Abstract
Background and objectives
IgA plays a key role in IgA nephropathy (IgAN) by forming immune complexes and depositing in the glomeruli, leading to an inflammatory response. However, the antigenic targets and functional characterization of IgA have been incompletely defined in this disease.
Design, setting, participants, & measurements
This study was performed in sera from patients who were studied as part of a prospective, observational study of IgAN. These patients (n=22) all had biopsy-proven IgAN within 3 years of study initiation, complete clinical data, annual urinary inulin clearance for GFRs, and at least 5 years of follow-up. Progression was defined as loss of >5 ml/min per 1.73 m2 per year of inulin clearance measured over at least 5 years. A protein microarray was used for detection of IgAN-specific IgA autoantibodies in blood across approximately 9000 human antigens to specifically identify the most immunogenic protein targets that drive IgA antibodies in IgAN (n=22), healthy controls (n=10), and non-IgAN glomerular diseases (n=17). Results were validated by ELISA assays in sera and by immunohistochemistry in IgAN kidney biopsies. IgA-specific antibodies were correlated with clinical and histologic variables to assess their effect on disease progression and prognosis.
Results
Fifty-four proteins mounted highly significant IgA antibody responses in patients with IgAN with a false discovery rate (q value) of ≤10%; 325 antibodies (P≤0.05) were increased overall. Antitissue transglutaminase IgA was significantly elevated in IgAN (P<0.001, q value of 0%). IgA antibodies to DDX4 (r=−0.55, P=0.01) and ZADH2 (r=−0.48, P=0.02) were significantly correlated with the decline of renal function. Specific IgA autoantibodies are elevated in IgAN compared with normal participants and those with other glomerular diseases.
Conclusions
In this preliminary study, IgA autoantibodies target novel proteins, highly expressed in the kidney glomerulus and tubules. These IgA autoantibodies may play important roles in the pathogenesis of IgAN.
Keywords: IgA nephropathy, nephritis, primary GN
Introduction
IgA nephropathy (IgAN) is the most common GN in the world and remains a common cause of ESRD (1,2). IgAN is diagnosed by kidney biopsy and defined by the deposition of IgA in the glomerulus with proliferation of the mesangium (3). The pathogenesis of IgAN is still poorly understood. There is likely underlying genetic predisposition as demonstrated by family, racial, and sex predisposition as well as several candidate genetic risk factors (4). Patients with IgAN generally demonstrate increased levels of galactose-deficient IgA1 (5). It has been demonstrated that immune complexes formed from IgA or IgG autoantibodies that recognize galactose-deficient IgA may precipitate in the glomerulus and cause injury (6–9). The prognosis of IgAN is also incompletely determined by typically measured clinical and pathologic features. Patients with greater degrees of proteinuria, renal dysfunction, or pathologic changes (proliferative, necrotic, or sclerotic) have higher risks of progressive disease (10,11). However, greater precision in risk assessment afforded by further biomarkers could aid in determining which patients to observe more closely, treat more aggressively, or include/stratify in treatment trials.
Our group and others previously demonstrated that autoantibodies may be important in the pathogenesis of IgAN (12,13). However, the specificities of IgA antibodies in IgAN have still not been broadly defined. These specificities could help to better understand the pathogenesis of this disease and may provide more refined biomarkers to prognosticate and guide therapy.
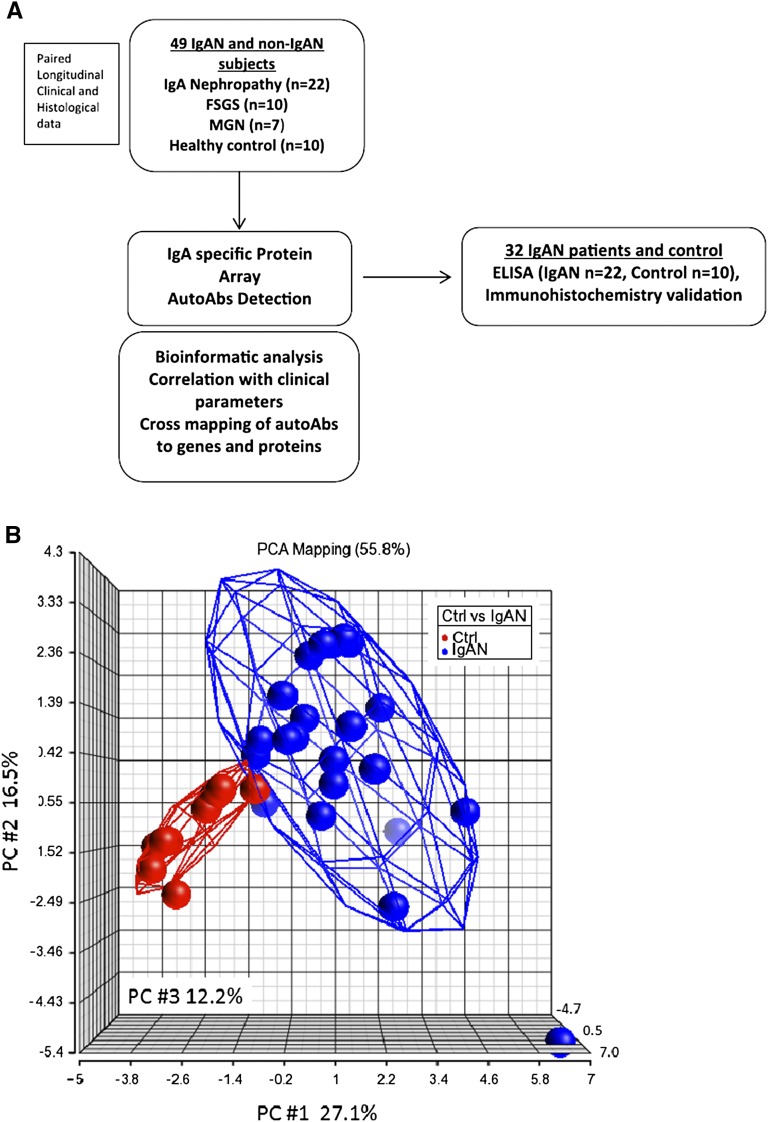
High-density protein microarrays have been used to screen serum antibodies (14) to >8000 individual human protein targets and have yielded findings of clinical interest (15). To characterize IgA and its antigenic targets, we used protein arrays to detect and describe IgA autoantibodies in patients with IgAN. We applied an integrative genomics approach to cross-map the IgA antibodies that are highly expressed in patients with IgAN with protein targets to assess their specificity to the kidney as well as other organs. Potential candidates can then be validated in biopsy tissue by immunohistochemistry and in serum samples by ELISA. The overall study approach is summarized in Figure 1A.
Figure 1.
Work summary and PCA of top 20 IgA autoantibodies. (A) Work flow of IgA autoantibody discovery in IgAN. (B) PCA of top 20 IgA autoantibodies with a cut-off threshold >1.65-fold increase in IgAN with P value <0.05 shows the comparison between IgAN and control participants. The axes represent principle components (PC1, PC2, and PC3). autoAb, autoantibody; Ctrl, control; IgAN, IgA nephropathy; MGN, membranous nephropathy; PCA, principal component analysis; PC, principle component.
Materials and Methods
Patients
Entry criteria for this study was biopsy-confirmed IgAN, and each enrolled patient with IgAN had to have at least 5 years of follow-up with complete clinical data, annual sera and urine sampling, and annually measured GFRs for absolute quantification of renal function. Given these stringent selection criteria, we could enroll 22 patients with IgAN that met all entry criteria.
A total of 49 participants were enrolled for this study: the 22 patients with biopsy-confirmed IgAN who met all above study entry criteria, 17 patients with biopsy-proven non-IgA causes of renal disease, and 10 age- and sex-matched healthy controls.
Within the cohort of IgAN participants, the rate of decline of measured GFR over the 5 years of follow-up was used to subclassify them into two groups. Patients with IgAN were classified as progressors (n=7) if the rate of measured GFR decline (ΔGFR) was >5.0 ml/min per 1.73m2 per year. Patients with a ΔGFR of <5.0 ml/min per 1.73 m2 per year were classified as nonprogressors. Blood (5 ml serum) and urine (50 ml) samples were collected annually over the 5-year follow-up from each patient with IgAN. GFR was measured using the urinary clearance of inulin, as previously described (16). The demographics of these patients are shown in Tables 1 and 2. The characteristics of seven patients with membranous nephropathy (MGN) include male sex (n=4), female sex (n=3), mean age (50.7 years), race (white, 43%; Hispanic, 43%; and Asian, 14%), serum creatinine (0.88 mg/dl), proteinuria (7.6 g/d). The characteristics of 10 patients with FSGS include male sex (n=7), female sex (n=3), mean age (50.7 years), race (white, 50%; Hispanic, 40%; and black, 10%), serum creatinine (2.0 mg/dl), and proteinuria (4.8 g/d). None of our participants were treated with immunosuppression (steroids, cytotoxics) at the time of their blood draws.
Table 1.
Demographics of patients with IgAN and healthy controls
| Demographic | Patients with IgAN | Controls (Non-IgAN) | P Value |
|---|---|---|---|
| Age (yr) | 38.0±10.2 | 29.9±10.4 | 0.05 |
| Sex (men/women) | 12/10 | 5/5 | 0.82 |
| Race (%) | |||
| White | 59 | 56 | |
| Asian | 32 | 33 | |
| Hispanic | 9 | ||
| Black | 11 | ||
| Mean systolic BP (mmHg) | 131.5±9.8 | 127.1±1 | 0.30 |
| Mean diastolic BP(mmHg) | 79.0±13.2 | 73.6±5.9 | 0.16 |
| Serum creatinine (mg/dl) | 1.0±0.31 | 0.94±0.18 | 0.20 |
| GFR (ml/min per 1.73 m2) | 72.0±21.7 | 88.2±12.6 | 0.10 |
| Biopsy features | |||
| Volume fraction of mesangium | 0.22±0.07 | — | — |
| Globally sclerotic glomeruli (%) | 15.8 (0–36) | — | — |
| Fractional interstitial area (%) | 25.1±6.7 | — | — |
Values are presented as the mean±SD unless otherwise indicated. Pathologic values as reported previously. Median and range normal values in our laboratory are volume fraction of mesangium 0.14±0.04, globally sclerotic glomeruli 0, (0–15)%, fractional interstitial area 14.9%±4.8% (26). IgAN, IgA nephropathy.
Table 2.
Demographics of IgAN progressors versus IgAN nonprogressors
| Demographic | IgAN Progressors | IgAN Nonprogressors | P Value |
|---|---|---|---|
| Age (yr) | 39.7±7.3 | 37.6±11.5 | 0.67 |
| Sex (men/women) | 5/2 | 7/8 | 0.30 |
| Mean systolic BP (mmHg) | 131.5±9.8 | 130.3±12.4 | 0.82 |
| Mean diastolic BP (mmHg) | 82.2±9.1 | 77.9±14.8 | 0.49 |
| Serum creatinine (mg/dl) | 1.38±0.22 | 1.05±0.28 | 0.01 |
| GFR (ml/min per 1.73 m2) | 57±13.7 | 79±21.4 | 0.02 |
| ΔGFR (ml/min per 1.73 m2 per yr) | −16.4±15.3 | 0.19±4.7 | <0.001 |
| Proteinuria (g/d) | 3.87±2.63 | 1.28±0.93 | 0.04 |
Immune Response Measurement Using Protein Microarrays
ProtoArray Human Protein Microarray v5.0 (Invitrogen, Carlsbad, CA) was used to characterize the specificity of IgA-specific autoantibody response in IgAN. The relative fluorescence intensity (in relative fluorescence units [RFU]) was measured using GenePix pro 6.0 software (Molecular Devices, Sunnyvale, CA). Data normalization was done using ProtoArray Prospector 5.2. We used the quantile normalization method because there were no positive controls for IgA to apply Robust Linear Model for normalization (17). Fluorescence signal values were measured for each protein after adjusting for background correction so that Z factors for all of the corrected intensities of the human protein features could be calculated. The differentially increased antibody signal in IgAN was analyzed after based on our previous quality control experience (14,18). The minimum signal threshold was set at 500 RFU and signal difference required between IgAN and healthy controls for any antibody was at least 200 RFU, and a Z score value of >3.0 was used as a parameter to identify significant antibody signals. The IgA signal was normalized using quantile normalization. Pearson’s correlation coefficients between selected antibodies and the rate of renal function decline (Δ inulin GFR, in milliliters per year) were calculated after transforming antibody signal intensities with the use of base-2 logarithms. The list of autoantibodies highly expressed in both IgAN and non-IgAN groups were cross-mapped based on statistical significance (P<0.05).
The analysis was also done with SAS software (version 9.2, enterprise guide 4.2). The pathway analysis was performed using Ingenuity Pathway Analysis (http://www.ingenuity.com).
Validation of IgA Antibodies Based on ELISA
Validation of IgAN-specific IgA autoantibodies identified by protein arrays was performed using the MSD ELISA platform (Meso Scale Discovery, Gaithersburg, MD). Receiver operating characteristic (ROC) analysis was conducted on a combined panel of antibodies to predict IgAN with fitted logistic regression models using log-transformed ELISA levels of antibodies against SIX homeobox 2 (SIX2), membrane protein, palmitoylated 1 (MPP1), TEA domain family member 4 (TEAD4), transglutaminase 2 (T-TG), zinc binding alcohol dehydrogenase domain containing 2 (ZADH2), glutamate receptor, ionotropic, N-methyl-D-aspartate-like 1A (GRINL1A), and ariadne homolog 2 (ARIH2). For ROC analyses, we generated all corresponding sensitivities and specificities for each of cut-off point (theta).
Immunohistochemistry
To evaluate the presence of DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 (DDX4), ZADH2, and GRINL1A in kidney tissue, we performed immunohistochemical staining on formalin-fixed, paraffin-embedded kidney biopsy tissue using corresponding antibodies. We used a new set of IgAN kidney biopsy samples, new normal kidney control samples (obtained from the normal kidney region obtained from nephrectomy samples for renal tumor), and FSGS and MGN tissue to evaluate their expression in kidneys.
Results
IgA Autoantibody Profiling in IgAN
Using an innovative IgA profiling method that allowed us to profile IgA antibodies against approximately 9000 human antigens, we were able to detect IgA autoantibodies in human serum samples. We identified 325 (P≤0.05) IgA autoantibodies that were increased in the sera collected from patients with IgAN compared with healthy controls, of which 54 met stricter statistical criteria, because they were increased with a false discover rate (q value) of ≤10%. The 20 IgA autoantibody targets of highest significance are listed in Table 3. Principal component analysis of these top 20 antibodies compared with non-IgAN controls demonstrated a clear difference in the pattern of participants with IgAN versus controls (Figure 1B). The significant antigens were subjected to pathway analysis using Ingenuity Pathway Analysis (http://www.ingenuity.com; Ingenuity Systems) which identified cell-mediated immune response, cellular development, cellular function, and maintenance as the main biologic processes of these proteins. We used the GO-Elite Pathway Analysis Tool (http://www.genmapp.org/go_elite/go_elite.html) to look for enriched pathways based on the immunogenic epitopes for IgA reactivity. The most significantly enriched pathways included the G13 signaling pathway (P=0.001), DNA damage response (P=0.001), the MAPK signaling pathway (P=0.002), and the WNT signaling pathway (P=0.003).
Table 3.
Top 20 most significant antigenic targets for autoantibodies in IgAN
| No. | Protein Name | Gene Symbol | P Value for IgAN | Q Value | Fold Increase | Expressed in Glomerulus | Expressed in Tubules |
|---|---|---|---|---|---|---|---|
| 1 | Transglutaminase 2 | TGM2 | <0.01 | 0.0 | 2.3 | Y | Y |
| 2 | TEA domain family member 4 | TEAD4 | <0.01 | 0.0 | 3.1 | NA | NA |
| 3 | Four and a half LIM domains protein 3 | FHL3 | <0.01 | 4.8 | 1.8 | Y | N |
| 4 | Homeobox C8 | HOXC8 | <0.01 | 4.8 | 1.9 | Y | Y |
| 5 | Membrane protein, palmitoylated 1 | MPP1 | <0.01 | 4.8 | 2.8 | N | Y |
| 6 | Polyadenylate- interacting protein 2 | PAIP2 | <0.01 | 4.8 | 2.5 | Y | Y |
| 7 | Ariadne homolog 2 | ARIH2 | <0.01 | 4.8 | 1.7 | NA | NA |
| 8 | LUC7-like 2 | LUC7L2 | 0.02 | 4.8 | 1.9 | NA | NA |
| 9 | Tandem C2 domains, nuclear | MTAC2D1 | 0.02 | 4.8 | 1.7 | N | Y |
| 10 | Protein phosphatase 2 | PPP2R5C | 0.01 | 5.9 | 2.0 | Y | Y |
| 11 | Pescadillo homolog 1 | PES1 | <0.01 | 7.6 | 2.6 | NA | NA |
| 12 | Leucine rich repeat containing 48 | LRRC48 | <0.0 | 7.6 | 1.9 | Y | Y |
| 13 | Developmental pluripotency- protein | DPPA4 | <0.01 | 7.6 | 2.1 | Y | Y |
| 14 | ATM/ATR-Substrate Chk2-Interacting Zn2+-finger protein | ASCIZ | 0.01 | 9.1 | 3.4 | NA | NA |
| 15 | Nuclear respiratory factor 1 | NRF1 | <0.01 | 7.6 | 2.2 | Y | Y |
| 16 | Hypothetical protein FLJ23356 | FLJ23356 | 0.01 | 7.6 | 1.9 | NA | NA |
| 17 | G protein-coupled receptor kinase | GIT2 | 0.01 | 7.6 | 2.2 | Y | Y |
| 18 | TANK-binding kinase 1 | TBK1 | 0.01 | 7.6 | 1.9 | NA | NA |
| 19 | Golgi SNAP receptor complex | GOSR1 | 0.01 | 7.6 | 1.9 | N | Y |
| 20 | Protein MYLE | DEXI | 0.02 | 7.6 | 2.4 | Y | Y |
Source: www.proteinatlas.org. Y, yes; NA, data not available; N, no.
IgA Immune Response Profiling in Patients with Progressive IgAN versus Nonprogressive IgAN
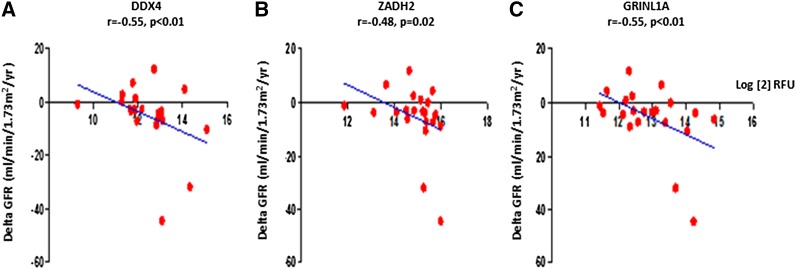
When we compared the IgA autoantibody profiles of patients with IgAN with progressive kidney disease versus those who did not have progressive disease but stable renal function, 109 autoantibodies were increased (P<0.05) and the top 20 most significant antigens are shown in Table 4. We performed a correlation analysis of each of these antibody levels with the change in GFR over a 5-year duration, as determined by annual measurements of inulin GFR. IgA antibodies to DDX4 showed a significant correlation to the decline of renal function (r=−0.55, P=0.01). DDX4 is a putative RNA helicase, with different spliced forms, and is involved in the alteration of RNA secondary structure, and mitochondrial splicing (19). IgA antibodies to GRINL1A and ZADH 2 also showed a significant correlation with the decline of renal function (GRINL1A ΔGFR, r=−0.44, P=0.04; ZADH2 ΔGFR, r=−0.48, P=0.02) (Figure 2). GRINL1A is the central component of the basal RNA polymerase II transcription machinery and has multiple alternatively spliced variants (20). ZADH2 also has four alternatively spliced mRNAs and plays a role in the redox reactions of cells, localizing primarily in the mitochondria. It is associated with a congenital aural atresia phenotype in individuals with the 18q deletion syndrome (21).
Table 4.
Top 20 most significant antigenic targets for autoantibodies in progressive IgAN
| No. | Protein Name | P Value |
|---|---|---|
| 1 | LIM and senescent cell antigen-like-containing domain protein 1 | <0.01 |
| 2 | Glial fibrillary acidic protein (GFAP) | <0.01 |
| 3 | Ubiquitin-conjugating enzyme E2E 2 (UBC4/5 homolog, yeast) (UBE2E2) | <0.01 |
| 4 | Regulator of G-protein signaling 3 (RGS3), transcript variant 4 | <0.01 |
| 5 | MAGUK p55 subfamily member 5 | <0.01 |
| 6 | Membrane protein, palmitoylated 5 (MAGUK p55 subfamily member 5) (MPP5) | <0.01 |
| 7 | Piccolo (presynaptic cytomatrix protein) (PCLO) | <0.01 |
| 8 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 (DDX4)a | <0.01 |
| 9 | Putative uncharacterized protein C14orf177 | <0.01 |
| 10 | RWD domain containing 2B (RWDD2B) | <0.01 |
| 11 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 17 (DDX17), transcript variant 2 | 0.01 |
| 12 | Rho GTPase-activating protein 15 | 0.01 |
| 13 | Glutamate receptor, N-methyl D-aspartate-like 1A (GRINL1A)a | 0.01 |
| 14 | Tropomodulin 4 (muscle) (TMOD4) | 0.01 |
| 15 | p21(CDKN1A)-activated kinase 4 (PAK4), transcript variant 1 | 0.01 |
| 16 | Branched chain ketoacid dehydrogenase kinase (BCKDK) | 0.01 |
| 17 | TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa (TAF6), transcript variant 1 | 0.01 |
| 18 | Kinesin family member 6 (KIF6) | 0.01 |
| 19 | Tropomodulin 1 (TMOD1) | 0.01 |
| 20 | Zinc binding alcohol dehydrogenase, domain containing 2 (ZADH2)a | 0.01 |
Autoantibodies of antigens correlated with renal function decline (Figure 2).
Figure 2.
Correlation of DDX4, ZADH2, and GRINL1A with renal function decline. DDX4 (A), ZADH2 (B), and GRINL1A (C) showed significant correlations with the decline of renal function (ΔGFR). Log[2] RFU, relative fluorescent units.
Specificity of IgA Autoantibodies
To ascertain whether the relative specificity of the IgA autoantibody profile in patients with IgAN was specific to this etiology and not just to glomerular injury, 23 additional protein arrays were conducted on sera samples on an additional group of participants with glomerular diseases (10 with FSGS and seven with MGN) and were evaluated together with six additional normal controls. The autoantibody panel increased in IgA was found to be largely distinct from these other glomerular diseases. Among 93 IgA autoantibodies that were significantly increased in IgAN (P<0.01), only five autoantibodies were also increased in FSGS and one autoantibody was also increased in MGN. The five autoantibodies increased in both IgAN and FSGS included CREB regulated transcription coactivator 2 (CRTC2), polyadenylate-binding protein-interacting protein 2, ARIH2, ankyrin repeat family A 2 (ANKRA2), and ninjurin 2 (NINJ2). Possibly of interest, ANKRA2 was increased in all three diseases. ANKRA2 is a protein that interacts with the cytoplasmic tail of megalin, which is a multiligand scavenger protein in proximal renal tubules (22).
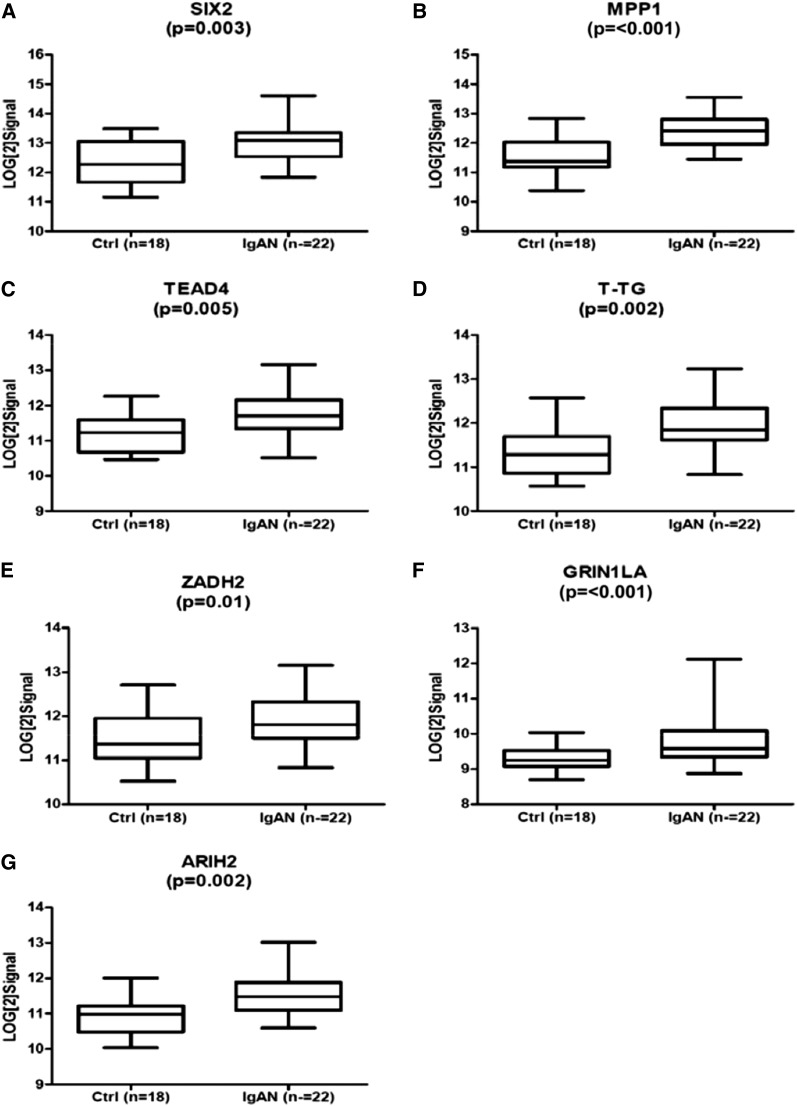
Customized ELISA Verification of IgAN-Specific Antibodies
To confirm the findings of the protein array analysis, we selected seven autoantibodies for further ELISA validation based on both the confirmed expression of the corresponding protein (antigen) in the kidney (as previously demonstrated by immunohistochemistry) and strong statistical significance (measured by the fold increase and q value <10%). These autoantibodies included IgA against SIX2, membrane protein, MPP1, ZADH2, GRINL1A, TEAD4, T-TG (also known as TGM2), and ARIH2. As demonstrated in Figure 3, each of these autoantibodies was confirmed to be significantly increased in IgAN compared with controls (P<0.01). The difference in these IgA antibodies remained significant when normalized against total IgA to test for possible effect of varying IgA levels in these samples: MPP1 (P=0.001), TEAD4 (P=0.01), T-TG (P=0.01), ZADH2 (P=0.03), GRIN1LA (P=0.03), and ARIH2 (P=0.01).
Figure 3.
ELISA verification of IgA antibodies. Seven antibodies were subjected for verification by ELISA assay. This included IgA against SIX homeobox 2 (SIX2) (A), membrane protein, palmitoylated 1 (MPP1) (B), zinc binding alcohol dehydrogenase domain containing 2 (ZADH2) (C), glutamate receptor, ionotropic, N-methyl D-aspartate-like 1A (GRINL-1A) (D), TEA domain family member 4 (TEAD4) (E), Transglutaminase 2 (T-TG) (F), and ariadne homolog 2 (ARIH2) (G).
Prediction of IgAN by Autoantibodies
The diagnosis of IgAN was strongly associated with seven autoantibodies by ROC curve analysis. The individual area under the curve (AUC) values for each of the autoantibodies were as follows: 0.84 for MPP1, 0.79 for T-TG, 0.79 for ARIH2, 0.78 for SIX2, 0.77 for GRIN1LA, 0.76 for TEAD4, and 0.74 for ZADH2. The significance of IgAN-specific antibodies was demonstrated by comparative ROC analyses that was performed on four IgAN-specific IgG antibodies against MATN2, UBE2W, DDX17, and PRKD1 (13) and six IgAN-specific IgA antibodies against GRINL1A, ZADH2, TEAD4, MPP1, TGM2, and SIX2. As evident from AUC from the two ROC analyses, combined performance of six IgAN-specific IgA antibodies was more discriminatory (AUC=1) compared with IgAN-specific IgG antibodies (AUC=0.86) as described previously (13). Of note, anti–T-TG IgA antibody has been used for the diagnosis of celiac disease (23), wherein there appears to be an increased risk of IgAN (24).
Immunohistochemistry
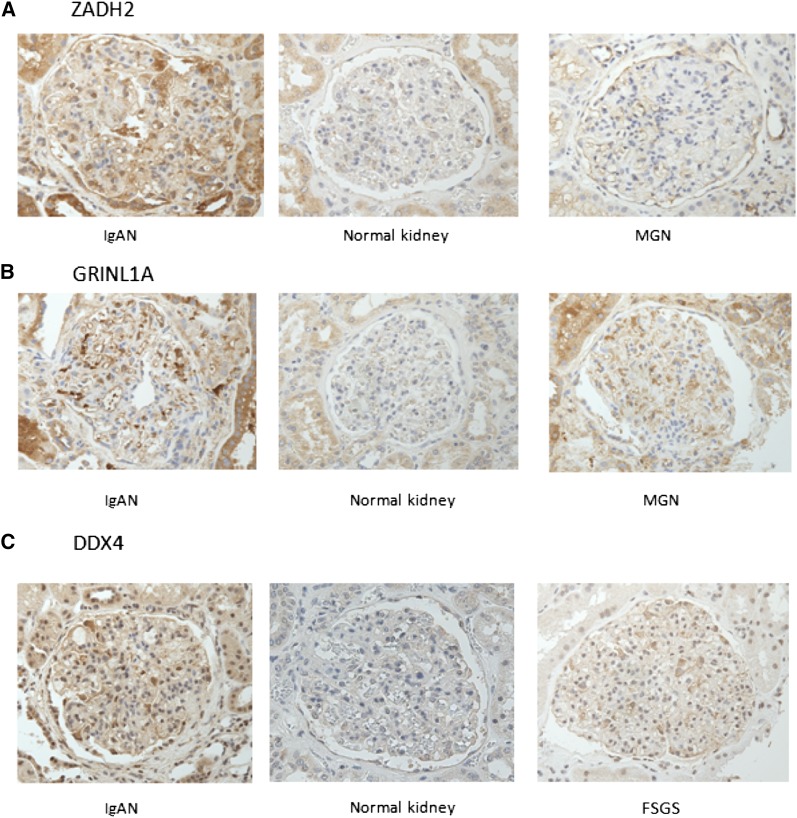
For those IgA autoantibodies that significantly associated with decline of renal function in IgAN (ZADH2, GRINL1A, and DDX4), we performed immunohistochemistry for their corresponding proteins/antigens on kidney biopsy samples from patients with the histologic diagnosis of IgAN, non-IgAN glomerular diseases (FSGS, membranous), and normal control kidney tissue (from microdissected kidney biopsy samples of normal kidney from resected renal tumor specimens). As demonstrated in Figure 4, immune staining for these antigens in the podocytes and tubular epithelium of participants with IgAN was present or relatively increased compared with normal control tissue or other glomerular diseases. This may suggest that increased or altered tissue expression of the corresponding antigen may underlie the increased autoantibody response to these renal proteins in IgAN.
Figure 4.
Immunohistological assessment of IgA specific antigens ZADH2, GRINL1A, and DDX4 in kidneys. (A) ZADH2 staining illustrates increased staining in podocytes in IgAN compared with normal kidney and MGN. (B) GRINL1A staining shows staining in IgAN and MGN, but normal kidneys did not show significant staining. (C) DDX4 staining shows cytoplasmic staining in glom and FSGS shows 1+ staining in cytoplasm.
Discussion
This study was carried out to further the understanding of IgAN by examining the antigen specificity of IgA among patients with IgAN. Utilizing protoarray technology, we were able to analyze each individual’s repertoire of autoantibodies to >9000 human antigens. We found a substantial difference in the levels of many antigen-specific IgA antibodies in participants with IgAN compared with healthy controls. When compared by principal component analysis, the patterns were highly distinct between controls and patients with disease. The results of the protoarray analysis were subsequently confirmed by ELISA for each of the autoantibodies that were tested, selected for the magnitude of difference from controls and for their possible biologic relevance. Because our patients had been observed and characterized for an average of 5 years, autoantibody levels could be compared in a group of patients with progression toward dialysis versus those without progression. Several of the autoantibodies were significantly higher among the progressor group, and a group of these antibodies correlated significantly with the loss of renal function, defined by the decline in measured GFR observed during the study. Comparison of autoantibody levels in the patients with IgAN to another control group of normal participants and patients with other progressive, proteinuric renal diseases suggests that the autoantibody patterns are distinct among different glomerular diseases with very little overlap. Finally, in an analysis of the described functions of the antigens targeted by IgA, potential mechanisms of disease are revealed, such as apoptosis, fibrosis, and inflammation. For example, we find potential for activation of the G13 pathway, which has been associated with podocyte collagen activation, proteinuria, and glomerulosclerosis (25). Likewise, the antibodies interacted with proteins that interact with the WNT signaling pathway and there has been evidence that changes in WNT signaling may be important in IgAN (26). These findings suggest that IgA could potentially be pathogenic in this disease through stimulating these mechanisms. If confirmed in larger cohorts, these IgA autoantibodies could serve as a biomarker or prognostic measure and may be proven to play a role in the progression of renal injury.
The pathogenesis of IgAN continues to emerge. Over recent years, there has been substantial focus on the role of systemic IgA in subsequent renal injury. Many studies have documented increased levels of galactose-deficient IgA among patients with IgAN (27). However, levels do not necessarily predict disease or disease severity among individuals. Further work has established the existence of autoantibodies to galactose-deficient IgA1. The levels of IgG and IgA specific to galactose-deficient IgA1 seem to be more predictive of disease and of disease severity (28). However, given that these autoantibodies are also found in healthy individuals, and that some patients with relatively low levels can manifest severe disease, other mechanisms and measures are still being sought. This study could not evaluate IgA autoantibodies to galactose-deficient IgA because specific hinge region components are not included in the antigen array.
We previously suggested that there are a wide number of IgG autoantibodies to various antigens, including many found in the kidney, increased among patients with IgAN (13). In this study, we find that IgA autoantibodies to various antigens, again including many that are found in the kidney itself, occur in a distinct pattern in IgAN and correlate with progression. However, many of the autoantibodies most specific for IgAN diagnosis were not strongly associated with the actual progression of renal disease. First, this observation may be due to the limited number of participants in this study who experienced disease progression. Inclusion of a larger cohort may show that some of these autoantibodies are significant in renal disease progression. Second, the most significant autoantibodies associated with the progression of the renal disease in this study may be nonspecific to disease etiology, in a way similar to proteinuria, irrespective of the initiating pathogenic mechanisms. Because we do not have serial GFR measurements in the non-IgAN cohort, we cannot test this hypothesis. This could be addressed by validation of these autoantibodies in prospective studies of progressive renal injury. In addition, although the exact functions of these autoantibodies are uncertain, some like anti T-Tg, found in celiac disease, may point to common pathways of autoimmune injury as a trigger for many renal diseases.
A distinct strength of this study is its highly select sample group, with detailed demographics, quantitative renal function data, and extended follow-up to evaluate the natural history of the disease with outcomes of disease progression. The bioinformatics analyses allowed for robust selection of highly significant IgA autoantibodies that segregated with IgA diagnosis and IgA-related progressive renal injury, which was verified and validated by the customized reverse ELISA assays. The defined panel of autoantibodies appears to have substantial accuracy for noninvasive detection of IgA disease and prediction of IgA renal progression.
We previously reported that IgG autoantibodies may play a role in IgAN through the recognition of kidney antigens and activation of immune response. The present, more robust results may suggest that circulating IgA has more refined specificities to antigens within the kidney and elsewhere, which may prove helpful in the diagnosis and prognosis of IgAN. It appears possible that some of these autoantibodies could be mediators of injury and potential targets of therapy. The possibility that both IgG and IgA contribute to disease activity is consistent with the hypothesis of others, who have demonstrated this for antibodies to galactose-deficient IgA (28). Although mesangial deposits of IgA immune complex have been known to be a main pathogenic feature of IgAN, it has also been shown that interstitial expansion and podocytopenia are strong predictors of the progression of IgAN (1,29). This study suggests that autoantibodies to specific antigenic targets in podocytes and tubules may be pathogenic for IgAN as they correlate with disease progression.
Nevertheless, there are significant limitations of this study. We did not have the ability to measure antibodies to galactose-deficient IgA antibodies or to measure glycation status of antigens. Therefore, we cannot comment on the potential contribution of galactose-deficient IgA antibodies to these findings. Furthermore, to allow for robust discovery across the large number of antigens screened, we used very stringent criteria for patient selection to limit false discovery rates, which limited the number of the participants that were available for inclusion in the discovery set in this study. Finally, because of the high cost associated with using high-density protein arrays, we have not been able to test a larger cohort of patients without IgAN. To overcome these limitations, the predefined antibody panels that correlated with IgAN diagnosis and progression might be used for independent validation in larger studies of patients with IgAN and other glomerular diseases, using lower-cost ELISA-based assays of the predefined antigenic targets. The validity of our studies and these conclusions cannot be well established until this is completed.
Disclosures
A patent related to the IgAN biomarker is pending. M.S. serves as a consultant for Bristol Meyers, Immucor, and ISIS.
Acknowledgments
The authors thank Dr. Bryan Myers for contributing precious plasma/serum samples and clinical histories on patients, as well as Dr. Neeraja Kambham for her reviewing and grading of the pathology for immunohistochemistry. The authors also thank the Stanford Functional Genomics Center at Stanford University for the Axon scanner, Dr. Marianne Delville for her support with the MSD ELISA, and Sarwal laboratory members for their help.
This work was supported by the Sobrato Research Fund (to S.H.W and R.A.L.).
Footnotes
Published online ahead of print. Publication date available at www.cjasn.org.
References
- 1.Lemley KV, Lafayette RA, Derby G, Blouch KL, Anderson L, Efron B, Myers BD: Prediction of early progression in recently diagnosed IgA nephropathy. Nephrol Dial Transplant 23: 213–222, 2008 [DOI] [PubMed] [Google Scholar]
- 2.Berger J, Hinglais N: [Intercapillary deposits of IgA-IgG]. J Urol Nephrol (Paris) 74: 694–695, 1968 [PubMed] [Google Scholar]
- 3.Cattran DC, Coppo R, Cook HT, Feehally J, Roberts IS, Troyanov S, Alpers CE, Amore A, Barratt J, Berthoux F, Bonsib S, Bruijn JA, D’Agati V, D’Amico G, Emancipator S, Emma F, Ferrario F, Fervenza FC, Florquin S, Fogo A, Geddes CC, Groene HJ, Haas M, Herzenberg AM, Hill PA, Hogg RJ, Hsu SI, Jennette JC, Joh K, Julian BA, Kawamura T, Lai FM, Leung CB, Li LS, Li PK, Liu ZH, Mackinnon B, Mezzano S, Schena FP, Tomino Y, Walker PD, Wang H, Weening JJ, Yoshikawa N, Zhang H, Working Group of the International IgA Nephropathy Network and the Renal Pathology Society : The Oxford classification of IgA nephropathy: Rationale, clinicopathological correlations, and classification. Kidney Int 76: 534–545, 2009 [DOI] [PubMed] [Google Scholar]
- 4.Kiryluk K, Julian BA, Wyatt RJ, Scolari F, Zhang H, Novak J, Gharavi AG: Genetic studies of IgA nephropathy: Past, present, and future. Pediatr Nephrol 25: 2257–2268, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allen AC, Bailey EM, Brenchley PE, Buck KS, Barratt J, Feehally J: Mesangial IgA1 in IgA nephropathy exhibits aberrant O-glycosylation: Observations in three patients. Kidney Int 60: 969–973, 2001 [DOI] [PubMed] [Google Scholar]
- 6.Galla JH: IgA nephropathy. Kidney Int 47: 377–387, 1995 [DOI] [PubMed] [Google Scholar]
- 7.Coppo R, Amore A, Hogg R, Emancipator S: Idiopathic nephropathy with IgA deposits. Pediatr Nephrol 15: 139–150, 2000 [DOI] [PubMed] [Google Scholar]
- 8.Coppo R, Troyanov S, Camilla R, Hogg RJ, Cattran DC, Cook HT, Feehally J, Roberts IS, Amore A, Alpers CE, Barratt J, Berthoux F, Bonsib S, Bruijn JA, D’Agati V, D’Amico G, Emancipator SN, Emma F, Ferrario F, Fervenza FC, Florquin S, Fogo AB, Geddes CC, Groene HJ, Haas M, Herzenberg AM, Hill PA, Hsu SI, Jennette JC, Joh K, Julian BA, Kawamura T, Lai FM, Li LS, Li PK, Liu ZH, Mezzano S, Schena FP, Tomino Y, Walker PD, Wang H, Weening JJ, Yoshikawa N, Zhang H, Working Group of the International IgA Nephropathy Network and the Renal Pathology Society : The Oxford IgA nephropathy clinicopathological classification is valid for children as well as adults. Kidney Int 77: 921–927, 2010 [DOI] [PubMed] [Google Scholar]
- 9.D’Amico G: Natural history of idiopathic IgA nephropathy: role of clinical and histological prognostic factors. Am J Kidney Dis 36: 227–237, 2000 [DOI] [PubMed] [Google Scholar]
- 10.Berthoux F, Mohey H, Laurent B, Mariat C, Afiani A, Thibaudin L: Predicting the risk for dialysis or death in IgA nephropathy. J Am Soc Nephrol 22: 752–761, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roberts IS, Cook HT, Troyanov S, Alpers CE, Amore A, Barratt J, Berthoux F, Bonsib S, Bruijn JA, Cattran DC, Coppo R, D’Agati V, D’Amico G, Emancipator S, Emma F, Feehally J, Ferrario F, Fervenza FC, Florquin S, Fogo A, Geddes CC, Groene HJ, Haas M, Herzenberg AM, Hill PA, Hogg RJ, Hsu SI, Jennette JC, Joh K, Julian BA, Kawamura T, Lai FM, Li LS, Li PK, Liu ZH, Mackinnon B, Mezzano S, Schena FP, Tomino Y, Walker PD, Wang H, Weening JJ, Yoshikawa N, Zhang H, Working Group of the International IgA Nephropathy Network and the Renal Pathology Society : The Oxford classification of IgA nephropathy: Pathology definitions, correlations, and reproducibility. Kidney Int 76: 546–556, 2009 [DOI] [PubMed] [Google Scholar]
- 12.Matsiota P, Dosquet P, Louzir H, Druet E, Druet P, Avrameas S: IgA polyspecific autoantibodies in IgA nephropathy. Clin Exp Immunol 79: 361–366, 1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sigdel TK, Woo SH, Dai H, Khatri P, Li L, Myers B, Sarwal MM, Lafayette RA: Profiling of autoantibodies in IgA nephropathy, an integrative antibiomics approach. Clin J Am Soc Nephrol 6: 2775–2784, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li L, Wadia P, Chen R, Kambham N, Naesens M, Sigdel TK, Miklos DB, Sarwal MM, Butte AJ: Identifying compartment-specific non-HLA targets after renal transplantation by integrating transcriptome and “antibodyome” measures. Proc Natl Acad Sci U S A 106: 4148–4153, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mattoon D, Michaud G, Merkel J, Schweitzer B: Biomarker discovery using protein microarray technology platforms: Antibody-antigen complex profiling. Expert Rev Proteomics 2: 879–889, 2005 [DOI] [PubMed] [Google Scholar]
- 16.Squarer A, Lemley KV, Ambalavanan S, Kristal B, Deen WM, Sibley R, Anderson L, Myers BD: Mechanisms of progressive glomerular injury in membranous nephropathy. J Am Soc Nephrol 9: 1389–1398, 1998 [DOI] [PubMed] [Google Scholar]
- 17.Sboner A, Karpikov A, Chen G, Smith M, Mattoon D, Freeman-Cook L, Schweitzer B, Gerstein MB: Robust-linear-model normalization to reduce technical variability in functional protein microarrays. J Proteome Res 8: 5451–5464, 2009 [DOI] [PubMed] [Google Scholar]
- 18.Sutherland SM, Li L, Sigdel TK, Wadia PP, Miklos DB, Butte AJ, Sarwal MM: Protein microarrays identify antibodies to protein kinase Czeta that are associated with a greater risk of allograft loss in pediatric renal transplant recipients. Kidney Int 76: 1277–1283, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hickford DE, Frankenberg S, Pask AJ, Shaw G, Renfree MB: DDX4 (VASA) is conserved in germ cell development in marsupials and monotremes. Biol Reprod 85: 733–743, 2011 [DOI] [PubMed] [Google Scholar]
- 20.Roginski RS, Mohan Raj BK, Birditt B, Rowen L: The human GRINL1A gene defines a complex transcription unit, an unusual form of gene organization in eukaryotes. Genomics 84: 265–276, 2004 [DOI] [PubMed] [Google Scholar]
- 21.Dostal A, Nemeckova J, Gaillyova R, Vranova V, Zezulkova D, Lejska M, Slapak I, Dostalova Z, Kuglik P: Identification of 2.3-Mb gene locus for congenital aural atresia in 18q22.3 deletion: A case report analyzed by comparative genomic hybridization. Otol Neurotol 27: 427–432, 2006 [DOI] [PubMed] [Google Scholar]
- 22.Rader K, Orlando RA, Lou X, Farquhar MG: Characterization of ANKRA, a novel ankyrin repeat protein that interacts with the cytoplasmic domain of megalin. J Am Soc Nephrol 11: 2167–2178, 2000 [DOI] [PubMed] [Google Scholar]
- 23.Baudon JJ, Johanet C, Absalon YB, Morgant G, Cabrol S, Mougenot JF: Diagnosing celiac disease: A comparison of human tissue transglutaminase antibodies with antigliadin and antiendomysium antibodies. Arch Pediatr Adolesc Med 158: 584–588, 2004 [DOI] [PubMed] [Google Scholar]
- 24.Welander A, Sundelin B, Fored M, Ludvigsson JF: Increased risk of IgA nephropathy among individuals with celiac disease. J Clin Gastroenterol 47: 678–683, 2013 [DOI] [PubMed] [Google Scholar]
- 25.Boucher I, Yu W, Beaudry S, Negoro H, Tran M, Pollak MR, Henderson JM, Denker BM: Gα12 activation in podocytes leads to cumulative changes in glomerular collagen expression, proteinuria and glomerulosclerosis. Lab Invest 92: 662–675, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cox SN, Sallustio F, Serino G, Pontrelli P, Verrienti R, Pesce F, Torres DD, Ancona N, Stifanelli P, Zaza G, Schena FP: Altered modulation of WNT-beta-catenin and PI3K/Akt pathways in IgA nephropathy. Kidney Int 78: 396–407, 2010 [DOI] [PubMed] [Google Scholar]
- 27.Suzuki H, Fan R, Zhang Z, Brown R, Hall S, Julian BA, Chatham WW, Suzuki Y, Wyatt RJ, Moldoveanu Z, Lee JY, Robinson J, Tomana M, Tomino Y, Mestecky J, Novak J: Aberrantly glycosylated IgA1 in IgA nephropathy patients is recognized by IgG antibodies with restricted heterogeneity. J Clin Invest 119: 1668–1677, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berthoux F, Suzuki H, Thibaudin L, Yanagawa H, Maillard N, Mariat C, Tomino Y, Julian BA, Novak J: Autoantibodies targeting galactose-deficient IgA1 associate with progression of IgA nephropathy. J Am Soc Nephrol 23: 1579–1587, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lemley KV, Lafayette RA, Safai M, Derby G, Blouch K, Squarer A, Myers BD: Podocytopenia and disease severity in IgA nephropathy. Kidney Int 61: 1475–1485, 2002 [DOI] [PubMed] [Google Scholar]