Abstract
Aims
Personal genomic testing (PGT) for common disease risk is becoming increasingly frequent, but little is known about people's array of emotional reactions to learning their genomic risk profiles and the psychological harms/benefits of PGT. We conducted a study of post-PGT affect, including positive, neutral, and negative states that may arise after testing.
Methods
Two hundred twenty-eight healthy adults received PGT for common disease variants and completed a semi-structured research interview within two weeks of disclosure. Study participants reported how PGT results made them feel in their own words. Using an iterative coding process, responses were organized into three broad affective categories (Negative, Neutral, and Positive affect).
Results
Neutral affect was the most prevalent response (53.9%), followed by Positive affect (26.9%) and Negative affect (19.2%). We found no differences by gender, race or education.
Conclusions
While <20% of participants reported negative affect in response to learning their genomic risk profile for common disease, a majority experience either neutral or positive emotions. These findings contribute to the growing evidence that PGT does not impose significant psychological harms. Moreover, they point to a need to better link theories and assessments in both emotional and cognitive processing to capitalize on PGT information for healthy behavior change.
Keywords: Genetic testing, Health psychology, Psychological impact
Introduction
Several systematic reviews of the psychological implications for adults participating in predictive genetic testing for rare, hereditary diseases (e.g., certain cancers, neurodegenerative conditions) have concluded that learning one's personal risk profile can lead to mildly elevated, short-term psychological distress that dissipates over time for most individuals [1-4]. For these rare conditions, genotype:phenotype associations tend to be strong. If a risk allele is highly penetrant, the associated disease almost always occurs. Penetrance is attenuated when some individuals do not go on to develop the disease, even though they carry the allele.
Personal genomic testing (PGT) for common diseases that are more prevalent in the general population (e.g., diabetes, heart disease) is distinguished by identifying multiple low penetrance genes or gene variants that only sometimes result in disease and can be influenced by other factors (e.g., the environment). Studies to date on the psychological implications of PGT suggest that this testing does not produce significant mental health burdens [5-7].
Data about the absence of psychological harm surrounding predictive genetic testing more generally, and PGT in particular, have set the stage for the proliferation of such tests in the consumer marketplace—and often without the benefit of consultation by genetic/genomic health care professionals [8]. However, the absence of psychological harm is a rather narrow lens through which to view emotional responses to a phenomena as complex as learning about genetic determinants of one's health, and might not sufficiently attend to the broader meanings ascribed to this information in everyday life [7]. For instance, individuals could respond more neutrally to information about their higher chance of developing a certain disease (e.g., diabetes) if they expected this finding based on their personal or family health history (e.g., being overweight, having a biological parent with diabetes). Moreover, individuals could even respond positively or favorably to knowing this information, since early identification of health risk could lead to preventive or risk-reducing treatments. For instance, a systematic review of at-risk individuals tested for predisposition to hereditary hemochromatosis found decreases in distress and increases in quality of life after such testing [9-11]. More relevant to PGT for common disease risk and risk factors, a recent study identified relief from self-blame following testing for a common obesity gene variant [12]. This and other studies that have conveyed genetic risk for obesity using either actual testing or hypothetical vignettes [13] note that those who are tested derived “personal” psychological benefits from testing. Indeed, the concept of “personal utility” of genetic risk information in a growing area of exploration [6,14]. Apart from distressing emotions, neutral and positive affect can also have implications for processing health information [15,16]. More positive affective associations with multiple health behavior domains, including diet and physical activity, have been linked to greater levels of engagement in these behaviors [17-20]. Positive emotions of happiness and relief have been included in measures of the psychological reaction to receiving genetic test results related to hereditary cancers [21]. However, the range of responses and the potential to capitalize on these as motivators of behavior change in the context of PGT has gone largely unexamined.
To help move the field of PGT forward, and with greater attention to a fuller range of its psychological implications, we believe it is appropriate to take a step back and reconsider the complete spectrum of emotional reactions that persons may experience in response to genetic testing for common disease risk. Gaining a complete understanding of the true nature and relative balances among positive, neutral, and negative affective responses of individuals to PGT is necessary in determining if there are additional patient education and counseling objectives that must be met and in fulfilling the promise of PGT as a means to improve health. As these are predicated on the notion that information derived from PGT can be incorporated into people's lives in impactful ways (i.e., motivating them to change their lifestyle behaviors, undergo preventive interventions, adhere to treatments, etc.), then affective responses may be an under-recognized opportunity to learn what matters most to individuals when considering future health threats. We also should consider whether these affective responses vary by important demographic factors, such as gender, race or educational attainment.
Study Purpose
In this report, we set out to characterize individuals' affective responses to their receipt of PGT results for common disease risk. Our data were drawn from the National Human Genome Research Institute's (NHGRI) Multiplex Initiative [22], a population-based study of healthy adults offered genetic testing for common disease risk. All participants underwent PGT and received their genetic test results prior to data collection. As part of a semi-structured research interview, these individuals were afforded the opportunity to report to us, in their own words, how they felt following the receipt of their genetic health risk profile. Our primary hypothesis was that individuals would report a broad array of affective, emotional outcomes to their test results that included but were not limited to negative affect. Moreover, we expected that negative affect, when reported by participants, would constitute a minority of responses. Finally, we also assessed whether responses varied by gender, race or education.
Method
Study Design
The NHGRI Multiplex Initiative has previously been described in detail elsewhere [23,24]. Briefly, the multiplex genetic susceptibility test used in this study included 15 genetic variants associated with increases in risk for 8 common chronic health conditions (diabetes, heart disease, high cholesterol, high blood pressure, lung cancer, colon cancer, skin cancer, and osteoporosis) [25].
Participants
Participants in the Multiplex Initiative were selected from a large health maintenance organization. Selection criteria included being enrolled in the plan for at least 2 years, being age 25-40 years, and not having the health conditions assayed through the multiplex test. A random sample of the members meeting these criteria was drawn, oversampling for male gender, African American race, and lower educational status (based on census tract information associated with current address; “lower” being ≤ high school). Study recruitment occurred from February, 2007 to May, 2008 [23]. All procedures were approved by the Institutional Review Boards at the NHGRI and the Henry Ford Health System. Data for the present analysis are based on the 228 patient participants who agreed to the multiplex test, completed a baseline telephone assessment, and a post-disclosure follow-up telephone call to complete a semi-structured research interview with a research educator within 2 weeks after receiving their results by mail.
Post-disclosure Call
As described elsewhere [26], the post-disclosure call was scripted and data collection assessments were conducted via a semi-structured research interview along with discussion of test results. Calls were recorded for quality assurance by a study manager. Study materials included contact information for a medical geneticist or genetic counselor, but no participants requested this. Development and implementation of the mailed feedback and education call relied on best practices in clear public health communication and health literacy [26-28]. The call focused on ensuring that participants had an accurate understanding of the meaning of the test results. A mixed-methods interview was embedded in this call. Importantly, the call did not include discussion of the psychological implications of test results, as would be typical within the context of genetic counseling [29]. Research educators were instructed not to probe for further clarification in order to keep the interview highly structured across participants.
Data and Measures
Baseline data
Sociodemographic information was collected at baseline and prior to the provision of a DNA sample for analysis. This included participant age, gender, race/ethnicity, marital status, and educational status. Educational status was indexed by the highest grade or year in school completed, later trichotomized as ≤ high school, some college/vocational school, and ≥ college degree.
Genetic risk profile
A complete description of the multiplex test feedback protocol is described elsewhere [26]. Briefly, participants received a folder by mail containing their results, as well as three supplementary one-page enclosures describing important caveats about the results and behavioral strategies to reduce disease risk, and preparing them for the research educator follow-up telephone call (Figure 1). Feedback was provided about health conditions (vs. genes and risk genes) due to greater familiarity with diseases.
Figure 1. Example of Multiplex test results feedback received by mail.

Patient-reported outcomes
A trained research educator attempted to contact participants by telephone within 10 days of receipt of mailed multiplex test results. Data for the current analysis are drawn from participants' responses to the open-ended question probe eliciting information about emotional reactions to learning their PGT results: “People can have different reactions to genetic test results. In your own words, how did you feel about getting your test results?” All responses were recorded verbatim and all interviews were audio recorded for quality control purposes.
Data Analysis
We first generated frequencies and descriptive statistics to determine participants' sociodemographic and genomic risk profile information. All semi-structured interview data were read, content coded, and analyzed by a team of experts (including C.B. and C.M.M.) using a study-specific coding scheme based on Grounded Theory [30,31]. These data were analyzed in two phases. In the first phase of data analysis, the coding scheme initially applied emotion-focused adjectives and other affect terms commonly encountered in scales of emotion, such as those included in the Positive and Negative Affect Schedule (PANAS) [32] (e.g., nervous, determined). Next, the team expanded its initial coding scheme as needed based on the frequency of participant responses falling outside of these parameters, adding additional affect codes and other codes that were nonspecific to affect (i.e., cognitions). This process ultimately resulted in 27 discrete codes, with an additional code used to capture more general comments that were neither affective nor cognitive in nature. Two independent coders (including C.M.B.) then applied this coding scheme to the available study data: inter-coder reliability across the 27 codes was high (Cohen's Kappa = 0.83) [33]. Discrepancies were resolved by consensus. Each response was assigned one or more codes depending on verbalization and context—all codes were applied equally. The frequency of each code was then computed, representing the number of times each code was applied to the dataset. These frequencies were then summed to provide an overall count of the total number of coded responses, and each code's percentage of the overall total was computed (individual code frequency/total code frequency).
In the second phase of data analysis, 2 additional independent coders (S.C.O., K.P.T.) sorted these 27 codes along a thematic continuum of affect ranging from negative to positive emotions, resulting in 3 broad thematic categories (Neutral [e.g., “Indifferent”], Positive [e.g., “Relieved”], and Negative [e.g., “Afraid”]). Four of these codes captured responses that were not affective, but rather, reflected responses to the education session itself. These were not considered further in this analysis, leaving 23 remaining codes. The team next applied Q-sort methodology (see Lilienfeld [34] for review) with 12 genetic health research assistants who rank-ordered each of the 23 codes into the 3 categories, based on their perceptions of the code's thematic likelihood of membership (from most to least likely). Codes were assigned to one of the three categories as determined by absolute majority of ≥ 51% endorsement. (The ‘next most likely’ membership ranks were used in the event of ties and to help resolve discrepant responses by consensus.) The results of our Q-sort, and sample responses offered by participants across the 23 codes, are presented in Table 1. Full consensus by overall majority of our 12 coders was reached for thematically categorizing all but one of the 23 codes. With respect to Negative affect and emotion, 10 codes were applied, seven were applied as Neutral, and six codes were applied to Positive affect and emotion.
Table 1. Thematic Coding and Q-Sort Results.
| Q-Sort Category of Emotion | ||||
|---|---|---|---|---|
|
| ||||
| Thematic Category of Emotion/Affect Code (frequency) | Neutral | Positive | Negative | Sample Response |
| Neutral Emotion (7) | ||||
| Not Surprised (50) | 12 | 0 | 0 | Not surprised. It didn't bother me. |
| Surprised (38) | 8 | 1 | 2 | Somewhat surprised with a few results. |
| Okay (31) | 8 | 3 | 0 | I just thought ‘okay’. |
| Indifferent (27) | 10 | 0 | 2 | I don't know, indifferent. |
| Not Concerned (22) | 6 | 5 | 1 | I didn't feel anything unusual, not scared. |
| Mixed Emotions (15) | 6 | 0 | 3 | Um, kind of a mix of emotions. |
| Just Information (6) | 8 | 0 | 0 | I guess it was just information. |
|
| ||||
| Positive Emotion (6) | ||||
| Interesting (32) | 1 | 9 | 0 | I was interested…to see what it said. |
| Excited (24) | 0 | 12 | 0 | I was definitely excited to get them. |
| Happy (16) | 0 | 12 | 0 | I had fewer predispositions than average so I was happy. |
| Curious (14) | 2 | 5 | 0 | Curious…(not) life changing. |
| Hopeful (4) | 0 | 12 | 0 | I was hopeful (about) getting the results. |
| Relieved (4) | 0 | 12 | 0 | Relieved, actually. |
|
| ||||
| Negative Emotion (10) | ||||
| Nervous (23) | 0 | 0 | 12 | Nervous at first. |
| Afraid (9) | 0 | 0 | 12 | It was kind of scary. |
| Concerned (7) | 2 | 1 | 7 | I was concerned, “freaked out”. |
| Shocked (7) | 3 | 0 | 8 | It was kind of a shock…(some results) I didn't expect. |
| Horrible/Disappointed (6) | 0 | 0 | 12 | It was horrible. I got all the genes. |
| Uninformative (5) | 3 | 0 | 7 | It does not say a lot to me. |
| Misinformation (4) | 0 | 0 | 9 | I didn't agree with them. |
| Skeptical (3) | 1 | 0 | 7 | I don't think that they were accurate. |
| Not understandable (2) | 1 | 0 | 10 | I didn't understand some of the numbers. |
| Vague (1) | 4 | 0 | 5 | I thought it was vague, I didn't have much reaction. |
The number of codes assigned to each category was then tallied, followed by computing each code's within-category percentage (individual code frequency/total code frequency for category). Each thematic category's relative frequency was then computed, followed by its relative percentage of the overall total (total code frequency for category/total code frequency). Finally, we performed t-tests to determine whether there are significant differences in the overall frequency of responses within each of the four categories by gender, race (White/non-White) and education (< college/≥ college).
Results
Sociodemographics
Participants who sought multiplex testing were, on average, 34 years old and just over half (57%) were female (Table 2). Sixty-two percent were non-Hispanic white and 27% were African American. Just over half (52%) were college educated. A majority of our participants were in a partnered relationship. Participants carried at least 1 variant associated with increased risk for 6 (M = 5.9, SD = 0.9) of the 8 health conditions; they carried an average of 9.2 risk alleles (SD = 1.7) out of the possible 15.
Table 2. Participant Sociodemographic Characteristics (N = 228).
| Variable | % | Mean (SD) |
|---|---|---|
| Demographics | ||
| Age (years) | 34.89 (4.17) | |
| Gender (% female) | 56.58% | |
| Race/ethnicity | ||
| Non-Hispanic white | 62.28% | |
| African American | 27.63% | |
| Other | 10.09% | |
| Education | ||
| ≤ High school | 10.53% | |
| Some college | 37.72% | |
| ≥ College | 51.75% | |
| Partnered | 67.11% | |
| Carry at least one genetic variant associated with increased risk | ||
| Diabetes | 98.25% | |
| Osteoporosis | 98.25% | |
| Heart disease | 97.37% | |
| High cholesterol | 85.53% | |
| Hypertension | 33.77% | |
| Lung cancer | 58.77% | |
| Colon cancer | 93.42% | |
| Skin cancer | 19.74% | |
Frequencies of Responses Generated
Responses generated by the 228 participants produced a total of 351 codes across 23 categories. Each participant's response received a mean of 1.88 codes (range 1-3). For example, “I guess not super concerned because I kind of figured I would get some sort of cancer and the diabetes and all that…so I kind of figured I'd have something…” generated one code (Not Surprised), while “I was eager to get them because I was curious what the results would be - I kind of figured I would be predisposed to many of the things that were tested. Skin cancer surprised me.” generated 3 codes (Nervous, Surprised, and Curious).
Reported Responses to Test Results
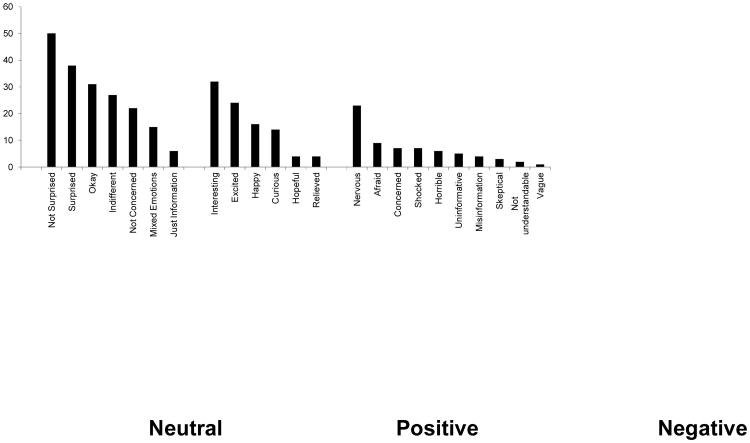
Coded frequencies within each category are presented in Figure 2. We present these data along a continuum from Neutral to Positive to Negative affect responses. Our participants displayed a full range of emotional responses following the receipt of their test results. Along this continuum, responses in the Neutral category were, by far, the most common, capturing over half of all responses (53.9%). Neutral responses were followed by responses in the Positive category, capturing just over a quarter of responses (26.9%). Reponses capturing Negative affect were least common, representing only 19.2% of all responses coded.
Figure 2. Frequency Distribution of Coded Affect Responses within Thematic Categories.
When we examined the most frequently coded responses within each category and across all categories, the most commonly encountered responses were “Not Surprised” (14.2%) and “Surprised” (10.8%) within the Neutral emotion category. In the Positive category, “Interesting” (9.1%) was the most frequent. “Nervous” (6.5%) was the most frequent response within the Negative emotion category.
Group Differences in Affective Responses
We examined differences in the overall frequency of Positive, Neutral, and Negative responses by gender, race and education. We found no significant group differences within any of the three response categories. In addition, there were no significant differences in the overall frequency of responses across these groups.
Discussion
Last year, the Food and Drug Administration (FDA) ordered the largest provider of direct-to-consumer genetic testing, 23andMe, to discontinue marketing of its health-related PGT. Concerns cited include that consumers may not adequately understand test results and as a consequence, might respond to test results in ways that would be counter to medical advice [35]. However, Green and Farahany [36] noted that while more systematic research is needed regarding the outcomes of consumer genomic testing, the growing number of studies to date do not point to significant harm as a result of this testing. Our findings contribute to this growing body of evidence that PGT for common disease risk does not pose significant psychological harms to participants, nor did these responses vary in a way to suggest that certain groups, such as those with less formal education, would be more likely to report these responses. Indeed, our work points to a need to better link theories and assessments in emotional and cognitive processing to capitalize on the range of responses individuals have to PGT information during patient education and counseling encounters. This could include several potential measurement methodologies, including existing scales, continued use of open-ended prompts such as the one used here, and more direct measurement of the affective experience. This future research agenda would benefit from a robust opportunity to offer PGT to individuals and carefully assess their understanding and application of this information to their health [36].
Future work should seek to consider the impact of positive affective experiences on the processing of genomic risk information. For instance, “Interesting” held the most responses in the Positive category. A growing body of literature [15,37,38] now suggests that positive affect can improve individuals' information processing and the quality of their health decision making. Interventions aimed at capturing positive affect and engagement associated with the receipt of PGT could enhance the processing of health information incorporated into interventions and produce greater behavior change and salience.
Affect varies not only in valence (positive/neutral/negative) but also strength [39-41]. While our method was most appropriate for capturing affective valence, many responses within the Neutral category, such as “OK,” “Indifferent,” and “Just Information” speak to an affective response that is not particularly strong. Further, even when asked for an emotional response, many participants did not report an affective reaction at all. Hay and colleagues [42] provided an early theoretical synthesis designed to anticipate the direction of psychological and behavioral research in PGT. They underscored the importance of affective engagement with genomic risk information. They postulated both positive and negative affective responses to genomic risk information, and state that engagement (or in other words, the strength of the affective response) is crucial in how individuals ultimately will apply this information to their health. Future work should measure a range of affective valence and strength as one way to capture levels of engagement with this health information. This work should also include development of novel measures that capture responses to PGT outside of the domain of distress and negative affect [43-46]. Our work can serve as a first step in this work by providing domains for item development and further measurement refinement.
Finally, given the most frequent response was “Not Surprised” followed by “Surprised,” one must consider the implications of information that either confirms or disconfirms the individual's expectations and the mental schema they hold regarding their health risks. Such findings are likely to increase in frequency in the era of whole genome and whole exome sequencing that takes the field beyond targeted testing in high-risk samples [47-50]. Limited research on patient perspectives of this approach has been done on this topic to date. Participants in one cohort expressed strong intentions for learning their sequence results, generally speaking, with the motivation of applying these to disease prevention [51]. In a more recent study of patients receiving diagnostic exome sequencing, 93.5% of these opted to learn incidental deleterious findings [52]. These studies suggest that, regardless of guidelines, many patients would seek to learn their sequence and interpretation to the degree possible using the literature to date. Therefore, patients' responses to deleterious findings in the absence of a significant family history of disease [53] should be the focus of research moving forward.
The participants in this study were part of a large, diverse, population-based study of healthy adults. Despite this, there are several limitations. While the initial sampling frame for this study began with oversampling for male, African-American and less educated participants, groups who have been less represented in genomic research to date, those who sought testing were more likely to be female, White and college educated. Therefore, while we found no group differences in the present analysis, the responses captured by the present work may not generalize to the population at large. Also, our data were generated from response to a single open-ended question. Employment of longer qualitative interviews with successive probes could have yielded additional data and provided more insight into the affective responses of these participants.
Despite these limitations, the present findings contribute to our understanding of the range of affective responses that healthy adults have to PGT. As PGT continues to reach greater segments of general public, it is critical to understand how patients might respond to this information, with or without the benefit of a health care practitioner. Our results suggest that when drawing from a large, diverse population of young, healthy adults, those who elect to be tested report a range of affective responses. Future research should seek to better understand these responses and determine how to capitalize on them to enhance public health. This research should include the development of measures that better capture the range of responses that we have identified in our study. Item development could follow this work, as well as scale development and validation in samples receiving PGT in diverse settings.
Acknowledgments
This work was supported by the Intramural Research Program of the National Human Genome Research Institute (NHGRI). However, the proposed research was made possible by collaboration with the Cancer Research Network funded by the National Cancer Institute (CA079689). Additional resources were provided by Group Health Research Institute and Henry Ford Hospital. Genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University (HHSN268200782096C). We thank the non-author members of the NHGRI Multiplex Initiative Steering Committee (Drs. Lawrence Brody, Robert Reid, Eric Larson, Andreas Baxevanis, and Sharon Kardia) who provided critical review of this report. Our thanks also go to the study participants who were all members of the Henry Ford Health System. Manuscript preparation was supported, in part, by grants from the American Cancer Society (10-110-01-CPPB) (to S.C.O.), the Office of the Director, National Institutes of Health (HG006754) (to K.P.T.) and the National Cancer Institute (P30051008). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
References
- 1.Claes E, Renson M, Delespesse A, De H, Haelterman HV, Kartheuser G, Van CE. Psychological implications of living with familial adenomatous polyposis. Acta Gastroenterol Belig. 2011;74:438–444. [PubMed] [Google Scholar]
- 2.Crotser CB, Boehmke M. Survivorship considerations in adults with hereditary breast and ovarian cancer syndrome: state of the science. J Cancer Surviv. 2009;3:21–42. doi: 10.1007/s11764-008-0077-7. [DOI] [PubMed] [Google Scholar]
- 3.Hilgart JS, Coles B, Iredale R. Cancer genetic risk assessment for individuals at risk of familial breast cancer. Cochrane Database Syst Rev. 2012;2:CD003721. doi: 10.1002/14651858.CD003721.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paulsen JS, Nance M, Kim JI, Carlozzi NE, Panegyres PK, Erwin C, Goh A, McCusker E, Williams JK. A review of quality of life after predictive testing for and earlier identification of neurodegenerative diseases. Prog Neurobiol. 2013;110:2–28. doi: 10.1016/j.pneurobio.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bloss CS, Schork NJ, Topol EJ. Effect of direct-to-consumer genomewide profiling to assess disease risk. N Engl J Med. 2011;364:524–534. doi: 10.1056/NEJMoa1011893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bloss CS, Wineinger NE, Darst BF, Schork NJ, Topol EJ. Impact of direct-to-consumer genomic testing at long term follow-up. J Med Genet. 2013;50:393–400. doi: 10.1136/jmedgenet-2012-101207. [DOI] [PubMed] [Google Scholar]
- 7.Roberts JS, Ostergren J. Direct-to-consumer genetic testing and personal genomics services: a review of recent empirical studies. Curr Genet Med Rep. 2013;1:182–200. doi: 10.1007/s40142-013-0018-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caulfield T, McGuire AL. Direct-to-consumer genetic testing: perceptions, problems, and policy responses. Annu Rev Med. 2012;63:23–33. doi: 10.1146/annurev-med-062110-123753. [DOI] [PubMed] [Google Scholar]
- 9.Anderson RT, Wenzel L, Walker AP, Ruggiero A, Acton RT, Hall MA, Tucker DC, Thomson E, Harrison B, Howe E, III, Holup J, Leiendecker-Foster C, Power T, Adams P. Impact of hemochromatosis screening in patients with indeterminate results: the hemochromatosis and iron overload screening study. Genet Med. 2006;8:681–687. doi: 10.1097/01.gim.0000245631.07117.ac. [DOI] [PubMed] [Google Scholar]
- 10.Meiser B, Dunn S, Dixon J, Powell LW. Psychological adjustment and knowledge about hereditary hemochromatosis in a clinic-based sample: a prospective study. J Genet Couns. 2005;14:453–463. doi: 10.1007/s10897-005-6192-y. [DOI] [PubMed] [Google Scholar]
- 11.Picot J, Bryant J, Cooper K, Clegg A, Roderick P, Rosenberg W, Patch C. Psychosocial aspects of DNA testing for hereditary hemochromatosis in at-risk individuals: a systematic review. Genet Test Mol Biomarkers. 2009;13:7–14. doi: 10.1089/gtmb.2008.0064. [DOI] [PubMed] [Google Scholar]
- 12.Meisel SF, Wardle J. ‘Battling my biology’: psychological effects of genetic testing for risk of weight gain. J Genet Couns. 2014;23:179–186. doi: 10.1007/s10897-013-9628-9. [DOI] [PubMed] [Google Scholar]
- 13.Meisel SF, Walker C, Wardle J. Psychological responses to genetic testing for weight gain: a vignette study. Obesity (Silver Spring) 2012;20:540–546. doi: 10.1038/oby.2011.324. [DOI] [PubMed] [Google Scholar]
- 14.Bunnik EM, Janssens AC, Schermer MH. Personal utility in genomic testing: is there such a thing? J Med Ethics. 2014 doi: 10.1136/medethics-2013-101887. May 28, Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 15.Fredrickson BL, Branigan C. Positive emotions broaden the scope of attention and thought-action repertoires. Cogn Emot. 2005;19:313–332. doi: 10.1080/02699930441000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friedman RS, Forster J. Implicit affective cues and attentional tuning: an integrative review. Psychol Bull. 2010;136:875–893. doi: 10.1037/a0020495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conner M, Rhodes RE, Morris B, McEachan R, Lawton R. Changing exercise through targeting affective or cognitive attitudes. Psychol Health. 2011;26:133–149. doi: 10.1080/08870446.2011.531570. [DOI] [PubMed] [Google Scholar]
- 18.Kiviniemi MT, Voss-Humke AM, Seifert AL. How do I feel about the behavior? The interplay of affective associations with behaviors and cognitive beliefs as influences on physical activity behavior. Health Psychol. 2007;26:152–158. doi: 10.1037/0278-6133.26.2.152. [DOI] [PubMed] [Google Scholar]
- 19.Kiviniemi MT, Duangdao KM. Affective associations mediate the influence of cost-benefit beliefs on fruit and vegetable consumption. Appetite. 2009;52:771–775. doi: 10.1016/j.appet.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walsh EM, Kiviniemi MT. Changing how I feel about the food: experimentally manipulated affective associations with fruits change fruit choice behaviors. J Behav Med. 2014;37:322–331. doi: 10.1007/s10865-012-9490-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cella D, Hughes C, Peterman A, Chang CH, Peshkin BN, Schwartz MD, Wenzel L, Lemke A, Marcus AC, Lerman C. A brief assessment of concerns associated with genetic testing for cancer: the Multidimensional Impact of Cancer Risk Assessment (MICRA) questionnaire. Health Psychol. 2002;21:564–572. [PubMed] [Google Scholar]
- 22.McBride CM, Alford SH, Reid RJ, Larson EB, Baxevanis AD, Brody LC. Characteristics of users of online personalized genomic risk assessments: implications for physician-patient interactions. Genet Med. 2009;11:582–587. doi: 10.1097/GIM.0b013e3181b22c3a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hensley Alford S, McBride CM, Reid RJ, Larson EB, Baxevanis AD, Brody LC. Participation in genetic testing research varies by social group. Public Health Genomics. 2011;14:85–93. doi: 10.1159/000294277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McBride CM, Alford SH, Reid RJ, Larson EB, Baxevanis AD, Brody LC. Putting science over supposition in the arena of personalized genomics. Nat Genet. 2008;40:939–942. doi: 10.1038/ng0808-939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wade CH, McBride CM, Kardia SL, Brody LC. Considerations for designing a prototype genetic test for use in translational research. Public Health Genomics. 2010;13:155–165. doi: 10.1159/000236061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaphingst KA, McBride CM, Wade C, Alford SH, Reid R, Larson E, Baxevanis AD, Brody LC. Patients' understanding of and responses to multiplex genetic susceptibility test results. Genet Med. 2012;14:681–687. doi: 10.1038/gim.2012.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doak CC, Doak LG, Root JH. Teaching Patients with Low Literacy Skills. 2nd. Philadelphia, PA: J.B. Lippincott Company; 1996. [Google Scholar]
- 28.National Cancer Institute. Clear & Simple: Developing Effective Print Materials for Low-Literate Readers. 2003 http://www.cancer.gov/aboutnci/oc/clear-and-simple.
- 29.Resta R, Biesecker BB, Bennett RL, Blum S, Hahn SE, Strecker MN, Williams JL. A new definition of Genetic Counseling: National Society of Genetic Counselors' Task Force report. J Genet Couns. 2006;15:77–83. doi: 10.1007/s10897-005-9014-3. [DOI] [PubMed] [Google Scholar]
- 30.Creswell JW, Plano Clark VL. Designing and Conducting Mixed Methods Research. 2nd. Thousand Oaks, CA: Sage Publications, Inc.; 2011. [Google Scholar]
- 31.Miles MB, Huberman AM. Qualitative Data Analysis: An Expanded Sourcebook. 2nd. Thousand Oaks, CA: Sage Publications, Inc.; 1994. [Google Scholar]
- 32.Watson D, Clark LA, Tellegen A. Development and validation of brief measures of positive and negative affect: the PANAS scales. J Pers Soc Psychol. 1988;54:1063–1070. doi: 10.1037//0022-3514.54.6.1063. [DOI] [PubMed] [Google Scholar]
- 33.Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33:159–174. [PubMed] [Google Scholar]
- 34.Lilienfeld SO. Methodological advances and developments in the assessment of psychopathy. Behav Res Ther. 1998;36:99–125. doi: 10.1016/s0005-7967(97)10021-3. [DOI] [PubMed] [Google Scholar]
- 35.Food and Drug Administration. Warning Letter. 23andMe, Inc; Inspections, Compliance, Enforcement, and Criminal Investigations. 11/22/13. http://www.fda.gov/iceci/enforcementactions/warningletters/2013/ucm376296.htm. [Google Scholar]
- 36.Green RC, Farahany NA. Regulation: The FDA is overcautious on consumer genomics. Nature. 2014;505:286–287. doi: 10.1038/505286a. [DOI] [PubMed] [Google Scholar]
- 37.Carpenter SM, Peters E, Vastfjall D, Isen AM. Positive feelings facilitate working memory and complex decision making among older adults. Cogn Emot. 2013;27:184–192. doi: 10.1080/02699931.2012.698251. [DOI] [PubMed] [Google Scholar]
- 38.Fredrickson BL. What good are positive emotions? Rev Gen Psychol. 1998;2:300–319. doi: 10.1037/1089-2680.2.3.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cacioppo JT, Gardner WL, Berntson GG. Beyond bipolar conceptualizations and measures: the case of attitudes and evaluative space. Pers Soc Psychol Rev. 1997;1:3–25. doi: 10.1207/s15327957pspr0101_2. [DOI] [PubMed] [Google Scholar]
- 40.Cacioppo JT, Gardner WL. Emotion Annu Rev Psychol. 1999;50:191–214. doi: 10.1146/annurev.psych.50.1.191. [DOI] [PubMed] [Google Scholar]
- 41.Harmon-Jones E, Gable PA, Price TF. Toward an understanding of the influence of affective states on attentional tuning: comment on Friedman and Forster (2010) Psychol Bull. 2011;137:508–512. doi: 10.1037/a0022744. [DOI] [PubMed] [Google Scholar]
- 42.Hay JL, Meischke HW, Bowen DJ, Mayer J, Shoveller J, Press N, Asgari M, Berwick M, Burke W. Anticipating dissemination of cancer genomics in public health: a theoretical approach to psychosocial and behavioral challenges. Ann Behav Med. 2007;34:275–286. doi: 10.1007/BF02874552. [DOI] [PubMed] [Google Scholar]
- 43.Bennette CS, Trinidad SB, Fullerton SM, Patrick D, Amendola L, Burke W, Hisama FM, Jarvik GP, Regier DA, Veenstra DL. Return of incidental findings in genomic medicine: measuring what patients value--development of an instrument to measure preferences for information from next-generation testing (IMPRINT) Genet Med. 2013;15:873–881. doi: 10.1038/gim.2013.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Esplen MJ, Stuckless N, Hunter J, Liede A, Metcalfe K, Glendon G, Narod S, Butler K, Scott J, Irwin E. The BRCA Self-Concept Scale: a new instrument to measure self-concept in BRCA1/2 mutation carriers. Psychooncology. 2009;18:1216–1229. doi: 10.1002/pon.1498. [DOI] [PubMed] [Google Scholar]
- 45.Esplen MJ, Stuckless N, Gallinger S, Aronson M, Rothenmund H, Semotiuk K, Stokes J, Way C, Green J, Butler K, Petersen HV, Wong J. Development and validation of an instrument to measure the impact of genetic testing on self-concept in Lynch syndrome. Clin Genet. 2011;80:415–423. doi: 10.1111/j.1399-0004.2011.01770.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Phelps C, Bennett P, Jones H, Hood K, Brain K, Murray A. The development of a cancer genetic-specific measure of coping: the GRACE. Psychooncology. 2010;19:847–854. doi: 10.1002/pon.1629. [DOI] [PubMed] [Google Scholar]
- 47.Allyse M, Michie M. Not-so-incidental findings: the ACMG recommendations on the reporting of incidental findings in clinical whole genome and whole exome sequencing. Trends Biotechnol. 2013;31:439–441. doi: 10.1016/j.tibtech.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Burke W, Matheny Antommaria AH, Bennett R, Botkin J, Clayton EW, Henderson GE, Holm IA, Jarvik GP, Khoury MJ, Knoppers BM, Press NA, Ross LF, Rothstein MA, Saal H, Uhlmann WR, Wilfond B, Wolf SM, Zimmern R. Recommendations for returning genomic incidental findings? We need to talk! Genet Med. 2013;15:854–859. doi: 10.1038/gim.2013.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Green RC, Berg JS, Grody WW, Kalia SS, Korf BR, Martin CL, McGuire AL, Nussbaum RL, O'Daniel JM, Ormond KE, Rehm HL, Watson MS, Williams MS, Biesecker LG. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med. 2013;15:565–574. doi: 10.1038/gim.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rosenblatt DS. Who's on first in exome and whole genome sequencing? Is it the patient or the incidental findings? Mol Genet Metab. 2013;110:1–2. doi: 10.1016/j.ymgme.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 51.Facio FM, Eidem H, Fisher T, Brooks S, Linn A, Kaphingst KA, Biesecker LG, Biesecker BB. Intentions to receive individual results from whole-genome sequencing among participants in the ClinSeq study. Eur J Human Genet. 2013;21:261–265. doi: 10.1038/ejhg.2012.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shahmirzadi L, Chao EC, Palmaer E, Parra MC, Tang S, Gonzalez KD. Patient decisions for disclosure of secondary findings among the first 200 individuals undergoing clinical diagnostic exome sequencing. Genet Med. 2014;16:395–399. doi: 10.1038/gim.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johnston JJ, Rubinstein WS, Facio FM, Ng D, Singh LN, Teer JK, Mullikin JC, Biesecker LG. Secondary variants in individuals undergoing exome sequencing: screening of 572 individuals identifies high-penetrance mutations in cancer-susceptibility genes. Am J Hum Genet. 2012;91:97–108. doi: 10.1016/j.ajhg.2012.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]