Abstract
The two major melanoma histologic subtypes, superficial spreading and nodular melanomas, differ in their speed of dermal invasion but converge biologically once they invade and metastasize. Herein, we tested the hypothesis that distinct molecular alterations arising in primary melanoma cells might persist as these tumors progress to invasion and metastasis. Ribosomal protein S6 kinase, 90 kDa, polypeptide 1 (RSK1; official name RPS6KA1) was significantly hyperactivated in human melanoma lines and metastatic tissues derived from nodular compared with superficial spreading melanoma. RSK1 was constitutively phosphorylated at Ser-380 in nodular but not superficial spreading melanoma and did not directly correlate with BRAF or MEK activation. Nodular melanoma cells were more sensitive to RSK1 inhibition using siRNA and the pharmacological inhibitor BI-D1870 compared with superficial spreading cells. Gene expression microarray analyses revealed that RSK1 orchestrated a program of gene expression that promoted cell motility and invasion. Differential overexpression of the prometastatic matrix metalloproteinase 8 and tissue inhibitor of metalloproteinases 1 in metastatic nodular compared with metastatic superficial spreading melanoma was observed. Finally, using an in vivo zebrafish model, constitutive RSK1 activation increased melanoma invasion. Together, these data reveal a novel role for activated RSK1 in the progression of nodular melanoma and suggest that melanoma originating from different histologic subtypes may be biologically distinct and that these differences are maintained as the tumors invade and metastasize.
Superficial spreading melanoma (SSM) and nodular melanoma (NM) represent the two most common primary melanoma histologic subtypes, accounting for 70% and 15% to 20% of cases, respectively.1,2 SSM is characterized by a radial growth phase (RGP) consisting of an intraepidermal component. Whereas SSM can proceed from a RGP to a vertical growth phase (VGP) and finally to distant metastases, NM grows rapidly in a vertical manner (VGP), with no horizontal growth phase.3
To date, the prognostic and therapeutic impact of melanoma histologic subtypes has been relatively limited. The American Joint Committee on Cancer staging system uses tumor thickness, ulceration, mitotic index, and lymph node status, but not histologic subtype, in the recurrence/metastasis risk assessment of patients with primary localized melanoma.4 This is, in part, due to the current linear model of melanoma progression, which dictates that melanoma begins with the transformation of epidermal melanocytes and an initial RGP, followed by a subsequent transition to a VGP and distant metastasis.5–7 Hence, it is generally accepted that the speed of dermal invasion is the only aspect that differentiates the NM and SSM subtypes.
Recent discoveries in other solid tumor types emphasize the potential role of histology-driven molecular characterization to assist in the diagnosis and treatment of cancer.8–11 Indeed, the utility of histologic classification in melanoma has been demonstrated with acral lentiginous melanoma, which composes approximately 10% of primary melanomas. The prevalence of molecular alterations in the c-KIT oncogene in this histologic melanoma subtype has defined acral lentiginous melanoma as a distinct and useful subclassification of melanoma, and a phase 2 trial of the c-KIT inhibitor imatinib validated the rationale of subtype-specific therapy for this group of melanoma patients.12 In contrast, the clinical relevance of the SSM and NM subtypes has been largely overlooked.
Recent reports by our groups and others suggest that primary SSM and NM might be distinct biological entities.13–19 Unbiased, high-throughput genetic techniques, such as comparative genomic hybridization, single nucleotide polymorphism arrays, and microarrays, have revealed the presence of recurrent SSM-specific deletions that are present or even amplified in NM and, thus, cannot be reconciled with the linear progression model, even when epigenetic modifications are taken into account. Similarly, significant alterations in mRNA and miRNA gene expression are observed when comparing SSM with nevi and NM, and these alterations cannot be explained by the existing stepwise (linear) model.16,17 Together, these findings suggest that distinct molecular alterations between SSM and NM might underlie the biological differences between these subtypes. However, it is unclear whether differences that exist between primary SSM and NM are retained during progression to invasion and metastasis.
Herein, we tested the hypothesis that subtype-specific differences between SSM and NM persist throughout progression of primary melanoma to metastatic disease. We used a combination of human melanoma cell lines representing SSM and NM, human tissues from metastatic SSM and NM, and a zebrafish model of melanoma to demonstrate the role of protein S6 kinase, 90 kDa, polypeptide 1 (RSK1; official name RPS6KA1) in melanoma invasion in vitro and in vivo. These data suggest that metastatic melanoma originating from different histologic subtypes might be biologically distinct and reveal differences that are maintained from the primary tumor to metastatic disease.
Materials and Methods
Cell Culture
Melanoma cell lines representing RGP (WM 35, WM 1552c, WM 1575, and WM 3211) and VGP (WM 98.1, WM 115, WM 278, WM 793c, WM 853.2, WM 902, WM 983e, WM 1361, WM 1366, and WM 3248) melanoma were purchased commercially from The Wistar Institute (Philadelphia, PA). Primary cell lines were cultured in MCDB153/L15 medium (4/1 v/v) supplemented with 2% fetal bovine serum, 5 μg/mL insulin, 15 μg/mL bovine pituitary extract, 1.68 mmol/L calcium chloride, 5 ng/mL epidermal growth factor, and 1% penicillin/streptomycin. Human epidermal melanocytes were cultured in DermaLife M melanocyte culture medium (Lifeline Cell Technology, Frederick, MD).
Western Blot Analysis and Antibodies
Cells and human tissues were harvested in extraction buffer, and Western blot analysis was performed as previously described.13 Phospho-RSK (p-RSK) (Ser-363; #9344), phospho-RSK (Ser-380; #9335), total RSK (#9335), RSK1 (#8408), and RSK2 (#9340) were purchased from Cell Signaling Technology Inc. (Beverly, MA). Anti-Ran (sc-1156) antibody was purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Cell Proliferation Assay
Specified cell lines were seeded at 5 to 10 × 103 cells per well in a 96-well plate in complete medium. The next day, cells were treated with equal amounts of dimethyl sulfoxide (DMSO) or the indicated concentration of BI-D1870 (purchased from the Division of Signal Transduction Therapy, University of Dundee, Scotland, UK) and were cultured for 72 hours. Cells were fixed with cold 1% glutaraldehyde, washed with phosphate-buffered saline, and stained with 0.1% crystal violet and 20% methanol. Cells were destained with 15% acetic acid, and absorbance was read at 595 nm using a FlexStation 3 benchtop multi-mode microplate reader (Molecular Devices, Sunnyvale, CA). Half maximal inhibitory concentrations were calculated using GraphPad Prism software version 6 (GraphPad Software Inc., La Jolla, CA).
RNA Extraction and Quantitative RT-PCR
RNA extraction and quantitative RT-PCR were used to verify siRNA target knockdown and to validate the mRNA array. Total RNA was isolated from cells or tumors using a QIAshredder and RNeasy mini kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. For mRNA detection, 1 μg of DNAse-treated RNA (DNA-free kit; Invitrogen, Carlsbad, CA) was reverse transcribed using a RETROscript kit (Invitrogen), and quantitative RT-PCRs were performed using gene-specific primers and a MyiQ Real-time PCR system (Bio-Rad Laboratories, Hercules, CA). Quantitative normalization was performed on the expression of glyceraldehyde-3-phosphate dehydrogenase. The relative expression levels between samples were calculated using the 2−ΔΔCT method, with a control sample as the reference point.14 Primer sequences are summarized in Table 1.
Table 1.
Primers Used for Quantitative RT-PCR
| Gene | Forward primers | Reverse primers |
|---|---|---|
| RPS6KA1 | 5′-TGAAGGTGCTGAAGAAGGCA-3′ | 5′-CAGCTTCACCACGAATGGGT-3′ |
| RPS6KA3 | 5′-AACCTATGGGAGAGGAGGAGA-3′ | 5′-AGGATCTGCCTTTTCATGTCC-3′ |
| GAPDH | 5′-GAGCCACATCGCTCAGACAC-3′ | 5′-CATGTAGTTGAGGTCAATGAAGG-3′ |
| MMP2 | 5′-AAGCCACTGACCAGCCTGGGA-3′ | 5′-AGGCATCTGCGATGAGCTTGGG-3′ |
| CCL2 | 5′-AGCAAGTGTCCCAAAGAAGCTGTG-3′ | 5′-AGCTGCAGATTCTTGGGTTGTGGA-3′ |
| ATF1 | 5′-ACGGCGCCCATCTTACAGAAAAA-3′ | 5′-ACTGTAAGGCTCCATTTGGGGCAA-3′ |
| TIMP3 | 5′-GGTCTACACCATCAAGCAGAT-3′ | 5′-AGCCCCGTGTACATCTTGCCATCATA-3′ |
| IL8 | 5′-TTTCAGAGACAGCAGAGCACACAAGC-3′ | 5′-CACCTTCACACAGAGCTGCAGAAA-3′ |
| CXCL1 | 5′-TATTTCTGAGGAGCCTGCAACATGCC-3′ | 5′-GCACATACATTCCCCTGCCTTCACAAT-3′ |
Serum Starvation
Cells (4 × 105) of specified cell lines were seeded in three 6-cm plates. The next day, growth medium in two-thirds of the plates was replaced with medium containing MCDB153/L15 medium (4/1 v/v), 1% primary melanoma cell medium, and 1% penicillin/streptomycin. After 24 hours, one serum-starved plate was stimulated for 10 minutes with complete primary melanoma cell medium. Plates were then trypsinized, washed with phosphate-buffered saline, and harvested for Western blot analysis.
Transfection of siRNAs
Cells were plated in 12-well plates at 30% to 50% confluency and transfected 24 hours later with 20 pmol/L siRNA using Lipofectamine 2000 reagent (Invitrogen Life Technologies, Grand Island, NY), according to the manufacturer's instructions. RSK1 (#S12275), RSK2 (#S12279), and negative control (#4390843) siRNA were purchased from Invitrogen Life Technologies. mRNA knockdown was quantified 48 hours later by quantitative RT-PCR.
Gene Expression Array
For expression profiling, we used the Affymetrix GeneChip system (Affymetrix Inc., Santa Clara, CA). The RNA quality and quantity were determined using a 2100 Bioanalyzer system (Agilent Technologies Inc., Santa Clara, CA) and a NanoDrop ND-1000 spectrophotometer (Thermo Scientific, Wilmington, DE). Total RNA (100 ng) extracted from WM 278 or WM 3248 cells treated for 24 or 48 hours with equal amounts of 5 μmol/L BI-D1870 or DMSO in duplicate were used to prepare cRNA following the 3′ IVT express kit (Affymetrix Inc.) labeling protocol and standardized array processing procedures recommended by Affymetrix Inc., including hybridization, fluidics processing, and scanning of the Affymetrix HG-U133 plus 2.0 arrays (Affymetrix Inc.). The raw data (Affymetrix CEL files) were normalized using the Robust Multichip Average algorithm in the GeneSpring GX software version 11 (Agilent Technologies Inc.).15 Differentially expressed genes between BI-D1870 cells and DMSO-treated cells were determined by Pavlidis Template Matching at P < 0.005, thresholded for fold change of 33%.12 The resulting genes were used for interpretation and validation analyses.
Statistical Analysis
Unless otherwise noted, data are presented as means ± SEM, and the two-tailed Student's t-test was used for comparison, with P < 0.05, P < 0.01, and P < 0.001 considered to be statistically significant.
Protein Pathway Array
A protein pathway array was performed as previously described.20–22 RGP (WM 35, WM 1552c, and WM 1575) and VGP (WM 98.1, WM 983e, and WM 3248) cells were seeded at 1.0 × 106 cells per 10-cm dish and grown to 80% confluence before harvesting. Total proteins were extracted from the cells using 1× cell lysis buffer (Cell Signaling Technology Inc., Danvers, MA) in the presence of 1× protease inhibitor and 1× phosphatase inhibitor cocktails (Roche Applied Science, Indianapolis, IN). Protein lysate (300 μg) was loaded in 1 well across the entire width of a 10% SDS-PAGE polyacrylamide gel, separated by electrophoresis, and blotted with 141 protein-specific or phosphorylation site–specific antibodies as described elsewhere.20–22 All the antibodies were validated independently before inclusion in the pathway array. For protein pathway array data analysis, the signal intensity of each protein band was determined by densitometric scanning (Quantity One software package; Bio-Rad Laboratories). The background was locally subtracted from the raw protein signal, and the background-subtracted intensity was normalized by a global median subtraction normalization method to reduce the variations arising from different runs (such as transferring and blotting efficiency, total protein loading amount, and exposure density). The normalized data were used in subsequent statistical analyses. Differentially expressed proteins were identified by Pavlidis Template Matching at P < 0.05.23
Cell Migration and Invasion Assays
Cells (1 × 106) were seeded in 10-cm dishes and were treated the next day for 24 hours with equal amounts of either DMSO or 5 μmol/L of BI-D1870. Cells (5 × 104 for migration assay or 9 × 104 for invasion assay) in serum-free medium containing equal amounts of DMSO or 5 μmol/L BI-D1870 were seeded in triplicate into the upper chamber of a porous 8-μm polyethylene terephthalate insert (BD Biosciences, Franklin Lakes, NJ) in a 24-well plate containing complete medium (chemoattractant). For invasion assays, the insert was coated with Matrigel (BD Biosciences). After 6 hours for migration assay or 20 hours for invasion assay, cells remaining in the upper chamber were carefully removed using a cotton swab. Cells adhering to the bottom of the filter were fixed with cold 1% glutaraldehyde and stained with 0.1% crystal violet and 20% methanol. Five high-power fields were photographed for each insert using an Axiovert 10 inverted microscope (Carl Zeiss, Oberkochen, Germany) and were counted.
Matrix Metalloproteinase Antibody Array
Metastatic samples from SSM and NM patients (n = 4 for each group) were analyzed for the expression of matrix metalloproteinases (MMPs) and tissue inhibitor of metalloproteinases (TIMPs) using a human MMP antibody array I (RayBiotech Inc., Norcross GA), according to the manufacturer's instructions. Total protein was extracted using lysis buffer and was quantified using the BCA protein assay (Thermo Scientific). Samples were incubated on the array membranes for 2 hours before washing the membranes and incubation with biotin-conjugated primary antibodies. The expression of MMPs and TIMPs was determined using chemiluminescence imaging and was quantified as pixel values. Mean values for duplicate samples were calculated and normalized using the positive controls.
Immunohistochemical Analysis
Immunohistochemical analysis was performed using formalin-fixed, paraffin-embedded metastatic samples from SSM and NM patients (n = 9 and n = 10, respectively). Tissues were evaluated for MMP-8 expression using an MMP-8–specific rabbit polyclonal antibody (catalog #ab78423, diluted 1:500; Abcam Inc., Cambridge, MA). An attending pathologist [Farbod Darvishian (New York, NY)] scored the expression of MMP-8 in each sample on a scale from 0 to 2.
Zebrafish Studies
MiniCoopR vectors were created using Gateway recombination (Invitrogen) as previously described.24 A constitutively active human RSK1 was cloned by adding an N-terminal myristoylation sequence (Myr-RSK1).25 For embryo assays, we performed microinjection of zebrafish embryos from an incross of nacre [nacre has a missense mutation in mitf (C417T/C417T) and results in loss of function], roy, and p53(M214K/M214K). Individual embryos were photographed using a Zeiss V8 stereoscope (Carl Zeiss) under transmitted light. For adult melanoma assays, we performed microinjection of zebrafish embryos from an incross of Tg[mitfa:BRAF(V600E);p53(M214K/M214K);nacre]. We raised melanocyte-rescued animals to adulthood and performed weekly observations for melanoma onset as previously described.26
Results
p-RSK1 Is Differentially Expressed in Human Melanoma Lines and Tissues Derived from Different Histologic Subtypes
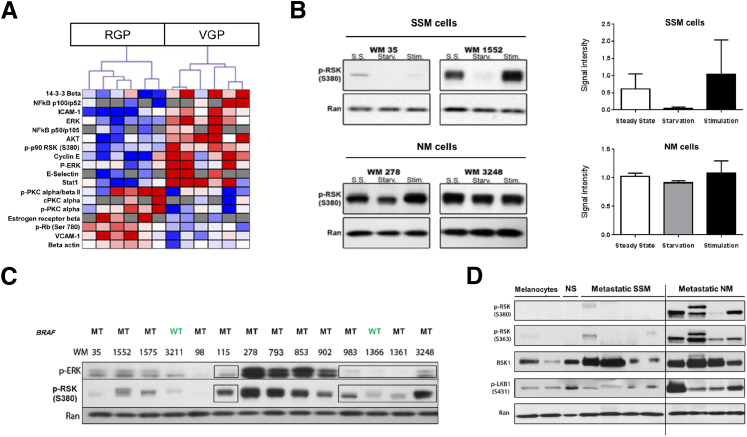
To identify signaling pathways that were differentially expressed or activated between RGP and VGP melanoma, we used a protein pathway array with duplicate protein samples using 141 protein- and phospho-specific antibodies (Supplemental Table S1) with protein extracts from three RGP and three VGP melanoma cell lines (RGP: WM 35, WM 1552c, and WM 1575; VGP: WM 983e, WM 98.1, and WM 3248), and we used the Pavlidis Template Matching algorithm23 (P < 0.05) in TIGR MultiExperiment Viewer (Dana-Farber Cancer Institute, Boston, MA) to identify 18 proteins or phospho-proteins (12.8%) that were differentially expressed between the two groups (Figure 1A and Supplemental Table S2). Of particular interest, the phosphorylated form of p90 ribosomal S6 kinase (RSK) was overexpressed in two independent experiments (P = 0.02 and P = 0.01) in VGP cell lines compared with RGP-derived cells. The vertebrate 90-kDa ribosomal S6 kinase (RSK1) family consists of four isoforms in humans, RSK1 to RSK4, which share 73% to 80% sequence homology at the amino acid level and are located downstream of extracellular signal-regulated (ERK) 1/2 in the mitogen-activated protein kinase (MAPK)/ERK signaling pathway.27 In addition to it belonging to the MAPK/ERK pathway, we chose to focus on RSK1 in this study for several reasons: we found p-RSK1 to be significantly differentially expressed between the two types of cell lines in two independent runs; deregulated RSK1 activity has been previously associated with other types of cancer,28,29 including melanoma.30 The present data extend these observations by suggesting that there may be preferential up-regulation of RSK1 activity in VGP melanoma.
Figure 1.
Differential expression/phosphorylation of tumorigenic pathways in radial (RGP) versus vertical (VGP) growth phase melanoma. A: Heat map generated from a protein pathway array demonstrating differential expression/phosphorylation of tumorigenic proteins in three RGP (WM 35, WM 1552c, and WM 1575) and three VGP (WM 98.1, WM 983e, and WM 3248) cell lines in duplicate (P < 0.05). Blue bars indicate proteins or phospho-proteins that were down-regulated; red bars, proteins or phospho-proteins that were up-regulated. B: Protein S6 kinase, 90 kDa, polypeptide 1 (RSK1) phosphorylation was serum dependent in superficial spreading melanoma (SSM) cells (WM 35 and WM 1552), whereas RSK1 was constitutively phosphorylated in a serum-independent manner in nodular melanoma (NM) cells (WM 3248 and WM 278). C: Phospho-RSK1 (p-RSK1) up-regulation cannot be exclusively explained by BRAF mutation status or extracellular signal-regulated (ERK) activation. Western blot analysis on an expanded panel of 14 primary melanoma cell lines. Boxes highlight cell lines in which phospho-ERK (p-ERK) and p-RSK levels did not directly correlate. D: p-RSK1 protein expression in human metastatic melanoma tissues by Western blot analysis in two melanocyte cell lines (HEM-377 and HEM-475), one human normal skin sample (NS), four human SSM metastatic tissues, and four human NM metastatic tissues. RSK was overexpressed and hyperphosphorylated at Ser-363 and Ser-380 in metastatic tissues, whereas RSK1 phosphorylation was absent in melanocytes and normal skin. Phospho-liver kinase B1 (p-LKB1), a known downstream effector of RSK1, was used as a control for p-RSK1 activation. Data are given as means ± SEM. MT, mutation; S.S., steady state; Starv., serum starved for 24 hours; Stim., serum starved for 24 hours, then stimulated for 10 minutes with serum; WT, wild type.
To establish whether RSK1 activation was correlated with the NM histologic subtype, we first investigated whether there was constitutive activation of RSK1 (p-RSK1) in VGP (NM-derived) melanoma cells. We considered an exclusively RGP cell line to be of SSM origin as, by definition, NM cannot have an RGP component. In contrast, VGP melanoma may be derived from NM or from SSM that progressed from RGP to acquire a VGP. Therefore, we equated VGP cell lines as NM only when there was published evidence stating that the tumor of origin was from the NM histologic subtype. We tested whether mitogenic (growth factor) stimulation was required to induce RSK1 phosphorylation in SSM versus NM cells. We, therefore, compared two primary NM cell lines (WM 278 and WM 3248) with two primary SSM cell lines (WM 35 and WM 1552c).
After 24 hours of serum starvation, we observed that RSK1 phosphorylation at Ser-380 was suppressed in SSM melanoma cell lines (WM 35 and WM 1552c) and then induced in these cells after 10 minutes of serum stimulation (Figure 1B). One SSM cell line (WM 1552) showed a higher level of baseline p-RSK1. However, we observed complete suppression of p-RSK1 on serum starvation of this cell line, suggesting that the SSM subtype may not be reliant on constitutively active RSK1. In contrast, NM cells (WM 278 and WM 3248) maintained RSK1 phosphorylation at Ser-380 after 24 hours of serum starvation and on serum stimulation (Figure 1B), suggesting that RSK1 may be constitutively phosphorylated at Ser-380 in NM irrespective of the presence of mitogenic stimuli.
Because RSK1 is an established downstream effector of the MAPK pathway,31,32 we reasoned that RSK1 hyperactivation might be due to activating BRAF mutations, the most common somatic MAPK pathway alteration in melanoma.33–35 We investigated this possibility by evaluating the relationship between p-RSK1 levels and activation of the MAPK pathway in an expanded panel of 14 melanoma cell lines (12 BRAF mutants and two BRAF wild types). Whereas four cell lines had higher expression of p-ERK than p-RSK1 (WM 278, WM 793, WM 853, and WM 902) by Western blot analysis, five cell lines showed low or no detectable expression of p-ERK (WM 115, WM 983, WM 1366, WM 1361, and WM 3248). For the latter cell lines, we observed high p-RSK1 expression despite the absence of p-ERK, suggesting that in some tumors RSK1 activation might occur independently of upstream MAPK activation or BRAF mutation (Figure 1C and Supplemental Figure S1). Although the four cell lines shown in Figure 1B all harbored a BRAF mutation, we observed a distinct p-RSK1 response to starvation, suggesting the independency of this event from MAPK activation. Together, these data suggest that RSK1 activation in melanoma cells can occur independently of upstream MAPK pathway activation.
To confirm that RSK1 is constitutively activated in human melanoma in a subtype-specific manner, we used Western blot analysis to analyze p-RSK1 expression in tissues from the New York University melanoma clinicopathologic biospecimen database.36 We observed little or no detectable expression of p-RSK1 in melanocytes, normal skin, or metastatic samples from SSM patients, whereas p-RSK1 was strongly expressed in human metastatic tissues originating from NM (Figure 1D). In one metastatic sample originating from primary NM, we observed a second, upper p-RSK1 band that we hypothesize might relate to multiple phosphorylation states of RSK1 in this specimen. Indeed, besides its ERK1/2 binding domain (Ser-363), RSK1 contains several domains involved in kinase activation (Ser-221 and Ser-732) or autophosphorylation (Ser-380) that are responsible for its complete activation. To further confirm the overexpression of p-RSK1 in the metastatic NM subtype, we also assessed the activation status of the protein kinase liver kinase B1 (also known as serine/threonine kinase 11), a known RSK1 substrate,37 in metastatic melanoma tissues and observed elevated phosphorylation of liver kinase B1 at Ser-431 specifically in metastatic tissues that originated from NM (Figure 1D). Taken together, these findings in melanoma cell lines and patient tissues indicate that RSK1 is preferentially activated in human primary and metastatic NM compared with SSM.
RSK1 Inhibition Decreases Cell Proliferation in NM but Not SSM Cell Lines
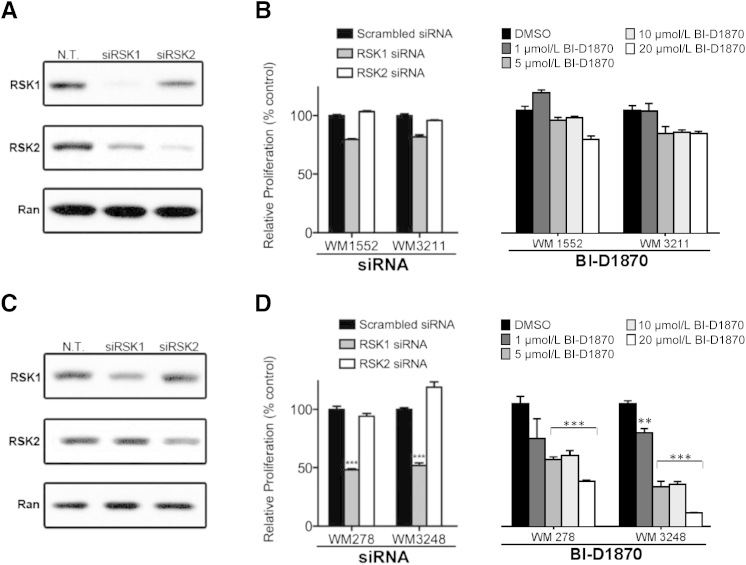
RSK1 and RSK2 have been previously implicated in cancer,28,29,38 including melanoma,30 where they regulate nuclear signaling, cell-cycle progression, and cell proliferation, survival, and migration, as well as nuclear signaling and protein synthesis. Therefore, we sought to determine the relative dependence of SSM and NM melanoma cell lines on RSK1 and RSK2. We used two SSM cell lines and two NM cell lines that were either transfected with RSK1- or RSK2-targeting siRNA or, alternatively, treated with a potent small-molecule RSK inhibitor, BI-D1870, that is specific for all four RSK isoforms and acts by reversibly competing with ATP binding to the N-terminal kinase domain ATP-interacting sequence.39 Notably, when tested against a panel of >50 other kinases, BI-D1870 did not block their activity even at concentrations >100-fold higher than that needed to block RSK phosphorylation.39
RNA interference knockdown of RSK1 in SSM cells (WM 1552 and WM 3211) (Figure 2A) induced a modest decrease in cell proliferation compared with nontargeting siRNA (Figure 2B), whereas transfection of NM cell lines with RSK2-targeting siRNA (Figure 2A) did not produce a change in cell proliferation (Figure 2B). Consistent with these findings, treating SSM cells with BI-D1870 for 72 hours induced a minimal decrease in proliferation (Figure 2B). In contrast, RNA interference–mediated knockdown of RSK1 in NM cell lines (WM 278 and WM 3248) (Figure 2C) resulted in a significant reduction in cell proliferation (51.8% reduction for WM 278 cells and 48.2% reduction for WM 3248 cells; P < 0.001) (Figure 2D). To confirm the effects of RSK1 knockdown, we tested two different siRNA molecules that were designed to specifically target RSK1 (Supplemental Figure S2). We selected the one displaying the most significant level of suppression (siRNA #1). By comparison, RSK2 knockdown (Figure 2C) induced a modest, nonsignificant reduction in WM 278 proliferation and a 19.0% increase (P < 0.05) in WM 3248 proliferation (Figure 2D). Treating NM cells with the small-molecule RSK inhibitor BI-D1870 for 72 hours induced a marked dose-dependent reduction in cell proliferation, with half maximal inhibitory concentrations of 9.5 and 3.1 μmol/L in WM 278 and WM 3248 cells, respectively (Figure 2D). To confirm this result, we treated an additional NM melanoma cell line with BI-D1870 and observed a similar dose-dependent reduction in cell proliferation (WM 1366; half maximal inhibitory concentration, 2.6 μmol/L) (Supplemental Figure S3). The RNA interference data are consistent with the BI-D1870 inhibitor data and demonstrate that pharmacological or genetic inhibition of RSK1 but not RSK2 can inhibit the growth of NM cells, supporting the postulate that NM cells depend on activated RSK1 to sustain their proliferation.
Figure 2.
Genetic or pharmacological inhibition of protein S6 kinase, 90 kDa, polypeptide 1 (RSK1) inhibits nodular melanoma (NM) but not superficial spreading melanoma (SSM) cell proliferation. A: Western blot analysis showing RSK1 levels after transfection with nontargeting (N.T.), RSK1-targeting, or RSK2-targeting siRNA in WM 1552 melanoma cells (SSM). B: Crystal violet proliferation assay of SSM cells 48 hours after transient transfection with RSK1 or RSK2 siRNA or 72 hours after treatment with escalating doses of the RSK inhibitor BI-D1870. Data were normalized by setting N.T. siRNA-transfected or dimethyl sulfoxide (DMSO)–treated cells to 100%. C: Same as in A but using WM 3248 melanoma cells (NM). D: Same as in B but using NM cell lines. Data are given as means ± SEM. ∗∗P < 0.01, ∗∗∗P < 0.001 compared with N.T. siRNA-transfected cells.
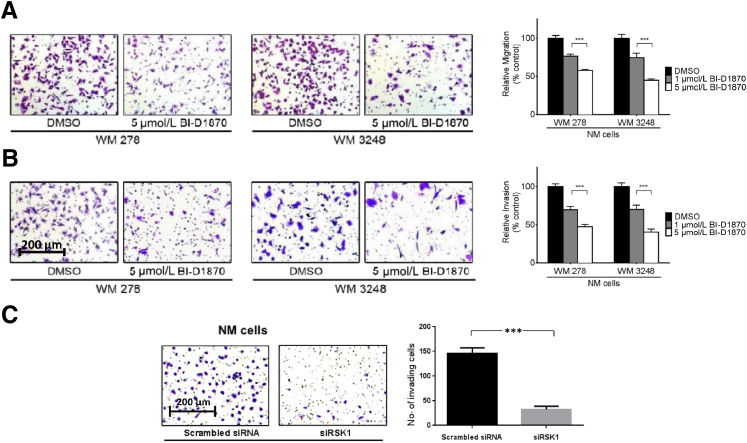
RSK1 Activation Promotes Migration and Invasion of NM-Derived Cells
Elevated cell migration and invasion are hallmarks of melanoma, and these processes support and promote metastatic progression of the disease and, ultimately, the death of patients. Several recent reports support a promigratory and proinvasive role for RSK family proteins in cancer.40,41 Therefore, we sought to determine the contribution of constitutively active RSK1 to cell migration and invasion in the NM histologic subtype. We treated NM cells with BI-D1870 and performed a Transwell cell migration assay. After 6 hours, the migration of BI-D1870–treated WM 278 cells was reduced by 42.7% compared with that of DMSO-treated cells (P < 0.001) (Figure 3A). Similarly, RSK inhibition with BI-D1870 decreased the number of WM 3248 cells that traversed the migration chamber by 55.6% compared with DMSO-treated cells (P < 0.001) (Figure 3A).
Figure 3.
p90 ribosomal S6 kinase (RSK) inhibition decreases nodular melanoma (NM) cell migration and invasion. A: WM 278 and WM 3248 cells treated with equal amounts of dimethyl sulfoxide (DMSO) or 5 μmol/L BI-D1870 migrated through 8-μm migration chambers for 6 hours and then were fixed and stained with crystal violet. Five high-power fields from each chamber were photographed, and cells were then counted. Images shown are representative of one high-power field from each condition. B: Same as A but chamber coated with Matrigel and cells allowed to invade for 20 hours. For A and B, data were normalized by setting DMSO-treated cells to 100%. C: Same as B but using siRNA against RSK1. Data are given as means ± SEM. ∗∗∗P < 0.001 compared with DMSO-treated cells (A and B) or with scrambled siRNA–treated cells (C).
In parallel experiments, we assessed whether RSK activation altered the invasive potential of NM by pretreating WM 278 and WM 3248 VGP cells with 5 μmol/L BI-D1870 for 20 hours and measuring their invasion through an extracellular matrix (Matrigel). WM 278 and WM 3248 cells that had been pretreated with BI-D1870 demonstrated a 55.8% and 57.3% reduction in cell invasion, respectively, compared with vehicle-treated controls (P < 0.001) (Figure 3B). Crucially, we confirmed that the RSK1 isoform was critical to melanoma cell invasion by performing an invasion assay with WM 3248 cells in which RSK1 expression was depleted after transfection with RSK1-specific siRNA and observed a 77.8% reduction in cell invasion after RSK1 knockdown compared with nontargeting siRNA-transfected cells (P < 0.0001) (Figure 3C). Taken together, these data indicate that RSK1 may contribute to the invasive NM phenotype by promoting cell migration and invasion.
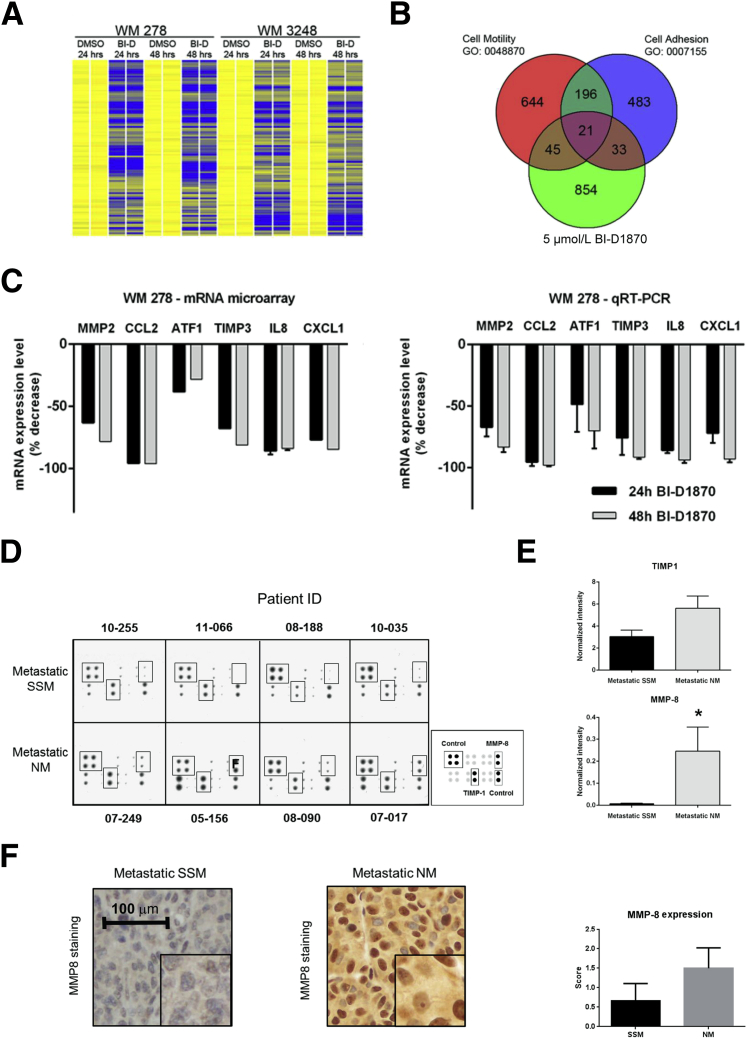
RSK1 Activation Orchestrates a Program of Gene Expression that Promotes Cell Motility and Adhesion and Differential Expression of MMP-8 and TIMP-1 in NM-Derived Cell Lines and Tissues
To elucidate the mechanisms by which RSK inhibition modulates melanoma cell migration and invasion, we performed unbiased mRNA array profiling using total mRNA isolated from WM 278 and WM 3248 VGP cells that were treated in duplicate with vehicle (DMSO) or 5 μmol/L BI-D1870 for 24 or 48 hours. To confirm that BI-D1870 leads to decreased p-RSK1 protein levels, we assessed the expression of p-RSK1 (S380) in an NM-derived cell line (WM 278); we observed that p-RSK1 expression was decreased by a factor of 2.4 (data not shown).
We reasoned that RSK inhibition would repress expression of genes implicated in invasion and migration; thus, we focused on analysis of those genes that were significantly down-regulated (P < 0.005) in both cell lines after treatment with BI-D1870 at 24 and 48 hours compared with the equivalent DMSO-treated cells. This identified 1398 mRNA probes (Figure 4A and Supplemental Table S3), for which the corresponding genes were annotated using the National Institute of Allergy and Infectious Diseases/NIH Database for Annotation, Visualization, and Integrated Discovery (DAVID) Bioinformatics Resource 6.7. Gene Ontology analysis of this data set revealed significant enrichment for Gene Ontology biological process terms related to cell motility (enrichment score, 3.22; P < 0.001) and cell adhesion (enrichment score, 2.37; P = 0.0025). We confirmed these findings using the AmiGO database (http://amigo1.geneontology.org/cgi-bin/amigo/go.cgi, last accessed October 23, 2014) focusing on 974 unique genes that correspond to the 1398 down-regulated mRNA probes; 66 genes belong to the functional class cell motility (GO: 0048870), and 54 genes belong to the functional class cell adhesion (GO: 2000145) (Figure 4B). Twenty-one genes overlapped and belonged to both functional sets and the BI-D1870 down-regulated gene set. In the BI-D1870 down-regulated gene set, we noted enrichment for several MMPs, which are known to be involved in the digestion and turnover of the extracellular matrix42 and have previously been implicated in melanoma invasion, including MMP-2.43 Furthermore, there was a significant concomitant reduction (P < 0.005) in the expression of genes that are known to induce MMP-2 expression/activation, including ATF1, CCL2, CXCL1, TIMP3, and CXCL8 (IL-8), a finding that we confirmed in BI-D1870–treated WM 278 cells using quantitative RT-PCR (Figure 4C).
Figure 4.
p90 ribosomal S6 kinase (RSK) inhibition suppresses a promotile transcriptional program. A: Heat map demonstrating down-regulation of 200 mRNAs involved in cell motility and/or cell adhesion after RSK inhibition. Yellow bars indicate the basal mRNA expression level. Blue bars indicate mRNA down-regulated ≥1.33 fold (P < 0.005) compared with dimethyl sulfoxide (DMSO)–treated cells. B: Venn diagram classifying 200 mRNAs that were down-regulated after RSK inhibition and functionally annotated with biological process terms related to cell motility and/or cell adhesion. C: Results from mRNA array demonstrating down-regulated expression of matrix metalloproteinase 2 (MMP-2) and five other related transcripts after 24 or 48 hours of treatment with BI-D1870. Data were normalized by setting DMSO-treated cells to 100%, and microarray was confirmed using quantitative RT-PCR (RT-qPCR). D: Human MMP antibody array comparing four superficial spreading melanoma (SSM) and four nodular melanoma (NM) metastatic samples. Boxes indicated correspond to tissue inhibitor of metalloproteinases 1 (TIMP-1) and MMP-8 as indicated in the key. E: Quantitative spot density analysis of the array shown in D. The intensity of each spot was normalized to the positive control, and duplicate values were evaluated as a mean. F: Validation of the array using immunohistochemical analysis. NM metastatic tissue showed higher expression of MMP-8 relative to SSM metastatic tissue. Samples were scored based on intensity (0 to 2) (mean of nine metastatic SSM samples and 10 metastatic NM samples). Data are given as means ± SEM. ∗P < 0.05. Original magnification: ×20 (F); ×40 (insets, F).
To investigate the relevance of the decreased expression of metalloproteinases after RSK inhibition, we used a commercially available antibody array to compare the expression of various MMPs and TIMPs between metastatic melanoma samples from patients with primary SSM and primary NM (n = 4 for each subtype) (Figure 4D). Quantitative analysis of the array spot densities when comparing metastatic samples from SSM patients with metastatic samples from NM patients revealed significant increases in the protein levels of MMP-8 and TIMP-1 (P = 0.03 and P = 0.05, respectively) in tissues that originated from the NM subtype (Figure 4E). Because MMP-8 was the most strikingly overexpressed, we performed immunohistochemical analysis using representative formalin-fixed, paraffin-embedded metastatic samples from SSM and NM patients (n = 9 and n = 10, respectively) with an MMP-8–specific antibody. These experiments revealed considerably higher cytoplasmic staining (2.3-fold increase) of MMP-8 in NM metastatic tissues compared with the SSM tissues (Figure 4F). Consistent with this finding, we observed a significant decrease in MMP-8 expression in the two NM cell lines on RSK1 inhibition in the mRNA array analysis (P < 0.005; fold reduction of 4.85 and 4.95 in WM 278 and WM 3248, respectively) (Supplemental Table S3).
In addition, we performed an invasion assay on two NM cell lines (WM 278 and WM 3248) that had been transfected with siRNA against MMP8 or TIMP1 or cotransfected with both. We observed a slight decrease (14%) in the invasion of NM cells on MMP-8 inhibition only compared with cells transfected with scrambled siRNA (data not shown). However, when cells were cotransfected with si-MMP-8 and si-TIMP-1, we observed a significant decrease in invasion of both NM cell lines (WM 278 and WM 3248; a mean 2.89-fold decrease; P = 0.0005; data not shown). These results suggest that the invasive phenotype of the NM subtype might not rely on a single MMP but on a concomitant and cooperative effect of several MMPs/TIMPs. Together, these data suggest that RSK1 drives a transcriptional program that promotes melanoma cell migration, invasion, and adhesion and that RSK1 might promote MMP-mediated invasion in NM, thus contributing to the distinctive biology of metastasis in this histologic subset of melanoma.
Constitutive RSK1 Activation Is Associated with Increased Melanoma Progression and Invasion in Zebrafish
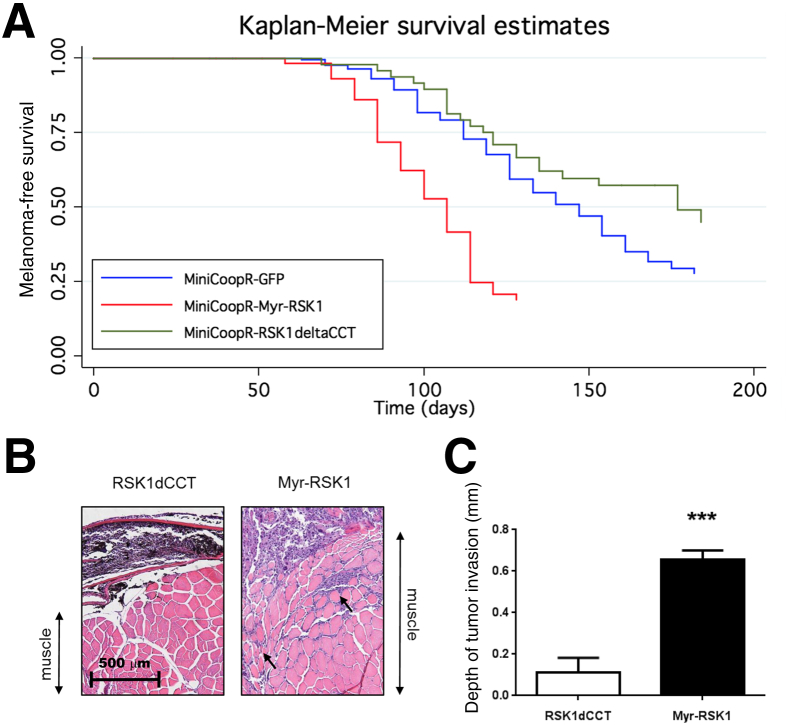
To confirm the role of constitutive RSK1 activation in the development and progression of melanoma in vivo, we used a high-throughput zebrafish model Tg[mitfa:BRAF(V600E), p53M214K/M214K]24 in which we used miniCoopR, a tol2-based transposon vector, to express a constitutively active,25 myristoylated human RSK1 specifically in melanocytes. In this model system, cooperating genes can be tested for the ability to accelerate or slow melanoma initiation. There is no mouse model of NM or SSM that we could use as an alternative approach. As experimental controls, we also expressed green fluorescent protein or an inactive RSK1 mutant (RSK1dCCT, generated by deletion of the last 11 amino acids, which encode an ERK docking site and, thus, prevent binding of ERK). We found that melanocyte-specific expression of constitutively active RSK1 (Myr-RSK1) significantly accelerated melanoma onset compared with green fluorescent protein (P = 2.92 × 10−10) (Figure 5A) or RSK1 mutant-expressing animals (P = 1.81 × 10−7) (Figure 5A). These data indicate that constitutively active RSK1 can cooperate with oncogenic BRAF to accelerate tumor onset in vivo.
Figure 5.
Constitutive activation of protein S6 kinase, 90 kDa, polypeptide 1 (RSK1) accelerates melanoma tumor onset and invasion in zebrafish. A: Melanoma-free survival curve of miniCoopR-green fluorescent protein (GFP)–, miniCoopR-Myr-RSK1–, and miniCoopR-RSK-deltaCCT–injected zebrafish. B: Histologic sections of zebrafish RSK1dCCT- and Myr-RSK1–expressing melanomas were stained with hematoxylin and eosin to assess tumor invasion depth. Arrows indicate muscular invasion by melanoma cells. C: Depth of invasion was measured for each tumor and was quantified by averaging the depth of invasion of four tumors for each group (RSK1dCCT and Myr-RSK1). Data are given as means ± SEM. ∗∗∗P < 0.001 compared with dominant-negative RSK1-expressing tumors.
To extend this finding, we analyzed tumor invasion in vivo, comparing the depth of invasion between green fluorescent protein and Myr-RSK1 tumors. The depth of invasion was measured on four tumors from each group (Myr-RSK1– and RSK1dCCT-expressing tumors), and we found that tumors expressing constitutively active RSK1 invaded significantly deeper into muscle compared with tumors expressing the dominant-negative form of RSK1 (Figure 5B) (an average of 0.11 mm for the RSK1dCCT-expressing tumors versus 0.66 mm for Myr-RSK1–expressing tumors; P = 0.0002) (Figure 5C). This finding suggests that activated RSK1 may contribute to the invasive phenotype observed in NM tumors.
Discussion
Herein, we investigated the hypothesis that subtype-specific differences between NM and SSM histologic subtypes persist throughout the progression of primary melanoma to metastatic disease and may account for the distinct biological behavior of each subtype. We identified preferential activation of RSK1 in VGP cell lines and demonstrated constitutive phosphorylation of RSK1 in VGP cell lines and metastatic NM patient tissues, suggesting that RSK1 activation is maintained throughout the progression of NM from primary to metastatic disease. In addition, we provide in vitro and in vivo evidence that supports a role for RSK1 activation in promoting NM growth and invasion, suggesting that it plays a driving role in the aggressive phenotype of this subtype.
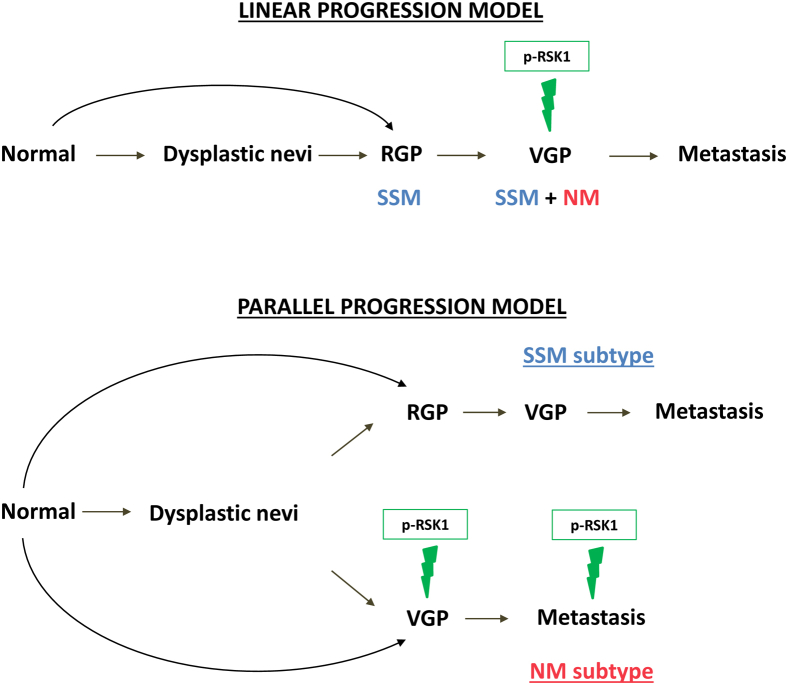
A growing body of clinical, pathologic, and molecular data supports the concept that primary NM and SSM evolve as distinct biological entities.44 For example, we and others have identified evidence supporting distinct pathways of development from transformed melanocytes to invasive SSM and NM, such as the presence of recurrent SSM-specific deletions that are not present in nevi or NM,17 distinct subtype-specific miRNA and mRNA expression patterns between SSM and NM that were independent of thickness,15,16,18 and, more recently, recurrent subtype-specific activating mutations in the GTPase RAC1.45 These data argue against a model in which SSM and NM develop along a linear pathway of progression that begins with the transformation of epidermal melanocytes and for which the difference between subtypes is limited to the speed with which the transformed melanocytes invade the dermis through a VGP (Figure 6).
Figure 6.
Proposed role of protein S6 kinase, 90 kDa, polypeptide 1 (RSK1) activation in a parallel progression model of melanoma. Schematic depicting the linear progression model of melanoma and the parallel progression model that we propose. Linear progression model: Superficial spreading melanoma (SSM) and nodular melanoma (NM) grow according to a linear model of progression, as malignant or normal melanocytes spread radially and then invade vertically. Parallel progression model: SSM and NM develop along divergent pathways from the transformed melanocyte to the primary melanoma, and these differences are maintained as the tumors invade and metastasize. p-RSK1, phospho-RSK1; RGP, radial growth phase; VGP, vertical growth phase.
We identify RSK1 activation as a feature of metastatic melanoma derived from NM, where it is preferentially maintained throughout the progression to metastasis. This distinction between metastatic NM and metastatic SSM is consistent with the study by Haqq et al,46 who compared gene expression signatures at different stages of malignant melanoma progression and identified two dichotomous gene expression patterns in metastases, reflecting those seen in VGP or RGP cells of primary melanoma. However, the present analysis is unique given the comparison of the expression and activation of signal transduction proteins between melanoma subtypes rather than contrasting mRNA expression profiles or copy number alterations. Together, these findings suggest that the differences that arise between primary melanoma subtypes persist throughout the invasive and metastatic processes. In addition to explaining, in part, the inherent biological differences between melanoma histologic subtypes, a revised model of melanoma progression may have implications for the subtype-specific treatment of metastatic melanoma.
We prioritized studying activated RSK1 as a specific feature of NM of the 18 significantly different proteins identified on the protein array for several reasons. First, we reasoned that activated RSK1 might play a key role in melanoma progression because it is situated in two signaling pathways that are cumulatively dysregulated in >90% of melanomas: the RAS-RAF-MEK-ERK and the PI3K/Akt pathways.47–49 Second, RSK signaling has been shown to facilitate signaling from activated BRAF to the mammalian target of rapamycin in melanoma and to promote tumor growth.30 We did not observe a correlation between activation of BRAF and RSK1 in NM cells, suggesting that RSK1 hyperactivation in this subtype can also result from BRAF-independent stimuli. In this regard, we did not find evidence to suggest that there are activating mutations in RSK1 in melanoma when we analyzed data from The Cancer Genome Atlas and our own RSK1 melanoma sequencing (data not shown).
The present data support a role for RSK1 hyperactivation in melanoma progression and a role for RSK1 in promoting the migration and invasion of NM cells. First, treatment of melanoma cells with BI-D1870, a small-molecule pan-RSK inhibitor,50 caused a significant reduction in melanoma cell migration and invasion in vitro. We used BI-D1870 in cell culture experiments at a final concentration of 5 μmol/L, which is half the concentration used in other published studies.51 Second, the zebrafish melanoma model indicated that RSK1 activation significantly accelerated melanoma development and invasion in vivo. To our knowledge, there is no genetic mouse model of SSM and NM. Third, BI-D1870 treatment of melanoma cell lines was associated with altered expression of genes associated with cell motility and cell adhesion and, in particular, with reduced levels of several MMPs, which, together with their tissue inhibitors (TIMPs), have been implicated in tumor progression and metastasis.52,53
Comparing MMP and TIMP expression in metastatic tissues from SSM and NM patients, we observed overexpression of MMP-8 and TIMP-1 exclusively in tumors derived from the NM subtype, suggesting that these proteins may have a role in the pathogenesis of NM metastasis.
In the mRNA microarray we performed on NM cells on RSK1 inhibition, we observed a significant concordant decrease in MMP-8 expression (4.85- and 4.95-fold reductions in WM 278 and WM 3248 cells, respectively; P < 0.005), and cotransfection of siRNA against MMP-8 and TIMP-1 produced a significant reduction in the invasive phenotype of NM cells. RSK1 has been shown to be overexpressed in breast and prostate cancers28,29 and to stimulate cell motility and invasion by activating a transcriptional program that modulates the extracellular matrix, the intracellular motility apparatus, and receptors that mediate communication between these two compartments.54 Furthermore, a genome-wide RNA interference screen demonstrated that multiple tumorigenic signaling pathways converge on RSK to stimulate cell migration and invasion and accelerate metastasis.41 Last, in vitro studies suggest that novel inhibitors of RSK function may hold promise for the treatment of a subset of cancers with RSK dependence.55 Further work is required to determine whether RSK inhibition might be an effective therapeutic strategy for the treatment of metastatic NM. A recent report showed that the RSK isoforms RSK3 and RSK4 mediate resistance to PI3K inhibitors in breast cancer,56 an interesting finding given that combined MEK/ERK and PI3K pathway blockade represents a promising approach to inhibit the growth of metastatic melanoma with activating BRAF or NRAS mutations.57,58
In summary, the present data indicate that RSK1 activation is a feature of the NM subtype and that it contributes to the invasion and progression of this disease. We demonstrate persistent RSK1 activation in the metastases of NM patients, suggesting that different subtypes of primary melanoma might use distinct biological pathways to invade and metastasize, a finding with implications for the development of subtype-specific therapies for the treatment of metastatic melanoma. This finding emphasizes the inherent biological variability of primary and metastatic melanoma histologic subtypes.
Acknowledgment
We thank Dr. Farbod Darvishian (New York University School of Medicine) for technical assistance in performing the scoring of the formalin-fixed, paraffin-embedded staining.
Footnotes
Supported by the NIH Cancer Center Support Grant to the New York University Perlmutter Cancer Center (P30 CA016087), The Chemotherapy Foundation grant 15-C0300-82115 (I.O.), the American Skin Association grant 12-00009 (J.A.F.), The Skin Cancer Foundation (J.A.F.), and the Marc Jacobs Campaign for Melanoma Research (I.O. and E.H.-M.).
A.S. and J.A.F. contributed equally to this work.
Disclosures: None declared.
Supplemental Data
Relative intensity of phospho–extracellular signal-regulated (p-ERK) and phospho–protein S6 kinase, 90 kDa, polypeptide 1 (p-RSK1) expression in vertical growth phase (VGP) cell lines. Relationship between p-RSK1 levels and activation of the mitogen-activated protein kinase pathway in an expanded panel of nine VGP melanoma cell lines. Whereas four cell lines have higher expression of p-ERK than p-RSK1 (WM 278, WM 793, WM 853, and WM 902) by Western blot analysis, five cell lines show low or no detectable expression of p-ERK (WM 115, WM 983, WM 1366, WM 1361, and WM 3248).
Test of two siRNA candidates targeting protein S6 kinase, 90 kDa, polypeptide 1 (RSK1). Western blot analysis of protein extracts from siRNA-treated cells using two different siRNA candidates. p-RSK1, phospho-RSK1. Ran was used as a loading control.
Nodular melanoma cell proliferation is more sensitive to protein S6 kinase, 90 kDa, polypeptide 1 knockdown/inhibition than is superficial spreading melanoma cell proliferation. Treating an additional nodular melanoma cell line, WM 1366, with BI-D1870 induces a dose-dependent reduction in cell proliferation. DMSO, dimethyl sulfoxide.
References
- 1.Clark W.H., Jr., From L., Bernardino E.A., Mihm M.C. The histogenesis and biologic behavior of primary human malignant melanomas of the skin. Cancer Res. 1969;29:705–727. [PubMed] [Google Scholar]
- 2.McGovern V.J., Mihm M.C., Jr., Bailly C., Booth J.C., Clark W.H., Jr., Cochran A.J., Hardy E.G., Hicks J.D., Levene A., Lewis M.G., Little J.H., Milton G.W. The classification of malignant melanoma and its histologic reporting. Cancer. 1973;32:1446–1457. doi: 10.1002/1097-0142(197312)32:6<1446::aid-cncr2820320623>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 3.Liu W., Dowling J.P., Murray W.K., McArthur G.A., Thompson J.F., Wolfe R., Kelly J.W. Rate of growth in melanomas: characteristics and associations of rapidly growing melanomas. Arch Dermatol. 2006;142:1551–1558. doi: 10.1001/archderm.142.12.1551. [DOI] [PubMed] [Google Scholar]
- 4.Balch C.M., Gershenwald J.E., Soong S.J., Thompson J.F., Atkins M.B., Byrd D.R., Buzaid A.C., Cochran A.J., Coit D.G., Ding S., Eggermont A.M., Flaherty K.T., Gimotty P.A., Kirkwood J.M., McMasters K.M., Mihm M.C., Jr., Morton D.L., Ross M.I., Sober A.J., Sondak V.K. Final version of 2009 AJCC melanoma staging and classification. J Clin Oncol. 2009;27:6199–6206. doi: 10.1200/JCO.2009.23.4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kwong L., Chin L., Wagner S.N. Growth factors and oncogenes as targets in melanoma: lost in translation? Adv Dermatol. 2007;23:99–129. doi: 10.1016/j.yadr.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ackerman A.B. Malignant melanoma. a unifying concept. Am J Dermatopathol. 1980;2:309–313. doi: 10.1097/00000372-198000240-00004. [DOI] [PubMed] [Google Scholar]
- 7.Barnhill R.L., Mihm M.C., Jr. The histopathology of cutaneous malignant melanoma. Semin Diagn Pathol. 1993;10:47–75. [PubMed] [Google Scholar]
- 8.Kim K.B., Eton O., Davis D.W., Frazier M.L., McConkey D.J., Diwan A.H., Papadopoulos N.E., Bedikian A.Y., Camacho L.H., Ross M.I., Cormier J.N., Gershenwald J.E., Lee J.E., Mansfield P.F., Billings L.A., Ng C.S., Charnsangavej C., Bar-Eli M., Johnson M.M., Murgo A.J., Prieto V.G. Phase II trial of imatinib mesylate in patients with metastatic melanoma. Br J Cancer. 2008;99:734–740. doi: 10.1038/sj.bjc.6604482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Motoi N., Szoke J., Riely G.J., Seshan V.E., Kris M.G., Rusch V.W., Gerald W.L., Travis W.D. Lung adenocarcinoma: modification of the 2004 WHO mixed subtype to include the major histologic subtype suggests correlations between papillary and micropapillary adenocarcinoma subtypes, EGFR mutations and gene expression analysis. Am J Surg Pathol. 2008;32:810–827. doi: 10.1097/PAS.0b013e31815cb162. [DOI] [PubMed] [Google Scholar]
- 10.Weigelt B., Reis-Filho J.S. Histological and molecular types of breast cancer: is there a unifying taxonomy? Nat Rev Clin Oncol. 2009;6:718–730. doi: 10.1038/nrclinonc.2009.166. [DOI] [PubMed] [Google Scholar]
- 11.Wyman K., Atkins M.B., Prieto V., Eton O., McDermott D.F., Hubbard F., Byrnes C., Sanders K., Sosman J.A. Multicenter Phase II trial of high-dose imatinib mesylate in metastatic melanoma: significant toxicity with no clinical efficacy. Cancer. 2006;106:2005–2011. doi: 10.1002/cncr.21834. [DOI] [PubMed] [Google Scholar]
- 12.Hodi F.S., Corless C.L., Giobbie-Hurder A., Fletcher J.A., Zhu M., Marino-Enriquez A., Friedlander P., Gonzalez R., Weber J.S., Gajewski T.F., O'Day S.J., Kim K.B., Lawrence D., Flaherty K.T., Luke J.J., Collichio F.A., Ernstoff M.S., Heinrich M.C., Beadling C., Zukotynski K.A., Yap J.T., Van den Abbeele A.D., Demetri G.D., Fisher D.E. Imatinib for melanomas harboring mutationally activated or amplified KIT arising on mucosal, acral, and chronically sun-damaged skin. J Clin Oncol. 2013;31:3182–3190. doi: 10.1200/JCO.2012.47.7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crocetti E., Caldarella A., Chiarugi A., Nardini P., Zappa M. The thickness of melanomas has decreased in central Italy, but only for thin melanomas, while thick melanomas are as thick as in the past. Melanoma Res. 2010;20:422–426. doi: 10.1097/CMR.0b013e32833d9d36. [DOI] [PubMed] [Google Scholar]
- 14.Geller A.C., Elwood M., Swetter S.M., Brooks D.R., Aitken J., Youl P.H., Demierre M.F., Baade P.D. Factors related to the presentation of thin and thick nodular melanoma from a population-based cancer registry in Queensland Australia. Cancer. 2009;115:1318–1327. doi: 10.1002/cncr.24162. [DOI] [PubMed] [Google Scholar]
- 15.Jaeger J., Koczan D., Thiesen H.J., Ibrahim S.M., Gross G., Spang R., Kunz M. Gene expression signatures for tumor progression, tumor subtype, and tumor thickness in laser-microdissected melanoma tissues. Clin Cancer Res. 2007;13:806–815. doi: 10.1158/1078-0432.CCR-06-1820. [DOI] [PubMed] [Google Scholar]
- 16.Poliseno L., Haimovic A., Segura M.F., Hanniford D., Christos P.J., Darvishian F., Wang J., Shapiro R.L., Pavlick A.C., Berman R.S., Hernando E., Zavadil J., Osman I. Histology-specific microRNA alterations in melanoma. J Invest Dermatol. 2012;132:1860–1868. doi: 10.1038/jid.2011.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rose A.E., Poliseno L., Wang J., Clark M., Pearlman A., Wang G., Vega YSdM E.C., Medicherla R., Christos P.J., Shapiro R., Pavlick A., Darvishian F., Zavadil J., Polsky D., Hernando E., Ostrer H., Osman I. Integrative genomics identifies molecular alterations that challenge the linear model of melanoma progression. Cancer Res. 2011;71:2561–2571. doi: 10.1158/0008-5472.CAN-10-2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scatolini M., Grand M.M., Grosso E., Venesio T., Pisacane A., Balsamo A., Sirovich R., Risio M., Chiorino G. Altered molecular pathways in melanocytic lesions. Int J Cancer. 2010;126:1869–1881. doi: 10.1002/ijc.24899. [DOI] [PubMed] [Google Scholar]
- 19.Warycha M.A., Christos P.J., Mazumdar M., Darvishian F., Shapiro R.L., Berman R.S., Pavlick A.C., Kopf A.W., Polsky D., Osman I. Changes in the presentation of nodular and superficial spreading melanomas over 35 years. Cancer. 2008;113:3341–3348. doi: 10.1002/cncr.23955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Che Y., Ye F., Xu R., Qing H., Wang X., Yin F., Cui M., Burstein D., Jiang B., Zhang D.Y. Co-expression of XIAP and cyclin D1 complex correlates with a poor prognosis in patients with hepatocellular carcinoma. Am J Pathol. 2012;180:1798–1807. doi: 10.1016/j.ajpath.2012.01.016. [DOI] [PubMed] [Google Scholar]
- 21.Wang H., Gillis A., Zhao C., Lee E., Wu J., Zhang F., Ye F., Zhang D.Y. Crocidolite asbestos-induced signal pathway dysregulation in mesothelial cells. Mutat Res. 2011;723:171–176. doi: 10.1016/j.mrgentox.2011.04.008. [DOI] [PubMed] [Google Scholar]
- 22.Ye F., Che Y., McMillen E., Gorski J., Brodman D., Saw D., Jiang B., Zhang D.Y. The effect of Scutellaria baicalensis on the signaling network in hepatocellular carcinoma cells. Nutr Cancer. 2009;61:530–537. doi: 10.1080/01635580902803719. [DOI] [PubMed] [Google Scholar]
- 23.Pavlidis P., Noble W.S. Analysis of strain and regional variation in gene expression in mouse brain. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-10-research0042. RESEARCH0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ceol C.J., Houvras Y., Jane-Valbuena J., Bilodeau S., Orlando D.A., Battisti V., Fritsch L., Lin W.M., Hollmann T.J., Ferre F., Bourque C., Burke C.J., Turner L., Uong A., Johnson L.A., Beroukhim R., Mermel C.H., Loda M., Ait-Si-Ali S., Garraway L.A., Young R.A., Zon L.I. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature. 2011;471:513–517. doi: 10.1038/nature09806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shimamura A., Ballif B.A., Richards S.A., Blenis J. Rsk1 mediates a MEK-MAP kinase cell survival signal. Curr Biol. 2000;10:127–135. doi: 10.1016/s0960-9822(00)00310-9. [DOI] [PubMed] [Google Scholar]
- 26.Iyengar S., Houvras Y., Ceol C.J. Screening for melanoma modifiers using a zebrafish autochthonous tumor model. J Vis Exp. 2012:e50086. doi: 10.3791/50086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lara R., Seckl M.J., Pardo O.E. The p90 RSK family members: common functions and isoform specificity. Cancer Res. 2013;73:5301–5308. doi: 10.1158/0008-5472.CAN-12-4448. [DOI] [PubMed] [Google Scholar]
- 28.Clark D.E., Errington T.M., Smith J.A., Frierson H.F., Jr., Weber M.J., Lannigan D.A. The serine/threonine protein kinase, p90 ribosomal S6 kinase, is an important regulator of prostate cancer cell proliferation. Cancer Res. 2005;65:3108–3116. doi: 10.1158/0008-5472.CAN-04-3151. [DOI] [PubMed] [Google Scholar]
- 29.Smith J.A., Poteet-Smith C.E., Xu Y., Errington T.M., Hecht S.M., Lannigan D.A. Identification of the first specific inhibitor of p90 ribosomal S6 kinase (RSK) reveals an unexpected role for RSK in cancer cell proliferation. Cancer Res. 2005;65:1027–1034. [PubMed] [Google Scholar]
- 30.Romeo Y., Moreau J., Zindy P.J., Saba-El-Leil M., Lavoie G., Dandachi F., Baptissart M., Borden K.L., Meloche S., Roux P.P. RSK regulates activated BRAF signalling to mTORC1 and promotes melanoma growth. Oncogene. 2013;32:2917–2926. doi: 10.1038/onc.2012.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anjum R., Blenis J. The RSK family of kinases: emerging roles in cellular signalling. Nat Rev Mol Cell Biol. 2008;9:747–758. doi: 10.1038/nrm2509. [DOI] [PubMed] [Google Scholar]
- 32.Roberts P.J., Der C.J. Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene. 2007;26:3291–3310. doi: 10.1038/sj.onc.1210422. [DOI] [PubMed] [Google Scholar]
- 33.Davies H., Bignell G.R., Cox C., Stephens P., Edkins S., Clegg S. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 34.Edlundh-Rose E., Egyhazi S., Omholt K., Mansson-Brahme E., Platz A., Hansson J., Lundeberg J. NRAS and BRAF mutations in melanoma tumours in relation to clinical characteristics: a study based on mutation screening by pyrosequencing. Melanoma Res. 2006;16:471–478. doi: 10.1097/01.cmr.0000232300.22032.86. [DOI] [PubMed] [Google Scholar]
- 35.Goel V.K., Lazar A.J., Warneke C.L., Redston M.S., Haluska F.G. Examination of mutations in BRAF, NRAS, and PTEN in primary cutaneous melanoma. J Invest Dermatol. 2006;126:154–160. doi: 10.1038/sj.jid.5700026. [DOI] [PubMed] [Google Scholar]
- 36.Wich L.G., Hamilton H.K., Shapiro R.L., Pavlick A., Berman R.S., Polsky D., Goldberg J.D., Hernando E., Manga P., Krogsgaard M., Kamino H., Darvishian F., Lee P., Orlow S.J., Ostrer H., Bhardwaj N., Osman I. Developing a multidisciplinary prospective melanoma biospecimen repository to advance translational research. Am J Transl Res. 2009;1:35–43. [PMC free article] [PubMed] [Google Scholar]
- 37.Sapkota G.P., Kieloch A., Lizcano J.M., Lain S., Arthur J.S., Williams M.R., Morrice N., Deak M., Alessi D.R. Phosphorylation of the protein kinase mutated in Peutz-Jeghers cancer syndrome, LKB1/STK11, at Ser431 by p90(RSK) and cAMP-dependent protein kinase, but not its farnesylation at Cys(433), is essential for LKB1 to suppress cell growth. J Biol Chem. 2001;276:19469–19482. doi: 10.1074/jbc.M009953200. [DOI] [PubMed] [Google Scholar]
- 38.Bignone P.A., Lee K.Y., Liu Y., Emilion G., Finch J., Soosay A.E., Charnock F.M., Beck S., Dunham I., Mungall A.J., Ganesan T.S. RPS6KA2, a putative tumour suppressor gene at 6q27 in sporadic epithelial ovarian cancer. Oncogene. 2007;26:683–700. doi: 10.1038/sj.onc.1209827. [DOI] [PubMed] [Google Scholar]
- 39.Sapkota G.P., Cummings L., Newell F.S., Armstrong C., Bain J., Frodin M., Grauert M., Hoffmann M., Schnapp G., Steegmaier M., Cohen P., Alessi D.R. BI-D1870 is a specific inhibitor of the p90 RSK (ribosomal S6 kinase) isoforms in vitro and in vivo. Biochem J. 2007;401:29–38. doi: 10.1042/BJ20061088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kang S.M., Elf S., Lythgoe K., Hitosugi T., Taunton J., Zhou W., Xiong L., Wang D.S., Muller S., Fan S.Q., Sun S.Y., Marcus A.I., Gu T.L., Polakiewicz R.D., Chen Z., Khuri F.R., Shin D.M., Chen J. p90 ribosomal S6 kinase 2 promotes invasion and metastasis of human head and neck squamous cell carcinoma cells. J Clin Invest. 2010;120:1165–1177. doi: 10.1172/JCI40582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smolen G.A., Zhang J., Zubrowski M.J., Edelman E.J., Luo B., Yu M., Ng L.W., Scherber C.M., Schott B.J., Ramaswamy S., Irimia D., Root D.E., Haber D.A. A genome-wide RNAi screen identifies multiple RSK-dependent regulators of cell migration. Genes Dev. 2010;24:2654–2665. doi: 10.1101/gad.1989110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rundhaug J.E. Matrix metalloproteinases, angiogenesis, and cancer: commentary re: A. C. Lockhart et al., Reduction of wound angiogenesis in patients treated with BMS-275291, a broad spectrum matrix metalloproteinase inhibitor. Clin. Cancer Res., 9: 00–00, 2003. Clin Cancer Res. 2003;9:551–554. [PubMed] [Google Scholar]
- 43.Godefroy E., Manches O., Dreno B., Hochman T., Rolnitzky L., Labarriere N., Guilloux Y., Goldberg J., Jotereau F., Bhardwaj N. Matrix metalloproteinase-2 conditions human dendritic cells to prime inflammatory T(H)2 cells via an IL-12-and OX40L-dependent pathway. Cancer Cell. 2011;19:333–346. doi: 10.1016/j.ccr.2011.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Greenwald H.S., Friedman E.B., Osman I. Superficial spreading and nodular melanoma are distinct biological entities: a challenge to the linear progression model. Melanoma Res. 2012;22:1–8. doi: 10.1097/CMR.0b013e32834e6aa0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mar V.J., Wong S.Q., Logan A., Nguyen T., Cebon J., Kelly J., Wolfe R., Dobrovic A., McLean C., McArthur G.A. Clinical and pathological associations of the activating RAC1 P29S mutation in primary cutaneous melanoma. Pigment Cell Melanoma Res. 2014;27:1117–1125. doi: 10.1111/pcmr.12295. [DOI] [PubMed] [Google Scholar]
- 46.Haqq C., Nosrati M., Sudilovsky D., Crothers J., Khodabakhsh D., Pulliam B.L., Federman S., Miller J.R., III, Allen R.E., Singer M.I., Leong S.P., Ljung B.M., Sagebiel R.W., Kashani-Sabet M. The gene expression signatures of melanoma progression. Proc Natl Acad Sci U S A. 2005;102:6092–6097. doi: 10.1073/pnas.0501564102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cohen C., Zavala-Pompa A., Sequeira J.H., Shoji M., Sexton D.G., Cotsonis G., Cerimele F., Govindarajan B., Macaron N., Arbiser J.L. Mitogen-actived protein kinase activation is an early event in melanoma progression. Clin Cancer Res. 2002;8:3728–3733. [PubMed] [Google Scholar]
- 48.Michailidou C., Jones M., Walker P., Kamarashev J., Kelly A., Hurlstone A.F. Dissecting the roles of Raf-and PI3K-signalling pathways in melanoma formation and progression in a zebrafish model. Dis Model Mech. 2009;2:399–411. doi: 10.1242/dmm.001149. [DOI] [PubMed] [Google Scholar]
- 49.Wu H., Goel V., Haluska F.G. PTEN signaling pathways in melanoma. Oncogene. 2003;22:3113–3122. doi: 10.1038/sj.onc.1206451. [DOI] [PubMed] [Google Scholar]
- 50.Bain J., Plater L., Elliott M., Shpiro N., Hastie C.J., McLauchlan H., Klevernic I., Arthur J.S., Alessi D.R., Cohen P. The selectivity of protein kinase inhibitors: a further update. Biochem J. 2007;408:297–315. doi: 10.1042/BJ20070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Galan J.A., Geraghty K.M., Lavoie G., Kanshin E., Tcherkezian J., Calabrese V., Jeschke G.R., Turk B.E., Ballif B.A., Blenis J., Thibault P., Roux P.P. Phosphoproteomic analysis identifies the tumor suppressor PDCD4 as a RSK substrate negatively regulated by 14-3-3. Proc Natl Acad Sci U S A. 2014;111:E2918–E2927. doi: 10.1073/pnas.1405601111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brooks P.C., Stromblad S., Sanders L.C., von Schalscha T.L., Aimes R.T., Stetler-Stevenson W.G., Quigley J.P., Cheresh D.A. Localization of matrix metalloproteinase MMP-2 to the surface of invasive cells by interaction with integrin alpha v beta 3. Cell. 1996;85:683–693. doi: 10.1016/s0092-8674(00)81235-0. [DOI] [PubMed] [Google Scholar]
- 53.Kawasaki K., Kawakami T., Watabe H., Itoh F., Mizoguchi M., Soma Y. Expression of matrilysin (matrix metalloproteinase-7) in primary cutaneous and metastatic melanoma. Br J Dermatol. 2007;156:613–619. doi: 10.1111/j.1365-2133.2006.07678.x. [DOI] [PubMed] [Google Scholar]
- 54.Doehn U., Hauge C., Frank S.R., Jensen C.J., Duda K., Nielsen J.V., Cohen M.S., Johansen J.V., Winther B.R., Lund L.R., Winther O., Taunton J., Hansen S.H., Frodin M. RSK is a principal effector of the RAS-ERK pathway for eliciting a coordinate promotile/invasive gene program and phenotype in epithelial cells. Mol Cell. 2009;35:511–522. doi: 10.1016/j.molcel.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aronchik I., Appleton B.A., Basham S.E., Crawford K., Del Rosario M., Doyle L.V., Estacio W.F., Lan J., Lindvall M.K., Luu C.A., Ornelas E., Venetsanakos E., Shafer C.M., Jefferson A.B. Novel potent and selective inhibitors of p90 ribosomal S6 kinase reveal the heterogeneity of RSK function in MAPK-driven cancers. Mol Cancer Res. 2014;12:803–812. doi: 10.1158/1541-7786.MCR-13-0595. [DOI] [PubMed] [Google Scholar]
- 56.Serra V., Eichhorn P.J., Garcia-Garcia C., Ibrahim Y.H., Prudkin L., Sanchez G., Rodriguez O., Anton P., Parra J.L., Marlow S., Scaltriti M., Perez-Garcia J., Prat A., Arribas J., Hahn W.C., Kim S.Y., Baselga J. RSK3/4 mediate resistance to PI3K pathway inhibitors in breast cancer. J Clin Invest. 2013;123:2551–2563. doi: 10.1172/JCI66343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Posch C., Moslehi H., Feeney L., Green G.A., Ebaee A., Feichtenschlager V., Chong K., Peng L., Dimon M.T., Phillips T., Daud A.I., McCalmont T.H., LeBoit P.E., Ortiz-Urda S. Combined targeting of MEK and PI3K/mTOR effector pathways is necessary to effectively inhibit NRAS mutant melanoma in vitro and in vivo. Proc Natl Acad Sci U S A. 2013;110:4015–4020. doi: 10.1073/pnas.1216013110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vredeveld L.C., Possik P.A., Smit M.A., Meissl K., Michaloglou C., Horlings H.M., Ajouaou A., Kortman P.C., Dankort D., McMahon M., Mooi W.J., Peeper D.S. Abrogation of BRAFV600E-induced senescence by PI3K pathway activation contributes to melanomagenesis. Genes Dev. 2012;26:1055–1069. doi: 10.1101/gad.187252.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Relative intensity of phospho–extracellular signal-regulated (p-ERK) and phospho–protein S6 kinase, 90 kDa, polypeptide 1 (p-RSK1) expression in vertical growth phase (VGP) cell lines. Relationship between p-RSK1 levels and activation of the mitogen-activated protein kinase pathway in an expanded panel of nine VGP melanoma cell lines. Whereas four cell lines have higher expression of p-ERK than p-RSK1 (WM 278, WM 793, WM 853, and WM 902) by Western blot analysis, five cell lines show low or no detectable expression of p-ERK (WM 115, WM 983, WM 1366, WM 1361, and WM 3248).
Test of two siRNA candidates targeting protein S6 kinase, 90 kDa, polypeptide 1 (RSK1). Western blot analysis of protein extracts from siRNA-treated cells using two different siRNA candidates. p-RSK1, phospho-RSK1. Ran was used as a loading control.
Nodular melanoma cell proliferation is more sensitive to protein S6 kinase, 90 kDa, polypeptide 1 knockdown/inhibition than is superficial spreading melanoma cell proliferation. Treating an additional nodular melanoma cell line, WM 1366, with BI-D1870 induces a dose-dependent reduction in cell proliferation. DMSO, dimethyl sulfoxide.