Abstract
Francisella tularensis, the causative agent of tularemia, presents a significant biological threat and is a Category A priority pathogen due to its potential for weaponization. The bacterial FASII pathway is a viable target for the development of novel antibacterial agents treating Gram-negative infections. Here we report the advancement of a promising series of benzimidazole FabI (enoyl-ACP reductase) inhibitors to a second-generation using a systematic, structure-guided lead optimization strategy, and the determination of several co-crystal structures that confirm the binding mode of designed inhibitors. These compounds display an improved low nanomolar enzymatic activity as well as promising low microgram/mL antibacterial activity against both F. tularensis and S. aureus and its methicillin-resistant strain (MRSA). The improvements in activity accompanying structural modifications lead to a better understanding of the relationship between the chemical structure and biological activity that encompasses both enzymatic and whole-cell activity.
Keywords: Enoyl reductase, benzimidazole scaffold, F. tularensis, FabI inhibitor, S. aureus, MRSA
Tularemia is a relatively rare infectious disease that spreads to humans through insect bites or from direct exposure to infected animals.1 Francisella tularensis, the causative agent of tularemia, is a Gram-negative bacterium that can invade macrophages and evade the immune system.2 The disease has a very low infectious dose, high mortality rate, and is categorized as a Category A priority pathogen due to its strong potential for weaponization and high risk to national security.2 The current recommended treatment is less than ideal in the event of a mass exposure due to the requirement for intravenous administration (aminoglycosides), or contraindication in pregnant women and children (fluoroquinolones and tetracyclines). This highlights a clear need for the development of novel therapeutics targeting tularemia.
Bacterial type II fatty acid synthesis, or FAS II, is a well-known pathway that has been targeted by many drug discovery groups.3–7 The essentiality of this pathway has been debated as some Gram-positive organisms have been shown to overcome FAS II inhibition by their ability to uptake and utilize fatty acids from the host environment.8–10 However, it has recently been shown that, while some Gram-positive organisms, such as Streptococci, possess a feedback regulatory mechanism that suppresses the endogenous pathway when exogenous fatty acids are available, other Gram-positive species such as S. aureus are unable to do so and hence remain susceptible to FAS II inhibition.11 However, the susceptibility of Gram-negative organisms such as F. tularensis to FAS II inhibition is not in question as they require β-hydroxy fatty acids to assemble the lipid A component of outer membrane lipopolysaccharides, and exogenous fatty acids cannot support lipid A biosynthesis.12 A recent study has experimentally confirmed the essentiality of the FAS II pathway specifically in F. tularensis.13
Enoyl-acyl carrier protein (ACP) reductase I, FabI, a key enzyme in the FAS II pathway, catalyzes a rate-limiting step in bacterial fatty acid synthesis and is an attractive antibacterial drug target. The aforementioned F. tularensis essentiality study specifically demonstrated that F. tularensis FabI (FtFabI) was essential for growth even in the presence of exogenous long chain fatty acids.13 These studies, along with the low sequence and structural similarity of FabI to its mammalian counterpart in the FAS I pathway, provide strong biochemical justification for the continued investigation of F. tularensis FabI as an antibacterial target in F. tularensis. Recently, our laboratory reported the identification and structural characterization of a novel series of benzimidazole FabI inhibitors as a new chemical scaffold with promising enzyme and antibacterial activity.14, 15 We now report the structural and biological evaluation of several second generation benzimidazole compounds with low nanomolar enzyme inhibition and promising antibacterial activity, not only against F. tularensis, but also against the more prevalent pathogen, S. aureus and MRSA. Our co-crystal structures demonstrate the binding modes of these second generation inhibitors in FtFabI and lay a solid foundation for analyzing strategies to improve pharmacokinetic properties while maintaining FabI inhibition.
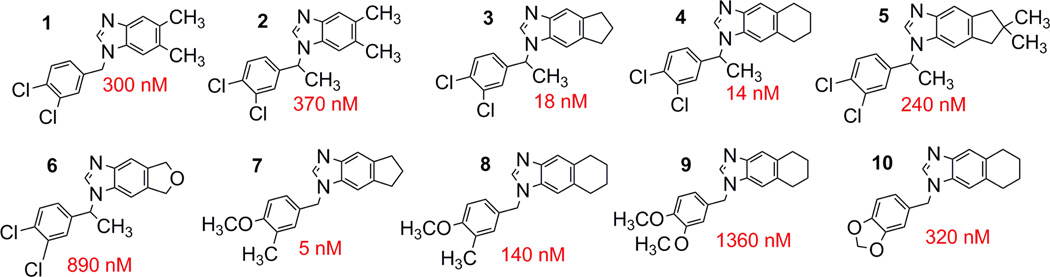
In our prior studies reporting hit identification, structural and enzymatic analyses of the first-generation benzimidazole FabI inhibitors,14, 15 the initial SAR was constructed primarily by testing commercially available benzimidazole analogs, resulting in a limited understanding of the structure-activity relationship. We now report activities from synthetic analogs of our prior best hit, compound 1 (Figure 1), and find that the second generation compounds display enhanced enzymatic inhibitory activity, along with significantly improved antibacterial activity. The most promising 2nd generation compounds are presented in Figure 1.
Figure 1. Structures of benzimidazole inhibitors.
Structures of the tested benzimidazole analogs with IC50 values displayed alongside.
The addition of a methyl group to the methylene linker, as in 2, does not significantly alter the inhibitory activity. Larger groups at this position were not tested, as the crystal structure of 1 bound to FtFabI (PDB ID 3UIC)15 demonstrates that large groups cannot be accommodated at this position. The addition of a methyl group at this position results in a chiral center, but only the racemic mixture of 2 has been tested to date.
We noted a significant improvement in activity upon the replacement of the 5 and 6 position methyl groups in 2 (IC50 of 370 nM) with a cyclopentyl ring system (3, IC50 of 18 nM) or a cyclohexyl ring (4, IC50 of 14 nM). There is little preference for the cyclopentyl vs cyclohexyl ring fused to the benzimidazole scaffold. Further substitutions to the cyclopentane ring, such as the dimethyl substitutions in 5 (IC50 of 240 nM), or replacement of the cyclopentane ring with a tetrahydrofuran ring fused to the benzimidazole ring in 6 (IC50 of 890 nM) resulted in weaker enzyme inhibitory activity relative to 3.
With the 1st generation benzimidazole compounds, we initially focused on halogen substituents to the N1 phenyl group, principally due to a known halogen bond interaction between FabI and triclosan, the stereotypical FabI inhibitor,14 which suggested that the halogen-substituted phenyl group could make a similar interaction. However our structure of 1 bound to FtFabI demonstrates this not to be the case.15 We now investigated the replacement of halogen substituents with other small, lipophilic groups, including methyl and methoxy groups. Compound 7, substituted with a meta-methyl and para-methoxy group, demonstrated that the activity is not dependent on halogen substitution at these positions, as the inhibitory activity was retained relative to the other compounds. Additionally, the replacement of the 5 and 6 position methyl groups in 1 with a cyclopentane ring system in 7 resulted in our most enzymatically potent 2nd generation compound, with an IC50 of 5 nM. Compound 7 has better enzyme inhibitory activity than 8 (IC50 =140 nM). The reason for this is not clear since no difference was observed between compounds 3 and 4, which also differed only in the cyclopentyl vs. cyclohexyl rings. Replacement of the meta-methyl group with a second methoxy group, as in 9, resulted in additional activity loss with an IC50 of 1360 nM. Replacement of the methyl and methoxy groups with a methylenedioxy group in these analogs yielded 10 (IC50 = 320 nM), with an improvement in enzyme inhibitory activity relative to dimethoxy-substituted 9, but reduced inhibitory activity relative to the methyl and methoxy substituted analog, 8, with these changes explained by the crystal structures described below.
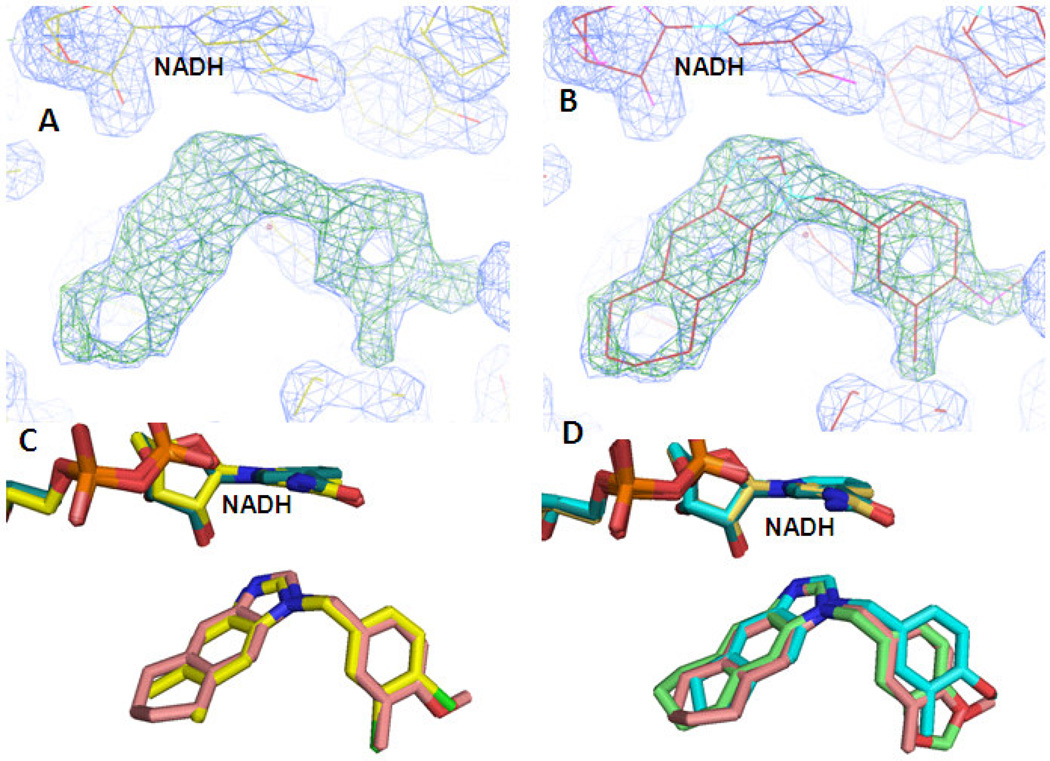
The co-crystal structures of 7, 8, and 10 bound to FtFabI were solved to resolutions of 2.45 Å, 1.85 Å, and 2.34 Å, respectively. The asymmetric unit of the 7 co-crystal structure contains two chains as a homodimer, while those of the 8 and 10 co-crystal structures contain eight chains as 4 homodimers. Data collection and refinement statistics for these structures are given in Table 1. As seen with our previous FtFabI co-crystal structure with 115, these inhibitors bind in the presence of NADH, and both the NADH and the inhibitor are present in each monomer. As the structures of the different chains in each of the enzyme/compound complexes were geometrically restrained to be identical during refinement, we focus our further discussion on the features of chain A in each complex. A superimposition of the backbone atoms of chain A from all three ternary complexes resulted in a calculated RMSD of < 0.25 Ǻ, indicating that these structures are nearly identical. Clear electron densities for both NADH and the inhibitor compounds are visible in all three ternary complexes and allow for the unambiguous positioning of the ligands. This is illustrated in Figure 2A with 8, where the structure was refined without the compound in the active site to generate an omit map that confirms the binding mode of the inhibitor (Figure 2B) and validates our previously published binding mode of 1 (Figure 2C).15 Figure 2D shows the overlap of the three compounds as they are in the co-crystal structures, with the binding mode being essentially identical.
Table 1.
Data collection and Refinement Statistics (data collection statistics in the highest resolution bin are indicated in brackets). PDB IDs are listed in parentheses in the first row.
| Data Collection: | Compound 7 + FtFabI (4J1N) |
Compound 8 + FtFabI (4J3F) |
Compound 10 + FtFabI (4J4T) |
|---|---|---|---|
| Space group | P21 212 | P21 2121 | P21 2121 |
| Unit cell parameters: | |||
| a,b,c (Å) | a = 85.2, | a = 85.4, | a = 85.4, |
| b = 123.3 | b = 123.3 | b = 123.3 | |
| c = 50.6 | c = 202.7 | c = 202.7 | |
| Resolution (Å) | 2.45 (2.5−2.45) | 1.85 (1.96−1.85) | 2.34 (2.48−2.34) |
| No. reflections | 96626 (7093) | 2473815 (228739) | 1204544 (146383) |
| No. averaged reflections (unique) | 20119 (1450) | 178885 (26126) | 90198 (13545) |
| Redundancy | 4.8 (4.9) | 13.8 (8.7) | 13.4 (10.8) |
| Rmerge (%) | 11.0 (67.5) | 6.6 (19.5) | 13.8 (49.4) |
| I/σI | 12.2 (2.1) | 29.5 (9.9) | 13.5 (4.9) |
| Completeness % | 99.7 (99.8) | 98.4 (90.6) | 98.9 (94.3) |
|
Refinement | |||
| Resolution range (Å) | 20.0 – 2.45 | 20.0 – 1.85 | 20.0 – 2.34 |
| no. reflections in working set | 19058 | 169807 | 85571 |
| No. of free reflections | 1023 | 8974 | 4518 |
| Rcrys (%) | 24.2 | 15.5 | 19.9 |
| Rfree (%) | 28.0 | 18.2 | 24.1 |
| Figure of merit | 0.76 | 0.88 | 0.81 |
| average B-factor (Å2) (protein) | 39.3 | 13.7 | 23.4 |
| No. of protein molecules in asymmetric unit | 2 | 8 | 8 |
| RMSD from ideal geometry: | |||
| Bond lengths (Å) | 0.006 | 0.010 | 0.011 |
| Bond angles (deg) | 1.099 | 1.363 | 1.474 |
| Ramachandran plot favored (%) | 92.4 | 95.5 | 96 |
| outliers (%) | 0.6 | 0.7 | 0.4 |
Figure 2. Crystal structures of the benzimidazole inhibitors in the active site of FtFabI.
(A) Electron density of the ligand in the FabI active site. This difference map was generated without the ligand in the active site. The Fo-Fc map (green grid lines) is contoured at 5 sigma while the 2Fo-Fc map (blue grid lines) is contoured at 1.5 sigma. (B) The density for the ligand in the active site allows for unambiguous positioning of 8. (C) Overlap of 8 (salmon backbone) and our previously published 1 or CB-7725253 (yellow backbone) confirms the binding mode. (D) Overlap of all three compounds: 7 (cyan backbone), 8 (salmon backbone) and 10 (green backbone) in the active site of FtFabI.
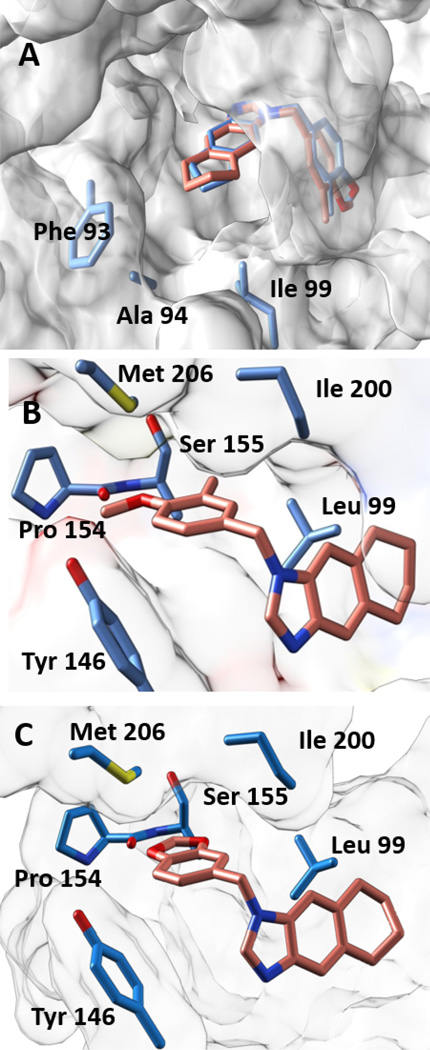
In the co-crystal structures of 7, 8, and 10, the N3 atom of the benzimidazole ring system is positioned to engage either the hydroxyl group of Tyr156 or the 2’-ribose hydroxyl group of the NADH cofactor in a hydrogen bond interaction. This interaction is one of the most conserved features observed in published FabI inhibitors.15 In the co-crystal structures of 7 and 8, the cyclopentyl and cyclohexyl rings are well accommodated in the active site and face the fairly lipophilic active site opening that is lined by Phe 93, Ala94, and Leu99, as indicated in Figure 3A. As noted above, we observe a preference for a meta-methyl substituent on the phenyl group over a methoxy at the same position. Analysis of the structure of the ternary complex with 8 reveals the structural basis for this observation. The meta-methyl group of the benzimidazole inhibitor falls in a lipophilic pocket bordered by Ile200, Leu99, and the backbone (primary and secondary carbon) atoms of Ser155. Figure 3B demonstrates that modifying the methyl group to a methoxy group would be less energetically favorable due to both its increased steric bulk and electrostatic potential. However, the para-methoxy group is observed to occupy an area near the solvent accessible active site opening, and is bordered by residues positioned near enough to engage in electrostatic interactions, including Pro154 (backbone carbonyl), Met206, and Tyr146. Similarly, the methylenedioxy analog in 10 showed decreased activity relative to the meta-methyl, para-methoxy substituted analogs, which may also reflect intolerance to the increased electrostatic potential of the oxygen atom at the meta position that is preferentially occupied by a meta-methyl group (Figure 3C). Sterically, this substitution is not as restrictive as the replacement of the meta-methyl group with the methoxy group, which could explain the improvement in activity seen with methylenedioxy analogs over their dimethoxy counterparts.
Figure 3. Co-crystal Structures.
(A) The cyclopentyl ring of 7 (blue backbone) and the cyclohexyl ring in 8 (salmon backbone) are well accommodated in the active site and face the fairly lipophilic active site opening that is lined by Phe 93, Ala94. (B) Positioning of the methyl and methoxy groups in 8 in the FtFabI active site. The methyl group at the meta position is in a very lipophilic environment. The methoxy group is surrounded by the backbone carbonyl of Pro154 (3.5 Ǻ away), the −OH of Tyr146 (4.8 Ǻ away) and −S of Met206 (3.5 Ǻ away). (C) Positioning of 10 in the active site of FtFabI. The region lined by Leu99 and Ile200 is fairly lipophilic and can accommodate fairly bulky modifications. The region lined by Met206, backbone of Pro154, and Tyr146 is more hydrophilic and restrictive but presents the possibility of exploiting hydrogen bonding in this region.
We have previously demonstrated that our inhibitors display competitive inhibition with respect to the substrate and uncompetitive inhibition with respect to the cofactor NADH.15 We have determined the Ki values for several of our inhibitors through rigorous experiments where concentrations of the substrate crotonyl-CoA and inhibitors were respectively varied as a function of each other at a saturating concentration of 200 µM of NADH, and the results are shown in Table 2. We found the experimental Ki values to be very close to the IC50 values. The Ki values for these inhibitors calculated using the Cheng-Prusoff equation16 were likewise similar. Burlingham and Wedlanski have noted that in the case of competitive inhibitors, the IC50 values approximate Ki when the substrate concentration used is much lower than the Km17, as it is in our case where our substrate concentration is 200 µM, while the Km is 830 µM.15 Although a rigorous experimental determination of the Ki for each compound wasn’t practical due to the limited availability and high price of the crotonyl-CoA substrate, these results indicate that the calculated Ki values for the remaining compounds are likely to closely approximate the true Ki values and can be estimated to be the IC50 value. Thus, the trend in our IC50 values can be assumed to reflect the trend in Ki values.
Table 2.
Enzymatic Activity: Experimental Ki values were determined for the listed compounds and found to be very close to both the IC50 and the calculated Ki values.
Experimental Ki
Calculated Ki using the Cheng-Prusoff Equation: where [S] = 200 µM and Km = 830 µM.15
We previously reported that 1 displayed a minimum inhibitory concentration (MIC) of 7.8 µg/mL against F. tularensis and 9.4 µg/mL against B. anthracis.14 The MICs of compounds shown in Figure 1 are presented in Table 3. The addition of the methyl group on the linker region in 2 resulted in an MIC >12.5 µg/mL against F. tularensis. However, the MIC values against S. aureus and MRSA were 2 to 4-fold better with 2 compared to 1 (12.5 µg/mL compared to 25 and 50 µg/mL for 1 against S. aureus and MRSA respectively). We speculate that this is likely due to an improvement in cell penetration with the Gram-positive organism resulting from an increase in lipophilicity of 2 over 1, and a decrease in F. tularensis cell penetration for the same reason. It is well known that Gram-negative organisms favor decreased lipophilicity for cell penetration due to porin passage, while Gram-positives favor increased lipophilicity due to transmembrane passage.18
Table 3.
MIC values. MIC is the minimum inhibitory concentration while MBC is the minimum bactericidal concentration. MBCs were not determined for compounds with MICs ≥ 12.5 µg/mL. E. coli TolC- is the E. coli TolC efflux pump knockout mutant. MIC values reported here are the mean of at least two runs. MICs for 1 have been previously published14 but are included here for reference. NT = Not tested
| MIC / MBC (both in µg/mL) | |||||
|---|---|---|---|---|---|
| Compound | F. tularensis | S. aureus | MRSA | E. coli | E. coli TolC- |
| 1 | 7.8 | 25 | 50 | >200 | 6.25 |
| 2 | >12.5 | 12.5 | 12.5 | >12.5 | 6.3 / 12.5 |
| 3 | 3.9 / 9.4 | 5.5 / > 25.0 | 6.3 / 12.5 | >12.5 | 3.1 / 12.5 |
| 4 | 3.1 / 25.0 | 3.1 / 25.0 | 3.1 / 25.0 | >12.5 | 3.1 / >25.0 |
| 5 | >12.5 | 4.7 / >12.5 | 4.7 / >12.5 | >12.5 | >12.5 |
| 6 | >12.5 | >12.5 | >12.5 | >12.5 | >12.5 |
| 7 | 5.5 / 12.5 | > 12.5 | > 12.5 | >12.5 | 12.5 |
| 8 | 3.1 / 6.3 | 6.3 / 25.0 | 12.5 | >12.5 | 4.7 / 9.4 |
| 9 | >12.5 | >12.5 | >12.5 | >12.5 | >12.5 |
| 10 | 4.7 / 18.8 | 4.7 / >25.0 | 6.3 / >25.0 | >12.5 | 9.4 / 25.0 |
We obtained excellent antibacterial activity of 3.1–6.3 µg/mL with compounds 3 and 4 against both the Gram-negative F. tularensis and the Gram-positive S. aureus, including MRSA. Similar activity is seen in the E. coli TolC-mutant suggesting that these compounds are significantly effluxed in the wild type E. coli strain, producing no observable activity at ≤12.5 µg/mL. Promising antibacterial activity is also observed with 7, 8, and 10. Generally we observe that the more enzymatically potent compounds have better antibacterial activity.
Compounds that were active against the two tested bacterial species also displayed promising antibacterial activity against the E. coli TolC- mutant with MIC values in a range similar to that observed for both F. tularensis and S. aureus. No antibacterial activity was observed for any of the compounds tested at ≤12.5 µg/mL against the wild type E. coli. This demonstrates that the target is intracellular and TolC mediated efflux is an issue for this species. However, as TolC transporters, or homologs thereof, are present in the other organisms tested, it is possible that these compounds are susceptible to efflux in these organisms as well, but are still able to achieve intracellular concentrations high enough to inhibit bacterial growth.
Thus, in the studies described here, we have utilized a systematic lead optimization strategy to progress a promising chemical series of enoyl-reductase inhibitors, with high nanomolar enzyme activity to a second-generation series with low nanomolar enzyme activity against F. tularensis FabI. These second generation benzimidazoles also display very promising, low microgram/mL antibacterial activity against not only F. tularensis, but also S. aureus, MRSA, and TolC-E. coli. This indicates that our benzimidazoles have the potential to display broad spectrum activity in species for which FabI is an essential enzyme.
Supplementary Material
Acknowledgements
This work was supported in part by National Institutes of Health Grants U01-AI077949 and R41AI110090. K.E.H. was supported during a portion of this work by NIDCR 5T32-DE018381, UIC College of Dentistry, MOST Program. P.C.S was supported by 13PRE14800030, a predoctoral fellowship from the American Heart Association, the Mid-West Affiliate. B.D.S was supported in part by the UIC Center for Clinical and Translational Science through grant UL1TR000050 from the National Institutes of Health. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor for the support of this research program (Grant 085P1000817). ChemDraw was used for drawing and displaying chemical structures (ChemDraw v13.0, CambridgeSoft Corp.). Molecular graphics and analyses were performed with the UCSF Chimera package. Chimera is developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIGMS P41-GM103311).
We hank Dr. Joseph Brunzelle for support with x-ray data collection and initial data processing at LS-CAT.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
PDB Accession Codes: 4J1N: FtFabI with Compound 7; 4J3F: FtFabI with Compound 8; 4J4T: FtFabI with Compound 10.
Experimental procedures including compound details, assay conditions, MIC determinations, and crystallization conditions and structure solution are available in the Supplementary Section.
References and Notes
- 1.Weber IB, Turabelidze G, Patrick S, Griffith KS, Kugeler KJ, Mead PS. Clin. Infect. Dis. 2012;55:1283. doi: 10.1093/cid/cis706. [DOI] [PubMed] [Google Scholar]
- 2.Oyston PC, Sjostedt A, Titball RW. Nat. Rev. Microbiol. 2004;2:967. doi: 10.1038/nrmicro1045. [DOI] [PubMed] [Google Scholar]
- 3.Seefeld MA, Miller WH, Newlander KA, Burgess WJ, DeWolf WE, Jr, Elkins PA, Head MS, Jakas DR, Janson CA, Keller PM, Manley PJ, Moore TD, Payne DJ, Pearson S, Polizzi BJ, Qiu X, Rittenhouse SF, Uzinskas IN, Wallis NG, Huffman WF. J. Med. Chem. 2003;46:1627. doi: 10.1021/jm0204035. [DOI] [PubMed] [Google Scholar]
- 4.Kitagawa H, Kumura K, Takahata S, Iida M, Atsumi K. Biorg. Med. Chem. 2007;15:1106. doi: 10.1016/j.bmc.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 5.Freundlich JS, Yu M, Lucumi E, Kuo M, Tsai HC, Valderramos JC, Karagyozov L, Jacobs WR, Jr, Schiehser GA, Fidock DA, Jacobus DP, Sacchettini JC. Bioorg. Med. Chem. Lett. 2006;16:2163. doi: 10.1016/j.bmcl.2006.01.051. [DOI] [PubMed] [Google Scholar]
- 6.Moir DT. Curr. Drug Targets Infect. Disord. 2005;5:297. doi: 10.2174/1568005054880154. [DOI] [PubMed] [Google Scholar]
- 7.England K, am Ende C, Lu H, Sullivan TJ, Marlenee NL, Bowen RA, Knudson SE, Knudson DL, Tonge PJ, Slayden RA, Antimicrob J. Chemother. 2009;64:1052. doi: 10.1093/jac/dkp307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brinster S, Lamberet G, Staels B, Trieu-Cuot P, Gruss A, Poyart C. Nature. 2009;458:83. doi: 10.1038/nature07772. [DOI] [PubMed] [Google Scholar]
- 9.Balemans W, Lounis N, Gilissen R, Guillemont J, Simmen K, Andries K, Koul A. Nature. 2010;463:E3. doi: 10.1038/nature08667. [DOI] [PubMed] [Google Scholar]
- 10.Brinster S, Lamberet G, Staels B, Trieu-Cuot P, Gruss A, Poyart C, Brinster, et al. Nature. 2010;463:E4. doi: 10.1038/nature07772. reply, [DOI] [PubMed] [Google Scholar]
- 11.Parsons JB, Frank MW, Subramanian C, Saenkham P, Rock CO. Proc. Natl. Acad. Sci. U. S. A. 2011;108:15378. doi: 10.1073/pnas.1109208108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cronan JE, Rock CO. Eco-Sal-Escherichia coli and Salmonella typhimurium. In: BöckI ea., editor. Cell. Mol. Biol. Washington, DC: American Society for Microbiology; 2008. [Google Scholar]
- 13.Kingry LC, Cummings JE, Brookman KW, Bommineni GR, Tonge PJ, Slayden RA. J Bacteriol. 2013;195:351. doi: 10.1128/JB.01957-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hevener KE, Mehboob S, Su PC, Truong K, Boci T, Deng J, Ghassemi M, Cook JL, Johnson ME. J Med Chem. 2012;55:268. doi: 10.1021/jm201168g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mehboob S, Hevener KE, Truong K, Boci T, Santarsiero BD, Johnson ME. J Med Chem. 2012;55:5933. doi: 10.1021/jm300489v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng Y, Prusoff WH. Biochem. Pharmacol. 1973;22:3099. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 17.Burlingham BT, Widlanski TS. J. Chem. Educ. 80(2003):214. [Google Scholar]
- 18.O'Shea R, Moser HE. J Med Chem. 2008;51:2871. doi: 10.1021/jm700967e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.