and carries activating mutations in most GISTs(2). The majority of GISTs with wild type KIT have activating mutations in the receptor tyrosine kinase platelet-derived growth factor receptor alpha (PDGFRA)(3,4). Activation of the phosphatidyl-inositol-3-kinase (PI3K) pathway downstream of mutant KIT/PDGFRA is essential for GIST cell growth and survival(5). In addition, mitogen-activated protein kinase (MAPK) pathway signaling is activated downstream of KIT, and plays a pivotal role in tumorigenesis through the stabilization of the transcription factor ETV1 and activation of an oncogenic transcriptional program(6). The introduction of targeted tyrosine kinase inhibitor (TKI) therapy has revolutionized the clinical management of GIST and exemplifies the success of targeted therapy in solid tumors, where 80–90% of GIST patients with unresectable or disseminated disease initially attain at least disease stabilization, or complete or partial response to imatinib mesylate(7). However, nearly 50% of GIST cases treated with imatinib develop secondary resistance in the first 2 years(8). Most frequently, secondary resistance is due to acquisition of additional mutations in KIT or PDGFRA that decrease the binding affinity for imatinib(9). However, another mechanism that is likely to account for acquired resistance in a subset of GISTs is activation of pathways other than KIT and PDGFRA, thereby bypassing the inhibitory effects of KIT/PDGFRA-targeted small molecules.
Receptor tyrosine kinases are tightly regulated in normal cells, but frequently acquire transforming functions due to mutation(s), overexpression and autocrine paracrine stimulation in human cancers. Selective tyrosine kinase inhibitors can block this activity and constitute a promising approach for molecularly guided therapeutics. For example, the FGF signaling network is deregulated in several human cancers, including breast, bladder, prostate, endometrial, and non-small cell lung cancer(10). Receptors may be aberrantly activated through mutations(11,12), amplifications(13), or fusions(14). The ligands for FGF receptors (FGFs) have also shown aberrant activity in a variety of cancers. High expression of FGF3, FGF8, and FGF10 has been reported in breast cancer(15), and correlates with malignant behavior. In prostate cancer(16), FGF2 expressed by stromal cells promotes tumor progression(17). Activation of the FGF signaling axis by FGF8, FGF9, and FGF10 over-expression is also associated with an aggressive clinical phenotype(18). In addition, FGF2 has recently been shown to mediate resistance to chemotherapy, and, as laid out in this paper, may also provide intrinsic protection of tumor cells in the presence of small-molecule kinase inhibitors.
Materials and Methods
siRNA and Kinase Inhibitors
The RAPID siRNA library has been previously described(19–21). All siRNAs were from Thermo Fisher Scientific Dharmacon RNAi Technologies. Each well contained a pool of 4 siRNAs. Cells were aliquoted at 66ul per well in a 96-well plate and 34 ul of siRNA/OptiMEM/siRNA mixture was added to each well. Oligofectamine and siRNA were used at a ratio of 1:6. For assessment of cell viability and proliferation, cells were subjected to the MTS assay after 96 h. PD173074, AZD-6244, and PI-103 were purchased from Selleck; imatinib and CHIR-258 were purchased from LC Labs.
Immunoblotting
All immunoblotting was performed using standard protocols. Data was analyzed with ImageJ.
GIST Tissue Samples
All patient specimens were obtained with informed consent of the patients on protocols approved by the Institutional Review Board of Oregon Health & Science University. To prepare Fresh frozen GIST tissue samples for immunoblotting, tissue was dissected on dry ice using a razor blade. 4 shavings per sample were sonicated 3 times for 3 seconds in 2× Cell Signaling Lysis Buffer.
Statistical Analyses
For cell proliferation assays, a Student t test was carried out for each treatment condition compared to untreated cells or appropriate controls. The P values for the t tests are indicated by asterisks (*): *.01 ≤ P < .05; **.001 ≤ P < .01; ***P < .001. To determine the significance of combination indices to indicate synergy, upper and lower confidence limits were calculated. Data points for combinations with upper confidence limits below 1 were considered synergistic.
For further experimental details, see Supplemental Materials and Methods.
Results
siRNA-mediated knockdown identifies FGFR3 dependence in GIST cell lines
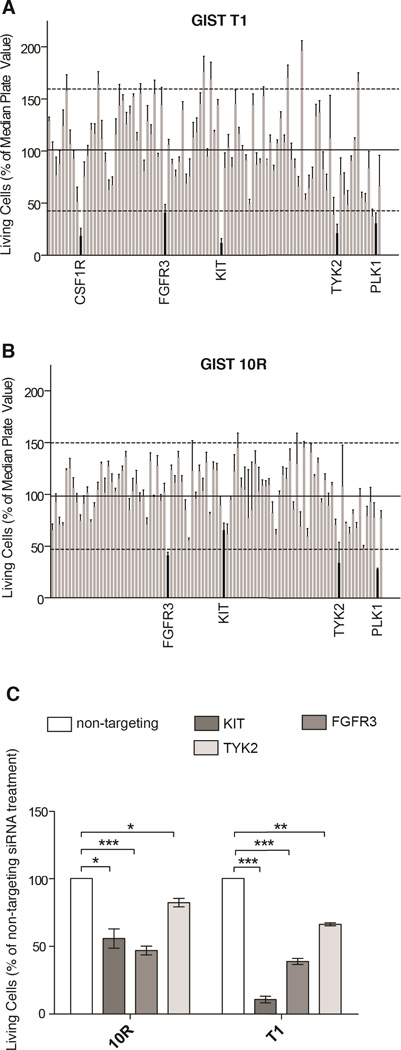
GIST T1 cells harbor a heterozygous deletion of KIT exon 11 and consequently exhibit high sensitivity to imatinib with potent suppression of cell proliferation at concentrations ranging from 100 to 1,000 nM. The GIST 10R cell line grew out as a colony after two months treatment of GIST T1 cells with 10,000 nM imatinib. Consequently, GIST 10R cells exhibit no IC50 at concentrations of imatinib up to 10,000 nM, although an IC25 is still apparent at 100 nM (Figure S1A). Interestingly, GIST 10R cells do exhibit reduced phosphorylation of KIT and its downstream signaling molecules after exposure to 1,000 nM imatinib (Figure S1B), and no secondary mutations were found in KIT, indicating that drug resistance in GIST 10R cells must be due to alternative mechanisms. The fact that inhibition is equal at equal concentrations of imatinib is likely due to the fact that GIST 10R does not carry additional mutations in KIT that should render this primary drug target less susceptible to inhibition. In addition, comparative RNA sequencing between GIST T1 and 10R revealed no point mutations or remarkable changes in gene expression that would explain drug resistance in these cells. To investigate possible alternative therapeutic targets in these cells, we transfected GIST T1 and 10R cells with a panel of siRNAs that collectively target the entire tyrosine kinase gene family in addition to NRAS and KRAS (93 genes total)(19,20). As expected, siRNA against KIT significantly reduced the relative number of proliferating GIST T1 cells (Figure 1A). We also observed a significant reduction in proliferation after silencing of colony stimulating factor 1 receptor (CSF1R), tyrosine kinase 2 (TYK2), and fibroblast growth factor receptor 3 (FGFR3). Polo-like kinase 1 (PLK1) plays a critical role in mitosis in all cells and was used as a positive control for effective siRNA-mediated silencing. Interestingly, GIST 10R cells retained residual sensitivity to KIT silencing (Figure 1B), however, silencing of TYK2 and FGFR3 reduced growth more significantly. To confirm reproducible and significant sensitivity to silencing of these genes, we independently assessed cell proliferation after silencing of KIT, FGFR3, and TYK2 compared with non-targeting siRNA. We confirmed the differential effect of KIT siRNA on GIST 10R and T1 cells (Figure 1C). KIT silencing significantly impacted both cell lines, but this impact was much less pronounced in GIST 10R cells than in GIST T1 cells. We also confirmed that both cell lines were sensitive to FGFR3 silencing to a comparable degree. For reference, FGFR3 phosphorylation and expression levels are compared in all cell line models in Figure S1C. Expression of FGFR3 after treatment of GIST 10R and T1 cells with 4 individual siRNA duplexes also resulted in reductions of GIST cell viability (Figure S2). To determine whether this impact on relative number of viable cells was predominantly an effect on cell growth or an induction of apoptosis, we also stained cells with Annexin V after silencing of KIT, FGFR3, or TYK2. While we observed minor increases in Annexin V staining after silencing of each gene, these changes were markedly less than the reduction observed in overall numbers of viable cells, indicating that reduced cell growth is largely responsible for the observed phenotype (Figure S1D).
Figure 1. GIST cell sensitivity to siRNA-mediated knockdown of the receptor tyrosine kinome, and target validation.
A, The cell line GIST T1 was transfected with siRNA pools individually targeting each member of the receptor tyrosine kinome. The cell viability was calculated by normalizing the cell proliferation (as determined by MTS assay) after 96h of treatment to the median plate value. B, The imatinib-resistant cell line GIST 10R was treated and analyzed analogous to GIST T1 in A. C, Target validation comparing the effect of KIT, FGFR3, and TYK2 silencing on proliferation of GIST cells (as determined by MTS assay). The bars represent the mean ± s.e.m. between independent experiments, each containing 3 replicates (n=3). The P values for the t tests are indicated by asterisks (*): *.01 ≤ P < .05; **.001 ≤ P < .01; ***P < .001. Viability measures for all tested siRNA constructs can be found in Table S1.
Inhibition and silencing of FGFR3 and KIT reveals crosstalk
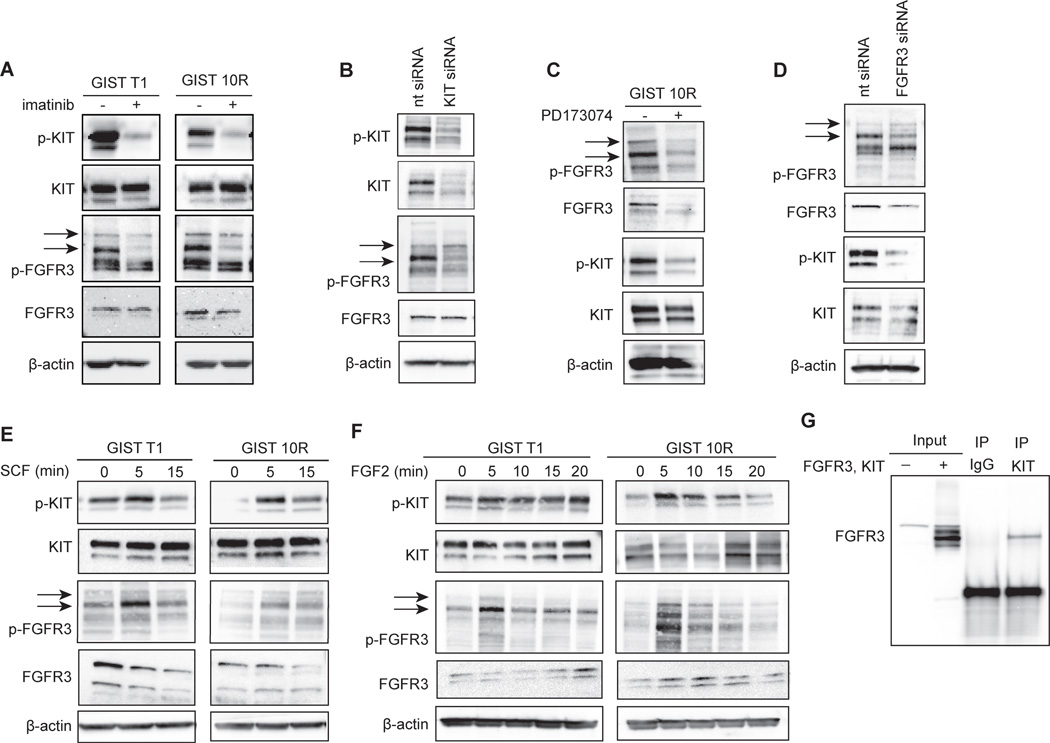
We next sought to understand the role of FGFR3 in maintenance of GIST T1/10R cell growth. First, we treated both cell lines with 1,000 nM imatinib for 2h and then detected the active, phosphorylated forms of KIT and FGFR3, as well as total protein, on an immunoblot (Figure 2A). Surprisingly, in both cell lines we observed a reduction in phospho-KIT and phospho-FGFR3 after imatinib treatment. To confirm that the reduction in phospho-FGFR3 was not due to an off-target effect of imatinib, we silenced KIT in GIST 10R and again assessed FGFR3 phosphorylation by immunoblot (Figure 2B). Phosphorylation was again reduced, indicating that FGFR3 activation in GIST cells is dependent on KIT activity. Next, we asked whether the connection between KIT and FGFR3 was reciprocal, so that FGFR3 inhibition or silencing would affect KIT activity. We treated GIST 10R cells with the FGFR inhibitor PD173074 at 1,000 nM for 2h and performed immunoblots (Figure 2C). Phospho-FGFR3 and total FGFR3 protein levels were markedly reduced after treatment with PD173074. The reduction in total protein may be due to degradation of the receptor after inhibition (Figure S3). Importantly, KIT is not a reported target of PD173074, yet phospho-KIT was reduced after treatment in both cell lines. To rule out direct inhibition of KIT as the source for reduced phosphorylation, we silenced FGFR3 using siRNA in GIST 10R cells and assessed KIT phosphorylation by immunoblot (Figure 2D). Again, phospho-KIT was reduced after FGFR3 silencing, suggesting that reciprocal crosstalk exists between KIT and FGFR3 in GIST cells. In addition, we stimulated KIT and FGFR3 by the addition of ligand for each receptor (SCF and FGF2, respectively) and observed increased phosphorylation for both KIT and FGFR3 in GIST 10R and, to a lesser degree, in GIST T1. (Figure 2E, F). Although FGF1 is the prototypic ligand of FGFR3, FGF2 also binds and activates FGFR3(22). We observed a similar increase in phosphorylation of FGFR3 and KIT in response to FGF1 (Figure S4A), however, FGF2 elicited a more dramatic protective effect after imatinib treatment (Figures 3 A–C) compared to FGF1 (Figure S4B). We thus chose to conduct subsequent experiments using FGF2.
Figure 2. Crosstalk between KIT-and FGFR3-signaling after inhibition or stimulation.
A, GIST cell lines were treated with 1uM imatinib (+) or medium (−) for 2 hours. Levels of total and phospho-KIT and FGFR3 as well as β-actin were assessed by immunoblot analysis. Phospho-FGFR3 bands are indicated by arrows. B, GIST cell lines were treated with siRNA against KIT or a non-targeting control pool (nt siRNA) for 96 h and cell lysates were subjected to immunoblot analysis. C, GIST cell lines were treated with 1uM of the FGFR-inhibitor PD173074 (+) or media (−) for 2 hours. Levels of total and phospho-KIT and FGFR3 as well as β-actin were assessed by immunoblot analysis. D, GIST cell lines were treated with siRNA against FGFR3 or a non-targeting control pool (nt siRNA) for 96 h and cell lysates were subjected to immunoblot analysis. E, GIST T1 and 10R cell lines were stimulated with 100 ng/ml SCF for 0, 5, or 15 minutes and cell lysates were subjected to immunoblot analysis. F, GIST T1 and 10R cell lines were stimulated with 100 ng/ml FGF2 for 0, 5, 10, 15, or 20 minutes and cell lysates were subjected to immunoblot analysis. G, HEK 293 cells were transfected with plasmids for the overexpression of KIT and FGFR3. After 48 hours, cell lysates were immunoprecipitated with IgG isotype control or antibody against KIT. Whole cell lysates as well as immunoprecipitates were subjected to immunoblot analysis using an antibody specific for FGFR3.
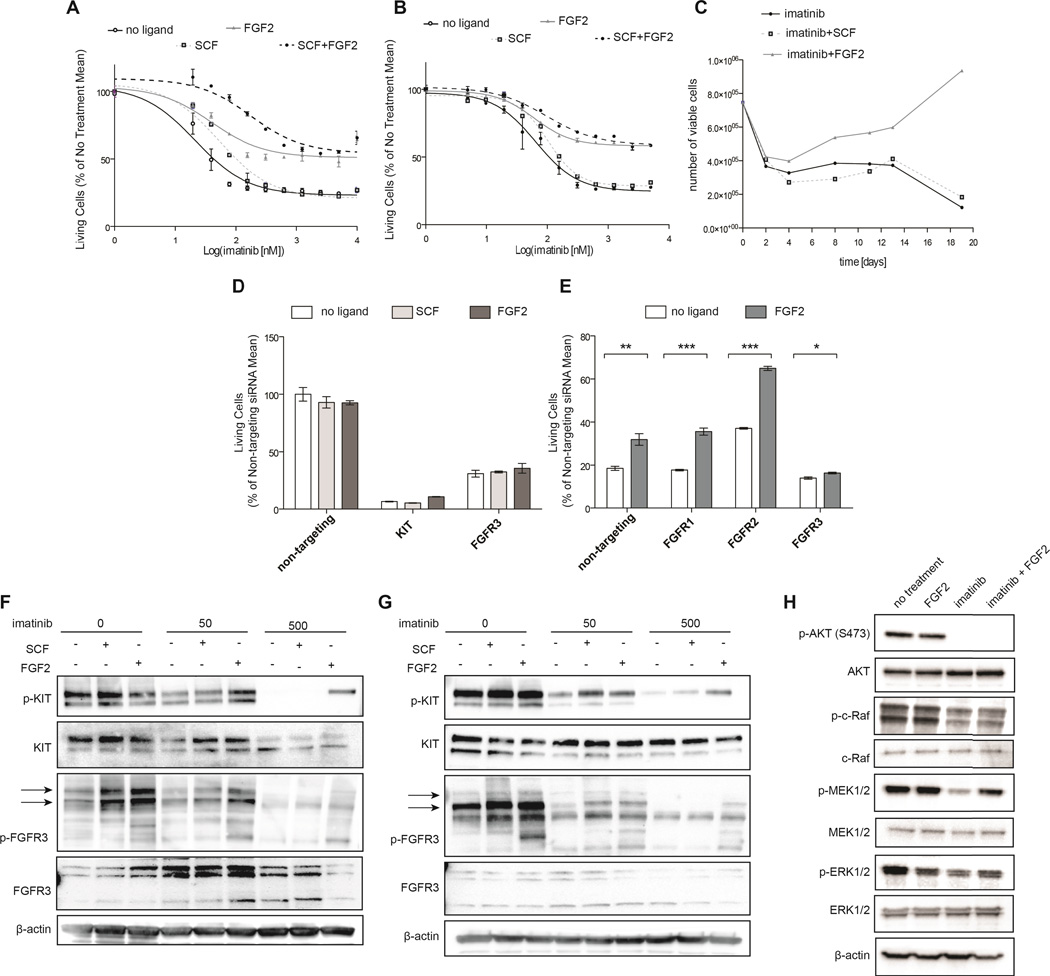
Figure 3. FGF2 rescues GIST cell lines from KIT inhibition in an FGFR3-dependent manner.
A, GIST T1 cells were treated with a dose gradient of imatinib in the presence of 10ng/ml SCF, FGF2, SCF+FGF2, or media (no ligand). After 48 h, viability was assessed by MTS assay and normalized to ligand treatment in the absence of drug. B, GIST 882 cells were treated and analyzed analogous to GIST T1 in A. C, Resistance in the presence of FGF2 leads to outgrowth of GIST T1 cells in a 20-day culture with imatinib. Cells were cultured with 1uM imatinib and 10ng/ml ligand. Viable cell counts were obtained by flow cytometry using PI exclusion. Data is representative of 3 experiments. D, GIST T1 cells were transfected with non-targeting, KIT, or FGFR3 siRNA and cultured for 48 h before the addition of 10ng/ml FGF2. After an additional 48 h, viability was assessed by MTS assay and normalized to no treatment. E, GIST T1 cells were transfected with siRNA targeting members FGFR1, FGFR2, or FGFR3 or with non-targeting siRNA, and cultured for 48 h. Cells were treated with imatinib (1000 nM) in the presence of FGF2 (10 ng/ml) and cell viability was assessed after an additional 48 h. F, GIST T1 cells were treated with a dose gradient of imatinib for 2 hours and then stimulated with media alone or media containing SCF (100 ng/ml) or FGF2 (100 ng/ml) for 5 minutes. Cell lysates were subjected to immunoblot analysis. G, GIST 10R cells were treated with a dose gradient of imatinib for 2 hours and then stimulated with media alone or media containing SCF (100 ng/ml) or FGF-1 (100 ng/ml) for 5 minutes. Cell lysates were subjected to immunoblot analysis. Phospho-FGFR3 bands are indicated by arrows. H, GIST T1 cells were treated with imatinib for 24h, and stimulated with FGF2 (100 ng/ml) for 5 minutes. Phosphorylation was assessed using antibodies specific for total and phospho AKT, MEK1/2, ERK1/2, and β-actin. The bars represent the mean ± s.e.m. between replicates (n=3). The P values for the t tests are indicated by asterisks (*): *.01 ≤ P < .05; **.001 ≤ P < .01; ***P < .001.
Co-immunoprecipitation in HEK 293 cells indicates direct crosstalk between KIT and FGFR3
Reciprocal crosstalk between two receptor tyrosine kinases may result from indirect interaction (mediated by a common downstream kinase), or direct interaction through the formation of heterodimers or other receptor clustering. To test the hypothesis that the crosstalk between KIT and FGFR3 is direct, we co-transfected HEK 293 cells with plasmids containing KIT and FGFR3 wild type cDNA. We performed immunoprecipitation with a KIT-specific antibody, followed by immunoblotting for FGFR3 (Figure 2G). We observed co-immunoprecipitation of KIT and FGFR3, suggesting a direct interaction between these receptor tyrosine kinases. The reverse co-immunoprecipitation was not successful, likely because there is no antibody suitable for immunoprecipitation of FGFR3 available to us.
Signaling through FGFR3 desensitizes GIST cells to imatinib treatment
We reasoned that presence of ligand, in particular FGF2, in the cell culture media might dampen the response of GIST T1 cells to imatinib. Consequently, we treated GIST T1 cells with a gradient of imatinib concentrations with or without a constant concentration of ligand (Figure 3A). In the absence of ligand, GIST T1 cells were extremely sensitive to imatinib, with an IC50 of 40 nM. In the presence of SCF, the IC50 was increased slightly, however, at higher concentrations of imatinib, cell proliferation was not improved. In contrast, the addition of FGF2 increased cell proliferation in the presence of imatinib dramatically, with no IC50 even at concentrations as high as 10,000 nM. Of note, the combination of FGF2 and SCF conferred a further increase in viability at low concentrations of imatinib. Similar results were observed using a different, KIT-sensitive GIST-derived cell line, GIST 882 (Figure 3B). We also cultured GIST T1 cells with 1,000 nM imatinib in the presence or absence of 10 ng/ml SCF or FGF2 (Figure 3C). Viable cells were counted every 2–3 days for 19 days. As expected, culture with SCF did not confer any growth advantage over cells cultured with imatinib and a vehicle control. The addition of FGF2, however, increased the number of viable cells starting at day 4. Importantly, cells not only persisted in the culture, but, after a lag phase, continued to divide and exceeded the number initially plated on day 0. We hypothesized that the desensitization of GIST cells to imatinib is indeed mediated through the interaction between KIT and FGFR3, and not the result of an alternative survival pathway replacing KIT signaling. To this end, we measured GIST T1 cell growth after KIT silencing in the presence or absence of FGF2 with the hypothesis that presence of KIT protein would be required for FGF2 rescue (Figure 3D). As predicted, rescue of cell growth by FGF2 was ineffective after KIT knockdown, indicating that FGF2 rescue requires presence of both KIT and FGFR3. We also showed that inhibition of FGFR3 by PD173074, which inhibits GIST T1 cell proliferation with an IC50 of 300 nM, can be partially reversed by the addition of SCF (Figure S5) and that SCF rescue is ineffective after silencing of FGFR3 (Figure 3D). To test whether desensitization to imatinib is, indeed, mediated by FGFR3, we performed siRNA knockdown of FGFR1, FGFR2, and FGFR3 in GIST T1, and subsequently treated with imatinib and FGF2 (Figure 3E). Knockdown of FGF receptors was confirmed via real time RT-PCR (Figure S6). FGF2 rescue of imatinib sensitivity remained effective after FGFR1 and FGFR2 silencing, however, FGFR3 silencing ablated the response to FGF2, implicating FGFR3 but not FGFR1 or FGFR2 in FGF2-mediated drug resistance. We next treated GIST T1 cells with 0, 50, or 500 nM imatinib and stimulated cells with SCF or FGF2. At baseline, KIT phosphorylation was responsive to both SCF and FGF2 stimulation. The response to SCF was conserved in the context of 50 nM imatinib treatment, but was markedly decreased after treatment with 500 nM imatinib. In contrast, FGF2 still restored KIT phosphorylation at 500 nM (Figure 3F). To ensure that this observation was not specific to the cell line GIST T1, we repeated this experiment in the cell line GIST 882 (Figure 3G). Again, we observed that KIT phosphorylation in GIST 882 was completely ablated at 500 nM imatinib without FGF2 stimulation, but could be partially restored by the addition of FGF2. We subsequently sought to determine the effects of FGF2 stimulation on downstream signaling in the setting of imatinib treatment (Figure 3H). Analysis of AKT and MAPK pathway activation revealed that both pathways are inhibited after imatinib treatment. However, while AKT phosphorylation remained inhibited after the addition of FGF2, to MEK1/2 and ERK1/2 phosphorylation were restored.
Combined inhibition of KIT and FGFR3 is highly synergistic in imatinib-resistant GIST cells
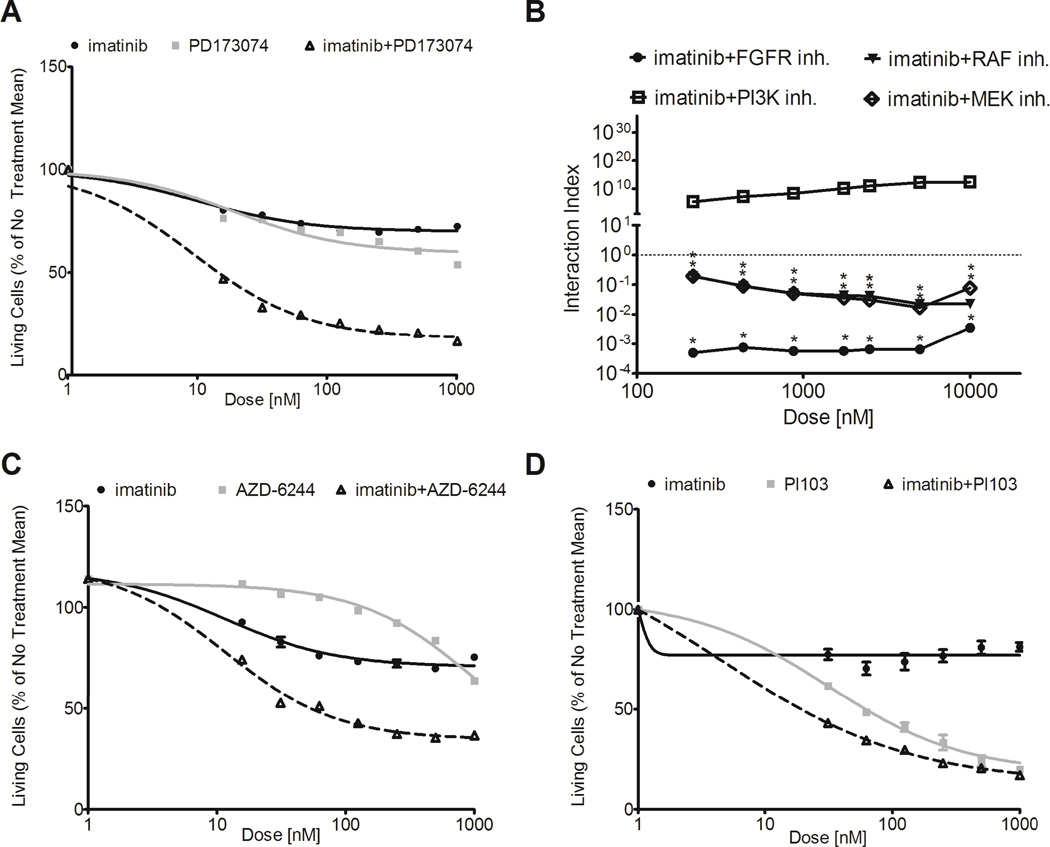
We hypothesized that combination treatment using imatinib and the selective FGFR inhibitor PD173074 may restore imatinib sensitivity in resistant GIST cells. Accordingly, we performed dose-response curves in the imatinib-resistant GIST 10R cells using imatinib and PD173074 alone as well as a constant, equimolar ratio combination of the two drugs (Figure 4A). We then determined the combination index (CI) for each dose point using the Chou-Talalay method to quantify synergy. Figure 4B shows CI values plotted against the log of drug dose. Over the entire dosing curve, the CI values ranged from 0.0005 to 0.004, indicating an extremely high degree of synergy between the two drugs.
Figure 4. Combination of FGFR-inhibitor or MAPK-pathway inhibitors with imatinib restores sensitivity in GIST 10R cells.
A, GIST 10R cells were treated with combinations of imatinib with PD173074 (FGFR inhibitor; 1:1 ratio). Cells were cultured in drug dilutions for 48 h and viability was quantified by MTS assay. B, Combination indices were calculated for each concentration point of each drug curve. Asterisks mark combinations that show significant synergy (upper limit of interaction index below 1). C, GIST 10R cells were treated with combinations of imatinib with AZD-6244 (MEK inhibitor; 1:1 ratio). Cells were cultured in drug dilutions for 48 h and viability was quantified by MTS assay. D, GIST 10R cells were treated with combinations of imatinib with PI103 (PI3K inhibitor; 1:10 ratio). Cells were cultured in drug dilutions for 48 h and viability was quantified by MTS assay.
We next wanted to determine whether signaling pathways activated downstream of FGFR3 might also exhibit synergy with KIT inhibition. To this end, we treated GIST 10R cells with combinations of imatinib and PLX-4720, a B-Raf inhibitor (Figure S7), AZD-6244, a MAPK inhibitor (Figure 4C), and PI103, a PI3K inhibitor (Figure 4D). Combination of imatinib with inhibitors of the RAF/MAPK pathway exhibited significant synergy at all concentrations tested. In contrast, combination of imatinib with inhibitors of the PI3K-AKT pathway did not result in synergy with imatinib. Combined treatment with imatinib and AZD-6244, in particular, led to a decrease in cell growth similar to that observed after imatinib and PD173074 treatment, suggesting that the MAPK pathway is a key mediator of imatinib resistance in GIST 10R cells.
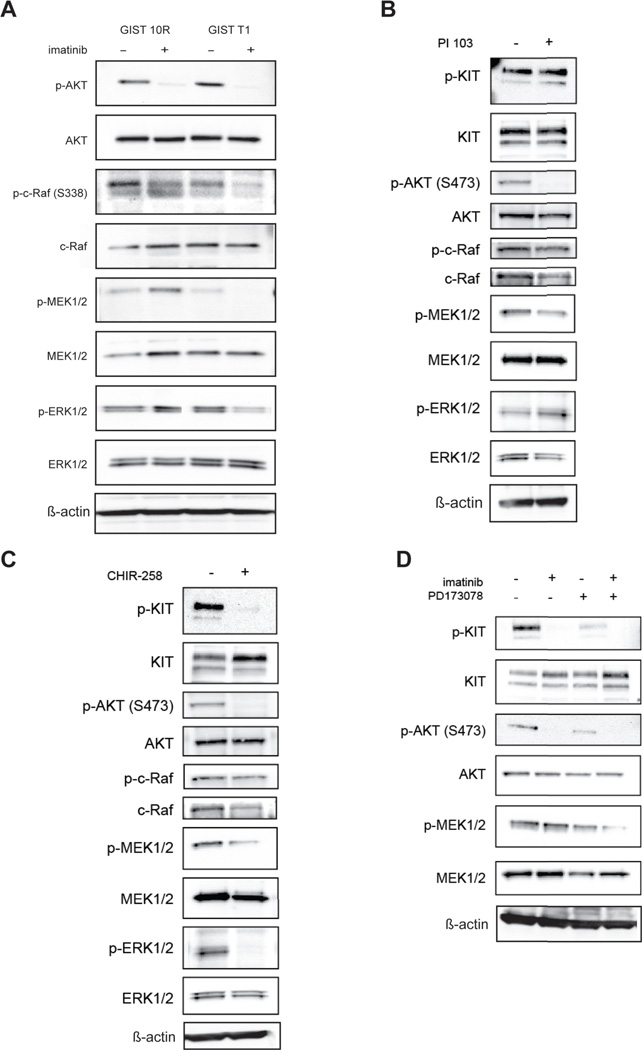
The MAPK signaling pathway is activated in GIST 10R cells in response to imatinib
We performed immunoblot analysis to assess phosphorylation of AKT, c-Raf, MEK, and ERK in GIST 10R and T1 cells after 2h treatment with 1,000 nM imatinib (Figure 5A). In both cell lines, AKT phosphorylation was equivalently reduced after treatment. However, we observed divergent effects on MAPK-pathway activation. Phosphorylation of c-Raf was reduced less markedly in GIST 10R compared to GIST T1 cells. Even more strikingly, phosphorylation of MEK and ERK was abolished in GIST T1 after imatinib treatment, but increased in GIST 10R. To determine whether this feedback mechanism could be solely regulated at the receptor level, or whether it might also be regulated after direct inactivation of the downstream PI3K signaling, we treated GIST 10R cells with the PI3K inhibitor PI103 and asked whether the same increased phosphorylation of MAPK signaling resulted (Figure 5B). No increase in c-Raf, MEK, or ERK phosphorylation was observed after inhibition of PI3K. To assess whether FGF receptors are involved in mediating this MAPK feedback mechanism, we inhibited KIT and FGFRs, either by the dual inhibitor CHIR-258 (dovitinib) (Figure 5C), or by a combination of imatinib and PD173074 (Figure 5D). We found that MAPK activation was abrogated in both cases, providing a mechanistic basis for the synergy observed in Figure 4.
Figure 5. The MAPK pathway is upregulated downstream of FGFRs in resistant GIST cells in response to imatinib.
A, GIST T1 and GIST 10R cells were treated with imatinib for 2 hours and cell lysates were subjected to immunoblot analysis using antibodies specific for total or phospho-AKT, C-RAF, MEK1/2, ERK1/2, or β-actin. B, GIST 10R cells were treated with the PI3K inhibitor PI103, C, the multi-kinase inhibitor CHIR-258 for 2 hours and cell lysates were subjected to immunoblot analysis, or D, a combination of imatinib and PD173074.
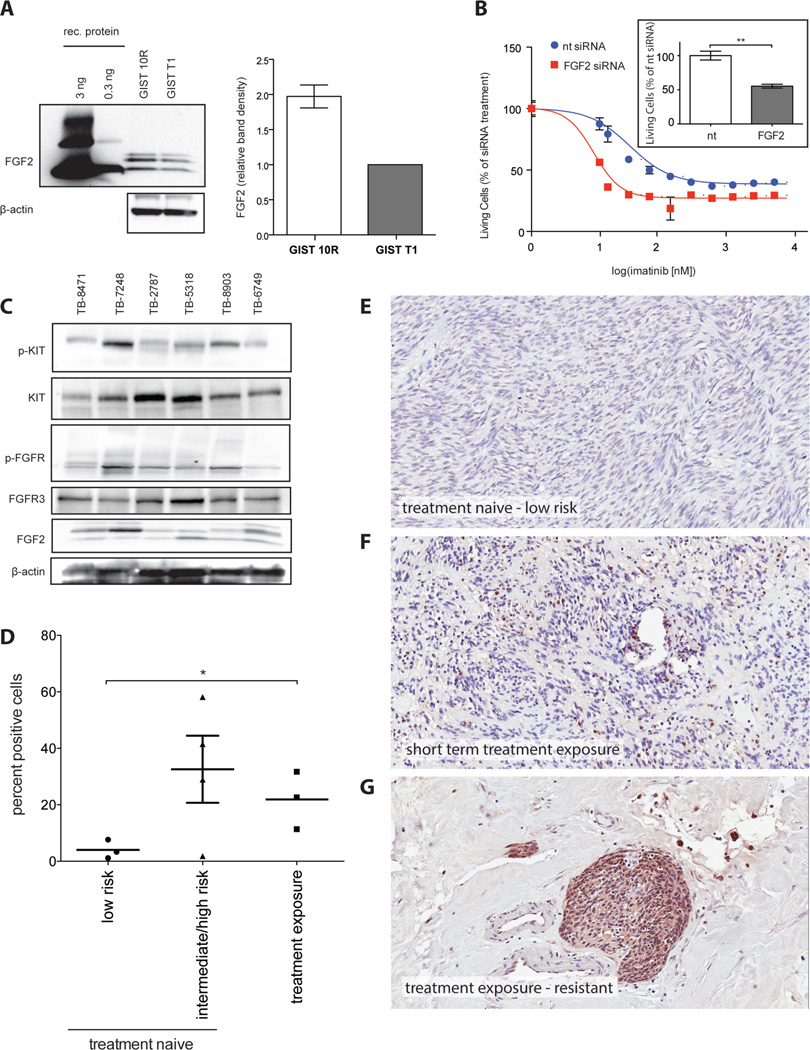
FGF2 is overexpressed in GIST 10R cells and is increased in tumor tissue post-imatinib treatment
Since our data suggest that FGF2 can promote imatinib resistance in GIST cells, we wanted to determine whether FGF2 levels are higher in GIST 10R than GIST T1 cells and whether this may underlie the resistance of GIST 10R cells to imatinib. It is well recognized that FGF2 associates with heparan sulfate in the extracellular matrix. We were thus unable to detect it in supernatant but found it present in cell lysate. Evaluation of FGF2 protein levels did reveal increased levels of FGF2 in GIST 10R cells compared to GIST T1 (Figure 6A). Subsequently, we investigated whether silencing of FGF2 in GIST 10R cells changes cell growth when compared to treatment with non-targeting siRNA. Indeed, we observed a significant decrease in cell growth after GIST 10R cells were treated with FGF2 siRNA (insert in Figure 6B). We next asked whether siRNA-mediated knockdown of FGF2 would re-sensitize these cells to imatinib. Silencing of FGF2 dramatically shifted the IC50 for imatinib in GIST 10R cells (Figure 6B), indicating that FGF2 contributes to imatinib-resistance in GIST 10R and that the inhibition of FGF-signaling restores the response to imatinib in a resistant GIST cell line. Knockdown of FGF2 expression was confirmed via real time RT-PCR (Figure S8). We sought to examine whether increased levels of FGF2 correlated with KIT activation in GIST patient samples. We obtained a panel of 6 frozen tissue specimens from GI stromal tumors with confirmed activating mutations in KIT exon 11. Five of the tumors were treatment-naïve at the time of biopsy, while one patient had received 4–5 weeks of imatinib prior to the tumor being removed. The treatment was discontinued 3–4 days before surgery. We performed immunoblots for activated KIT and FGFR, as well as FGF2 levels in these samples (Figure 6C) and observed increased FGF2 in specimen TB-7248, which had been exposed to imatinib. The pan-phospho-FGFR antibody yielded immunoblots with lower background and was thus chosen here for use on tumor lysates. Concurrent with high FGF2 levels, we observed a high degree of phosphorylated KIT and FGFR. This observation is consistent with our model of FGF2-mediated re-activation of KIT. We sought to validate these findings in a larger number of GIST samples by performing immunohistochemical staining for FGF2 on 10 sections of formalin fixed paraffin-embedded tissue (Figure 6D). An illustration of the quantification process is provided in Figure S9. Samples not previously exposed to treatment were segregated by mitotic rate and tumor size according to the Fletcher risk table into low risk and intermediate/ high risk categories. Figures 6E–G provide examples of immunohistochemical staining in treatment naïve, low risk disease, as well as two cases of imatinib exposed tumor samples. Overall, low risk was associated with low FGF2 staining, while intermediate/ high risk and treatment exposure or resistance were associated with elevated levels, warranting further study of FGF2 levels in even larger cohorts of primary GIST patient specimens in the future.
Figure 6. FGF2 is overexpressed in GIST 10R cells and a pre-treated patient sample.
A, Cell lysates from untreated GIST 10R and GIST T1 cells were subjected to immunoblot analysis using antibodies specific for FGF2 or β-actin. Recombinant FGF2 is included for comparison. Sample blot is shown. FGF levels on immunoblots were quantified as bioluminescence units (BLU) relative to actin and normalized to levels in GIST T1 cells for comparison. B, GIST 10R cells were transfected with non-targeting or FGF2 siRNA and cultured for 48 h. An imatinib gradient was added to cells and viability was assessed after another 48 h. For comparison, viability was normalized to the effect of the respective siRNA alone. The effect of siRNA alone on viable cell number is shown in the boxed inlay. The bars represent the mean ± s.e.m. between replicates (n=3). GIST 10R cells were transfected with non-targeting or FGF2 siRNA and cultured for 96 hours. Viability was determined by MTS assay. The P value is indicated by asterisks **.001 ≤ P < .01 C, Frozen GIST tissue samples were prepared for immunoblot analysis and probed with antibodies specific for total and phospho KIT, FGFR, FGF2, or β-actin. D, GIST tumor FFPE samples were subjected to immunohistochemical staining for FGF2. Staining was quantified using ImageScope software. *.01 ≤ P < .05 E, Example of immunohistochemical staining for FGF2 on a treatment naïve, low risk tumor sample. F, Immunohistochemical staining for FGF2 on a tumor sample after short-term exposure to imatinib. G, Example of immunohistochemical staining for FGF2 on an imatinib exposed, resistant tumor sample.
Discussion
Our study describes for the first time a functional cooperation between FGFR3 and KIT in human GIST and analyzes the molecular mechanisms that underlie this cooperation. The findings provide insight into the protective potential of FGF signaling in GIST and into the signal transduction pathways that mediate resistance to small molecule inhibitors of KIT in these tumors. Moreover, this represents a new mechanism of resistance in this setting that can account for GIST patients progressing on imatinib in the absence of a secondary resistance mutation in KIT.
In addition to the paracrine action of cancer cell-secreted FGF2 on endothelial cells, FGF2 can provide autocrine pro-survival and mitogenic signals, and confer resistance to chemotherapeutic drugs. FGF2 (over)expression has been observed after chemoradiation and is correlated with recurrence risk in some cancers. FGF2 modulates the response of a wide range of tumor types, such as neuroblastoma, breast cancer, melanoma and non small cell lung cancer (NSCLC) to chemotherapy or radiation(23–26). There are also accounts of FGF signaling involvement in resistance to targeted therapy, for example through de-repression of FGFR2 and FGFR3 in NSCLC cell lines, which leads to resistance to EGFR inhibitor therapy(27). Although the mechanism is not clear in every setting, FGF2 protects both non small cell lung cancer and endothelial cells from apoptosis in a Raf-dependent manner after chemotherapy or VEGFR-inhibition, respectively(28,29). FGF2 overexpression is not only associated with resistance to chemo- and radiotherapy, but also correlates with an increased risk of recurrence and reduced overall survival. The array of tumor types and treatments to which FGF2 is connected suggest a global protective role for this ligand, which is in line with FGF2’s role in normal tissues during injury and inflammation.
Here, we present one of the first accounts of FGF2 mediating resistance to targeted therapy. Recently, activation of FGFR3 and the downstream MEK/ERK pathway was also implicated in resistance to a targeted agent, the B-RAF inhibitor vemurafenib, in melanoma(25). The protective effect of FGF2 in this setting is mediated by the MAPK pathway and downstream activation of ribosomal protein S6 kinase 2 (S6K2)(30,31). Similarly, an autocrine signaling loop was identified in non-small-cell lung cancer, where FGFRs and their ligands were co-expressed and provided an alternative pathway to EGFR signaling in cells treated with gefitinib(32). This correlates well with our finding that MAPK pathway members are preferentially activated after FGF2 stimulation in the presence of imatinib in GIST T1 cells. Similarly, this pathway remained active in GIST 10R cells during imatinib treatment, and these cells could be re-sensitized by combined imatinib and MAPK-inhibitor treatment. The Raf/MEK/ERK signaling pathway is already recognized as an important driver of cell proliferation, survival, and angiogenesis in GIST, as evidenced by an ongoing phase II multicenter trial of the Raf inhibitor sorafenib in imatinib- and sunitinib-resistant GIST. Selective inhibition of this pathway also inhibited proliferation and induced apoptosis and cell cycle arrest in patient-derived GIST xenograft lines(33). Taken together with our observations, this underscores the potential of the Raf/MEK/ERK signaling pathway as potential future targets of molecular therapy in GIST.
We propose that, in addition to preferential activation of the MAPK pathway, FGFR3 activation partially restores KIT activity. In the setting of overexpression of both receptors, we demonstrated an interaction between the two receptors. Although this data is supportive of a direct interaction between KIT and FGFR3, this is only one mechanism potentially underlying the crosstalk observed in GIST cells, and other possibilities such as the involvement of downstream mediators should not be discarded. Receptor crosstalk and heterodimerization to circumvent targeted therapy has been explored extensively in the setting of EGFR inhibition. EGFR can interact with other RTKs such as MET, ERBB2, and IGF-1R[38]. These mechanisms were identified at the clinical and pre-clinical level in non-small-cell lung cancer and breast cancer. Overexpression and activation of EGFR can promote transphosphorylation of MET in lung and epidermal carcinoma cell lines. Activation of MET occurs in an EGFR-ligand dependent manner in the setting of EGFR overexpression, or independently of ligand in glioblastomas expressing a constitutively active EGFR variant. Xenograft models of glioblastoma require targeting of both EGFR and MET to achieve growth inhibition(34). The crosstalk between KIT and FGFR3 we present in this paper involves two hitherto unconnected signaling pathways, which are highly relevant to human cancers. Our findings are consistent with the biology reported for crosstalk in other systems. Although no previous reports exist of transactivation between KIT and FGFR3, there is evidence that FGFRs can crosstalk with other RTKs. For example, the cytoplasmic domains of FGF receptors and EphA4 can interact and transphosphorylate each other(35).
In light of the propensity of FGF2 to desensitize GIST cells to the short- and long-term effects of imatinib, we suggest that FGF2 expression in treated tumors provides the basis for the development of resistance. Future studies should determine the mechanism of FGF2 upregulation and examine the dynamics of FGF2 expression with imatinib treatment. This would provide valuable information to identify patient populations who may be at risk of FGF-mediated resistance, either by constitutive overexpression or by sustained upregulation in response to therapy. Our observations are an added incentive to pursue targeted treatment that combines inhibition of KIT with antagonism of protective signaling from autocrine loops or the tumor stroma. This strategy could be especially powerful with screening to identify patients at risk for microenvironment-induced resistance.
In summary, we show for the first time that the FGFR3 pathway crosstalks with KIT, and that FGF2 mediates survival and outgrowth of GIST cells during imatinib treatment. We further elucidate the molecular mechanisms of FGF2-mediated drug rescue. Our data suggest that incorporation of FGFR3 inhibitors to combination therapeutic regimens may be beneficial in overcoming clinical resistance to targeted therapies for some patients with GIST.
Supplementary Material
Acknowledgements
N.J.-S. is supported by the Oregon Clinical and Translational Research Institute (OCTRI), grant number TL1 RR024159 from the National Center for Advancing Translational Sciences (NCATS), a component of the National Institutes of Health (NIH), and NIH Roadmap for Medical Research. B.J.D. is an investigator of the Howard Hughes Medical Institute. J.W.T. is supported by grants from the V Foundation for Cancer Research, The Leukemia & Lymphoma Society, the Gabrielle’s Angel Foundation for Cancer Research, and the National Cancer Institute (5R00CA151457-04; 1R01CA183974-01). BPR and AG are supported by the Life Raft Group.
M.C.H. consults for and has equity interest in Molecular MD and is a consultant for Novartis, Pfizer, Ariad, and Onyx; M.C.H. has intellectual property on patenting of the use of imatinib to treat GIST (licensed by OHSU to Novartis); C.L.C. consults for Novartis; B.P.R. is on the speaker’s bureau and is an advisory board participant for Novartis; B.J.D serves as a consultant to Molecular MD, Blueprint Medicines, Gilead Sciences, Cell Therapeutics, Inc., AstraZeneca, Cylene Pharmaceuticals and Lorus Therapeutics. OHSU on behalf of B.J.D. is recipient of a subcontract from Oncotide’s NIH STTR 1R41CA165570-01. OHSU patents from which B.J.D. has or will receive royalties as an inventor: #843 Mutated ABL Kinase Domains (has been licensed to ARIAD Pharmaceuticals, Inc.; Array BioPharma, Inc.; Curis, Inc.; Molecular MD Corporation; Pfizer, Inc.; Piramal Health Care, Ltd.; Praecis Pharmaceuticals, Inc.; SGX Pharmaceuticals, Inc.; The Translational Genomics Research Institute; Vertex Pharmaceuticals, Inc.; and others; #0996 Detection of Gleevec Resistant Mutations (licensed to Molecular MD); #0606 Treatment of Gastrointestinal Stromal Tumors (exclusively licensed to Novartis).
Footnotes
Conflict of Interest Disclosure Statement: OHSU has clinical trial contracts with Novartis, Bristol-Myers Squibb and ARIAD to pay for patient costs, nurse and data manager salaries, and institutional overhead. B.J.D. does not derive salary, nor does his laboratory receive funds, from these contracts. OHSU and B.J.D. have a financial interest in Molecular MD. This potential individual and institutional conflict of interest has been reviewed and managed by OHSU.
References
- 1.Miettinen M, Lasota J. Gastrointestinal Stromal Tumors: Review on Morphology. Molecular Pathology, Prognosis, and Differential Diagnosis. 2009 Oct 1; doi: 10.5858/2006-130-1466-GSTROM. [DOI] [PubMed] [Google Scholar]
- 2.Hirota S, Nishida T, Isozaki K, Taniguchi M, Nakamura J, Okazaki T, et al. Gain-of-function mutation at the extracellular domain of KIT in gastrointestinal stromal tumours. J Pathol. 2001:505–510. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH818>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 3.Hirota S, Ohashi A, Nishida T, Isozaki K, Kinoshita K, Shinomura Y, et al. Gain-of-function mutations of platelet-derived growth factor receptor α gene in gastrointestinal stromal tumors. Gastroenterology. 2003 Sep;:660–667. doi: 10.1016/s0016-5085(03)01046-1. [DOI] [PubMed] [Google Scholar]
- 4.Heinrich MC, Corless CL, Duensing a, McGreevey L, Chen CJ, Joseph N, et al. PDGFRA activating mutations in gastrointestinal stromal tumors. Science (80-) 2003 Jan 31;:708–710. doi: 10.1126/science.1079666. [DOI] [PubMed] [Google Scholar]
- 5.Bauer S, Duensing a, Demetri GD, Fletcher Ja. KIT oncogenic signaling mechanisms in imatinib-resistant gastrointestinal stromal tumor: PI3-kinase/AKT is a crucial survival pathway. Oncogene. 2007 Nov 29;:7560–7568. doi: 10.1038/sj.onc.1210558. [DOI] [PubMed] [Google Scholar]
- 6.Chi P, Chen Y, Zhang L, Guo X, Wongvipat J, Shamu T, et al. ETV1 is a lineage survival factor that cooperates with KIT in gastrointestinal stromal tumours. Nature. 2010 Oct 14;:849–853. doi: 10.1038/nature09409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schlemmer M, Bauer S, Schütte R, Hartmann J, Bokemeyer C, Hosius C, et al. Activity and side effects of imatinib in patients with gastrointestinal stromal tumors: data from a german multicenter trial. Eur J Med Res. 2011 May;:206. doi: 10.1186/2047-783X-16-5-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blanke CD, Rankin C, Demetri GD, Ryan CW, von Mehren M, Benjamin RS, et al. Phase III randomized, intergroup trial assessing imatinib mesylate at two dose levels in patients with unresectable or metastatic gastrointestinal stromal tumors expressing the kit receptor tyrosine kinase: S0033. J Clin Oncol. 2008 Feb 1;:626–632. doi: 10.1200/JCO.2007.13.4452. [DOI] [PubMed] [Google Scholar]
- 9.Liegl B, Kepten I, Le C, Zhu M, Demetri GD, Heinrich MC, et al. Heterogeneity of kinase inhibitor resistance mechanisms in GIST. 2008:64–74. doi: 10.1002/path. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greulich H, Pollock PM. Targeting mutant fibroblast growth factor receptors in cancer. Trends Mol Med. 2011 May;:283–292. doi: 10.1016/j.molmed.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Di Martino E, Tomlinson DC, Knowles Ma. A Decade of FGF Receptor Research in Bladder Cancer: Past, Present, and Future Challenges. Adv Urol. 2012 Jan;:429213. doi: 10.1155/2012/429213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tomlinson DC, Baldo O, Harnden P, Knowles Ma. FGFR3 protein expression and its relationship to mutation status and prognostic variables in bladder cancer. J Pathol. 2007:91–98. doi: 10.1002/path.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Onwuazor ON, Wen X-Y, Wang D-Y, Zhuang L, Masih-Khan E, Claudio J, et al. Mutation, SNP, and isoform analysis of fibroblast growth factor receptor 3 (FGFR3) in 150 newly diagnosed multiple myeloma patients. Blood. 2003 Jul 15;:772–773. doi: 10.1182/blood-2003-04-1204. [DOI] [PubMed] [Google Scholar]
- 14.Li Z. The myeloma-associated oncogene fibroblast growth factor receptor 3 is transforming in hematopoietic cells. Blood. 2001 Apr 15;:2413–2419. doi: 10.1182/blood.v97.8.2413. [DOI] [PubMed] [Google Scholar]
- 15.Tomlinson DC, Knowles Ma, Speirs V. Mechanisms of FGFR3 actions in endocrine resistant breast cancer. Int J Cancer. 2012 Jun 15;:2857–2866. doi: 10.1002/ijc.26304. [DOI] [PubMed] [Google Scholar]
- 16.Kwabi-Addo B, Ozen M, Ittmann M. The role of fibroblast growth factors and their receptors in prostate cancer. Endocr Relat Cancer. 2004 Dec;:709–724. doi: 10.1677/erc.1.00535. [DOI] [PubMed] [Google Scholar]
- 17.Yang F, Strand DW, Rowley DR. Fibroblast growth factor-2 mediates transforming growth factor-beta action in prostate cancer reactive stroma. Oncogene. 2008 Jan 17;:450–459. doi: 10.1038/sj.onc.1210663. [DOI] [PubMed] [Google Scholar]
- 18.Murphy T, Darby S, Mathers ME, Gnanapragasam VJ. Evidence for distinct alterations in the FGF axis in prostate cancer progression to an aggressive clinical phenotype. J Pathol. 2010:452–460. doi: 10.1002/path.2657. [DOI] [PubMed] [Google Scholar]
- 19.Tyner JW, Walters DK, Willis SG, Luttropp M, Oost J, Loriaux M, et al. RNAi screening of the tyrosine kinome identifies therapeutic targets in acute myeloid leukemia. Blood. 2008 Feb 15;:2238–2245. doi: 10.1182/blood-2007-06-097253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tyner JW, Deininger MW, Loriaux MM, Chang BH, Gotlib JR, Willis SG, et al. RNAi screen for rapid therapeutic target identification in leukemia patients. Proc Natl Acad Sci U S A. 2009 May 26;:8695–8700. doi: 10.1073/pnas.0903233106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bicocca VT, Chang BH, Masouleh BK, Muschen M, Loriaux MM, Druker BJ, et al. Crosstalk between ROR1 and the Pre-B cell receptor promotes survival of t(1;19) acute lymphoblastic leukemia. Cancer Cell. 2012 Nov 13;:656–667. doi: 10.1016/j.ccr.2012.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eswarakumar VP, Lax I, Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine growth factor Rev. 2005 Apr;:139–149. doi: 10.1016/j.cytogfr.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 23.Johnson MD, O’Connell MJ, Pilcher W, Reeder JE. Fibroblast growth factor receptor-3 expression in meningiomas with stimulation of proliferation by the phosphoinositide 3 kinase-Akt pathway. J Neurosurg. 2010 May;:934–939. doi: 10.3171/2009.7.JNS09726. [DOI] [PubMed] [Google Scholar]
- 24.Tomlinson D, Knowles M, Speirs V. Mechanisms of FGFR3 actions in endocrine resistant breast cancer. Int J Cancer. 2011 Jul;:1–10. doi: 10.1002/ijc.26304. [DOI] [PubMed] [Google Scholar]
- 25.Yadav V, Zhang X, Liu J, Estrem S, Li S, Gong X-Q, et al. Reactivation of mitogen-activated protein kinase (MAPK) pathway by FGF receptor 3 (FGFR3)/Ras mediates resistance to vemurafenib in human B-RAF V600E mutant melanoma. J Biol Chem. 2012 Aug 10;:28087–28098. doi: 10.1074/jbc.M112.377218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wesche J, Haglund K, Haugsten EM. Fibroblast growth factors and their receptors in cancer. Biochem J. 2011 Jul 15;:199–213. doi: 10.1042/BJ20101603. [DOI] [PubMed] [Google Scholar]
- 27.Ware KE, Marshall ME, Heasley LR, Marek L, Hinz TK, Hercule P, et al. Rapidly acquired resistance to EGFR tyrosine kinase inhibitors in NSCLC cell lines through de-repression of FGFR2 and FGFR3 expression. PLoS One. 2010 Jan;:e14117. doi: 10.1371/journal.pone.0014117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Semrad TJ, Mack PC. Fibroblast Growth Factor Signaling in Non–Small-Cell Lung Cancer. Clin Lung Cancer. 2012:90–95. doi: 10.1016/j.cllc.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 29.Ware KE, Hinz TK, Kleczko E, Singleton KR, Marek LA, Helfrich BA, et al. A mechanism of resistance to gefitinib mediated by cellular reprogramming and the acquisition of an FGF2-FGFR1 autocrine growth loop. Oncogenesis. 2013 Jan;:e39. doi: 10.1038/oncsis.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salm F, Cwiek P, Ghosal A, Lucia Buccarello A, Largey F, Wotzkow C, et al. RNA interference screening identifies a novel role for autocrine fibroblast growth factor signaling in neuroblastoma chemoresistance. Oncogene. 2013 Aug 22;:3944–3953. doi: 10.1038/onc.2012.416. [DOI] [PubMed] [Google Scholar]
- 31.Pardo OE, Wellbrock C, Khanzada UK, Aubert M, Arozarena I, Davidson S, et al. FGF-2 protects small cell lung cancer cells from apoptosis through a complex involving PKCepsilon, B-Raf and S6K2. EMBO J. 2006 Jul 12;:3078–3088. doi: 10.1038/sj.emboj.7601198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marek L, Ware KE, Fritzsche A, Hercule P, Helton WR, Smith JE, et al. Fibroblast Growth Factor (FGF) and FGF Receptor-Mediated Autocrine Signaling in Non – Small-Cell Lung Cancer Cells. Cell Prolif. 2009:196–207. doi: 10.1124/mol.108.049544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huynh H, Lee JWJ, Chow PKH, Ngo VC, Bin Lew G, Lam IWL, et al. Sorafenib induces growth suppression in mouse models of gastrointestinal stromal tumor. Mol Cancer Ther. 2009 Jan 1;:152–159. doi: 10.1158/1535-7163.MCT-08-0553. [DOI] [PubMed] [Google Scholar]
- 34.Karamouzis MV, Konstantinopoulos PA, Papavassiliou AG. Targeting MET as a strategy to overcome crosstalk-related resistance to EGFR inhibitors. Lancet Oncol. 2009 Jul;:709–717. doi: 10.1016/S1470-2045(09)70137-8. [DOI] [PubMed] [Google Scholar]
- 35.Yokote H, Fujita K, Jing X, Sawada T, Liang S, Yao L, et al. Trans-activation of EphA4 and FGF receptors mediated by direct interactions between their cytoplasmic domains. Proc Natl Acad Sci U S A. 2005:18866–18871. doi: 10.1073/pnas.0509741102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.