Abstract
We report two different unique HIV-1 recombinant viruses from two HIV-positive men who have sex with men (MSM) in Beijing, China. Phylogenetic analysis of near full-length genomes (NFLG) showed that the unique recombinant forms (URFs) were comprised of gene regions from two circulating recombinant forms, CRF01_AE and CRF07_BC, both common in China. The parental CRF01_AE region of the recombinants clustered together with a previously described cluster 4 lineage of CRF01_AE. The CRF07_BC regions of both the recombinants clustered within the CRF07_BC radiation, but were distinct from other CRF07_BC reference sequences. The two recombinant forms had two breakpoints in common. The emergence of the two URFs indicates the ongoing generation of recombinant viruses involving CRF01_AE and CRF07_BC, and may provide insight into our understanding of the dynamics and complexity of the HIV-1 epidemic in China.
HIV-1 group M strains play a major role in the global HIV-1 epidemic and are further divided into nine subtypes [A (A1, A2), B, C, D, F (F1, F2), G, H, J, K]. Furthermore, at least 72 circulating recombinant forms (CRFs) and innumerable unique recombinant forms (URFs) have been reported, especially in regions where multiple subtypes and/or CRFs cocirculate.1
Beijing is a metropolitan area with a large number of men who have sex with men (MSM). The HIV prevalence among MSM in Beijing has increased from 3.1% in 20022 to 7.8% in 2010, and is projected to be >20% by 2020 if there are no enhanced HIV interventions,3 and the prevalence among MSM in Beijing is much higher than those in most other Chinese cities (1.3–1.6%).4–6 HIV-1 genotype distribution among MSM in China was first studied in Beijing in 2005–2006. Subtype B was predominant among MSM (71.1%), followed by CRF01_AE (24.4%) and CRF07_BC (4.4%). Follow-up surveys of Beijing MSM revealed that subtype B infections decreased to 41.9% in 2007 and to 30.8% in 2010. In contrast, CRF01_AE increased from 3.7% in 2005 to 56.0% in 2010 and CRF07_BC increased from <5% in 2005 to 12.6% in 2010.7
The cocirculation of viruses from different subtypes and/or CRFs in the same region and risk groups fosters the emergence of new intersubtype recombinant viruses. Two HIV-1 URFs composed of gene regions from CRF01_AE and subtype B have been reported in Beijing among MSM.8 In a current study to characterize HIV sequences in Beijing MSM (2013–2014), two new HIV-1 URFs consisting of gene regions from CRF01_AE and CRF07_BC were detected.
This study was approved by the China CDC Institutional Ethics Committee, and written informed consent was obtained from study participants. Plasma from two of the MSM samples yielded different URFs. Detailed information concerning the two participants is given in Table 1. Both were most likely infected through MSM, and were confirmed as HIV-1 seropositive in July 2013 for BJMP3002B and in May 2013 for BJMP3026B, respectively. Participant BJMP3002B had more than 500 MSM sexual partners in his lifetime and more than 20 in the past 3 months. Subject BJMP3026B had 50 male sexual contacts but only 2 in the past 3 months. Both of them reported having used illicit drugs in their lifetime; illicit drug use was comparatively rare in the MSM cohort as a whole (1.6%, 59/3,618, unpublished data). CD4+ T cell counts were 387 cells/μl and 531 cells/μl and viral loads were 2,000 copies/ml and 25,000 copies/ml for BJMP3002B and BJMP3026B, respectively. They are not sexual partners.
Table 1.
Demographics, HIV-Related Behaviors, and Disease Status of Two HIV-Infected Study Participants
| Variable | BJMP3002B | BJMP3026B |
|---|---|---|
| Age (year) | 43 | 24 |
| Education | Elementary school | College |
| Marital status | Divorced | Single |
| Predominant role in anal sex | Receptive | Receptive |
| Number of lifetime male sex partners | >500 | 50 |
| Number of male sexual partners in the past 3 months | 20 | 2 |
| Ever used illicit drugs | Yes | Yes |
| Viral loads (copies/ml) | 2,000 | 25,000 |
| CD4+ T cell count (cells/μl) | 387 | 531 |
For near full-length genome (NFLG) amplification and sequencing, RNA was extracted from the participant's plasma sample using the QIAamp Viral Mini Kit (QIAGEN, Germany). RNA was then transcribed into cDNA using the Superscript III First-strand synthesis system (Invitrogen, USA). With the near-endpoint diluted cDNA template, the NFLGs were amplified with TaKaRa LA Taq (TaKaRa, Dalian, China) using the same nested polymerase chain reaction (PCR) amplification conditions in both rounds, as described previously.9 The positive PCR products were purified using the QIAquick Gel Extraction Kit (QIAGEN, Germany) and sequenced by ABI 3730XL sequencer using BigDye terminators (Applied Biosystems, Foster City, CA). The chromatogram data were edited manually and assembled using Sequencher v. 5.1 (Gene Codes Corporation, Ann Arbor, MI).
To detect potential laboratory cross-contamination, we conducted a BLAST search during which the NFLG sequences were queried against all sequences previously generated in our laboratory. The NFLG sequences were then aligned against standard Los Alamos National Laboratory (www.hiv.lanl.gov/content/sequence/HIV.html) subtype reference sequences, which includes subtypes A1, A2, B, C, D, F, G, H, J, P, CRF01_AE, CRF07_BC, and CRF08_BC, and edited manually using BioEdit v. 7.1.10 Subgenomic phylogenetic analyses were performed using the neighbor-joining method based on the Kimura two-parameter model implemented in MEGA v. 5.05.11 To analyze the recombinant structure of the two strains, the sequences were submitted to the recombination identification program (RIP) (www.hiv.lanl.gov/content/sequence/RIP/RIP.html) using all default settings except a window size of 300, and a similarity plot analysis was performed using SimPlot (version 3.5.1).12
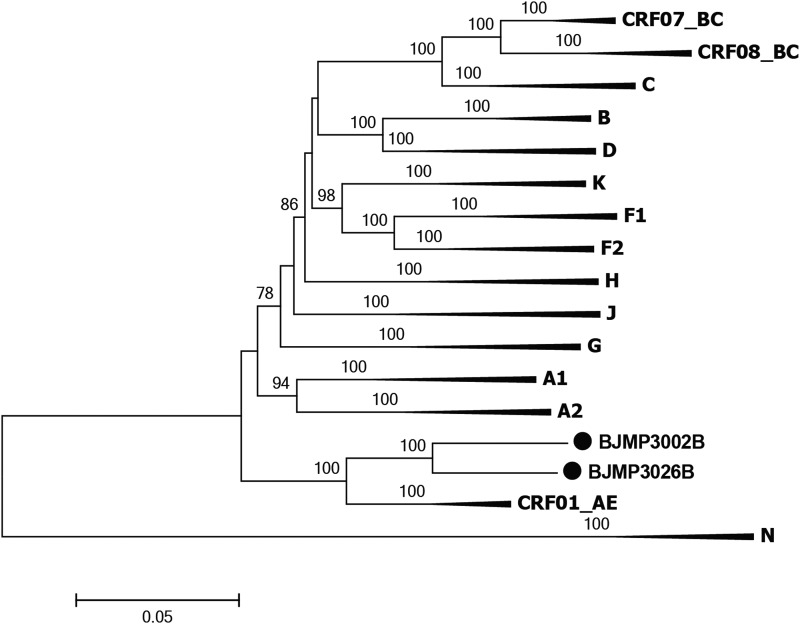
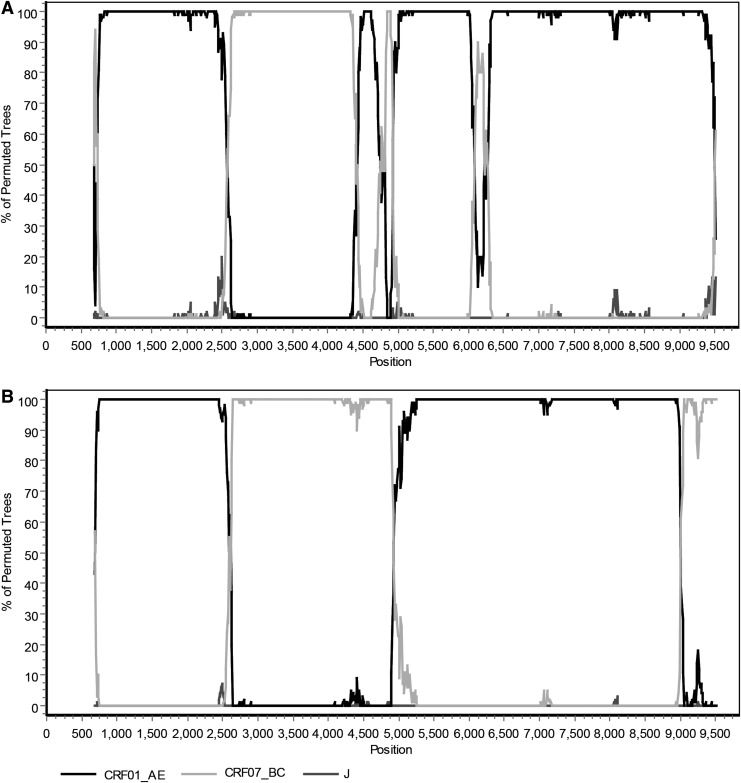
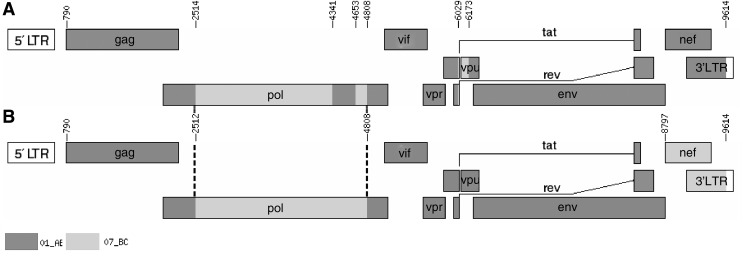
In the phylogenetic trees reconstructed with full genome reference sequences (Fig. 1), the two URFs clustered with CRF01_AE reference sequences with a high bootstrap value (100%), yet branched independently of them, initially suggesting they may have recombinant structures. The result of subgenomic phylogenetic analyses revealed a CRF01_AE parental backbone with three CRF07_BC fragments inserted in the pol and vpu regions (BJMP3002B, Fig. 2A and Fig. 3A) and two CRF07_BC fragments inserted in pol and nef (BJMP3026B, Fig. 2B and Fig. 3B), respectively. We used the Map-Draw Tool available at the Los Alamos HIV sequence database to map the breakpoints and illustrate the genomic structure of the two new HIV URFs (www.hiv.lanl.gov/content/sequence/DRAW_CRF/recom_mapper.html) (Fig. 3).
FIG. 1.
Neighbor-joining phylogenetic tree of BJMP3002B and BJMP3026B. The tree was constructed using MEGA 6.0. Solid circle (●) marks the indicated sequences. The stability of each node was confirmed by bootstrapping with 1,000 replicates and only significant bootstrap values ≥70% were shown at the corresponding nodes. The scale bar represents 5% genetic distance.
FIG. 2.
Bootscan analysis of the near full-length nucleotide sequences of BJMP3002B (A) and BJMP3026B (B). Bootscan analysis was performed using CRF01_AE, CRF07_BC, and subtype J reference sequences. The bootscan window was 300 bp with a step size of 10 bp using SimPlot 3.5.1 software. The x-axis is the nucleotide position in the HIV genomic sequence; the y-axis indicates the percentage supporting the clustering with reference sequences.
FIG. 3.
Genetic map of BJMP3002B (A) and BJMP3026B (B). Map-draw tool is available at the Los Alamos HIV sequence database (www.hiv.lanl.gov/content/sequence/DRAW_CRF/recom_mapper.html). (A) In BJMP3002B, three segments of CRF07_BC were inserted into the genome of the subtype CRF01_AE backbone from nucleotide 2514 to 4341, 4653 to 4808, and 6029 to 6173, respectively, as referenced to the HXB2 complete genome. (B) In BJMP3026B, two segments of CRF07_BC were inserted into the genome of subtype CRF01_AE from nucleotide 2512 to 4808 and from 8797 to 9614, respectively. The common breakpoints are shown by dotted lines.
The phylogenetic trees of the individual gene regions confirmed the breakpoints of the two NFLG sequences as follows (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/aid): I (790–2,513 nt) CRF01_AE, II (2,514–4,340 nt) CRF07_BC, III (4,341–4,652 nt) CRF01_AE, IV (4,653–4,807 nt) CRF07_BC, V (4,808–6,028 nt) CRF01_AE, VI (6,029–6,172 nt) CRF07_BC, and VII (6,173–9,614 nt) CRF01_AE (BJMP3002B); I (790–2,511 nt) CRF01_AE, II (2,512–4,807 nt) CRF07_BC, III (4,808–8,796 nt) CRF01_AE, and IV (8,797–9,614 nt) CRF07_BC (BJMP3026B). Bootscan analysis of the two NFLG sequences revealed that they shared two identical breakpoints (nt 2,513±1 and 4,808), using HXB2 as a reference, as shown in the genomic map (Fig. 3).
Subregion tree analysis indicates that the parental origin of all CRF01_AE regions of the two NFLGs were from the MSM-related CRF01_AE cluster 4 lineage13 (Supplementary Fig. S1), which is circulating primarily among MSM in Beijing. The CRF07_BC regions in BJMP3002B (II, IV, VI) and BJMP3026B (II, IV) clustered with CRF07_BC reference sequences. In conclusion, the parental origins of the two novel second-generation URFs were CRF01_AE cluster 4 lineage and CRF07_BC.
Increasing genetic diversity is a characteristic of HIV-1 and is a product of both the accumulation of point mutations and recombination. HIV-1 recombination is an ongoing event and can significantly contribute to the epidemics in different regions.14–18 While URFs are not strains of epidemic importance, they hold the potential to enter high-risk transmission networks and become new CRFs. It is also of importance to note that recombination requires coinfection or superinfection of viral strains within an individual. The identification of numerous CRFs and even more numerous URFs indicates that coinfection and/or superinfection occurs at an alarming rate. Therefore, it is important to identify the range of CRFs and URFs to monitor the dynamics and complexity of the HIV epidemic and determine their importance to vaccine development.
Sequence Data
The NFLG sequences of BJMP3002B and BJMP3026B have been deposited in GenBank with accession numbers KM982719 and KM982720, respectively.
Supplementary Material
Acknowledgments
This study was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award R01AI094562, the Chinese National Science and Technology Major Projects for Infectious Diseases Control and Prevention (2012ZX1001-002), and the Chinese National Natural Science Foundation of China (81261120393, 81471962). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Hemelaar J: The origin and diversity of the HIV-1 pandemic. Trends Mol Med 2012;18(3):182–192 [DOI] [PubMed] [Google Scholar]
- 2.Choi KH, Liu H, Guo Y, et al. : Emerging HIV-1 epidemic in China in men who have sex with men. Lancet 2003;361(9375):2125–2126 [DOI] [PubMed] [Google Scholar]
- 3.Lou J, Blevins M, Ruan Y, et al. : Modeling the impact on HIV incidence of combination prevention strategies among men who have sex with men in Beijing, China. PloS One 2014;9(3):e90985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cai WD, Tan JG, Chen L, et al. : A survey of the characteristics and STD/HIV infection of homosexuality in Shenzhen. Modern Prevent Med 2005;32:328–330 [Google Scholar]
- 5.Choi KH, Ning Z, Gregorich SE, and Pan QC: The influence of social and sexual networks in the spread of HIV and syphilis among men who have sex with men in Shanghai, China. J Acquir Immune Defic Syndr 2007;45(1):77–84 [DOI] [PubMed] [Google Scholar]
- 6.Gu YQP, Xu L, Luo M, et al. : Survey of knowledge, attitude, behavior and practices related to STI/HIV among male homosexuality in Shenyang. Chin J Publ Health 2004;20:573–574 [Google Scholar]
- 7.Li L, Han N, Lu J, et al. : Genetic characterization and transmitted drug resistance of the HIV type 1 epidemic in men who have sex with men in Beijing, China. AIDS Res Hum Retroviruses 2013;29(3):633–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang W, Meng Z, Zhou M, et al. : Near full-length sequence analysis of two new HIV type 1 unique (CRF01_AE/B) recombinant forms among men who have sex with men in China. AIDS Res Hum Retroviruses 2012;28(4):411–417 [DOI] [PubMed] [Google Scholar]
- 9.Rousseau CM, Birditt BA, McKay AR, et al. : Large-scale amplification, cloning and sequencing of near full-length HIV-1 subtype C genomes. J Virol Methods 2006;136(1–2):118–125 [DOI] [PubMed] [Google Scholar]
- 10.Hall TA: Bioedit: A user-friendly biological sequence alignment editor and analysis program for windows 95/98/nt. Paper presented at the Nucleic Acids Symposium Series1999 [Google Scholar]
- 11.Tamura K, Dudley J, Nei M, and Kumar S: Mega4: Molecular evolutionary genetics analysis (mega) software version 4.0. Mol Biol Evol 2007;24(8):1596–1599 [DOI] [PubMed] [Google Scholar]
- 12.Lole KS, Bollinger RC, Paranjape RS, et al. : Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol 1999;73(1):152–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng Y, He X, Hsi JH, et al. : The rapidly expanding CRF01_AE epidemic in China is driven by multiple lineages of HIV-1 viruses introduced in the 1990s. AIDS 2013;27(11):1793–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han X, An M, Zhang M, et al. : Identification of 3 distinct HIV-1 founding strains responsible for expanding epidemic among men who have sex with men in 9 Chinese cities. J Acquir Immune Defic Syndr 2013;64(1):16–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li X, Ning C, He X, et al. : Near full-length genome sequence of a novel HIV type 1 second-generation recombinant form (CRF01_AE/CRF07_BC) identified among men who have sex with men in Jilin, China. AIDS Res Hum Retroviruses 2013;29(12):1604–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei H, Su L, Feng Y, et al. : Near full-length genomic characterization of a novel HIV type 1 CRF07_ BC/01_AE recombinant in men who have sex with men from Sichuan, China. AIDS Res Hum Retroviruses 2013;29(8):1173–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo H, Guo D, Wei JF, et al. : First detection of a novel HIV type 1 CRF01_AE/07_BC recombinant among an epidemiologically linked cohort of IDUs in Jiangsu, China. AIDS Res Hum Retroviruses 2009;25(4):463–467 [DOI] [PubMed] [Google Scholar]
- 18.He X, Xing H, Ruan Y, et al. : A comprehensive mapping of HIV-1 genotypes in various risk groups and regions across China based on a nationwide molecular epidemiologic survey. PloS One 2012;7(10):e47289. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.