Abstract
KPC-producing Klebsiella pneumoniae isolates have emerged as important pathogens of nosocomial infections, and tigecycline is one of the antibiotics recommended for severe infections caused by KPC-producing K. pneumoniae. To identify the susceptibility profile of KPC-producing K. pneumoniae to tigecycline and investigate the role of efflux pumps in tigecycline resistance, a total of 215 KPC-producing K. pneumoniae isolates were collected. The minimum inhibitory concentration (MIC) of tigecycline was determined by standard broth microdilution tests. Isolates showing resistance to tigecycline underwent susceptibility test with efflux pump inhibitors. Expression levels of efflux pump genes (acrB and oqxB) and their regulators (ramA, marA, soxS and rarA) were examined by real-time PCR, and the correlation between tigecycline MICs and gene expression levels were analysed. Our results show that the tigecycline resistance rate in these isolates was 11.2%. Exposure of the tigecycline-resistant isolates to the efflux pump inhibitor NMP resulted in an obvious decrease in MICs and restored susceptibility to tigecycline in 91.7% of the isolates. A statistically significant association between acrB expression and tigecycline MICs was observed, and overexpression of ramA was found in three tigecycline-resistant isolates, further analysis confirmed ramR mutations existed in these isolates. Transformation of one mutant with wild-type ramR restored susceptibility to tigecycline and repressed overexpression of ramA and acrB. These data indicate that efflux pump AcrAB, which can be up-regulated by ramR mutations and subsequent ramA activation, contributed to tigecycline resistance in K. pneumoniae clinical isolates.
Introduction
Klebsiella pneumoniae has emerged worldwide as an important pathogen of nosocomial infections that causes a variety of infections, including pneumonia, liver abscesses, urinary-tract infections and bacteraemia. Carbapenems are often the last resort for treating infections due to the emergence of multidrug-resistant K. pneumoniae [1]. However, the acquisition of carbapenemase has contributed to resistance to all β-lactams including carbapenem antibiotics. Carbapenem-hydrolysing Klebsiella pneumoniae carbapenemase (KPC)-type enzymes have been identified mostly in K. pneumoniae. In fact, most KPC carbapenemase-producing K. pneumoniae show resistance to almost all antibiotics except colistin and tigecycline.
Tigecycline, one type of glycylcycline, is a novel expanded-spectrum antibiotic. It is a derivative of minocycline, which inhibits the initial codon recognition step of tRNA accommodation and prevents rescue by the tetracycline resistance protein TetM [2, 3]. Tigecycline is effective against most carbapenemase-producing bacteria including K. pneumoniae and has been approved for clinical use in China during recent years. K. pneumoniae has previously been reported to be non-susceptible to tigecycline in other countries [4, 5]. The resistance rate to tigecycline in multidrug-resistant K. pneumoniae in the USA was approximately 9.2% (MIC≥8 mg/L, FDA) [6], while the resistance rate in ESBL-producing isolates was approximately 33.3% in Spain (MIC >2 mg/L, EUCAST) [7].
The mechanism of tigecycline resistance has not yet been clearly elucidated. It has been reported that the increased expression of efflux pumps such as AcrAB and OqxAB is one of the possible mechanisms [4, 8, 9]. Expression of the acrAB operon is controlled by its local repressor AcrR [10]. Several global transcriptional regulators of the AraC family, RamA, MarA, SoxS, and RarA, may participate in tigecycline resistance via efflux pump activation [5, 11, 12]. ramR, marR and soxR are repressors of ramA, marA and soxS, respectively. RamA is also regulated by the Lon protease [13]. Mutation in these genes might be responsible for ramA, marA and soxS overexpression that subsequently leads to upregulation of the efflux pumps [14–16].
In This study, a total of 215 KPC-producing K. pneumoniae were collected from four hospitals in three provinces in China. We identified the tigecycline susceptibility profiles of these isolates. Furthermore, we investigated the role of efflux pumps and the function of regulators in tigecycline resistance.
Material and Methods
Bacterial isolates
A total of 215 KPC-producing K. pneumoniae isolates were collected between Jan. 2010 and Dec. 2013 from the following centres in China: First Affiliated Hospital, School of Medicine, Zhejiang University (ZJF); Sir Run Run Shaw Hospital, School of Medicine, Zhejiang University (ZJS); The First Affiliated Hospital of Kunming Medical University (KM); The First Affiliated Hospital of Zhengzhou University (ZZ). All isolates were identified using the VITEK 2 system (bioMérieux, France). The bla KPC gene was amplified to confirm the KPC-producing K. pneumoniae isolates [17].
Antimicrobial susceptibility test
The MIC of tigecycline was determined using standard broth microdilution tests with fresh (<12 h) ISO-Sensitest broth (Oxoid LTD, Basingstoke, Hampshire, England). MIC results were interpreted according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) clinical breakpoints (for tigecycline, ≤1.0 mg/L is susceptible, 2.0 mg/L is intermediate, and >2.0 mg/L is resistant) [18]. Escherichia coli ATCC 25922 was used for quality control in the susceptibility assays. Isolates that showed resistance to tigecycline also underwent susceptibility testing for tigecycline by adding efflux pump inhibitors 1-(1-naphthylmethyl)-piperazine (NMP), phenylalanine arginine β-naphthylamide (PAβN) or carbonyl cyanide m-chlorophenylhydrazone (CCCP) to the medium [19]. The MICs of other antimicrobial agents were determined using the agar dilution method or Etest method.
PFGE analysis
Genomic DNA was digested with restriction enzyme XbaI (TaKaRa, Dalian, China), and DNA fragments were separated by electrophoresis in 1% agarose III (Sangon, Shanghai, China) in 0.5× TBE (45 mM Tris, 45 mM boric acid, 1.0 mM EDTA; pH 8.0) buffer with a CHEF apparatus (CHEF Mapper XA, Bio-Rad, USA) at 14°C and 6 V/cm and with alternating pulses at a 120° angle in a 6 to 36 s pulse time gradient for 22 h. The results of PFGE were analysed using BioNumerics 7.0 (Applied Maths, Austin, TX, USA) software.
Real-time PCR
mRNA expression levels of efflux pump genes (acrB and oqxB) and regulators (ramA, marA, soxS and rarA) were examined by real-time PCR. Overnight bacterial cultures were diluted 1/100 in LB broth (Sangon, Shanghai, China) and grown to log phase at 37°C with vigorous shaking (230 rpm). RNase-free DNase (TaKaRa, Dalian, China)-treated RNA was harvested using the Purelink RNA Mini Kit (Ambion, Carlsbad, USA). The yield and quality of RNA were determined using a Nanodrop 2000C (Thermo, USA). Two micrograms of total RNA were reverse transcribed into cDNA using the PrimeScript RT Reagent kit (TaKaRa, Dalian, China). Real-time quantitative RT-PCR was run on a LightCycler 480 II (Roche, Germany) with 40 cycles of 5 s at 95°C, 30 s at 54°C, and 30 s at 72°C. SYBR Premix Ex Taq (TaKaRa, Dalian, China) was used to quantify the expression of the target gene. The reactions were performed in a volume of 20 μL. All experiments were performed in triplicate. The primers used in these experiments are listed in Table 1. Expression of each gene was normalised to that of a housekeeping gene (rpoB). Relative expression of each target gene was then calibrated against the corresponding expression of a tigecycline-susceptible isolate K134 (expression = 1), which served as the control. Data were analysed by using the 2-ΔΔCT method.
Table 1. Primers used for real-time PCR studies and PCR amplification.
| Primers for this study (5’-3’) | Usage | Reference | ||
|---|---|---|---|---|
| rpoB | rpoB-F | CCGTATCTACGCTGTGCT | RT-PCR | This study |
| rpoB-R | TGTTACCGTGACGACCTG | RT-PCR | This study | |
| acrB | acrB-F | CGATAACCTGATGTACATGTCC | RT-PCR | [27] |
| acrB-R | CCGACAACCATCAGGAAGCT | RT-PCR | [27] | |
| oqxB | oqxB-F | CGAAGAAAGACCTCCCTACCC | RT-PCR | This study |
| oqxB-R | CGCCGCCAATGAGATACA | RT-PCR | This study | |
| ramA | ramA-F | GCATCAACCGCTGCGTATT | RT-PCR | This study |
| ramA-R | GGGTAAAGGTCTGTTGCGAAT | RT-PCR | This study | |
| marA | marA-F | TAATGACGCCATCACTATCCA | RT-PCR | This study |
| marA-R | ATGTACTGGCCGAGGGAATG | RT-PCR | This study | |
| soxS | soxS-F | TAGTCGCCAGAAAGTCAGGAT | RT-PCR | This study |
| soxS-R | AGAAGGTTTGCTGCGAGACG | RT-PCR | This study | |
| rarA | rarA-F | GTTTGTTGACGAAGTGCA | RT-PCR | This study |
| rarA-R | GCCATCATTTCCAGGGTA | RT-PCR | This study | |
| ramR | ramR-F | GATGGCGACCACGCTAAA | Amplification | This study |
| ramR-R | GCTCGGTAAACGGGTAGGT | Amplification | This study | |
| lon | lon-F | TCCCGCCGTTGAATGTGTGG | Amplification | This study |
| lon-R | ACTTACCAGCCCTATTTTTAT | Amplification | This study | |
| marR | MarR-F | TAATGTTGACTTATGATTGCCT | Amplification | This study |
| MarR-R | ACATCATCTTACCTCTTCTT | Amplification | This study | |
| soxR | SoxR-F | TTTTGTCTGCGGGCGAGTAT | Amplification | This study |
| SoxR-R | GCGAGATAATGCGAAAGACA | Amplification | This study | |
Statistical analysis
The association between MICs and gene expression levels was analysed using the SPSS Statistics 17.0 software. According to the normality test and homogeneity of variances test, expression levels of soxS, marA, rarA and oqxB appear to be a normal distribution with equal variance, so an analysis of variance (ANOVA) statistical test was adopted. The expression levels of acrB and ramA appear to be a normal distribution with unequal variance, so a Kruskal-Wallis Test was adopted. Statistical significance was established by using a conventional level of P < 0.05.
Mutation analysis of acrR, ramR, marR, soxR and lon
acrR, ramR, marR, soxR and lon, were amplified and sequenced to identify mutations within these genes. The primers designed for these studies are listed in Table 1.
ramR plasmid construction and transformation
A DNA fragment carrying the wild-type ramR gene was amplified from a tigecycline-susceptible isolate (K134) with the primers listed in Table 1. After amplification, the amplimer was cloned into pCR-Blunt II -TOPO. The mutant strain S21 (kanamycin-susceptible) was used for transformation. The influence of the ramR mutation on the tigecycline MIC and transcriptional expression levels of ramA and acrB was examined using standard broth microdilution tests and real-time RT-PCR.
Results
Tigecycline resistance and MICs distribution
Of the 215 KPC-producing K. pneumoniae isolates, 24 isolates were resistant to tigecycline (MIC>2 mg/L). The MIC distribution is presented in Fig. 1. The tigecycline resistance rate for these strains was 11.2% (MIC>2 mg/L, EUCAST). The range of tigecycline MICs was 0.25–8 mg/L. The MIC50 and the MIC90 were 1 and 4 mg/L, respectively.
Fig 1. Tigecycline MIC distribution for 215 KPC-producing K. pneumoniae clinical isolates.
The results of antimicrobial susceptibility testing of tigecycline-resistant isolates are presented in Table 2. All isolates were resistant to multiple antimicrobial agents. Exposure of tigecycline-resistant isolates to the efflux pump inhibitor NMP resulted in an obvious decrease (4 to 16-fold) in the MICs of tigecycline and restored susceptibility for all except two strains (S21 and K23), while susceptible isolate MICs declined slightly from 0.75 to 0.125 mg/L. The effects of PAβN and CCCP were not significant (Table 2).
Table 2. Susceptibilities of 24 tigecycline-resistant isolates to 12 antimicrobial agents and MIC values of tigecycline in the presence of efflux pump inhibitors NMP, PAβN or CCCP.
| Isolate | Clonal group | Hospital | MIC (mg/L) a | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TZP b | CAZ b | FEP b | SCF b | IPM b | MEM b | AK c | CIP c | TS c | MC c | CO c | TGC | TGC+NMP | TGC+PAβN | TGC+CCCP | |||
| K22 | A | ZJF | >256 | 128 | 64 | >256 | 32 | 16 | >256 | >32 | >32 | 8 | 0.25 | 4 | 0.25 | 1 | 1 |
| K23 | B | ZJF | 256 | >256 | 32 | 64 | 16 | 8 | >256 | >32 | >32 | 32 | 0.25 | 8 | 4 | 2 | 8 |
| K83 | C | ZJF | >256 | 128 | 128 | >256 | 128 | 64 | 0.25 | >32 | >32 | 32 | 0.5 | 4 | 1 | 4 | 2 |
| Y13 | D | ZJF | >256 | >256 | >256 | >256 | 128 | 256 | >256 | >32 | 0.25 | 6 | 0.5 | 4 | 1 | 2 | 4 |
| Y17 | E | ZJF | >256 | >256 | >256 | >256 | 256 | >256 | >256 | >32 | 1 | 8 | 0.5 | 8 | 1 | 2 | 4 |
| S21 | F | ZJS | >256 | 256 | 64 | >256 | 8 | 128 | 1.5 | 4 | >32 | >256 | 0.5 | 8 | 2 | 4 | 8 |
| H65 | G | KM | >256 | 256 | 32 | >256 | 64 | 128 | >256 | >32 | 1 | 8 | 0.5 | 8 | 1 | 2 | 8 |
| Q4 | H | ZZ | >256 | >256 | 64 | >256 | 64 | 256 | >256 | >32 | >32 | 4 | 0.5 | 4 | 1 | 2 | 4 |
| Q5 | H | ZZ | >256 | >256 | 256 | >256 | 64 | 128 | >256 | >32 | >32 | 4 | 0.5 | 4 | 1 | 2 | 4 |
| Q6 | H | ZZ | >256 | >256 | 256 | >256 | 64 | 64 | >256 | >32 | >32 | 4 | 0.5 | 4 | 0.5 | 4 | 2 |
| Q8 | H | ZZ | >256 | >256 | 256 | >256 | 64 | 128 | >256 | >32 | >32 | 8 | 0.5 | 4 | 1 | 2 | 2 |
| Q10 | H | ZZ | 256 | >256 | 256 | >256 | 64 | 128 | >256 | >32 | >32 | 8 | 0.5 | 4 | 1 | 4 | 2 |
| Q11 | H | ZZ | >256 | >256 | 256 | >256 | 64 | 128 | >256 | >32 | >32 | 4 | 0.5 | 4 | 0.5 | 2 | 2 |
| Q12 | H | ZZ | >256 | >256 | 256 | >256 | 64 | 64 | >256 | >32 | >32 | 8 | 0.5 | 8 | 0.5 | 4 | 4 |
| Q14 | H | ZZ | >256 | >256 | 256 | >256 | 64 | 128 | >256 | >32 | >32 | 8 | 0.5 | 8 | 0.5 | 4 | 4 |
| Q15 | H | ZZ | >256 | >256 | 256 | >256 | 64 | 64 | >256 | >32 | >32 | 4 | 0.5 | 4 | 0.5 | 4 | 2 |
| Q17 | H | ZZ | >256 | >256 | >256 | >256 | 64 | >256 | 0.5 | >32 | >32 | 4 | 0.5 | 4 | 0.5 | 4 | 2 |
| Q20 | H | ZZ | >256 | >256 | 256 | 256 | 64 | 128 | >256 | >32 | >32 | 4 | 0.5 | 8 | 1 | 4 | 4 |
| Q22 | H | ZZ | >256 | >256 | 256 | >256 | 128 | 256 | >256 | >32 | >32 | 4 | 0.5 | 4 | 0.5 | 2 | 2 |
| Q28 | I | ZZ | >256 | 64 | 256 | >256 | 128 | 256 | 1 | >32 | >32 | 8 | 0.5 | 8 | 0.5 | 4 | 2 |
| Q30 | H | ZZ | >256 | >256 | 64 | >256 | 128 | 256 | 1 | >32 | >32 | 4 | 0.5 | 4 | 0.5 | 4 | 4 |
| Q38 | H | ZZ | >256 | >256 | >256 | >256 | 128 | 128 | >256 | >32 | >32 | 4 | 0.5 | 8 | 0.5 | 4 | 4 |
| Q39 | H | ZZ | >256 | >256 | >256 | >256 | 128 | 256 | >256 | >32 | >32 | 4 | 0.5 | 8 | 0.5 | 4 | 4 |
| Q40 | H | ZZ | >256 | >256 | >256 | >256 | 128 | 128 | 0.5 | >32 | >32 | 4 | 0.5 | 4 | 0.5 | 2 | 4 |
aAbbreviations: TZP, piperacillin/tazobatam; CAZ, ceftazidime; FEP, cefepime; SCF, cefoperazone/sulbactam; IPM, imipenem; MEM, meropenem; AK, amikacin; CIP, ciprofloxacin; TS, trimethoprim/sulfamethoxazole; MC, minocycline; CO, colistin; TGC, tigecycline; TGC+NMP, tigecycline with NMP; TGC+PAβN, tigecycline with PAβN, TGC+CCCP, tigecycline with CCCP
bTested by agar dilution method
cTested by Etest method
PFGE analysis
Three groups of isolates were selected for PFGE analysis; the 24 tigecycline-resistant isolates were designated as group one, a random selection of 24 isolates (equal to the number of tigecycline-resistant isolates) with a MIC = 1 mg/L isolates were designated as group two, and seven total isolates with a MIC = 0.25 mg/L were designated as group three. PFGE analysis revealed that isolates in group one could be divided into nine clonal groups (Fig. 2), group two isolates could be divided into 13 clonal groups (S1 Fig.), and group three isolates could be divided into seven clonal groups (S2 Fig.).
Fig 2. Phylogenetic clone analysis of 24 tigecycline-resistant isolates.
. These isolates were divided into 9 clonal groups (group A to I).
Among the tigecycline-resistant isolates, all five isolates from ZJF hospital are clonally distinct (group A—E). One isolate from ZJS hospital belongs to an individual clonal group (group F), and the isolate from KM hospital also belongs to an individual clonal group (group G). Clone dissemination was observed in ZZ hospital isolates. Sixteen of 17 isolates were clustered together with similar patterns belonging to clone group H, and the other isolate belonged to an individual clonal group (group I).
Gene expression analysis and relationship with tigecycline MICs
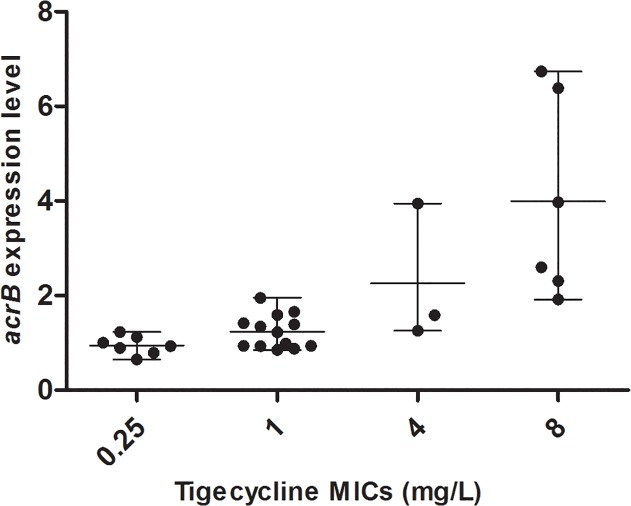
Clonally distinct isolates from each MIC group were selected for gene expression analysis. The gene expression levels of different MIC groups are presented in Table 3. According to the results of the Kruskal-Wallis Test (Table 4), there was a statistically significant association between acrB expression and tigecycline MICs (P<0.05). The Kruskal-Wallis Test indicates that acrB expression level in isolates with MICs of ≥4 mg/L is statistically significantly different from those with MICs of 1 mg/L and 0.25 mg/L and that acrB expression in isolates with MICs of 1 mg/L is also statistically significantly different from MICs of 0.25 mg/L. From these results, we found a trend for higher acrB expression as the tigecycline MIC increases (Fig. 3).
Table 3. Expression of acrB, ramA, soxS, marA, rarA, oqxB and mutation of acrR, ramR in different MIC groups of K. pneumoniae clinical isolates.
| Isolate | MIC(mg/L) c | Relative expression a | Mutation | ||||||
|---|---|---|---|---|---|---|---|---|---|
| acrB | ramA | soxS | marA | rarA | oqxB | acrR | ramR | ||
| K134 | 0.25 | 1 | 1 | 1 | 1 | 1 | 1 | - | - |
| K27 | 0.25 | 0.93±0.09 | 1.56±0.66 | 2.00±1.08 | 4.50±1.36 | 1.65±0.56 | 2.04±1.20 | - | - |
| K32 | 0.25 | 0.89±0.16 | 1.53±0.42 | 1.83±0.86 | 5.05±1.15 | 1.58±1.02 | 2.29±1.18 | - | - |
| K72 | 0.25 | 1.12±0.23 | 0.97±0.37 | 1.65±0.37 | 3.28±1.80 | 1.58±0.51 | 2.47±0.97 | - | - |
| K82 | 0.25 | 0.79±0.07 | 1.00±0.08 | 1.13±0.33 | 3.43±0.69 | 1.05±0.14 | 1.23±0.34 | - | - |
| K135 | 0.25 | 0.65±0.12 | 0.71±0.27 | 0.90±0.44 | 2.79±0.46 | 0.75±0.22 | 0.73±0.33 | - | - |
| K148 | 0.25 | 1.23±0.10 | ND b | 1.92±0.76 | 5.62±1.66 | 1.26±0.16 | 3.55±2.89 | - | - |
| K16 | 1 | 0.98±0.39 | 1.33±0.60 | 1.70±0.76 | 6.01±3.39 | 0.96±0.26 | 1.77±0.76 | - | - |
| K25 | 1 | 0.88±0.18 | 1.77±0.94 | 1.64±1.02 | 4.98±4.09 | 0.92±0.55 | 1.63±1.10 | - | - |
| K29 | 1 | 0.94±0.19 | 1.06±0.50 | 1.36±0.74 | 4.21±1.87 | 1.28±0.92 | 1.54±1.10 | - | - |
| K49 | 1 | 0.85±0.19 | 0.73±0.41 | 0.79±0.50 | 2.64±1.38 | 0.62±0.23 | ND b | - | - |
| K76 | 1 | 0.93±0.17 | 1.49±0.49 | 1.74±0.58 | 5.51±1.95 | 1.30±0.33 | 2.50±0.19 | - | - |
| K101 | 1 | 1.42±0.21 | 0.98±0.51 | 1.09±0.67 | 3.38±1.87 | 0.98±0.43 | 1.17±0.56 | IS5 d | - |
| K128 | 1 | 0.94±0.28 | ND b | 1.52±0.95 | 4.99±3.83 | 0.98±0.28 | 1.68±0.80 | - | - |
| K155 | 1 | 1.22±0.34 | 0.96±0.57 | 1.12±0.59 | 3.93±2.33 | 0.68±0.27 | ND b | IS5 d | - |
| Y8 | 1 | 1.39±0.25 | ND b | 1.50±1.31 | 4.62±4.25 | 0.98±0.52 | ND b | IS5 d | - |
| H33 | 1 | 1.34±0.42 | 1.14±0.68 | 1.34±0.72 | 4.23±2.51 | 0.87±0.42 | ND b | IS5 d | - |
| S7 | 1 | 1.95±0.15 | 2.07±0.18 | 2.33±0.11 | 6.86±0.84 | 1.51±0.03 | 2.91±0.34 | IS5 d | - |
| S10 | 1 | 1.66±0.16 | 1.38±0.41 | 1.53±0.50 | 4.39±1.96 | 1.30±0.20 | ND b | IS5 d | - |
| S17 | 1 | 1.59±0.18 | ND b | 1.00±0.42 | 3.24±1.43 | 0.95±0.22 | ND b | IS5 d | - |
| K22 | ≥4 | 1.26±0.21 | 1.10±0.39 | 0.99±0.43 | 3.54±1.11 | 0.76±0.11 | 1.00±0.28 | A20D | - |
| K23 | ≥4 | 1.92±0.40 | 0.29±0.12 | 0.17±0.06 | 1.02±0.13 | 0.37±0.11 | 3.04±0.87 | A20D | - |
| K83 | ≥4 | 1.58±0.15 | 2.71±0.41 | 2.24±0.64 | 11.48±1.76 | 1.56±0.35 | 3.35±0.29 | - | - |
| Y13 | ≥4 | 3.94±0.30 | 1.93±0.50 | 1.02±0.21 | 3.89±0.98 | 0.82±0.07 | ND b | IS5 d | - |
| Y17 | ≥4 | 6.38±2.64 | 9.43±3.89 | 0.67±0.48 | 2.11±1.08 | 0.70±0.22 | 1.24±0.48 | IS5 d | E113K |
| H65 | ≥4 | 6.74±1.06 | 7.82±2.17 | 1.17±0.24 | 4.49±1.76 | 1.06±0.08 | ND b | IS5 d | I106F |
| S21 | ≥4 | 3.97±0.49 | 13.77±2.90 | 1.94±0.74 | 7.19±2.25 | 1.88±0.71 | 2.11±0.94 | - | Q122Stop |
| Q28 | ≥4 | 2.31±0.28 | 0.73±0.08 | 1.01±0.22 | 2.30±0.68 | 0.67±0.09 | ND b | IS5 d | - |
| Q38 | ≥4 | 2.60±0.43 | 1.67±0.42 | 1.84±0.66 | 5.27±1.67 | 1.79±1.17 | 2.84±1.73 | IS5 d | - |
a Relative expression compared with K134 (expression = 1). Results are means of 3 runs ± standard deviation.
b ND, not determined. Mutations in primer region or gene deletion may have affected expression.
c MIC of tigecycline.
d IS5 insertion element into the nucleotide position 276–277 of the acrR gene.
Table 4. Kruskal-Wallis Test of acrB and ramA expression on the tigecycline MICs.
| Gene | Number of clonally distinct isolates | MIC | x̅±s | χ2 | P |
|---|---|---|---|---|---|
| acrB | 7 | 0.25 | 0.94±0.20 | 16.201 | 0.001 |
| 13 | 1 | 1.24±0.35 | |||
| 9 | ≥4 | 3.41±2.02 | |||
| ramA | 6 a | 0.25 | 1.13±0.34 | 3.345 | 0.188 |
| 10 b | 1 | 1.29±0.41 | |||
| 9 | ≥4 | 4.38±4.78 |
a One isolate was not determined.
b Three isolate were not determined.
Fig 3. Relationship between acrB expression levels and tigecycline MICs.
The relationship between oqxB expression and tigecycline MICs was not evident (Table 5). High expression of oqxB was found both in tigecycline resistant and susceptible isolates (Table 3). The role of OqxAB pump in tigecycline resistance was uncertain.
Table 5. ANOVA of marA, soxS, rarA and oqxB expression on the tigecycline MICs.
| Gene | Number of clonally distinct isolates a | MIC | x̅±s | F | P |
|---|---|---|---|---|---|
| marA | 7 | 0.25 | 3.67±1.56 | 0.495 | 0.615 |
| 13 | 1 | 4.54±1.16 | |||
| 9 | ≥4 | 4.59±3.17 | |||
| soxS | 7 | 0.25 | 1.49±0.47 | 0.650 | 0.530 |
| 13 | 1 | 1.44±0.39 | |||
| 9 | ≥4 | 1.23±0.66 | |||
| rarA | 7 | 0.25 | 1.27±0.35 | 0.930 | 0.407 |
| 13 | 1 | 1.03±0.26 | |||
| 9 | ≥4 | 1.07±0.54 | |||
| oqxB | 7 | 0.25 | 1.90±0.99 | 0.382 | 0.688 |
| 7 a | 1 | 1.89±0.60 | |||
| 6 b | ≥4 | 2.26±0.98 |
a Five isolates were not determined.
b Three isolates were not determined.
Overexpression of ramA were found in tigecycline resistant isolates (Y17, H65, S21) which also with high expression level of acrB (Table 3). Overexpression of ramA could contribute to the up-regulation of acrB in these isolates. The difference of marA, soxS, and rarA expression between tigecycline resistant isolates and susceptible isolates were not significant.
Mutation analysis of acrR, ramR, marR, soxR and lon
The sequences of the lon, marR and soxR regions of tigecycline-resistant isolates are identical to those of the susceptible isolates, and the sequences also align with the reference sequence of tigecycline-susceptible isolate K. pneumoniae subsp. pneumoniae MGH 78578 (GenBank accession no. CP000647) [15].
Mutations in the acrR gene were observed in 14 isolates (Table 3), and 12 isolates have IS5 insertion element into the nucleotide position 276–277 of the gene and two isolates harboured point mutations. The expression level of the acrB gene in the isolates harboring mutant acrR genes were higher than the control strain K134 which with a wild-type acrR gene, but not significant.
Mutations in the ramR gene were observed in three resistant isolates (Y17, H65, S21). One isolate (S21) harboured a point mutation leading to a premature stop codon, which resulted in a predicted truncated RamR protein that was most likely non-functional, and two isolates (Y17, H65) harboured point mutations leading to amino acid exchanges in the coding region of ramR (Table 3). Overexpressing of ramA could be due to the mutations of ramR in these strains.
ramR mutations contribute to tigecycline resistance in clinically isolated Klebsiella pneumoniae
Transformation of S21 with wild-type ramR K134 (S21/ramR K134) lowered the MIC for tigecycline from 8 mg/L to 1 mg/L. The influence of ramR mutations on the transcript expression levels of ramA and acrB in S21 was analysed by real-time PCR. Transformation of wild-type ramR (from K134) into S21 (S21/ramR K134) resulted in strongly repressed ramA expression (14.20-fold), and acrB expression was also downregulated (4.73-fold) (Table 6). No change was noted when transformed with the empty pCR-BluntII -TOPO vector (S21/ pCR-BluntII -TOPO). These data indicate that ramR mutations via ramA activation subsequently resulted in the up-regulation of efflux pump acrAB contribute to tigecycline resistance in K. pneumoniae clinical isolates.
Table 6. Tigecycline MIC and relative expressions of ramA and acrB when complemented with wild-type ramR in S21.
| Isolates | MIC (mg/L) b | ramR mutations | Relative expression a | |
|---|---|---|---|---|
| ramA | acrB | |||
| S21 | 8 | Q122Stop | 13.77±2.90 | 3.97±0.49 |
| S21/ramR K134 | 1 | 0.97±0.22 | 0.84±0.14 | |
| S21/ pCR-BluntⅡ-TOPO | 8 | 12.91±1.77 | 3.55±0.41 | |
| K134 | 0.25 | 1 | 1 | |
a Relative expression compared with K134 (expression = 1). Results are means of 3 runs ± standard deviation.
b MIC of tigecycline.
Discussion
KPC-producing K. pneumoniae isolates have emerged as important pathogens of nosocomial infections. These strains often show resistance to almost all antibiotics, and their worldwide spread usually causes a great threat to public health. Tigecycline is one of the antibiotics recommended for severe infections caused by KPC-producing K. pneumoniae [20]. In This study, we identified the susceptibility profile of tigecycline in KPC-producing K. pneumoniae in China. The MIC range and MIC90 in these isolates are identical to previous findings in the USA in which the activity of tigecycline was determined in multidrug-resistant K. pneumoniae [6]. According to US FDA criteria, the resistance rate in these isolates was 4.7% (MIC ≥8 mg/L), and the susceptibility rate was 88.8% (MIC≤2 mg/L). It appears that tigecycline retains good activity against KPC-producing K. pneumoniae in vitro.
Tigecycline resistance occurring in K. pneumoniae during therapy has recently been reported in many cases [21–24]. The mechanism of resistance is not yet clear. However, it is becoming apparent that the development of resistance to tigecycline is rather complicated, and more than one mechanism may be involved. In This study, 24 isolates belong to 9 clonal groups show resistance to tigecycline. Exposure of these isolates to the efflux pump inhibitor NMP resulted in an obvious decrease in the MICs of tigecycline and restored susceptibility to tigecycline in 91.7% of the isolates. These data suggests that efflux pumps are involved in decreased tigecycline susceptibility. However, the effects of PAβN and CCCP were not significant. Kern WV et al. deem that different antibiotics may have different binding sites on the pump with which the EPIs might interfere in a variable manner [25]. The different effect of the three EPIs on tigecycline MICs might be due to the different action mode of the EPIs and the particular binding sites of tigecycline. AcrAB-TolC, an RND-type efflux pump, has been linked to the non-susceptibility of tigecycline in a variety of Enterobacteriaceae [4, 8, 15, 16, 26]. Our results support the hypothesis that increased expression of the AcrAB pump is associated with increased MICs of tigecycline in K. pneumoniae. However, in two strains (K22, K83), the expression level of acrB was relatively low, suggesting that efflux pumps other than AcrAB may take effect in these strains. We also examined the expression level of oqxB in This study, but the role of OqxAB pump in tigecycline resistance was uncertain.
Transcriptional activators RamA, MarA, SoxS, and RarA have been linked to efflux pump-mediated resistance to tigecycline [5, 11, 12, 16]. However, the difference of marA, soxS, and rarA expression between tigecycline resistant and susceptible isolates were not significant, which indicating marA, soxS, and rarA may not in the domination position in tigecycline resistance of K. pneumoniae.
Mutations in the acrR gene were observed in some isolates. The expression level of the acrB gene in the isolates harboring mutant acrR genes were higher than those with a wild-type acrR gene, but not significant. The acrR gene mutations may partly contribute to AcrAB pump-mediated tigecycline resistance in K. pneumoniae. Mutations in the ramR gene were observed in three tigecycline resistant isolates (Y17, H65, S21). The high expression of acrB in isolate S21 could be due to mutations in ramR which led to the up-regulation of ramA. Moreover, isolates Y17 and H65 both have acrR mutation and ramR mutation, and the two mutations may together contribute to the overexpression of acrB.
For the one tigecycline-resistant strain (K23) that is insensitive to efflux pump inhibitors and had low expression of all genes examined in This study, it is likely that other mechanisms are involved in the development of tigecycline resistance.
Supporting Information
These isolates were divided into 13 clonal groups.
(TIF)
These isolates were divided into 7 clonal groups.
(TIF)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by research grants from the National Natural Science Foundation of China (No. NSFC81230039 and No. NSFC81000757). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Qi Y, Wei Z, Ji S, Du X, Shen P, Yu Y. ST11, the dominant clone of KPC-producing Klebsiella pneumoniae in China. J Antimicrob Chemother. 2011;66: 307–312. 10.1093/jac/dkq431 [DOI] [PubMed] [Google Scholar]
- 2. Pankey GA. Tigecycline. J Antimicrob Chemother. 2005;56: 470–480. [DOI] [PubMed] [Google Scholar]
- 3. Jenner L, Starosta AL, Terry DS, Mikolajka A, Filonava L, Yusupov M, et al. Structural basis for potent inhibitory activity of the antibiotic tigecycline during protein synthesis. Proc Natl Acad Sci U S A. 2013;110: 3812–3816. 10.1073/pnas.1216691110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ruzin A, Visalli MA, Keeney D, Bradford PA. Influence of transcriptional activator RamA on expression of multidrug efflux pump AcrAB and tigecycline susceptibility in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2005;49: 1017–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ruzin A, Immermann FW, Bradford PA. Real-time PCR and statistical analyses of acrAB and ramA expression in clinical isolates of Klebsiella pneumoniae. Antimicrob Agents Chemother. 2008;52: 3430–3432. 10.1128/AAC.00591-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. DiPersio JR, Dowzicky MJ. Regional variations in multidrug resistance among Enterobacteriaceae in the USA and comparative activity of tigecycline, a new glycylcycline antimicrobial. Int J Antimicrob Agents. 2007;29: 518–527. [DOI] [PubMed] [Google Scholar]
- 7. Vazquez MF, Romero ED, Garcia MI, Rodriguez JA, Bellido JL. Comparative in vitro activity of tigecycline against enterobacteria producing two or more extended-spectrum beta-lactamases. Int J Antimicrob Agents. 2008;32: 541–543. 10.1016/j.ijantimicag.2008.06.024 [DOI] [PubMed] [Google Scholar]
- 8. Roy S, Datta S, Viswanathan R, Singh AK, Basu S. Tigecycline susceptibility in Klebsiella pneumoniae and Escherichia coli causing neonatal septicaemia (2007–10) and role of an efflux pump in tigecycline non-susceptibility. J Antimicrob Chemother. 2013;68: 1036–1042. 10.1093/jac/dks535 [DOI] [PubMed] [Google Scholar]
- 9. De Majumdar S, Veleba M, Finn S, Fanning S, Schneiders T. Elucidating the regulon of multidrug resistance regulator RarA in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2013;57: 1603–1609. 10.1128/AAC.01998-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schneiders T, Amyes SG, Levy SB. Role of AcrR and ramA in fluoroquinolone resistance in clinical Klebsiella pneumoniae isolates from Singapore. Antimicrob Agents Chemother. 2003;47: 2831–2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bratu S, Landman D, George A, Salvani J, Quale J. Correlation of the expression of acrB and the regulatory genes marA, soxS and ramA with antimicrobial resistance in clinical isolates of Klebsiella pneumoniae endemic to New York City. J Antimicrob Chemother. 2009;64: 278–283. 10.1093/jac/dkp186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Veleba M, Schneiders T. Tigecycline resistance can occur independently of the ramA gene in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2012;56: 4466–4467. 10.1128/AAC.06224-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ricci V, Blair JM, Piddock LJ. RamA, which controls expression of the MDR efflux pump AcrAB-TolC, is regulated by the Lon protease. J Antimicrob Chemother. 2014;69: 643–650. 10.1093/jac/dkt432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bialek-Davenet S, Marcon E, Leflon-Guibout V, Lavigne JP, Bert F, Moreau R, et al. In vitro selection of ramR and soxR mutants overexpressing efflux systems by fluoroquinolones as well as cefoxitin in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2011;55: 2795–2802. 10.1128/AAC.00156-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hentschke M, Wolters M, Sobottka I, Rohde H, Aepfelbacher M. ramR mutations in clinical isolates of Klebsiella pneumoniae with reduced susceptibility to tigecycline. Antimicrob Agents Chemother. 2010;54: 2720–2723. 10.1128/AAC.00085-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Keeney D, Ruzin A, McAleese F, Murphy E, Bradford PA. MarA-mediated overexpression of the AcrAB efflux pump results in decreased susceptibility to tigecycline in Escherichia coli. J Antimicrob Chemother. 2008;61: 46–53. [DOI] [PubMed] [Google Scholar]
- 17. Shen P, Wei Z, Jiang Y, Du X, Ji S, Yu Y, et al. Novel genetic environment of the carbapenem-hydrolyzing beta-lactamase KPC-2 among Enterobacteriaceae in China. Antimicrob Agents Chemother. 2009;53: 4333–4338. 10.1128/AAC.00260-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.EUCAST. Breakpoint tables for interpretation of MICs and zone diameters. Version 3.1. 2013.
- 19. Schumacher A, Steinke P, Bohnert JA, Akova M, Jonas D, Kern WV. Effect of 1-(1-naphthylmethyl)-piperazine, a novel putative efflux pump inhibitor, on antimicrobial drug susceptibility in clinical isolates of Enterobacteriaceae other than Escherichia coli. J Antimicrob Chemother. 2006;57: 344–348. [DOI] [PubMed] [Google Scholar]
- 20. Peleg AY, Hooper DC. Hospital-acquired infections due to gram-negative bacteria. N Engl J Med. 2010;362: 1804–1813. 10.1056/NEJMra0904124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rodriguez-Avial C, Rodriguez-Avial I, Merino P, Picazo JJ. Klebsiella pneumoniae: development of a mixed population of carbapenem and tigecycline resistance during antimicrobial therapy in a kidney transplant patient. Clin Microbiol Infect. 2012;18: 61–66. 10.1111/j.1469-0691.2011.03482.x [DOI] [PubMed] [Google Scholar]
- 22. Tsai HY, Liao CH, Cheng A, Liu CY, Huang YT, Sheng WH, et al. Emergence of tigecycline-resistant Klebsiella pneumoniae after tigecycline therapy for complicated urinary tract infection caused by carbapenem-resistant Escherichia coli. J Infect. 2012;65: 584–586. 10.1016/j.jinf.2012.09.007 [DOI] [PubMed] [Google Scholar]
- 23. Nigo M, Cevallos CS, Woods K, Flores VM, Francis G, Perlman DC, et al. Nested case-control study of the emergence of tigecycline resistance in multidrug-resistant Klebsiella pneumoniae. Antimicrob Agents Chemother. 2013;57: 5743–5746. 10.1128/AAC.00827-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Duin D, Cober E, Richter SS, Perez F, Cline M, Kaye KS, et al. Tigecycline Therapy for Carbapenem-Resistant Klebsiella pneumoniae (CRKP) Bacteriuria Leads to Tigecycline Resistance. Clin Microbiol Infect. 2014. [DOI] [PMC free article] [PubMed]
- 25. Kern WV, Steinke P, Schumacher A, Schuster S, von Baum H, Bohnert JA. Effect of 1-(1-naphthylmethyl)-piperazine, a novel putative efflux pump inhibitor, on antimicrobial drug susceptibility in clinical isolates of Escherichia coli. J Antimicrob Chemother. 2006;57: 339–343. [DOI] [PubMed] [Google Scholar]
- 26. Keeney D, Ruzin A, Bradford PA. RamA, a transcriptional regulator, and AcrAB, an RND-type efflux pump, are associated with decreased susceptibility to tigecycline in Enterobacter cloacae. Microb Drug Resist. 2007;13: 1–6. [DOI] [PubMed] [Google Scholar]
- 27. Doumith M, Ellington MJ, Livermore DM, Woodford N. Molecular mechanisms disrupting porin expression in ertapenem-resistant Klebsiella and Enterobacter spp. clinical isolates from the UK. J Antimicrob Chemother. 2009;63: 659–667. 10.1093/jac/dkp029 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
These isolates were divided into 13 clonal groups.
(TIF)
These isolates were divided into 7 clonal groups.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.