Abstract
High-risk human papillomavirus (HPV) is a causative agent for an increasing subset of oropharyngeal squamous cell carcinomas (OPSCCs), and current evidence supports these tumors as having identifiable risk factors and improved response to therapy. However, the biochemical and molecular alterations underlying the pathobiology of HPV-associated OPSCC (designated HPV+ OPSCC) remain unclear. Herein, we profile miRNA expression patterns in HPV+ OPSCC to provide a more detailed understanding of pathologic molecular events and to identify biomarkers that may have applicability for early diagnosis, improved staging, and prognostic stratification. Differentially expressed miRNAs were identified in RNA isolated from an initial clinical cohort of HPV+/− OPSCC tumors by quantitative PCR–based miRNA profiling. This oncogenic miRNA panel was validated using miRNA sequencing and clinical data from The Cancer Genome Atlas and miRNA in situ hybridization. The HPV-associated oncogenic miRNA panel has potential utility in diagnosis and disease stratification and in mechanistic elucidation of molecular factors that contribute to OPSCC development, progression, and differential response to therapy.
Head and neck squamous cell carcinoma (HNSCC) is a leading cause of cancer death worldwide. High-risk human papillomavirus (HPV), particularly HPV16, is a causative agent for an increasing subset of oropharyngeal SCCs (OPSCCs). Current evidence supports these tumors (designated HPV+ OPSCC) as having unique biology because they have relatively low mutational density, wild-type cellular tumor antigen p53, and p16 protein overexpression.1 HPV+ OPSCC is already well established as a distinct clinical entity with identifiable epidemiologic characteristics and risk factors.2 However, a detailed understanding of the biology and the translation of that understanding into clinically applicable tools to decrease mortality and to lessen morbidity from treatment is clearly lacking. Although clinical trials assessing differing management strategies for the two diseases are under way, currently, HPV+ patients are treated within the standard of care guidelines for OPSCC, in the same manner as HPV− patients. However, HPV+ tumors of the oropharynx are associated with distinct survival strata because HPV+ patients have prolonged progression-free responses and often, with appropriate surgical management, chemotherapy, and irradiation, experience complete responses that seem durable.3 Thus, early identification and characterization of this patient cohort is necessary to tailor care to the unique biology of their tumors. As such, characterization of an HPV-driven OPSCC tumor is itself a biomarker that demands management strategies distinct from those associated with traditional HNSCC.
Owing to public health efforts regarding smoking cessation, the overall incidence of most HNSCC has decreased in recent years. In contrast, US incidence rates of OPSCC have increased significantly yearly between 1973 and 2004 for base of tongue (1.3%) and tonsillar (0.6%) carcinomas, corresponding to a 225% increase in the population-level incidence of HPV+ OPSCC between 1984 and 2004.4,5 Among individuals aged 12 to 69 years, oral HPV infection has an overall prevalence of@∼7%; however, men have a significantly higher prevalence compared with women (10.1% versus 3.6%).6 These numbers are heavily influenced by sexual behavior and smoking history: prevalence seems to be as high as 20% in heavy smokers or individuals with >20 lifetime sexual partners. These trends are globally relevant because OPSCC is increasing in several countries, particularly in younger men.7 Furthermore, cervical cancer incidence is declining in developed countries, whereas HPV-associated orophargyneal cancers are projected to surpass annual numbers of cervical cancers in the United States by 2020, making HPV+ OPSCC the dominant HPV-associated cancer in the United States.5 Therefore, prevention strategies for HPV+ OPSCC are urgently needed, and developing clinically useful biomarkers that are closely tied to the ability of the virus to transform host cells and the subsequent neoplastic and metastatic progressions are critical.
Expression profiling of cancers and functional studies performed in cancer cell lines and murine models have revealed provocative patterns implicating the importance of miRNA-mRNA dysregulation in tumor development and progression. Somewhat independent of miRNA biology, miRNA profiles offer diagnostic and prognostic value in cancer and other diseases that may guide treatment.8,9 Our aim was to profile HPV+ versus HPV− OPSCC to provide a more detailed understanding of pathologic molecular events and to identify biomarkers that may have applicability for early diagnosis, improved staging, and prognostic stratification. We identify an oncogenic miRNA panel that represents the host response to an oncogenic HPV infection and validate this panel in additional clinical cohorts using publicly available sequencing and clinical data from The Cancer Genome Atlas (TCGA; Bethesda, MD) and miRNA in situ hybridization (ISH) analysis of arrayed human OPSCC tissues. This molecular signature may help differentiate oropharyngeal tumors with different prognoses and, thus, distinct management strategies and facilitate mechanistic elucidation of molecular factors that contribute to OPSCC development, progression, and response to therapy.
Materials and Methods
Patient Samples for miRNA Profiling
Tissues for the initial study cohort were obtained from the University of Missouri (Columbia, MO) surgical pathology archives (2006 to 2011) with Institutional Review Board approval and represent histologically confirmed tonsillar or base of tongue SCC (OPSCC). Tissue for study was identified by staining for p1610 according to the manufacturer's instructions (CINtec histology kit and E6H4 clone; Ventana Medical Systems Inc., a member of the Roche Group, Tucson, AZ) and was evaluated using a binary rating system, with positive representing extensive (>50%) tumor cell–specific cytoplasmic and nuclear staining. Negative staining represented sparse or absent tumor-specific staining. Focal staining patterns, in the presence of mostly negative staining, were interpreted as negative. All cases included for laser capture microdissection represented unambiguous staining patterns.
Laser Capture Microdissection
Using p16 staining, we identified cases from surgical excisions that were strongly positive (>90% immunoreactivity for p16 with minimal criteria ≥70%) or completely negative. Of the 109 cases stained, 53 had sufficient primary tumor tissue available for laser capture, performed using the ArcturusXT system (Invitrogen, Carlsbad, CA). For each case, a minimum of 8000 cancer cells were dissected, and caps were stored at −80°C. RNA purification used the miRNeasy formalin-fixed, paraffin-embedded (FFPE) kit (Qiagen Inc., Valencia, CA). All the samples were assessed using a NanoDrop spectrophotometer (Thermo Scientific, Wilmington, DE) and Agilent 2100 bioanalyzer (Agilent Technologies Inc., Santa Clara, CA). Additionally, a limited number of samples were run on an miRNA QC PCR array (Qiagen Inc.) as a means of positive control to assess the stability of the small RNA fraction.
Real-Time PCR-Based miRNA Profiling
Qiagen's miRNome global real-time quantitative PCR (qPCR) array (Qiagen Inc.), which includes assays for 1008 individual miRNA species, was used for profiling. Each array consisted of three 384-well plates such that each patient's sample was run on three separate plates. The 1008 sequences profiled represent all the annotated miRNA sequences (miRNAseqs) in the human miRNA genome (miRBase release 16). cDNA was synthesized using the miScript II reverse transcription kit (Qiagen Inc.), and qPCR was performed using the LightCycler 480 system (Roche Applied Science, Indianapolis, IN). Each assay was begun with hot start activation and included 45 cycles of amplification and a melt curve analysis step for determining the specificity of each probe (by calculating melting temperature). Absolute quantitation was used to generate threshold values using the second derivative maximum algorithm unique to the LightCycler 480 system (auto baseline). Statistical analysis was performed using the geNorm algorithm. To improve the threshold of detection, preamplification was performed on cDNA synthesized using the miScript II reverse transcription kit using the miScript PreAMP PCR kit and miScript PreAMP primer mix (Qiagen Inc.). Amplified cDNA was added to the miScript SYBR mix, water, and miScript universal primer and was dispensed to miRNome plates. A test miRNome array was first used to determine whether the dilution for real-time PCR resulted in a high percentage of call rates. Preamplified samples, including p16+ (n = 15) and p16− (n = 9) samples, were loaded on the miRNome plates (15 ng of cDNA per sample). Subsequent analysis was performed as described above using CT ≤ 30 as a cutoff point for inclusion.
FFPE-Based miRNA Cohort Bioinformatics
Raw data from the RT-PCR arrays were subjected to extensive quality control analyses based on specialized internal controls on the arrays including positive PCR controls, which test the efficiency of the PCR itself, and reverse transcription controls to detect any impurities that inhibited the reverse transcription phase of the procedure. We also calculated mean, SD, and coefficient of variation values and compared them with values published on FFPE cancer samples for these arrays.11 Any sample that did not fall within the acceptable range of metrics as defined by Qiagen Inc. was excluded from the analysis; one of the 24 samples failed this step. Next, miRNAs with CT values >30 (or 0) for preamplifed samples were considered not reliably detected and were excluded from analysis by replacing that CT with NA to indicate missing. Reference gene determination for normalization was performed on a plate-by-plate basis according to the geNorm algorithm using the Bioconductor package SLqPCR version 1.26.0.12–16 To test which miRNAs were differentially expressed based on HPV status, we used the Bioconductor limma package version 3.16.7. To focus on widely expressed miRNAs and to increase power, nonspecific filtering of miRNAs was performed as follows: only miRNAs that were detected in >90% of samples were carried forward for subsequent analysis. Moderated t statistics were applied to each miRNA using an empirical Bayes approach in which the standard errors are shrunk toward a common value.17 The comparison of interest was HPV+ versus HPV−, expressed in terms of fold change (HPV+/HPV−). The comparison was made after adjusting for smoking, smoking and HPV interaction, and age to account for potential confounding effects. In the event of a significant interaction between smoking and HPV for a specific miRNA, the interpretation of the effect of either smoking or HPV in isolation was made with caution.
Validation of the Oncogenic miRNA Panel Using Patient Cohorts from TCGA
Patient consent/enrollment and use of data were conducted in accordance with TCGA Human Subjects Protection and Data Access Policies. Clinical data were first downloaded for HNSCC patients (458 cases), HPV ISH or HPV p16 testing was assessed, and then patients with a definitive status (positive or negative) were selected, resulting in 66 cases. Of these, 11 cases were positive and 11 were definitively HPV– according to p16 and/or HPV ISH. Annotated RNA sequencing data were then downloaded from TCGA Data Portal in September and October 2013. Because the comparison of interest was HPV+ versus HPV− cancer, individual normalized expression values for a particular miRNA were compared between groups of HPV+ patients (n = 11) and HPV− patients (n = 11) by using Student's t-test, and the results were sorted according to the level of significance. For miRNAseq, the relative abundance of a particular miRNA is represented by the absolute number of sequence reads.
Validation by miRNA ISH in HNSCC TMAs
HNSCC tissue microarrays (TMAs) were assembled from cases available in the Department of Pathology and Immunology, Washington University School of Medicine (St. Louis, MO), using tissues obtained with approval of the Human Research Protection Office. The TMA included 357 cases of HNSCC, with two tumor tissue cores per case. Immunohistochemical analysis was performed for p16 on a full FFPE section using a Ventana BenchMark automated immunostainer (Ventana Medical Systems Inc.) according to standard protocols with a known p16-expressing SCC case and normal tonsils as positive and negative controls, respectively. Antigen retrieval used Ventana CC1, EDTA-Tris, pH 8.0, solution. Staining was read by one study pathologist (J.S.L.), and all the positive cases demonstrated nuclear and cytoplasmic staining. Staining was graded in a quartile manner as follows: 0 = no staining, 1+ = 1% to 25% staining; 2+ = 26% to 50%; 3+ = 51% to 75%, and 4+ = 76% to 100%. Cases were then classified as positive (any convincing expression) versus negative, and then separately as strong positive staining (3+ or 4+) versus negative or weak staining (0, 1+, or 2+). More than 90% of cases were either strongly and diffusely positive or completely negative. The TMAs were then processed for RNA ISH for HPV or miRNA ISH for miR-9. ISH for high-risk HPV E6/E7 RNA was performed using the RNAscope HPV kit (Advanced Cell Diagnostics Inc., Hayward, CA) according to the manufacturer's instructions and was classified by the study pathologist (J.S.L.) as either positive or negative. Positive cases had granular cytoplasmic and/or nuclear brown staining that was above the signal on the negative control slide.
ISH for miR-9 was performed at the Center for RNA Interference and Non-Coding RNAs at MD Anderson Cancer Center (Houston, TX). Double digoxigenin–labeled (5′ and 3′) miRCURY LNA probes were obtained from Exiqon A/S (Vedbaek, Denmark). For visualization, the digoxigenins were detected with a polyclonal antidigoxigenin antibody and alkaline phosphatase–conjugated secondary antibody using 5-bromo-4-chloro-3-indolyl-phosphate/nitro blue tetrazolium. In these experiments, the full-length mature miRNAseq for miR-9 was used for specific LNA probes: 5′-TCATACAGCTAGATAACCAAAGA-3′. All the tumor tissues stained for miR-9 were counterstained with nuclear fast red. One entire TMA was stained with LNA U6 snRNA probe as positive controls without counterstain. Each TMA contained normal reference tissues that served as orientation and as negative controls (liver, thyroid, or small bowel). All the resulting slides were assessed in a blinded manner (D.L.M. and/or J.S.L.) as to HPV status, hematoxylin and eosin morphology, and all other clinicopathologic parameters. The stained slides were read and scored in a systematic manner according to the following three parameters: i) proportion of tumor cells with identifiable ISH signal (0 = < 10%, 1 = 10% to 24%, 2 = 25% to 49%, 3 = 50% to 74%, and 4 = ≥75%); ii) strength of ISH signal (weak, weak/moderate, moderate, and strong); and iii) staining pattern (punctate, diffuse, or punctate + diffuse). The staining results were tabulated and then unblinded and combined with data regarding HPV status and additional clinical data. The subsequent logistic regression analysis was performed separately for each binary outcome (HPV status measured by p16 and ISH), with the following for predictors: staining proportion, strength of ISH signal, and staining pattern. Model selection criteria were used to guide the selection of the final variables included in the model. Internal cross-validation was reported to indicate the performance of the model.
Evaluation of E-Cadherin and miR-9 in Cell Lines
OPSCC cell lines of known HPV status, a gift from Dr. Susanne M. Gollin (University of Pittsburgh, Pittsburgh, PA), were cultured in M-10 medium containing l-glutamine, gentamicin, and nonessential amino acids. For Western blot analysis, 20 μg of protein from cell lysates was electrophoresed on SDS-PAGE and electroblotted to polyvinylidene difluoride membranes. Blots were probed with anti–E-cadherin primary antibody (BD #610181, clone 36 recognizing the C-terminus, 1:100 dilution; BD Biosciences, San Jose, CA) and a rabbit secondary antibody (Sigma A-6667, 1:4000 dilution; Sigma-Aldrich, St. Louis, MO). Loading controls were probed with rabbit polyclonal anti–glyceraldehyde-3-phosphate dehydrogenase (Abcam #ab9485, 1:2000 dilution; Abcam Inc., Cambridge, MA) as a primary antibody. To assess levels of miR-9 in cell lines, RNA was extracted in technical triplicates using the miRCURY RNA isolation kit (Exiqon A/S) according to the manufacturer's instructions, polyadenylated, and reverse transcribed into cDNA in a single reaction with the Universal cDNA synthesis kit II (Exiqon A/S). A synthetic RNA spike-in, UniSp6, was added to each sample to monitor reverse transcription efficiency and reproducibility. The LNATM primer sets for miR-9, four candidate reference genes, and two positive control primer sets were obtained from Exiqon A/S. cDNA from the reverse transcription reactions were diluted 100× with nuclease-free water combined 1:1 with 2× PCR master mix (Exiqon A/S) and 0.05 ng of total RNA per reaction. Real-time PCR amplification and melting curve analysis was performed using an ABI StepOnePlus system (Applied Biosystems, Foster City, CA) in a standard (2-hour) run according to the following cycles: polymerase activation/denaturation at 95°C for 10 minutes, amplification for 40 cycles at 95°C for 10 seconds, followed by 60°C for 1 minute at a ramp rate of 1.6 degrees centigrade per second, and melting curve analysis as specified by the StepOnePlus system.
Results
Characterization of Oropharyngeal Tissues
Because national and international rates of HPV-driven OPSCC have been called epidemic in scale,18 we asked whether patients previously diagnosed as having OPSCC at the University of Missouri reflect concordant epidemiologic proportions. Of the cases (n = 109) stained for p16 expression, 58% were positive (Figure 1A). Multi-institutional US studies estimate that 65% to 70% of OPSCC is caused by HPV.5 The average ages in the cohorts under study were 56.49 and 61.00 years for p16+ and p16−, respectively (Figure 1B and Supplemental Table S1). HPV/OPSCC has a male predominance and no consistent association with smoking. Disease staging also has been associated with differences between HPV+ and HPV− OPSCC, with HPV+ disease more commonly associated with stage IV disease and frequent lymph node metastases at initial presentation. Overall, we interpret these clinical data as a positive signal that the p16+ and p16− cohorts reflect true disease trends.
Figure 1.
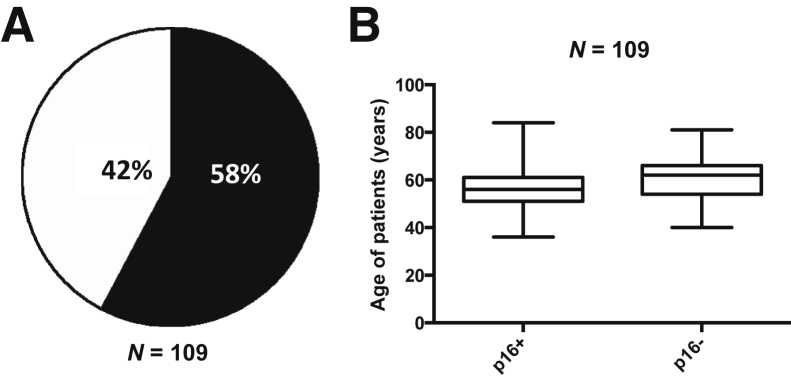
Human papillomavirus (HPV) prevalence in formalin-fixed, paraffin-embedded (FFPE) cases. A: The clinical history for patients diagnosed between 2006 to 2011 was reviewed, archived FFPE tissue blocks were assessed for available tissue, and available hematoxylin and eosin slides were reviewed. After an Institutional Review Board–approved protocol, tissues were sectioned, stained for p16 protein expression, and scored as positive or negative as described. The results showed that 58% of cases were HPV+ as defined in Materials and Methods. B: The average ages in the p16+ and p16− cohorts under study were 56.49 and 61.00 years, respectively. In the boxplot, the median is denoted by the solid horizontal line within the rectangle. The top boundary is at 75th percentile and the bottom boundary is at the 25th percentile. The whiskers extending from the rectangle are set at 1.5× the interquartile range.
HPV+ and HPV− Tumors Have Distinct miRNA Profiles
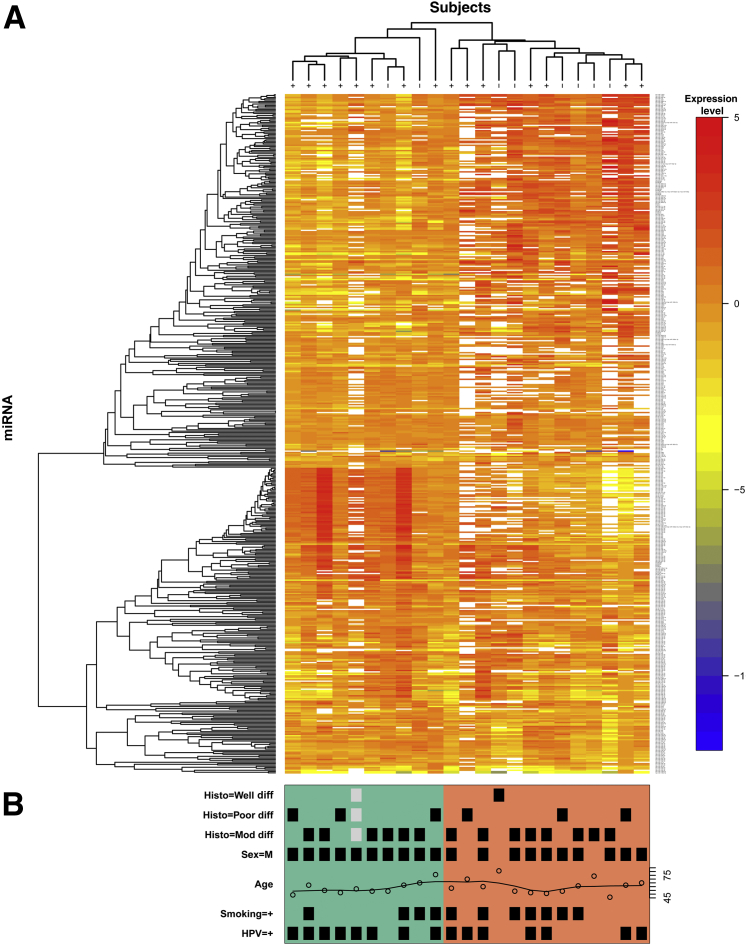
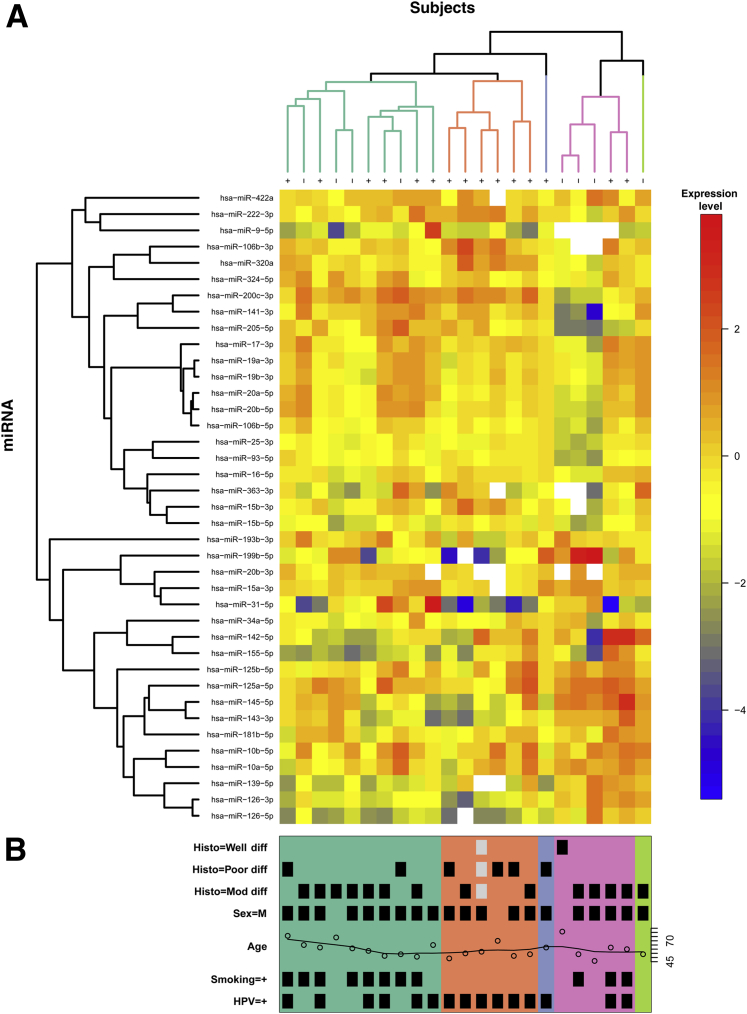
To identify a distinct miRNA signature that can differentiate HPV+ from HPV− OPSCC, we performed PCR-based miRNA profiling using a minimum of ten 10-μm sections from each of 24 cases. After preamplification, improved signal detection was evident (Supplemental Figure S1). One case was excluded based on quality control measures. Before nonspecific filtering there were 511 miRNAs; afterward, 276 miRNAs remained and were used for modeling. Results from the linear model showed that three individual miRNAseqs were significantly up-regulated in HPV+ patients: miR-320a, miR-222-3p, and miR-93-5p. The most statistically significant down-regulated miRNAs included six sequences representing four unique mature miRNAs: miR-199a-3p//miR-199b-3p, miR-143, miR-145, and miR-126a (Figure 2 and Table 1). The top 10 miRNAs that were most affected by age or smoking status do not show fold changes of the magnitude we found associated with HPV status (Supplemental Table S2). Three miRNAs that showed a significant HPV effect also had a significant smoking × HPV effect (miR-320a, miR-126, and miR-143; results not shown). The full expression matrix (511 miRNAs, 23 samples) was log2 transformed and clustered using unsupervised hierarchical clustering based on average agglomeration with as the distance measure. The results are presented as a heat map, with dendrograms indicating the clustering of patient samples and miRNAs (Figure 2). Additional unsupervised hierarchical clustering was performed using a list of 39 miRNAs implicated by the present data set and those from the literature (Figure 3). These analyses resulted in data that strongly support the hypothesis that HPV+ oropharyngeal tumors display distinct miRNA profiles and that groups of miRNAs can be associated with an oncogenic HPV infection.
Figure 2.
Profiling of miRNA expression on formalin-fixed, paraffin-embedded samples by quantitative RT-PCR. Tumors from 23 patients, including 15 p16+ and eight p16− samples, were profiled as described in Materials and Methods. A: An unsupervised hierarchical clustering heat map of normalized data (before nonspecific filtering or testing) representing 511 miRNAs is shown, with red indicating greater levels of relative expression, blue lesser levels, and white unreliable data (CT > 30 or CT = 0). The dendrogram at the top of the heat map illustrates which patient samples had the most similar miRNA profiles, whereas the dendrogram on the left (y axis) illustrates which miRNAs had similar profiles across patients. Items that were most similar were linked sooner to each other than items that were less similar. B: Clinical information is shown for each sample, with a black square marking the presence of the indicated variable (gray indicates missing data); green [8 of 10 human papillomavirus (HPV) positive] and salmon (6 of 13 HPV−) shading indicate how the samples cluster into two groups. Diff, differentiated.
Table 1.
Oncogenic miRNA Profile from FFPE HPV+ versus HPV− OPSCC Cases
| miRNA | Fold change | Log2FC | P value |
|---|---|---|---|
| hsa-miR-320a | 2.83 | 1.5 | 2.02 × 10−3 |
| hsa-miR-93-5p | 2.36 | 1.24 | 5.41 × 10−3 |
| hsa-miR-222-3p | 2.31 | 1.21 | 4.58 × 10−3 |
| hsa-miR-199a-3p//hsa-miR-199b-3p | 0.14 | −2.87 | 5.32 × 10−3 |
| hsa-miR-199b-5p | 0.14 | −2.85 | 8.80 × 10−3 |
| hsa-miR-145-5p | 0.20 | −2.30 | 5.91 × 10−3 |
| hsa-miR-143-3p | 0.21 | −2.28 | 2.50 × 10−3 |
| hsa-miR-126-5p | 0.23 | −2.14 | 5.48 × 10−3 |
| hs-miR-126-3p | 0.26 | −1.94 | 6.94 × 10−3 |
Fold change (FC) and log2FC are shown for the nine most statistically significant miRNA sequences, representing seven distinct miRNAs.
FFPE, formalin-fixed, paraffin-embedded; HPV, human papillomavirus; OPSCC, oropharyngeal squamous cell carcinoma.
Figure 3.
Unsupervised hierarchical clustering heat map of miRNA expression in formalin-fixed, paraffin-embedded samples. A: An unsupervised hierarchical clustering heat map of normalized data representing 39 selected miRNAs is shown, with red indicating greater levels of relative expression, blue lesser levels, and white unreliable data (CT > 30 or CT = 0). The dendogram at the top of the heat map illustrates which patient samples had the most similar miRNA profiles, whereas the dendrogram on the left (y axis) illustrates which miRNAs had similar profiles across patients. Items that were most similar were linked sooner to each other than items that were less similar. B: Clinical information is shown for each sample, with a black square marking the presence of the indicated variable (gray indicates missing data); the samples sort into five distinct clusters, with most samples falling into three groups: green [mostly smokers regardless of human papillomavirus (HPV) status], salmon and blue (all HPV+ nonsmokers), and pink and bright green (mixed). Diff, differentiated.
Correlation of miRNA Expression with HPV Expression from TCGA Database Validates the PCR-Based HPV-Associated miRNA Profile
To further assess changes in the miRNA expression profile associated with HPV+ OPSCC, we used publically available clinical and miRNAseq data from TCGA and identified 11 primary OPSCC cases (tonsil or base of tongue) positive for either p16 IHC or HPV ISH that were paired with 11 HPV− cases, which we designated as TCGA cohort 1 (Supplemental Table S3). In TCGA cohort 1, we found 84 miRNAs to be significant [P < 0.05 and mean reads per kilobase of transcript per million reads mapped (RPKM) >10 in both groups]. Using the average RPKM for each group as general estimations of expression level, fold changes were calculated and cutoff values of log2 fold changes of ±1.0 were used, yielding a list of 36 miRNAs (Supplemental Table S4).
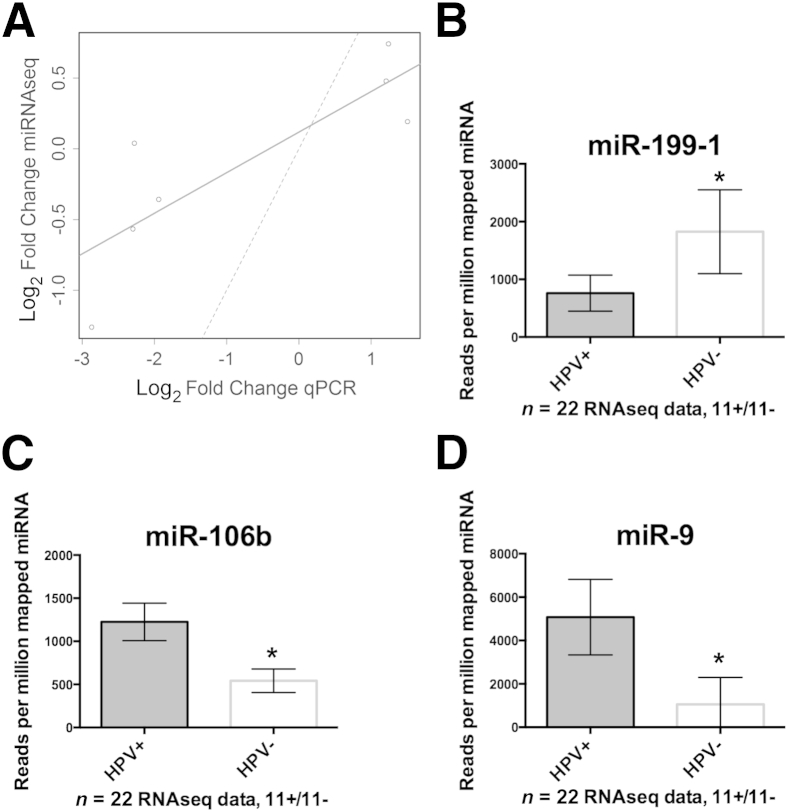
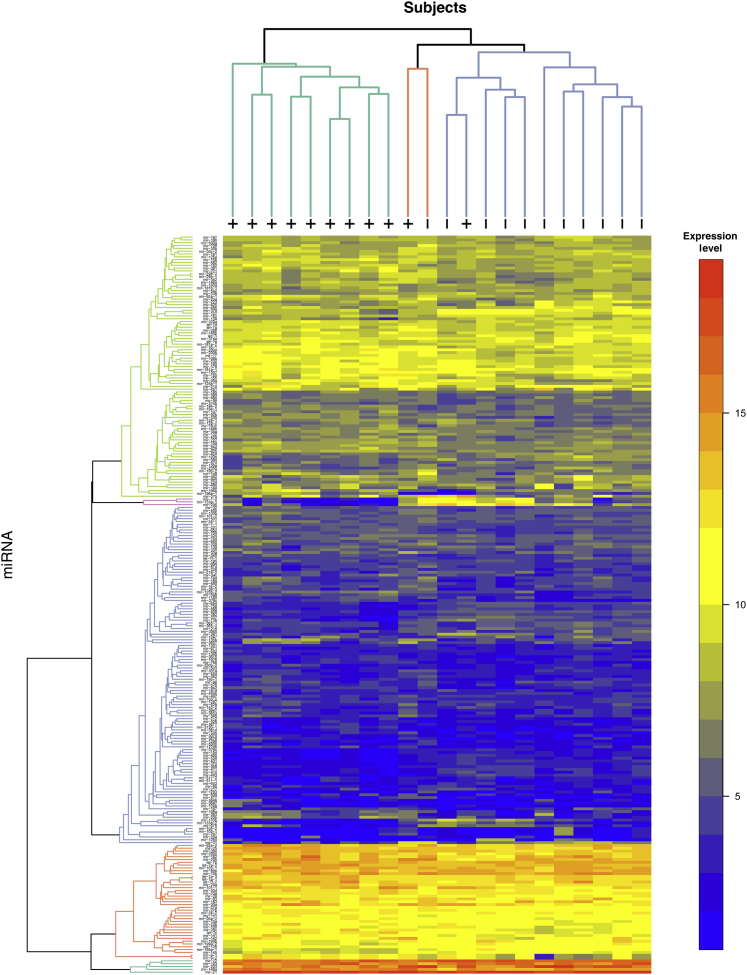
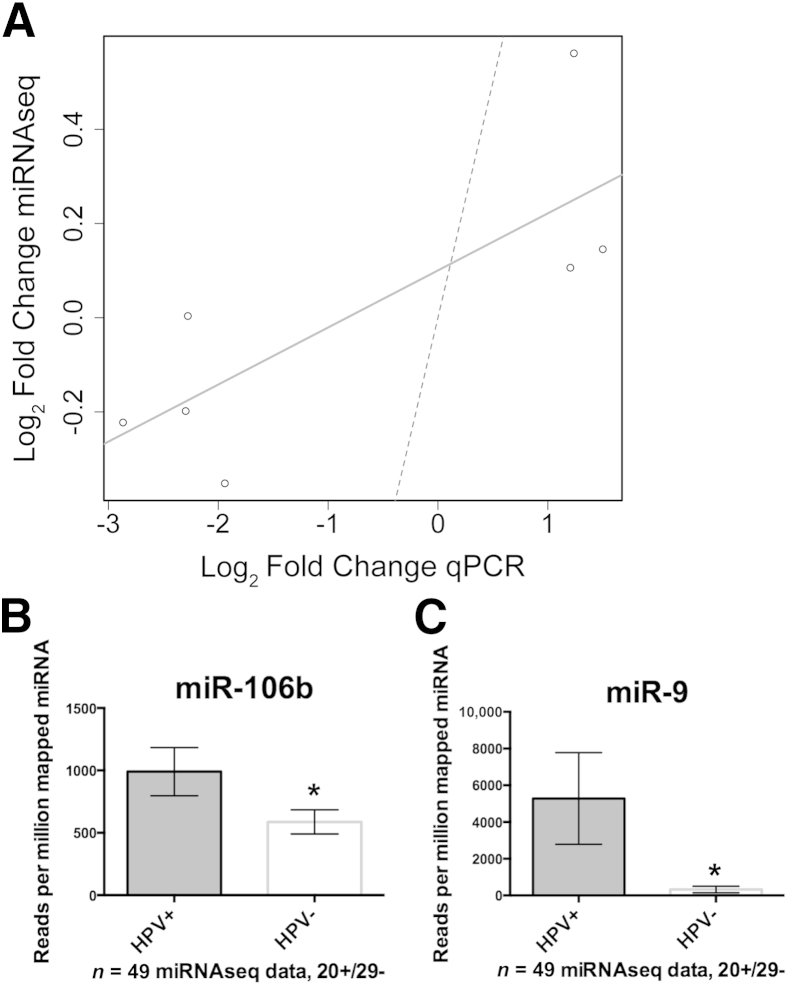
The results from TCGA cohort 1 were compared with the panel of miRNAs identified from the PCR profiling of FFPE tissue. The fold changes of these seven miRNAs between HPV+ and HPV− patients are concordant between the two data sets based on the Spearman rank correlation (rho = 0.85; P = 0.02) and the Pearson correlation (r = 0.83; P = 0.02; 95% CI, 0.21–0.97) (Figure 4A). Among the most statistically significant miRNAs from TCGA cohort 1, 16 of which are shown in Table 2, miR-199-1, miR-106b, and miR-9 were highly correlated to patients who are HPV+ irrespective of other covariates, such as age or smoking status (Figure 4, B–D). miR-9 was up-regulated 1.84-fold (P = 0.14) in the FFPE qPCR cohort, and it has been identified by two independent published studies as an HPV-associated miRNA in OPSCC. Therefore, the up-regulation of miR-9-1 and miR-9-2 in TCGA cohort 1 miRNAseq data served as strong validation that this is an HPV-associated miRNA in OPSCC. Unsupervised clustering performed on the full miRNAseq data indicated that, in this data set, HPV+ disease is associated with distinct miRNA profiles (Figure 5). Notably, a group of miRNAs that are highly expressed in HPV+ and HPV− disease, including miR-21, miR-203, and miR-22, clustered at the bottom of this heat map, implicating these miRNAs as potentially important in squamous differentiation of HNSCC regardless of HPV status.
Figure 4.
Analysis of The Cancer Genome Atlas (TCGA) cohort 1 miRNA sequencing (miRNAseq) data. A: Patients composing TCGA cohort 1 were identified as described. The graph shows comparison of the seven differentially expressed miRNAs identified by PCR profiling of microdissected formalin-fixed, paraffin-embedded (fold changes in log2 scale for symmetry) versus results obtained from analysis of TCGA cohort 1 miRNAseq data. Strong concordance between the data sets was obtained based on the Spearman rank correlation (rho = 0.85; P = 0.02) and the Pearson product moment correlation (r = 0.83; P = 0.02; 95% CI, 0.21–0.97). A best-fit line (solid) indicated this relative concordance, whereas a 45° reference line (dashed) indicated that there was not perfect absolute agreement between the data sets and the assay technique. B: Mean normalized miRNA read counts showing miR-199-1 as a validated human papillomavirus (HPV)–associated miRNA. C: Mean normalized miRNA read counts showed up-regulation of miR-106b in HPV+ tumors. D: Mean normalized read counts showed that miR-9 was significantly up-regulated in HPV+ tumors. All the error bars indicate 95% CIs. ∗P < 0.05. qPCR, real-time quantitative PCR.
Table 2.
Analysis of TCGA Cohorts 1 and 2
| Data source | miRNA | Mean RPKM |
Fold change | Log2FC | P value | |
|---|---|---|---|---|---|---|
| HPV+ | HPV− | |||||
| TCGA cohort 1 | hsa-miR-106b | 1225 | 542 | 2.26 | 1.18 | 1.7 × 10−5 |
| hsa-miR-148a | 54,501 | 20,645 | 2.64 | 1.40 | 4.8 × 10−5 | |
| hsa-miR-625 | 306 | 111 | 2.76 | 1.46 | 8.8 × 10−5 | |
| hsa-miR-335 | 113 | 41 | 2.71 | 1.44 | 5.4 × 10−4 | |
| hsa-miR-9-1 | 5078 | 1054 | 4.81 | 2.27 | 5.4 × 10−4 | |
| hsa-miR-9-2 | 5091 | 1055 | 4.82 | 2.27 | 5.8 × 10−4 | |
| hsa-miR-214 | 13 | 46 | 0.30 | −1.75 | 8.9 × 10−4 | |
| hsa-miR-337 | 10 | 36 | 0.29 | −1.79 | 1.3 × 10−3 | |
| hsa-miR-378c | 24 | 7 | 3.49 | 1.80 | 1.7 × 10−3 | |
| hsa-miR-29c | 4456 | 1204 | 3.70 | 1.89 | 2.5 × 10−3 | |
| hsa-miR-598 | 38 | 14 | 2.68 | 1.42 | 3.3 × 10−3 | |
| hsa-miR-107 | 133 | 52 | 2.55 | 1.35 | 5.2 × 10−3 | |
| hsa-miR-150 | 2796 | 746 | 3.75 | 1.91 | 5.6 × 10−3 | |
| hsa-miR-106a | 55 | 18 | 3.05 | 1.61 | 6.6 × 10−3 | |
| hsa-miR-378 | 2154 | 742 | 2.90 | 1.54 | 6.7 × 10−3 | |
| hsa-miR-20b | 152 | 9 | 16.61 | 4.05 | 7.4 × 10−3 | |
| TCGA cohort 2 | hsa-miR-20b | 47 | 7 | 6.79 | 2.76 | 2.6 × 10−4 |
| hsa-miR-9-2 | 5287 | 325 | 16.26 | 4.02 | 5.4 × 10−4 | |
| hsa-miR-9-1 | 5282 | 326 | 16.18 | 4.02 | 5.4 × 10−4 | |
| hsa-miR-106b | 990 | 587 | 1.69 | 0.75 | 5.4 × 10−4 | |
| hsa-miR-574 | 44 | 69 | 0.63 | −0.66 | 6.8 × 10−4 | |
| hsa-miR-193b | 101 | 256 | 0.40 | −1.34 | 9.0 × 10−4 | |
| hsa-miR-363 | 30 | 7 | 4.38 | 2.13 | 9.3 × 10−4 | |
| hsa-miR-16-2 | 23 | 11 | 1.99 | 0.99 | 1.7 × 10−3 | |
| hsa-miR-15b | 521 | 269 | 1.93 | 0.95 | 1.7 × 10−3 | |
| hsa-miR-25 | 11,787 | 7377 | 1.60 | 0.68 | 1.8 × 10−3 | |
Fold change (FC) and log2FC are shown for the most statistically significant miRNA sequences from each cohort.
HPV, human papillomavirus; RPKM, reads per kilobase of transcript per million reads mapped; TCGA, The Cancer Genome Atlas.
Figure 5.
Heat map of normalized expression data from The Cancer Genome Atlas (TCGA) cohort 1. The color scale represents greater (red) or lesser (blue) levels of relative expression across TCGA head and neck squamous cell carcinoma samples (columns) and miRNAs (rows). Unsupervised hierarchical clustering of the samples and the miRNAs was performed using Euclidean distance and the average linkage methods, and the resulting dendrograms are shown in the margins, where + or − indicates human papillomavirus (HPV) status. Items that are most similar cluster lower in the dendrogram. There seem to be two distinct clusters of samples, one entirely HPV+ and the other mostly HPV−, as well as four distinct clusters of miRNAs. As an example, reflecting the color scale, the bottom rows show miRNA expressed across all samples at levels of 15,000 to 320,000 reads per million miRNA mapped (Supplemental Table S4).
Further Validation Studies Using TCGA and miRNA ISH
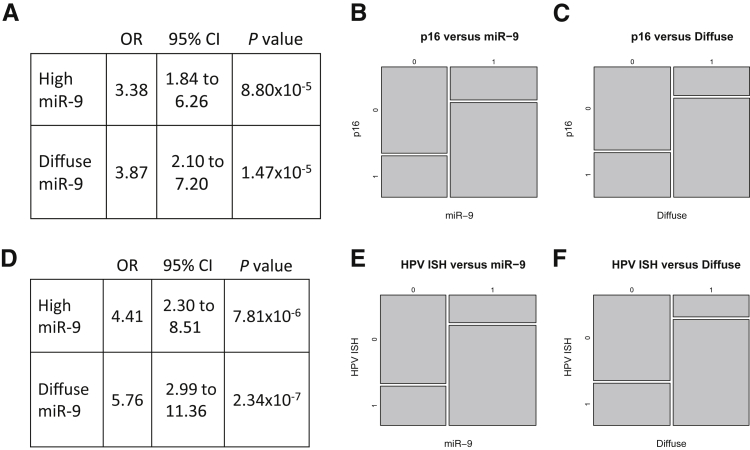
Based on a recent analysis of mRNAseq data from TCGA across 3775 malignancies for the presence of viral sequences, a second expanded cohort of patients from TCGA with HPV-associated HNSCC was identified19 that comprised 20 cases that lacked available clinical data regarding p16 or HPV ISH and were, therefore, not identified in the original analyses. However, these 20 cases were shown to express various viral transcripts (18 cases expressed transcripts from HPV16 and two from HPV33) and to have HPV DNA integrated into the host genome and were, thus, treated as HPV+ and were compared with an additional set of 29 HPV− cases (identified based on available p16 and/or HPV ISH data), herein designated TCGA cohort 2 (Supplemental Table S5). After extracting miRNAs that were expressed at suitable levels in both cohorts (>10 mean RPKM), a list of 43 miRNAs was generated (Supplemental Table S4), the top 10 of which are shown in Table 2. The fold changes of the seven-miRNA panel from FFPE studies show concordance between the two data sets based on the Spearman rank correlation (rho = 0.75; P = 0.06) and the Pearson correlation (r = 0.78; P = 0.03) (Figure 6). miR-9 and miR-106b were again among the most statistically significant miRNAs. miR-9-1 and miR-9-2 were expressed at RPKM of approximately 5000 in patients whose tumors express HPV transcripts. This is in stark contrast to patients whose tumors were negative for p16 IHC and/or HPV ISH, where, on average, miR-9 levels were approximately 16-fold lower. The tissue level of miR-9 expression was also assessed via ISH on HNSCC TMAs containing 357 cases, including 270 OPSCC cases with known p16 status and 226 cases with known HPV RNA ISH results. Representative positive and negative punctate and diffuse staining is shown in Figure 7. The odds that high-tumor miR-9 expression occurs in the setting of p16+ disease were more than three times greater [odds ratio (OR) = 3.38; P < 0.001; 95% CI, 1.84–6.26] than the odds of low-tumor miR-9 expression; similarly, the odds of diffuse miR-9 ISH were nearly four times greater (OR = 3.87; P < 0.001; 95% CI, 2.10–7.20) than the odds of nondiffuse miR-9 ISH (Figure 8). When the OR calculations were based on HPV mRNA ISH, the ORs increased (OR = 4.41; P < 0.001; 95% CI, 2.30–8.51 and OR = 5.76; P < 0.001; 95% CI, 2.99–11.36) for both miR-9 positivity and a diffuse cytoplasmic/nuclear pattern for miR-9 ISH signal (Figure 8). As has been shown in analyses of FFPE samples and analysis of TCGA cohort 1 (p16/ISH-confirmed cases), HPV-associated tumors are characterized by up-regulation of miRNAs belonging to the miR-106b∼25 cluster (miR-106b, miR-93, miR-25). The validation studies using TCGA cohort 2 strongly support up-regulation of members of this cluster in association with HPV status. Furthermore, the expression levels of the closely related miR-106a∼363 cluster (miR-20b and miR-363) are tightly correlated with HPV status, although the expression levels of these miRNAs are low, making it difficult to interpret the potential biological significance (Table 2). Note that others have reported up-regulation of miR-20b and miR-363 based on qPCR profiling and microarray analyses of FFPE samples and cell lines, respectively (Supplemental Table S6).20,21
Figure 6.
Analysis of The Cancer Genome Atlas (TCGA) cohort 2 miRNA sequencing (miRNAseq) data. A: Patients in TCGA cohort 2 were identified as described in Materials and Methods. The graph shows comparison of the seven differentially expressed miRNAs identified by PCR profiling of microdissected formalin-fixed, paraffin-embedded (fold changes in log2 scale) versus results obtained from analysis of TCGA cohort 2 miRNAseq data. A strong concordance between data sets was obtained based on the Spearman rank correlation (rho = 0.75; P = 0.06) and the Pearson product moment correlation (r = 0.78; P = 0.03; 95% CI, 0.07–0.96). A best-fit line (solid) indicates this relative concordance, whereas a 45° reference line (dashed) indicates that there is not perfect absolute agreement between the data sets and the assay technique. B: Mean normalized miRNA read counts showing miR-106b as a validated human papillomavirus (HPV)–associated miRNA. C: Mean normalized miRNA read counts showing miR-9 as a validated HPV-associated miRNA. All the error bars indicate 95% CIs. ∗P < 0.05. qPCR, real-time quantitative PCR.
Figure 7.
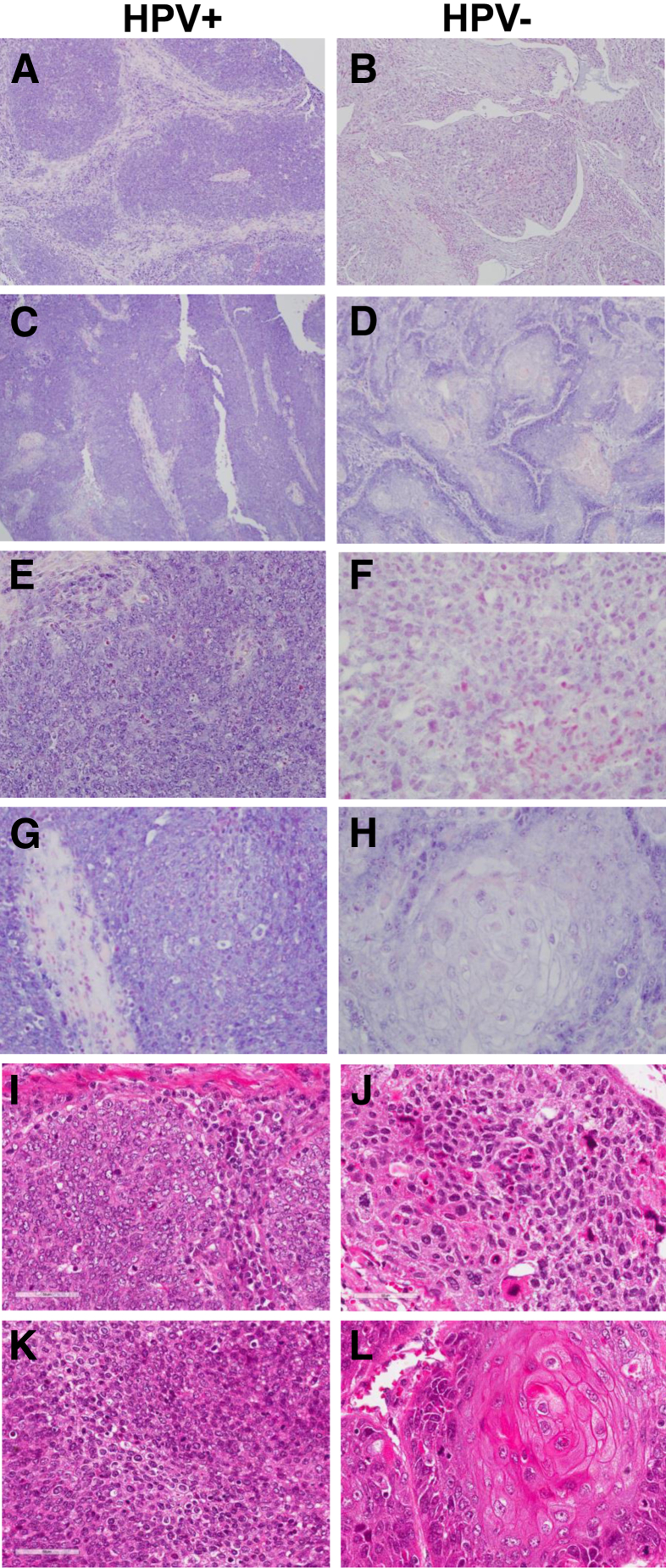
In situ hybridization (ISH) analysis of miR-9 expression in head and neck squamous cell carcinoma. A–D: Low-power images depicting ISH patterns for miR-9 in four oropharyngeal squamous cell carcinoma tissue cores. The ISH signal is represented by a deep purple color. The counterstain for miRNA-probed tissues sections was nuclear fast red; therefore, purple coloration represents ISH signal and pink/red is the counterstain. A and C: Human papillomavirus (HPV)–positive tumors showed a strong ISH signal. B and D: HPV− tumors had a weak or absent ISH signal. E–H: High-power images depicting ISH patterns for miR-9 in four oropharyngeal squamous cell carcinoma tissue cores. Staining in HPV+ cores was punctate (E) or diffuse (G), whereas HPV– tumors had weak or absent signal. I–L: Corresponding hematoxylin and eosin images. Original magnification: ×100 (A–D); ×400 (E–L).
Figure 8.
Odds ratios (ORs) and mosaic plots of high-tumor miR-9 occurring in relation to p16 status. A–C: The odds that high-tumor miR-9 expression occurs in the setting of p16+ disease were more than three times greater (OR = 3.38; P < 0.001; 95% CI, 1.84–6.26) than the odds of low-tumor miR-9 expression in p16+; similarly, the odds of diffuse miR-9 in situ hybridization (ISH) were nearly four times greater in p16+ (OR = 3.87; P < 0.001; 95% CI, 2.10–7.20) than the odds of nondiffuse miR-9 ISH in p16+. D–F: The ORs increased when the outcome was validated human papillomavirus (HPV) mRNA ISH. The model prediction based on HPV mRNA ISH had sensitivity = 0.62 and specificity = 0.875.
Discussion
HPV-Associated miRNA Profiling
The incidence rates for HPV+ OPSCC increased dramatically from 1988 to 2004, from 0.8 to 2.6 per 100,000, an increase of 225%.5 In striking contrast, HPV-negative cancers have declined 50%. These trends are also apparent internationally.7 Thus, it is of great interest to identify specific up-regulated miRNAs characteristic of an oncogenic HPV infection of the head and neck, as these miRNAs may represent novel diagnostic/prognostic biomarkers and may provide a more detailed understanding of the molecular pathogenesis of the disease. We provided miRNA profiles of three independent cohorts of patients (n = 94) with SCC of the aerodigestive tract performed with PCR arrays and next-generation deep sequencing. Comparative analysis of these cohorts strongly supports HPV-associated up-regulation of miR-9 and members of the miR-106b∼25 cluster and down-regulation of miR-199-1.
The fundamental mechanism associated with HPV+ disease is a disruption of cellular differentiation induced by the virus, wherein the cell acquires resistance to growth inhibition, immune evasion, subversion of apoptosis, genomic instability, and, ultimately, dysregulated proliferation such that viral DNA can replicate in synchrony with chromosomal DNA. Thus, HPV may causally modulate miRNAs that then act as central nodes in affecting numerous genes important in progression and metastatic spread, resulting in divergent gene regulation. In contrast to cervical cancer, viral integration is not necessary for the initiation of oncogenesis in OPSCC, and episomal HPV DNA seems to be a frequent occurrence in tonsillar carcinomas.22 However, similar to cervical cancer, transcriptionally active HPV is linked to the differentiation state of the host cell and in some circumstances leads to aberrant cell proliferation and genomic instability that is due, in part, to mitotic spindle defects that result in cytogenetic abnormalities.23 Alternatively, miRNAs could be suppressed owing to aberrant DNA methylation or chromosomal disruption.24 Because an individual miRNA can affect hundreds to thousands of genes, many of these often lie in the same biological pathway.25 Thus, specific cellular pathways important for immune surveillance, mitogenic signaling, or metabolism, which are integral for HPV infection, could also lead to typified miRNA expression.
Several studies have examined miRNA expression in the context of HPV and head and neck cancer, with heretofore a lack of consensus among reports.20,21,26–28 This may be due, in part, to the biological redundancy in miRNA function, the inherent genomic instability characteristic of HPV+ tumors, and the use of differing profiling methods and sample sizes. Nevertheless, emerging common themes among the various data sets are shown in Supplemental Table S6, which summarizes miRNAs identified in at least two independent studies. Note that miR-20b shares significant sequence homology with miR-106b and miR-93, these miRNAs share the same seed sequence, and they are closely related to miR-363. It has been reported that members of the miR-106b∼25 cluster are up-regulated independent of HPV status relative to normal tonsillar epithelium (n = 88 versus n = 7).21 This is in contrast to the present study showing up-regulation of members of the miR-106b∼25 cluster in HPV+ OPSCC relative to HPV− as determined in the FFPE PCR-based miRNA profile, TCGA cohort 1, and TCGA cohort 2. Recent in vitro experiments that sought to identify miRNAs altered as a function of HPV transfection used deep sequencing on organotypic raft cultures derived from normal or HPV31-transfected human foreskin keratinocytes isolated from the same donor, demonstrating two- to three-fold up-regulation of miR-106b and miR-25 in the HPV-transfected cultures relative to normal.29 In the context of the current human tumor data, this study strongly supports that these miRNAs are altered as a function of HPV oncoproteins.
TCGA deep sequencing data sets analyzed herein add another dimension to available data on the expression of miRNAs shown to be statistically significant between HPV+ and HPV− disease, as normalized read counts provide information on the relative abundance of a specific miRNA. Thus, it is interesting to note that miR-363, which has been shown to be up-regulated via microarray analysis in HPV+ disease by two independent studies, shows relatively low expression overall (<50 RPM in HPV+ and HPV− cohorts), yet the fold change for this miRNA is quite high. Similar in this regard is miR-20b, part of the same transcriptional unit and also previously reported21 as up-regulated in HPV+ disease. In the present study, we observed up-regulation of miR-20b (16- and 7-fold in TCGA cohorts 1 and 2, respectively); however, it is also expressed at relatively low levels. In contrast, miR-106b and members of the miR-106b∼25 cluster are expressed at levels approximately 30-fold higher. Because the biological significance of these fold differences are unknown, the relative importance of the two clusters in HPV+ OPSCC should be addressed experimentally because it is unclear whether expression of these miRNAs at very low levels may nevertheless regulate important biological functions.
Up-Regulated miRNAs
miRNA families are determined based on common seed sequence and are predicted to target overlapping sets of genes. miR-106b∼25 and miR-106a∼363 are genomic paralogs of the miR-17∼92 cluster, one of the best-characterized groups of miRNAs in human cancer, with oncogenic function in lymphoma, multiple myeloma, medulloblastoma, lung cancer, and colorectal cancer.30,31 miR-106b, miR-93, and miR-20b are members of the miR-17 family; as such, their seed sequence AAAGUG is identical to that of miR-17, miR-20a, miR-20b, miR-106a, and miR-106b, and they are predicted to have redundant function.32 However, the biological implications of intracluster redundancy are not entirely understood, as clustering of microRNAs with similar seed sequences is highly conserved, which suggests that members of the same cluster with identical seed sequences may have functional importance.
Members of the miR-106b∼25 and/or miR-106a∼363 cluster seem to be sensitive for differentiating HPV+ from HPV− HNSCC; however, the mechanistic basis for up-regulation is not entirely clear. HPV+ HNSCC and cervical cancers are characterized by up-regulated expression of distinct and larger subsets of cell cycle and DNA replication genes,33,34 and transcription of miR-106b∼25 is concurrent with the protein coding gene MCM7.31 Indeed, MCM7 is a known RB/E2F target gene, and E2F family transcription factors may be paramount in mediating miR-106b∼25 and MCM7 transcription.35 The MCM7 promoter has RB/E2F binding sites, and mRNA expression correlates with miR-106b expression.36 This is provocative because MCM7 has been proposed as a protein biomarker that distinguishes between HPV+ and HPV− HNSCC.37 Thus, the miR-106b∼25 cluster members may be sensitive for differentiating HPV+ from HPV− HNSCC as relatively increased expression of this cluster is highly characteristic of HPV+ tumors.
There is also considerable evidence that miR-106b∼25 and its paralogs can act as bona fide oncogenes (oncomirs) with defined roles in overcoming transforming growth factor β–mediated growth suppression, enhancing transforming growth factor β signaling, cell-cycle promotion, and increased cell survival.38 Repression of MCM7 together with miR-106b∼25 expression is associated with induction of p21 and phosphatase and tensin homolog in breast and prostate cancer.35 In neuroblastoma, miR-17∼92 expression induced potent inhibition of key transforming growth factor β signaling effectors and direct inhibition of transforming growth factor β–responsive genes.39 Increased expression of miR-106b∼25 and miR-17∼92 in retinoblastoma was also reported,40 leading to the hypothesis that in the context of genetic Rb loss, high miR-17∼92 expression may circumvent the need for the large numbers of genetic hits required for tumorigenesis. These results highlight the importance of context in miRNA oncogenic function and suggest the hypothesis that the current finding of miR-93, miR-106b, miR-20b, and miR-363 overexpression in OPSCC tumors supports the oncogenic activity instigated by the main viral oncoproteins: E6, E7, and E5.
In addition to the miR106b∼25 cluster, strong up-regulation of miR-9 is also observed in HPV+ OPSCC. miR-9 has been linked to metastatic potential via modulation of E-cadherin, acting to prime breast cancer cells for an epithelial-mesenchymal transition and stimulate angiogenesis.41 Because lymph node metastasis is extremely common in patients with HPV+ OPSCC, and increased miR-9 levels in HPV+ relative to HPV− OPSCC were observed in the present study and others,21,27 we asked whether there was a relationship between miR-9 and E-cadherin expression in OPSCC using RT-qPCR and Western blot analysis of OPSCC cell lines of known HPV status (Supplemental Figure S2); however, no relationship between miR-9 and E-cadherin was observed. Expression of E-cadherin is generally inversely correlated with prognosis in HNSCC, with early studies showing reduced E-cadherin expression as an independent prognostic marker for metastasis and local recurrence.42 A recent study (n = 152) reported that the expression of E-cadherin is extensive in OPSCC, with no correlation between E-cadherin and HPV, nodal or distant metastasis, histopathologic type, or survival.43 A second analysis (n = 102) suggested that HPV+/p16+ tumors from the oropharynx express high E-cadherin and β-catenin levels.44 Together, these results indicate that high miR-9 in HPV+ OPSCC is not directly altering E-cadherin expression, suggesting that other functionally important miR-9 targets should be explored. Interestingly, miR-9 may modulate the microenvironment in HPV+ OPSCC, as there is compelling evidence that miR-9 is packaged into microvesicles or may function as a tumor suppressor.45,46 Physiologic miR-9 expression may temper innate immune responses.47 In cancer, there is limited evidence that miR-9 may be involved in modulating immunoregulatory genes, including major histocompatibility complex class I– and interferon-regulated genes.48 Data surrounding miR-9 and adaptive immune response are also lacking. Nonetheless, the biology associated with adaptive immune response in HPV+ OPSCC is of particular interest and clinically relevant as these tumors are being considered for immune checkpoint inhibitor therapy with anti–programmed cell death 1 or programmed cell death ligand 1 antibodies and adoptive cell transfer therapy.49 It has also been shown that most transcriptionally active HPV+ OPSCC tumors express programmed cell death ligand 1 (also called B7-H1), although the significance of this and its relevance to patient survival may be limited.49,50 Thus, in light of strong up-regulation of miR-9 in HPV+ OPSCC, future studies will correlate miR-9 expression and the presence of immune infiltrates.
Down-Regulated miRNAs
Down-regulation of miR-143, miR-145, miR-199a-3p, miR-199b-3p, miR-199b-5p, and miR-126 has been observed in HPV+ OPSCC and cervical disease.26,28 Similar results were obtained in an organotypic keratinocyte raft culture model, showing significant down-regulation of miR-199a-5p and miR-145, suggesting a potential mechanistic link to early stages of the HPV life cycle.29 The present analyses on nonpreamplified patient samples offered preliminary suggestions that miR-199 and miR-145 were also down-regulated in patient samples (data not shown). After analyzing amplified samples and calculating relative expression of miRNAs based on the geNorm algorithm, an extremely robust and informatics-intensive method for analyzing RT-qPCR data, the data set confidently supports an HPV-specific down-regulation of miR-145. The seed sequence of miR-145 is present in the E1 open reading frame of a variety of papillomaviruses, and E1 has been shown to be a bona fide miR-145–reactive element.29 Moreover, miR-145 plays a role in modulating the viral life cycle, with a differentiation-dependent reduction in levels of this miRNA in HPV-transfected compared with control keratinocyte raft cultures that seems to depend on the function of viral E7 protein. Lentiviral forced expression of miR-145 plays a role in controlling genome amplification with a significant reduction of episomal viral DNA in undifferentiated cells. These in vitro data should be interpreted as having true biological significance because other studies evaluating miRNA profiles in HNSCC and cervical cancer corroborate an HPV-associated down-regulation of miR-145 and miR-143.26 Taken together, these data indicate that HPV may down-regulate miR-145 to allow for differentiation-specific genome amplification and that this miRNA remains repressed in human tumors. In this context, we can understand miR-145 as a putative tumor-suppressive miRNA in HPV disease.
Conclusions
Most HPV+ HNSCC tumors arise deep in the crypts of the tonsils of the oropharynx. These tumors are often obscured from routine gross visualization in dental examinations and, as such, most patients present with lymph node metastases. However, relative to patients with HPV− OPSCC, HPV+ status is positively correlated with a two- to three-fold increase in overall survival,3 indicating that the time is rapidly approaching whereupon HPV status will dictate therapy. The HPV-associated oncogenic miRNA panel identified herein may be incorporated into a multitarget diagnostic platform that can contribute to early detection and/or disease stratification to aid in differentiating oropharyngeal tumors with different prognoses and, thus, distinct management strategies. Furthermore, the oncogenic miRNA panel will facilitate the mechanistic elucidation of molecular factors that contribute to OPSCC development, progression, and response to therapy.
Acknowledgment
OPSCC cell lines of known HPV status were a gift from Dr. Susanne M. Gollin (University of Pittsburgh, Pittsburgh, PA).
Footnotes
Supported by NIH RL Kirschstein National Research Service Award F31 DE021926 (D.L.M.), NIH/National Cancer Institute research grant RO1 CA085870 (M.S.S.), and a Walther Cancer Foundation Advancing Basic Cancer Research Grant (M.S.S.).
Disclosures: None declared.
Supplemental Data
Signal detection for PCR plates. Four bars represent the relative proportions of CT values for miRNAs detected on the three 384-well PCR plates. Approximately 45% are not expressed (CT of 30 to 34), and only approximately 15% are detected at CT values <30. The proportions of miRNAs falling in detectable ranges are greatly improved by preamplification.
Expression of E-cadherin and miR-9 in oropharyngeal squamous cell carcinoma (OPSCC) cell lines. OPSCC cell lines of known human papillomavirus (HPV) status were cultured in M-10 medium containing l-glutamine, gentamicin, and nonessential amino acids. A: Western blot analysis for E-cadherin. Protein (20 μg) from cell lysates was electrophoresed on SDS-PAGE and electroblotted to polyvinylidene difluoride membranes and probed with anti–E-cadherin primary antibody and rabbit secondary antibody. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a loading control. B: Quantitative RT-PCR for miR-9. RNA was extracted in technical triplicates from cell lines, as indicated, polyadenylated, and reverse transcribed into cDNA in a single reaction. A synthetic RNA spike-in, UniSp6, was added to each sample to monitor reverse transcription efficiency and reproducibility. No concordance is observed between miR-9 and E-cadherin expression.
References
- 1.Miller D.L., Puricelli M.D., Stack M.S. Virology and molecular pathogenesis of HPV (human papillomavirus)-associated oropharyngeal squamous cell carcinoma. Biochem J. 2012;443:339–353. doi: 10.1042/BJ20112017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gillison M.L., D'Souza G., Westra W., Sugar E., Xiao W., Begum S., Viscidi R. Distinct risk factor profiles for human papillomavirus type 16-positive and human papillomavirus type 16-negative head and neck cancers. J Natl Cancer Inst. 2008;100:407–420. doi: 10.1093/jnci/djn025. [DOI] [PubMed] [Google Scholar]
- 3.Ang K.K., Harris J., Wheeler R., Weber R., Rosenthal D.I., Nguyen-Tân P.F., Westra W.H., Chung C.H., Jordan R.C., Lu C., Kim H., Axelrod R., Silverman C.C., Redmond K.P., Gillison M.L. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363:24–35. doi: 10.1056/NEJMoa0912217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marur S., D'Souza G., Westra W.H., Forastiere A.A. HPV-associated head and neck cancer: a virus-related cancer epidemic. Lancet Oncol. 2010;11:781–789. doi: 10.1016/S1470-2045(10)70017-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaturvedi A.K., Engels E.A., Pfeiffer R.M., Hernandez B.Y., Xiao W., Kim E., Jiang B., Goodman M.T., Sibug-Saber M., Cozen W., Liu L., Lynch C.F., Wentzensen N., Jordan R.C., Altekruse S., Anderson W.F., Rosenberg P.S., Gillison M.L. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol. 2011;29:4294–4301. doi: 10.1200/JCO.2011.36.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gillison M.L., Broutian T., Pickard R.K., Tong Z.Y., Xiao W., Kahle L., Graubard B.I., Chaturvedi A.K. Prevalence of oral HPV infection in the United States, 2009-2010. JAMA. 2012;307:693–703. doi: 10.1001/jama.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaturvedi A.K., Anderson W.F., Lortet-Tieulent J., Curado M.P., Ferlay J., Franceschi S., Rosenberg P.S., Bray F., Gillison M.L. Worldwide trends in incidence rates for oral cavity and oropharyngeal cancers. J Clin Oncol. 2013;31:4550–4559. doi: 10.1200/JCO.2013.50.3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schetter A.J., Leung S.Y., Sohn J.J., Zanetti K.A., Bowman E.D., Yanaihara N., Yuen S.T., Chan T.L., Kwong D.L., Au G.K., Liu C.G., Calin G.A., Croce C.M., Harris C.C. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schultz N.A., Dehlendorff C., Jensen B.V., Bjerregaard J.K., Nielsen K.R., Bojesen S.E., Calatayud D., Nielsen S.E., Yilmaz M., Holländer N.H., Andersen K.K., Johansen J.S. MicroRNA biomarkers in whole blood for detection of pancreatic cancer. JAMA. 2014;311:392–404. doi: 10.1001/jama.2013.284664. [DOI] [PubMed] [Google Scholar]
- 10.Lewis J.S., Jr. p16 Immunohistochemistry as a standalone test for risk stratification in oropharyngeal squamous cell carcinoma. Head Neck Pathol. 2012;(Suppl 1):S75–S82. doi: 10.1007/s12105-012-0369-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Philippidou D., Schmitt M., Moser D., Margue C., Nazarov P.V., Muller A., Vallar L., Nashan D., Behrmann I., Kreis S. Signatures of microRNAs and selected microRNA target genes in human melanoma. Cancer Res. 2010;70:4163–4173. doi: 10.1158/0008-5472.CAN-09-4512. [DOI] [PubMed] [Google Scholar]
- 12.Peltier H.J., Latham G.J. Normalization of microRNA expression levels in quantitative RT-PCR assays: identification of suitable reference RNA targets in normal and cancerous human solid tissues. RNA. 2008;14:844–852. doi: 10.1261/rna.939908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Appaiah H.N., Goswami C.P., Mina L.A., Badve S., Sledge G.W., Jr., Liu Y., Nakshatri H. Persistent upregulation of U6:SNORD44 small RNA ratio in the serum of breast cancer patients. Breast Cancer Res. 2011;13:R86. doi: 10.1186/bcr2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., Hornik K., Hothorn T., Huber W., Iacus S., Irizarry R., Leisch F., Li C., Maechler M., Rossini A.J., Sawitzki G., Smith C., Smyth G., Tierney L., Yang J.Y., Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schaefer A., Jung M., Miller K., Lein M., Kristiansen G., Erbersdobler A., Jung K. Suitable reference genes for relative quantification of miRNA expression in prostate cancer. Exp Mol Med. 2010;42:749–758. doi: 10.3858/emm.2010.42.11.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smyth G.K. Limma: linear models for microarray data. In: Gentleman R., Carey V., Dudoit S., Irizarry R., Huber W., editors. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Springer; New York: 2005. pp. 397–420. [Google Scholar]
- 18.Mroz E.A., Forastiere A.A., Rocco J.W. Implications of the oropharyngeal cancer epidemic. J Clin Oncol. 2011;29:4222–4223. doi: 10.1200/JCO.2011.37.8893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khoury J.D., Tannir N.M., Williams M.D., Chen Y., Yao H., Zhang J., Thompson E.J., TCGA Network, Meric-Bernstam F., Medeiros L.J., Weinstein J.N., Su X. Landscape of DNA virus associations across human malignant cancers: analysis of 3,775 cases using RNA-Seq. J Virol. 2013;87:8916–8926. doi: 10.1128/JVI.00340-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wald A.I., Hoskins E.E., Wells S.I., Ferris R.L., Khan S.A. Alteration of microRNA profiles in squamous cell carcinoma of the head and neck cell lines by human papillomavirus. Head Neck. 2011;33:504–512. doi: 10.1002/hed.21475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hui A.B., Lin A., Xu W., Waldron L., Perez-Ordonez B., Weinreb I., Shi W., Bruce J., Huang S.H., O'Sullivan B., Waldron J., Gullane P., Irish J.C., Chan K., Liu F.F. Potentially prognostic miRNAs in HPV-associated oropharyngeal cancer. Clin Cancer Res. 2013;19:2154–2162. doi: 10.1158/1078-0432.CCR-12-3572. [DOI] [PubMed] [Google Scholar]
- 22.Syrjänen S. HPV infections and tonsillar carcinoma. J Clin Pathol. 2004;57:449–455. doi: 10.1136/jcp.2003.008656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duensing A., Spardy N., Chatterjee P., Zheng L., Parry J., Cuevas R., Korzeniewski N., Duensing S. Centrosome overduplication, chromosomal instability, and human papillomavirus oncoproteins. Environ Mol Mutagen. 2009;50:741–747. doi: 10.1002/em.20478. [DOI] [PubMed] [Google Scholar]
- 24.Johannsen E., Lambert P.F. Epigenetics of human papillomaviruses. Virology. 2013;445:205–212. doi: 10.1016/j.virol.2013.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lal A., Thomas M.P., Altschuler G., Navarro F., O'Day E., Li X.L., Concepcion C., Han Y.C., Thiery J., Rajani D.K., Deutsch A., Hofmann O., Ventura A., Hide W., Lieberman J. Capture of microRNA-bound mRNAs identifies the tumor suppressor miR-34a as a regulator of growth factor signaling. PLoS Genet. 2011;7:e1002363. doi: 10.1371/journal.pgen.1002363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lajer C.B., Nielsen F.C., Friis-Hansen L., Norrild B., Borup R., Garnæs E., Rossing M., Specht L., Therkildsen M.H., Nauntofte B., Dabelsteen S., von Buchwald C. Different miRNA signatures of oral and pharyngeal squamous cell carcinomas: a prospective translational study. Br J Cancer. 2011;104:830–840. doi: 10.1038/bjc.2011.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao G., Gay H.A., Chernock R.D., Zhang T.R., Luo J., Thorstad W.L., Lewis J.S.J., Wang X. A microRNA expression signature for the prognosis of oropharyngeal squamous cell carcinoma. Cancer. 2013;119:72–80. doi: 10.1002/cncr.27696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lajer C.B., Garnaes E., Friis-Hansen L., Norrild B., Therkildsen M.H., Glud M., Rossing M., Lajer H., Svane D., Skotte L., Specht L., Buchwald C., Nielsen F.C. The role of miRNAs in human papilloma virus (HPV)-associated cancers: bridging between HPV-related head and neck cancer and cervical cancer. Br J Cancer. 2012;106:1526–1534. doi: 10.1038/bjc.2012.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gunasekharan V., Laimins L.A. Human papillomaviruses modulate microRNA 145 expression to directly control genome amplification. J Virol. 2013;87:6037–6043. doi: 10.1128/JVI.00153-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ventura A., Young A.G., Winslow M.M., Lintault L., Meissner A., Erkeland S.J., Newman J., Bronson R.T., Crowley D., Stone J.R., Jaenisch R., Sharp P.A., Jacks T. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mendell J.T. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133:217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Concepcion C.P., Bonetti C., Ventura A. The microRNA-17-92 family of microRNA clusters in development and disease. Cancer J. 2012;18:262–267. doi: 10.1097/PPO.0b013e318258b60a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pyeon D., Newton M.A., Lambert P.F., den Boon J.A., Sengupta S., Marsit C.J., Woodworth C.D., Connor J.P., Haugen T.H., Smith E.M., Kelsey K.T., Turek L.P., Ahlquist P. Fundamental differences in cell cycle deregulation in human papillomavirus-positive and human papillomavirus-negative head/neck and cervical cancers. Cancer Res. 2007;67:4605–4619. doi: 10.1158/0008-5472.CAN-06-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Slebos R.J., Jehmlich N., Brown B., Yin Z., Chung C.H., Yarbrough W.G., Liebler D.C. Proteomic analysis of oropharyngeal carcinomas reveals novel HPV-associated biological pathways. Int J Cancer. 2013;132:568–579. doi: 10.1002/ijc.27699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thangavel C., Boopathi E., Ertel A., Lim M., Addya S., Fortina P., Witkiewicz A.K., Knudsen E.S. Regulation of miR106b cluster through the RB pathway: mechanism and functional targets. Cell Cycle. 2013;12:98–111. doi: 10.4161/cc.23029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Markey M.P., Angus S.P., Strobeck M.W., Williams S.L., Gunawardena R.W., Aronow B.J., Knudsen E.S. Unbiased Analysis of RB-mediated transcriptional repression identifies novel targets and distinctions from E2F action cancer. Cancer Res. 2002;62:6587–6597. [PubMed] [Google Scholar]
- 37.Strati K., Pitot H.C., Lambert P.F. Identification of biomarkers that distinguish human papillomavirus (HPV)-positive versus HPV-negative head and neck cancers in a mouse model. Proc Natl Acad Sci U S A. 2006;103:14152–14157. doi: 10.1073/pnas.0606698103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petrocca F., Visone R., Onelli M.R., Shah M.H., Nicoloso M.S., de Martino I., Iliopoulos D., Pilozzi E., Liu C.G., Negrini M., Cavazzini L., Volinia S., Alder H., Ruco L.P., Baldassarre G., Croce C.M., Vecchione A. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13:272–286. doi: 10.1016/j.ccr.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 39.Mestdagh P., Boström A.K., Impens F., Fredlund E., Van Peer G., De Antonellis P., von Stedingk K., Ghesquière B., Schulte S., Dews M., Thomas-Tikhonenko A., Schulte J.H., Zollo M., Schramm A., Gevaert K., Axelson H., Speleman F., Vandesompele J. The miR-17-92 microRNA cluster regulates multiple components of the TGF-beta pathway in neuroblastoma. Mol Cell. 2010;40:762–773. doi: 10.1016/j.molcel.2010.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Conkrite K., Sundby M., Mukai S., Thomson J.M., Mu D., Hammond S.M., MacPherson D. miR-17∼92 cooperates with RB pathway mutations to promote retinoblastoma. Genes Dev. 2011;25:1734–1745. doi: 10.1101/gad.17027411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma L., Young J., Prabhala H., Pan E., Mestdagh P., Muth D., Teruya-Feldstein J., Reinhardt F., Onder T.T., Valastyan S., Westermann F., Speleman F., Vandesompele J., Weinberg R.A. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol. 2010;12:247–256. doi: 10.1038/ncb2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muller S., Su L., Tighiouart M., Saba N., Zhang H., Shin D.M., Chen Z. Distinctive E-cadherin and epidermal growth factor receptor expression in metastatic and nonmetastatic head and neck squamous cell carcinoma. Cancer. 2008;113:97–107. doi: 10.1002/cncr.23557. [DOI] [PubMed] [Google Scholar]
- 43.Ukpo O.C., Thorstad W.L., Zhang Q., Lewis J.S., Jr. Lack of association of cadherin expression and histopathologic type, metastasis, or patient outcome in oropharyngeal squamous cell carcinoma: a tissue microarray study. Head Neck Pathol. 2012;6:38–47. doi: 10.1007/s12105-011-0306-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rampias T., Pectasides E., Prasad M., Sasaki C., Gouveris P., Dimou A., Kountourakis P., Perisanidis C., Burtness B., Zaramboukas T., Rimm D., Fountzilas G., Psyrri A. Molecular profile of head and neck squamous cell carcinomas bearing p16 high phenotype. Ann Oncol. 2013;24:2124–2131. doi: 10.1093/annonc/mdt013. [DOI] [PubMed] [Google Scholar]
- 45.Zhuang G., Wu X., Jiang Z., Kasman I., Yao J., Guan Y., Oeh J., Modrusan Z., Bais C., Sampath D., Ferrara N. Tumour secreted miR-9 promotes endothelial cell migration and angiogenesis by activating the JAK-STAT pathway. EMBO J. 2012;31:3513–3523. doi: 10.1038/emboj.2012.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsai K.W., Liao Y.L., Wu C.W., Hu L.Y., Li S.C., Chan W.C., Ho M.R., Lai C.H., Kao H.W., Fang W.L., Huang K.H., Lin W.C. Aberrant hypermethylation of miR-9 genes in gastric cancer. Epigenetics. 2011;6:1189–1197. doi: 10.4161/epi.6.10.16535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bazzoni F., Rossato M., Fabbri M., Gaudiosi D., Mirolo M., Mori L., Tamassia N., Mantovani A., Cassatella M.A., Locati M. Induction and regulatory function of miR-9 in human monocytes and neutrophils exposed to proinflammatory signals. Proc Natl Acad Sci U S A. 2009;106:5282–5287. doi: 10.1073/pnas.0810909106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gao F., Zhao Z.L., Zhao W.T., Fan Q.R., Wang S.C., Li J., Zhang Y.Q., Shi J.W., Lin X.L., Yang S., Xie R.Y., Liu W., Zhang T.T., Sun Y.L., Xu K., Yao K.T., Xiao D. miR-9 modulates the expression of interferon-regulated genes and MHC class I molecules in human nasopharyngeal carcinoma cells. Biochem Biophys Res Commun. 2013;431:610–616. doi: 10.1016/j.bbrc.2012.12.097. [DOI] [PubMed] [Google Scholar]
- 49.Lyford-Pike S., Peng S., Young G.D., Taube J.M., Westra W.H., Akpeng B., Bruno T.C., Richmon J.D., Wang H., Bishop J.A., Chen L., Drake C.G., Topalian S.L., Pardoll D.M., Pai S.I. Evidence for a role of the PD-1:PD-L1 pathway in immune resistance of HPV-associated head and neck squamous cell carcinoma. Cancer Res. 2013;73:1733–1741. doi: 10.1158/0008-5472.CAN-12-2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ukpo O.C., Thorstad W.L., Lewis J.S., Jr. B7-H1 expression model for immune evasion in human papillomavirus-related oropharyngeal squamous cell carcinoma. Head Neck Pathol. 2013;7:113–121. doi: 10.1007/s12105-012-0406-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Signal detection for PCR plates. Four bars represent the relative proportions of CT values for miRNAs detected on the three 384-well PCR plates. Approximately 45% are not expressed (CT of 30 to 34), and only approximately 15% are detected at CT values <30. The proportions of miRNAs falling in detectable ranges are greatly improved by preamplification.
Expression of E-cadherin and miR-9 in oropharyngeal squamous cell carcinoma (OPSCC) cell lines. OPSCC cell lines of known human papillomavirus (HPV) status were cultured in M-10 medium containing l-glutamine, gentamicin, and nonessential amino acids. A: Western blot analysis for E-cadherin. Protein (20 μg) from cell lysates was electrophoresed on SDS-PAGE and electroblotted to polyvinylidene difluoride membranes and probed with anti–E-cadherin primary antibody and rabbit secondary antibody. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a loading control. B: Quantitative RT-PCR for miR-9. RNA was extracted in technical triplicates from cell lines, as indicated, polyadenylated, and reverse transcribed into cDNA in a single reaction. A synthetic RNA spike-in, UniSp6, was added to each sample to monitor reverse transcription efficiency and reproducibility. No concordance is observed between miR-9 and E-cadherin expression.