Abstract
Mycobacterium tuberculosis (MTB) is a highly successful pathogen because of its ability to persist in human lungs for long periods of time. MTB modulates several aspects of the host immune response. Lymphocyte-activation gene 3 (LAG3) is a protein with a high affinity for the CD4 receptor and is expressed mainly by regulatory T cells with immunomodulatory functions. To understand the function of LAG3 during MTB infection, a nonhuman primate model of tuberculosis, which recapitulates key aspects of natural human infection in rhesus macaques (Macaca mulatta), was used. We show that the expression of LAG3 is highly induced in the lungs and particularly in the granulomatous lesions of macaques experimentally infected with MTB. Furthermore, we show that LAG3 expression is not induced in the lungs and lung granulomas of animals exhibiting latent tuberculosis infection. However, simian immunodeficiency virus–induced reactivation of latent tuberculosis infection results in an increased expression of LAG3 in the lungs. This response is not observed in nonhuman primates infected with non-MTB bacterial pathogens, nor with simian immunodeficiency virus alone. Our data show that LAG3 was expressed primarily on CD4+ T cells, presumably by regulatory T cells but also by natural killer cells. The expression of LAG3 coincides with high bacterial burdens and changes in the host type 1 helper T-cell response.
Mycobacterium tuberculosis (MTB), the causative agent of tuberculosis (TB), is thought to have infected more than one third of the world's current population.1 MTB is responsible for approximately 1.3 million deaths a year, meaning that this pathogen results in greater mortality than any other infectious bacterium.2,3 Each year, approximately 9 million people become newly infected with MTB.1 Of those individuals infected with MTB, approximately 90% remain latently infected, with an asymptomatic infection that is not cleared by the immune response.4 Only 5% to 10% of MTB-infected individuals progress to active disease, where a breakdown of MTB containment and clinical symptoms of TB take place.5 During active TB, the release of MTB bacilli occurs, resulting in individuals who are highly infectious, leading to the spread of MTB.
The granuloma is crucial to determining the progression or control of MTB infection.6 In human pulmonary TB, the structure of the lung granuloma within the host is well organized and composed mainly of immune cells. A classic-type MTB-induced lung granuloma consists of a necrotic central region encircled by monocyte-derived cells, including infected macrophages, epithelioid macrophages, multinucleated giant cells, and foamy macrophages, which is then surrounded by an outer ring of mostly T and B lymphoid-type cells and fibroblasts.6,7 The most accepted view is that the formation of this lung granuloma is the host's attempt to contain and control the growth of MTB bacilli, yet it has been suggested that granuloma formation might inadvertently assist in the persistence of infection.8,9 These two views hint at the constant struggle that occurs between host and pathogen to gain advantage during infection.
Immunomodulation within the host is critical for successful containment of the bacilli within the lung granuloma. A suppressed immune response will prevent granuloma formation and maintenance, and an overactive response will result in excessive inflammation, causing immunopathogenesis and allowing for the proliferation and spread of MTB.10,11 MTB has previously been shown to use certain immunomodulatory proteins of the host to regulate the immune response in its favor; this has been observed with the up-regulation of IL-10, as well as potentially with the increased presence of indoleamine 2,3-dioxygenase.12–14 Moreover, previous studies have illustrated that infected macrophages produce the immunosuppressive cytokine IL-10 that can inhibit the production of IL-12, thus controlling T-cell differentiation.12,15 Expression of IL-10 in regulatory T cells (Tregs) during early MTB infection has been observed in virulent MTB strains, resulting in the dampening of the type 1 helper T-cell (Th1) response.12 Another method through which MTB is able to control the host response to infection is through the suppression of dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin (DC-SIGN) within dendritic cells, which causes decreased dendritic cell function and maturation.16
Lymphocyte-activation gene 3 (LAG3) protein is expressed on populations of activated T cells, such as Tregs and natural killer (NK) cells, and some monocyte-derived cell populations.17,18 LAG3 is a negative costimulatory receptor that is homologous to CD4, yet has a 2-log higher affinity for major histocompatibility complex II.17 This molecule dampens the immune response through the activation and resulting proliferation of Tregs, as well as via the inhibition of monocyte differentiation, both of which have deleterious downstream effects on Th1 effector T-cell proliferation and function, and are essential for an adequate host response to control MTB infection.17–20 LAG3 up-regulation has already been shown to be detrimental to the host response in certain chronic infections, such as hepatitis B virus and Plasmodium falciparum.21,22 The blockade of LAG3 with monoclonal antibodies has resulted in an enhanced ability of antigen-presenting cells to generate a Th1 response, with increased levels of interferon (IFN)-γ being present.23 Building on these facts, it appears as though LAG3 and IL-10 could play similar roles during an MTB infection in both the inhibition of the Th1 immune response, as well as with presentation of antigen.
Our group has previously shown that the granuloma-rich lung tissue from actively MTB-infected rhesus macaques has a 25-fold higher expression of LAG3 RNA than that of naïve and latently infected animals.24 We propose that LAG3 plays a role in modulating the local lung immune response to MTB to attempt to contain infection through dampening the immune response to reduce host-mediated immunopathogenesis. We believe that LAG3 up-regulation correlates with active TB due to a diminished immune response and may be functionally linked to IL-10.12 As part of the current study, we have tested this hypothesis in the rhesus macaque (Macaca mulatta) model. This system was used because of its ability to reproducibly emulate the progression of MTB infection, where the clinical signs experienced by nonhuman primates (NHPs) during active infection and MTB-induced lung granuloma structure closely mirror what is experienced in humans.25 Furthermore, NHPs are able to recapitulate latency, as well as reactivation when infected with simian immunodeficiency virus (SIV).5,26 Our findings show that LAG3 expression is localized to the lungs of animals experiencing active TB. Furthermore, we show that LAG3 is expressed within the outer periphery of MTB-induced lung granulomas. The cells expressing LAG3 are believed to be regulatory T cells and NK cells.
Materials and Methods
Human Tissue Samples
Archived paraffinized lung tissue biopsy samples from humans diagnosed with TB were collected in accordance with a protocol approved by the Ethics Committee of the National Institute of Respiratory Diseases (Mexico City, Mexico), as previously described.27
Animals
Adult male Indian origin rhesus macaques acquired from the Tulane National Primate Research Center (Covington, LA) breeding colony were used for our studies. These animals were quarantined for 90 days and were tested for previous exposure to MTB with the tuberculin skin test (TST) and an NHP IFN-γ release assay (Primagam; Life Technologies-Thermo Fisher Scientific, Waltham, MA).28 MTB-infected animals were housed in Biosafety Level 3 conditions. Blood draws, bronchoalveolar lavage (BAL), and all other physical data collection were performed as previously described.28,29 Samples were collected for this study from animals with active TB as well as latent TB infection (LTBI) and reactivation, which had been described in detail earlier.14,24 The Tulane National Primate Research Center Institutional Animal Care and Use Committee and the Institutional Biosafety Committee approved all procedures.
Infections
The experimental design for MTB infection via inhalation and SIV coinfection has been previously illustrated.24 Herein, we used 32 Indian rhesus macaques (Table 1). For the study of active TB, 1000 colony-forming units (CFUs) of MTB CDC1551 was deposited into the lungs of 10 animals via the head-only aerosol method, as previously described.24 For the study of LTBI, 22 remaining animals were infected through the same method to deposit 50 CFUs of MTB CDC1551.24 MTB infection was confirmed by conversion to positive TST and PRIMAGAM. Over a period of 9 weeks, the animals were bled weekly for complete blood cell count, serum chemistry analysis, and serum C-reactive protein (CRP) assays. BAL collection was also performed at weeks 3 and 7 to determine MTB levels. In a subset of animals infected with the lower dose of MTB and with no signs of disease, SIV infections were performed with 300 samples of 50% tissue culture–infective dose of SIVmac239 virus diluted in RPMI 1640 medium (Sigma-Aldrich, St. Louis, MO) and injected i.v. at week 9 after MTB infection. The criteria to determine the onset of active TB, LTBI, and reactivation of LTBI included the following: TST and PRIMAGAM-IFN-γ release assay positivity, serum CRP levels, and bacterial CFUs in the BAL of these animals over the course of infection. All procedures had been described in detail in our prior publications.14,24,25,28 Plasma SIV levels were measured using an SIV assay developed by the Tulane National Primate Research Center Pathogen Quantification and Detection Core.15 Animals with acute brucellosis were infected with Brucella melitensis via aerosol with between 8.5 × 105 and 1.3 × 106 CFUs, as previously described.30 Animals infected with SIV alone were administered the virus i.v. with 100 animal infectious dose.31 Humane end points were predefined in this protocol and applied as a measure of reduction of discomfort.
Table 1.
Classification of TB Status in MTB-Infected Rhesus Macaques
| Identification code | Infection/treatment | Classification | Age at infection (years) | Mamu subtype | TST (−2 weeks) | TST (3 weeks) | TST (7 weeks) | E or EN | Time to necropsy (days) |
|---|---|---|---|---|---|---|---|---|---|
| HD0001 | MTB | Active TB | 9.5 | NA | NNN | PPP | ND | E | 21 |
| HD0002 | MTB | Active TB | 10.3 | NA | NNN | PPP | ND | E | 25 |
| HD0003 | MTB | Active TB | 2.4 | A-11 and DR2011 | NNN | PPP | ND | E | 25 |
| HD0004 | MTB | Active TB | 3.5 | A-02, A-11, and B-01 | NNN | PPP | ND | E | 34 |
| HD0005 | MTB | Active TB | 6.6 | A-11 and B-17 | NNN | PPP | ND | E | 34 |
| HD0006 | MTB | Active TB | 3.7 | A-08 and B-01 | NNN | PPP | ND | E | 35 |
| HD0007 | MTB | Active TB | 7.8 | A-02 | NNN | PPP | ND | E | 38 |
| HD0008 | MTB | Active TB | 13.8 | A-02 and B-01 | NNN | PPP | ND | E | 51 |
| HD0009 | MTB | Active TB | 13.4 | A-02 and B-01 | NNN | PPP | ND | E | 52 |
| HD0010 | MTB | Active TB | 9.4 | A-02, A-08, B01, and DR2011 | NNN | PPP | ND | E | 61 |
| LD0001 | MTB | Active TB | 9.3 | A-02 and DR2011 | NNN | ND | PPP | E | 105 |
| LD0002 | MTB | Active TB | 6.5 | NA | NNN | ND | PPP | E | 133 |
| LD0003 | MTB | Active TB | 3.5 | NA | NNN | PPP | ND | E | 46 |
| LD0004 | MTB | Active TB | 3.7 | A08 | NNN | PPP | ND | E | 43 |
| LD0005 | MTB | LTBI | 9.4 | A-02 and B-17 | NNN | ND | PPP | EN | 126 |
| LD0006 | MTB | LTBI | 9.4 | A-08 | NNN | ND | PPP | EN | 136 |
| LD0007 | MTB | LTBI | 11.6 | NA | NNN | ND | PPP | EN | 53 |
| LD0008 | MTB | LTBI | 12.5 | A-01 and DR2011 | NNN | ND | PPP | EN | 52 |
| LD0009 | MTB | LTBI | 4.5 | DR2011 | NNN | ND | PPP | EN | 127 |
| LD0010 | MTB | LTBI | 3.4 | A-01 and A-08 | NNN | ND | PPP | EN | 120 |
| LD0011 | MTB | LTBI | 6.4 | NA | NNN | PPP | PPP | EN | 190 |
| LD0012 | MTB or SIV | Reactivation | 3.1 | A08 | NNN | ND | PPP | E | 147 |
| LD0013 | MTB or SIV | Reactivation | 3.5 | A-01, A-02, and DR2011 | NNN | ND | PPP | E | 114 |
| LD0014 | MTB or SIV | Reactivation | 3.5 | B-01 | NNN | ND | PPP | E | 112 |
| LD0015 | MTB or SIV | Reactivation | 3.3 | A-01 and B-01 | NNN | ND | PPP | E | 127 |
| LD0016 | MTB or SIV | Reactivation | 3.4 | A-01 and B-01 | NNN | ND | PPP | E | 167 |
| LD0017 | MTB or SIV | Reactivation | 8.5 | A-11 | NNN | ND | PPP | E | 153 |
| LD0018 | MTB or SIV | Reactivation | 3.4 | A-02 and B-01 | NNN | ND | PPP | E | 104 |
| LD0019 | MTB or SIV | Reactivation | 2.2 | NA | NNN | ND | PPP | E | 167 |
| LD0020 | MTB or SIV | LTBI | 4.5 | A-01 and B-17 | NNN | ND | PPP | EN | 154 |
| LD0021 | MTB or SIV | LTBI | 8.2 | A-11 | NNN | ND | PPP | EN | 126 |
| LD0022 | MTB or SIV | LTBI | 3.1 | A-01 and A-08 | NNN | ND | PPP | EN | 137 |
Animals were divided into two subsets on the basis of high- and low-dose MTB infection. TST data were obtained 24, 48, and 72 hours after mammalian tuberculin injection. Negative results for the TST were represented with an N, and a P showed positive results. SIV coinfection was performed 63 days after MTB infection.
E, euthanasia; EN, experimental necropsy; HD, animals that received high-dose MTB infection when included in animal identification code; LD, animals that received low-dose MTB infection when included in identification code; LTBI, latent tuberculosis infection; MTB, Mycobacterium tuberculosis; N, a single negative; NA, data not available; ND, not done; P, a single positive test result at each time of reading; SIV, simian immunodeficiency virus; TB, tuberculosis; TST, tuberculin skin test.
RNA Extraction and Quantitative RT-PCR
RNA was extracted from lung tissue and BAL that had been stored at −80°C. Lung, spleen, bronchial lymph node, and BAL were placed in TRIzol (Life Technologies, Grand Island, NY) before RNA extraction. Tissue was then homogenized in M tubes (Miltenyi Biotec, Auburn, CA) with a GentleMACS Dissociator (Miltenyi Biotec) using the RNA_02 setting. All samples were run through QIAshredders (Qiagen, Valencia, CA) to ensure cell lysis. RNA was then extracted with a TRIzol/RNeasy Kit (Qiagen) hybrid protocol. DNA was removed from the RNA samples using a TURBO DNA-free Kit (Life Technologies). RNA was quantified with the NanoDrop 2000 (Thermo Scientific, Wilmington, DE). Because of low RNA yield, BAL samples were amplified with MessageAmp aRNA Amplification Kit (Life Technologies) and concentrated with a Vacufuge (Eppendorf, Hauppauge, NY). cDNA synthesis was performed with the High Capacity RNA-to-cDNA Kit (Life Technologies). Primers were as follows: LAG3, 5′-TCTTTCCTTACTGCCAAGTGGGCT-3′ (forward) and 5′-AATGTGACAGTGGCATTGAGCTGC-3′ (reverse); IL-10, 5′-TGAGAACCACGACCCAGACATCAA-3′ (forward) and 5′-AAAGGCATTCTTCACCTGCTCCAC-3′ (reverse); and β-actin, 5′-TCGTCCACCGCAAATGC-3′ (forward) and 5′-TCAAGAAAGGGTGTAACGCAACT-3′ (reverse). These primers were designed and optimized (Integrated DNA Technologies, Coralville, IA). The reactions were performed on an ABI 7900 RT-PCR machine (Applied Biosystems, Carlsbad, CA) with SYBR Green (Life Technologies) as a detector. Samples were performed in duplicate, and both positive and negative controls (nontemplate controls) were used. The expression levels were calculated using the 2E−ΔΔCT method, where samples were normalized against β-actin expression and uninfected controls.
Transforming Growth Factor β Cytokine Assay
Lung tissue was homogenized in M tubes with a GentleMACS Dissociator using the Protien_01 setting with Tissue Extraction Reagent I (Invitrogen, Life Technologies, Grand Island, NY) supplemented with Protease Inhibitor Cocktail (Sigma-Aldrich). Supernatant was filtered with a 0.2-μm sterile polyethersulfone filter (VWR, Radnor, PA). Samples were then acid treated (the pH was <3.0 and then neutralized). Milliplex TGFβ Magnetic Bead 3 Plex Kit (Millipore, Billerica, MA) was used to detect presence of cytokine using the Bio-Plex 200 array reader (Bio-Rad, Hercules, CA) and analyzed with Bio-Plex Manager software version 6.1 (Bio-Rad).
Flow Cytometry
Flow cytometry staining was performed on blood and BAL, as described earlier,24 as well as granuloma-rich lung tissue. Lung was processed into a single-cell suspension by digestion with collagenase type IV (Sigma-Aldrich) and DNase I (Sigma-Aldrich), followed by filtration through sterile 40-μm cell strainers (BD Biosciences, San Jose, CA).32 BAL was pelleted before flow cytometry staining. All samples were stained with the extracellular antibodies and were then treated with BD FACS Lysing Solution (BD Biosciences) to remove erythrocytes. After staining, all samples were treated with BD Stabilizing Fixative (BD Biosciences). The cell samples were labeled in stain buffer (phosphate-buffered saline, 1% bovine serum albumin, and 0.5% sodium azide) with the following fluorochrome-conjugated antibodies: CD3 (PacBlue; BD Biosciences), CD4 (APC-H7; BD Biosciences), CD8 (R-phycoerythrin-TxR; Invitrogen), and LAG3 (R-phycoerythrin; R&D Systems, Minneapolis, MN). Samples were read using a BD LSR II flow cytometer (BD Biosciences). Flow cytometric analysis was performed with FlowJo software version 8 (Treestar, Ashland, OR). Before gating for cell populations fluorescing positive for antibodies, lymphocytes were gated on the basis of their forward and side scatter characteristics, and doublets were excluded by plotting forward scatter area versus forward scatter height (Supplemental Figure S1).
Fluorescent Immunohistochemistry and Confocal Microscopy
Lung tissue sections were placed in a stimulation solution containing phytohaemagglutinin (Invitrogen), 4β-phorbol 12-myristate 13-acetate (Sigma-Aldrich), calcium ionophore (Sigma-Aldrich), lipopolysaccharide (Sigma-Aldrich), and brefeldin A (Sigma-Aldrich) before being frozen within optimal cutting temperature embedding matrix. Tissues were then cut into sections (15 μm thick). To retrieve antigen, the slides were boiled in Antigen Unmasking Solution (Vector, Olean, NY); human paraffin sections were deparaffinized before the antigen retrieval process. Primary antibodies against the following proteins were used: CD3 [1:20, mouse IgG1 (Dako, Carpinteria, CA), or neat, rabbit (Dako)], forkhead box P3 (FOXP3) [1:200, rabbit (Abcam, Cambridge, MA)], granzyme B [1:200, mouse IgG2a (BD Pharmingen, San Jose, CA)], Ham56 [1:50, mouse IgM (Dako)], IL-10 [1:25, mouse IgG2b (R&D)], LAG3 fluorescein isothiocyanate conjugated [1:50, mouse IgG1 (Lifespan Bioscience, Seattle, WA)], and TO-PRO-3 nuclear stain [1:2000 (Molecular Probes, Life Technologies)]. The above primary antibodies were conjugated with the following secondary antibodies from Molecular Probes at a 1:1000 concentration derived from goat: Alexa Fluor 488 anti-rabbit, Alexa Fluor 568 anti-mouse IgG1, Alexa Fluor 568 anti-mouse IgG2a, Alexa Fluor 568 anti-mouse IgG2b, Alexa Fluor 568 anti-mouse IgM, Alexa Fluor 568 anti-rabbit, Alexa Fluor 647 anti-mouse IgG1, and Alexa Fluor 647 anti-rabbit. Imaging was performed with a Leica True Confocal Laser Scanning Microscope SP2 laser scanning confocal microscope (Leica, Buffalo Grove, IL), and the images were analyzed with Volocity 3D image analysis software version 6.3 (Perkin Elmore, Waltham, MA).
Statistical Analysis
Statistical analyses were performed with GraphPad Prism 6 (San Diego, CA). For comparison between two groups, the unpaired, two-tailed, Student's t-test was used to show statistical significance. For greater numbers of variables, significance was calculated using analysis of variance, followed by the post hoc Bonferroni test for significance between experimental groups. P < 0.05 was considered significant, and P < 0.01 and P < 0.001 are indicated.
Results
Progression of MTB Infection
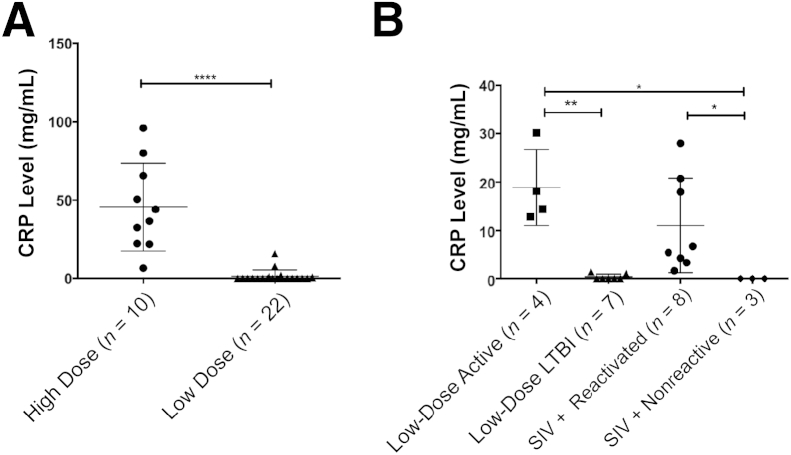
We monitored the progression of MTB infection in animals infected with the following: i) high-dose MTB (10 Indian rhesus macaques), ii) low dose of MTB (22 Indian rhesus macaques), or iii) low-dose of MTB followed by i.v. coinfection with SIV (a subset of 11 Indian rhesus macaques from ii) (Table 1). Although all animals infected with MTB developed TST positivity, all 10 animals infected with a high dose of MTB developed acute pulmonary TB, requiring humane euthanasia. We previously showed that serum CRP levels accurately predict the advent of active (or reactivation) TB in macaques.28 Thus, animals exposed to a high dose of MTB exhibited high peak serum CRP values after infection, between weeks 0 and 9 (Figure 1A). These values were significantly higher than those for the animals exposed to a low dose of MTB, because many of these animals did not exhibit serum CRP values above baseline in a comparable time period (Figure 1A). In fact, repeated serum CRP levels remained undetectable for 20 of 22 animals in this group.
Figure 1.
Serum C-reactive protein (CRP) levels discriminate between various infection outcomes in nonhuman primates infected with Mycobacterium tuberculosis (MTB). A: Peak serum CRP levels after MTB infection up to week 9 in a group of 10 animals that received a high dose of MTB CDC1551 (circles) and a group of 22 animals that received a low dose of the same strain (triangles). Statistical significance was determined using a two-tailed Student's t-test in GraphPad Prism 6. B: Of the 22 animals, which exhibited latent tuberculosis infection (LTBI), 11 were followed up long-term, whereas the other 11 were coinfected with simian immunodeficiency virus (SIV) at week 9. Peak serum CRP levels between weeks 9 and 24 are shown for animals that reactivated spontaneously (squares), due to SIV (circles), or exhibited LTBI despite SIV coinfection (diamonds). Animals that exhibited long-term LTBI are shown in triangles. Statistical significance was derived by using a one-way analysis of variance in Prism, with Sidak's multiple-comparison test. Each point represents one data point, and the means ± SD are represented by horizontal bars. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗∗P < 0.0001.
After SIV coinfection, 8 of 11 animals exhibited signs of clinical reactivation, whereas 3 remained recalcitrant to reactivation, where no clinical signs of TB were observed (Table 1). Similarly, of the 11 animals that were not SIV coinfected, 7 continued to exhibit LTBI, whereas 4 gradually developed active TB. We, therefore, compared the peak serum CRP values in animals infected with a low dose of MTB with and without SIV coinfection 9 weeks after MTB infection (the time at which SIV was administered) through the end of the experiment (Figure 1B). Again, the levels of serum CRP were significantly higher in the animals with reactivation of LTBI, whether spontaneous or SIV induced, relative to animals that maintained a latent MTB infection.
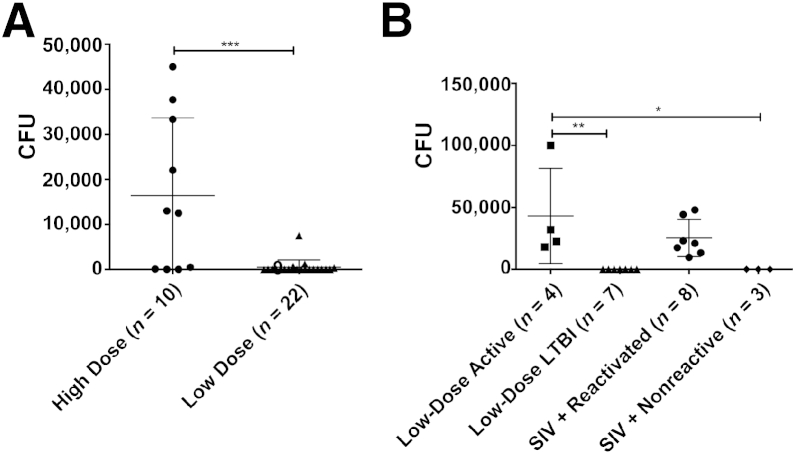
To verify that serum CRP values could accurately predict the outcome of mycobacterial disease in the 32 animals, we also studied the total bacterial burden in BAL of these animals. BAL bacillary burden is somewhat comparable to sputum MTB levels in clinical patients.33 Absence of MTB in the BAL accurately predicts LTBI, whereas detectable bacterial levels in BAL highly correlate with active TB disease. BAL CFUs mirrored serum CRP levels (Figure 2A). In high-dose MTB infections, 6 of 10 animals exhibited detectable BAL bacterial burdens, whereas the lavage of almost every low-dose animal was devoid of culturable MTB between weeks 0 and 9. Of the animals that had previously established LTBI and were followed up long-term, four with elevated CRP levels exhibited a spike in BAL MTB levels at different time points, thus illustrating reactivation of LTBI (Figure 2B). Other long-term animals remained free of detectable MTB in BAL, except for eight animals that received SIV i.v. (Figure 2B). Hence, from this point onward, we studied three groups of animals: i) active TB (10 high-dose animals and 4 low-dose animals), ii) LTBI (7 low-dose animals and 3 low-dose animals with SIV coinfection), and iii) reactivation (8 low-dose animals with SIV coinfection). The combination of both an immunological marker (serum CRP) and direct microbiological detection (BAL Mtb levels) allowed us to distinctly classify animals into these wanted groups.
Figure 2.
Bacillary load in bronchoalveolar lavage (BAL) distinguishes between different infection outcomes in Mycobacterium tuberculosis (MTB)–infected animals. A: Peak BAL colony-forming units (CFUs) after MTB infection up to week 9 in a group of 10 animals that received a high dose of MTB CDC1551 (circles) and a group of 22 animals that received a low dose of the same strain of MTB (triangles). Statistical significance was determined using a two-tailed Student's t-test. B: Of the 22 animals that exhibited latent tuberculosis infection (LTBI), 11 were followed up long-term, whereas the other 11 were coinfected with simian immunodeficiency virus (SIV) at week 9. Peak BAL CFUs between weeks 9 and 24 are shown for animals that reactivated spontaneously (squares), due to SIV (circles), or exhibited LTBI despite SIV coinfection (diamonds). Animals that exhibited long-term LTBI are shown in triangles. Statistical significance was derived by using a one-way analysis of variance in GraphPad Prism 6, with Sidak's multiple-comparison test. Each point represents one data point, and the means ± SD are represented by horizontal bars. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001.
LAG3 Transcript Levels in the BAL and Lungs of Macaques Infected with MTB
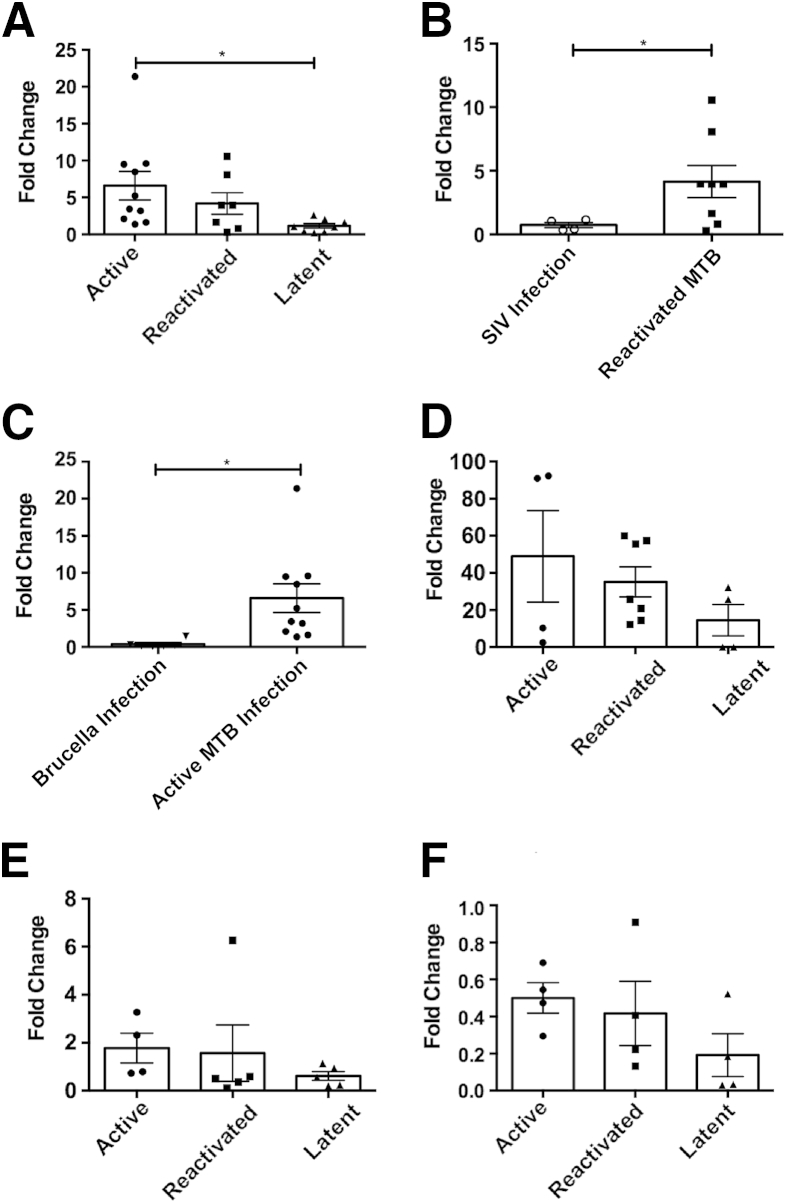
We next confirmed the results from our previous transcriptomics screen, which had revealed that the LAG3 transcript was one of the most abundant in macaque granulomatous lung during active TB disease.24 Although the relationship between LAG3 and MTB infections has not been previously explored in detail, LAG3 is a key immunosuppressive molecule present on subsets of Tregs with the aim of modulating excessive proinflammatory immune responses. Granuloma samples were retrieved from the lungs of MTB-infected (or MTB/SIV coinfected) animals at euthanasia due to either active TB or SIV-induced reactivation or at experimental necropsy of animals with LTBI. RNA was isolated from granuloma-rich lung tissue, and LAG3 expression was determined by quantitative RT-PCR. As expected, the expression of LAG3 was strongly induced in animals with active TB (with a fold change of 6.59 relative to normal lung) (Figure 3A). These values were significantly higher than those observed in lung granulomas from LTBI animals (a fold change of 1.15 relative to normal lung). Next, we assessed the lung granuloma lesions from animals experiencing reactivation of LTBI due to SIV coinfection. The expression of the LAG3 transcript was induced in these reactivation samples to an extent comparable to animals with active TB (a fold change of 4.18 relative to normal lung) (Figure 3A). However, these levels were not statistically different from LAG3 expression levels derived from LTBI granulomas (Figure 3A). Thus, our results suggest that the expression of LAG3 in the lungs of primates exposed to MTB correlates with the extent of TB disease and bacterial burden.
Figure 3.
Lymphocyte-activation gene 3 (LAG3) transcription levels in Mycobacterium tuberculosis (MTB)–infected nonhuman primates undergoing activation of tuberculosis (TB). The fold change of LAG3 mRNA expression was measured using quantitative RT-PCR through the calculation of the 2−ΔΔCT, where normalization was calculated with β-actin and corresponding uninfected tissue. LAG3 transcription levels were compared between lung tissue of active (n = 10), reactivated (n = 7), and latent tuberculosis infection (n = 8) MTB infections (A); active MTB (n = 10) and Brucella infections (n = 6) (B); and reactivated MTB infections (n = 7) and simian immunodeficiency virus (SIV) infections (n = 4) (C); BAL of active (n = 4), reactivated (n = 7), and LTBI (n = 4) MTB infections (D); spleen of active (n = 4), reactivated (n = 5), and LTBI (n = 5) MTB infections (E); and bronchial lymph node (BrLN) of active (n = 4), reactivated (n = 4), and LTBI (n = 4) MTB infections (F). Data points are categorized by infection type, where circles indicate active TB; squares, reactivated TB; triangles, LTBI; clear circles, SIV alone; and upside-down triangles, Brucella infections. Each point represents one data point, and the means ± SD are represented by horizontal bars. Statistical significance was determined by either a one-way analysis of variance (A and D–F) or a two-tailed Student's t-test (B and C) in GraphPad Prism 6. ∗P < 0.05.
To determine whether the expression of LAG3 was specifically linked to MTB infection, we performed two additional investigations. First, we studied the expression of LAG3 in the lungs of macaques infected with SIVmac239 alone during acute to chronic disease at 12 to 20 weeks after infection. These lung samples were obtained at comparable times to the MTB/SIV coinfected lung samples in relation to the time period between SIV infection and euthanasia. SIV infection alone failed to result in the induction of LAG3 levels in the lungs of macaques (Figure 3B). These expression levels were significantly lower than those in samples derived from MTB/SIV coinfected animals. Second, we studied the expression of LAG3 transcript in the lungs of macaques acutely infected with B. melitensis via aerosol exposure. Brucella infection resulted in virtually no change in lung LAG3 expression, and these values were significantly lower than those for active MTB infection (Figure 3C). This shows that the induction of LAG3 in the lungs of primates is most likely specific to TB and not just a generalized response to SIV infection or infection of aerosolized intracellular bacteria. Furthermore, the expression of LAG3 was not found to be induced in the lungs of animals infected with MTB:Δ-sigH mutant, which causes a nonpathogenic infection,26 and in animals infected with various mutants in the dormancy survival regulon pathway.
Having established that LAG3 up-regulation in the lungs of infected macaques was specific to MTB infection, we next assessed LAG3 levels in the BAL. LAG3 levels were examined by RT-PCR in samples that were collected from animals immediately before euthanasia. There was no statistical difference between the three groups of animals, with the active animals having a mean of 48.98-fold elevation of the LAG3 transcript, relative to BAL from uninfected macaques (Figure 3D). MTB/SIV coinfected animals showed an increase of 35.15-fold, when compared to BAL from naïve animals. Although the differences were not statistically significant, in both groups of animals that succumbed to MTB infection (MTB and MTB/SIV coinfection), a >35-fold increase in LAG3 expression took place compared to normal BAL (the baseline).
Finally, to ensure that significant LAG3 up-regulation was specific to the lung of active and reactivated TB animals, LAG3 expression was examined in both the spleen and bronchial lymph node. Up-regulation did not occur in either active or reactivated animals when compared to latent animals (Figure 3, E and F). This suggests that not only is the up-regulation of LAG3 levels specific for active rather than latent infection, it is also highly specific to MTB infection within the lung.
IL-10 Transcript Levels in the BAL and Lungs of Macaques Infected with MTB
IL-10 is a key anti-inflammatory cytokine, and its expression is known to correlate with loss of effective Th1 responses during MTB infection.12,15 It is increasingly being discovered that LAG3 is highly expressed on specific populations of Tregs that are primary producers of IL-10, thus mediating key anti-inflammatory functions.34,35 Consequently, we decided to study the expression of IL-10 in macaques in parallel with LAG3.
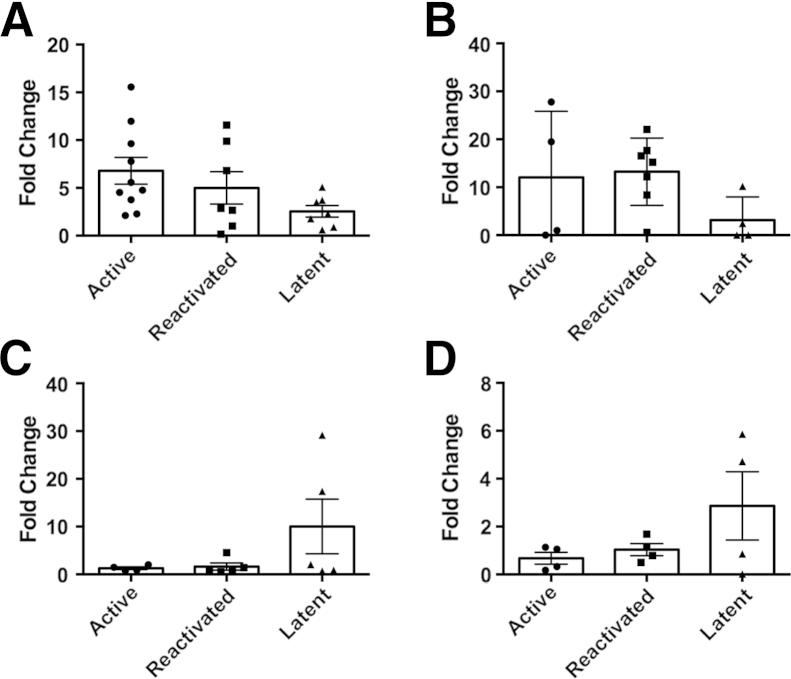
The expression of IL-10 mirrored that of LAG3 in the three groups of NHPs. Thus, animals with active TB exhibited an average fold change of 6.80 in lung granulomas, which was higher than the observed value for animals with LTBI (a fold change of 2.55) (Figure 4A). Similarly, SIV-induced reactivation also exhibited higher levels of IL-10 expression, and these values (an average fold change of 5.0) were higher than those for the LTBI cohort, but they were not statistically different when compared to the active TB data (Figure 4A).
Figure 4.
IL10 transcription levels in Mycobacterium tuberculosis (MTB)–infected nonhuman primates undergoing activation of tuberculosis (TB). Quantitative RT-PCR was used to measure the fold change of IL-10 mRNA expression through the calculation of the 2−ΔΔCT, where normalization was calculated with β-actin and uninfected tissue. IL10 transcription levels were compared between lung tissue of active (n = 10), reactivated (n = 7), and latent tuberculosis infection (n = 8) MTB infections (A); BAL of active (n = 4), reactivated (n = 7), and LTBI (n = 4) MTB infections (B); spleen of active (n = 4), reactivated (n = 5), and LTBI (n = 5) MTB infections (C); and bronchial lymph node of active (n = 4), reactivated (n = 4), and LTBI (n = 4) MTB infections (D). Data points are categorized by infection type, where circles indicate active TB; squares, reactivated TB; and triangles, LTBI. Each point represents one data point, and the means ± SD are represented by horizontal bars. Statistical significance was determined by a one-way analysis of variance in Prism version 6.
We then analyzed the expression of IL-10 in the BAL of the different groups of macaques (Figure 4B). These findings were similar to what was observed in the LAG3 expression data above. Hence, animals with LTBI expressed the lowest levels of IL-10 when compared to the active and reactivation TB groups. Although no statistical difference was observed between the three groups, there was greater expression of IL-10 in both the active group, with a 12.06-fold increase, and the MTB/LTBI group, with a 13.24-fold increase, when compared to normal lung tissue. The LTBI animals only experienced a 3.16-fold increase in IL-10. One interesting observation in the active animal group was that there appeared to be a division in IL-10 expression; half of the animals had IL-10 levels similar to what was present in the latent animals. Also, IL-10 levels did not appear to be significantly increased in the spleen and bronchial lymph node of active MTB-infected animals (Figure 4, C and D), respectively.
Detection of LAG3 Protein in Peripheral Blood and Lungs of Animals with Active TB, LTBI, and SIV-Induced Reactivation
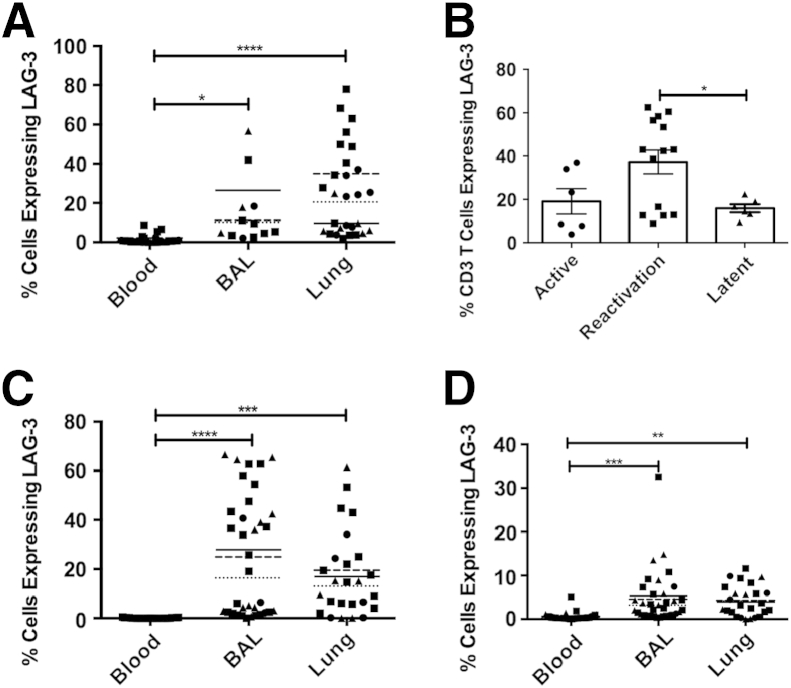
To study the frequency of cells expressing LAG3 in the peripheral blood, lung, and BAL at the protein level, we used an anti-LAG3 antibody in combination with flow cytometry. The LAG3+ populations were gated for as previously described in Supplemental Figure S1. Virtually no cells expressed LAG3 in the peripheral blood (Figure 5). However, an average of 27% of T cells derived from the lung granuloma expressed LAG3 (Figure 5A). Similarly, 11% of all BAL T cells were positive for LAG3 expression. Thus, the expression of the LAG3 protein, like its cognate transcript, was limited to the lung during MTB infection in macaques. We then studied if the expression of LAG3 on T cells differed depending on the status of the MTB infection. Reactivation TB animals exhibited a significantly greater frequency of T cells expressing LAG3 (Figure 5B).
Figure 5.
Lymphocyte-activation gene 3 (LAG3) protein expression levels in lymphocytes from peripheral blood, bronchoalveolar lavage (BAL), and lung of Mycobacterium tuberculosis (MTB)–infected nonhuman primates. LAG3+ cells were compared between CD3+ T cells in blood (n = 53), BAL (n = 24), and lung (n = 22) (A); CD3+ T cells in lung tissue of active (n = 6), reactivated (n = 14), and latent tuberculosis infection (LTBI) (n = 6) infections (B); CD3+CD4+ T cells in blood (n = 53), BAL (n = 53), and lung (n = 22) (C); and CD3+CD4+ T cells in blood (n = 53), BAL (n = 53), and lung (n = 22) (D). Data points are categorized by infection type, where circles indicate active TB; squares, reactivated TB; and triangles, LTBI. The mean for each infection category is represented by a dotted line for active TB, a dashed line for reactivated TB, and a solid line for LTBI (A, C, and D). Each point represents one data point, and the means are represented by horizontal bars. Statistical significance was determined by a one-way analysis of variance in GraphPad Prism. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, and ∗∗∗∗P < 0.0001.
Furthermore, LAG3 expression in CD3+CD4+ and CD3+CD8+ populations (Figure 5, C and D), respectively, was significantly higher in both the lung granuloma and BAL samples of MTB-infected animals relative to the peripheral blood. The frequency of LAG3-expressing CD8+ T cells was 0.5% in blood, whereas in BAL and lung, the frequency was significantly greater at 4.7% and 3.8%, respectively. CD4+ T cells expressing LAG3 occurred at an increased rate, with a mean of 18.7% T cells expressing LAG3 in BAL and 17.1% in the lung. However, in these populations, there was no significant difference between the frequencies of LAG3-expressing cells of the TB infection groups. We believe that the lack of significance can be explained by the up-regulation of LAG3 in certain populations of cells, yet not necessarily through an increase in the number of cells expressing LAG3.
Identification of LAG3-Expressing Cells Using Immunofluorescence
Having conclusively shown that LAG3 transcript and protein are highly induced during active TB and SIV-induced reactivation TB within the lung, we sought to determine the cell phenotypes that express LAG3 within the granuloma-rich lung samples that were obtained at euthanasia in NHPs that succumbed to active TB.
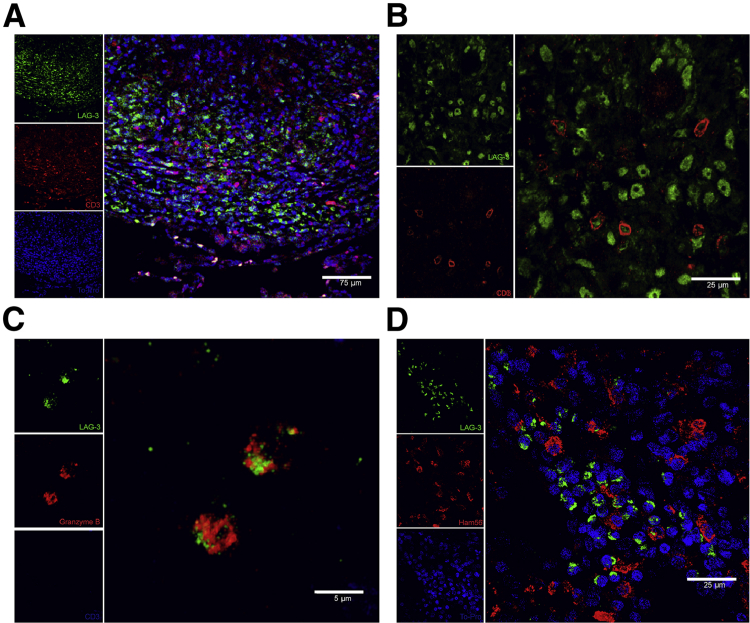
To achieve this, we used multicolor confocal immunofluorescence. We studied if as shown in the flow data, T cells were the source for LAG3 expression in lung lesions. The outer periphery of the lung granuloma was the location where most LAG3 expression was shown to be occurring (Figure 6A); a significant number of LAG3-expressing cells were also CD3 surface positive (Figure 6B), thus confirming T cells as a major source of lung-derived LAG3. However, approximately 50% of all strongly LAG3+ cells in the lung were not CD3+. In size and morphology, these cells were similar to lymphocytes. We, therefore, hypothesized that these cells could potentially be NK cells. Because we were unable to optimize an antibody that could definitively identify NK cells in rhesus macaques, we used a negative selection strategy instead. Sections were stained for CD3, LAG3, and granzyme B. We surmised that CD3−granzyme B+ cells are highly likely to be NK cells. Our results clearly showed that LAG3 is highly expressed in this subset after the onset of active MTB infection (Figure 6C). Because of the imperative role of lung macrophages during an MTB infection, we also wanted to verify if such cells in the lung compartment expressed this immunosuppressive protein. We accomplished this by using the pan–macrophage-specific marker HAM56.36 Although there were LAG3-expressing cells in close vicinity, it was apparent that none of the macrophages were expressing LAG3 (Figure 6D).
Figure 6.
Identification of cellular subsets expressing lymphocyte-activation gene 3 (LAG3) in the lung granulomas of macaques with active tuberculosis. Optimal cutting temperature–frozen and stimulated lung tissue sections were assessed using immunofluorescence staining and confocal microscopy imaging. A and B: Images were stained for T cells with LAG3 (green), CD3 (red), and TO-PRO (blue) nuclear stain at low and high magnification. C: Natural killer cells were stained using a negative selection strategy with LAG3 (green), granzyme B (red), and CD3 (blue). D: Alveolar macrophages do not express LAG3 [using markers for LAG3 (green), Ham56 (red), and TO-PRO (blue)].
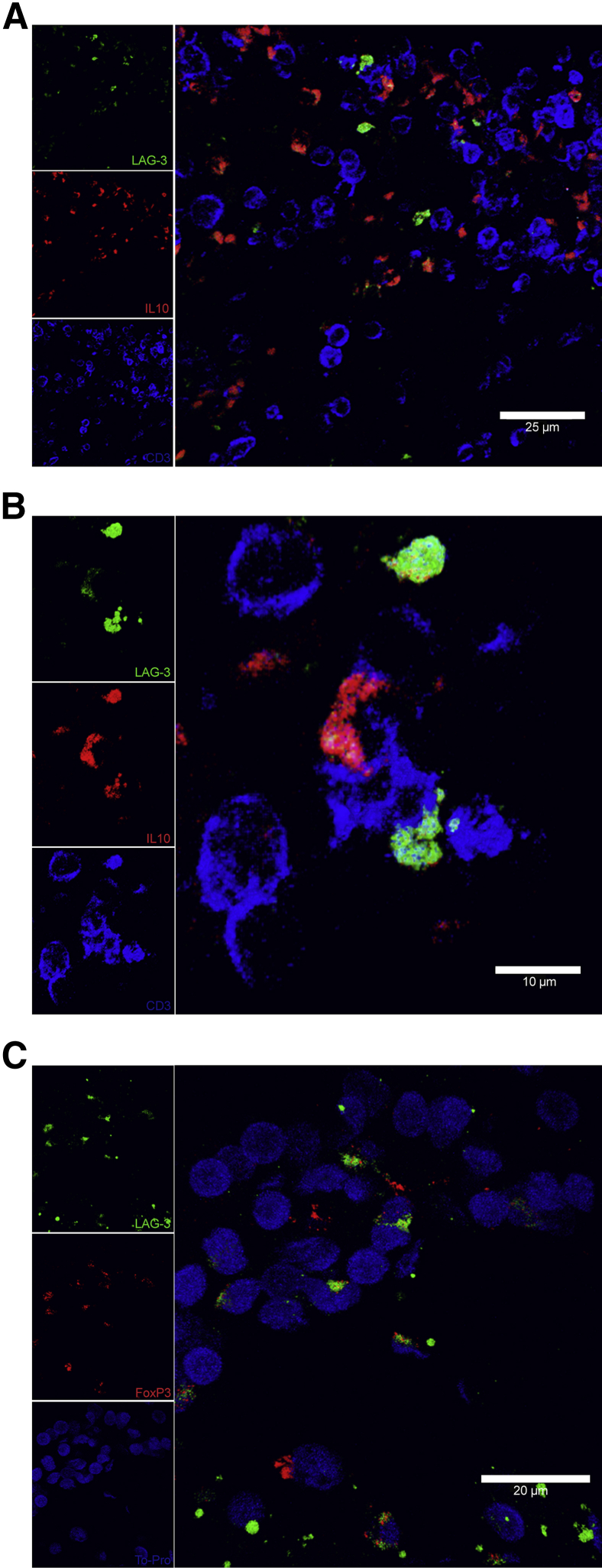
Because previous research had confirmed that Tregs are a subpopulation of T cells that express LAG3,18 we investigated whether these LAG3+ Tregs are present within the MTB-infected lung granuloma. We first used IL-10, a well-documented cytokine produced by Tregs, in combination with CD3 as markers to define Tregs. We observed that within the lung granuloma, there were cells that expressed both CD3 and IL-10 in addition to LAG3 (Figure 7, A and B). It has been recently suggested that LAG3 expression on CD4+ T cells marks a subpopulation of CD4+CD25−LAG3+ Tregs, which express the FOXP3 transcription factor. Therefore, we next examined if the LAG3 signal on macaque lung granulomas colocalized with the FOXP3 signal. FOXP3 was present within certain CD3+ T cells in combination with LAG3, thus showing increased evidence that LAG3-expressing Tregs are located within the MTB-infected lung granuloma (Figure 7C). Interestingly, although LAG3-expressing Tregs were found within the lungs of these actively infected animals, the same animals did not possess elevated levels of transforming growth factor β when compared to the lungs of uninfected and latently infected animals (Supplemental Figure S2). This might suggest that expansion and maintenance of Tregs between infection groups may not actually differ significantly.
Figure 7.
Regulatory T cells (Tregs) express lymphocyte-activation gene 3 (LAG3). Optimal cutting temperature–frozen and stimulated lung tissue sections were assessed using immunofluorescence staining and confocal microscopy imaging. A and B: Low- and high-magnification images showing colocalization of LAG3 and IL-10 in T cells with the following markers: LAG3 (green), IL-10 (red), and CD3 (blue). C: Tregs expressing LAG3 were stained for with LAG3 (green), FOXP3 (red), and TO-PRO (blue).
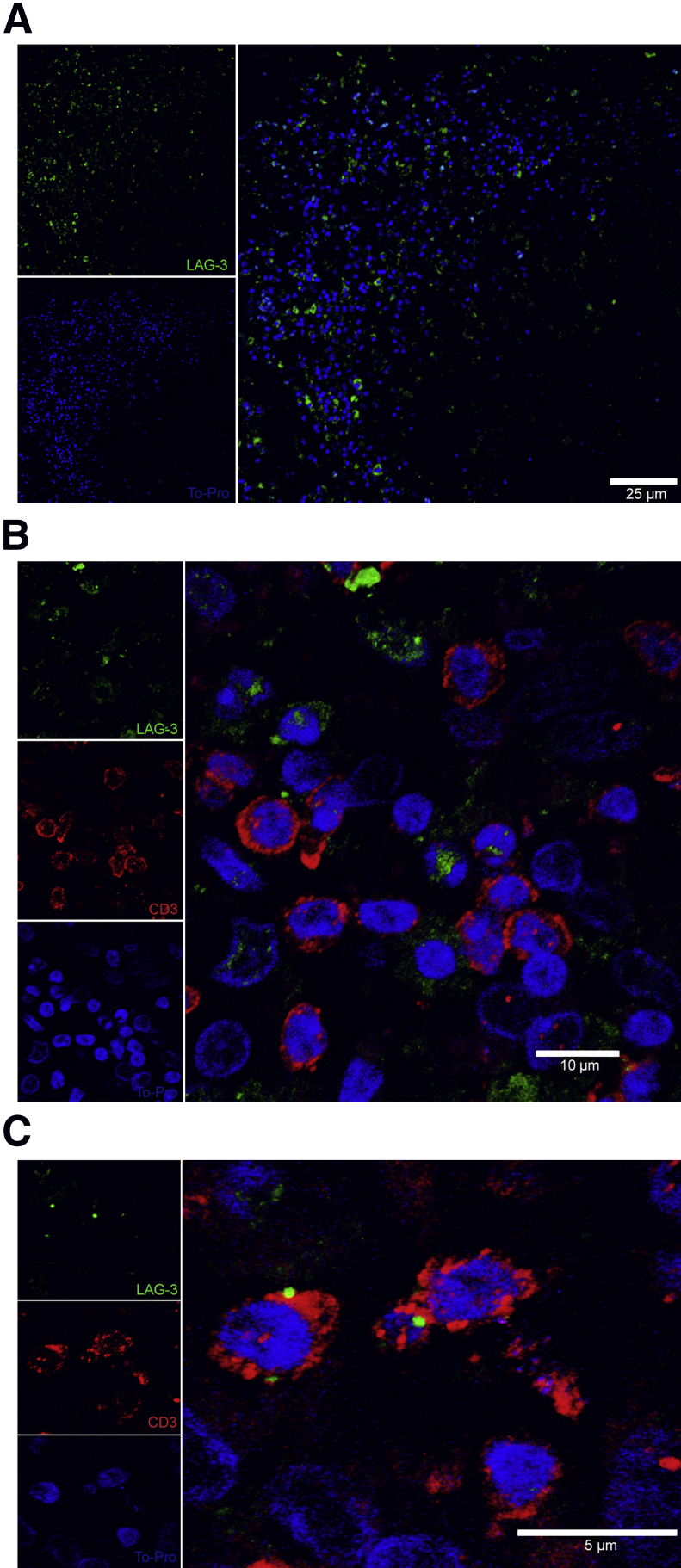
To verify the significance of LAG3 expression in NHP TB lesions, and to demonstrate that a similar expression pattern is observed in humans with active TB, we stained human lung tissue sections containing MTB-induced granulomas using fluorescent immunohistochemistry. We observed that LAG3 expression mainly occurs within the outer periphery of the lung granuloma (Figure 8A), similar to what was shown to occur in actively infected rhesus macaques. In addition, many of the LAG3-expressing cells within the periphery of the granuloma were coexpressing CD3, thus enforcing the fact that many of the LAG3-expressing cells are within lung granulomas and are T cells (Figure 8, B and C). These images illustrate that the similarities observed between LAG3 expression in MTB-induced lung granulomas in humans and NHPs are great, and imply that our previous findings in NHPs can be applied to active TB cases involving humans (Figures 6 and 8).
Figure 8.
A: Paraffinized Mycobacterium tuberculosis (MTB)–infected human lung tissue shows a similar presence and distribution of lymphocyte-activation gene 3 (LAG3) within the MTB containing lung granuloma with LAG3 (green) and TO-PRO (blue). B and C: Low and high magnification of LAG3 within T cells with markers for LAG3 (green), CD3 (red), and TO-PRO (blue).
Discussion
MTB is one of the most successful pathogens of humankind; emerging data indicate that this remarkable pathogen coevolved with humanity as it emerged from Africa 50,000 years ago.37 MTB has infected humans for such a long time with a high degree of penetration; it causes chronic, rather than acute, infections, illustrating that MTB infections are able to be extremely well managed.38 Thus, numerous examples exist in the literature, which point to the fact that MTB successfully engages with and modulates the protective response mounted by the host.39,40 The IFN-γ network is central to immunity against TB.41 Thus, MTB interferes with IFN-γ signaling pathways at multiple levels, by repressing the expression of proinflammatory chemokines and by down-regulating the expression of the IFN-γ receptor.31,42–44 In addition, MTB is able to alter the effects of other host immune pathways aimed at its eradication. Thus, mutants in sigH and sigE, two related inducible transcription factors of MTB that allow the pathogen to respond to and survive against a battery of stress conditions such as phagocytosis,45 oxidation reduction stress,46 enduring hypoxia,47 and in vivo infection,26,48,49 allow for the generation of host chemokine responses that were suppressed by the parental wild-type MTB strains.50,51 Furthermore, MTB is reported to use matrix metalloproteases52,53 and cAMP54 to modulate the host response to infection. It may be the result of these interventions that the effective, antigen-specific, CD4+ T-cell response to MTB is delayed relative to other infections.55 Moreover, although MTB has the ability to actively subvert and delay the host response mounted to clear the infection, the effectiveness of this initial immune response itself is questionable. Thus, it is established that the response is only able to plateau or control, rather than sterilize, the infection, both in experimental models and in humans.
Korf et al56 first reported that mycolic acid components from MTB result in an increased expression of Treg markers in the lung, causing an increased tolerance to experimental asthma. As to how the immunosuppression is regulated during an ongoing mycobacterial infection is not totally understood. Although the activity of Treg cells appears to be diminished in certain diseases,57 whether directly or indirectly, MTB appears to manipulate Treg activity in a manner that assists in its own persistence. Thus, Tregs were shown to suppress anti-MTB immune function in patients.58,59 In a mouse model of vaccination, ablation of Treg responses resulted in more potent IFN-γ activation, which, in turn, resulted in significantly reduced bacterial burdens.60 Such FOXP3+CD25− T cells have been shown to deplete pathogen-specific IFN-γ, CD4, and CD8 degranulation responses.61 In the macaque model, active TB is characterized by greater recruitment of these immunosuppressive cells.62
Herein, we characterized the role of LAG3 and the cells expressing it during active, latent, and reactivated MTB infections using the highly tractable and faithful rhesus macaque model. The animals progressed to active TB because of either a high-burden infection with MTB or a low-dose infection over a prolonged period. Reactivation was modeled by coinfection with SIVmac239. Expectedly, significantly more animals exhibited SIV-mediated, rather than spontaneous, reactivation.
Our results show that LAG3 expression was highly induced in the lungs of primates with active, but not latent, TB infections (Figures 3 and 5). Furthermore, LAG3 expression was elevated in animals in which LTBI was reactivated because of coinfection with SIV, modeling MTB/HIV coinfection in humans (Figures 3 and 5). Our results substantiate that LAG3 levels are elevated in the lungs of animals with active TB at both the transcript and the protein level. Interestingly, levels of LAG3 were not increased in the peripheral blood. These results, taken with the fact that other pulmonary infections do not appear to generate comparable LAG3 induction (Figure 3, B and C), indicate that LAG3 may be a potential suitable host-derived biomarker for active TB. Thus, assays may be optimized in the future on the basis of LAG3 levels to predict the progression of active TB or HIV-induced reactivation TB in a population of LTBI patients. The fact that LAG3 induction is not detected in peripheral blood means that such future assays will need to sample cells from the lung. However, the fact that LAG3 is readily detected in BAL, which predominantly samples cells from the alveolus, means that noninvasive assays using patient sputum may someday be developed. In a field with few leads on effective biomarkers, whether pathogen or host derived, our results represent a substantial growth of the field. More interesting are the results obtained when LAG3 transcripts were sampled in the BAL of MTB/SIV coinfected animals. Although most animals in this group exhibited reactivation of LTBI, characterized by high serum CRP levels (Figure 1) and high BAL-MTB burdens (Figure 2), a few of these animals remained recalcitrant to reactivation, at least until the time (13 to 17 weeks after infection) when they were sent to experimental necropsy. Although LAG3 levels remained low in the lungs of these animals, consistent with the low serum CRP and MTB levels, the BAL of these animals, performed just before necropsy, exhibited higher levels of LAG3. This was the only instance in which RNA levels of LAG3 in the lung lesions did not correlate tightly with those from BAL. On the other hand, BAL from LTBI animals infected with a low dose of MTB continued to exhibit low LAG3 RNA levels, as late as 24 weeks after infection. Although the results are not significant because of the smaller sample size of the group, which remained recalcitrant to reactivation after SIV coinfection, they indicate that these coinfected animals may have been in the process of reactivating and that LAG3 levels in BAL may provide early indication of the potential to reactivate.
Another key finding of this report is that the elevation in LAG3 levels observed in both BAL and lung granuloma primarily occurred in T cells (Figure 5, A and B). In fact, the frequency of both CD3+CD4+ (Figure 5C) and CD3+CD8+ (Figure 5D) T cells expressing LAG3 was slightly higher in BAL samples than in lungs. One unexpected result was a significantly greater percentage of LAG3+ T cells from the reactivated animals expressing LAG3 when compared to the latent animal group, yet the same was not observed in the active infection group (Figure 5B). It is possible that this increased frequency of LAG3 in the reactivated animals could be partially due to CD4 T-cell depletion from the SIV infection. In addition, although the presence of LAG3-expressing cells is no different between the active and latent infection groups, the transcription levels on individual cells could actually be significantly up-regulated, thus explaining the variation in LAG3 presence observed (Figures 3A and 5B). These results underlie our contention that LAG3+ Tregs are recruited to the lungs of infected animals with TB, perhaps to contain inflammation, and eventually home into lung lesions, and that increased transcription of LAG3 is indicative of conversion to active infection.
Our confocal microscopy studies (Figure 6) shed light on the type of cells that express LAG3 in response to MTB infection and their potential function. Colocalization of the CD3 and the LAG3 signal in the lung granulomas of animals with active TB adds further evidence, along with the flow cytometry data, that most of the LAG3 signal can be attributed to Tregs (Figure 6, A and B). However, we also observed that in addition to CD3+ cells, LAG3 signal was associated with other small cells in the granulomas (Figure 6B). We suspected that these could be NK cells, because LAG3 is known to be expressed on this cell type.63 However, despite trying several clones, we were unable to optimize any human NK cell antibody in rhesus macaques. We, therefore, devised a strategy to identify NK cells on the basis of negative selection (Figure 6C) and showed that NK cells express a portion of the lung LAG3 signal present in the lung granuloma. This strategy was on the basis of identifying small cells that were CD3− and expressing granzyme B.
Finally, we showed that LAG3+ Tregs are recruited to the lungs of macaques with active TB (Figure 7) and that these cells express high levels of IL-10 (Figure 7, A and B). This is a critical observation in light of several recent publications, which show that different subsets of Tregs may exist, with LAG3 marking the more activated, functional type.64,65 Because of similar antibody isotypes, we were unable to perform fluorescent immunohistochemistry showing one image containing CD3, FOXP3, and LAG3 expression. However, we were able to show in two images that Tregs expressing FOXP3 are found within the lung granuloma (Figure 7C), and that most LAG3+ T cells in the lung samples expressed high levels of IL-10 (Figure 7, A and B), indicating that these cells were phenotypically functional Tregs. We studied IL-10 levels at both the transcriptional and protein level in peripheral blood, BAL, and end-point lung samples of the infected animals and found that IL-10 expression levels mirrored those of LAG3.
In addition, we examined LAG3 expression in lung tissue containing MTB-induced granulomas from a human with active TB (Figure 8). We found that the presence and location of LAG3 within these structures (Figure 8A) are similar to what we observed within the rhesus macaque samples (Figure 6A). Furthermore, many of these LAG3-positive cells were also shown to be CD3+. The similarities observed between the human and NHP lung granuloma samples point to the fact that the NHP data are highly translatable to humans. These strong similarities confirm an important role for LAG3 in the modulation of intragranulomatous immune responses in human lungs. Thus, LAG3 expression was not only enhanced in NHP lesions with experimental MTB infection over a few weeks to months, but is also potentially induced in humans with active TB, in whom infection occurred several months to years ago. This suggests that MTB potentially survives in TB granulomas characterized by high LAG3 expression, which, in turn, suggests that the human TB granuloma environment is immunosuppressed in the months after initial infection, an observation recapitulated in our NHP model.26
Hence, our results indicate that LAG3 marks a subpopulation of Tregs that are highly active and produce high levels of the cytokine IL-10, which are recruited to the lungs of primates with uncontrolled MTB replication. Although it is likely that this response is able to mitigate uncontrolled inflammation, it may also assist the pathogen in its persistence. These results illustrate crucial implications for host-directed therapies against TB. Furthermore, a high level of LAG3 induction in response to MTB replication (in active and reactivation TB) and not expressed during latent or initial infection strongly suggests that this molecule can serve as a biomarker to differentiate between vaccinated and latently infected individuals on one end of the spectrum and people with a high potential for loss of immune control at the other.
Acknowledgments
We thank the members of B.L.P.'s doctoral thesis committee, including Drs. Andrew Lackner, Lucy Freytag, and James McLachlan (Tulane University, New Orleans, LA) and Dr. Alistair Ramsay (Louisiana State University, Baton Rouge, LA) for helpful suggestions; Nadia Abraham for excellent technical assistance and program management; and Drs. Chad Roy and Marcelo Kuroda for Brucella- and SIV-infected macaque tissue, respectively.
B.L.P. designed and performed experiments, performed data analysis, and wrote the manuscript; S.M. and M.H.A. performed research; M.S. and S.A.K. provided human clinical tuberculosis samples and insight; and D.K. designed experiments, performed data analysis, wrote the manuscript, and provided funding.
Footnotes
Supported by NIH grants HL106790, AI089323, AI091457, RR026006, RR020159, OD011104, and AI058609; the Tulane National Primate Research Center (TNPRC) Office of the Director; the TNPRC Pilot Projects Program; the Louisiana Vaccine Center; the Tulane Center for Infectious Diseases; and the Tulane Office of Vice-President for Research Bridge Fund (all to D.K.).
Disclosures: None declared.
Supplemental Data
Flow cytometry gating strategy where lymphocyte-activation gene 3 (LAG3) protein expression was detected in lymphocyte populations derived from peripheral blood, bronchoalveolar lavage, and lung of Mycobacterium tuberculosis–infected nonhuman primates. FSC-A, forward scatter area; SSC-A, side scatter area.
Transforming growth factor β (TGF-β) levels within the lung of uninfected (n = 4), active (n = 4), reactivated (n = 4), and latent (n = 4) tuberculosis animals. Both protein levels of active TGF-β1 (A) and transcript levels of TGF-β normalized against uninfected lung tissue (B) were investigated.
References
- 1.WHO: Global Tuberculosis Report 2013. World Health Organization; Geneva, Switzerland: 2013. [Google Scholar]
- 2.Raviglione M.C. The new Stop TB Strategy and the Global Plan to Stop TB, 2006-2015. Bull World Health Organ. 2007;85:327. doi: 10.2471/06.038513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Philips J.A., Ernst J.D. Tuberculosis pathogenesis and immunity. Annu Rev Pathol. 2012;7:353–384. doi: 10.1146/annurev-pathol-011811-132458. [DOI] [PubMed] [Google Scholar]
- 4.Dye C., Scheele S., Dolin P., Pathania V., Raviglione M.C. Consensus statement. Global burden of tuberculosis: estimated incidence, prevalence, and mortality by country. WHO Global Surveillance and Monitoring Project. JAMA. 1999;282:677–686. doi: 10.1001/jama.282.7.677. [DOI] [PubMed] [Google Scholar]
- 5.Bloom B.R., Murray C.J. Tuberculosis: commentary on a reemergent killer. Science. 1992;257:1055–1064. doi: 10.1126/science.257.5073.1055. [DOI] [PubMed] [Google Scholar]
- 6.Russell D.G. Who puts the tubercle in tuberculosis? Nat Rev Microbiol. 2007;5:39–47. doi: 10.1038/nrmicro1538. [DOI] [PubMed] [Google Scholar]
- 7.Ramakrishnan L. Revisiting the role of the granuloma in tuberculosis. Nat Rev Immunol. 2012;12:352–366. doi: 10.1038/nri3211. [DOI] [PubMed] [Google Scholar]
- 8.Ehlers S., Schaible U.E. The granuloma in tuberculosis: dynamics of a host-pathogen collusion. Front Immunol. 2012;3:411. doi: 10.3389/fimmu.2012.00411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guirado E., Schlesinger L.S. Modeling the Mycobacterium tuberculosis granuloma: the critical battlefield in host immunity and disease. Front Immunol. 2013;4:98. doi: 10.3389/fimmu.2013.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin P.L., Myers A., Smith L., Bigbee C., Bigbee M., Fuhrman C., Grieser H., Chiosea I., Voitenek N.N., Capuano S.V., Klein E., Flynn J.L. Tumor necrosis factor neutralization results in disseminated disease in acute and latent Mycobacterium tuberculosis infection with normal granuloma structure in a cynomolgus macaque model. Arthritis Rheum. 2010;62:340–350. doi: 10.1002/art.27271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fenhalls G., Stevens L., Bezuidenhout J., Amphlett G.E., Duncan K., Bardin P., Lukey P.T. Distribution of IFN-gamma, IL-4 and TNF-alpha protein and CD8 T cells producing IL-12p40 mRNA in human lung tuberculous granulomas. Immunology. 2002;105:325–335. doi: 10.1046/j.1365-2567.2002.01378.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Redford P.S., Murray P.J., O'Garra A. The role of IL-10 in immune regulation during M. tuberculosis infection. Mucosal Immunol. 2011;4:261–270. doi: 10.1038/mi.2011.7. [DOI] [PubMed] [Google Scholar]
- 13.Cooper A.M. Cell-mediated immune responses in tuberculosis. Annu Rev Immunol. 2009;27:393–422. doi: 10.1146/annurev.immunol.021908.132703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mehra S., Alvarez X., Didier P.J., Doyle L.A., Blanchard J.L., Lackner A.A., Kaushal D. Granuloma correlates of protection against tuberculosis and mechanisms of immune modulation by Mycobacterium tuberculosis. J Infect Dis. 2013;207:1115–1127. doi: 10.1093/infdis/jis778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giacomini E., Iona E., Ferroni L., Miettinen M., Fattorini L., Orefici G., Julkunen I., Coccia E.M. Infection of human macrophages and dendritic cells with Mycobacterium tuberculosis induces a differential cytokine gene expression that modulates T cell response. J Immunol. 2001;166:7033–7041. doi: 10.4049/jimmunol.166.12.7033. [DOI] [PubMed] [Google Scholar]
- 16.Geijtenbeek T.B., Van Vliet S.J., Koppel E.A., Sanchez-Hernandez M., Vandenbroucke-Grauls C.M., Appelmelk B., Van Kooyk Y. Mycobacteria target DC-SIGN to suppress dendritic cell function. J Exp Med. 2003;197:7–17. doi: 10.1084/jem.20021229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poirier N., Haudebourg T., Brignone C., Dilek N., Hervouet J., Minault D., Coulon F., de Silly R.V., Triebel F., Blancho G., Vanhove B. Antibody-mediated depletion of lymphocyte-activation gene-3 (LAG-3(+) )-activated T lymphocytes prevents delayed-type hypersensitivity in non-human primates. Clin Exp Immunol. 2011;164:265–274. doi: 10.1111/j.1365-2249.2011.04329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Macon-Lemaitre L., Triebel F. The negative regulatory function of the lymphocyte-activation gene-3 co-receptor (CD223) on human T cells. Immunology. 2005;115:170–178. doi: 10.1111/j.1365-2567.2005.02145.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li N., Wang Y., Forbes K., Vignali K.M., Heale B.S., Saftig P., Hartmann D., Black R.A., Rossi J.J., Blobel C.P., Dempsey P.J., Workman C.J., Vignali D.A. Metalloproteases regulate T-cell proliferation and effector function via LAG-3. EMBO J. 2007;26:494–504. doi: 10.1038/sj.emboj.7601520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Workman C.J., Wang Y., El Kasmi K.C., Pardoll D.M., Murray P.J., Drake C.G., Vignali D.A. LAG-3 regulates plasmacytoid dendritic cell homeostasis. J Immunol. 2009;182:1885–1891. doi: 10.4049/jimmunol.0800185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li F.J., Zhang Y., Jin G.X., Yao L., Wu D.Q. Expression of LAG-3 is coincident with the impaired effector function of HBV-specific CD8(+) T cell in HCC patients. Immunol Lett. 2013;150:116–122. doi: 10.1016/j.imlet.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 22.Illingworth J., Butler N.S., Roetynck S., Mwacharo J., Pierce S.K., Bejon P., Crompton P.D., Marsh K., Ndungu F.M. Chronic exposure to Plasmodium falciparum is associated with phenotypic evidence of B and T cell exhaustion. J Immunol. 2013;190:1038–1047. doi: 10.4049/jimmunol.1202438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andreae S., Piras F., Burdin N., Triebel F. Maturation and activation of dendritic cells induced by lymphocyte activation gene-3 (CD223) J Immunol. 2002;168:3874–3880. doi: 10.4049/jimmunol.168.8.3874. [DOI] [PubMed] [Google Scholar]
- 24.Mehra S., Golden N.A., Dutta N.K., Midkiff C.C., Alvarez X., Doyle L.A., Asher M., Russell-Lodrigue K., Monjure C., Roy C.J., Blanchard J.L., Didier P.J., Veazey R.S., Lackner A.A., Kaushal D. Reactivation of latent tuberculosis in rhesus macaques by coinfection with simian immunodeficiency virus. J Med Primatol. 2011;40:233–243. doi: 10.1111/j.1600-0684.2011.00485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaushal D., Mehra S., Didier P.J., Lackner A.A. The non-human primate model of tuberculosis. J Med Primatol. 2012;41:191–201. doi: 10.1111/j.1600-0684.2012.00536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mehra S., Golden N.A., Stuckey K., Didier P.J., Doyle L.A., Russell-Lodrigue K.E., Sugimoto C., Hasegawa A., Sivasubramani S.K., Roy C.J., Alvarez X., Kuroda M.J., Blanchard J.L., Lackner A.A., Kaushal D. The Mycobacterium tuberculosis stress response factor SigH is required for bacterial burden as well as immunopathology in primate lungs. J Infect Dis. 2012;205:1203–1213. doi: 10.1093/infdis/jis102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slight S.R., Rangel-Moreno J., Gopal R., Lin Y., Fallert Junecko B.A., Mehra S., Selman M., Becerril-Villanueva E., Baquera-Heredia J., Pavon L., Kaushal D., Reinhart T.A., Randall T.D., Khader S.A. CXCR5(+) T helper cells mediate protective immunity against tuberculosis. J Clin Invest. 2013;123:712–726. doi: 10.1172/JCI65728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dutta N.K., Mehra S., Didier P.J., Roy C.J., Doyle L.A., Alvarez X., Ratterree M., Be N.A., Lamichhane G., Jain S.K., Lacey M.R., Lackner A.A., Kaushal D. Genetic requirements for the survival of tubercle bacilli in primates. J Infect Dis. 2010;201:1743–1752. doi: 10.1086/652497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gormus B.J., Blanchard J.L., Alvarez X.H., Didier P.J. Evidence for a rhesus monkey model of asymptomatic tuberculosis. J Med Primatol. 2004;33:134–145. doi: 10.1111/j.1600-0684.2004.00062.x. [DOI] [PubMed] [Google Scholar]
- 30.Lee K.M., Chiu K.B., Sansing H.A., Didier P.J., Ficht T.A., Arenas-Gamboa A.M., Roy C.J., Maclean A.G. Aerosol-induced brucellosis increases TLR-2 expression and increased complexity in the microanatomy of astroglia in rhesus macaques. Front Cell Infect Microbiol. 2013;3:86. doi: 10.3389/fcimb.2013.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kincaid E.Z., Ernst J.D. Mycobacterium tuberculosis exerts gene-selective inhibition of transcriptional responses to IFN-gamma without inhibiting STAT1 function. J Immunol. 2003;171:2042–2049. doi: 10.4049/jimmunol.171.4.2042. [DOI] [PubMed] [Google Scholar]
- 32.Cai Y., Sugimoto C., Arainga M., Alvarez X., Didier E.S., Kuroda M.J. In vivo characterization of alveolar and interstitial lung macrophages in rhesus macaques: implications for understanding lung disease in humans. J Immunol. 2014;192:2821–2829. doi: 10.4049/jimmunol.1302269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anderson C., Inhaber N., Menzies D. Comparison of sputum induction with fiber-optic bronchoscopy in the diagnosis of tuberculosis. Am J Respir Crit Care Med. 1995;152:1570–1574. doi: 10.1164/ajrccm.152.5.7582296. [DOI] [PubMed] [Google Scholar]
- 34.Okamura T., Fujio K., Shibuya M., Sumitomo S., Shoda H., Sakaguchi S., Yamamoto K. CD4+CD25-LAG3+ regulatory T cells controlled by the transcription factor Egr-2. Proc Natl Acad Sci U S A. 2009;106:13974–13979. doi: 10.1073/pnas.0906872106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sumitomo S., Yamamoto K. CD4+CD25-LAG-3+ T cells in mouse and human. Nihon Rinsho Meneki Gakkai Kaishi. 2010;33:92–98. doi: 10.2177/jsci.33.92. [DOI] [PubMed] [Google Scholar]
- 36.Adams C.W., Poston R.N. Macrophage histology in paraffin-embedded multiple sclerosis plaques is demonstrated by the monoclonal pan-macrophage marker HAM-56: correlation with chronicity of the lesion. Acta Neuropathol. 1990;80:208–211. doi: 10.1007/BF00308926. [DOI] [PubMed] [Google Scholar]
- 37.Hershberg R., Lipatov M., Small P.M., Sheffer H., Niemann S., Homolka S., Roach J.C., Kremer K., Petrov D.A., Feldman M.W., Gagneux S. High functional diversity in Mycobacterium tuberculosis driven by genetic drift and human demography. PLoS Biol. 2008;6:e311. doi: 10.1371/journal.pbio.0060311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Russell D.G. The evolutionary pressures that have molded Mycobacterium tuberculosis into an infectious adjuvant. Curr Opin Microbiol. 2013;16:78–84. doi: 10.1016/j.mib.2012.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paige C., Bishai W.R. Penitentiary or penthouse condo: the tuberculous granuloma from the microbe's point of view. Cell Microbiol. 2010;12:301–309. doi: 10.1111/j.1462-5822.2009.01424.x. [DOI] [PubMed] [Google Scholar]
- 40.Flynn J.L., Chan J., Triebold K.J., Dalton D.K., Stewart T.A., Bloom B.R. An essential role for interferon gamma in resistance to Mycobacterium tuberculosis infection. J Exp Med. 1993;178:2249–2254. doi: 10.1084/jem.178.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cooper A.M., Dalton D.K., Stewart T.A., Griffin J.P., Russell D.G., Orme I.M. Disseminated tuberculosis in interferon gamma gene-disrupted mice. J Exp Med. 1993;178:2243–2247. doi: 10.1084/jem.178.6.2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fortune S.M., Solache A., Jaeger A., Hill P.J., Belisle J.T., Bloom B.R., Rubin E.J., Ernst J.D. Mycobacterium tuberculosis inhibits macrophage responses to IFN-gamma through myeloid differentiation factor 88-dependent and -independent mechanisms. J Immunol. 2004;172:6272–6280. doi: 10.4049/jimmunol.172.10.6272. [DOI] [PubMed] [Google Scholar]
- 43.Nagabhushanam V., Solache A., Ting L.M., Escaron C.J., Zhang J.Y., Ernst J.D. Innate inhibition of adaptive immunity: mycobacterium tuberculosis-induced IL-6 inhibits macrophage responses to IFN-gamma. J Immunol. 2003;171:4750–4757. doi: 10.4049/jimmunol.171.9.4750. [DOI] [PubMed] [Google Scholar]
- 44.Martinez A.N., Mehra S., Kaushal D. Role of interleukin 6 in innate immunity to Mycobacterium tuberculosis infection. J Infect Dis. 2013;207:1253–1261. doi: 10.1093/infdis/jit037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Graham J.E., Clark-Curtiss J.E. Identification of Mycobacterium tuberculosis RNAs synthesized in response to phagocytosis by human macrophages by selective capture of transcribed sequences (SCOTS) Proc Natl Acad Sci U S A. 1999;96:11554–11559. doi: 10.1073/pnas.96.20.11554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mehra S., Kaushal D. Functional genomics reveals extended roles of the Mycobacterium tuberculosis stress response factor sigmaH. J Bacteriol. 2009;191:3965–3980. doi: 10.1128/JB.00064-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rustad T.R., Harrell M.I., Liao R., Sherman D.R. The enduring hypoxic response of Mycobacterium tuberculosis. PLoS One. 2008;3:e1502. doi: 10.1371/journal.pone.0001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaushal D., Schroeder B.G., Tyagi S., Yoshimatsu T., Scott C., Ko C., Carpenter L., Mehrotra J., Manabe Y.C., Fleischmann R.D., Bishai W.R. Reduced immunopathology and mortality despite tissue persistence in a Mycobacterium tuberculosis mutant lacking alternative sigma factor, SigH. Proc Natl Acad Sci U S A. 2002;99:8330–8335. doi: 10.1073/pnas.102055799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Manganelli R., Voskuil M.I., Schoolnik G.K., Dubnau E., Gomez M., Smith I. Role of the extracytoplasmic-function sigma factor sigma(H) in Mycobacterium tuberculosis global gene expression. Mol Microbiol. 2002;45:365–374. doi: 10.1046/j.1365-2958.2002.03005.x. [DOI] [PubMed] [Google Scholar]
- 50.Fontan P.A., Aris V., Alvarez M.E., Ghanny S., Cheng J., Soteropoulos P., Trevani A., Pine R., Smith I. Mycobacterium tuberculosis sigma factor E regulon modulates the host inflammatory response. J Infect Dis. 2008;198:877–885. doi: 10.1086/591098. [DOI] [PubMed] [Google Scholar]
- 51.Dutta N.K., Mehra S., Martinez A.N., Alvarez X., Renner N.A., Morici L.A., Pahar B., Maclean A.G., Lackner A.A., Kaushal D. The stress-response factor SigH modulates the interaction between Mycobacterium tuberculosis and host phagocytes. PLoS One. 2012;7:e28958. doi: 10.1371/journal.pone.0028958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Volkman H.E., Pozos T.C., Zheng J., Davis J.M., Rawls J.F., Ramakrishnan L. Tuberculous granuloma induction via interaction of a bacterial secreted protein with host epithelium. Science. 2010;327:466–469. doi: 10.1126/science.1179663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Elkington P.T., Ugarte-Gil C.A., Friedland J.S. Matrix metalloproteinases in tuberculosis. Eur Respir J. 2011;38:456–464. doi: 10.1183/09031936.00015411. [DOI] [PubMed] [Google Scholar]
- 54.Agarwal N., Bishai W.R. cAMP signaling in Mycobacterium tuberculosis. Indian J Exp Biol. 2009;47:393–400. [PubMed] [Google Scholar]
- 55.Subbian S., Tsenova L., Yang G., O'Brien P., Parsons S., Peixoto B., Taylor L., Fallows D., Kaplan G. Chronic pulmonary cavitary tuberculosis in rabbits: a failed host immune response. Open Biol. 2011;1:110016. doi: 10.1098/rsob.110016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Korf J.E., Pynaert G., Tournoy K., Boonefaes T., Van Oosterhout A., Ginneberge D., Haegeman A., Verschoor J.A., De Baetselier P., Grooten J. Macrophage reprogramming by mycolic acid promotes a tolerogenic response in experimental asthma. Am J Respir Crit Care Med. 2006;174:152–160. doi: 10.1164/rccm.200507-1175OC. [DOI] [PubMed] [Google Scholar]
- 57.Cools N., Ponsaerts P., Van Tendeloo V.F., Berneman Z.N. Regulatory T cells and human disease. Clin Dev Immunol. 2007;2007:89195. doi: 10.1155/2007/89195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen X., Zhou B., Li M., Deng Q., Wu X., Le X., Wu C., Larmonier N., Zhang W., Zhang H., Wang H., Katsanis E. CD4(+)CD25(+)FoxP3(+) regulatory T cells suppress Mycobacterium tuberculosis immunity in patients with active disease. Clin Immunol. 2007;123:50–59. doi: 10.1016/j.clim.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 59.Hougardy J.M., Place S., Hildebrand M., Drowart A., Debrie A.S., Locht C., Mascart F. Regulatory T cells depress immune responses to protective antigens in active tuberculosis. Am J Respir Crit Care Med. 2007;176:409–416. doi: 10.1164/rccm.200701-084OC. [DOI] [PubMed] [Google Scholar]
- 60.Majlessi L., Brodin P., Brosch R., Rojas M.J., Khun H., Huerre M., Cole S.T., Leclerc C. Influence of ESAT-6 secretion system 1 (RD1) of Mycobacterium tuberculosis on the interaction between mycobacteria and the host immune system. J Immunol. 2005;174:3570–3579. doi: 10.4049/jimmunol.174.6.3570. [DOI] [PubMed] [Google Scholar]
- 61.Geffner L., Basile J.I., Yokobori N., Sabio Y.G.C., Musella R., Castagnino J., Sasiain M.C., de la Barrera S. CD4(+) CD25(high) forkhead box protein 3(+) regulatory T lymphocytes suppress interferon-gamma and CD107 expression in CD4(+) and CD8(+) T cells from tuberculous pleural effusions. Clin Exp Immunol. 2014;175:235–245. doi: 10.1111/cei.12227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Green A.M., Mattila J.T., Bigbee C.L., Bongers K.S., Lin P.L., Flynn J.L. CD4(+) regulatory T cells in a cynomolgus macaque model of Mycobacterium tuberculosis infection. J Infect Dis. 2010;202:533–541. doi: 10.1086/654896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kisielow M., Kisielow J., Capoferri-Sollami G., Karjalainen K. Expression of lymphocyte activation gene 3 (LAG-3) on B cells is induced by T cells. Eur J Immunol. 2005;35:2081–2088. doi: 10.1002/eji.200526090. [DOI] [PubMed] [Google Scholar]
- 64.Okamura T., Fujio K., Sumitomo S., Yamamoto K. Roles of LAG3 and EGR2 in regulatory T cells. Ann Rheum Dis. 2012;71(Suppl 2):i96–i100. doi: 10.1136/annrheumdis-2011-200588. [DOI] [PubMed] [Google Scholar]
- 65.Workman C.J., Vignali D.A. Negative regulation of T cell homeostasis by lymphocyte activation gene-3 (CD223) J Immunol. 2005;174:688–695. doi: 10.4049/jimmunol.174.2.688. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Flow cytometry gating strategy where lymphocyte-activation gene 3 (LAG3) protein expression was detected in lymphocyte populations derived from peripheral blood, bronchoalveolar lavage, and lung of Mycobacterium tuberculosis–infected nonhuman primates. FSC-A, forward scatter area; SSC-A, side scatter area.
Transforming growth factor β (TGF-β) levels within the lung of uninfected (n = 4), active (n = 4), reactivated (n = 4), and latent (n = 4) tuberculosis animals. Both protein levels of active TGF-β1 (A) and transcript levels of TGF-β normalized against uninfected lung tissue (B) were investigated.