Interaction with a protein phosphatase and dephosphorylation affects the polar localization and endocytic trafficking of an auxin-related membrane protein and its impact on cell pattern formation.
Abstract
In plants, cell morphogenesis is dependent on intercellular auxin accumulation. The polar subcellular localization of the PIN-FORMED (PIN) protein is crucial for this process. Previous studies have shown that the protein kinase PINOID (PID) and protein phosphatase6-type phosphatase holoenzyme regulate the phosphorylation status of PIN1 in root tips and shoot apices. Here, we show that a type-one protein phosphatase, TOPP4, is essential for the formation of interdigitated pavement cell (PC) pattern in Arabidopsis (Arabidopsis thaliana) leaf. The dominant-negative mutant topp4-1 showed severely inhibited interdigitated PC growth. Expression of topp4-1 gene in wild-type plants recapitulated the PC defects in the mutant. Genetic analyses suggested that TOPP4 and PIN1 likely function in the same pathway to regulate PC morphogenesis. Furthermore, colocalization, in vitro and in vivo protein interaction studies, and dephosphorylation assays revealed that TOPP4 mediated PIN1 polar localization and endocytic trafficking in PCs by acting antagonistically with PID to modulate the phosphorylation status of PIN1. In addition, TOPP4 affects the cytoskeleton pattern through the Rho of Plant GTPase-dependent auxin-signaling pathway. Therefore, we conclude that TOPP4-regulated PIN1 polar targeting through direct dephosphorylation is crucial for PC morphogenesis in the Arabidopsis leaf.
Polarity is a fundamental characteristic underlying various cellular processes. In plants, cell polarity and morphogenesis are closely linked to the development and function of certain cells in their corresponding tissues and organs. Arabidopsis (Arabidopsis thaliana) leaf pavement cells (PCs), which have a jigsaw puzzle shape, provide a representative model to study the mechanisms of plant cell polarity and morphogenesis (Yang, 2008; Qian et al., 2009). The interdigitated lobes and indentations of PCs result from intercalary growth (Fu et al., 2005). he cell polarity and morphogenesis are dependent on cell adhesion, signaling networks, the cytoskeleton, and protein transport (Falbel et al., 2003; Mathur et al., 2003; Fu et al., 2005; Wang et al., 2007; Chary et al., 2008; Xu et al., 2010; Li et al., 2013). A well-known mechanism that controls interdigitated growth of PCs is Rho of Plant (ROP) GTPase-mediated cytoskeletal reorganization in Arabidopsis (Fu et al., 2002, 2005, 2009). The cytoskeleton is thought to be critical for the PC shape. Cortical filamentous actins (F-actins) are localized to lobe sites and promote lobe initiation and outgrowth, whereas highly ordered cortical microtubules (MTs) are arranged transversely in the neck regions and restrict cell expansion (Fu et al., 2005). In PC lobes, activated ROP2 and ROP4 act redundantly to promote lobe growth via activation of ROP-interactive Cdc42- and Rac-interactive binding motif-containing protein4 (RIC4)-mediated assembly of cortical diffuse F-actins, and also by suppressing RIC1-mediated organization of cortical MT arrays (Fu et al., 2005). In the indentations of PCs, activated ROP6 interacts with RIC1 to promote well-ordered MTs and suppress lateral expansion (Fu et al., 2009). Recently, it was reported that katanin (KATANIN1) is a downstream component of RIC1, which promotes MT-severing activity by interacting with RIC1 in Arabidopsis PCs (Lin et al., 2013).
Auxin is a fundamental plant hormone that regulates many aspects of plant development and mediates various cellular responses (Wu et al., 2011). Treatment of plants with the synthetic auxin naphthalene acetic acid (NAA) promotes PC lobe formation, while quadruple yucca mutants with defects in auxin biosynthesis exhibit reduced PC interdigitation (Xu et al., 2010; Li et al., 2011). More recent studies have revealed that both a plasma membrane-localized transmembrane kinase (TMK) and an auxin binding protein1 (ABP1) form an auxin-sensing complex at cell surface to activate ROP GTPase signaling (Xu et al., 2014). The auxin gradient modulates the ROP-RIC pathway through the ABP1-TMK complex in PCs (Xu et al., 2010, 2014). Auxin is transported from the sites of synthesis to the sites of action via polarized transporters. Polar subcellular localization of the PIN-FORMED (PIN) family proteins directs auxin flow and establishes an auxin gradient (Petrásek et al., 2006; Wisniewska et al., 2006). Reversible protein phosphorylation and dephosphorylation mediated by protein kinases and phosphatases play an essential role in regulating PIN1 polarity and trafficking (Friml et al., 2004; Michniewicz et al., 2007; Kleine-Vehn et al., 2009; Huang et al., 2010). In root and shoot regions, PINOID (PID), a protein Ser/Thr kinase, directly phosphorylates PIN proteins and regulates their polar targeting (Benjamins et al., 2001; Michniewicz et al., 2007). A pid loss-of-function mutant causes an apical-to-basal shift in PIN polarity in shoot meristem, whereas the gain-of-function PID results in the opposite PIN polarity shift (basal to apical). The type-A regulatory subunit of protein phosphatase2A (PP2A) and PID act antagonistically on PIN phosphorylation and polarity (Michniewicz et al., 2007). Recently, it was found that the corresponding PP6-type heterotrimeric holoenzyme complex, consisting of PP2AA, SAL (for Sit4-associated protein [SAP] subunits domain-like), and FyPP1 (phytochrome-associated Ser/Thr protein phosphatase), directly regulates the phosphorylation status of PIN proteins and consequently affects their polarity (Dai et al., 2012). In addition, PP2A and PID influence brefeldin A (BFA)-independent PIN1 endocytosis in roots (Kleine-Vehn et al., 2009). In Arabidopsis PCs, PIN1 is preferentially localized to the plasma membrane of the lobe regions and is essential for PC morphogenesis (Xu et al., 2010; Li et al., 2011). Overexpression of PIN1 promotes lobe formation, whereas a pin1 mutant produces PCs devoid of lobes (Xu et al., 2010). Recent report shows that the polar localization of PIN1 modulates PC interdigitation, and PID- and FyPP1-dependent PIN1 phosphorylation affects PIN1 polarity in PCs (Li et al., 2011). However, how the phosphorylated PIN1 in PCs is dephosphorylated remains elusive.
In mammals, PP1 is a major Ser/Thr phosphatase that regulates multiple cellular processes, including cell division, cytoskeletal reorganization, metabolism, synaptic plasticity, transcription, and translation (Shi, 2009). A critical role of PP1 in epithelial cell polarity is regulating the phosphorylation status of a polarity scaffold Partitioning defective3 (Traweger et al., 2008). The catalytic subunit of PP1 is highly conserved, with approximately 70% or greater protein sequence identity among all eukaryotes (Shi, 2009). It is likely that PP1 is also required for plant cell polarity and morphogenesis. In Arabidopsis, nine type-one protein phosphatase (TOPP) genes encoding the catalytic subunits of PP1 have been cloned (Smith and Walker, 1993; Lin et al., 1998). However, their functions are still poorly understood. In this study, we characterized a unique function of TOPP4 in regulating PC morphogenesis in Arabidopsis. Genetic data suggest that TOPP4 acts antagonistically with PID in regulating PC development. Biochemical analyses demonstrated that TOPP4 directly interacts with and dephosphorylates PIN1, which is crucial for PIN1 polar localization and endocytic trafficking.
RESULTS
The topp4-1 Mutant Displays Severe PC Defects
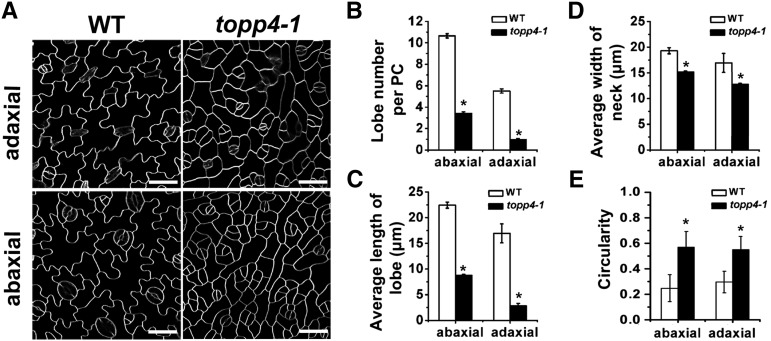
In our previous work, we identified a severely dwarfed dominant-negative mutant topp4-1 and found that TOPP4 acts as a positive regulator in GA signaling through dephosphorylating DELLA proteins (Qin et al., 2014). The topp4-1 mutant was also found to show PC defects. Analyses of the tissue-specific expression using TOPP4:TOPP4-GFP plants showed that TOPP4 is also expressed in leaf PCs (Supplemental Fig. S1). When the mutant was backcrossed to ecotype Columbia, the PC shape of F2 population resulting from self-pollination had a segregation ratio of 94:247:109 (normal PC:moderate PC defect:serious PC defect), which was close to the expected 1:2:1 segregation ratio for a semidominant single locus. The rosette leaves of topp4-1 mutant were small, narrow, and curly (Supplemental Fig. S2, A and D). PCs, stomata, and trichomes were all present in the topp4-1 mutant, suggesting that the mutation did not affect cell differentiation in leaves. PCs on abaxial and adaxial leaf surfaces in the wild type are shaped like a jigsaw puzzle with lobes and necks that interlock with those of adjacent cells (Fig. 1A). In the topp4-1 mutant, the PC shape in cotyledons and the first and second true leaves was similar to those in the wild-type plants (Supplemental Fig. S3). However, after the third leaves, the PCs had either no lobes or less pronounced lobes in the mutant (Fig. 1A). The outlines of PCs in topp4-1 were straighter than those in the wild-type plants and similar to those in rop4-1 ROP2 RNA interference (R2i-34) or RIC1-OVEREXPRESSION (OX) mutants (Fu et al., 2005). The lobe number and length as well as the neck width of PCs in topp4-1 were reduced dramatically (Fig. 1, B–D). The average PC area was also dramatically reduced in topp4-1 compared with that of the wild type (Supplemental Fig. S2E). Circularity has been used as a key parameter to characterize PC geometry, which is independent of its size (Dewitte et al., 2003). Using the geometric analytical method (Guo et al., 2013), we found larger values of circularity in topp4-1 PCs (Fig. 1E), indicating that the reduced lobe length of PCs in topp4-1 was not the result of a decrease in cell size. Transverse sections of mature rosette leaves indicated that more cells were produced in topp4-1, the palisade cell size was smaller, and the shapes of palisade and sponge mesophyll cells were indistinguishable (Supplemental Fig. S2, B and C).
Figure 1.
The topp4-1 mutant shows aberrant PC shape. A, PCs on the abaxial and adaxial leaf sides in the wild type (WT) and topp4-1. Bars = 50 µm. B, The average lobe number per PC in topp4-1 was fewer than the wild type. C, The average lobe length of topp4-1 PCs was significantly reduced compared with that of the wild type. D, The average neck width of topp4-1 PCs was significantly reduced compared with that of the wild type. E, The circularity values of PCs in the wild type and topp4-1. Asterisks in B to E represent statistical differences from the wild type based on Student’s t test with P < 0.01. Error bars represent se (n = 100).
Next, we performed a time course analysis of PC development in the topp4-1 mutant. The development of Arabidopsis leaf PCs is separated into three stages (Fu et al., 2002). In the first phase, wild-type PCs were mostly square, rectangular, or pentagonal, but some cells were slightly more expanded (Supplemental Fig. S4A). PCs of the topp4-1 mutant were similar to those of the wild-type plants at this stage (Supplemental Fig. S4D). At stage II, wild-type PCs had apparent lobes and one or more necks along the long axis, whereas topp4-1 PCs continued to expand and had no visible lobes (Supplemental Fig. S4, B and E). Subsequently, stage II wild-type PCs expanded largely in the direction of the PC long axis, resulting in highly lobed interlocking stage III cells (Supplemental Fig. S4C). However, at stage III, lobes appeared to form in topp4-1 PCs, but they were much less obvious than that in wild-type PCs (Supplemental Fig. S4F). These results showed that TOPP4 might be involved in the lobe formation and outgrowth of PCs.
Overexpression of TOPP4 in the topp4-1 Mutant Rescues the PC Defects, and Its Overexpression in Wild-Type Plants Promotes PC Interdigitated Growth
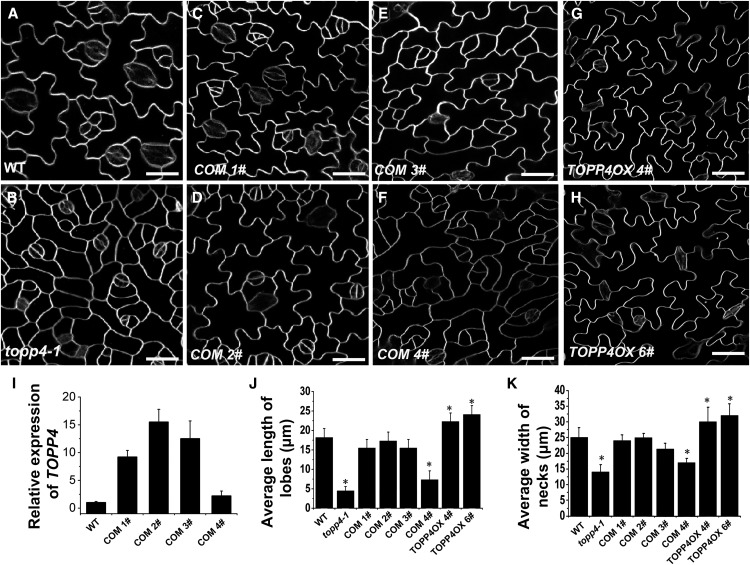
To confirm whether the single nucleotide substitution in TOPP4 is responsible for the defective PC phenotype, we observed four independent 35S:TOPP4 topp4-1 T1-transformed lines (Qin et al., 2014). Three lines (complement [COM] 1#–COM 3#) showed a completely recovered PC phenotype (Fig. 2, A–E). One line (COM 4#) had a weak complemented phenotype (Fig. 2F). Quantitative PCR analysis indicated that COM 4# had a lower overexpression level of TOPP4 than the other three lines (Fig. 2I). Therefore, it appeared that the recovery effect on PCs was positively correlated with the expression level of TOPP4 gene in topp4-1. We also analyzed PC morphology of five individual TOPP4:TOPP4 topp4-1-transformed lines (Qin et al., 2014). They all showed a slightly recovered PC phenotype (two lines are showed in Supplemental Fig. S5) with a deeper and larger number of lobes than the topp4-1 mutant, but the PCs in the transformed lines still displayed shallower lobes than the wild type (Supplemental Fig. S5). Taken together, these results demonstrated that TOPP4 is responsible for the leaf PC defect in the topp4-1 mutant.
Figure 2.
Overexpression of TOPP4 in topp4-1 complements the mutant PC defect, while its overexpression promotes interdigitated PC growth. A to H, PCs on the adaxial leaf side in the wild type (WT), topp4-1, four complemented lines (COM 1#–COM 4#), and two TOPP4 overexpression lines (TOPP4OX 4# and TOPP4OX 6#). Bars = 25 µm. I, The relative expression levels of TOPP4 in four complemented lines shown in C to F. The expression level of TOPP4 in the wild type was set to 1.0. J and K, Quantitative analyses of lobe length (J) and neck width (K) in the wild type, topp4-1, four complemented lines (COM 1#–COM 4#), and two TOPP4 overexpression lines (TOPP4OX 4# and TOPP4OX 6#). Asterisks indicate significant difference from the wild type (P < 0.01 by Student’s t test). Error bars represent se (n = 100).
In addition, two transfer DNA insertion lines of TOPP4, SALK_090980, and N466328 did not show any obvious PC defects (Qin et al., 2014; Supplemental Fig. S6), possibly due to the high expression level of TOPP4 in these two lines or functional redundancy among TOPP genes.
To further investigate the function of TOPP4 in PC morphology, two representative TOPP4-overexpressing transgenic lines with the highest expression level of TOPP4 (lines 4# and 6#) were selected for further phenotypic analyses (Qin et al., 2014; Supplemental Fig. S7A). These two lines exhibited longer and more frequent lobes as well as a wider neck than the wild-type plants (Fig. 2, G, H, J, and K), confirming that TOPP4 promotes lobe outgrowth and lateral expansion of PCs. The representative transgenic lines with the highest expression level of TOPP4 (6#) were selected for further analysis and renamed as 35S:TOPP4.
Expression of topp4-1 in Wild-Type Plants Recapitulates the PC Defects of the topp4-1 Mutant, and Inhibition of PP1 Hinders Interdigitated Growth of PCs
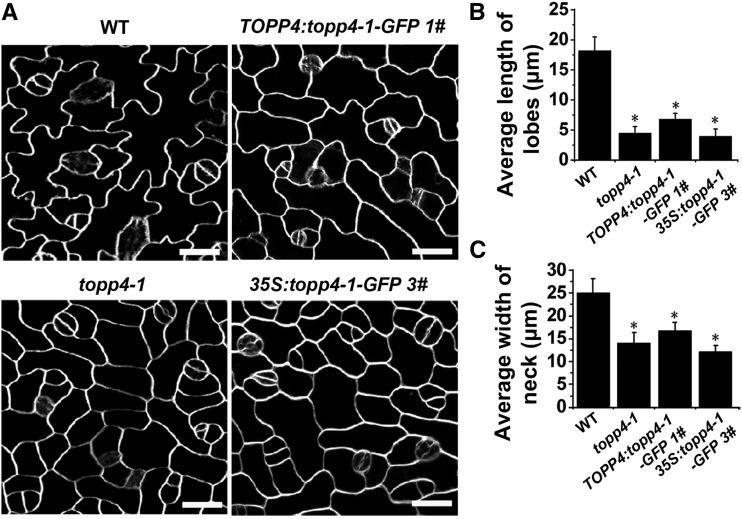
To verify whether the mutant topp4-1 protein had a dominant-negative effect on PC growth, we first analyzed three TOPP4:topp4-1-GFP lines (Qin et al., 2014). Constitutive expression of topp4-1 in these lines was confirmed by quantitative reverse transcription (qRT)-PCR (Supplemental Fig. S7B). The representative transgenic line (1#) with the highest expression level was selected for further phenotypic analyses, which recapitulated the PC defects of topp4-1, as characterized by a reduced lobe length and neck width (Fig. 3). We also observed the phenotypes of the 35S:topp4-1-GFP transgenic lines (Qin et al., 2014). The PCs of all overexpression lines were highly reminiscent of the topp4-1 mutant (line 3# with the highest expression level was displayed in Supplemental Fig. S7C and Fig. 3). These results suggest that topp4-1 affects PC development in a dominant-negative fashion.
Figure 3.
Overexpression of topp4-1 gene in wild-type (WT) plants recapitulates PC phenotype of topp4-1. A, PCs on the adaxial leaf side in wild-type, topp4-1, TOPP4:topp4-1-GFP 1#, and 35S:topp4-1-GFP 3# plants. Bars = 25 µm. B and C, Quantitative analyses of lobe length (B) and neck width (C) of PCs in wild-type, topp4-1, TOPP4:topp4-1-GFP 1#, and 35S:topp4-1-GFP 3# plants. Asterisks represent statistical differences from the wild type based on Student’s t test with P < 0.01. Error bars represent se (n = 100).
It has been reported that tautomycin is a specific inhibitor of PP1, because it inhibits PP2A with 10-fold lower sensitivity in vitro (Favre et al., 1997; Takemiya et al., 2006). Similar to mammalian PP1 phosphatases, Arabidopsis TOPP phosphatases are potently inhibited by tautomycin (Stubbs et al., 2001). We therefore investigated the effects of tautomycin on the PC morphology of wild-type plants. Treatment with 0.1 µm tautomycin inhibited the lobe growth and formation of PCs in true leaves (Supplemental Fig. S8), and a higher concentration of tautomycin (1.0 µm) strongly inhibited PC growth (Supplemental Fig. S8). These results suggest that TOPP phosphatases are involved in the interdigitated PC growth.
The topp4-1 Mutant Displays Auxin-Related Phenotypes, and Exogenously Applied Auxin Dose Not Rescue PC Lobe Formation
The phenotypes of the topp4-1 mutant (i.e. the PC interdigitation defect, reduced apical dominance, increased number of branches, and reduced root length) resemble those in mutants of auxin synthesis, transport, and response (Supplemental Fig. S9, A–C). These defects may be related to the action of auxin. Therefore, we first analyzed local auxin distribution in topp4-1 using the reporter of auxin-responsive promoter DR5 (Ulmasov et al., 1997). In wild-type seedlings, the GUS signal was restricted to the tips of the leaves (Supplemental Fig. S9D). However, GUS-stained areas in topp4-1 were greatly increased in the leaf tips, and these areas extended up to the leaf margins (Supplemental Fig. S9D). By contrast, the activity of the DR5 reporter was less at the primary root tip of topp4-1, but it was significantly higher in 35S:TOPP4 roots than in the wild-type plant roots (Supplemental Fig. S9E). This result indicated that DR5 activity is affected in the topp4-1 mutant.
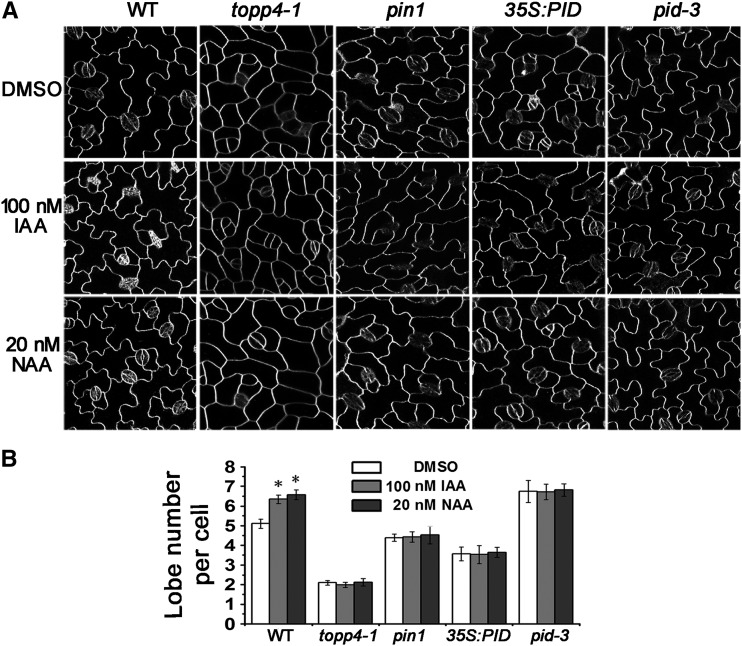
Next, we examined the effect of exogenous auxin on the degree of PC interdigitation. In wild-type plants, the lobe number of PCs was significantly increased by application of exogenous auxin (Fig. 4). The mean number of lobes was increased from 5.14 to approximately 6.5 lobes per cell after 100 nm indole-3-acetic acid (IAA) or 20 nm NAA treatment. However, neither IAA nor NAA treatment rescued PC lobe formation in the topp4-1 mutant (Fig. 4); similar results were obtained in pin1, 35S:PID, and pid-3 mutants (Fig. 4; Xu et al., 2010). Thus, we hypothesized that TOPP4 is involved in the auxin-promoted PC interdigitation.
Figure 4.
The PC interdigitation in topp4-1 is not rescued by exogenous auxin. A, PCs on the adaxial leaf side in wild-type (WT), topp4-1, pin1, 35S:PID, and pid-3 plants treated with DMSO, 100 nm IAA, or 20 nm NAA. Bars = 50 µm. B, Analyses of lobe number per PC in wild-type, topp4-1, pin1, 35S:PID, and pid-3 plants treated with DMSO, 100 nm IAA, or 20 nm NAA. Asterisk represents statistical difference from the untreated wild type based on Student’s t test with P < 0.01. Error bars represent se (n = 300).
TOPP4 Genetically Interacts with PIN1 and PID in PC Interdigitation and Plant Development
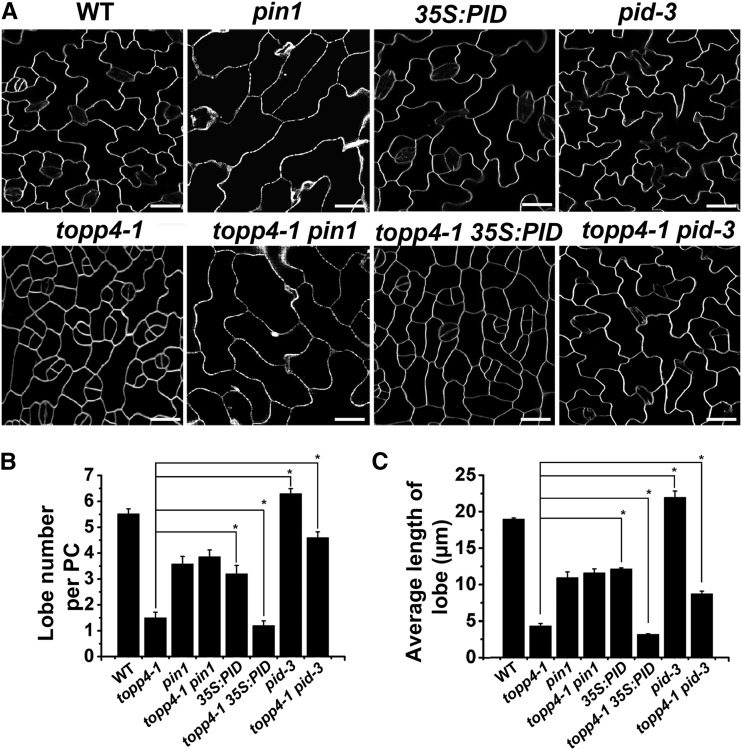
A pin1 mutant, which is defective for auxin transport, has abnormal PC interdigitation in cotyledons and true leaves (Xu et al., 2010). Considering that the PC shape in topp4-1 mutant is similar to that in the pin1 mutant, we analyzed the phenotype of topp4-1 pin1 double mutants to determine whether both TOPP4 and PIN1 function genetically in the same pathway. All double mutants generated from self-pollination of topp4–/– pin1+/– plants exhibited pin1 phenotypes (Supplemental Fig. S10). Among the progeny of pin1+/–, 12.4% (n = 646) of seedlings was defective for the formation of cotyledons and true leaves. However, the overall frequency of aberrant leaf phenotypes was higher in topp4–/– pin1+/– progeny, with up to 17.5% (n = 426) of seedlings. These results indicate that topp4-1 enhances pin1 loss-of-function phenotypes, suggesting that TOPP4 is involved in PIN1-mediated auxin transport. Moreover, most of the adaxial leaf PCs of topp4-1 pin1 double mutants resembled those of the pin1 mutant (Fig. 5). Consistently, the PC shape of the pin1 mutant treated with tautomycin resembled that of the pin1 mutant (Supplemental Fig. S11). These data indicate that both TOPP4 and PIN1 likely function genetically in the same pathway to regulate PC interdigitation and that PIN1 is located downstream of TOPP4.
Figure 5.
TOPP4 acts antagonistically with PID to regulate leaf PC morphogenesis. A, PCs on the adaxial leaf side in the wild type (WT), topp4-1, pin1, topp4-1 pin1, 35S:PID, topp4-1 35S:PID, pid-3, and topp4-1 pid-3 double mutants. Bars = 25 µm. B and C, Quantitative analyses of lobe number (B) and lobe length (C) of PCs. Asterisks represent statistical difference from topp4-1 based on Student’s t test with P < 0.05. Error bars represent se (n = 100).
Compared with the PCs of the wild type, those of the PID gain-of-function mutant (35S:PID) were significantly decreased in lobe number and length, whereas PCs of the PID loss-of-function mutant (pid-3) exhibited more frequent and deeper lobes (Fig. 5). The PC defects of the topp4-1 mutant are similar to those of 35S:PID, but opposite to those of the pid-3 mutant (Fig. 5), suggesting that TOPP4 and PID may have a genetic interaction. To test this hypothesis, we constructed topp4-1 35S:PID and topp4-1 pid-3 double mutants. The topp4-1 35S:PID double mutants showed more severe defects in plant development than each single mutant (Supplemental Fig. S12). Some topp4-1 35S:PID plants failed to form true leaves and showed arrested growth as seedlings. By contrast, the PC defects of pid-3 were partially suppressed by topp4-1 (Fig. 5). The topp4-1 pid-3 PCs had fewer and shorter lobes than those of pid-3. The length of inflorescence stems in the topp4-1 pid-3 double mutant decreased compared with the pid-3 single mutant (Supplemental Fig. S13). Additionally, the PC interdigitated growth of 35S:PID and pid-3 mutants was partially inhibited by tautomycin treatment, which is similar to what was observed in topp4-1 35S:PID and topp4-1 pid-3 double mutants (Supplemental Fig. S11). These genetic analyses suggest that TOPP4 may act antagonistically with PID to regulate PC morphogenesis.
TOPP4 Directly Interacts with PIN1 in Vitro and in Vivo
To reveal the subcellular localization of TOPP4 protein, 35S:TOPP4-GFP was expressed in wild-type Arabidopsis. We found that TOPP4 protein was mainly localized to the nucleus and plasma membrane, and some TOPP4 proteins were found in the cytoplasm (Fig. 6A; Qin et al., 2014). Analyses of TOPP4-GFP expression under the control of its native promoter in the wild-type plants confirmed the subcellular distribution of TOPP4 protein (Supplemental Fig. S1).
Figure 6.
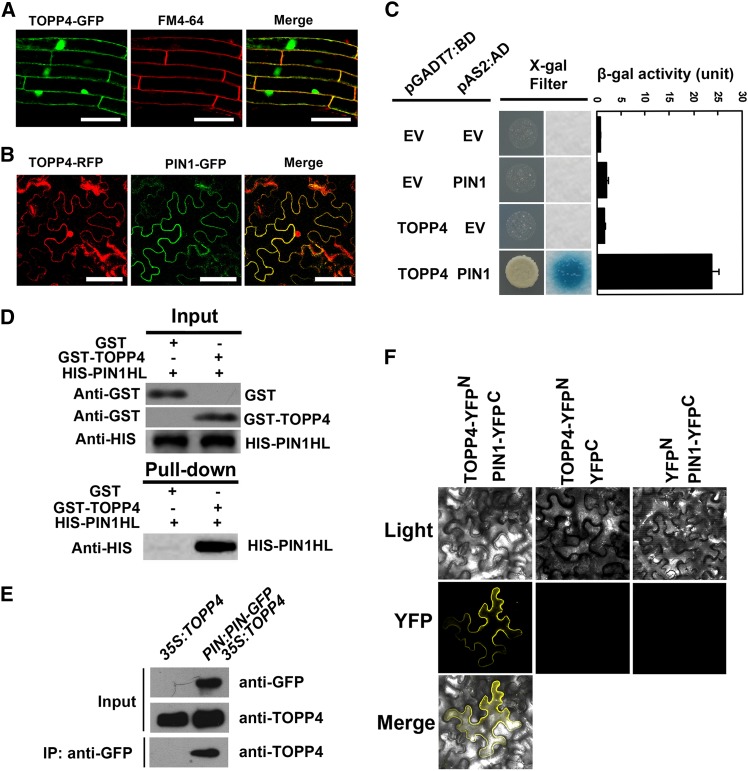
TOPP4 physically interacts with PIN1 protein in vitro and in vivo. A, Subcellular localization of TOPP4-GFP and colocalization of TOPP4-GFP and FM4-64 in Arabidopsis roots. Bars = 100 µm. B, Colocalization of TOPP4-YFP and PIN1-GFP at the plasma membrane of N. benthamiana leaf epidermal cells. Bars = 100 µm. C, Yeast two-hybrid assay was used to determine the interactions between TOPP4 and PIN1. Quantitative measurements of β-gal activities are shown on the right. Error bars represent se (n = 3). X-Gal, 5-Bromo-4-chloro-3-indolyl-β-d-galactopyranoside acid. D, Pull-down assay between GST-TOPP4 and HIS-PIN1. Precipitated HIS-PIN1 was detected by anti-HIS antibody. E, co-IP of TOPP4 with PIN1-GFP. Membrane extracts of PIN1:PIN1-GFP 35S:TOPP4 plants were immunoprecipitated with an anti-GFP antibody and detected by immunoblotting using an anti-TOPP4. Immunoprecipitation (IP) by the anti- GFP in 35S:TOPP4 was used as a negative control. F, BiFC assays showed that TOPP4 interacts with PIN1 at plasma membrane. TOPP4-YFPN and PIN1-YFPC fusion proteins were expressed in N. benthamiana epidermal cells. No YFP signal was detected in negative controls in which either TOPP4-YFPN or PIN1-YFPC was coexpressed with the corresponding empty vector (EV). Light indicates bright field, YFP indicates YFP fluorescence, and merge indicates merged view of the light and YFP images.
At the subcellular level, PIN1 protein is localized to the plasma membrane (Gälweiler et al., 1998; Michniewicz et al., 2007). Our results showed colocalization of a fraction of TOPP4 with FM4-64 and PIN1 at the plasma membrane (Fig. 6, A and B). Next, we determined whether TOPP4 interacts with PIN1 directly. We examined the interactions of TOPP4 and PIN1 by a series of biochemical approaches. Using a yeast (Saccharomyces cerevisiae) two-hybrid assay, TOPP4 was expressed as a DNA-binding domain (BD) fusion protein, and PIN1 was expressed as a transactivation domain (AD) fusion protein in the yeast strain Y190. Interactions of TOPP4-BD and PIN1-AD were confirmed by β-galactosidase (β-gal) activity (Fig. 6C). Recombinant His (HIS)-PIN1 hydrophilic loop (PIN1HL) and glutathione S-transferase (GST)-TOPP4 were purified from Escherichia coli and followed by pull-down experiments. HIS-PIN1HL was pulled down together with GST-TOPP4 using glutathione Sepharose 4B resin (Fig. 6D).
Furthermore, to test the interaction of TOPP4 and PIN1 in vivo, we performed coimmunoprecipitation (co-IP) and bimolecular fluorescence complementation (BiFC) assays. We crossed PIN1:PIN1-GFP with 35S:TOPP4 to generate PIN1:PIN1-GFP 35S:TOPP4 for use in co-IP assays. 35S:TOPP4 was used as a negative control. PIN1 protein was immunoprecipitated with an anti-GFP antibody, and PIN1-bound proteins were subjected to immunoblot analysis. As a result, TOPP4 was coimmunoprecipitated with PIN1 in PIN1:PIN1-GFP 35S:TOPP4, but not in 35S:TOPP4 (Fig. 6E). When TOPP4-N-terminal yellow fluorescent protein and PIN1-C-terminal yellow fluorescent protein were transiently coexpressed in the leaves of Nicotiana benthamiana, strong yellow fluorescent signal was clearly detected at the plasma membrane of PCs (Fig. 6F). Taken together, the above experiments demonstrated that TOPP4 directly interacts with PIN1 in vitro and in vivo.
TOPP4 Directly Dephosphorylates PIN1
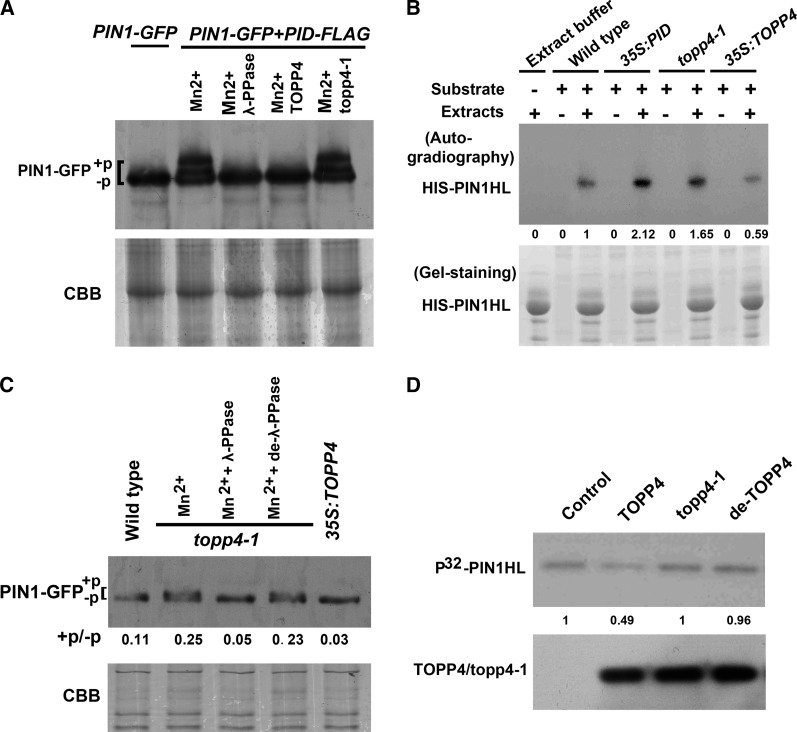
Next, we determined whether TOPP4 dephosphorylated PIN1 directly. A previous study has shown that higher Mr bands of endogenous PIN1 appear when PID is overexpressed (Michniewicz et al., 2007). Therefore, we isolated wild-type protoplasts and transfected them with 35S:PIN1-GFP and/or 35S:PID-FLAG. PIN1-GFP was observed predominantly at the protoplast plasma membrane by a confocal microscopy (Supplemental Fig. S14A). PID-FLAG was detected on protein blots with the expected Mr (Supplemental Fig. S14B). We observed additional signals of PIN1-GFP with a high Mr in protein extracts of protoplasts cotransfected with 35S:PIN1-GFP and 35S:PID-FLAG (Fig. 7A). To test whether the observed electrophoretic mobility shift corresponded to PIN1 phosphorylation, we incubated protein extracts from protoplasts that were transiently transfected with the two constructs with λ-phosphatase. The appearance of additional PIN1 bands was sensitive to λ-phosphatase treatment (Fig. 7A), indicating that these additional PIN1-GFP signals with reduced mobility were due to phosphorylation. We also incubated the protein extracts of TOPP4 or mutant topp4-1, which were immunoprecipitated from wild-type or topp4-1 plants, respectively, with anti-TOPP4 antibody. Consistent with the results obtained by λ-phosphatase treatment, the additional PIN1 bands disappeared when treated with TOPP4, but not mutant topp4-1 (Fig. 7A), indicating that TOPP4 can dephosphorylate PIN1.
Figure 7.
TOPP4 directly dephosphorylates PIN1. A, Immunoblot assay of PIN1-GFP protein incubated with λ protein phosphatase (λ-PPase), TOPP4, or mutant topp4-1. Western blot demonstrating PIN1-GFP expression in protoplasts transfected with 35S:PIN1-GFP and/or 35S:PID-FLAG. A high Mr of PIN1-GFP signals appeared in cotransfected protoplasts. The appearance of additional PIN1-GFP bands was sensitive to λ-PPase and TOPP4 treatments but stable in the presence of topp4-1. B, Increased phosphorylation of the HIS-PIN1 by total protein extracts of 35S:PID and topp4-1 compared with those of the wild type and 35S:TOPP4. Numbers under lanes indicate relative band intensities that were quantified and normalized for each section. C, Increased accumulation of higher Mr of PIN1-GFP in topp4-1 compared with the wild type on SDS-PAGE, whereas the retarded bands did not appear in 35S-TOPP4. These bands were sensitive to λ-PPase treatment but stable in the presence of denatured λ-PPase (de-λ-PPase). D, In the top row, HIS-PIN1HL residues were phosphorylated by GST-PID using [γ-32P] ATP and then incubated with TOPP4, mutant topp4-1, or denatured TOPP4. Numbers under lanes indicate relative band intensities that were quantified and normalized for each section. In the bottom row, western blot of the precipitated TOPP4/topp4-1 proteins using anti-TOPP4 antibody. +P indicates phosphorylated status, and −P indicates dephosphorylated status. +P/–P indicates blot value ratios of phosphorylated PIN1 to unphosphorylated one quantified by ImageJ. CBB, Coomassie Brilliant Blue.
Furthermore, we used an in vitro phosphorylation assay to examine the ability of total protein extracts derived from plant materials to phosphorylate HIS-PIN1HL (Michniewicz et al., 2007; Dhonukshe et al., 2010; Dai et al., 2012). We used the hydrophilic loop of PIN1 as the substrate. Equal amounts of recombinant HIS-PIN1HL residues expressed in E. coli were coincubated with equal amounts of extracts prepared from wild-type, 35S:PID, topp4-1, or 35S:TOPP4 plants. Consistent with the results of previous reports (Michniewicz et al., 2007; Dai et al., 2012), autoradiograph results in this study showed that the amounts of phosphorylated HIS-PIN1HL residues were significantly higher upon incubation with 35S:PID protein extracts than that observed after incubation with the wild-type protein extracts (Fig. 7B). Similarly, the amounts of phosphorylated HIS-PIN1HL were evidently higher in protein extracts derived from topp4-1 plant, but lower in 35S:TOPP4 protein extracts, than in the protein extracts from the wild-type plants (Fig. 7B). These results suggest that the topp4-1 mutation might reduce PIN1 dephosphorylation.
To test whether PIN1 can be dephosphorylated by TOPP4 in planta, we first compared a migration of PIN1-GFP from wild-type, topp4-1, and 35S:TOPP4 plants expressing PIN1-GFP by SDS-PAGE. Immunoblot analysis using an anti-GFP antibody produced a single band of the expected size for PIN1-GFP in all plants (Supplemental Fig. S15). A previous report has shown that higher Mr bands of PIN1 appear on SDS-PAGE (Abas and Luschnig, 2010). Therefore, PIN1-GFP proteins were immunoprecipitated with the anti-GFP antibody from wild-type, topp4-1, and 35S:TOPP4 plants expressing PIN1-GFP and then run on SDS-PAGE for immunoblot analysis. A higher accumulation of the slowly migrating forms of PIN1-GFP was observed in topp4-1 but not in 35S:TOPP4 (Fig. 7C). Moreover, the retarded bands in topp4-1 were sensitive to λ-phosphatase treatment but stable in the presence of denatured λ-phosphatase (Fig. 7C). These data support the notion that TOPP4 is required for PIN1 dephosphorylation.
To further confirm that TOPP4 dephosphorylates PIN1 directly, we first obtained the phosphorylated HIS-PIN1HL residues by incubating HIS-PIN1HL with GST-PID in an in vitro phosphorylation reaction. Equal amounts of phosphorylated PIN1HL residues were then treated with TOPP4, mutant topp4-1, or denatured TOPP4, which were immunopurified from plants. Autoradiographic results showed that phosphorylated HIS-PIN1HL was dephosphorylated by TOPP4, but not by mutant topp4-1, or denatured TOPP4 (Fig. 7D). These data support the conclusion that TOPP4 acts antagonistically with PID to dephosphorylate PIN1 protein directly.
TOPP4 Is Required for PIN1 Polarity Maintenance
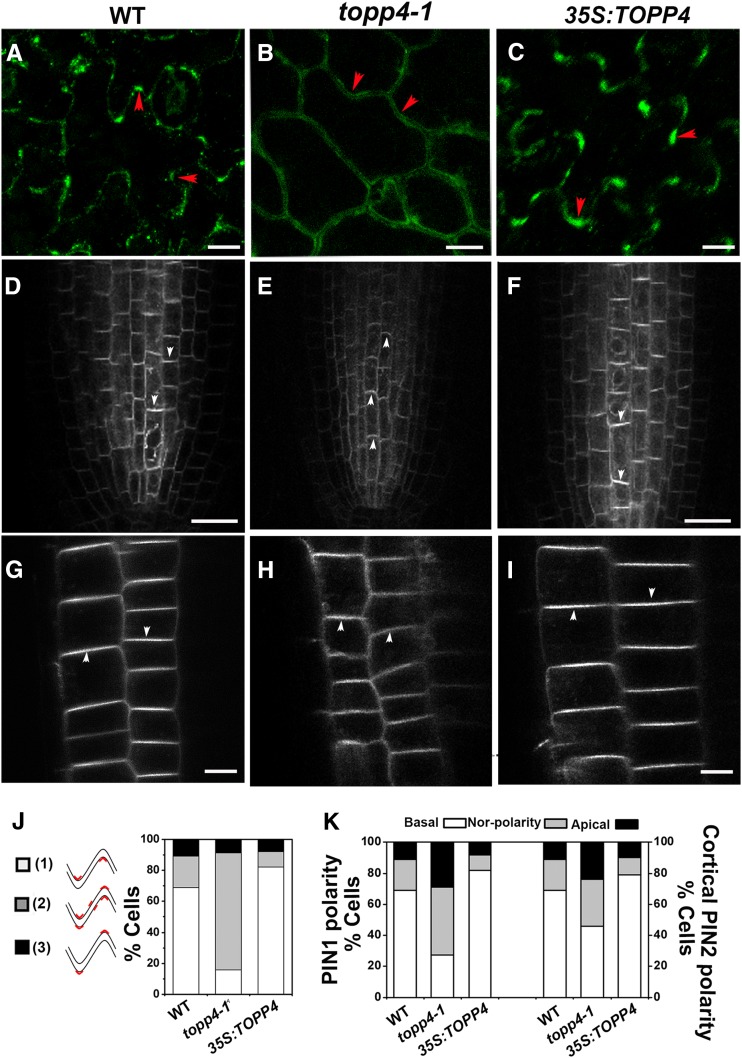
In PCs, PIN1 protein is preferentially localized to the plasma membrane of the lobe regions, and the PIN1 polarity switch between lobes and indentations is controlled by protein phosphorylation (Xu et al., 2010; Li et al., 2011). PIN1 polarity shifts from lobes to indentations in 35S:PID and FYPP1 (a catalytic subunit gene of PP6) loss-of-function mutants (Li et al., 2011). To test the effect of TOPP4-mediated PIN1 dephosphorylation on PIN1 localization, we performed an immunolocalization assay in wild-type, topp4-1, and 35S:TOPP4 PCs. In wild-type PCs, approximately 70% of the cells had preferential PIN1 localization to the plasma membrane of lobe regions (Fig. 8, A and J), consistent with previous observations (Li et al., 2011). However, in topp4-1 PCs, the polar localization of PIN1 was much less pronounced, and a significant proportion of PIN1 shifted from the lobes to indentations or nonlobing regions (Fig. 8, B and J). The polar localization of PIN1 was almost unaltered in 35S:TOPP4 PCs, but the number of PCs with preferential localization of PIN1 to lobe regions in 35S:TOPP4 was higher than that of the wild-type PCs (Fig. 8, C and J).
Figure 8.
Polar localization of PIN1 and PIN2. A to C, Subcellular localization of PIN1 in PCs was detected by immunofluorescence with the anti-PIN1 antibody. Bars = 25 µm. Red arrowheads indicate the accumulation of PIN1 at lobe regions. D to I, Live images of GFP-tagged PIN1 (D–F) and PIN2 (G–I) in the roots. Bars = 50 µm. White arrowheads indicate polarity of PIN localization. J, Quantitative analysis of PIN1 localization patterns in PCs. 1, PIN1 preferentially localized only to lobe sides. 2, PIN1 localized to lobe and indentation sides or nonlobing regions. 3, PIN1 localized to indentation sides. The percentage of three types of PIN1 localization patterns of PCs is shown on the right. K, Quantification of PIN1 and PIN2 polarity defects in the roots. The PIN localization pattern was quantified from more than 30 randomly chosen cells. WT, Wild type.
To investigate whether the polar localization of PIN was also affected in the roots of topp4-1 and 35S:TOPP4 plants, we crossed them with PIN1:PIN1-GFP and PIN2:PIN2-GFP marker lines. In wild-type roots, PIN1 was localized preferentially to the basal side of stele cells, while PIN2 was localized at the apical side of epidermis cells and the basal side of cortex cells (Fig. 8, D and G; Blilou et al., 2005). In topp4-1 stele cells, the basal polarity of PIN1 was much less pronounced, and the number of cells with nonpolar and apical PIN1 was increased significantly (Fig. 8, E and K). In addition, the basal PIN2 polarity in the cortex cells of topp4-1 was much less pronounced, but the apical PIN2 localization in epidermis cells was not visibly affected (Fig. 8, H and K). Compared with PID and PP2A mutations, the polar PIN1 and PIN2 shifts in topp4-1 appeared to be more gradual. In 35S:TOPP4, there was an increase in the proportion of cells with normal PIN1 or PIN2 localization, while the proportion of cells with nonpolar PIN1 or PIN2 decreased (Fig. 8, F, I, and K). Taken together, these observations indicate that TOPP4-mediated PIN dephosphorylation is required for PIN polarity in leaf PCs and root cells.
TOPP4 Is Required for Endocytic Trafficking of PIN1 in PCs
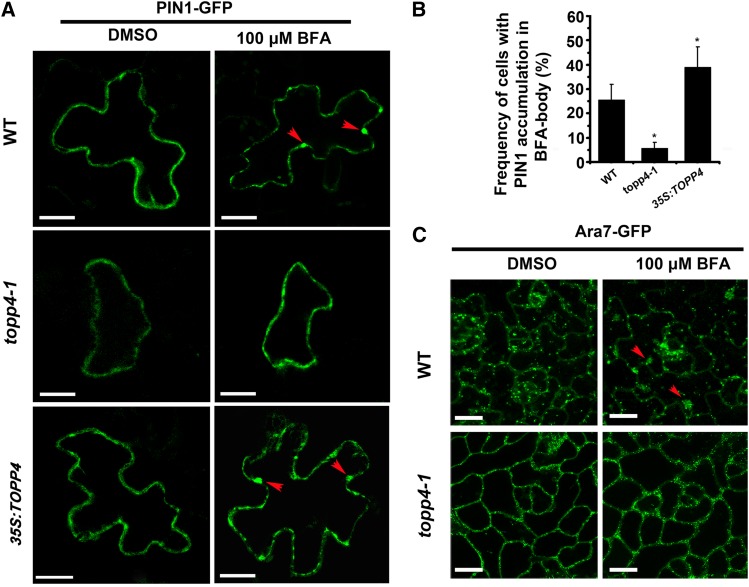
The fungal toxin BFA is useful to investigate early endocytic trafficking (Geldner et al., 2001; Kleine-Vehn et al., 2008). Recent studies have shown that PIN1-GFP is preferentially internalized in the indentation regions of PCs, and BFA treatment induces large aggregations of PIN1-GFP into BFA bodies (Nagawa et al., 2012). In wild-type root cells, BFA induced PIN1 internalization and formed large aggregates (Kleine-Vehn et al., 2008; Supplemental Fig. S16). However, the effect of BFA on PIN1 localization was severely impaired in topp4-1, as manifested by the significantly reduced BFA-induced PIN1 internalization and PIN1 retention at the plasma membrane (Supplemental Fig. S16). In 35S:TOPP4 root cells, BFA rapidly induced PIN1 internalization and formed larger BFA compartments compared with the wild type (Supplemental Fig. S16). To test the effect of TOPP4-dependent PIN1 dephosphorylation on the endocytic trafficking in PCs, we first monitored BFA-induced intracellular accumulation of PIN1 of topp4-1 and 35S:TOPP4 PCs. 35S:PIN1-GFP was transiently expressed in wild-type, topp4-1, and 35S:TOPP4 PCs by the ballistics-mediated method (Fu et al., 2002). After treatment with 100 µm BFA, the plasma membrane-localized PIN1-GFP was rapidly internalized from the plasma membrane into BFA compartments in wild-type and 35S:TOPP4 PCs. However, in topp4-1 PCs, intracellular accumulation of PIN1 was quite rare, with most PIN1-GFP remaining at the plasma membrane (Fig. 9A). Quantitative analysis showed that the frequency of cells with accumulated PIN1-GFP in BFA bodies was significantly lower in topp4-1, but higher in 35S:TOPP4 than in the wild type (Fig. 9B). This finding suggests that the BFA-induced PIN1 internalization is severely compromised in the topp4-1 mutant, and TOPP4 overexpression in the wild type enhances BFA-induced PIN1 internalization.
Figure 9.
PIN1-GFP and Ara7-GFP internalization in PCs after treatment with BFA. A, 35S:PIN1-GFP was transiently expressed for 24 h in leaf PCs, and then these leaves were treated with DMSO or 100 µm BFA. Red arrowheads represent the PIN1 accumulation in BFA bodies. Bars = 20 µm. B, Quantitative analyses of intracellular accumulation of PIN1 in BFA-treated PCs. Asterisks indicate significant difference from the wild type (WT; P < 0.01 by Student’s t test). Error bars represent se (n = 20). C, Distribution of the endocytic marker Ara7-GFP in wild-type and topp4-1 PCs. The leaves of wild-type and topp4-1 plants labeled with Ara7-GFP treated with DMSO or 100 µm BFA. Red arrowheads represent the Ara7-GFP accumulation in BFA bodies. Bars = 50 µm.
Next, we examined localization of the endocytic marker Ara7-GFP, which resides in an endosomal compartment and is necessary for targeting PIN1 to vacuoles or recycling to the plasma membrane (Lee et al., 2004). In wild-type PCs, Ara7-GFP was localized to endosomal compartments (Fig. 9C). After treatment with 100 µm BFA for 90 min, Ara7-GFP had accumulated greatly in intracellular endosomal aggregations (Fig. 9C). However, the localization of Ara7-GFP was affected in the topp4-1 mutant by mainly residing in the plasma membrane (Fig. 9C). In addition, BFA-induced Ara7-GFP aggregations were rarely observed in topp4-1 PCs (Fig. 9C). Taken together, these results suggest that TOPP4-mediated PIN1 dephosphorylation is required for endocytic trafficking-dependent polar targeting of PIN1.
TOPP4 Is Involved in Regulation of ROP GTPase-Mediated Cytoskeletal Distribution
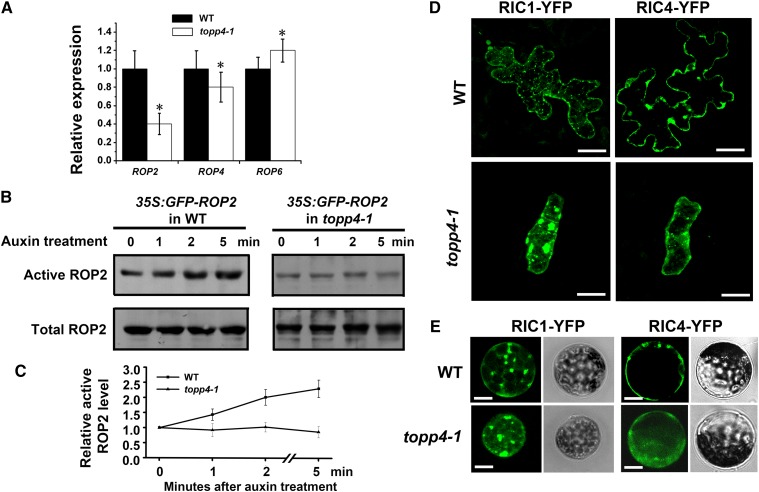
The reduced lobe expansion and narrow necks in the topp4-1 mutant were similar to those in rop2-1 rop4-1 and ROP6- or RIC1-overexpressing plants (Fu et al., 2002, 2005, 2009). To determine whether TOPP4 is required for the ROP GTPase-dependent auxin-signaling pathway, we first examined the expression of ROP genes in the topp4-1 mutant by qRT-PCR. Compared with the wild type, ROP2 and ROP4 expression levels were lower, while the ROP6 expression level was higher in topp4-1 (Fig. 10A). Subsequently, we examined the effect of auxin on ROP2 activity in topp4-1 stably expressing 35S:GFP-ROP2. Because RIC1 can specifically bind to the active-form ROP2 but not the inactive-form ROP2 both in vivo and in vitro, RIC1 has been used to measure ROP2 activity (Xu et al., 2010). Measurements of GFP-bound ROP2 showed that the activity of ROP2 was rapidly increased by auxin in wild-type plants (Fig. 10, B and C). However, auxin-stimulated ROP2 activity was abolished in the topp4-1 mutant (Fig. 10, B and C). Furthermore, if TOPP4 positively regulates the activation of ROP2, the distributions of RIC1-GFP and RIC4-GFP should be disrupted in the topp4-1 mutant. To verify this hypothesis, we first transiently expressed 35S:RIC1/RIC4-GFP in the leaves of the wild-type plants and topp4-1 mutant by the ballistics-mediated method. In wild-type PCs, RIC1-GFP often formed numerous dot-like structures along cortical MTs. However, in topp4-1 PCs, some RIC1-GFP had accumulated in very large structures (Fig. 10D). RIC4-GFP was preferentially localized to the lobe tips and plasma membrane of wild-type PCs (Fig. 10D), but its localization to the plasma membrane was replaced by diffuse distribution in the cytosols of topp4-1 PCs (Fig. 10D). We also transiently expressed 35S:RIC1/RIC4-GFP in the protoplasts of the wild type and topp4-1. Similarly, we observed that the localization of RIC1-GFP and RIC4-GFP was compromised in topp4-1 protoplasts (Fig. 10E). These results indicate that the ROP2 GTPase-dependent auxin-signaling pathway is disrupted in topp4-1.
Figure 10.
TOPP4 is required for the auxin-activated ROP-RIC pathway. A, The relative expression of ROP2, ROP4, and ROP6 in topp4-1. Asterisks represent statistical differences from the wild type (WT) based on Student’s t test with P < 0.01. B, Measurements of GTP-bound ROP2 activity in protoplasts isolated from the wild type and topp4-1 stably expressing 35S:GFP-ROP2 by co-IP assay. C, Quantitative analyses of the relative active ROP2 activity. Error bars represent se (n = 3). D, The distribution patterns of RIC1-GFP and RIC4-GFP in the PCs of the wild type and topp4-1. Bars = 20 µm. E, Transiently expressed RIC1-GFP and RIC4-GFP in protoplasts isolated from wild-type and topp4-1 plants. Right section shows the bright field images of corresponding protoplasts. Scale bars = 20 µm.
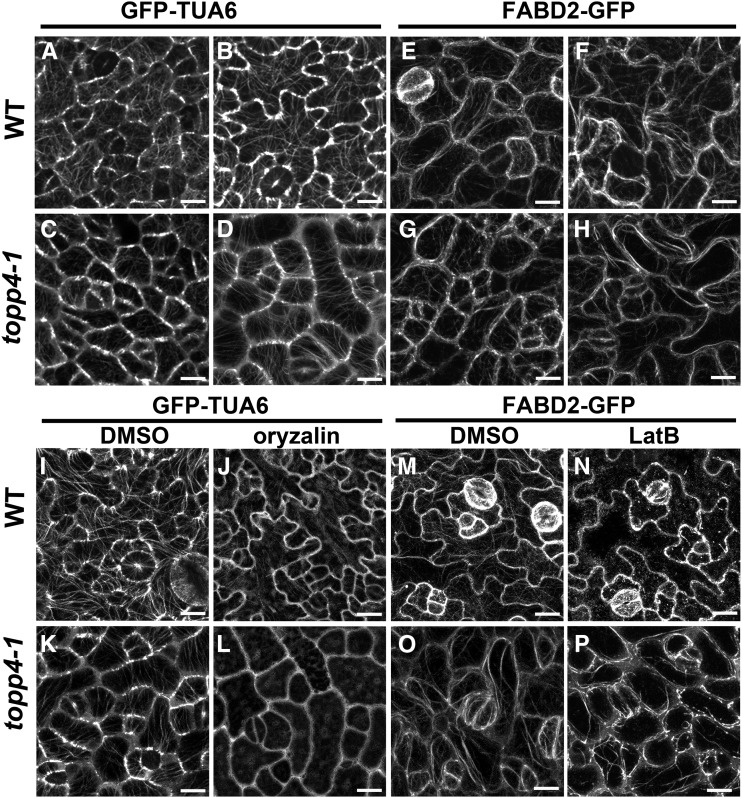
Considering that the ROP-RIC pathway is defective in topp4-1, RIC1-associated cortical MTs and RIC4-associated F-actins may be disrupted in the mutant. We observed the cortical MT distribution in topp4-1 PCs using GFP-tagged tubulin (Ueda et al., 1999). At the early development stage of PCs, cortical MTs were oriented randomly in both the wild type and topp4-1, but the number of MTs in topp4-1 was lower than that of the wild type (Fig. 11, A and C). At the late developmental stage of PCs, cortical MTs formed a fine and complex network with random orientations, and some transverse cortical MTs were present in the neck regions of the wild type (Fig. 11B). However, in topp4-1, the cortical MTs were highly ordered, and the MTs of most PCs were oriented perpendicularly to the long cellular axis (Fig. 11D). We also observed the actin distribution in topp4-1 PCs using F-actins labeled with the second actin-binding domain of Arabidopsis fimbrin (FABD2; Voigt et al., 2005). At the early developmental stage of wild-type and topp4-1 PCs, F-actins were found throughout the cell cortex, and strong fluorescence signals were associated with lobe formation sites (Fig. 11, E and G). At the late developmental stage of PCs, cortical F-actins had become more intense in the expanding lobes, and the F-actin cables formed a fine network in the wild type (Fig. 11F). Conversely, in topp4-1, cortical F-actin cables appeared to lose their fine network organization and formed some thicker actin cables along the plasma membrane of PCs (Fig. 11H).
Figure 11.
The organization of cortical MTs and F-actins is altered in topp4-1, and MTs are hypersensitive to oryzalin treatment, but F-actins are more resistant to LatB treatment in topp4-1. A to D, The cortical MT organization in PCs on the adaxial leaf side in wild-type (WT) and topp4-1 plants labeled with GFP-Tubulin α-6 (TUA6). A and C, The early development stage of PCs. B and D, The late development stage of PCs. Bars = 25 µm. E to H, The F-actin organization in PCs on the adaxial leaf side in the wild type and topp4-1 labeled with FABD2-GFP. E and G, The early development stage of PCs. F and H, The late development stage of PCs. Bars = 25 µm. I to L, The cortical MT organization in PCs in wild-type and topp4-1 plants labeled with GFP-TUA6 treated with or without 20 µm oryzalin for 45 min. Bars = 25 µm. M to P, The F-actin organization in PCs in wild-type and topp4-1 plants labeled with FABD2-GFP treated with or without 800 nm LatB for 30 min. Bars = 25 µm.
To determine whether the observed MT disorganization in topp4-1 was accompanied by changes in its stability, we compared the sensitivity of MTs to oryzalin (an MT-disrupting drug that disrupts MTs by binding to α-tubulin; Nakamura et al., 2004) in wild-type and topp4-1 PCs. In topp4-1 PCs, most MTs were completely disassembled after treatment with 20 μm oryzalin for 45 min, while MTs in most wild-type PCs remained intact (Fig. 11, I–L). To assess the effect of TOPP4 on actin polymerization, we infiltrated wild-type and topp4-1 seedlings with 800 nm latrunculin B (LatB) that inhibits actin polymerization by binding to monomeric actin and preventing its assembly on filament ends (Sampathkumar et al., 2011). After 30 min of LatB treatment, the F-actin bundles had disappeared completely and were replaced by diffuse green fluorescence in most wild-type PCs (Fig. 11, M and N). However, F-actin bundles remained largely in most of the topp4-1 PCs (Fig. 11, O and P). These observations indicate that the topp4-1 mutation enhances the sensitivity of MTs to oryzalin but reduces the sensitivity of F-actins to LatB in PCs. Taken together, these results suggest that TOPP4 affects PC cytoskeletal organization by coordinating MT stabilization and F-actin polymerization via the ROP GTPase-dependent auxin-signaling pathway.
DISCUSSION
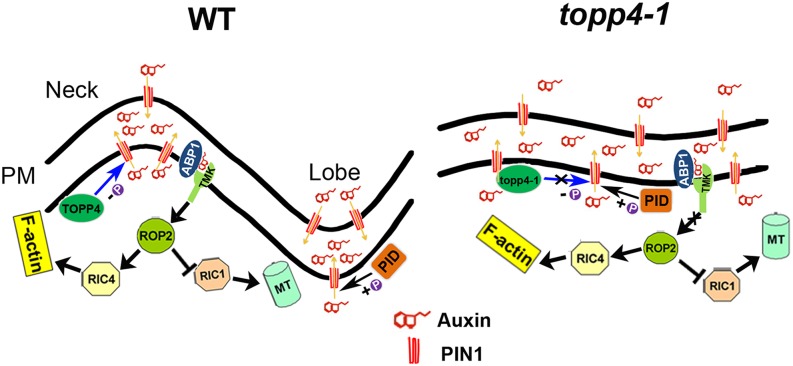
Cell polarity and morphogenesis, which are critical for plant organ development, are modulated by numerous cytoskeleton-associated proteins and various developmental and environmental signals (Fu et al., 2005; Hamant et al., 2008; Kleine-Vehn et al., 2008; Xu et al., 2010; Li et al., 2013). Cell polarity is often presented as a polar distribution of molecules within the cell. In Arabidopsis, the PIN family of auxin efflux carriers is asymmetrically targeted to the plasma membrane of cells, which is crucial for cell polarity and auxin distribution-dependent organogenesis (Benková et al., 2003). Previous studies indicate that the phosphorylation status of PIN proteins determine its polarity in embryos, inflorescences, and roots, mediated by PID, PP2A, and PP6 (Friml et al., 2004; Michniewicz et al., 2007; Kleine-Vehn et al., 2009; Dhonukshe et al., 2010; Huang et al., 2010; Dai et al., 2012). A recent study shows that PIN1 is polarly localized to the lobes of PCs, which is altered in a fypp1 mutant (Xu et al., 2010; Li et al., 2011). Although it is known that phosphorylated PIN1 regulates PC interdigitation formation, how PIN1 is dephosphorylated in this process is still poorly understood. Here, we provide our extensive genetic and biochemical data to demonstrate that TOPP4 plays an antagonistic role with PID in regulating the polar localization and endocytosis of PIN1 in auxin-mediated PC morphogenesis via a reversible phosphorylation and dephosphorylation process (Fig. 12). We also found that TOPP4-dependent PIN1 dephosphorylation is involved in the coordination of ROP GTPase-mediated cytoskeletal distribution (Fig. 12).
Figure 12.
A current model for TOPP4 controlling interdigitated PC growth. In wild-type (WT) PCs, PIN kinase and TOPP4 phosphatase regulate PIN1 polarity in the lobes and indentations of PCs. The PIN1 polarity determines auxin flow and establishes auxin gradients in two adjacent PCs. This extracellular auxin in turn activates ROP GTPase signaling through the ABP1-TMK complex to control PC morphogenesis. By contrast, in topp4-1 PCs, dephosphorylation of PIN1 is diminished, causing PIN1 to localize to indentation or nonlobing regions. Thus, the nonpolarity PIN1 reduces auxin accumulation in lobes and neighboring indentations of two adjacent PCs, disturbing the ROP GTPase signaling and causing the PC interdigitation defect (Pietra and Grebe, 2010; Chen and Yang, 2014). +P indicates phosphorylated status, and −P indicates dephosphorylated status. PM, Plasma membrane.
A previous study showed that the PIN phosphorylation status, mediated by PID and PP2A, controls the polarity switch of PIN proteins by affecting their endocytic recycling in root cells (Geldner et al., 2001; Kleine-Vehn et al., 2009). In this study, our genetic analyses showed that TOPP4 functions antagonistically with PID in PIN1-mediated auxin promotion of PC interdigitation (Figs. 4 and 5). Biochemical analyses demonstrated that TOPP4 directly interacts with and dephosphorylates phosphorylated PIN1 protein in vitro and in vivo (Figs. 6 and 7). Further studies showed that the topp4-1 mutation leads to alterations in PIN1 polarity (Fig. 8), which resembles the PIN1 polarity distribution of fypp1 PCs and 35S:PID or pp2aa1 pp2aa3 root cells (Michniewicz et al., 2007; Li et al., 2011). It is well known that polar PIN localization is dynamic, and PIN proteins constitutively undergo cycles of exocytosis and endocytosis to and from the plasma membrane (Dhonukshe et al., 2007). Ara7-dependent endocytic trafficking is crucial for the generation of cell polarity in roots via regulating PIN polar localization (Tanaka et al., 2013). We showed that PIN1 dephosphorylation mediated by TOPP4 regulated its polar localization in PCs via affecting Ara7-dependent PIN1 internalization (Fig. 9C). The topp4-1 mutation decreased BFA-induced PIN internalization in PCs, whereas TOPP4 overexpression in the wild-type plants enhanced the sensitivity of PIN1 internalization to BFA (Fig. 9, A and B). These results are consistent with those from a previous report showing that 35S:PID and pp2aa1 pp2aa3 plants exhibit a reduction of BFA-induced PIN1 internalization in roots (Kleine-Vehn et al., 2009).
In addition, PIN1 endocytosis is increased in dominant-negative mutant for ROP and loss-of-function rop4 R2i-34 mutants (Nagawa et al., 2012). In topp4-1, although the activity of ROP2 was reduced (Fig. 10), BFA-induced PIN1 internalization was inhibited (Fig. 9). These results suggest that the compromised localization of PIN1 in topp4-1 PCs is not the effect of reduced ROP2 activity but rather due to the defect in PIN1 dephosphorylation caused by the TOPP4 mutation, which is similar to the function of PP2A in roots (Kleine-Vehn et al., 2009).
Many studies have shown that MTs and F-actins play important roles in PC shape (Wang et al., 2007). It is well known that the ROP GTPase-signaling pathway regulates the organization of both cortical MTs and F-actins (Fu et al., 2002, 2005, 2009). Our results indicated that auxin-stimulated ROP2 activity was greatly disturbed in topp4-1 (Fig. 10). Moreover, similar to pin1 and tmk mutants (Xu et al., 2010, 2014), plasma membrane-localized RIC4-GFP was reduced and RIC1-GFP within cortical MTs was abolished in topp4-1, supporting a critical role of TOPP4 in localized ROP2 activation. The organization of RIC1-associated cortical MTs and RIC4-associated F-actins was also disrupted in topp4-1 (Fig. 11), which is similar to that observed in rop4 R2i and RIC1-OX plants (Fu et al., 2005, 2009). Therefore, we concluded that TOPP4 is required for ROP GTPase-mediated cytoskeletal distribution in PCs.
Cell size is tightly correlated with its shape (Zhang et al., 2011). We found that the PC size of topp4-1 was smaller than that of the wild type. To rule out the effect of cell size on cell shape, we analyzed auxin and ROP-RIC signaling and cytoskeleton organization in cyclin D-type3;1 (CYCD3;1) overexpression plants, in which the PC size was also dramatically reduced compared with the wild type (Zhou et al., 2003; Supplemental Fig. S17). The circularity value of PCs in 35S:CYCD3;1 was similar to that of the wild type (Supplemental Fig. S17), indicating that the PC shape of 35S:CYCD3;1 was not altered. Furthermore, similar to wild-type plants, auxin and ROP-RIC signaling were normal in 35S:CYCD3;1 plants (Supplemental Fig. S18). The cortical MTs and F-actins also showed normal distribution in 35S:CYCD3;1 PCs compared with wild-type PCs (Supplemental Fig. S18). These results suggest that the abnormal PC shape in topp4-1 is not caused by the small cell size effects on auxin signaling.
PIN1-mediated auxin efflux may lead to a localized accumulation of extracellular auxin in the indenting region of neighboring cells, activating complementary ROP2 and ROP6 pathways in two adjacent cells through a cell surface auxin perception complex, ABP1-TMK (Xu et al., 2010, 2014; Li et al., 2011). Therefore, based on the aforementioned results, we propose that the ROP GTPase signaling may be at the downstream of TOPP4. In wild-type PCs, hypophosphorylated PIN1, mediated by TOPP4, localizes preferentially to the plasma membrane of lobe regions, promoting the auxin flow from lobes to neighboring indentations and forming a localized accumulation of extracellular auxin in two adjacent cells. This extracellular auxin in turn activates ROP signaling via its perception complex to control PC morphogenesis by regulating the organization of cortical MTs and F-actins. In topp4-1 PCs, dephosphorylation of PIN1 is abolished, causing hyperphosphorylated PIN1 to localize to indentation or nonlobing regions. This nonpolarity of PIN1 distribution reduces auxin accumulation in lobes and neighboring indentations, thus inactivating ROP2 and disturbing RIC-controlled cytoskeletal organization, resulting in abnormal PCs. In addition, as the PC defects in topp4-1 were more severe than those in auxin transport-defective mutants, such as pin1 and 35S:PID (Fig. 4), we could not completely exclude the possibility that TOPP4 also participates in another pathway to regulate PC morphogenesis.
The mechanism of TOPP4 in PP1 family modulating the PIN1 phosphorylation status is similar to the action of PP2A and PP6 (Michniewicz et al., 2007; Dai et al., 2012). The catalytic domains of Ser/Thr-specific phosphoprotein phosphatases are highly conserved among all eukaryotes (Farkas et al., 2007; Shi, 2009). As a catalytic subunit of protein phosphatase, TOPP4 must bind to different regulatory subunits to form a functional enzyme to participate in multiple signaling pathways (Tian and Wang, 2002). Because the regulatory subunits of phosphatases are dynamic at different developmental stages or in response to various environmental signals (Dai et al., 2013), PP1 and PP6 may respond to different developmental signals in different organs or tissues at various developmental stages. The topp4-1 mutant showed PC defects only in and after the third true leaves, whereas the fypp1 mutant has PC defects in cotyledons and true leaves (Li et al., 2011). Furthermore, TOPP4 was only detected in the stele cells of the root, but FyPP1 and FyPP3 were ubiquitously expressed in the root. Therefore, the root defect in the fypp mutant was more severe than that in topp4-1 (Dai et al., 2012). Thus, although PP1 and PP6 share similar substrates and catalytic action, they may perform different functions in different developmental organs and tissues through binding to specific regulatory subunits. Future investigation will focus on identifying the different regulatory subunits of TOPP4.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Plants were grown on either one-half-strength Murashige and Skoog (MS) medium or soil in greenhouse at 22°C ± 1°C, with a 16-h-light/8-h-dark photoperiod. The double mutants topp4-1 pin1, topp4-1 35S:PID, and topp4-1 pid-3 were generated from the separate crosses of topp4-1 with pin1 (SALK_047613, from the Arabidopsis Biological Resource Center), 35S:PID 21# (N9867, from the European Arabidopsis Stock Centre [NASC]), and pid-3 (N5219, from NASC), respectively. Double mutants were identified from the F2 progeny by PCR-based molecular analyses. Primers used for identifying homozygous lines are indicated in Supplemental Table S1. The topp4-1 crossed with DR5:GUS, DR5:GFP, PIN1-GFP (N23889, from NASC), or Ara7-GFP (Lee et al., 2004) lines and screened the homozygous lines from the F2 progeny by PCR-based analysis and GFP florescence observation.
Plasmid Construction
For transient expression in Arabidopsis (Arabidopsis thaliana) leaves or protoplasts, full-length complementary DNAs of PIN1, RIC1, and RIC4 were amplified and cloned into pA7-GFP vector. Primers used for plasmid construction are indicated in Supplemental Table S1.
Microscopic Analyses of PC Shape
The third true leaves from 21-d-old seedlings were used for phenotype analyses. The Arabidopsis leaves were stained with 10 µm FM4-64 dye (Sigma) for 30 min and imaged using a confocal microscopy (Olympus FluoView FV1000MPE). Both lobe length and neck width were measured with Image J, as described by Fu et al. (2002). The measurements were consistently conducted on PCs at the same developmental stage. Each piece of data was averaged from the measurements of 100 cells of five leaves. Data from different figures were obtained from independent experiments.
Chemical Treatments
To detect the effects of tautomycin (Merck) on the PC morphology, the seeds were grown on one-half-strength MS agar plates that were supplemented with 0.1 or 1 µm tautomycin. To examine the effect of exogenous auxin on the degree of PC interdigitation, NAA or IAA (Sigma) was dissolved in dimethyl sulfoxide (DMSO) and prepared as a stock solution of 100 µm, which was added into one-half-strength MS media to obtain a final concentration of 20 nm NAA or 100 nm IAA for seedling treatments. Analysis of PIN1-GFP internalization was performed as described by Nagawa et al. (2012). 35S:PIN1-GFP was transiently transfected in Arabidopsis leaves for 24 h by ballistics-mediated method, and then these leaves were treated with DMSO or 100 µm BFA for 90 min. PIN1-GFP signal was observed by the confocal microscopy. Each treatment was repeated at least three times with three independent seedlings each time.
Visualization of F-Actins and MTs and Drug Treatments
To visualize the cortical MTs and F-actins in leaf PCs, we crossed GFP-tagged α-tubulin (Ueda et al., 1999) or FABD2-GFP (Voigt et al., 2005) into topp4-1 and examined their distribution using the confocal microscope. For oryzalin (Sigma) treatments, 21-d-old seedlings expressing GFP-tagged α-tubulin were incubated in one-half-strength MS medium containing DMSO or 20 μm oryzalin for 45 min. For LatB (Sigma) treatments, 21-d-old seedlings expressing FABD2-GFP were incubated with one-half-strength MS medium containing DMSO or 800 nm LatB for 30 min. Each treatment was repeated at least three times.
Yeast Two-Hybrid Assay
The yeast (Saccharomyces cerevisiae) strain Y190 was used in our experiments. Yeast transformations were performed according to the MATCHMAKER two-hybrid system 3 (Clontech). Full-length TOPP4 gene fused to the DNA-binding domain of GAL4 was used as the bait protein, and PIN1HL fused to the transcriptional activation domain of GAL4 was used as the prey protein. Yeast clones containing the GAL4-BD-TOPP4 and GAL4-AD-PIN1HL constructs were plated on SD-His-Trp-Leu medium for 5 d at 30°C to assay for interaction. β-gal activity was performed according to the manufacturer’s protocol (Clontech). This experiment was repeated at least three times.
In Vitro Pull-Down and co-IP Assays
The GST-TOPP4 and HIS-PIN1HL proteins were expressed in Escherichia coli BL21. The recombinant proteins were coincubated in the presence of glutathione Sepharose 4B resin (GE), which was used to selectively bind the GST fusion proteins with phosphate-buffered saline (PBS) buffer (140 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, and 1.8 mm KH2PO4). The bound proteins were eluted with 1× SDS loading buffer and analyzed with anti-GST and anti-HIS antibodies. This experiment was repeated at least three times.
co-IP studies of TOPP4 and PIN1 were performed on 10-d-old seedlings of 35S:TOPP4 PIN1:PIN1-GFP and 35S:TOPP4. IP of PIN1 protein used an anti-GFP antibody (Invitrogen). Protein G agarose (GE) was used to precipitate the immunoprotein complexes with IP buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 5 mm EDTA, 0.2% [v/v] Triton X-100, and 1% [v/v] protease inhibitors). After IP, beads were washed four times with IP buffer. Proteins were then released and collected by boiling in 2× SDS loading buffer for 5 min. IP products were detected by immunoblot analysis with a TOPP4 antibody. This experiment was repeated at least three times.
BiFC Assay
The coding sequence of sequences of TOPP4 or PIN1 was amplified and cloned into pEearleygate201-YN or pEearleygate202-YC BiFC vectors to generate TOPP4-N-terminal yellow fluorescent protein or PIN1-C-terminal yellow fluorescent protein, respectively (Song et al., 2010). Agrobacterium tumefaciens strains containing the BiFC constructs and the p19-silencing plasmid were infiltrated into leaves of 4-week-old Nicotiana benthamiana plants (Feng et al., 2008). The YFP fluorescence was observed with confocal laser-scanning microscopy.
Immunofluorescence Microscopy
Whole-mount immunolocalization on Arabidopsis leaf PCs were performed as described (Sauer et al., 2006; Li et al., 2011). Briefly, true leaves were submerged into a fixation solution (PBS plus 4% paraformaldehyde) for 1 h. Fixed leaves were washed in PBS two to three times for 5 to 10 min each. PBS was removed, pure methanol was added, and materials were incubated for 10 min at 37°C; this was repeated two more times until chlorophyll was gone. The materials were transferred to wash buffer (1× PBS and 50 mm Gly) in Eppendorf tubes, and 3% bovine serum albumin was added for 1 h. The materials were incubated with a PIN1 antibody (NASC, 1:200) at 37°C for 3 h and then incubated with a second antibody (fluorescein isothiocyanate-conjugated anti-sheep IgG [1:200], KPL) for 1 to 3 h at 37°C. Stained tissues were observed under confocal microscope. Lobe and indentation regions containing PIN1 were quantified from over 30 cells from three independent experiments. Preferential localization of PIN1 to lobes or indentations and equal localization to both regions were determined by eyeballing of the confocal stacked images.
Antibodies and IP Assay
Anti-TOPP4 polyclonal antibodies were raised against the N terminus of TOPP4 (amino acids 1–150), which is specific for TOPP4 protein, in rabbits. Antiserum was isolated from rabbits immunized alternately with soluble recombinant HIS-TOPP4 N150aa protein. One-half gram of cyanogen bromide-activated Sepharose 4B (GE Healthcare) was incubated with 20 mg of HIS-TOPP4 N150aa for 1 h; the medium was transferred to 0.1 m Tris-HCl, pH 8.0, for 2 h and washed alternatively with 0.5 m NaCl, 0.1 m ethanoic acid, pH 4.0, 0.5 m NaCl, and 0.1 m Tris-HCl, pH 8.0; the Sepharose was balanced, mixed with antiserum slowly for 40 min, and washed with buffer (0.1 m citric acid, pH 2.0); and the anti-TOPP4 polyclonal antibodies were eluted with elution buffer (1 m Tris-HCl, pH 8.0).
For IP assay, 21-d-old seedlings were harvested and suspended in IP buffer (20 mm HEPES, pH 7.5, 40 mm KCl, 1 mm dithiothreitol [DTT], and 1% protease inhibitors [Sangon Biothch]). TOPP4 or topp4-1 was immunoprecipitated from wild-type or topp4-1 plants using the anti-TOPP4 antibody followed by protein A beads. The beads in column were washed by IP buffer containing 0.2% Triton X-100 three times. After IP, the beads were suspended in 50 µL of IP buffer containing 1% of the protease inhibitor cocktail. Immunoprecipitated proteins were detected by immunoblot analysis with the anti-TOPP4 at 1:400 dilutions.
In Vitro Phosphorylation Assays
Arabidopsis protoplasts were isolated from leaves of 4-week-old plants, transformed according to Meskiene et al. (2003) and harvested after 10 to 22 h. Cell pellets were lysed by freeze-thaw cycles followed by a Dounce-type homogenizer. The extraction buffer used was previously described (Michniewicz et al., 2007; Abas and Luschnig, 2010).
Membrane fractions were solubilized with 0.1% Brij35 (Sigma) and preheated at 65°C for 10 min to inactivate endogenous enzymes. After λ-phosphatase buffer (New England Labs) was added, four treatments were performed in a final volume of 30 mL: sample plus 3 mm MnCl2; sample plus 3 mm MnCl2 and 100 units of λ-phosphatase (New England Labs); and sample plus TOPP4 or topp4-1, which was immunoprecipitated with anti-TOPP4 antibody from wild-type or topp4-1 plants, respectively. All samples were incubated at 30°C for 30 min. These experiments were repeated at least three times.
Recombinant HIS-tagged PIN1 hydrophilic loop (HIS-PIN1HL) was expressed in E. coli and purified using N+-nitrilotriacetic acid resin (Invitrogen). Total proteins were extracted with 1× kinase buffer (25 mm Tris-HCl, pH 7.5, 1 mm DTT, and 5 mm MgCl2) plus 1× protease inhibitor and 1 mm phenylmethylsulfonyl fluoride. According to previous methods (Michniewicz et al., 2007; Dhonukshe et al., 2010; Dai et al., 2012), 1 μg of HIS:PIN1HL and 25 µg of plant seedling extracts were mixed in 1× kinase buffer and 1× ATP solution (100 µm ATP and 1 µCi [γ-32P] ATP) in a total volume of 25 µL. The reactions were incubated at 30°C for 30 min and then stopped by adding 5× loading buffer and boiling for 5 min, or approximately 1 µg of purified HIS-PIN1HL was phosphorylated by GST-PID using kinase reaction mix and then incubated at 30°C for 3 h with TOPP4, topp4-1, or denatured TOPP4, which were immunoprecipitated from the wild type or topp4-1. Products were separated by electrophoresis through 12.5% acrylamide gels, and the gels were stained, dried, and then visualized by exposure to x-ray films.
In Vivo Phosphorylation Assays
Arabidopsis seedlings expressing PIN1:PIN1-GFP in wild-type, topp4-1, and 35S:TOPP4 backgrounds were grown to 12 d, and then these seedlings were harvested. The membrane protein extraction was performed as previously described (Michniewicz et al., 2007; Abas and Luschnig, 2010). PIN1-GFP was immunoprecipitated by incubation with anti-GFP antibody-coupled protein A beads. Membrane fractions were subjected to λ-phosphatase treatment as described previously (Dai et al., 2012). Samples were separated as described (Abas and Luschnig, 2010) and probed with the anti-GFP antibody (1:1,000; Invitrogen).
Particle Bombardment-Mediated Transient Expression in Arabidopsis Leaves
For particle bombardment, all plasmids were amplified in E. coli strain DH5α and purified using plasmid midi or mini kits according to the manufacturer’s instructions (Qiagen). Expanding rosette leaves of 0.8 to 1.2 cm in length were collected from 3-week-old plants and were bombarded with gold particles coated with plasmids using a Bio-Rad PDS-1000/He particle delivery system. In all experiments, 0.5 g of constructs were used. The bombardment procedure was described previously for leaves (Fu et al., 2002). Bombarded leaves were incubated in water before observation with the confocal microscope.
ROP2 Activity Assay
GFP-tagged active ROP2 was pulled down by use of GST-RIC1 as described previously (Xu et al., 2010). Arabidopsis protoplasts were isolated from leaves of 3-week-old wild-type or topp4-1 plants transformed with 35S:GFP-ROP2 and harvested after 4 to 6 h. The protoplasts were treated with or without 100 nm NAA and frozen by liquid nitrogen. Total protein was extracted from 105 to 106 protoplasts by extraction buffer (25 mm HEPES, pH 7.4, 10 mm MgCl2, 10 mm KCl, 5 mm DTT, 5 mm Na3VO4, 5 mm NaF, 1 mm phenylmethylsulfonyl fluoride, 1% protease inhibitor, and 1% TritonX-100). A part of total proteins was used as control to determine the total amount of GFP-ROP2. A saturated amount of GST-RIC1-conjugated beads was added to the same amount of protoplast extracts, which were then gently shaken at 4°C for 2 h. Beads were washed in a washing buffer (25 mm HEPES, pH 7.4, 1 mm EDTA, 5 mm MgCl2, 1 mm DTT, and 0.5% TritonX-100) for three times at 4°C (5 min each). Western blotting with the anti-GFP antibody was used for analysis of the GTP-bound active form of GFP-ROP2 that was associated with the GST-RIC1 beads.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers TOPP4 (At2g39840), PIN1 (AT1G73590), PID (AT2G34650), ROP2 (AT1G20090), ROP4 (AT1G75840), ROP6 (AT4G35020), RIC1 (AT2G33460), and RIC4 (AT5G16490).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Tissue-specific expression of TOPP4 protein.
Supplemental Figure S2. The rosette leaf phenotypes of topp4-1.
Supplemental Figure S3. The cotyledon PCs show normal shape in topp4-1.
Supplemental Figure S4. Effect of TOPP4 mutation on PC development at different stages.
Supplemental Figure S5. Transformed TOPP4:TOPP4 into the topp4-1 mutant slightly rescues the mutant phenotype.
Supplemental Figure S6. Phenotype analysis of two transfer DNA insertion alleles of TOPP4.
Supplemental Figure S7. The TOPP4/topp4-1 expression levels of the 35S:TOPP4, TOPP4:topp4-1-GFP, or 35S:topp4-1-GFP transgenic plants.
Supplemental Figure S8. Inhibition of PP1 partially repressed interdigitated growth of PCs.
Supplemental Figure S9. The topp4-1 mutant shows auxin-related defects.
Supplemental Figure S10. The phenotype of topp4-1 pin1 double mutants.
Supplemental Figure S11. Effect of tautomycin on PC shape in wild-type, pin1, 35S:PID, and pid-3 plants.
Supplemental Figure S12. The phenotype of topp4-1 35S:PID double mutants.
Supplemental Figure S13. The phenotype of 2-month-old topp4-1 pid-3 double mutants.
Supplemental Figure S14. Transient expression of 35S:PIN1-GFP and 35S:PID-FLAG in Arabidopsis protoplasts.
Supplemental Figure S15. Immunoblot analyses of PIN1-GFP in wild-type, topp4-1, and 35S:TOPP4 plants expressing PIN1:PIN1-GFP.
Supplemental Figure S16. PIN1 internalization in root cells after BFA treatment.
Supplemental Figure S17. The effect of exogenous auxin on the PC lobe formation in 35S:CYCD3;1.
Supplemental Figure S18. The effect of auxin on ROP2 activity and the organization of cortical MTs and F-actins in 35S:CYCD3;1.
Supplemental Table S1. Primers used for plasmid construction and qRT-PCR.
Supplementary Material
Acknowledgments
We thank Xiaoping Gou, Yun Xiang, Longfeng Yan, and Qingxiang Gao for technical assistance, Yun Xiang for providing FABD2-GFP seeds, Ver⊚nica A. Grieneisen (Utrecht University) for providing Ara7-GFP seeds, and Yongming Zhou (Huazhong University) for providing 35S:CYCD3;1 seeds.
Glossary
- PC
pavement cell
- F-actin
filamentous actin
- MT
microtubule
- NAA
naphthalene acetic acid
- BFA
brefeldin A
- qRT
quantitative reverse transcription
- IAA
indole-3-acetic acid
- β-gal
β-galactosidase
- GST
glutathione S-transferase
- BiFC
bimolecular fluorescence complementation
- LatB
latrunculin B
- MS
Murashige and Skoog
- NASC
European Arabidopsis Stock Centre
- DMSO
dimethyl sulfoxide
- co-IP
coimmunoprecipitation
- PBS
phosphate-buffered saline
- DTT
dithiothreitol
Footnotes
This work was supported by the National Basic Research Program of China (grant nos. 2011CB915401 and 2009CB941501), the Natural Science Foundation of China (grant nos. 91017002, 31070247, and 31271460), the Ministry of Agriculture of the People’s Republic of China (grant no. 2013ZX08009–003–002), and the Fundamental Research Funds for the Central Universities (grant no. lzujbky–2013–240).
References
- Abas L, Luschnig C (2010) Maximum yields of microsomal-type membranes from small amounts of plant material without requiring ultracentrifugation. Anal Biochem 401: 217–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamins R, Quint A, Weijers D, Hooykaas P, Offringa R (2001) The PINOID protein kinase regulates organ development in Arabidopsis by enhancing polar auxin transport. Development 128: 4057–4067 [DOI] [PubMed] [Google Scholar]
- Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 [DOI] [PubMed] [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B (2005) The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44 [DOI] [PubMed] [Google Scholar]
- Chary SN, Hicks GR, Choi YG, Carter D, Raikhel NV (2008) Trehalose-6-phosphate synthase/phosphatase regulates cell shape and plant architecture in Arabidopsis. Plant Physiol 146: 97–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Yang Z (2014) Novel ABP1-TMK auxin sensing system controls ROP GTPase-mediated interdigitated cell expansion in Arabidopsis. Small GTPases 30: e29711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai M, Terzaghi W, Wang H (2013) Multifaceted roles of Arabidopsis PP6 phosphatase in regulating cellular signaling and plant development. Plant Signal Behav 8: e22508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai M, Zhang C, Kania U, Chen F, Xue Q, McCray T, Li G, Qin G, Wakeley M, Terzaghi W, et al. (2012) A PP6-type phosphatase holoenzyme directly regulates PIN phosphorylation and auxin efflux in Arabidopsis. Plant Cell 24: 2497–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewitte W, Riou-Khamlichi C, Scofield S, Healy JM, Jacqmard A, Kilby NJ, Murray JA (2003) Altered cell cycle distribution, hyperplasia, and inhibited differentiation in Arabidopsis caused by the D-type cyclin CYCD3. Plant Cell 15: 79–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhonukshe P, Aniento F, Hwang I, Robinson DG, Mravec J, Stierhof YD, Friml J (2007) Clathrin-mediated constitutive endocytosis of PIN auxin efflux carriers in Arabidopsis. Curr Biol 17: 520–527 [DOI] [PubMed] [Google Scholar]
- Dhonukshe P, Huang F, Galvan-Ampudia CS, Mähönen AP, Kleine-Vehn J, Xu J, Quint A, Prasad K, Friml J, Scheres B, et al. (2010) Plasma membrane-bound AGC3 kinases phosphorylate PIN auxin carriers at TPRXS(N/S) motifs to direct apical PIN recycling. Development 137: 3245–3255 [DOI] [PubMed] [Google Scholar]
- Falbel TG, Koch LM, Nadeau JA, Segui-Simarro JM, Sack FD, Bednarek SY (2003) SCD1 is required for cytokinesis and polarized cell expansion in Arabidopsis thaliana [corrected]. Development 130: 4011–4024 [DOI] [PubMed] [Google Scholar]
- Farkas I, Dombrádi V, Miskei M, Szabados L, Koncz C (2007) Arabidopsis PPP family of serine/threonine phosphatases. Trends Plant Sci 12: 169–176 [DOI] [PubMed] [Google Scholar]
- Favre B, Turowski P, Hemmings BA (1997) Differential inhibition and posttranslational modification of protein phosphatase 1 and 2A in MCF7 cells treated with calyculin-A, okadaic acid, and tautomycin. J Biol Chem 272: 13856–13863 [DOI] [PubMed] [Google Scholar]
- Friml J, Yang X, Michniewicz M, Weijers D, Quint A, Tietz O, Benjamins R, Ouwerkerk PB, Ljung K, Sandberg G, et al. (2004) A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science 306: 862–865 [DOI] [PubMed] [Google Scholar]
- Fu Y, Gu Y, Zheng Z, Wasteneys G, Yang Z (2005) Arabidopsis interdigitating cell growth requires two antagonistic pathways with opposing action on cell morphogenesis. Cell 120: 687–700 [DOI] [PubMed] [Google Scholar]
- Fu Y, Li H, Yang Z (2002) The ROP2 GTPase controls the formation of cortical fine F-actin and the early phase of directional cell expansion during Arabidopsis organogenesis. Plant Cell 14: 777–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Xu T, Zhu L, Wen M, Yang Z (2009) A ROP GTPase signaling pathway controls cortical microtubule ordering and cell expansion in Arabidopsis. Curr Biol 19: 1827–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gälweiler L, Guan C, Müller A, Wisman E, Mendgen K, Yephremov A, Palme K (1998) Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282: 2226–2230 [DOI] [PubMed] [Google Scholar]
- Geldner N, Friml J, Stierhof YD, Jürgens G, Palme K (2001) Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature 413: 425–428 [DOI] [PubMed] [Google Scholar]
- Guo X, Lu W, Ma Y, Qin Q, Hou S (2013) The BIG gene is required for auxin-mediated organ growth in Arabidopsis. Planta 237: 1135–1147 [DOI] [PubMed] [Google Scholar]
- Hamant O, Heisler MG, Jönsson H, Krupinski P, Uyttewaal M, Bokov P, Corson F, Sahlin P, Boudaoud A, Meyerowitz EM, et al. (2008) Developmental patterning by mechanical signals in Arabidopsis. Science 322: 1650–1655 [DOI] [PubMed] [Google Scholar]
- Huang F, Zago MK, Abas L, van Marion A, Galván-Ampudia CS, Offringa R (2010) Phosphorylation of conserved PIN motifs directs Arabidopsis PIN1 polarity and auxin transport. Plant Cell 22: 1129–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J, Dhonukshe P, Sauer M, Brewer PB, Wiśniewska J, Paciorek T, Benková E, Friml J (2008) ARF GEF-dependent transcytosis and polar delivery of PIN auxin carriers in Arabidopsis. Curr Biol 18: 526–531 [DOI] [PubMed] [Google Scholar]
- Kleine-Vehn J, Huang F, Naramoto S, Zhang J, Michniewicz M, Offringa R, Friml J (2009) PIN auxin efflux carrier polarity is regulated by PINOID kinase-mediated recruitment into GNOM-independent trafficking in Arabidopsis. Plant Cell 21: 3839–3849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GJ, Sohn EJ, Lee MH, Hwang I (2004) The Arabidopsis rab5 homologs rha1 and ara7 localize to the prevacuolar compartment. Plant Cell Physiol 45: 1211–1220 [DOI] [PubMed] [Google Scholar]
- Li H, Lin D, Dhonukshe P, Nagawa S, Chen D, Friml J, Scheres B, Guo H, Yang Z (2011) Phosphorylation switch modulates the interdigitated pattern of PIN1 localization and cell expansion in Arabidopsis leaf epidermis. Cell Res 21: 970–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Xu T, Lin D, Wen M, Xie M, Duclercq J, Bielach A, Kim J, Reddy GV, Zuo J, et al. (2013) Cytokinin signaling regulates pavement cell morphogenesis in Arabidopsis. Cell Res 23: 290–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D, Cao L, Zhou Z, Zhu L, Ehrhardt D, Yang Z, Fu Y (2013) Rho GTPase signaling activates microtubule severing to promote microtubule ordering in Arabidopsis. Curr Biol 23: 290–297 [DOI] [PubMed] [Google Scholar]
- Lin Q, Li J, Smith RD, Walker JC (1998) Molecular cloning and chromosomal mapping of type one serine/threonine protein phosphatases in Arabidopsis thaliana. Plant Mol Biol 37: 471–481 [DOI] [PubMed] [Google Scholar]
- Mathur J, Mathur N, Kernebeck B, Hülskamp M (2003) Mutations in actin-related proteins 2 and 3 affect cell shape development in Arabidopsis. Plant Cell 15: 1632–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meskiene I, Baudouin E, Schweighofer A, Liwosz A, Jonak C, Rodriguez PL, Jelinek H, Hirt H (2003) Stress-induced protein phosphatase 2C is a negative regulator of a mitogen-activated protein kinase. J Biol Chem 278: 18945–18952 [DOI] [PubMed] [Google Scholar]
- Michniewicz M, Zago MK, Abas L, Weijers D, Schweighofer A, Meskiene I, Heisler MG, Ohno C, Zhang J, Huang F, et al. (2007) Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell 130: 1044–1056 [DOI] [PubMed] [Google Scholar]
- Nagawa S, Xu T, Lin D, Dhonukshe P, Zhang X, Friml J, Scheres B, Fu Y, Yang Z (2012) ROP GTPase-dependent actin microfilaments promote PIN1 polarization by localized inhibition of clathrin-dependent endocytosis. PLoS Biol 10: e1001299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Naoi K, Shoji T, Hashimoto T (2004) Low concentrations of propyzamide and oryzalin alter microtubule dynamics in Arabidopsis epidermal cells. Plant Cell Physiol 45: 1330–1334 [DOI] [PubMed] [Google Scholar]
- Petrásek J, Mravec J, Bouchard R, Blakeslee JJ, Abas M, Seifertová D, Wisniewska J, Tadele Z, Kubes M, Covanová M, et al. (2006) PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 312: 914–918 [DOI] [PubMed] [Google Scholar]
- Pietra S, Grebe M (2010) Auxin paves the way for planar morphogenesis. Cell 143: 29–31 [DOI] [PubMed] [Google Scholar]
- Qian P, Hou S, Guo G (2009) Molecular mechanisms controlling pavement cell shape in Arabidopsis leaves. Plant Cell Rep 28: 1147–1157 [DOI] [PubMed] [Google Scholar]
- Qin Q, Wang W, Guo X, Yue J, Huang Y, Xu X, Li J, Hou S (2014) Arabidopsis DELLA protein degradation is controlled by a type-one protein phosphatase, TOPP4. PLoS Genet 10: e1004464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampathkumar A, Lindeboom JJ, Debolt S, Gutierrez R, Ehrhardt DW, Ketelaar T, Persson S (2011) Live cell imaging reveals structural associations between the actin and microtubule cytoskeleton in Arabidopsis. Plant Cell 23: 2302–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer M, Paciorek T, Benková E, Friml J (2006) Immunocytochemical techniques for whole-mount in situ protein localization in plants. Nat Protoc 1: 98–103 [DOI] [PubMed] [Google Scholar]
- Shi Y. (2009) Serine/threonine phosphatases: mechanism through structure. Cell 139: 468–484 [DOI] [PubMed] [Google Scholar]
- Smith RD, Walker JC (1993) Expression of multiple type 1 phosphoprotein phosphatases in Arabidopsis thaliana. Plant Mol Biol 21: 307–316 [DOI] [PubMed] [Google Scholar]
- Song Y, Wu K, Dhaubhadel S, An L, Tian L (2010) Arabidopsis DNA methyltransferase AtDNMT2 associates with histone deacetylase AtHD2s activity. Biochem Biophys Res Commun 396: 187–192 [DOI] [PubMed] [Google Scholar]
- Stubbs MD, Tran HT, Atwell AJ, Smith CS, Olson D, Moorhead GB (2001) Purification and properties of Arabidopsis thaliana type 1 protein phosphatase (PP1). Biochim Biophys Acta 1550: 52–63 [DOI] [PubMed] [Google Scholar]
- Takemiya A, Kinoshita T, Asanuma M, Shimazaki K (2006) Protein phosphatase 1 positively regulates stomatal opening in response to blue light in Vicia faba. Proc Natl Acad Sci USA 103: 13549–13554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Kitakura S, Rakusová H, Uemura T, Feraru MI, De Rycke R, Robert S, Kakimoto T, Friml J (2013) Cell polarity and patterning by PIN trafficking through early endosomal compartments in Arabidopsis thaliana. PLoS Genet 9: e1003540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Q, Wang J (2002) Role of serine/threonine protein phosphatase in Alzheimer’s disease. Neurosignals 11: 262–269 [DOI] [PubMed] [Google Scholar]
- Traweger A, Wiggin G, Taylor L, Tate SA, Metalnikov P, Pawson T (2008) Protein phosphatase 1 regulates the phosphorylation state of the polarity scaffold Par-3. Proc Natl Acad Sci USA 105: 10402–10407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda K, Matsuyama T, Hashimoto T (1999) Visualization of microtubules in living cells of transgenic Arabidopsis thaliana. Protoplasma 206: 201–206 [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt B, Timmers AC, Šamaj J, Müller J, Baluška F, Menzel D (2005) GFP-FABD2 fusion construct allows in vivo visualization of the dynamic actin cytoskeleton in all cells of Arabidopsis seedlings. Eur J Cell Biol 84: 595–608 [DOI] [PubMed] [Google Scholar]
- Wang X, Zhu L, Liu B, Wang C, Jin L, Zhao Q, Yuan M (2007) Arabidopsis MICROTUBULE-ASSOCIATED PROTEIN18 functions in directional cell growth by destabilizing cortical microtubules. Plant Cell 19: 877–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewska J, Xu J, Seifertová D, Brewer PB, Ruzicka K, Blilou I, Rouquié D, Benková E, Scheres B, Friml J (2006) Polar PIN localization directs auxin flow in plants. Science 312: 883. [DOI] [PubMed] [Google Scholar]
- Wu HM, Hazak O, Cheung AY, Yalovsky S (2011) RAC/ROP GTPases and auxin signaling. Plant Cell 23: 1208–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Dai N, Chen J, Nagawa S, Cao M, Li H, Zhou Z, Chen X, De Rycke R, Rakusová H (2014) Cell surface ABP1-TMK auxin-sensing complex activates ROP GTPase signaling. Science 343: 1025–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Wen M, Nagawa S, Fu Y, Chen JG, Wu MJ, Perrot-Rechenmann C, Friml J, Jones AM, Yang Z (2010) Cell surface- and rho GTPase-based auxin signaling controls cellular interdigitation in Arabidopsis. Cell 143: 99–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. (2008) Cell polarity signaling in Arabidopsis. Annu Rev Cell Dev Biol 24: 551–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Halsey LE, Szymanski DB (2011) The development and geometry of shape change in Arabidopsis thaliana cotyledon pavement cells. BMC Plant Biol 11: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Wang H, Gilmer S, Whitwill S, Fowke LC (2003) Effects of co-expressing the plant CDK inhibitor ICK1 and D-type cyclin genes on plant growth, cell size and ploidy in Arabidopsis thaliana. Planta 216: 604–613 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.