Abstract
OBJECTIVE
To examine the levels of Oxalobacter formigenes in probiotic supplements marketed by ™PRO Lab, Ltd, Toronto, Canada, and capsules of Oxalo™ purchased from Sanzyme Ltd, Hyderabad, India, and to measure the ability of these preparations to degrade oxalate in vitro.
METHODS
Probiotic supplements and pure cultures of O. formigenes were cultured in a number of media containing oxalate. OD595 was used to measure bacterial growth and ion chromatography was used to measure loss of oxalate in culture media. O. formigenes specific and degenerate Lactobacillus primers to the oxalate decarboxylase gene (oxc) were used in PCR.
RESULTS
Incubating probiotic supplements in different media did not result in growth of oxalate-degrading organisms. PCR indicated the absence of organisms harboring the oxc gene. Culture and 16S rRNA gene sequencing indicated the ™PRO Lab supplement contained viable Lactococcus lactis subsp. lactis (GenBank accession no. KJ095656.1), while Oxalo™ contained several Bacillus species and Lactobacillus plantarum.
CONCLUSION
The probiotic supplement sold over the internet by ™PRO Lab, Ltd and Sanzyme Ltd did not contain identifiable O. formigenes or viable oxalate-degrading organisms, and they are unlikely to be of benefit to calcium oxalate kidney stone patients.
Keywords: Oxalobacter formigenes, oxalate, probiotic, calcium oxalate kidney stone disease
INTRODUCTION
Calcium oxalate kidney stone patients may consider purchasing kidney stone probiotic supplements sold by ™PRO Lab, Ltd, Toronto, Canada, or Oxalo™, from Sanzyme, Ltd, Hyderabad, India. The ™PRO Lab, Ltd website implies the supplement contains the highly efficient, oxalate-degrading, intestinal bacterium, Oxalobacter formigenes, while Sanzyme Ltd directly states their Oxalo™ product contains O. formigenes. O. formigenes is a non-pathogenic, Gram-negative, obligate anaerobic bacterium that commonly inhabits the human gut and degrades oxalate as its major energy and carbon source.1 Because of the contribution of dietary oxalate to calcium oxalate stone disease, the potential relationship of this organism to intestinal oxalate balance, urinary oxalate excretion and calcium oxalate kidney stone formation has attracted considerable attention.2 Whether high oral doses of this organism can promote sufficient intestinal oxalate secretion to diminish the oxalate burden on the kidney in individuals with Primary Hyperoxaluria is currently being tested in a clinical trial financed by the biopharmaceutical company OxThera AB (http://www.oxthera.com/). The association between recurrent calcium oxalate stone disease and colonization with O. formigenes was assessed in a study of 247 calcium oxalate stone formers and 259 matched controls.3 The odds ratio for forming a recurrent stone when colonized was 0.3; i.e., a 70% reduction in stone risk. Controlled dietary studies have also indicated colonized individuals excrete lower levels of urinary oxalate4,5 and have lower levels of plasma oxalate.5 A review of the colonization frequencies conducted worldwide indicates that 38 – 77% of a normal population is colonized and it was consistently observed that the colonization frequency in calcium oxalate stone formers was about half that in normal subjects.3,6 Several studies have indicated that the intake of antibiotics can result in the loss of colonization6–8, and this is supported by lower prevalence of O. formigenes in both cystic fibrosis patients9, and calcium oxalate stone formers who are frequently prescribed antibiotics.8,10 It is also possible that a lower rate of colonization in stone formers is due to patients restricting dietary oxalate intake. To date, there has only been one study to examine factors that impact colonization, and in this study6 only a slight (non-significant) trend was observed between prevalence of colonization (simply whether or not a person was colonized with O. formigenes) in normal subjects and oxalate intake. The impact of oxalate deprivation on O. formigenes colonization warrants further examination.
O. formigenes colonization may prove to be an efficacious and inexpensive method for limiting calcium oxalate stone risk. The goals of this study were to evaluate the levels of O. formigenes in kidney stone probiotic supplements sold by ™PRO Lab, Ltd, and Oxalo™, purchased from Sanzyme, Ltd, and to measure the ability of these preparations to degrade oxalate in culture.
MATERIALS AND METHODS
Culture conditions
Pure cultures of O. formigenes, strain OxCC13, were grown at 37°C in either Schaedler broth (SBO, BD Biosciences), medium B1 (an undefined medium with minerals, metals, cysteine, carbonate buffering system, oxalate, acetate, and 0.1% yeast extract), or medium B with 0.5% yeast extract (OXMY). All media were supplemented with 100 mM sodium oxalate and 10 mM sodium acetate.
For determination of colony forming units (CFU) on solid plate medium, a variation of medium B was used that contained 40 mM sodium oxalate, 7 mM CaCl2, 0.5% yeast extract, 0.1% Na2CO3, and 2.0% Bacto agar (OXMC). Plates were incubated in anaerobic containers using GasPak EZ Anaerobe Container System with Indicator (BD Biosciences) sachets at 37°C.
OD595 measurements were performed using a Biotek Synergy HT plate reader and pureGrade BRANDplates®, 96 well, Transparent, F-bottom.
A single batch preparation of a 60-day supply of the dehydrated kidney stone probiotic supplement (catalog # 1246-87, Lot # 460-04) was purchased November 23, 2013, from ™PRO Lab (http://www.probiotic-lab.com). One 10 × 10 capsules box of Oxalo™ (http://www.nu-division.com/oxalo.html) was purchased April 26, 2014. The expiration date listed on the box of Oxalo™ was June 2015. There was no expiration date listed on the ™PRO Lab supplement. The Oxalo™ product claimed to contain no less than 2.5 billion cells per capsule of O. formigenes, Lactobacillus acidophilus, Bifidobacterium lactis, and Bacillus coagulans. All culture experiments were completed within three months of purchase date. Culture experiments were conducted in triplicate.
The ™PRO Lab supplement powder (5 mg/ml) was directly added to medium B, SBO, Brain Heart Infusion (BHI, BD Biosciences) and Lactobacilli MRS broth (BD Biosciences) supplemented with 20 mM sodium oxalate. As recommended by the company, 1 g powder was reconstituted overnight in 500 ml sterile water and then a 10% inoculum was added to BHI and MRS without added oxalate, and MRS broth, SBO, and medium B supplemented with 20 mM oxalate. Cultures grown in BHI and MRS were grown without shaking, and cultures grown in SBO, medium B, and MRS containing oxalate were grown in anaerobically sealed serum vials or Hungate tubes. Serial dilutions of overnight sterile water reconstituted cultures were also cultured onto BHI plates containing 2.0% Bacto agar, and L agar plates (1.0% tryptone, 0.5% yeast extract, 0.5% NaCl, and 1.5% Bacto agar), and incubated aerobically.
One Oxalo™ capsule (0.3 g) was cultured in 5 ml of anaerobic medium B containing 20 mM sodium oxalate. In some experiments following the addition of the Oxalo™ capsule to the medium, the initial enrichment culture was serially diluted (~10−1 to 10−6) in anaerobic 20 mM oxalate medium B. These cultures were incubated at 37°C for 14 days. In addition, a capsule was dissolved in 5 ml of BHI, and 0.5 ml of this culture was added to 10 ml of BHI, SBO, MRS, and these cultures were grown without shaking. A 5% inoculum from the BHI culture was also added to SBO, OXMY, and Schaedler broth containing no oxalate, and these cultures were grown in anaerobically sealed Hungate tubes. For culture on solid medium, the dissolved capsule was plated on a variation of medium B that contained 40 mM sodium oxalate, 0.5% yeast extract, 0.1% Na2CO3, and 2.0% Bacto agar. These plates were grown aerobically and in anaerobic containers using GasPak EZ Anaerobe Container System with Indicator sachets.
Oxalate Assay
Following incubation in various media containing 20 mM oxalate, the oxalate content was measured by ion chromatography using an AS22 column, as previously described.4
DNA extraction
DNA was extracted from pure cultures of O. formigenes using Promega's Wizard Genomic DNA Purification Kit according to the manufacturer’s protocol. DNA was extracted from the supplement powder or Oxalo™ capsule using Maxwell DNA Tissue Purification Kit (Promega) according to the manufacturer’s instructions.
PCR analysis and sequencing
Primers were designed to amplify a 1284-bp sequence of oxalyl coenzyme A decarboxylase (oxc) gene from O. formigenes strain OxCC13. The forward primer was oxc7 (5’-ATGTAGAGTTGACTGATGGC-3’), and the reverse primer was oxc4 (5’-AGCCCATACCAATACCCATAAC-3’). Degenerate primers, oxc9 and oxc8 (5’-ATGTATGGTGTTGTMGGYATT-3’ and 5’-TCMAMGTAAACACCACCTGGA-3’) were also used to amplify Lactobacillus oxc.11 The 16S rRNA gene was amplified with the universal bacterial primers 515F and 805R (5’-GTGCCAGCMGCCGCGGTAA-3’ and 5’-GACTACCAGGGTATCTAATC-3’). PCR was performed with GoTaq Green Master Mix (Promega). For sequencing, PCR products were purified using Wizard SV Gel and PCR Clean Up System (Promega) following the manufacturer’s protocol. Sequencing of the 16S rRNA gene was done using primer 515F. Sequencing was performed by the Heflin Center Genomics Core Facility (University of Alabama at Birmingham, Birmingham, Alabama).
RESULTS
Culture
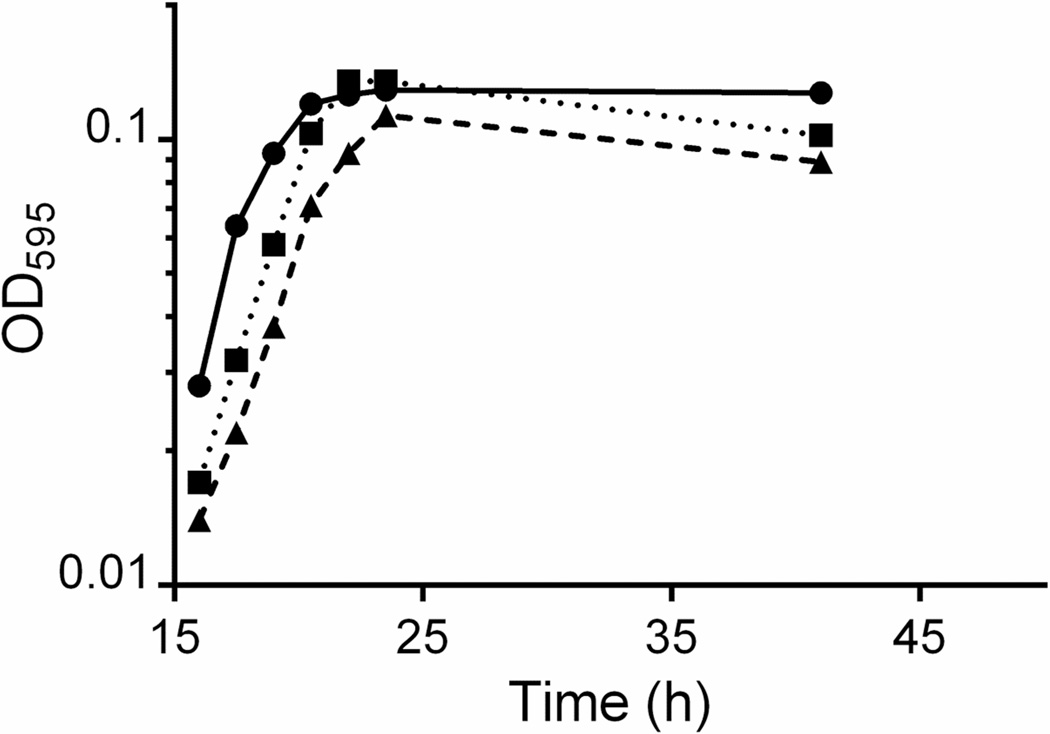
Growth curves of pure cultures of OxCC13 are shown in Fig. 1. OxCC13 grew to a higher density at any given time point in SBO compared to OXMY and medium B. OxCC13 growth in medium B reached a lower maximum OD than growth in SBO and OXMY. OxCC13 showed similar specific growth rates during log growth in all media, μ = 0.34 ± 0.01, equivalent to a doubling time of 2.1 ± 0.1 hours. Plating OxCC13 indicated an OD595 of 0.10, close to the end of logarithmic growth, was equivalent to 1.1 × 108 CFU/ml. OxCC13 appeared as translucent, smooth colonies on OXMC plates, and after 3 days the colonies grew to a uniform size of approximately 1 mm in diameter.
Figure 1.
Growth of O. formigenes OxCC13. Representative growth in SBO (●), medium B (- -▲- -), and medium B supplemented with 0.5% yeast (··■··). All media contained 100mM sodium oxalate and 10mM sodium acetate. Starter cultures were grown to end of log growth (OD595 0.1), and then diluted 3/10,000 into each medium to monitor growth over time.
To determine whether O. formigenes and other oxalate-degrading organisms were present in the probiotic batch preparation obtained from ™PRO Lab, Ltd, the supplement was either directly, or after overnight reconstitution in sterile water, incubated in SBO, BHI, MRS broth, and medium B containing 20 mM oxalate. No growth, as determined by OD595, was observed in any of the media to which powder was directly added, and no growth was observed in SBO and medium B to which overnight reconstituted supplement was added. In contrast, overnight reconstituted supplement incubated in BHI media and MRS broth with and without oxalate showed evidence of bacterial growth. However, IC measurement of media oxalate showed no loss of oxalate in any of the cultures. In contrast, less than 0.1 mM oxalate remained after culture with pure OxCC13 in SBO and medium B.
The overnight reconstituted powder was plated on BHI and L agar plates containing no oxalate. Colonies with similar size and morphology were observed on both plates. The average CFU/ml from three independent experiments plated onto BHI was 8.8 × 106 (standard error 3.1 × 103). The overnight supplement culture was also plated on OXMC, and after 14 days no colonies or clear zones, which are indicative of oxalate degradation, were visible.
All Oxalo™ medium B cultures (diluted or non-diluted) showed no loss of oxalate after 14 days incubation. Bacterial growth was evident in BHI, MRS, SBO, and OXMY; however, there was no loss of media oxalate.
PCR analysis and sequencing
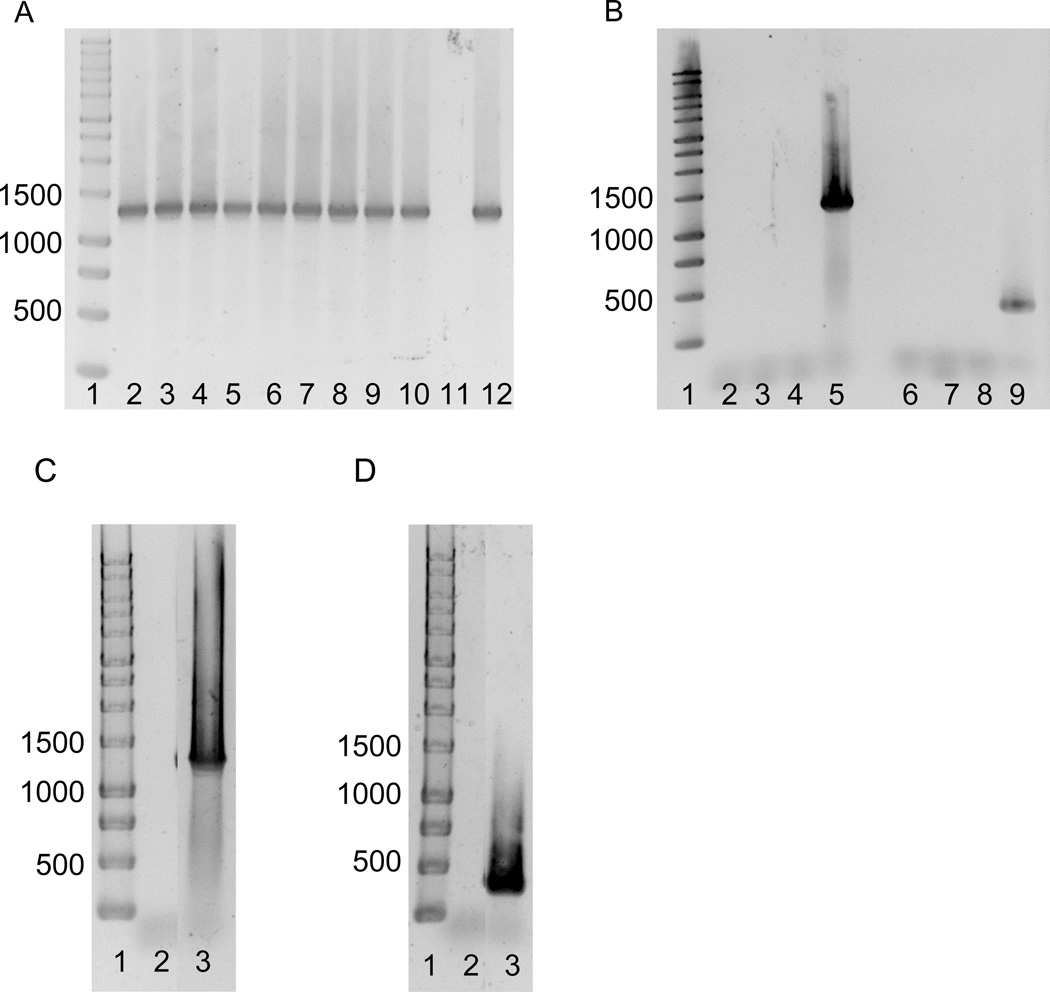
DNA was isolated from both probiotic supplements and a number of pure O. formigenes strains: OxB, BA1, Sox4, OxCC13, OxWR, HOxBLS, OxK, HC1, and OxGP. PCR was performed using both O. formigenes oxc-specific primers (Fig. 2A), and degenerate primers designed to detect oxc in Lactobacillus strains (Fig 2B). All O. formigenes strains tested with oxc-specific primers exhibited a band of the expected size (1284 bp). Strain OxCC13 was also tested with the degenerate Lactobacillus primers and exhibited a band of the expected size (419 bp). However, no PCR product was detected in either probiotic supplement using both O. formigenes specific and degenerate Lactobacillus oxc primers (Fig 2).
Figure 2.
Amplification of oxc.
(A) All Oxalobacter strains tested with oxc-specific primers exhibited a band of the expected size (1284 bp). Lanes: 1, molecular weight ladder (bp); 2, OxB; 3, BA-1; 4, Sox4; 5, OxCC13; 6, OxWR; 7, HOxBLS; 8, OxK; 9, HC1; 10, OxGP; 11, no DNA control; 12, purified DNA from CC13. (B) Amplification of oxc using oxc-specific primers (lanes 2–5) and degenerate oxc primers (lanes 6–9, expected size 419 bp) and DNA extracted from ™PRO Lab supplement powder, and BHI cultures of ™PRO Lab supplement. Lanes: 1, molecular weight ladder (bp); 2, BHI ™PRO Lab supplement DNA; 3, ™PRO Lab supplement DNA; 4, no DNA control; 5, purified DNA from CC13; 6, BHI ™PRO Lab DNA; 7, ™PRO Lab DNA; 8, no DNA control; 9, purified DNA from CC13. (C) Amplification of oxc using oxc-specific primers and DNA extracted from Oxalo™ capsule. Lanes: 1, molecular weight ladder (bp); 2, Oxalo™ capsule DNA; 3, purified DNA from OxCC13. (D) Amplification of oxc using degenerate oxc primers and DNA extracted from Oxalo™ capsule. Lanes: 1, molecular weight ladder (bp); 2, Oxalo™ capsule DNA; 3, purified DNA from OxCC13.
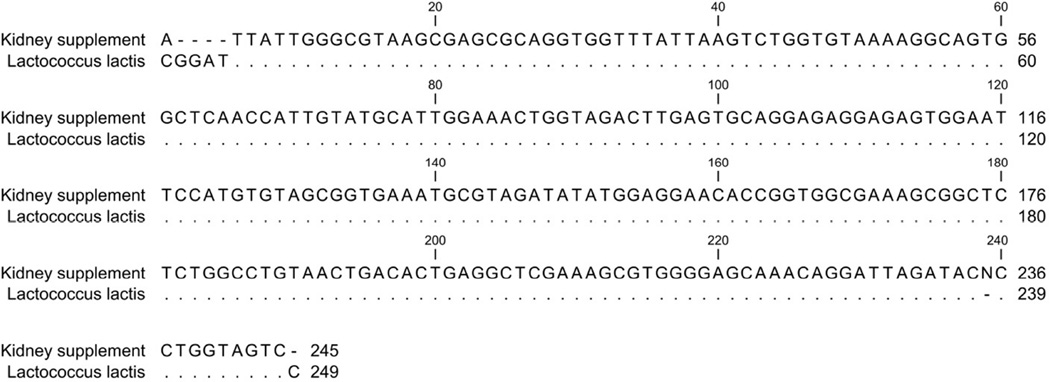
DNA was extracted from ™PRO Lab supplement, and the universal bacterial primers 515F and 805R were used to amplify the 16S rRNA gene. Sequencing of the reaction revealed a PCR product 98% identical to Lactococcus lactis subsp. lactis (Fig. 3, GenBank accession no. KJ095656.1). In addition, DNA was isolated from an overnight reconstituted supplement culture and from a supplement culture grown in BHI. Sequencing of 16S rRNA genes from these samples also indicated the major viable bacterium in the supplement was Lactococcus lactis subsp. lacti.
Figure 3.
Sequence alignment of 16S rRNA from ™PRO Lab supplement powder. This PCR product was 98% identical with GenBank accession no. KJ095656.1, Lactococcus lactis subsp. lactis.
DNA was extracted from an Oxalo™ capsule and from cultures grown in BHI, MRS, SBO, OXMY and Schaedler Broth. Sequencing of 16S rRNA gene indicated Lactobacillus plantarum strain Sha 7 was present (GenBank accession no. KF040094.1) Sequencing of 16S rRNA gene from the different cultures identified several Bacillus species (Table 1).
Table 1.
Organisms Present in the Probiotic Supplements, ™PRO Lab, “Kidney stone probiotic”, and Sanzyme, Ltd, Oxalo™.
| ™PRO Lab supplement | GenBank accession no. |
|---|---|
| Lactococcus lactis subsp. lacti | KJ095656.1 |
| Sanzyme Ltd Oxalo™ capsule | |
| Lactobacillus plantarum | KF040094.1 |
| Bacillus licheniformis | JQ353819.1 |
| Bacillus cereus | KF527826.1 |
| Bacillus coagulans | KJ466151.1 |
| Bacillus subtilis | JQ829568.1 |
COMMENT
Colonization of the intestine with the highly efficient, oxalate-degrading, intestinal bacterium Oxalobacter formigenes may reduce the risk of calcium oxalate stone disease. Probiotic supplements that may contain O. formigenes are available for purchase over the internet from ™PRO Lab, Ltd and Sanzyme Ltd. Such products would allow both researchers and physicians the opportunity to examine the impact of O. formigenes colonization on oxalate balance and recurrence of calcium oxalate stone disease. This study used culture and PCR methods to detect O. formigenes within a single batch preparation of each probiotic product. However, results from these approaches indicated the batch preparations contained no O. formigenes. Furthermore, cultures in a number of different media containing oxalate, and PCR using degenerate Lactobacillus primers that amplify the oxalate decarboxylase gene, both indicated that the preparations lacked bacteria that may have the capacity to degrade oxalate. Further analysis by 16S rRNA gene sequencing indicated the predominant organism in the ™PRO Lab preparation was Lactococcus lactis subsp. lacti. This organism has not been reported in the literature to have the oxc gene or degrade oxalate in culture. 16S rRNA gene sequencing identified several Bacillus species and Lactobacillus plantarum in the Oxalo™ capsule. Interestingly, of these species identified in Oxalo™, oxalate decarboxylase is a minor component of the spore coat of Bacillus subtilis, Bacillus lichenformis and Bacillus cereus.12 The genome of Bacillus lichenformis, Bacillus cereus, Bacillus coagulans contains the oxalate/formate antiporter central to ATP synthesis in O.formigenes.13 Of the 4 stated organisms listed on the Oxalo™ product only Bacillus coagulans was detected by 16S rRNA gene sequencing.
This study raises questions about the difficulty of culturing and preparing O. formigenes for probiotic use. For example, the procedure recommended by the company to first reconstitute the powder overnight in sterile water may compromise O. formigenes viability. A recent randomized, double-blind, placebo-controlled multicenter study showed that ingestion of a lyophilized enteric coated capsulated preparation of O. formigenes, Oxabact®, currently not available for purchase, did not result in a significant reduction in urinary oxalate excretion14, which the authors suggested could have been due to problems with bioavailability of the supplement or viability of O. formigenes in this formulation. Thus, studies examining the factors that impact O. formigenes survival and revival following processing for probiotic supplement preparation are warranted.
A limitation of this study is only one batch supply of each probiotic supplement was purchased and examined, and it is possible the preparation received was from a bad batch. Nevertheless, the study raises concerns about the efficacy of these products.
CONCLUSION
The probiotic supplements sold by ™PRO Lab, Ltd, and Sanzyme Ltd, claim to contain the oxalate-degrading intestinal bacterium Oxalobacter formigenes. However, both culture and PCR methods did not detect the presence of O. formigenes in a batch preparation of each product. In addition, the culture and PCR methods used indicated the preparations do not contain other oxalate-degrading organisms. It is our belief that the probiotic supplement sold by ™PRO Lab, Ltd and Oxalo™ purchased from Sanzyme Ltd, will be of little or no benefit to calcium oxalate kidney stone patients.
ACKNOWLEDGEMENTS
This research was supported by NIH grant RO1 DK087967.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Allison MJ, Dawson KA, Mayberry WR, et al. Oxalobacter formigenes gen. nov., sp. nov.: oxalate-degrading anaerobes that inhabit the gastrointestinal tract. Archives of microbiology. 1985;141:1–7. doi: 10.1007/BF00446731. [DOI] [PubMed] [Google Scholar]
- 2.Knight J, Deora R, Assimos DG, et al. The genetic composition of Oxalobacter formigenes and its relationship to colonization and calcium oxalate stone disease. Urolithiasis. 2013;41:187–196. doi: 10.1007/s00240-013-0566-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaufman DW, Kelly JP, Curhan GC, et al. Oxalobacter formigenes may reduce the risk of calcium oxalate kidney stones. J Am Soc Nephrol. 2008;19:1197–1203. doi: 10.1681/ASN.2007101058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang J, Knight J, Easter LH, et al. Impact of dietary calcium and oxalate, and Oxalobacter formigenes colonization on urinary oxalate excretion. J Urol. 2011;186:135–139. doi: 10.1016/j.juro.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siener R, Bangen U, Sidhu H, et al. The role of Oxalobacter formigenes colonization in calcium oxalate stone disease. Kidney international. 2013;83:1144–1149. doi: 10.1038/ki.2013.104. [DOI] [PubMed] [Google Scholar]
- 6.Kelly JP, Curhan GC, Cave DR, et al. Factors related to colonization with Oxalobacter formigenes in U.S. adults. Journal of endourology / Endourological Society. 2011;25:673–679. doi: 10.1089/end.2010.0462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kharlamb V, Schelker J, Francois F, et al. Oral antibiotic treatment of Helicobacter pylori leads to persistently reduced intestinal colonization rates with Oxalobacter formigenes. Journal of endourology/Endourological Society. 2011;25:1781–1785. doi: 10.1089/end.2011.0243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mittal RD, Kumar R, Bid HK, et al. Effect of antibiotics on Oxalobacter formigenes colonization of human gastrointestinal tract. Journal of endourology/Endourological Society. 2005;19:102–106. doi: 10.1089/end.2005.19.102. [DOI] [PubMed] [Google Scholar]
- 9.Sidhu H, Hoppe B, Hesse A, et al. Absence of Oxalobacter formigenes in cystic fibrosis patients: a risk factor for hyperoxaluria. Lancet. 1998;352:1026–1029. doi: 10.1016/S0140-6736(98)03038-4. [DOI] [PubMed] [Google Scholar]
- 10.Sidhu H, Schmidt ME, Cornelius TJG, et al. Direct correlation between hyperoxaluria/oxalate stone disease and the absence of the gastrointestinal tract dwelling bacterium Oxalobacter formigenes: Possible prevention by gut recolonization or enzyme replacement therapy. J Am Soc Nephrol. 1999;10:S334–S340. [PubMed] [Google Scholar]
- 11.Miller AW, Kohl KD, Dearing MD. The gastrointestinal tract of the white-throated Woodrat (Neotoma albigula) harbors distinct consortia of oxalatede-grading bacteria. Appl Environ Microbiol. 2014;80:1595–1601. doi: 10.1128/AEM.03742-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costa T, Steil L, Martins LO, et al. Assembly of an oxalate decarboxylase produced under sigmaK control into the Bacillus subtilis spore coat. Journal of bacteriology. 2004;186:1462–1474. doi: 10.1128/JB.186.5.1462-1474.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anantharam V, Allison MJ, Maloney PC. Oxalate:formate exchange. The basis for energy coupling in Oxalobacter. J Biol Chem. 1989;264:7244–7250. [PubMed] [Google Scholar]
- 14.Hoppe B, Groothoff JW, Hulton SA, et al. Efficacy and safety of Oxalobacter formigenes to reduce urinary oxalate in primary hyperoxaluria. Nephrol Dial Transplant. 2011;26:3609–3615. doi: 10.1093/ndt/gfr107. [DOI] [PubMed] [Google Scholar]