Abstract
Mannobiose-modified polyethylenimines (PEI) are used in gene therapy to generate nanoparticles of DNA that can be targeted to the antigen-presenting cells of the immune system. We report that the sugar modification alters the DNA organization within the nanoparticles from homogenous to shell-like packing. The depth-dependent packing of DNA within the nanoparticles was probed using AFM nano-indentation. Unmodified PEI/DNA nanoparticles display linear elastic properties and depth-independent mechanics, characteristic of homogenous materials. Mannobiose-modified nanoparticles, however, showed distinct force regimes that were dependent on indentation depth, with ‘buckling’-like response that is reproducible and not due to particle failure. By comparison with theoretical studies of spherical shell mechanics, the structure of mannobiosylated particles was deduced to be a thin shell with wall thickness in the order of few nanometers, and a fluid-filled core. The shell-core structure is also consistent with observations of nanoparticle denting in altered solution conditions, with measurements of nanoparticle water content from AFM images, and with images of DNA distribution in Transmission Electron Microscopy.
Keywords: DNA condensation, mannobiose, nanoparticles, polyethylenimine, Atomic force microscopy, indentation
1. INTRODUCTION
DNA is an anionic polymer, with a semirigid (persistence length ≈ 50 nm) and self-repelling backbone that occupies an expanded volume in solution. However in the presence of polymeric cations (having three positive charges or higher), the DNA backbone packs or condenses into less than one thousandth of its unperturbed volume 1 2, 3. There is a growing interest in using nanometer-size dispersed particles or nanoparticles of condensed DNA for delivering DNA to cells in both clinical gene therapy and basic molecular biology applications 4, 5. As a carrier for DNA, cationic polymers are relatively less toxic than deactivated viruses. Importantly, they can be functionalized for targeting specific cells and for achieving specific drug-delivery attributes 6.
Polyethyleneimine (PEI) is a widely used cationic DNA carrier for a number of reasons 5, 7-11. The close arrangement of positively charged amine groups on the PEI backbone makes the polymer effective at neutralizing DNA and condensing it into stable nanoparticles 11-13. Only about half of the PEI amines are protonated and charged at physiological pH. Additional protonation occurs at lower pH and the polymer functions like a buffer. The ‘proton-sponge’ character of PEI is critical for stabilizing DNA nanoparticles in the acidic environment of cell uptake vesicles, and for subsequent osmotic bursting of these vesicles to release the nanoparticles into the cytoplasm 14-16. Recent studies report that PEI-DNA nanoparticles are being actively transported to the vicinity of the cell nucleus 12.
Drug-delivery studies using DNA-PEI nanoparticles have focused on improving the morphological, cytotoxicity, and cell-targeting characteristics of the DNA/PEI complexes by linking compounds to the PEI backbone 17-22. Mannobiose is grafted to the PEI backbone to generate DNA particles that can be targeted to the Antigen-Presenting Cells (APCs) of the immune system 23. APCs like Dendritic and Langerhans cells have surface receptors for mannose and are present under the skin close to openings in the body. Their chief function is to intercept invading pathogens, and display the pathogenic antigens on their surface for activating/priming T-cells 17, 22-24. There is a lot of interest in developing clinical immunotherapies that load APCs with antigens against cancer and AIDS, to ensure a sustained and long-term immune response against these conditions. Mannobiosylated-PEI or PEIm has been used for delivering DNA to APCs in one form of clinical immunotherapy being developed against the HIV virus 17, 24-26.
It is not known how grafting of compounds onto the cationic polymer alters the physical packing of DNA within the nanoparticles. In this paper we study the effect of PEI mannobiosylation on the structure of DNA nanoparticles by indenting the particles using an Atomic Force Microscope (AFM). A pyramidal tip (about 8nm diameter) indents nanoparticles (~150 – 250 nm diameter) that are gently adsorbed on a mica surface in a fluid environment. The indentation maps depth-dependent changes in the elastic modulus of the particles (elastic tomography) due to inhomogeneities in DNA packing on a length scale that is larger than the tip contact diameter. The chief advantage of elastic tomography to study structural organization is that the nanoparticles are maintained in their native fluid environment with minimal distortion of the packing structure from staining and fixation. Indentation probing picked up significant differences in the DNA packing between mannobiosylated and unmodified nanoparticles. These mannobiosylated nanoparticles showed a shell-core architecture, which was confirmed by complementary observations of nanoparticle denting in altered solution conditions, non-uniform staining for DNA distribution in TEM, and a shell-like distribution of nanoparticle water content with nanoparticle size.
2. EXPERIMENTAL METHODS
2.1 Nanoparticle preparation
Nanoparticles were prepared with a low molecular weight (22 kDa) linear PEI that is known to interact with blood components to a lesser degree, and has reduced toxicity and high transfection efficiency in vivo 27-30. The polyethylenimine was modified with mannobiose according to the procedure in Toke et al 17. An unmodified 22 kDa linear polyethylenimine (PEI) was purchased from Polysciences Inc. (Warrington, PA). A 12.5 kb plasmid DNA used in this study 31. The particles were prepared from a solution of 1 ng/μL DNA, 1.36 mM PEIm/PEI and 50 μM NaCl. The PEI amine to DNA phosphate ratio in the solution was greater than 10. The condensate solution was incubated for 3 hours to form nanoparticles and centrifuged at 553 g for 1 min prior to experimental studies.
2.2 Atomic Force Microscopy
Dry Imaging
A commercial AFM instrument (Multimode/PicoForce system with NS-V controller, Bruker Nano-surfaces, Inc. Santa Barbara, CA) was used to image air-dried nanoparticles on freshly cleaved mica surface. Silicon Cantilevers (OMCL, ~300kHz, 42N/m by Olympus, Japan) were used to acquire height, amplitude error, and phase images of the nanoparticles. The phase and amplitude images produced the clearest 3D visualization of the nanoparticle surface. AFM images were analyzed using the particle-analysis toolbox of the NIH ImageJ software (available at http://rsb.info.nih.gov/ij, National Institutes of Health, Bethesda, MD) to measure the area footprint (A) and volume (V) of the surface-adsorbed nanoparticles. Particle diameters < 5 nm were not included in the image analysis. Two kinds of particle diameters were calculated from the measured A and V. The ‘area-equivalent’ diameter was calculated as the diameter of a circle having the same area footprint as the nanoparticle on the imaging surface, DA= (4A/π)½. The ‘volume-equivalent’ diameter was calculated as the diameter of a sphere having the same volume as the nanoparticle on the imaging surface, DV = (6V/π)1/3. (see Supplementary information S2).
Fluid imaging and Nano-indentation
A Bioscope Catalyst (NS-V controller, Bruker Nano-surfaces, Santa Barbara, CA) was used for imaging and indentation studies performed in fluid. The cantilevers were general-purpose silicon nitride, with square pyramidal tip and 15 nm nominal tip radius (MSCT, Bruker). The spring constant of each cantilever was determined using the thermal tune method and was in the range of 0.045 N/m. The Deflection Sensitivity of the cantilevers was determined by ramping on plain mica surface before each nanoparticle indentation experiment. The sample for testing was prepared by adsorbing 4 μL of the condensate solution on the mica surface confined within a fluid cell for 10 min, followed by rinsing with buffer. Indentation was performed across a grid on the nanoparticle maintained in fluid environment. The indentation ramp rate was 0.1 μm/sec. The height of the nanoparticle at different points on the grid was obtained from the contact point of the indentation force curve. Unless noted, the indentation curves shown in this paper are from the grid point with the tallest nanoparticle height. All average and distribution data shown were culled from three sample sets.
2.3 Dynamic Light Scattering
Dynamic Light Scattering (DLS) measurements were performed with a Precision Detector - Expert Laser Light Scattering DLS Workstation equipped with a HeNe laser (wavelength: 698 nm) on 200 μL of condensate solutions. The condensate solutions were filtered with a 400 nm polycarbonate filter (#11607 Nucleopore, Whatman) before DLS measurements to remove dust and large aggregates.
3. RESULTS AND DISCUSSION
3.1 Comparison of nanoparticle morphology and size distribution by dry AFM imaging
A 3μL solution of PEI/DNA and PEIm/DNA nanoparticles was air-dried on a clean mica surface and imaged with AFM (Fig.1). While air-drying and surface interactions can distort the appearance of adsorbed particles to different extents, there appeared to be a significant difference in the surface height and smoothness of the mannobiosylated and unmodified particles. The PEI/DNA nanoparticles appeared fluffy, with irregular heights and ruffled edges, whereas the PEIm/DNA nanoparticles appeared as well-formed spheres (see phase images in top inset) with smoothly changing surface and sharp particle outlines (Fig. 1). The field of PEIm/DNA nanoparticles was relatively uniformly distributed in size compared to the PEI/DNA nanoparticles (bottom inset in Figs. 1a and b). The volume- and area- equivalent diameters of the nanoparticles were calculated from the particle volume and area on the surface (see Methods). Figures 1c and d show the distribution of these diameters for both particle types. As apparent from the field images, the PEIm nanoparticles were larger and slightly less polydisperse than the PEI nanoparticles. The peak values for their area diameters were 125 nm and 85 nm, respectively, indicating that the PEIm nanoparticles were significantly larger than their unmannobiosylated counterpart. Also the peak values of the volume diameter, which is an estimate of the dried mass in each nanoparticle, were again modestly higher for the PEIm/DNA nanoparticles than the PEI/DNA nanoparticles (50 vs. 35 nm). The standard deviations for the diameter were calculated for the unimodal histogram corresponding to the peak values, and not for the entire distribution.
Fig.1.
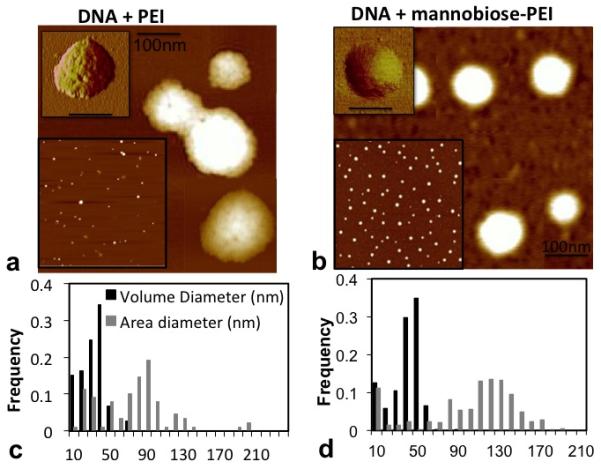
Analysis of air-dried PEI (left) and PEIm (right) DNA nanoparticle. The AFM height images of the particles are shown in figures a and b (scale is 100nm). The top insets in these figures are phase images of the nanoparticles (scale = 100nm). The bottom insets are the nanoparticle height images in a 6mmX6mm scan field. Figures c and d are plots of the volume- and area- equivalent diameters of the nanoparticles calculated from the AFM height images.
3.2 Fluid imaging and nano-indentation of the nanoparticles
The nanoparticles were adsorbed on the mica surface in their native fluid environment and imaged in a fluid cell. The AFM phase image and surface height profiles of representative PEI/DNA and PEIm/DNA nanoparticles are shown in Fig. 2. In both cases, the area diameter of the nanoparticles was about 120 nm and the peak height was 70 nm. Albeit slightly oblong, the PEI and PEIm nanoparticles retained their shape without collapsing when adsorbed onto the imaging surface in a fluid environment. Interestingly, in separate experiments we found that DNA nanoparticles with branched PEI did not hold their shape, but collapsed on the imaging surface (See Supplementary Information S3). The ability to hold the shape can be explained to some extent by the difference in packing densities for nanoparticles with linear polycations (~70% 1) and branched PEI (~10% 32).
Fig.2.
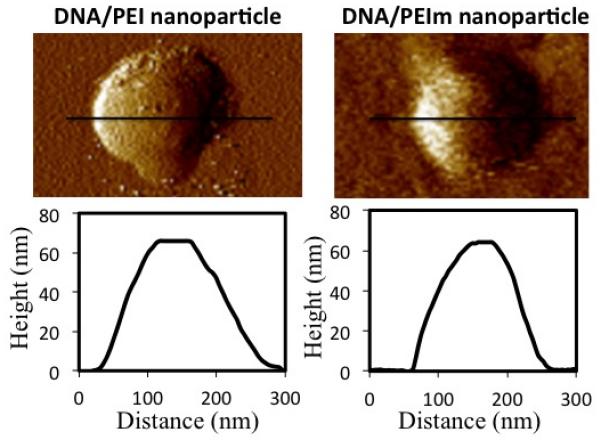
AFM images of PEI (left) and PEIm (right) DNA nanoparticles adsorbed on mica surface in fluid images. Shown below is height profile of along each nanoparticle.
Nanoparticles in the 150 nm median diameter range were chosen for testing with nanoindentation. At this size, the nanoparticle diameters are larger than the tip diameter and the indentation analysis models described below are applicable. Fig. 3a shows the deflection curves obtained for PEI and PEIm nanoparticles of similar size, along with the indentation curve on mica. The PEI/DNA nanoparticle response could be fitted by Hertzian contact mechanics of a sphere indenting into a homogenous elastic material33. The linear elasticity modulus calculated with this model33 was found to be ~2.8 MPa, which exceeds that of soft swollen gels34. In the model, the strain under the indenting sphere decays as 1/z, where z is the distance from the point of contact. Therefore, the force at ~10 nm indentation is the integrated effect of material resistance over a larger depth, indicating that PEI/DNA nanoparticles were uniformly filled with DNA on the length scale of the tip contact area.
Fig.3.
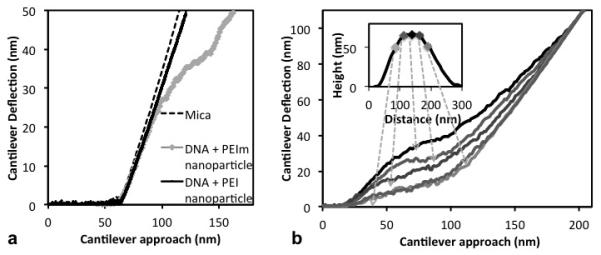
(a) Nano-indentation force curves at the center of PEI/DNA and PEIm/DNA nanoparticles, along with that on mica. (b) All the force curves at indentation points along the diameter of PEIm/DNA nanoparticles have region of reversed curvature followed by nonlinear stiffening.
PEIm/DNA nanoparticles, however, did not show depth-independent, homogenous mechanics. Initially, at small indentations, the response was elastic and similar to that of PEI nanoparticles. However, the slope of the force curve decreased significantly at intermediate indentations, and increased again at large depths. This depth-dependent indentation response, consisting of a region with reversed curvature followed by nonlinear stiffening, was observed at all points on the nanoparticle. An example is given in Fig. 3b where the nanoparticle was indented along a grid, with each point about 26 nm apart. The force curves at points along the particle diameter are only shown for clarity. The approximate locations of these indentations are given in the inset on the nanoparticle height profile. All the force curves display the region of reversed curvature followed by nonlinear stiffening. The depth of the reversed-curvature region is maximal at the particle center, and decreases on either side away from the center. The reproducible and symmetric pattern of the depth-dependent force response confirmed that it was a material property and not due to large-scale material failure. The reversed curvature and depth-dependence was not observed during the retraction phase of the force curves at all indentation points (See Supplementary Information S4).
3.3 Comparison of PEIm/DNA indentation mechanics against that of thin-walled fluid-filled shells
In Fig. 4 the force response is plotted as a function of indentation depth instead of cantilever approach. The indentation depth (Δz) was calculated from the contact point (z0), the cantilever approach (z) and the cantilever deflection (Δy) (Eq. 1). The indentation force was calculated from the spring constant of the cantilever (kc, which is ~ 0.045 N/nm) and from the cantilever deflection (Eq. 2).
| Eq.1 |
| Eq.2 |
Fig.4.
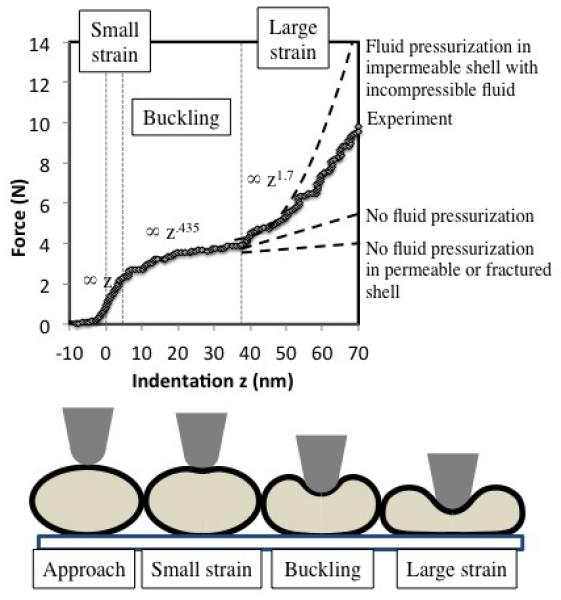
Indentation force curve of a PEIm/DNA nanoparticle is similar to that of a hollow fluid-filled shell and can be separated into three regimes with different deformation modes.
The shape of the force curve is remarkably similar to that derived by analytical 35, simulation 36, and experimental 37 models for the compression of thin walled hollow shells under point loads. A range of hollow shells has been studied by indentation analysis, including beach balls 36 and tennis balls 35, micro-scale polyelectrolyte capsules 38 and nanometer-scale viruses 39. A classic hollow shell shows reversed force curvature when its shell wall is less that 1/10th the radius of the nanoparticle, due to buckling of the shell wall 35. The indentation response of a thin shell can be separated into three regions: small-strain, buckling, and large-strain regimes, with different deformation modes and therefore different power-law dependence on indentation depth. The PEIm/DNA nanoparticle force curve is discussed below in the context of these three regimes. Interestingly, the power law dependence in the small-strain and large-strain regions is not affected by the shape of the shell (spherical, ellipsoidal, or cylindrical) 40.
Small strain region
At indentation depths that are on the order of the shell thickness, thin-walled shells show a linear force response where F∞∆z. In this region the indentation is resisted by the shell wall bending under the indenter; the bending deformation requiring less elastic energy than stretching of the shell wall (Fig. 4). The linear dependence between force and indentation can be derived using thin plate theory for small-strain bending, and it is unaffected by the internal pressure within the shell 40. The linear behavior was observed for up to 5 nm depth in the case of DNA/PEIm nanoparticles, indicating that the shell thickness was also of the same order (Fig. 4).
Buckling region
When a thin-walled shell is compressed to depths larger than the wall thickness, a region of decreasing stiffness appears in the force curve, which is coincident with the appearance of a dimple or inverted cap in the shell wall around the indenter (Fig. 4). The dimple forms as the shell wall buckles under the compressive load, storing most of its bending strain energy along the edge of the dimple 38. The shape and extent of the dimpled region depends on indenter shape and curvature, shell wall elasticity, and the indentation rate 41. In this region the force is expected to increase with depth as F∞∆z0.538. In cases where the shell wall has been observed fracture instead of buckling, example the virus capsids, the force decreases with indentation (negative power law). We obtained a power law dependence of about 0.435, which is in the range expected for a buckling shell wall. Interestingly this region extends to indentation depth of 30 nm into the nanoparticle.
Large strain region
In the large strain region the force curves develop differently depending on the pressurization and permeability of the shell. Tabor et al 35 calculated that the force response of the hollow and pressurized shells are similar till about 20% of the shell radius, since fluid pressure is relatively small at these compressions. Beyond the buckling region, the force increases to different extents in hollow and pressurized shells. In the case of an unpressurized shell, indentation is resisted by the bending and compression of the shell wall, and the force increases linearly with indentation 35, 36. In a pressurized shell, there is geometric stretching of the shell due to the internal fluid pressure, as the incompressible fluid within accommodates the volume and shape change of compression. Volume conservation principles dictate that the force in pressurized shells increases as a cubic power of indentation, F∞∆z3, in the large-strain region 38. In the case of the DNA/PEIm nanoparticles we measured a power dependency of 1.7 between force and indentation (Fig. 4). The reduced exponent can be explained by a number of factors that are not accounted in the analysis: the boundary conditions of surface adsorption, membrane pre-stress, and permeability of the shell wall. However the indentation response in this region strongly suggests the presence of a fluid-filled core, which is also supported by the observations of ramp rate dependence and the effect of solvent quality that are discussed below.
We note that the force response with reverse curvature has only been observed for thin shells that are strong enough that the tip does not penetrate through the shell layer. In the case of soft shells like liposomes, the force curve does not exhibit reversed curvature, but shows a discontinuity; it appears that the tip completely penetrates one wall and jumps onto the second. We did not observe indentation mechanics akin to extremely soft shells in the DNA nanoparticles.
Indentation rate dependence
Nanoparticles of similar size were chosen for this experiment. We observed that when the ramp rate of indentation was increased from 0.1μm/ sec to 1μm/sec, there was no change in the slope of the small strain region (Fig. 5). However buckling occurred later in the indentation cycle (~7nm) and developed rapidly. Beyond the buckling region, the force increased non-linearly and was again independent of the ramp rate. The pattern of ramp rate dependence suggests that the buckling phase involves fluid rearrangement that was retarded by the increased rate of indentation. The retardation of buckling rearrangement is again consistent with our observation that the core of the DNA/PEIm nanoparticle is filled with fluid. Further studies were performed on DNA/PEIm nanoparticles to confirm the shell-core structure, and are described below.
Fig.5.
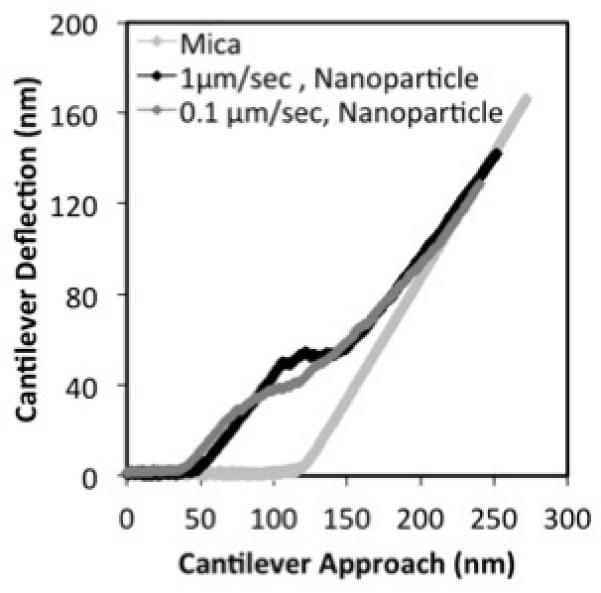
Ramp rate dependence of the indentation mechanics of a DNA/PEIm nanoparticle
3.4 Effect of solvent quality on the morphology of PEIm/DNA nanoparticles
The PEIm/DNA nanoparticles were concentrated and transferred to solution of 2% glycerol in phosphate buffered saline. Surface-adsorbed particles exposed to the new solvent conditions were air-dried on a mica surface and visualized with AFM. We observed that the area-diameters of these nanoparticles did not change significantly and still retained a peak value at ~120 nm (Fig. 6a), indicating that the nanoparticles did not appear to break up and re-form in the new solvent conditions. (data not shown). However all the nanoparticles had a single dent formed in the middle, clearly visible in the 2D and 3D height images (Fig. 6b). The exact dimensions of the dented region is expected to be smaller than that observed in the AFM image because of the scanning probe finite dimensions. The occurrence of denting in the nanometer-sized shell assemblies has also been reported by other researchers 42 and is consistent with the presence of a fluid-filled core. We note that the observed denting of air-dried DNA nanoparticles may be the consequence of glycerol’s plasticizing effect. It is known that the glass transition temperature of glycerol-plasticized polymers is significantly reduced 43.
Fig.6.
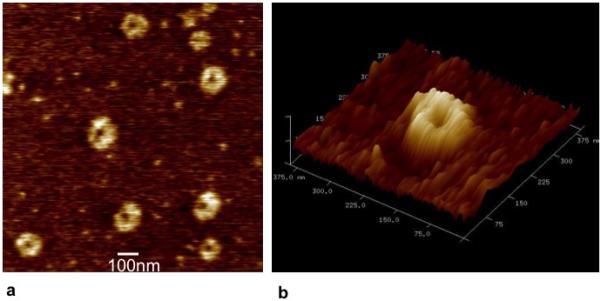
(a) AFM image of PEIm/DNA nanoparticles subject to osmotic deswelling and air-dried. (b) 3D rendering of the height image of a representative particle from (a) showing the dent in the middle.
3.5 Estimation of water content and shell wall thickness of DNA/PEIm nanoparticles
The hydrodynamic diameter of the PEIm/DNA nanoparticles was measured using Dynamic Light Scattering (Fig. 7a). The distribution of hydrodynamic diameters and its peak value of 150 nm were similar to that of the area-equivalent diameters of air-dried particles (Fig. 7a and inset). This finding implies that the diameter of the nanoparticles air-dried on the imaging surface was a close approximation to their diameters in solution. It appears that the nanoshells were drying on the surface without significant spreading.
Fig.7.
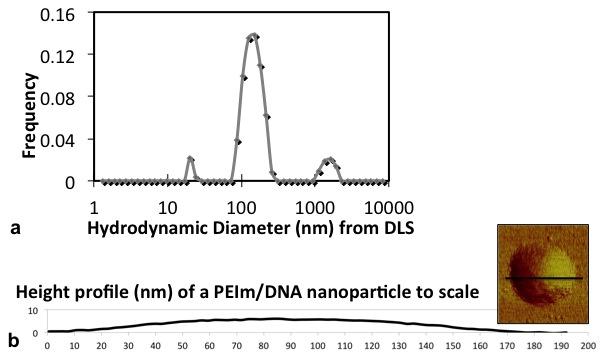
(a) Distribution of particle hydrodynamic diameters determined by Dynamic Light Scattering at 90° detection angle. The distribution of hydrodynamic diameters is similar to the distribution of area-equivalent diameters shown inset. (b) Height profile across a representative air-dried PEIm\DNA nanoparticle (left) showing the large diameter-height ratio produced by air-drying.
It was observed that the height of air-dried particles was significantly smaller (4 – 6 nm) than their area-equivalent diameter (average of 120 nm). An example of the height profile of an air-dried nanoparticle is shown to scale in Fig. 7b. There is a significant decrease in height during drying which can be attributed to the loss of water. However all the air-dried PEIm/DNA nanoparticles had a smooth surface (Fig. 7b) with no indications of water loss by shell damage. Therefore the water loss must be from permeation through the shell wall. However, the absence of shrinkage wrinkles in the air-dried particles may also reflect self-healing of the shell walls. Under these conditions, the volume of the nanoparticles after air-drying is a good measure of the volume of the solid component in each particle.
We estimated the water content of each air-dried nanoparticle by subtracting its dried volume from its swollen volume. The plot of water-fraction vs. particle diameter is shown in Fig. 8. The water fraction increases rapidly as a function of particle diameter and then becomes nearly uniform for diameters > 50 nm. The nearly constant water fraction at these diameters is ≈ 95%, which is much greater than water fractions for DNA nanoparticles reported in literature 1, 32. The shape of the water fraction curve is similar to that obtained from fluid-filled shells of different diameter but same wall thickness (Fig. 8). In fact the PEIm/DNA nanoparticles plot is within the limits calculated for fluid-filled shells with wall thickness ranging from 0.5 – 1.5 nm.
Fig.8.
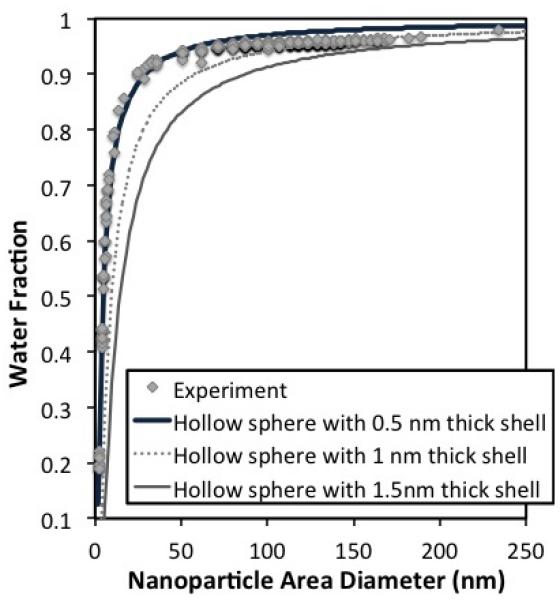
(a) Distribution of water content as a function of nanoparticle area-equivalent diameter. The water fraction of each particle was determined by subtracting the volumes corresponding to its area-equivalent and volume-equivalent diameters. In the diameter range of interest (> 80 nm), the water fraction is nearly constant at about 0.95. Shown alongside is the water-fraction estimate for water-filled hollow spheres of different diameters, for three shell thickness values: 0.5, 1, and 1.5 nm. The experimental data lie between the estimates for 1 nm and 0.5 nm shell thickness.
The water-fraction studies predict a wall thickness of 0.5 – 1.5nm for the DNA/PEIm nanoparticles, based on the assumption of a fluid-filled shell with constant wall thickness. This wall thickness appears to be lower than that of a double stranded DNA in solution, which is about ~2 nm, but in the typical range of heights measured for surface-adsorbed DNA using AFM 44. We also note that according to hollow shell indentation theory, the shell thickness is roughly equal to the indentation depth of the small-strain region. This puts the thickness estimate for swollen DNA/PEIm shells at ≈ 4 nm.
3.6 TEM scans of the PEIm/ and PEI/ DNA nanoparticles are consistent with nano-indentation measurements
The nanoparticles were positively stained with Uranyl Acetate and imaged with Transmission Electron Microscopy using standard protocols12 (Fig. 9a). While the staining of DNA/PEI nanoparticles was similar to that published in literature and was homogenous through the particles12 (inset, Fig. 9a), the DNA/PEIm particles stained non-homogenously with stronger staining on the circumference (Fig. 9a and b). Uranyl acetate binds DNA and the staining in TEM images is proportional to the DNA mass density in the nanoparticle in the path of the electron beam. A profile of the intensity of uranyl acetate staining across a representative DNA/PEIm particle is shown in Fig. 9b. The stain intensity increases sharply at the particle boundary and decreases towards the particle center, but is still higher than the background. Similar TEM staining profiles have been seen in hollow polymeric structures of polymerosomes 45.
Fig.9.
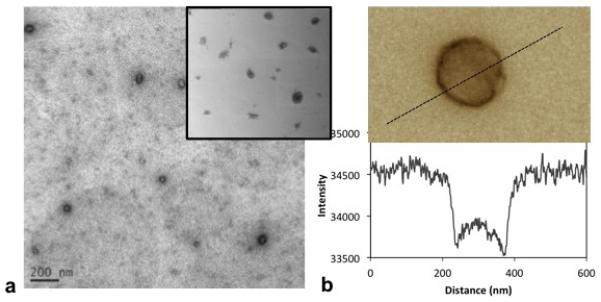
(a) TEM images of a field of PEIm/DNA nanoparticles compared with that of PEI/DNA nanoparticles (inset). (b) Profile of intensity of uranyl-acetate staining in a representative PEIm/DNA nanoparticle.
CONCLUSIONS
We report differences in the packing of DNA in nanoparticles that were assembled with PEI and mannobiosylated-PEI (PEIm) under the same preparation conditions. The depth-dependent packing of DNA within the nanoparticles was investigated with AFM nanoindentation. The particles were maintained in their native swollen state, and only changes in packing on length scales larger than the AFM tip were probed. PEI/DNA nanoparticles displayed a linear response well described by the Hertzian model of indentation into a homogenous linear-elastic material. PEIm/DNA nanoparticles, on the other hand, did not display homogenous packing. Nanoindentation analysis suggested that these particles exhibit thin-walled shell structure with a fluid-filled core. The particles buckled at intermediate indentation depths and stiffened at large strains probably due to fluid pressurization. The profile of DNA staining seen in TEM images was also consistent with a shell structure. Analysis of the water fraction revealed that the shell wall was about a few nanometers thick. The wall thickness is in the range predicted by nanoindentation data.
In summary, we report an interesting finding that mannobiosylated PEI alters the packing of DNA in nanoparticles from homogenous to that of a thin-walled shell with a fluid-rich core. Packing density and morphology in nanoparticle-based drug delivery systems are critically important. The drug should retain its biological activity until it reaches its target. Then it should be released at an optimal rate. The architecture of nanoparticles plays a crucial role in protecting the incorporated drug from degradation and preventing early release of the active ingredients. Advancing our ability to tailor nanoparticles opens new avenues to design and engineer novel biomaterials (e.g., nanomedicines. drug carriers, biosensors) for future biomedical applications, which combine multiple functionalities while maintaining the nanoscopic length scale.
Supplementary Material
ACKNOWLEDGEMENTS
This research was supported by the Intramural Research Program of the NICHD, NIH. The authors are grateful to Mr. Quentinn Roby and Mr. Ayele Guggsaa at Howard University for their assistance with the TEM imaging of PEI nanoparticles.
REFERENCES
- 1.Bloomfield VA. Biopolymers. 1991;31:1471–1481. doi: 10.1002/bip.360311305. [DOI] [PubMed] [Google Scholar]
- 2.Bloomfield VA. Current Opinion in Structural Biology. 1996;6:334–341. doi: 10.1016/s0959-440x(96)80052-2. [DOI] [PubMed] [Google Scholar]
- 3.Gosule LC, Schellman JA. Journal of Molecular Biology. 1978;121:311–326. doi: 10.1016/0022-2836(78)90366-2. [DOI] [PubMed] [Google Scholar]
- 4.Challa K, Josef H, Carola L, editors. Nanofabrication Towards Biomedical Applications. Springer; Netherlands: 2006. [Google Scholar]
- 5.Santhakumaran LM, Chen A, Pillai CKS, Thomas T, He H, Thomas TJ. Nanotechnology in Nonviral Gene Delivery. Wiley-VCH Verlag GmbH & Co. KGaA; 2005. [Google Scholar]
- 6.Lemkine G, Demeneix B. Current Opinion in Molecular Therapeutics. 2001;3:178–182. [PubMed] [Google Scholar]
- 7.Ahn HH, Lee JH, Kim KS, Lee JY, Kim MS, Khang G, Lee IW, Lee HB. Biomaterials. 2008;29:2415–2422. doi: 10.1016/j.biomaterials.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 8.Ahn HH, Lee MS, Cho MH, Shin YN, Lee JH, Kim KS, Kim MS, Khang G, Hwang KC, Lee IW, Diamond SL, Lee HB. Colloids and Surfaces A: Physicochemical and Engineering Aspects. 2008;313–314:116–120. [Google Scholar]
- 9.Choosakoonkriang S, Lobo BA, Koe GS, Koe JG, Middaugh CR. Journal of Pharmaceutical Sciences. 2003;92:1710–1722. doi: 10.1002/jps.10437. [DOI] [PubMed] [Google Scholar]
- 10.Iida T, Mori T, Katayama Y, Niidome T. Journal of Controlled Release. 2007;118:364–369. doi: 10.1016/j.jconrel.2006.12.027. [DOI] [PubMed] [Google Scholar]
- 11.Godbey WT, Wu KK, Mikos AG. Journal of Controlled Release. 1999;60:149–160. doi: 10.1016/s0168-3659(99)00090-5. [DOI] [PubMed] [Google Scholar]
- 12.Pollard H, Remy J-S, Loussouarn G, Demolombe S, Behr J-P, Escande D. Journal of Biological Chemistry. 1998;273:7507–7511. doi: 10.1074/jbc.273.13.7507. [DOI] [PubMed] [Google Scholar]
- 13.Goula D, Remy JS, Erbacher P, Wasowicz M, Levi G, Abdallah B, Demeneix BA. Anglais. 1998;5:712–717. doi: 10.1038/sj.gt.3300635. [DOI] [PubMed] [Google Scholar]
- 14.Demeneix B, Behr J, Leaf Huang M-CH, Ernst W. Advances in Genetics. Volume 53. Academic Press; 2005. pp. 215–230. [Google Scholar]
- 15.Dunlap DD, Maggi A, Soria MR, Monaco L. Nucleic Acids Research. 1997;25:3095–3101. doi: 10.1093/nar/25.15.3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boussif O, Lezoualc’h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr JP. Proceedings of the National Academy of Sciences. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toke ER, Lorincz O, Somogyi E, Lisziewicz J. International Journal of Pharmaceutics. 2010;392:261–267. doi: 10.1016/j.ijpharm.2010.03.048. [DOI] [PubMed] [Google Scholar]
- 18.Wilson RW, Bloomfield VA. Biochemistry. 1979;18:2192–2196. doi: 10.1021/bi00578a009. [DOI] [PubMed] [Google Scholar]
- 19.Dey D, Inayathullah M, Lee AS, LeMieux MC, Zhang X, Wu Y, Nag D, De Almeida PE, Han L, Rajadas J, Wu JC. Biomaterials. 2011;32:4647–4658. doi: 10.1016/j.biomaterials.2011.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang G, Min S-H, Oh E, Hahn S. Biotechnology and Bioprocess Engineering. 2007;12:684–689. [Google Scholar]
- 21.Kim N, Jiang D, Jacobi AM, Lennox KA, Rose SD, Behlke MA, Salem AK. International Journal of Pharmaceutics. 2012;147:123–133. doi: 10.1016/j.ijpharm.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diebold SS, Kursa M, Wagner E, Cotten M, Zenke M. Journal of Biological Chemistry. 1999;274:19087–19094. doi: 10.1074/jbc.274.27.19087. [DOI] [PubMed] [Google Scholar]
- 23.Irache JM, Salman HH, Gamazo C, Espuelas S. Expert Opinion on Drug Delivery. 2008;5:703–724. doi: 10.1517/17425247.5.6.703. [DOI] [PubMed] [Google Scholar]
- 24.Orsolya L, R. TE, Eszter S, Ferenc H, L. CP, F. DJ, János S, Julianna L. Nanomedicine : nanotechnology, biology, and medicine. 2012;8:497–506. doi: 10.1016/j.nano.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nátz E, Lisziewicz J. In: Gene Vaccines. Thalhamer J, Weiss R, Scheiblhofer S, editors. Springer; Vienna: 2012. pp. 127–143. [Google Scholar]
- 26.Lisziewicz J, Trocio J, Whitman L, Varga G, Xu J, Bakare N, Erbacher P, Fox C, Woodward R, Markham P, Arya S, Behr JP, Lori F. Journal of Investigative Dermatology. 2005;124:160–169. doi: 10.1111/j.0022-202X.2004.23535.x. [DOI] [PubMed] [Google Scholar]
- 27.Moghimi SM, Symonds P, Murray JC, Hunter AC, Debska G, Szewczyk A. Mol Ther. 2005;11:990–995. doi: 10.1016/j.ymthe.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 28.Goula D, Benoist C, Mantero S, Merlo G, Levi G, Demeneix BA. Gene Therapy. 1998;5:1291–1295. doi: 10.1038/sj.gt.3300717. [DOI] [PubMed] [Google Scholar]
- 29.Wightman L, Kircheis R, Rössler V, Carotta S, Ruzicka R, Kursa M, Wagner E. The Journal of Gene Medicine. 2001;3:362–372. doi: 10.1002/jgm.187. [DOI] [PubMed] [Google Scholar]
- 30.Jeong JH, Kim SW, Park TG. Progress in Polymer Science. 2007;32:1239–1274. [Google Scholar]
- 31.Somogyi E, Xu J, Gudics Á, Tóth J, Kovács AL, Lori F, Lisziewicz J. Vaccine. 2011;29:744–753. doi: 10.1016/j.vaccine.2010.11.019. [DOI] [PubMed] [Google Scholar]
- 32.Clamme JP, Azoulay J, MÈly Y. Biophysical Journal. 2003;84:1960–1968. doi: 10.1016/S0006-3495(03)75004-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin DC, Dimitriadis EK, Horkay F. Journal of Biomechanical Engineering. 2007;129:430–440. doi: 10.1115/1.2720924. [DOI] [PubMed] [Google Scholar]
- 34.Vijayanathan V, Thomas T, Thomas TJ. Biochemistry. 2002;41:14085–14094. doi: 10.1021/bi0203987. [DOI] [PubMed] [Google Scholar]
- 35.Taber L. Journal of Applied Mechanics. 1982;49:121–128. [Google Scholar]
- 36.Vella D, Ajdari A, Vaziri A, Boudaoud A. Journal of The Royal Society Interface. 2012;9:448–455. doi: 10.1098/rsif.2011.0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roos WH, Wuite GJ. Advanced Materials. 2009;21:1187–1192. [Google Scholar]
- 38.Zoldesi C, Ivanovska I, Quilliet C, Wuite G, Imhof A. Physical Review E. 2008;78:051401. doi: 10.1103/PhysRevE.78.051401. [DOI] [PubMed] [Google Scholar]
- 39.Ivanovska IL, Miranda R, Carrascosa JL, Wuite GJL, Schmidt CF. Proceedings of the National Academy of Sciences. 2011;108:12611–12616. doi: 10.1073/pnas.1105586108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vella D, Ajdari A, Vaziri A, Boudaoud A. Physical review letters. 2012;109:144302. doi: 10.1103/PhysRevLett.109.144302. [DOI] [PubMed] [Google Scholar]
- 41.Nasto A, Ajdari A, Lazarus A, Vaziri A, Reis PM. Soft Matter. 2013;9:6796–6803. [Google Scholar]
- 42.Gao C, Leporatti S, Moya S, Donath E, Möhwald H. Langmuir. 2001;17:3491–3495. [Google Scholar]
- 43.Domján A, Bajdik J, Pintye-Hódi K. Macromolecules. 2009;42:4667–4673. [Google Scholar]
- 44.Chandran P, Dimitriadis E, Horkay F. Probing structure of DNA nanoparticles with short DNA strands. Boston: 2012. [Google Scholar]
- 45.Urban P, Estelrich J, Adeva A, Cortes A, Fernandez-Busquets X. Nanoscale Research Letters. 2011;6:620–628. doi: 10.1186/1556-276X-6-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


