A negative regulator of strawberry fruit development and ripening in strawberry fruit ripening is a homolog of a well-characterized protein kinase.
Abstract
Whereas the regulatory mechanisms that direct fruit ripening have been studied extensively, little is known about the signaling mechanisms underlying this process, especially for nonclimacteric fruits. In this study, we demonstrated that a SUCROSE NONFERMENTING1-RELATED PROTEIN KINASE2, designated as FaSnRK2.6, is a negative regulator of fruit development and ripening in the nonclimacteric fruit strawberry (Fragaria × ananassa) and can also mediate temperature-modulated strawberry fruit ripening. FaSnRK2.6 was identified as an ortholog of OPEN STOMATA1. Levels of FaSnRK2.6 transcript rapidly decreased during strawberry fruit development and ripening. FaSnRK2.6 was found to be capable of physically interacting with strawberry ABSCISIC ACID INSENSITIVE1, a negative regulator in strawberry fruit ripening. RNA interference-induced silencing of FaSnRK2.6 significantly promoted fruit ripening. By contrast, overexpression of FaSnRK2.6 arrested fruit ripening. Strawberry fruit ripening is highly sensitive to temperature, with high temperatures promoting ripening and low temperatures delaying it. As the temperature increased, the level of FaSnRK2.6 expression declined. Furthermore, manipulating the level of FaSnRK2.6 expression altered the expression of a variety of temperature-responsive genes. Taken together, this study demonstrates that FaSnRK2.6 is a negative regulator of strawberry fruit development and ripening and, furthermore, that FaSnRK2.6 mediates temperature-modulated strawberry fruit ripening.
Ripening of fleshy fruits is a complex process that involves dramatic changes in color, texture, flavor, and aroma (Giovannoni, 2001; Seymour et al., 2013). Fleshy fruits are physiologically classified as climacteric or nonclimacteric. Whereas climacteric fruits exhibit an increase in respiration and a concomitant burst in ethylene production soon after the onset of ripening, nonclimacteric fruits do not (Nitsch, 1953; Coombe, 1976; Brady, 1987). Much of our knowledge of fruit ripening is based on climacteric fruits, such as tomato (Solanum lycopersicum; Klee and Giovannoni, 2011). However, the mechanism underlying the ripening of nonclimacteric fruits, such as strawberry (Fragaria × ananassa), is poorly understood.
Early theories on ripening emphasize the senescence-associated deteriorative aspects of this process; however, it is now widely accepted that ripening is a highly coordinated process determined by genetic programs (Brady, 1987; Fischer and Bennett, 1991; Giovannoni, 2001). Although the mechanisms underlying the molecular regulation of fruit ripening have been studied extensively, these studies have largely focused on the contribution of phytohormones and transcription factors to this process, and little is known about the signaling mechanisms that underlie fruit development and ripening (Fischer and Bennett, 1991; Giovannoni, 2001; Klee and Giovannoni, 2011; Seymour et al., 2013).
Protein kinase/phosphatase-catalyzed reversible phosphorylation plays a central role in cellular signal transduction (Bourret et al., 1991; Freeman and Gurdon, 2002; Ma et al., 2009). SUCROSE NONFERMENTING1-RELATED PROTEIN KINASES2 (SnRK2s) are a family of plant-specific protein kinases implicated in stress and abscisic acid (ABA) signaling (Yoshida et al., 2002; Boudsocq et al., 2004; Kobayashi et al., 2005; Umezawa et al., 2004; Fujii et al., 2007; Shukla and Mattoo, 2008; Fujita et al., 2009; Hirayama et al., 2010; Kulik et al., 2011; Hrabak et al., 2013). OPEN STOMATA1 (OST1), also designated as SnRK2E or SnRK2.6, is a member of the SnRK2 family that plays a critical role in stomatal movement, and in Arabidopsis (Arabidopsis thaliana), mutation of OST1 renders guard cells insensitive to ABA, such that stomata remain open in the presence of ABA (Schroeder et al., 2001; Mustilli et al., 2002; Assmann, 2003; Yoshida et al., 2006; Imes et al., 2013). OST1 and its orthologs in other plant species are thought to have important roles in various biological processes in addition to their involvement in stomatal regulation (Sirichandra et al., 2010; Zheng et al., 2010; Xue et al., 2011; Imes et al., 2013; Vilela et al., 2013; Wang et al., 2013a). Recently, ABA was shown to act as an internal cue that plays an important role in strawberry fruit ripening (Jia et al., 2011). Furthermore, it was reported that manipulating the expression of either an ABA receptor, strawberry PYRABACTIN RESISTANCE1 (FaPYR1), or its downstream signaling component, strawberry ABSCISIC ACID INSENSITIVE1 (FaABI1), affects strawberry fruit ripening (Chai et al., 2011; Jia et al., 2013a). Given that OST1 is a critical mediator of ABA signaling, it is possible that the strawberry ortholog of OST1 plays an important role in strawberry fruit development and ripening. However, this possibility has not been explored.
Fleshy fruit ripening is determined both by internal regulators, such as phytohormones and signaling proteins, and external environmental cues (Ulrich, 1958; Giovannoni, 2001; Llop-Tous et al., 2002; Owen et al., 2009; Giribaldi et al., 2010; Hirayama et al., 2010; Koyama et al., 2010; Gagné et al., 2011; Sun et al., 2012; Yang et al., 2012; Seymour et al., 2013). Temperature, photon flux density, and water availability affect both fruit ripening and quality, and temperature is particularly important for controlling the progression of fruit development and ripening during fruit production. While much attention has focused on the mechanisms by which internal cues regulate fruit ripening, little is known about the mechanisms by which external cues regulate fruit development and ripening. Given that fleshy fruit arose as a mechanism to facilitate seed dispersal and promote the survival of the plant in ever-changing environments (Coombe, 1976), the regulation of fruit ripening by external cues is of great importance in the plant’s adaptation to environmental stresses (Shulaev et al., 2008). Given that OST1 is a critical mediator of ABA signaling, and that SnRK2s are tightly associated with plant stress signaling (Yoshida et al., 2002; Boudsocq et al., 2004; Umezawa et al., 2004; Kobayashi et al., 2005; Fujii et al., 2007; Shukla and Mattoo, 2008; Fujita et al., 2009; Hirayama et al., 2010; Kulik et al., 2011; Hrabak et al., 2013), we sought to identify the ortholog of OST1 in strawberry and to evaluate whether this protein contributes to the regulation of strawberry fruit development and ripening, and particularly whether it modulates fruit ripening in response to temperature.
RESULTS
Genome-Wide Identification of FaSnRK2 Subfamily Members
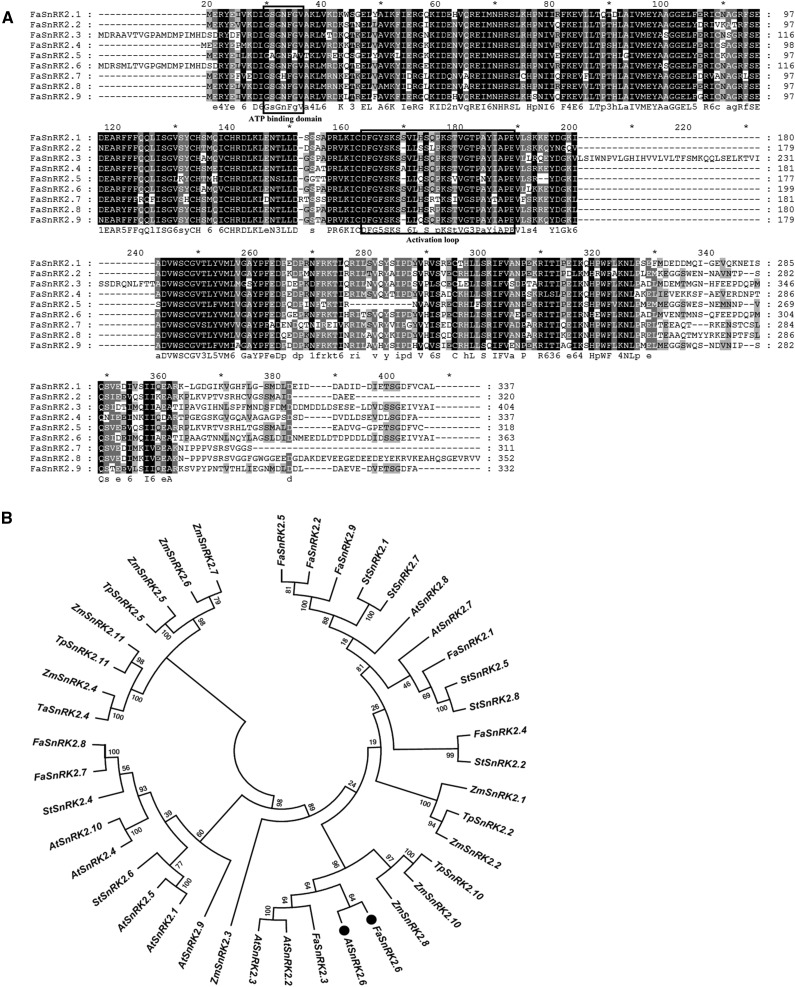
To identify the strawberry ortholog of OST1, we first searched for SnRK2 subfamily members in the strawberry genome. BLAST searches against the strawberry sequence database (https://strawberry.plantandfood.co.nz/) using the coding sequences of all members of the Arabidopsis SnRK subfamily as query (Boudsocq et al., 2004) detected nine nonredundant gene sequences. Protein sequence alignment showed that all nine members contained a conserved SnRK2 domain (Hrabak et al., 2003; Boudsocq et al., 2004); therefore, we named these proteins FaSnRK2.1 to FaSnRK2.9 (Fig. 1A). To compare FaSnRK2s with AtSnRK2s and other SnRK2s reported previously (Park et al., 1993; Mustilli et al., 2002; Hrabak et al., 2003; Mao et al., 2010; Bucholc et al., 2011), we constructed a phylogenetic tree based on the full-length complementary DNA (cDNA) sequences of the selected SnRK2s using the neighbor-joining method in MEGA6 software (Fig. 1B). The FaSnRK2s clustered into four clades. Among all of the sequences selected, OST1/AtSnRK2.6 shares the highest degree of similarity (95%) with FaSnRK2.6. Within the FaSnRK2 subfamily, FaSnRK2.6 and FaSnRK2.3 belong to the same clade and share only 80% sequence similarity. The high degree of similarity between FaSnRK2.6 and OST1 suggests that FaSnRK2.6 is an ortholog of OST1.
Figure 1.
Phylogenetic analysis of SnRK2.6 proteins. A, Sequence alignments of the deduced amino acid sequences of FaSnRK2s. The deduced amino acid sequences of FaSnRK2.1 to FaSnRK2.9 were aligned using ClustalX 2.0.12 software with default settings. The alignments were edited and marked using GeneDoc. Black and light-gray shading indicate identical and similar amino acid residues, respectively. The predicted functional domains are boxed. B, Phylogenic analysis of SnRK2 homologs from various plant species. The phylogenetic trees were constructed using the neighbor-joining method in MEGA6 software with 1,000 bootstrap replicates. The numbers at nodes represent bootstrap values. OST1 and its ortholog in strawberry are marked with black circles. Species name abbreviations are as follows: At, Arabidopsis; Np, Nicotiana plumbaginifolia; St, potato (Solanum tuberosum); Ta, wheat (Triticum aestivum); Tp, Triticum polonicum; and Zm, maize.
Temporospatial Expression of FaSnRK2.6
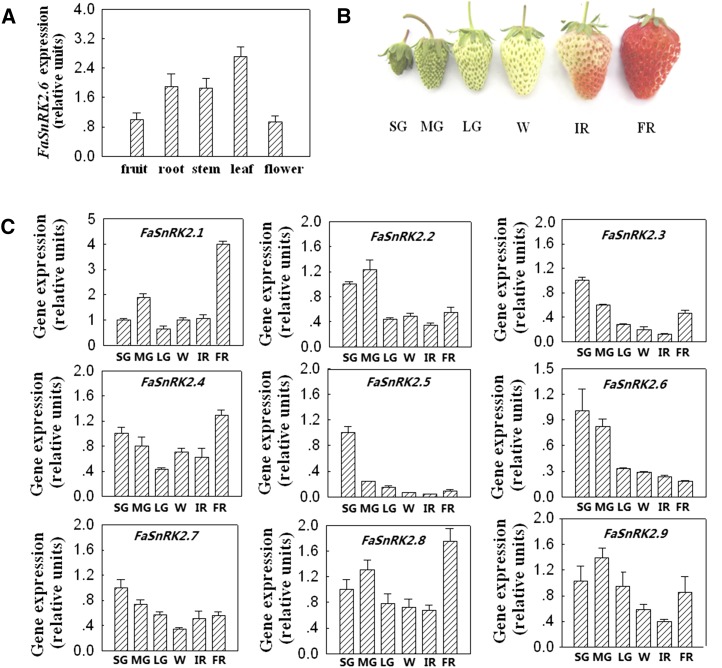
Using quantitative PCR, we showed that FaSnRK2.6 was ubiquitously expressed in strawberry plants, with the highest transcript level detected in the leaves (Fig. 2A). We were particularly interested in determining the expression pattern of FaSnRK2.6 during fruit development and ripening. The process of strawberry fruit development, from fruit set to ripening, can be divided into three major stages, the green fruit stage, white fruit stage, and red fruit stage, and the green and red fruit stages can be divided again into several substages, the small green fruit, middle green fruit, and large green fruit and initially reddening and fully reddening substages (Fig. 2B). As shown in Figure 2C, FaSnRK2.6 was strongly expressed in the small green fruit substage, declined sharply from the middle green fruit to large green fruit substages, and then decreased gradually until the fully reddening substage, suggesting that there is a tight negative correlation between FaSnRK2.6 expression and the progression of fruit development and ripening. As a comparison, we also examined the expression patterns of other members of the FaSnRK2 subfamily. While the expression of FaSnRK2.3 and FaSnRK2.5 also declined throughout fruit growth and development, no clear correlations were detected between the expression of other members and fruit ripening (Fig. 2C).
Figure 2.
Temporospatial pattern of FaSnRK2 expression in strawberry fruits. A, Quantitative reverse transcription (qRT)-PCR analysis of FaSnRK2.6 expression in different organs of the strawberry plant. B, Phenotypes showing different developmental stages of strawberry fruit: SG, small green fruit; MG, middle green fruit; LG, large green fruit; W, white fruit; IR, initial reddening; and FR, full reddening. C, Quantitative PCR analysis of FaSnRK2 expression in different developmental stages of strawberry fruit. Labels below bars denote the corresponding developmental stages as explained in B. For A and C, qRT-PCR was conducted using FaACTIN as an internal control. Values are means + sd of five biological replicates.
Expression of FaSnRK2.6 in Response to Internal Signals
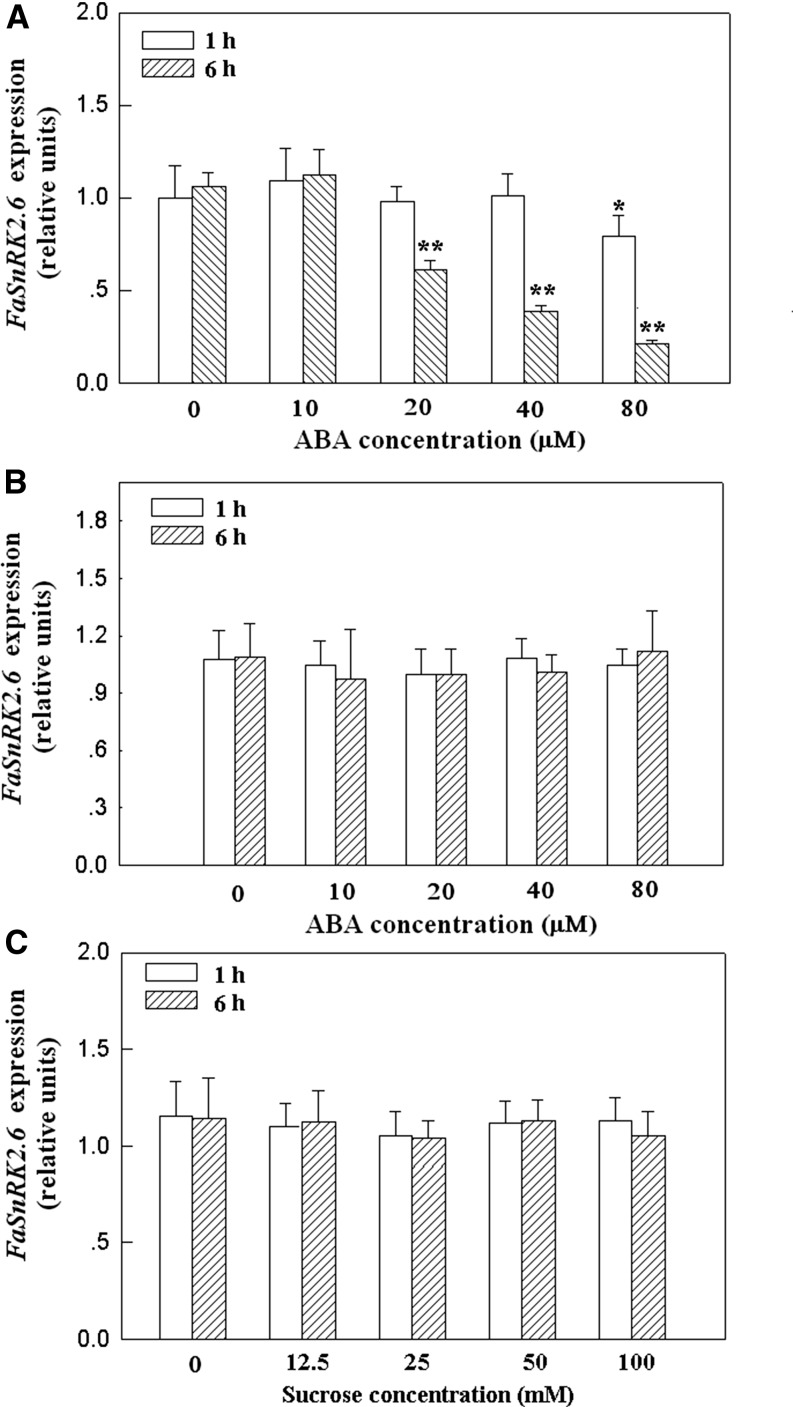
As ABA and Suc have been identified as internal signals that control strawberry fruit development and ripening (Jia et al., 2011, 2013b), we examined the expression of FaSnRK2.6 in response to ABA and Suc stimuli in fruits. As shown in Figure 3A, the level of FaSnRK2.6 expression declined upon ABA treatment in a concentration-dependent manner. Furthermore, FaSnRK2.6 expression was sensitive to ABA, as just 20 mm ABA significantly reduced FaSnRK2.6 expression. Based on the findings that OST1 regulates stomatal movement in leaves (Mustilli et al., 2002) and that FaSnRK2.6 is largely expressed in leaves (Fig. 2A), we further examined the response of FaSnRK2.6 expression to ABA in leaves. Unexpectedly, FaSnRK2.6 expression in the leaves was not affected by ABA treatment (Fig. 3B), suggesting that different mechanisms regulate FaSnRK2.6 expression in leaves and fruits. Additionally, Suc treatment had no effect on the level of FaSnRK2.6 expression in fruits, suggesting that FaSnRK2.6 is specifically involved in ABA signaling (Fig. 3C).
Figure 3.
qRT-PCR analysis of FaSnRK2.6 expression in response to ABA and Suc treatments in fruits and leaves. A, FaSnRK2.6 expression in response to ABA in fruits. Detached fruits in the large green fruit substage (i.e. corresponding to 18 DPA) were treated with various concentrations of ABA for 1 or 6 h, and then FaSnRK2.6 expression was examined. B, FaSnRK2.6 expression in response to ABA treatment in leaves. Young, fully expanded leaves were treated with various concentrations of ABA for 1 or 6 h, and then the FaSnRK2.6 expression level was examined. C, FaSnRK2.6 expression in response to Suc treatment in fruits. Detached fruits in the large green fruit substage were treated with various concentrations of Suc for 1 or 6 h, and then the FaSnRK2.6 expression level was examined. qRT-PCR was conducted using FaACTIN as an internal control. Values are means + sd of four biological replicates. Asterisks denote significant differences compared with the control sample (i.e. the 0 concentration) at *P < 0.05 and **P < 0.01, according to Student’s t test.
Manipulation of FaSnRK2.6 Expression Modulated Fruit Development and Ripening
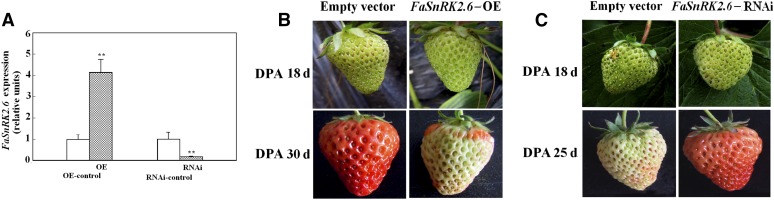
The close relationship between the pattern of FaSnRK2.6 expression and fruit development and ripening (Fig. 2) suggests that FaSnRK2.6 is an important regulator of strawberry fruit development and ripening. To test this possibility, we performed loss- and gain-of-function experiments using transient overexpression (OE) and RNA interference (RNAi) techniques (Jia et al., 2013b). As shown in Figure 4A, FaSnRK2.6-OE resulted in a more than 4-fold increase in the level of FaSnRK2.6 transcript, whereas FaSnRK2.6-RNAi resulted in a dramatic decrease in FaSnRK2.6 transcript level. Thus, the transient OE and RNAi techniques successfully manipulated FaSnRK2.6 expression in the strawberry fruits. Interestingly, FaSnRK2.6-OE greatly delayed the progression of strawberry fruit development and ripening. While the control fruits transfected with empty vector became red, the fruits of FaSnRK2.6-OE plants commonly remained in the white stage (Fig. 4B). By contrast, while the control fruits were still in the white stage, the fruits of FaSnRK2.6-RNAi plants commonly became red (Fig. 4C).
Figure 4.
Effect of FaSnRK2.6 OE and RNAi on the levels of FaSnRK2.6 transcript and strawberry fruit development and ripening. A, Effect of FaSnRK2.6-OE and FaSnRK2.6-RNAi on the levels of FaSnRK2.6 transcript. FaSnRK2.6 OE and RNAi were performed as described in “Materials and Methods.” The OE and RNAi constructs were injected into the fruits at 18 DPA. Levels of FaSnRK2.6 were examined at 30 DPA for the OE samples and at 25 DPA for the RNAi samples. Control samples were transfected with the empty vector (pCambia1304). qRT-PCR was conducted using FaACTIN as an internal control. Values are means + sd of four biological replicates. Asterisks denote significant difference at P < 0.01 using Student’s t test. B, Effect of FaSnRK2.6-OE on strawberry fruit development and ripening. The top row shows the phenotypes of fruits immediately after construct injection. Note the color change caused by the injection. In the bottom row, note that when the control fruit became red, the OE fruit was still in the white stage. C, Effect of FaSnRK2.6-RNAi on strawberry fruit development and ripening. Note that when the control fruit was at the white stage, the RNAi fruit had already become red.
Consistent with these phenotypic observations, FaSnRK2.6-OE and -RNAi strongly affected ripening-related physiological and biochemical metabolism and the related gene expression (Table I). Compared with the corresponding control, the FaSnRK2.6-OE and -RNAi lines exhibited a large increase and decrease in fruit firmness, respectively, and conversely, a large decrease and increase in anthocyanin content. FaSnRK2.6-OE and -RNAi also resulted in significant changes in major fruit quality parameters such as flavonoid and total phenol contents. Furthermore, FaSnRK2.6-OE and -RNAi led to an increase and decrease, respectively, in the contents of aldehyde and alcohol compounds (such as hexanal, 2-hexenal, octanal, etc.), and conversely, a decrease and increase in the contents of keton, furan, and ester compounds (such as 4H-pyran-4-one-2,3-dihydro-3,5-dihydroxy-6-methyl,hexanoic acid, methyl ester, etc.). Given that mature fruits contain greater concentrations of ketone, furan, and ester compounds and lower concentrations of aldehyde and alcohol compounds than immature fruit (El Hadi et al., 2013), the changes in aroma, pigment, and cell wall metabolism in the FaSnRK2.6-OE and -RNAi lines collectively suggested that the FaSnRK2.6-OE and -RNAi lines inhibited and promoted strawberry fruit ripening, respectively.
Table I. Effects of FaSnRK2.6-OE and FaSnRK2.6-RNAi on the major fruit ripening-related parameters.
The OE and RNAi constructs were transfected into fruits at 18 DPA, and 7 d (for RNAi) or 12 d (for OE) after transfection, fruits were detached and used for the analysis. Values are means ± sd of four samples (each sample includes five fruits). Asterisks denote significant differences compared with the control sample (i.e. OE-C or RNAi-C) at P < 0.01, using Student’s t test.
| Parameters | OE-C | OE | RNAi-C | RNAi | Notes |
|---|---|---|---|---|---|
| Firmness (kg cm−2) | 2.815 ± 0.262 | 6.990 ± 0.126** | 6.185 ± 0.117 | 2.155 ± 0.158** | Cell wall metabolism-related parameter |
| Anthocyanin content (mg g−1 fresh wt) | 0.110 ± 0.005 | 0.039 ± 0.002** | 0.040 ± 0.003 | 0.096 ± 0.004** | Pigment metabolism-related compounds |
| Flavonoid content (μg g−1 fresh wt) | 2.073 ± 0.057 | 3.142 ± 0.160** | 4.346 ± 0.095 | 2.790 ± 0.080** | |
| Total phenol content (μg g−1 fresh wt) | 3.795 ± 0.091 | 4.661 ± 0.081** | 6.367 ± 0.090 | 4.463 ± 0.064** | |
| Fru content (mg g−1 fresh wt) | 15.825 ± 1.163 | 16.105 ± 1.297 | 13.635 ± 1.277 | 14.348 ± 0.364 | Sugar metabolism-related compounds |
| Suc content (mg g−1 fresh wt) | 26.328 ± 1.225 | 25.659 ± 1.015 | 21.865 ± 1.365 | 21.118 ± 0.965 | |
| Glc content (mg g−1 fresh wt) | 17.921 ± 1.137 | 18.021 ± 0.129 | 16.011 ± 1.021 | 15.213 ± 0.361 | |
| Total titratable acid content (%) | 1.127 ± 0.036 | 1.125 ± 0.089 | 1.401 ± 0.021 | 1.263 ± 0.062 | Acid metabolism-related parameter |
| Hexanal | 0.437 ± 0.002 | 0.743 ± 0.005** | 1.510 ± 0.062 | 0.653 ± 0.011** | Aroma metabolism-related compounds (data are expressed as percentage of the total volatiles) |
| 2-Hexenal | 1.106 ± 0.042 | 2.585 ± 0.008** | 4.833 ± 0.009 | 1.800 ± 0.008** | |
| Octanal | 0.000 ± 0.000 | 1.520 ± 0.001** | 0.000 ± 0.000 | 0.000 ± 0.000 | |
| 1-Hexanol | 0.422 ± 0.016 | 0.731 ± 0.001** | 1.550 ± 0.003 | 0.717 ± 0.023** | |
| 1-Pentanol, 2-methyl | 0.000 ± 0.000 | 2.200 ± 0.018** | 3.227 ± 0.004 | 0.000 ± 0.000** | |
| 1,5-Pentanediol | 4.662 ± 0.142 | 5.881 ± 0.0.029** | 6.476 ± 0.015 | 5.420 ± 0.109** | |
| 2-Pentanol | 0.000 ± 0.000 | 2.230 ± 0.019** | 0.000 ± 0.000 | 0.000 ± 0.000 | |
| 1-Pentanol, 2-ethyl-4-methyl | 0.000 ± 0.000 | 0.720 ± 0.009** | 0.000 ± 0.000 | 0.000 ± 0.000 | |
| 1,3-Propanediol, 2-ethyl-2-(hydroxymethyl) | 5.520 ± 0.091 | 0.000 ± 0.000** | 0.000 ± 0.000 | 7.050 ± 0.017** | |
| 4H-Pyran-4-one, 2,3-dihydro-3,5-dihydroxy-6-methyl | 8.438 ± 0.089 | 1.600 ± 0.041** | 5.656 ± 0.593 | 9.672 ± 0.099** | |
| 2-Furancarboxaldehyde, 5-(hydroxymethyl) | 12.416 ± 0.123 | 2.350 ± 0.090** | 6.677 ± 0.311 | 15.542 ± 0.821** | |
| 2,4(1H,3H)-Pyrimidinedione, 5-methyl | 1.230 ± 0.030 | 1.370 ± 0.005 | 2.140 ± 0.048 | 2.160 ± 0.031 | |
| Hexanoic acid, methyl ester | 1.030 ± 0.006 | 0.000 ± 0.000** | 0.000 ± 0.000 | 1.020 ± 0.005** | |
| Acetic acid, hexyl ester | 0.653 ± 0.001 | 0.000 ± 0.000** | 0.000 ± 0.000 | 2.530 ± 0.050** | |
| Butanoic acid, 3-oxo, propyl ester | 1.800 ± 0.068 | 0.000 ± 0.000** | 0.000 ± 0.000 | 2.360 ± 0.013** | |
| 2-Hexen-1-ol | 3.692 ± 0.017 | 1.850 ± 0.099** | 1.542 ± 0.041 | 3.253 ± 0.007** | |
| Diethyl phthalate | 1.857 ± 0.009 | 0.680 ± 0.002** | 1.450 ± 0.011 | 2.071 ± 0.016** | |
| 1,2-Benzenedicarboxylic acid, butyl octyl ester | 3.337 ± 0.027 | 3.419 ± 0.015 | 2.928 ± 0.009 | 2.777 ± 0.039 | |
| 1,2-Benzenedicarboxylic acid, bis(2-methylpropyl) ester | 4.073 ± 0.092 | 2.363 ± 0.024** | 1.415 ± 0.004 | 2.219 ± 0.011** |
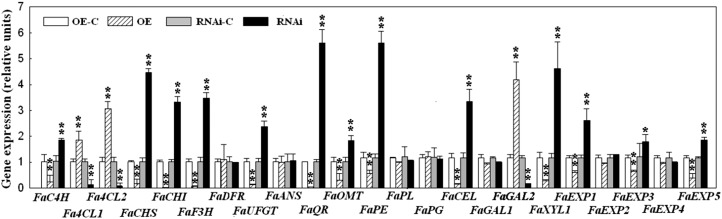
Consistent with the observations of the changed patterns of the ripening-related physiological parameters as described above, FaSnRK2.6-OE and -RNAi dramatically modulated the expression of a series of genes implicated in fruit color and aroma metabolism (Fig. 5), including FaCHS (chalcone synthase), FaCHI (chalcone isomerase), FaF3H (flavanone 3-hydroxylase), FaUFGT (UDP-glycose flavonoid 3-O-glycosyltransferase), Fa4CL1 (4-coumaroyl-CoA ligase1), and Fa4CL2 (4-coumaroyl-CoA ligase2), in a converse manner. Notably, FaSnRK2.6-OE almost completely blocked the expression of FaQR (quinine oxidoreductase), a key gene implicated in aroma metabolism, whereas FaSnRK2.6-RNAi resulted in a more than 5-fold increase in the level of FaQR transcript (Fig. 5A). Also in a converse manner, FaSnRK2.6-OE and -RNAi greatly affected the expression of a series of genes implicated in fruit softening, such as FaPE (pectinesterase), FaCEL (cellulose), FaGAL2 (β-galactosidase2), FaXYL1 (β-xylosidase1), and FaEXP1 (expansin1). Collectively, these results strongly indicate that FaSnRK2.6 plays a critical role in the regulation of strawberry fruit development and ripening.
Figure 5.
Effect of FaSnRK2.6 OE and RNAi on the expression of ripening-related genes. FaSnRK2.6-OE and FaSnRK2.6-RNAi fruits were generated as described in “Materials and Methods.” The OE and RNAi constructs were injected into fruits at 18 DPA, and gene expression was examined at 30 and 25 DPA, respectively, using qRT-PCR, with FaACTIN as the internal control. OE-C and RNAi-C denote controls for the OE or RNAi fruit (i.e. fruit transformed with the corresponding empty vector). C4H, Cinnamic acid 4-hydroxylase; DFR, dihydroflavonolreductase; OMT, O-methyltransferase; PL, pectatelyase; PG, polygalacturonase. Values are means + sd of four biological replicates. Asterisks denote significant differences compared with the control sample (i.e. OE-C or RNAi-C) at *P < 0.05 and **P < 0.01, using Student’s t test.
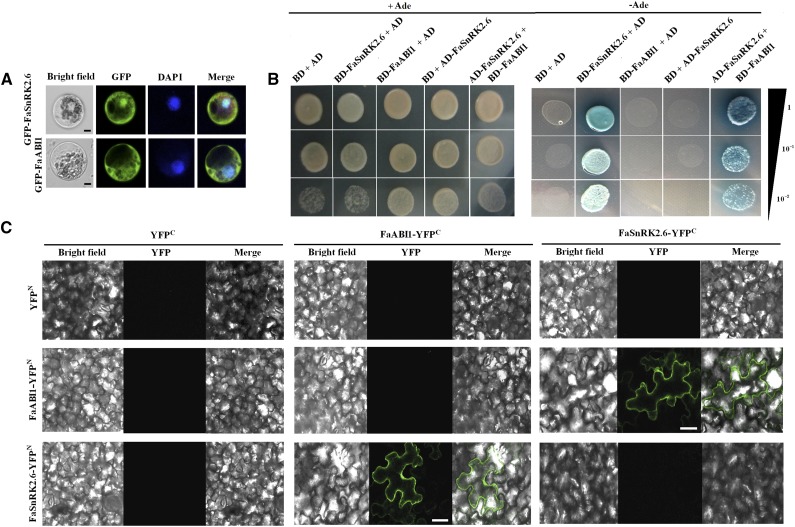
Subcellular Localization of FaSnRK2.6 and the Physical Interaction between FaSnRK2.6 and FaABI1
A previous study demonstrated that FaABI1, a key signal in the ABA signaling cascade, is a negative regulator of strawberry fruit development and ripening (Vlad et al., 2009; Jia et al., 2013a). We here examined the physical interaction between FaSnRK2.6 and FaABI1. FaSnRK2.6-GFP fluorescence indicated that FaSnRK2.6 was localized mainly to the nucleus and cytoplasm of maize (Zea mays) protoplasts (Fig. 6A). Yeast two-hybrid analysis indicated that FaSnRK2.6 undergoes autoactivation, because yeast (Saccharomyces cerevisiae AH109) cells continued to grow well on synthetic dextrose/−adenine,−Leu,−Trp,−His medium when harboring DNA binding domain (BD)-FaSnRK2.6 and activation domain (AD) empty plasmid but not when harboring AD-FaSnRK2.6 and BD empty plasmid. The clear positive result for the combination of BD-FaABI1 and AD-FaSnRK2.6 indicates that FaSnRK2.6 interacts physically with ABI1 (Fig. 6B). To provide further evidence for the interaction between FaSnRK2.6 and FaABI1, we performed a bimolecular fluorescence complementation (BiFC) assay. As shown in Figure 6C, fluorescence was clearly observed for both the FaABI1-YFPN and FaSnRK2.6-YFPC combination and the FaABI1-YFPC and FaSnRK2.6-YFPN combination. By contrast, no fluorescence was observed for the other combinations tested, strongly suggesting that FaSnRK2.6 interacts with FaABI1.
Figure 6.
Subcellular localization of FaSnRK2.6 and FaABI1 and physical interaction between FaSnRK2.6 and FaABI1. A, Subcellular localization of FaSnRK2.6 and FaABI1. pEZS-NL-Fasnrk2.6 and pEZS-NL-Faabi1 constructs were transformed into maize protoplasts, and fluorescence was observed by confocal microscopy as described in “Materials and Methods.” Bars = 8 µm. B, Yeast two-hybrid analysis of the physical interaction between FaSnRK2.6 and FaABI1. The protein interaction was examined using various combinations of prey and bait vectors. All tests were conducted on media containing adenine (+Ade; /−Leu/−Trp/+His/+Ade) or lacking adenine (−Ade; /−Leu/−Trp/−His/−Ade). Interactions were determined based on cell growth and were confirmed by an α-Gal assay on medium lacking adenine (/−Leu/−Trp/−His/−Ade). Dilutions (1, 10−1, and 10−2) of saturated cultures were spotted onto the plates. C, BiFC analysis of the physical interaction between FaSnRK2.6 and FaABI1. FaSnRK2.6 and FaABI1 were fused with either the C or N terminus of yellow fluorescent protein (YFP; designated as YFPc or YFPN, respectively). Different combinations of the fused constructs were cotransformed into tobacco (Nicotiana tabacum) cells, and the cells were then visualized using confocal microscopy as described in “Materials and Methods.” YFP and bright field were excited at 488 and 543 nm, respectively. Bars = 20 μm.
Temperature-Modulated Fruit Development and Ripening in Relation to FaSnRK2.6 Expression
Since temperature is a major environmental cue affecting strawberry development and ripening, we investigated a possible role for FaSnRK2.6 in temperature-modulated fruit ripening. To evaluate the effect of temperature on strawberry fruit development and ripening, plants were first grown under normal conditions until fruit set (i.e. day/night, 25°C/18°C) and then were divided into two groups and grown under a day/night regimen of either 32°C/20°C or 20°C/12°C. As shown in Figure 7, the two different temperature treatments resulted in great differences in the progression of strawberry fruit development and ripening, with fruit grown at the higher temperature ripening approximately 10 d before that grown at the lower temperature, indicating that strawberry fruit ripening is sensitive to temperature.
Figure 7.
Effect of temperature on strawberry fruit development and ripening. A, Photographs showing the progression of strawberry fruit development and ripening under two different temperatures. Strawberry plants were first grown under normal temperature conditions (i.e. 25°C/18°C, day/night) until fruit setting and then were subjected to one of two different temperature conditions (i.e. 32°C/20°C or 20°C/12°C, day/night), with other conditions left unchanged. Successive photographs show the developmental progression of a representative fruit, with the numbers indicating the duration (in days) of treatment. B, The number of fully ripened fruits was counted for a given number of plants subjected either to high-temperature (32°C/20°C) or low-temperature (20°C/12°C) treatments. The horizontal distance between the two lines represents the difference in the number of days to ripening under the two different temperature conditions.
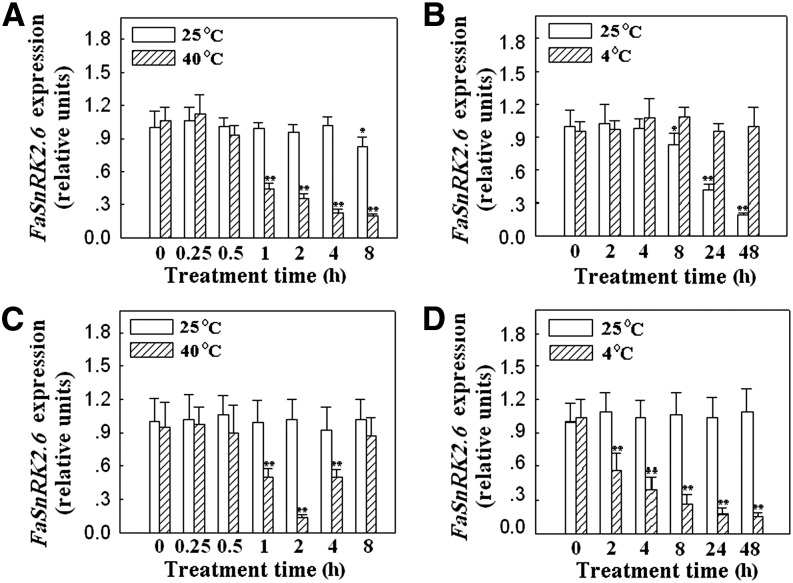
To further investigate whether FaSnRK2.6 might be associated with temperature-modulated fruit development and ripening, we examined the effect of temperature on FaSnRK2.6 expression. As shown in Figure 8A, transient exposure to high temperature greatly reduced the expression of FaSnRK2.6 (Fig. 8A), and by contrast, low temperature treatment completely blocked the fruit development-induced reduction in FaSnRK2.6 expression (Fig. 8B). These results suggest that the pattern of temperature-modulated FaSnRK2.6 expression was consistent with that of FaSnRK2.6 expression during the progression of strawberry fruit ripening and development. Together with the finding that strawberry fruit development and ripening could be manipulated by modulating the expression of FaSnRK2.6 (Figs. 4 and 5; Table I), these results suggest that FaSnRK2.6 mediates temperature-modulated fruit development and ripening.
Figure 8.
FaSnRK2.6 expression in response to temperature treatments in fruits and leaves. A and B, Fruits were detached at the large green fruit substage (corresponding to 18 DPA) and then treated with high (40°C) or low (4°C) temperature for the indicated periods of time. Samples were then subjected to qRT-PCR analysis to evaluate FaSnRK2.6 expression. For high-temperature treatment (A), the fruit samples were incubated at 40°C for 0 to 8 h, and for low-temperature treatment (B), the fruit samples were treated at 4°C for 0 to 48 h. C, High-temperature treatment in leaves. D, Low-temperature treatment in leaves. For C and D, young, fully expanded leaves were used, and the method for temperature treatment was the same as described in A and B. qRT-PCR was conducted using FaACTIN as an internal control. Values are means + sd of three biological replicates. Asterisks denote significant differences at *P < 0.05 and **P < 0.01, using Student’s t test, as compared with the control sample (i.e. 0 h, 25°C).
Since FaSnRK2.6 was largely expressed in leaves (Fig. 2A), and importantly, since its ortholog OST1 controls stomatal movement in leaves (Mustilli et al., 2002), we also examined FaSnRK2.6 expression in response to temperature stimuli in leaves (Fig. 8, C and D). Interestingly, in response to high-temperature treatment, FaSnRK2.6 expression initially declined but rapidly recovered to its original level (Fig. 8C). Moreover, in response to low-temperature treatment, FaSnRK2.6 expression decreased rather than increased in leaves, suggesting that the mechanism underlying the regulation of FaSnRK2.6 expression is distinctly different in fruits and leaves.
Roles of FaSnRK2.6 in Temperature-Modulated Gene Expression
To provide further evidence that FaSnRK2.6 is involved in temperature-modulated fruit development and ripening, we examined whether manipulating FaSnRK2.6 expression by OE or RNAi would alter the expression of temperature-responsive genes in fruits. To do so, we first transfected fruits at the large green fruit stage with FaSnRK2.6-OE, FaSnRK2.6-RNAi, or empty vector constructs and incubated the fruits for 5 d at room temperature (25°C). We then subjected these fruits to a transient treatment of high (40°C, 8 h) or low (4°C, 24 h) temperature and tested for an expressional induction of typical temperature-responsive genes, such as those implicated in color metabolism (e.g. FaCHS, FaCHI, and FaUFGT), cell wall metabolism (e.g. FaPE and FaXYL1), and aroma metabolism (e.g. FaQR).
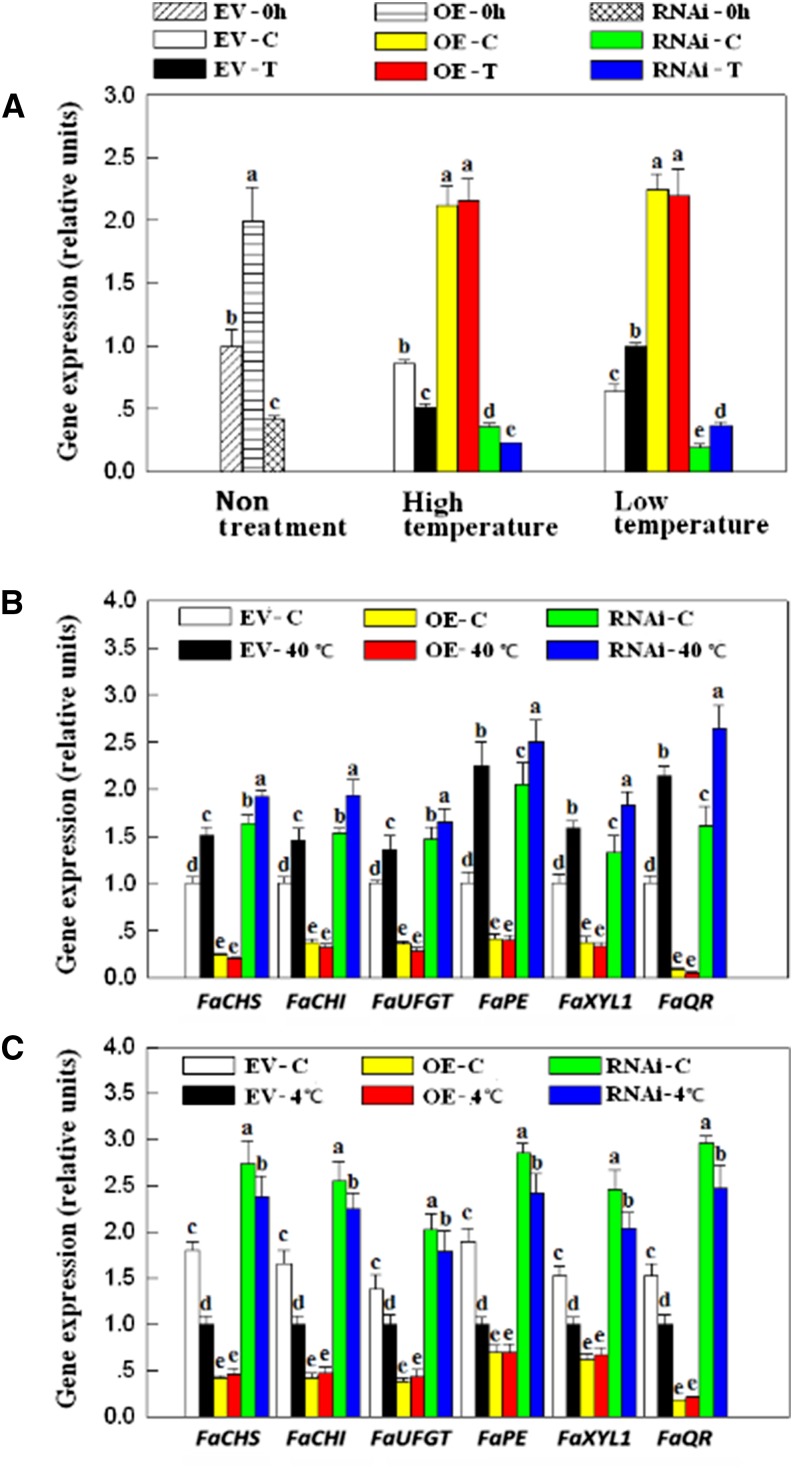
As shown in Figure 9A, transient transfection with FaSnRK2.6-OE and FaSnRK2.6-RNAi resulted in a marked increase and decrease, respectively, in the level of FaSnRK2.6 transcript. High- or low-temperature treatment resulted in a significant decrease or increase, respectively, in the level of FaSnRK2.6 transcript in the fruits transfected with the empty vector, whereas it had no significant effect on fruits transfected with FaSnRK2.6-OE. High or low temperature also resulted in a decrease or increase, respectively, in the level of FaSnRK2.6 transcript in FaSnRK2.6-RNAi fruits, but to a lesser extent than in fruits transfected with the empty vector.
Figure 9.
Effect of FaSnRK2.6 OE and RNAi on the expression of temperature-responsive genes. The FaSnRK2.6-OE and FaSnRK2.6-RNAi constructs were generated and transformed into fruits at 18 DPA, as described in “Materials and Methods.” Five days after transformation, the fruits were detached and used to analyze temperature-modulated gene expression. A, Effect of FaSnRK2.6-OE and FaSnRK2.6-RNAi on the level of FaSnRK2.6 transcript in fruits. High-temperature treatment was 40°C for 8 h, and low-temperature treatment was 4°C for 24 h. Non treatment indicates that gene expression was analyzed immediately after fruits were detached, and EV indicates empty vector; EV-0h, OE-0h, and RNAi-0h all indicate nontreatment. C, Control temperature (i.e. 25°C); T, high- or low-temperature treatment. B, Effects of FaSnRK2.6-OE and FaSnRK2.6-RNAi on FaSnRK2.6 expression in response to high-temperature treatment. C, Control temperature (i.e. 25°C); 40°C, high-temperature treatment for 8 h. C, Effects of FaSnRK2.6-OE and FaSnRK2.6-RNAi on FaSnRK2.6 expression in response to low-temperature treatment. C, Control temperature (i.e. 25°C); 4°C, low-temperature treatment for 24 h. qRT-PCR was conducted using FaACTIN as an internal control. Values are means + sd of four biological replicates. Different lowercase letters indicate significant differences according to Fisher’s lsd (P < 0.05).
As shown in Figure 9, B and C, while the overexpression or silencing of FaSnRK2.6 resulted in a large increase or decrease, respectively, in the level of FaSnRK2.6 transcript (Fig. 9A), they resulted in a large decrease or increase, respectively, in the transcript levels of the ripening-related genes relative to the empty vector control, regardless of the temperature treatment applied (Fig. 9, B and C). High or low temperature induced or suppressed, respectively, the expression of ripening-related genes in fruits transfected with empty vector (Fig. 9, B and C), but the induction or suppression of these genes was completely blocked in fruits transfected with FaSnRK2.6-OE. These findings suggest that the expression of the temperature-responsive genes can be modulated by manipulating the expression of FaSnRK2.6. Collectively, these results strongly suggest that (1) FaSnRK2.6 plays a critical role in temperature-modulated gene expression, and (2) the FaSnRK2.6-mediated regulation of the ripening-related genes in response to a temperature stimulus is largely achieved by regulating FaSnRK2.6 at the transcriptional level.
Methylation Analysis of the FaSnRK2.6 Promoter
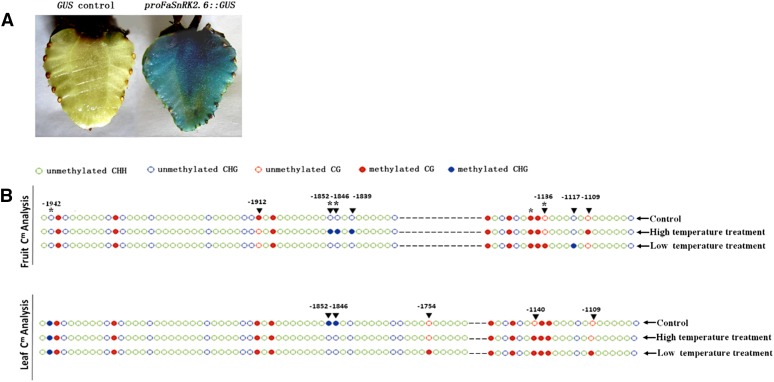
DNA methylation is a key mechanism by which the expression of a specific gene is regulated in different cells/tissues (Finnegan et al., 1998). Given that strawberry fruit development and ripening are largely controlled by the transcriptional regulation of FaSnRK2.6 and that temperature had a different effect on FaSnRK2.6 expression in fruits and leaves, we analyzed the DNA methylation status of the FaSnRK2.6 promoter in relation to its expression in fruits and leaves. The fragment 2,288 bp upstream of the FaSnRK2.6 transcriptional start site was isolated and demonstrated to have promoter activity in a GUS-mediated gene expression analysis (Fig. 10A). As expected, many sites within this fragment were differently methylated in DNA isolated from fruits and leaves. Under the control temperature (25°C), no methylation occurred at positions −1,942, −1,852, −1,846, and −1,136 bp relative to the transcriptional start site of FaSnRK2.6 in DNA isolated from fruits, whereas all of these sites were methylated in DNA from leaves. Interestingly, while high-temperature treatment resulted in methylation at positions −1,852, −1,846, −1,839, and −1,109 bp of FaSnRK2.6 relative to the transcriptional start site and demethylation at position −1,912 bp in DNA isolated from fruits, it resulted in demethylation at positions −1,852 and −1,846 bp and methylation at position −1,140 bp in leaves. Also, while low-temperature treatment resulted in methylation at positions −1,136 and −1,117 bp and demethylation at position −1,912 bp of FaSnRK2.6 in DNA isolated from fruits, it resulted in methylation at positions −1,754, −1,140, and −1,109 bp and demethylation at positions −1,852 and −1,846 bp in DNA isolated from leaves (Fig. 10B), suggesting that both high and low temperature have different effects on the methylation and demethylation of DNA isolated from fruit and leaves. Bioinformation analysis indicated that most of the sites involved in the regulation of DNA methylation were within the cis-elements of the FaSnRK2.6 promoter and that these sites had a variety of different functions (Supplemental Table S1). Collectively, these results suggest that the modification of promoter methylation is an important mechanism by which FaSnRK2.6 expression is uniquely regulated in fruits and leaves.
Figure 10.
Cytosine methylation analysis of proFaSnRK2.6 in fruit and leaf. A, Confirmation that proFaSnRK2.6 is active. proFaSnRK2.6 was inserted into the pBI121 vector to drive the GUS reporter gene. The construct was introduced into fruits (18 DPA), and proFaSnRK2.6 activity was estimated by GUS staining 6 d later. As a control, the pBI121 empty vector with the GUS promoter deleted was used. B, Cytosine methylation levels of proFaSnRK2.6 in fruits and leaves were estimated using Bisulfite Sequencing-PCR. A total of 500 ng of genomic DNA extracted from fruits (18 DPA) or leaves was used to perform bisulfite modification, and the product was used as a template to amplify proFaSnRK2.6 using eight primer pairs; 12 independent clones from each PCR were sequenced and analyzed using Kismeth (http://katahdin.mssm.edu/kismeth/revpage.pl). The control temperature was 25°C, high-temperature treatment was 40°C for 8 h, and low-temperature treatment was 4°C for 24 h. Asterisks indicate sites of difference between fruit and leaf, and black triangles indicate sites of difference caused by temperature treatment.
DISCUSSION
Previous studies demonstrated that ABA as well as PYR1 and FaABI1 were involved in the regulation of strawberry fruit development and ripening (Chai et al., 2011; Jia et al., 2011, 2013a). It is not surprising that PYR1 and ABI1 are implicated in strawberry fruit development and ripening, because molecular interaction between PYR and ABI1 is required to initiate ABA signaling. In this study, we identified a novel signal component, FaSnRK2.6, that functions downstream of PYR and ABI1 and serves as a negative regulator of fruit development and ripening. Interestingly, FaSnRK2.6 was the only ortholog of OST1 identified in the strawberry genome (Fig. 1), implying that FaSnRK2.6 may regulate both stomatal movement and fruit ripening. Given that FaSnRK2.6 is a negative regulator of strawberry fruit development and ripening, whereas OST1 is a positive regulator of stomatal movement, FaSnRK2.6 must operate via distinct mechanisms in these biological processes. Although SnRK2 protein kinases have been suggested to be downstream signal transducers of ABA (Kobayashi et al., 2005; Wang et al., 2013b) and although we found that SnRK2.6 interacts physically with FaABI1 (Fig. 6), the FaSnRK2.6-mediated regulation of strawberry fruit development and ripening likely does not occur via the posttranscriptional regulation of FaSnRK2.6 by the ABA/FaPYR1/FaABI1 signaling cascade, because this signaling cascade would be expected to activate rather than deactive FaSnRK2.6, based on our understanding of ABA signaling (Soon et al., 2012; Wang et al., 2013b). The most likely mechanism by which FaSnRK2.6 mediates ABA-regulated strawberry fruit ripening is the regulation of FaSnRK2.6 at the transcriptional level. Consistent with this notion, we found that FaSnRK2.6 expression could be sensitively inhibited by ABA in a concentration-dependent manner (Fig. 3A). These results, in combination with the previous observations that ABA content is substantially increased during strawberry fruit development and ripening (Jia et al., 2011), strongly suggest that FaSnRK2.6 mediates ABA-regulated fruit ripening mainly through the transcriptional regulation of FaSnRK2.6.
Fruit ripening is not only regulated by internal signals but also is affected by a variety of environmental factors, such as light, water availability, and temperature (Ulrich, 1958; Giovannoni, 2001; Llop-Tous et al., 2002; Yang et al., 2012; Seymour et al., 2013). In this study, we found that strawberry fruit development and ripening are indeed very sensitive to changes in temperature (Fig. 7). Consistent with the observation that high temperature dramatically accelerated the onset of fruit ripening (Fig. 7), we found that the expression of the ripening-related genes could be sensitively regulated by temperature (i.e. the expression of a series of ripening-related genes was rapidly up-regulated upon exposure to a high-temperature stimulus; Fig. 9B). By contrast, the expression of FaSnRK2.6 rapidly decreased upon exposure to a high-temperature stimulus (Fig. 8A). These results suggest that the temperature-modulated expression of ripening-related genes is correlated with the regulation of FaSnRK2.6 expression. Importantly, we found that overexpression of FaSnRK2.6 completely blocked high-temperature-induced gene expression, which suggests that the temperature-modulated expression of ripening-related genes is mediated by FaSnRK2.6. Furthermore, the transcript levels of ripening-related genes were always negatively correlated with the transcript level of FaSnRK2.6, regardless of the temperature treatment applied (Fig. 9, B and C), implying that the temperature-modulated expression of the ripening-related genes also largely occurs through the transcriptional regulation of FaSnRK2.6.
The SnRK2 family has been suggested to have evolved specifically for hyperosmotic stress signaling (Boudsocq et al., 2004; Umezawa et al., 2004; Kobayashi et al., 2005; Fujii et al., 2007; Shukla and Mattoo, 2008; Fujita et al., 2009). For example, Kobayashi et al. (2005) identified 10 SnRK2 protein kinases encoded in rice (Oryza sativa), and all of these SnRK2s were specifically activated by osmotic stress at both the transcriptional and the posttranscriptional levels. The involvement of the stress signal, FaSnRK2.6, in the regulation of fruit development and ripening may reasonably be explained from a plant ecological perspective. Seeds are known to be the most stress-tolerant organ, and hence, facilitating seed dispersal can be regarded as a strategy for plants to cope with stresses. From an evolutionary perspective, fleshy fruits arose to facilitate seed dispersal, thereby enabling plants to survive and thrive in ever-changing environments (Nitsch, 1953; Coombe, 1976; Brady, 1987). Accordingly, the involvement of FaSnRK2.6 in the regulation of fruit development and ripening may just be a consequence of its essential role in plant stress signaling. While mechanisms of plant stress signaling have been studied extensively, much less is known about the signaling mechanisms underlying fruit development and ripening. Knowledge of plant stress signaling may provide important clues for better understanding the molecular mechanisms underlying strawberry fruit development and ripening.
Ripening of fleshy fruits generally has been thought of as a distinct developmental stage, during which dramatic changes in a series of physiological and biochemical metabolic processes occur, including modification of cell wall ultrastructure and texture, increases/decreases in the levels of sugars/organic acids, and the biosynthesis and accumulation of aromatic volatiles (Seymour et al., 1993, 2013; Giovannoni, 2001). A key question regarding the ripening process is how an assortment of otherwise unrelated pathways and processes act coordinately during this stage of fruit development (Giovannoni, 2001). Basic cell biology studies have established that in both eukaryotic and prokaryotic organisms, coordinated cellular metabolism is essentially based on the regulation of cellular signaling (Bourret et al., 1991; Freeman and Gurdon, 2002; Bekaert et al., 2005; Anantharaman et al., 2007). Given the complexity of the metabolic processes occurring during fruit ripening, it is not difficult to imagine that fruit ripening must be tightly controlled through a complicated signaling network. In this study, we have found that suppression of FaSnRK2.6 accelerated the biosynthesis of anthocyanins and reduced fruit firmness, whereas it appeared to have no significant effect on the sugar and total titratable acid contents. It is likely that for each metabolic pathway there exists a distinct signaling cascade, such that a single signaling cascade or component is not enough to govern all of the ripening-related metabolic processes. Alternatively, FaSnRK2.6 alone may not be capable of controlling all of the ripening-related metabolic pathways, which implies that the different pathways of the ripening-related metabolic processes are temporospatially separated.
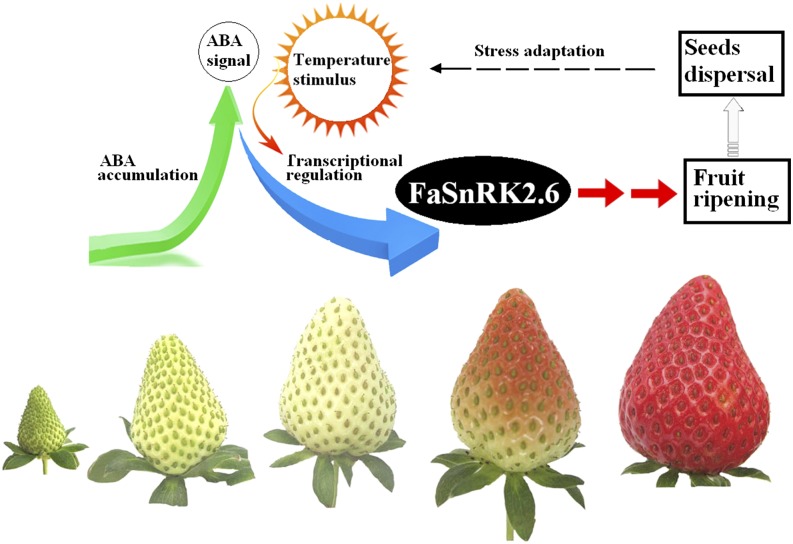
In summary (Fig. 11), we have identified a novel protein signal, FaSnRK2.6, that regulates strawberry fruit development and ripening. FaSnRK2.6 is a downstream signal of ABA, as indicated by its physical interaction with FaABI1. FaSnRK2.6 expression is not only negatively correlated with strawberry fruit development and ripening but also is negatively regulated by ABA and temperature, internal and external cues, respectively, that are known to modulate strawberry fruit development and ripening. Manipulating FaSnRK2.6 expression was capable of modulating strawberry fruit ripening as well as the expression of a variety of ripening-related genes, and moreover, it was also capable of modulating the expression of a variety of temperature-responsive genes. Collectively, these results demonstrate that FaSnRK2.6 is an important regulator of strawberry fruit development and ripening. The FaSnRK2.6-mediated signaling pathway implicated here in temperature-modulated fruit ripening embodies a wonderful strategy by which FaSnRK2.6 functions as a stress signal, from a plant ecological perspective. This study greatly enhances our understanding of the regulatory mechanism underlying the ripening of nonclimacteric fruits.
Figure 11.
Diagram of FaSnRK2.6 signaling in relation to the regulation of strawberry fruit development and ripening. Levels of FaSnRK2.6 transcript decrease during the progression of strawberry fruit development and ripening are shown. OE- and RNAi-mediated suppression of FaSnRK2.6 expression delay and accelerate, respectively, the onset of strawberry fruit ripening. ABA and high temperature down-regulate the expression of FaSnRK2.6, thereby promoting strawberry fruit ripening. As seed maturation and dispersal can be regarded as a strategy for plants to withstand environmental stresses, the regulation of strawberry fruit ripening by FaSnRK2.6 embodies a role of FaSnRK2.6 in stress signaling.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Strawberry plants (Fragaria × ananassa ‘Benihoppe’) were planted in pots (diameter, 230 mm; and depth, 230 mm) containing a mixture of nutrient soil, vermiculite, and organic fertilizer (7:2:1, v/v/v). The seedlings were grown in a controlled environment with the following conditions: 25°C/18°C (day/night), 60% humidity, and a 12-h photoperiod with a photosynthetic photon flux density of 450 µmol m−2 s−1.
Gene Isolation and Analysis
Total RNA was extracted from fruit receptacles using the SV Total RNA Isolation System (Promega) according to the manufacturer’s instructions. To identify SnRK2s from strawberry, the coding sequences of all members of the AtSnRK2 subfamily in Arabidopsis (Arabidopsis thaliana) were BLASTed in the Web site https://strawberry.plantandfood.co.nz/. Nine Sucrose Nonfermenting1-related protein kinases, which were designated as FaSnRK2.1 to FaSnRK2.9, were identified. The FaSnRK2s were cloned from cDNA by PCR under the following conditions: 98°C for 30 s, followed by 35 cycles of 98°C for 10 s, 54°C for 20 s, and 72°C for 2 min, with a final extension of 72°C for an additional 5 min (Q5 High-Fidelity DNA Polymerase; New England Biolabs). The PCR products were ligated into a pMD19-T simple vector and subsequently transformed into Escherichia coli DH5α. Positive colonies were selected and amplified and then sequenced by Invitrogen. The nine FaSnRK2 sequences were submitted to the National Center for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov/genbank/). Primer sequences and GenBank accession numbers are shown in Supplemental Table S2.
The deduced amino acid sequences of FaSnRK2.1 to FaSnRK2.9 were aligned using ClustalX 2.0.12 with default settings. The alignment results were edited and marked using GeneDoc.
Phylogenetic Analysis
The full-length SnRK2 cDNA sequences from Arabidopsis, maize (Zea mays), Triticum polonicum, potato (Solanum tuberosum), wheat (Triticum aestivum), and Nicotiana plumbaginifolia were obtained from the NCBI nucleotide database (http://www.ncbi.nlm.nih.gov/nucleotide/) as reported in previous studies (Park et al., 1993; Mustilli et al., 2002; Hrabak et al., 2003; Mao et al., 2010; Bucholc et al., 2011).
The DNA sequences of FaSnRKs were aligned with the homologous genes in Arabidopsis, maize, T. polonicum, potato, wheat, and N. plumbaginifolia using ClustalX 2.0.12 software (Thompson et al., 1994) with default settings. The phylogenetic trees were constructed using the neighbor-joining method in MEGA6 software with 1,000 bootstrap replicates for evaluating the reliability of different phylogenetic groups. Tree files were viewed and edited using MEGA6 software.
Temporospatial Expression of FaSnRK2.6
Fruit ripening stages were divided according to DPA and fruit development as follows: small green fruit (6 DPA), middle green fruit (12 DPA), large green fruit (18 DPA), white fruit (24 DPA), initially reddening (27 DPA), and fully reddening (30 DPA).
Ten fruits were harvested at each stage, and the root, stem, leaf, and flower used for organ-specific expression analysis were sampled from 10 plants grown under the same conditions. All fruits (without seeds) and samples were immediately frozen in liquid nitrogen, mixed, and stored at –80°C until further use. Quantitative PCR primers to detect FaSnRK2.1 to FaSnRK2.9 were designed using Primer3Plus (http://www.primer3plus.com/cgi-bin/dev/primer3plus.cgi). Primer sequences are shown in Supplemental Table S3.
qRT-PCR Analysis
Total RNA was extracted from fruits using the SV Total RNA Isolation System (Promega) according to the manufacturer’s instructions. To eliminate genomic DNA contamination, RNA was treated with DNase I (Takara) for 20 min. First-strand cDNA was synthesized from total RNA with the Takara RNA PCR Kit. qRT-PCR was performed in the ABI 7500 Real-Time PCR System (Applied Biosystems) following the manufacturer’s instructions and using SYBR Premix ExTaq (Perfect Real Time; Takara Bio). qRT-PCR was conducted with three biological replicates, and each sample was analyzed at least in triplicate and normalized using FaACTIN as an internal control. Transcription levels are presented in the form 2–ΔΔCT, where ΔCT represents the difference between the cycle threshold values of the target and the reference genes (Schmittgen et al., 2008). Primers used for the qRT-PCR analysis of ripening-related genes are provided in Supplemental Table S4.
ABA and Suc Treatment
Strawberry fruits and leaves were incubated in vitro with ABA as described (Berüter et al., 1995; Jiang and Zhang, 2001). After washing with distilled water, the freshly harvested fruits or leaves were dried carefully. Discs (10 mm in diameter and 1 mm in thickness) of fruits (with seeds removed) or leaves were prepared. The disc samples were first vacuum infiltrated for 30 min in equilibration buffer (Archbold, 1999) consisting of 50 mm MES-Tris (pH 5.5), 10 mm MgCl2, 10 mm EDTA, 5 mm CaCl2, 200 mm mannitol, and 5 mm vitamin C and then shaken for 1 or 6 h at 25°C in equilibration buffer containing various concentrations of ABA (Sigma-Aldrich; A1049) or Suc. After incubation, the samples were washed with double distilled water, frozen immediately in liquid nitrogen, and kept at −80°C until further use. These experiments were repeated three times.
Construction of FaSnRK2.6 OE and RNAi Plasmids
To generate the FaSnRK2.6 OE construct, full-length FaSnRK2.6 cDNA was obtained by reverse transcription-PCR using the following primers: forward, 5′-GTCGACATGGATCGGTCTATGCT-3′ (with a SalI restriction site); reverse, 5′-AAGCTTTCAAATTGCATACACTATTTCC-3′ (with a HindIII restriction site). The cDNA was cloned into the SalI and HindIII restriction sites of the plant expression vector pCambia1304. To generate an intron containing a hairpin RNAi fragment of FaSnRK2.6, a 720-bp fragment near the 5′ end of FaSnRK2.6 cDNA was PCR amplified using the primer pair 5′-GCATGCATGGATCGGTCTATGCTG-3′ (with a SphI restriction site; forward) and 5′-GTCGACGTACTGGACACTCGTTATACGATGT-3′ (with a SalI restriction site; reverse). The 720-bp fragment obtained was sequenced and forward cloned into the SphI and SalI restriction sites of pCambia1304. A 281-bp fragment complementary to the 3′ end of the 720-bp fragment described above was PCR amplified from strawberry fruit cDNA using the following primers: 5′-GCATGCACACATTGTTAGATGGAAGTC-3′ (with a SphI restriction site; forward) and 5′-AAGCTTGTACTGGACACTCGTTATACGATG-3′ (with a HindIII restriction site; reverse). The 281-bp fragment obtained was sequenced and forward cloned into the SphI and HindIII restriction sites of pCambia1304; by this means, a hairpin would be produced when this 281-bp fragment was exposed to its complementary sequence at the 3′ end of the 720-bp fragment.
Transfection of Strawberry by Agroinfiltration
Agrobacterium tumefaciens strain AGL0 (Lazo et al., 1991), containing pCambia1304, OE-pCambia1304-FaSnRK2.6, or RNAi-pCambia1304-FaSnRK2.6, was grown at 28°C in Luria-Bertani liquid medium containing 10 mm MES and 20 µm acetosyringone with appropriate antibiotics. When the culture reached an optical density at 600 nm of approximately 0.1, A. tumefaciens cells were harvested and resuspended in infection buffer (10 mm MgCl2, 10 mm MES, pH 5.6, and 200 µm acetosyringone) and shaken for 2 h at room temperature before being used for infiltration. For transfection, attached fruits with basically identical sizes and shapes at 18 DPA were selected, and the A. tumefaciens suspension was evenly injected into the fruits with a syringe until the whole fruit became hygrophanous (Jia et al., 2013b). The transfections were carried out in different batches according to different experimental aims, and in each batch, 40 fruits were injected, with 20 fruits being transformed with FaSnRK2.6-RNAi or FaSnRK2.6-OE and the other 20 used as the control (i.e. injected with the pCambia1304 empty vector). To examine the effect of FaSnRK2.6-RNAi and FaSnRK2.6-OE on the expression of ripening-related genes, the fruits were collected 7 and 12 d after transfection, respectively, for FaSnRK2.6-RNAi and FaSnRK2.6-OE. For the expressional analysis of ripening-related genes, five fruits were combined as an individual sample. Seeds of the fruit samples were removed, and the receptacles were frozen in liquid nitrogen and stored at –80°C until use. To examine the effect of FaSnRK2.6-RNAi and FaSnRK2.6-OE on temperature-modulated gene expression, the fruits were collected 5 d after transfection and then treated with different temperature regimens as described below.
Temperature Treatment Experiment
To observe the effect of temperature on the progression of fruit development and ripening, strawberry seedlings were first grown in pots until fruit set under normal conditions as described in “Plant Materials and Growth Conditions.” On the day before temperature treatment started, flower and fruit thinning was performed so that for each plant, only five fruits were retained. For temperature treatment, the plants were divided into two groups. One group was grown in a controlled environment under 32°C/20°C (day/night), 60% humidity, and a 12-h photoperiod with a photosynthetic photon flux density of 450 µmol m−2 s−1, and the other group was grown under 20°C/12°C (day/night), 60% humidity, and a 12-h photoperiod with a photosynthetic photon flux density of 450 µmol m−2 s−1. The numbers of mature fruits as judged by changes in fruit color were successively recorded until all fruits matured. To examine the effect of temperature on the expression of ripening-related genes, for each treatment, 15 detached fruits of 18 DPA were used, with five fruits combined into one sample, such that the 15 fruits were divided into three samples. The samples were individually used for the analysis of gene expression. The fruits were separated longitudinally into two even halves, and one half was treated with high or low temperature and the other at 25°C as the control. The high-temperature treatment was performed at 40°C for 0, 0.25, 0.5, 1, 2, 4, or 8 h, and the low-temperature treatment was performed at 4°C for 0, 2, 4, 8, 24, or 48 h. After the temperature treatment, the seeds were removed and the fruits were frozen in liquid nitrogen and stored at –80°C until further use. For temperature treatment of leaves, whole plants grown in pots were treated with different temperatures (as described above for fruits), and the young fully expanded leaves were sampled for analysis of gene expression.
To examine the effect of FaSnRK2.6-RNAi and FaSnRK2.6-OE on temperature-modulated gene expression, the fruits were first transfected with the FaSnRK2.6-RNAi or FaSnRK2.6-OE construct or the empty vector, and 5 d after the transfection, the fruits were detached and treated with different temperatures (i.e. 40°C for 8 h or 4°C for 24 h), as described above.
Determination of Fruit Firmness, Pigments, Soluble Sugar Content, Titratable Acidity, and Aroma Components
Pigment, anthocyanin, total phenol, and flavonoid contents of fruits were determined as described (Fuleki and Francis, 1968; Lees and Francis,1971). The soluble sugar content was determined as described (Jia et al., 2011). The total titratable acidity was determined using the acid-base titration method (Kafkas et al., 2007). Titratable acidity was calculated as malic acid. The firmness of fruits transfected by agroinfiltration was determined before freezing. Flesh firmness was measured after the removal of fruit skin on opposite sides of each fruit using a GY-4 fruit hardness tester (Zhejiang TOP Instrument). Aroma production by fruits was characterized by head space solid-phase microextraction and gas chromatography-mass spectrometry as described (Dong et al., 2013). The relative content of each aroma component was calculated using the peak areas and expressed as a percentage of the total volatiles.
Subcellular Localization
For subcellular localization of FaSnRK2.6 and FaABI1, the FaSnRK2.6 cDNA sequence was amplified using the forward primer 5′-CTCGAGATGGATCGGTCTATGCTG-3′ and the reverse primer 5′-GAATTCTAATTGCATACACTATTTCCCCG-3′. The PCR product was then cloned into XhoI/EcoRI-cleaved pEZS-NL transient expression vector (from David Ehrhardt, Carnegie Institution). The FaABI1 cDNA sequence was amplified using the forward primer 5′-CTCGAGATGGAGGAGATGTCACC-3′ and the reverse primer 5′-GGATCCTCTGTTTTACTTTTAAACTTC-3′, and the PCR product was then cloned into the XhoI/BamHI-cleaved pEZS-NL transient expression vector. Maize protoplast isolation and transformation were carried out according to a protocol described by Sheen et al. (1995), with minor modifications (Li et al., 2012). After pEZS-NL-Fasnrk2.6 or pEZS-NL-Faabi1 was transformed into maize protoplasts, the protoplasts were incubated for 16 h and then stained with 0.1 mg mL−1 4′,6-diamino-phenylindole (DAPI; Sigma-Aldrich) in phosphate-buffered saline for 10 min. FaSnRK2.6 and FaABI1 localization patterns were assessed by visualizing GFP fluorescence using a Nikon D-ECLIPSE C1 spectral confocal laser-scanning system, and the nuclei were visualized by DAPI fluorescence. GFP and DAPI were excited at 488 and 408 nm, respectively. Fluorescence analysis was performed using a Nikon ECLIPSE TE2000-E inverted fluorescence microscope.
Yeast Two-Hybrid Assays
Yeast two-hybrid assays were performed using the Matchmaker GAL4-based Two-Hybrid System 3 (Clontech), according to the manufacturer’s instructions. The full-length cDNA sequences of FaABI1 and FaSnRK2.6 were subcloned into the pGADT7 and pGBKT7 vectors, respectively. All constructs were transformed into yeast (Saccharomyces cerevisiae) strain AH109 by the lithium acetate method, and yeast cells were grown on a minimal medium/−Leu−Trp according to the manufacturer’s instructions (Clontech). Transformed colonies were plated onto a minimal medium/−Leu/−Trp/−His/−adenine containing 20 µg mL−1 5-bromo-4-chloro-3-indolyl-α-d-galactopyranoside to test for possible interactions. The primers used for yeast two-hybrid assays are listed in Supplemental Table S5.
BiFC Assays
To generate the BiFC constructs, the full-length cDNA sequences of FaABI1 and FaSnRK2.6 were subcloned into the pCambia1300-YFPN and pCambia1300-YFPC vectors. Coexpression was conducted in tobacco (Nicotiana tabacum) leaves as described by Schütze et al. (2009). The chimeric fluorescence of the expressed fusion proteins was detected 2 to 4 d after infiltration. Fluorescence images were acquired using a Nikon D-ECLIPSE C1 spectral confocal laser-scanning system. YFP and bright field were excited at 488 and 543 nm, respectively. Fluorescence analysis was performed using a Nikon ECLIPSE TE2000-E inverted fluorescence microscope. The primers used for BiFC assays are listed in Supplemental Table S6.
Isolation and Activity Analysis of FaSnRK2.6 Promoters
The promoter sequences of FaSnRK2.6 in fruit and leaf were isolated according to information provided at https://strawberry.plantandfood.co.nz/. The PCR primers were 5′-TTAAGCTAATCAGTTTGTTGG-3′ and 5′-CTCAGAAATGGTGGCTCCTATGAT-3′, and the products were ligated into the pMD19-T simple vector and subsequently transformed into E. coli DH5α. Positive colonies were selected and amplified and then sequenced by Invitrogen. The fruit and leaf shared the same FaSnRK2.6 promoters. The proFaSnRK2.6 sequence was submitted to the NCBI database (http://www.ncbi.nlm.nih.gov/genbank/) and assigned GenBank accession number KJ959615. Promoter analysis was performed using PLACE (http://www.dna.affrc.go.jp/PLACE/signalscan.html; Prestridge, 1991; Higo et al., 1999).
Transient expression assays were employed to test the activity of the promoter. The full-length FaSnRK2.6 promoter was obtained by reverse transcription-PCR using the following primers: forward, 5′-AAGCTTTTAAGCTAATCAGTTTGTTGG-3′ (with a HindIII restriction site); reverse, 5′-GGATCCCTCAGAAATGGTGGCTCCTATGAT-3′ (with a BamHI restriction site). The cDNA was cloned into the HindIII and BamHI restriction sites of the plant expression vector pBI121, generating the proFaSnRK2.6-GUS construct. The constructs were introduced into A. tumefaciens strain AGL0, and the A. tumefaciens was infiltrated into fruits at 18 DPA (Jia et al., 2013b). After 6 d, the fruits were treated with GUS solution (Gallagher, 1992).
Methylation Analysis of proFaSnRK2.6
BSP-PCR analysis was performed as described (Telias et al., 2011). Briefly, 500 ng of genomic DNA from fruit (18 DPA), leaf, fruit treated for 8 h at 40°C, leaf treated for 8 h at 40°C, fruit treated for 24 h at 4°C, and leaf treated for 24 h at 4°C was treated using the EZ DNA Methylation-Gold Kit (Zymo Research). Using the treated DNA as template, eight proFaSnRK2.6 fragments of each sample were amplified using Q5 High-Fidelity DNA polymerase (New England Biolabs), ligated into the PMD19-T simple vector (Takara), and then sequenced. Sequences of 12 independent clones of each fragment were analyzed using Kismeth (http://katahdin.mssm.edu/kismeth/revpage.pl), and the methylation level of each fragment was calculated. Three biological replicates were performed. Primer sequences are shown in Supplemental Table S7.
Genes and accession numbers are as follows: AtSnRK2.1, NM120946; AtSnRK2.2, AT3G50500.2; AtSnRK2.3, NM126087; AtSnRK2.4, AT1G10940.2; AtSnRK2.5, NM125760; AtSnRK2.6, NM119556; AtSnRK2.7, NM120165; AtSnRK2.8, AT1G78290.2; AtSnRK2.9, NM127867; AtSnRK2.10, NM104774; ZmSnRK2.1, EU676033; ZmSnRK2.2, EU676034; ZmSnRK2.3, EU676035; ZmSnRK2.4, EU676036; ZmSnRK2.5, EU676037; ZmSnRK2.6, EU676038; ZmSnRK2.7, EU676039; ZmSnRK2.8, EU676040; ZmSnRK2.10, EU676041; ZmSnRK2.11, EU676042; TpSnRK2.2, KF688097; TpSnRK2.5, KF688098; TpSnRK2.10, KF688099; TpSnRK2.11, KF688100; StSnRK2.1, JX280911; StSnRK2.2, JX280912; StSnRK2.3, JX280913; StSnRK2.4, JX280914; StSnRK2.5, JX280915; StSnRK2.6, JX280916; StSnRK2.7, JX280917; StSnRK2.8, JX280918; TaSnRK2.4, GQ384359; and NtSnRK2, FJ882981.
Supplemental Data
The following supplemental materials are available.
Supplemental Table S1. Bioinformation analysis of sites subjected to pro-FaSnRK2.6 DNA methylation.
Supplemental Table S2. Primers used to amplify FaSnRK2.1 to FaSnRK2.9 coding sequences and GenBank accession numbers.
Supplemental Table S3. Quantitative PCR primers used in the temporospatial analysis of FaSnRK2.1 to FaSnRK2.9.
Supplemental Table S4. Quantitative PCR primers used to analyze the expression of ripening-related marker genes in FaSnRK2.6 OE and RNAi fruits.
Supplemental Table S5. Primers used to amplify FaSnRK2.6 and FaABI1 in yeast two-hybrid assays.
Supplemental Table S6. Primers used for FaSnRK2.6 and FaABI1 in BiFC assays.
Supplemental Table S7. Primers used to amplify proFaSnRK2.6 in the BSP-PCR analysis.
Supplementary Material
Glossary
- ABA
abscisic acid
- cDNA
complementary DNA
- RNAi
RNA interference
- OE
overexpression
- BiFC
bimolecular fluorescence complementation
- NCBI
National Center for Biotechnology Information
- qRT
quantitative reverse transcription
- DAPI
4′,6-diamino-phenylindole
- YFP
yellow fluorescent protein
Footnotes
This work was supported by the National Natural Science Foundation (grant nos. 31101527, 31171921, and 31471851), the Research Fund for the Doctoral Program of Higher Education (grant no. 20110008120024), and the National High-Tech R&D Program of China (grant no. 2011AA100204).
Articles can be viewed without a subscription.
References
- Anantharaman V, Iyer LM, Aravind L (2007) Comparative genomics of protists: new insights into the evolution of eukaryotic signal transduction and gene regulation. Annu Rev Microbiol 61: 453–475 [DOI] [PubMed] [Google Scholar]
- Archbold DD. (1999) Carbohydrate availability modifies sorbitol dehydrogenase activity of apple fruit. Physiol Plant 105: 391–395 [Google Scholar]
- Assmann SM. (2003) OPEN STOMATA1 opens the door to ABA signaling in Arabidopsis guard cells. Trends Plant Sci 8: 151–153 [DOI] [PubMed] [Google Scholar]
- Bekaert M, Rousset JP (2005) An extended signal involved in eukaryotic-1 frameshifting operates through modification of the E site tRNA. Mol Cell 17: 61–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berüter J, Feusi MES (1995) Comparison of sorbitol transport in excised tissue discs and cortex tissue of intact apple fruit. J Plant Physiol 146: 95–102 [Google Scholar]
- Boudsocq M, Barbier-Brygoo H, Laurière C (2004) Identification of nine sucrose nonfermenting 1-related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana. J Biol Chem 279: 41758–41766 [DOI] [PubMed] [Google Scholar]
- Bourret RB, Borkovich KA, Simon MI (1991) Signal transduction pathways involving protein phosphorylation in prokaryotes. Annu Rev Biochem 60: 401–441 [DOI] [PubMed] [Google Scholar]
- Brady CJ. (1987) Fruit ripening. Annu Rev Plant Physiol 38: 155–178 [Google Scholar]
- Bucholc M, Ciesielski A, Goch G, Anielska-Mazur A, Kulik A, Krzywińska E, Dobrowolska G (2011) SNF1-related protein kinases 2 are negatively regulated by a plant-specific calcium sensor. J Biol Chem 286: 3429–3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai YM, Jia HF, Li CL, Dong QH, Shen YY (2011) FaPYR1 is involved in strawberry fruit ripening. J Exp Bot 62: 5079–5089 [DOI] [PubMed] [Google Scholar]
- Coombe BG. (1976) The development of fleshy fruits. Annu Rev Plant Physiol 27: 207–228 [Google Scholar]
- Dong J, Zhang YT, Tang XW, Jin WM, Han ZH (2013) Differences in volatile ester composition between Fragaria × ananassa and F. vesca and implications for strawberry aroma patterns. Sci Hortic (Amsterdam) 150: 47–53 [Google Scholar]
- El Hadi MA, Zhang FJ, Wu FF, Zhou CH, Tao J (2013) Advances in fruit aroma volatile research. Molecules 18: 8200–8229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan EJ, Genger RK, Peacock WJ, Dennis ES (1998) DNA methylation in plants. Annu Rev Plant Physiol Plant Mol Biol 49: 223–247 [DOI] [PubMed] [Google Scholar]
- Fischer RL, Bennett AB (1991) Role of cell wall hydrolases in fruit ripening. Annu Rev Plant Biol 42: 675–703 [Google Scholar]
- Freeman M, Gurdon JB (2002) Regulatory principles of developmental signaling. Annu Rev Cell Dev Biol 18: 515–539 [DOI] [PubMed] [Google Scholar]
- Fujii H, Verslues PE, Zhu JK (2007) Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell 19: 485–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Nakashima K, Yoshida T, Katagiri T, Kidokoro S, Kanamori N, Umezawa T, Fujita M, Maruyama K, Ishiyama K, et al. (2009) Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol 50: 2123–2132 [DOI] [PubMed] [Google Scholar]
- Fuleki T, Francis FJ (1968) Quantitative methods for anthocyanins. J Food Sci 33: 266–274 [Google Scholar]
- Gagné S, Cluzet S, Mérillon JM, Gény L (2011) ABA initiates anthocyanin production in grape cell cultures. J Plant Growth Regul 30: 1–10 [Google Scholar]
- Gallagher SR. (1992) GUS Protocols: Using the GUS Gene as a Reporter of Gene Expression. Academic Press, San Diego, CA [Google Scholar]
- Giovannoni J. (2001) Molecular biology of fruit maturation and ripening. Annu Rev Plant Physiol Plant Mol Biol 52: 725–749 [DOI] [PubMed] [Google Scholar]
- Giribaldi M, Gény L, Delrot S, Schubert A (2010) Proteomic analysis of the effects of ABA treatments on ripening Vitis vinifera berries. J Exp Bot 61: 2447–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama T, Shinozaki K (2010) Research on plant abiotic stress responses in the post-genome era: past, present and future. Plant J 61: 1041–1052 [DOI] [PubMed] [Google Scholar]
- Hrabak EM, Chan CW, Gribskov M, Harper JF, Choi JH, Halford N, Kudla J, Luan S, Nimmo HG, Sussman MR, et al. (2003) The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol 132: 666–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imes D, Mumm P, Böhm J, Al-Rasheid KA, Marten I, Geiger D, Hedrich R (2013) Open stomata 1 (OST1) kinase controls R-type anion channel QUAC1 in Arabidopsis guard cells. Plant J 74: 372–382 [DOI] [PubMed] [Google Scholar]
- Jia HF, Wang Y, Sun M, Li B, Han Y, Zhao Y, Li X, Ding N, Li C, Ji W, et al. (2013b) Sucrose functions as a signal involved in the regulation of strawberry fruit development and ripening. New Phytol 198: 453–465 [DOI] [PubMed] [Google Scholar]
- Jia HF, Chai YM, Li CL, Lu D, Luo JJ, Qin L, Shen YY (2011) Abscisic acid plays an important role in the regulation of strawberry fruit ripening. Plant Physiol 157: 188–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia HF, Lu D, Sun JH, Li CL, Xing Y, Qin L, Shen YY (2013a) Type 2C protein phosphatase ABI1 is a negative regulator of strawberry fruit ripening. J Exp Bot 64: 1677–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, Zhang J (2001) Effect of abscisic acid on active oxygen species, antioxidative defence system and oxidative damage in leaves of maize seedlings. Plant Cell Physiol 42: 1265–1273 [DOI] [PubMed] [Google Scholar]
- Kafkas E, Koşar M, Paydaş S, Kafkas S, Başer KHC (2007) Quality characteristics of strawberry genotypes at different maturation stages. Food Chem 100: 1229–1236 [DOI] [PubMed] [Google Scholar]
- Klee HJ, Giovannoni JJ (2011) Genetics and control of tomato fruit ripening and quality attributes. Annu Rev Genet 45: 41–59 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Murata M, Minami H, Yamamoto S, Kagaya Y, Hobo T, Yamamoto A, Hattori T (2005) Abscisic acid-activated SNRK2 protein kinases function in the gene-regulation pathway of ABA signal transduction by phosphorylating ABA response element-binding factors. Plant J 44: 939–949 [DOI] [PubMed] [Google Scholar]
- Koyama K, Sadamatsu K, Goto-Yamamoto N (2010) Abscisic acid stimulated ripening and gene expression in berry skins of the Cabernet Sauvignon grape. Funct Integr Genomics 10: 367–381 [DOI] [PubMed] [Google Scholar]
- Kulik A, Wawer I, Krzywińska E, Bucholc M, Dobrowolska G (2011) SnRK2 protein kinases: key regulators of plant response to abiotic stresses. Omics 15: 859–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazo GR, Stein PA, Ludwig RA (1991) A DNA transformation-competent Arabidopsis genomic library in Agrobacterium. Biotechnology (N Y) 9: 963–967 [DOI] [PubMed] [Google Scholar]
- Lees DH, Francis FJ (1971) Quantitative methods for anthocyanins. J Food Sci 36: 1056–1060 [Google Scholar]
- Li B, Zhao Y, Liang L, Ren H, Xing Y, Chen L, Sun M, Wang Y, Han Y, Jia H, et al. (2012) Purification and characterization of ZmRIP1, a novel reductant-inhibited protein tyrosine phosphatase from maize. Plant Physiol 159: 671–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llop-Tous I, Domínguez-Puigjaner E, Vendrell M (2002) Characterization of a strawberry cDNA clone homologous to calcium-dependent protein kinases that is expressed during fruit ripening and affected by low temperature. J Exp Bot 53: 2283–2285 [DOI] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E (2009) Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324: 1064–1068 [DOI] [PubMed] [Google Scholar]
- Mao X, Zhang H, Tian S, Chang X, Jing R (2010) TaSnRK2.4, an SNF1-type serine/threonine protein kinase of wheat (Triticum aestivum L.), confers enhanced multistress tolerance in Arabidopsis. J Exp Bot 61: 683–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustilli AC, Merlot S, Vavasseur A, Fenzi F, Giraudat J (2002) Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 14: 3089–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitsch JP. (1953) The physiology of fruit growth. Annu Rev Plant Physiol 4: 199–236 [Google Scholar]
- Owen SJ, Lafond MD, Bowen P, Bogdanoff C, Usher K, Abrams SR (2009) Profiles of abscisic acid and its catabolites in developing Merlot grape (Vitis vinifera) berries. Am J Enol Vitic 60: 277–284 [Google Scholar]
- Park YS, Hong SW, Oh SA, Kwak JM, Lee HH, Nam HG (1993) Two putative protein kinases from Arabidopsis thaliana contain highly acidic domains. Plant Mol Biol 22: 615–624 [DOI] [PubMed] [Google Scholar]
- Prestridge DS. (1991) SIGNAL SCAN: a computer program that scans DNA sequences for eukaryotic transcriptional elements. Comput Appl Biosci 7: 203–206 [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3: 1101–1108 [DOI] [PubMed] [Google Scholar]
- Schroeder JI, Allen GJ, Hugouvieux V, Kwak JM, Waner D (2001) Guard cell signal transduction. Annu Rev Plant Physiol Plant Mol Biol 52: 627–658 [DOI] [PubMed] [Google Scholar]
- Schütze K, Harter K, Chaban C (2009) Bimolecular fluorescence complementation (BiFC) to study protein-protein interactions in living plant cells. Methods Mol Biol 479: 189–202 [DOI] [PubMed] [Google Scholar]
- Seymour GB, Østergaard L, Chapman NH, Knapp S, Martin C (2013) Fruit development and ripening. Annu Rev Plant Biol 64: 219–241 [DOI] [PubMed] [Google Scholar]
- Seymour GB, Taylor JE, Tucker GA (1993) Biochemistry of Fruit Ripening. Chapman & Hall, London [Google Scholar]
- Sheen J, Hwang S, Niwa Y, Kobayashi H, Galbraith DW (1995) Green-fluorescent protein as a new vital marker in plant cells. Plant J 8: 777–784 [DOI] [PubMed] [Google Scholar]
- Shukla V, Mattoo AK (2008) Sucrose non-fermenting 1-related protein kinase 2 (SnRK2): a family of protein kinases involved in hyperosmotic stress signaling. Physiol Mol Biol Plants 14: 91–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulaev V, Cortes D, Miller G, Mittler R (2008) Metabolomics for plant stress response. Physiol Plant 132: 199–208 [DOI] [PubMed] [Google Scholar]
- Sirichandra C, Davanture M, Turk BE, Zivy M, Valot B, Leung J, Merlot S (2010) The Arabidopsis ABA-activated kinase OST1 phosphorylates the bZIP transcription factor ABF3 and creates a 14-3-3 binding site involved in its turnover. PLoS ONE 5: e13935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soon FF, Ng LM, Zhou XE, West GM, Kovach A, Tan MH, Suino-Powell KM, He Y, Xu Y, Chalmers MJ, et al. (2012) Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases. Science 335: 85–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Sun Y, Zhang M, Wang L, Ren J, Cui M, Wang Y, Ji K, Li P, Li Q, et al. (2012) Suppression of 9-cis-epoxycarotenoid dioxygenase, which encodes a key enzyme in abscisic acid biosynthesis, alters fruit texture in transgenic tomato. Plant Physiol 158: 283–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telias A, Lin-Wang K, Stevenson DE, Cooney JM, Hellens RP, Allan AC, Hoover EE, Bradeen JM (2011) Apple skin patterning is associated with differential expression of MYB10. BMC Plant Biol 11: 93–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich R. (1958) Postharvest physiology of fruits. Annu Rev Plant Physiol 9: 385–416 [Google Scholar]
- Umezawa T, Yoshida R, Maruyama K, Yamaguchi-Shinozaki K, Shinozaki K (2004) SRK2C, a SNF1-related protein kinase 2, improves drought tolerance by controlling stress-responsive gene expression in Arabidopsis thaliana. Proc Natl Acad Sci USA 101: 17306–17311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilela B, Moreno-Cortés A, Rabissi A, Leung J, Pagès M, Lumbreras V (2013) The maize OST1 kinase homolog phosphorylates and regulates the maize SNAC1-type transcription factor. PLoS ONE 8: e58105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlad F, Rubio S, Rodrigues A, Sirichandra C, Belin C, Robert N, Leung J, Rodriguez PL, Laurière C, Merlot S (2009) Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. Plant Cell 21: 3170–3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Yuan F, Hao H, Zhang Y, Zhao H, Guo A, Hu J, Zhou X, Xie CG (2013a) BolOST1, an ortholog of Open Stomata 1 with alternative splicing products in Brassica oleracea, positively modulates drought responses in plants. Biochem Biophys Res Commun 442: 214–220 [DOI] [PubMed] [Google Scholar]
- Wang P, Xue L, Batelli G, Lee S, Hou YJ, Van Oosten MJ, Zhang H, Tao WA, Zhu JK (2013b) Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effectors of abscisic acid action. Proc Natl Acad Sci USA 110: 11205–11210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue S, Hu H, Ries A, Merilo E, Kollist H, Schroeder JI (2011) Central functions of bicarbonate in S-type anion channel activation and OST1 protein kinase in CO2 signal transduction in guard cell. EMBO J 30: 1645–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Peng H, Whitaker BD, Conway WS (2012) Characterization of a calcium/calmodulin-regulated SR/CAMTA gene family during tomato fruit development and ripening. BMC Plant Biol 12: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida R, Hobo T, Ichimura K, Mizoguchi T, Takahashi F, Aronso J, Ecker JR, Shinozaki K (2002) ABA-activated SnRK2 protein kinase is required for dehydration stress signaling in Arabidopsis. Plant Cell Physiol 43: 1473–1483 [DOI] [PubMed] [Google Scholar]
- Yoshida T, Nishimura N, Kitahata N, Kuromori T, Ito T, Asami T, Shinozaki K, Hirayama T (2006) ABA-Hypersensitive Germination3 encodes a protein phosphatase 2C (AtPP2CA) that strongly regulates abscisic acid signaling during germination among Arabidopsis protein phosphatase 2Cs. Plant Physiol 140: 115–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Xu X, Crosley RA, Greenwalt SA, Sun Y, Blakeslee B, Wang L, Ni W, Sopko MS, Yao C, et al. (2010) The protein kinase SnRK2.6 mediates the regulation of sucrose metabolism and plant growth in Arabidopsis. Plant Physiol 153: 99–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.