A methyl-CpG-binding domain protein participates in active DNA demethylation and appears to act as an anti-silencing agent in Arabidopsis.
Abstract
Although researchers have established that DNA methylation and active demethylation are dynamically regulated in plant cells, the molecular mechanism for the regulation of active DNA demethylation is not well understood. By using an Arabidopsis (Arabidopsis thaliana) line expressing the Promoter RESPONSIVE TO DEHYDRATION 29A:LUCIFERASE (ProRD29A:LUC) and Promoter cauliflower mosaic virus 35S:NEOMYCIN PHOSPHOTRANSFERASE II (Pro35S:NPTII) transgenes, we isolated an mbd7 (for methyl-CpG-binding domain protein7) mutant. The mbd7 mutation causes an inactivation of the Pro35S:NPTII transgene but does not affect the expression of the ProRD29A:LUC transgene. The silencing of the Pro35S:NPTII reporter gene is associated with DNA hypermethylation of the reporter gene. MBD7 interacts physically with REPRESSOR OF SILENCING5/INCREASED DNA METHYLATION2, a protein in the small heat shock protein family. MBD7 prefers to target the genomic loci with high densities of DNA methylation around chromocenters. The Gypsy-type long terminal repeat retrotransposons mainly distributed around chromocenters are most affected by mbd7 in all transposons. Our results suggest that MBD7 is required for active DNA demethylation and antisilencing of the genomic loci with high densities of DNA methylation in Arabidopsis.
DNA methylation is an important epigenetic marker for genome stability and the regulation of gene expression in both plants and animals (Law and Jacobsen, 2010; He et al., 2011). In plants, the molecular mechanisms for DNA methylation have been well characterized by the use of powerful genetic screening systems (Bartee et al., 2001; Lindroth et al., 2001; Matzke et al., 2004; He et al., 2009). A transgene or an endogenous gene may be silenced because of DNA hypermethylation in the promoter region. Screenings for mutants with release of the silenced marker genes have identified many components that are involved in RNA-directed DNA methylation (RdDM) and in maintaining DNA methylation (Matzke and Birchler, 2005; Law and Jacobsen, 2009; He et al., 2011; Bender, 2012). DNA methylation is catalyzed by DNA methyltransferases including DNA METHYLTRANSFERASE1 (MET1) and CHROMOMETHYLASE3 (CMT3), which maintain symmetric CG and CHG methylation, respectively, during DNA replication, and DOMAINS REARRANGED METHYLASE2 (DRM2) and CMT2, which are required for establishing CHG and asymmetric CHH methylation during each cell cycle. DRM2 also catalyzes CG methylation (Law and Jacobsen, 2010; Haag and Pikaard, 2011; He et al., 2011; Zemach et al., 2013; Stroud et al., 2014). Twenty-four-nucleotide small RNAs produced through the RdDM pathway target genomic regions to guide the establishment of DNA methylation by DRM2 (Cao et al., 2003).
DNA methylation can be actively removed by a subfamily of bifunctional DNA glycosylases/lyases including REPRESSOR OF SILENCING1 (ROS1; Gong et al., 2002) and its paralogs DEMETER and DEMETER-LIKE2/3 (Gehring et al., 2006; Ortega-Galisteo et al., 2008). DNA methylation can also be passively lost during DNA replication when DNA methylation cannot be maintained (Zhu, 2009). Promoter RESPONSIVE TO DEHYDRATION 29A:LUCIFERASE (ProRD29A:LUC) in the ProRD29A:LUC/Promoter cauliflower mosaic virus 35S:NEOMYCIN PHOSPHOTRANSFERASE II (Pro35S:NPTII) transgenic Arabidopsis (Arabidopsis thaliana) line has been used as a marker to identify ros1 and ros3 mutants in which both ProRD29A:LUC and Pro35S:NPTII are silenced (Gong et al., 2002; Zheng et al., 2008). ROS3 is an RNA-binding protein that facilitates the function of ROS1 in active DNA demethylation at certain genomic loci. Using Pro35S:NPTII as a selection marker for kanamycin-sensitive mutants and the 35S-SUC2 transgene or a chop PCR marker for assaying DNA methylation at the 3′ region of At1g26400 from transfer DNA (T-DNA) insertion mutants, researchers recently identified two genes involved in active DNA demethylation: ROS4/INCREASED DNA METHYLATION1 (IDM1) and ROS5/IDM2 (Li et al., 2012; Qian et al., 2012, 2014; Zhao et al., 2014). ROS4/IDM1 is a plant homeodomain-finger domain-containing histone acetyltransferase that catalyzes histone H3 lysine18 (H3K18) and lysine23 (H3K23) acetylation (Li et al., 2012; Qian et al., 2012). ROS5/IDM2 is a member of the small heat shock protein family that interacts physically with ROS4/IDM1 for the regulation of active DNA demethylation. Genetic analysis indicates that ROS1, ROS4/IDM1, and ROS5/IDM2 are in the same genetic pathway and that ROS4/IDM1 and ROS5/IDM2 may form a protein complex for the regulation of active DNA demethylation (Qian et al., 2014; Zhao et al., 2014).
During the genetic screening for kanamycin-sensitive mutants using the ProRD29A:LUC/Pro35S:NPTII transgenic line in this study, we identified another mutant, mbd7, where the Pro35S:NPTII transgene is specifically silenced. MBD7 is a methyl-CpG-binding domain (MBD) protein containing three MBD motifs that bind in vitro to methylated symmetric CG sites. MBD7 localizes to all highly CpG-methylated chromocenters in vivo (Zemach and Grafi, 2003; Zemach et al., 2008). Recruitment of MBD7 to chromocenters is disrupted in decrease in DNA methylation1 (ddm1) and met1, two mutants with great reductions in DNA methylation, suggesting that DNA methylation is required for proper MBD7 localization (Zemach et al., 2005). In this study, we found that MBD7 interacts physically with ROS5/IDM2 and is required for the active DNA demethylation of certain genomic loci, especially for the Gypsy-type long terminal repeat (LTR) retrotransposons with high densities of DNA methylation around chromocenters in Arabidopsis.
RESULTS
MBD7 Is Required for Active DNA Demethylation at the 3′ Region of Pro35S:NPTII
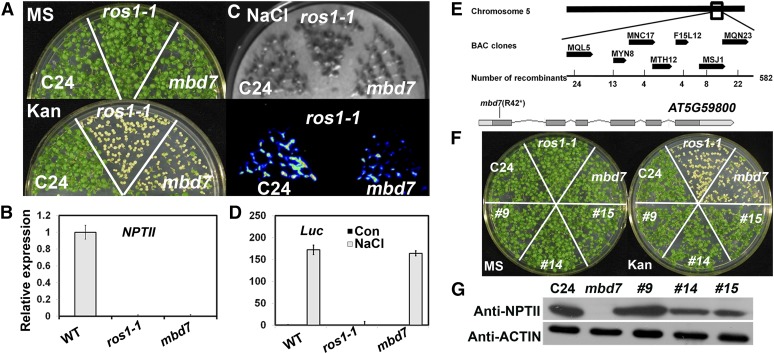
The C24 transgenic Arabidopsis line carries ProRD29A:LUC and Pro35S:NPTII transgenes, which are actively expressed (Gong et al., 2002). We used an ethyl methanesulfonate-mutagenized population of this line (as the wild type in this study) to screen for kanamycin-sensitive mutants (Li et al., 2012; Zhao et al., 2014). The kanamycin-sensitive mutant, mbd7, was isolated during the screening (Fig. 1A, left). Like NTPII expression in ros1-1, NTPII expression in mbd7 was only weakly detected but was clearly detected in the C24 wild type (Fig. 1B). mbd7 was backcrossed with the wild type at least three times to remove background mutations before further analysis. F1 seedlings of mbd7 crossed with the wild type exhibited the wild-type phenotype for kanamycin resistance, and F2 seedlings were about 1:3 kanamycin sensitive (52 seedlings) to kanamycin resistant (178 seedlings), indicating that mbd7 is caused by a recessive, single nuclear gene mutation. Like ros4 and ros5-1 mutants (Li et al., 2012; Zhao et al., 2014), the mbd7 mutant did not affect the expression of ProRD29A:LUC (Fig. 1, C, right, and D), but ros1-1 silenced both ProRD29A:LUC and Pro35S:NPTII (Fig. 1, A–D).
Figure 1.
Identification of MBD7 by map-based cloning. A, mbd7 mutants silence Pro35S:NPTII but do not affect ProRD29A:LUC. Seedlings were grown on Murashige and Skoog (MS) medium or MS medium supplemented with 50 mg L−1 kanamycin (Kan). The C24 accession was used as the wild type (WT). ros1-1, in which both Pro35S:NPTII and ProRD29A:LUC are silenced, was included for comparison. B, NPTII expression in C24, ros1-1, and mbd7 as determined by real-time PCR. Three independent experiments were done with similar results, each with three technical repeats. The results of one representative experiment are shown. Values are means ± se (n = 3). C, mbd7 does not affect the expression of ProRD29A:LUC. Seedlings of C24, mbd7, and ros1-1 were treated with 300 mm NaCl for 3 h before luminescence images were captured. D, Real-time PCR analyses of LUC transcripts from the samples in C. Three independent experiments were done with similar results, each with three technical repeats. The results of one representative experiment are shown. Values are means ± se (n = 3). E, Identification of MBD7 by map-based cloning. The location of MBD7 was narrowed to BAC clones MNC17 and F15L12 by use of simple sequence length polymorphism markers. A G-to-A mutation, which changes Arg-42 to a stop codon, was found in AT5G59800. F, The kanamycin-sensitive phenotype of mbd7 was complemented by the MBD7 gene as shown in three independent transgenic lines. G, NPTII protein levels in C24, mbd7, and three transgenic lines as indicated by western blot with NPTII antibodies. ACTIN was used as a loading control.
We used map-based cloning to isolate the MBD7 gene. The mbd7 mutant in the C24 accession was crossed with the wild-type Columbia accession. The 582 F2 plants that contained the NPTII gene (checked by PCR) but were kanamycin sensitive were isolated and used for mapping. MBD7 was narrowed to a region between bacterial artificial chromosome (BAC) clones MNC17 and F15L12 on chromosome 5 (Fig. 1E). Sequencing the candidate genes on the BAC clone MTH12 revealed a G-to-A point mutation in AT5G59800, which changes an Arg-42 to a putative stop codon. mbd7 should be a null mutant. To confirm that the point mutation in AT5G59800 leads to Pro35S:NPTII transgene silencing in mbd7, we transformed a wild-type genomic fragment containing the full-length AT5G59800, including the 2,643-bp promoter and the 278-bp 3′ region, into mbd7 mutant plants. As shown in Figure 1, G and F, three randomly selected, independent transgenic lines (lines 9, 14, and 15) complemented the kanamycin-sensitive phenotype with expression of the NPTII protein. Analysis of GUS staining in the transgenic plants expressing ProMBD7:GUS indicated that MBD7 is highly expressed at the early seedling stage; the timing of this expression overlaps with that of ROS4/IDM1 and ROS5/IDM2 (Supplemental Fig. S1; Li et al., 2012; Zhao et al., 2014).
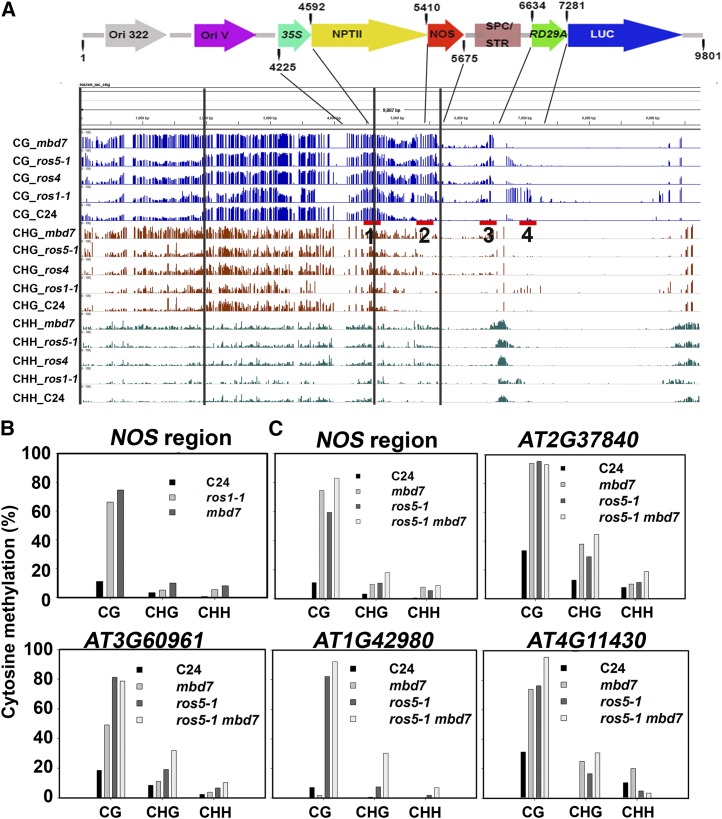
In previously isolated ros mutants, ros1-1 increased the DNA methylation in both the RD29A promoter and the 3′ NOPALINE SYNTHASE (NOS) terminator region, and ros4 and ros5-1 increased DNA methylation only in the 3′ NOS terminator (Zhao et al., 2014). Whole-genome bisulfite sequencing indicated that, like ros4 and ros5-1, mbd7 increased DNA methylation only in the 3′ NOS terminator region and did not change DNA methylation in the RD29A promoter (Fig. 2A); this is consistent with the lower expression level of NPTII but the similar expression level of LUC relative to the wild type (Fig. 1, B and D). Bisulfite sequencing confirmed the DNA hypermethylation in the 3′ NOS terminator region in mbd7 (Fig. 2B). The DNA methylation level in 3′ NOS was not greater in the ros5-1 mbd7 double mutant than in the ros5-1 or mbd7 single mutant. We further checked four loci (AT2G37840, AT3G60961, AT1G42980, and AT4G11430) in which DNA methylation was found to be increased by ros5-1 mutation in a previous study (Zhao et al., 2014). As was the case for 3′ NOS, the methylation level at the four loci was not greater in the ros5-1 mbd7 double mutant than in the ros5-1 or mbd7 single mutant. These results suggest that ROS5/IDM2 and MBD7 work in the same pathway to mediate DNA methylation. Treatment with 5′-aza-2′-deoxycytidine (a cytosine methylation inhibitor) consistently rescued the kanamycin-sensitive phenotype of mbd7 seedlings (Supplemental Fig. S2A) and induced a high level of NPTII expression (Supplemental Fig. S2B). The introduction of a DNA replication mutation, DNA replication factor c1-1 (rfc1-1), into mbd7 released the silencing of Pro35S:NPTII (Supplemental Fig. S2C), which is similar to previous reports for ros1-1, ros4, and ros5-1 mutants (Liu et al., 2010; Li et al., 2012; Zhao et al., 2014). Previous study indicates that mutations in the RdDM pathway greatly reduce the expression of ROS1. However, ROS1 expression was not affected by mbd7 (Supplemental Fig. S2D).
Figure 2.
MBD7 prevents DNA hypermethylation in the 3′ NOS region in ProRD29A:LUC/Pro35S:NTPII transgenic plants. A, Diagram of the whole T-DNA region and corresponding DNA methylation patterns in mbd7, ros5-1, ros4, ros1-1, and C24 (the wild type) as determined by whole-genome bisulfite sequencing. The red bars with numbers 1 to 4 indicate the fragments used for the ChIP assay in Figure 5A. Ori, Replication origin. B, Confirmation of DNA methylation in the NOS region in mbd7 and ros1-1 by bisulfite sequencing. C, DNA methylation levels in ros5-1 and mbd7 mutants are not additive in the 3′ NOS region or in AT2G37840, AT3G60961, AT1G42980, or AT4G11430 in the mbd7 ros5-1 double mutant.
MBD7 Interacts Physically with ROS5/IDM2
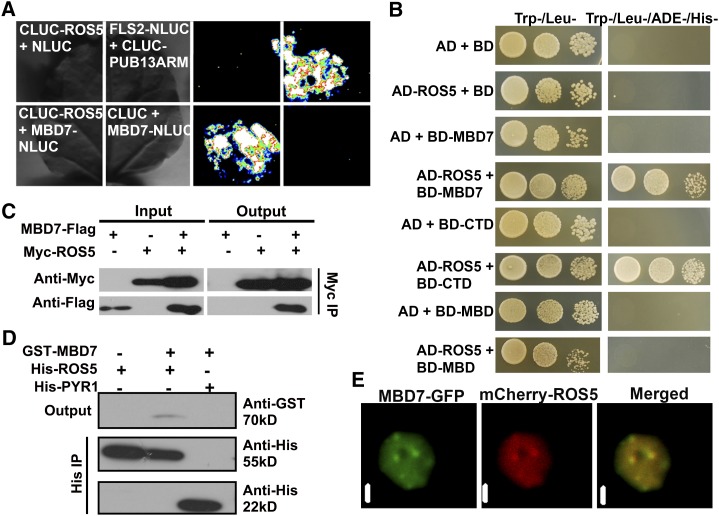
ROS5/IDM2 is a small, heat shock protein-like protein with a conserved α-crystallin domain that forms an approximately 16-mer complex (Zhao et al., 2014). ROS5 interacts physically with ROS4/IDM1 and is required for histone H3K18 acetylation by ROS4/IDM1 (Qian et al., 2014; Zhao et al., 2014). Because ROS1, ROS4, ROS5, and MBD7 are all required for active DNA demethylation in the 3′ NOS terminator region of 35S-NPTII, and because they are all identified by the same genetic screening, we wanted to know whether they operate in the same protein complex. We first used a firefly luciferase complementation imaging assay to determine whether MBD7 interacts with ROS1, ROS4, or ROS5 in tobacco (Nicotiana tabacum) leaves (Chen et al., 2008). We found that MBD7 interacts with ROS5 (Fig. 3A) but not with ROS4 or ROS1 (Supplemental Fig. S3A). The interaction of FLS2 with the PUB13-ARM domain was used as a positive control (Lu et al., 2011). The yeast (Saccharomyces cerevisiae) two-hybrid assay indicated that ROS5 interacts with the MBD7 C-terminal domain but not with the MBD7 N-terminal region, which contains three MBD domains (Fig. 3B). MBD7 did not interact with ROS1 or ROS4/IDM1 in the yeast two-hybrid assay (Supplemental Fig. S3B). An in vivo coimmunoprecipitation assay using proteins from Arabidopsis protoplasts transiently expressing different plasmids indicated that MBD7-Flag was coimmunoprecipitated with Myc-ROS5 (Fig. 3C). An in vitro protein pull-down assay, which used purified proteins expressed in E. coli, also showed that GLUTATHIONE S-TRANSFERASE (GST)-MBD7 pulled down His-ROS5 (Fig. 3D). We also analyzed the subnuclear localization of mCherry-ROS5 and MBD7-GFP in transiently coexpressing tobacco epidermal cells. As shown by the yellow signals in Figure 3E, mCherry-ROS5 was colocalized with MBD7-GFP. Because ROS4/IDM1 interacts with ROS5/IDM2, and because ROS1 is likely in the complex of ROS5/IDM1 (Zhao et al., 2014), these results suggest that ROS1, ROS4/IDM1, ROS5/IDM2, and MBD7 form a protein complex in the nucleus.
Figure 3.
MBD7 interacts with ROS5/IDM2. A, MBD7 interacts with ROS5/IDM2 as indicated by a firefly luciferase complementation imaging assay in tobacco leaves. The interaction of FLAGELLIN-SENSING2 (FLS2) with the C-terminal ARMADILLO (ARM) repeat domain of Plant U-box (PUB) E3 ubiquitin ligase was used as a positive control. B, MBD7 interacts with ROS5/IDM2 in a yeast two-hybrid assay. The full-length, C-terminal domain (CTD) and MBD domain of MBD7 were analyzed by yeast two-hybrid assay for interaction with ROS5. AD, GAL4 activation domain in the pGADT7 plasmid; BD, GAL4 binding domain in the pGBKT7 plasmid. C, Interaction of MBD7 with ROS5 as indicated by coimmunoprecipitation (IP) in a protoplast transient assay. Three independent experiments were done with similar results. D, Interaction of MBD7 with ROS5 as indicated by protein pull-down assay using proteins expressed from Escherichia coli. E, Colocalization of MBD7-GFP with mCherry-ROS5 as indicated by a transient assay in tobacco epidermal cells. MBD7 was fused to the N terminus of GFP, and mCherry was fused to the N terminus of ROS5. Agrobacterium tumefaciens carrying MBD7-GFP and mCherry-ROS5 was coinjected into tobacco epidermis cells. Bars = 1 μm.
MBD7 Prevents DNA Methylation Mainly in the Genomic Regions around Chromocenters
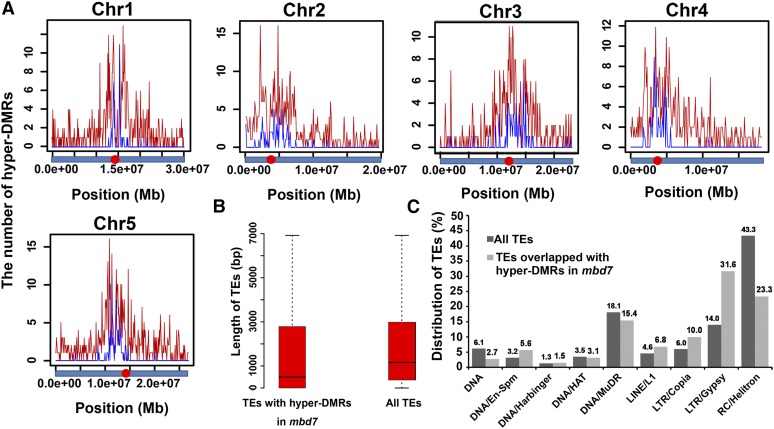
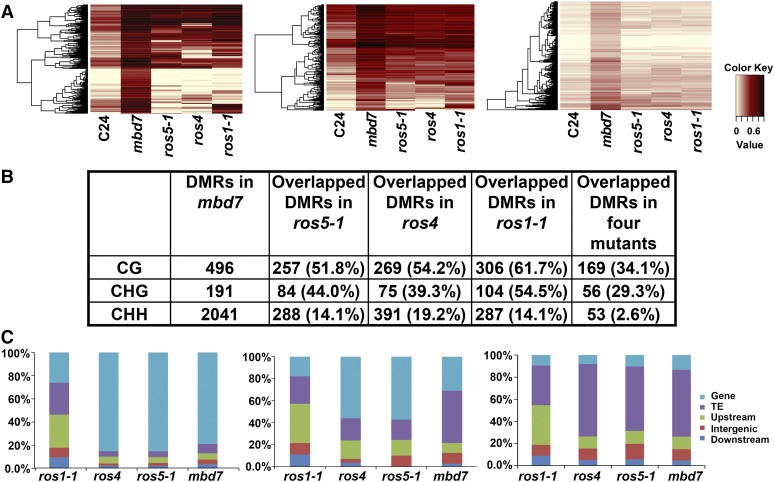
To determine whether MBD7 plays a role in active DNA demethylation at the whole-genome level, we compared the genome-wide DNA methylation profiles of mbd7 and the C24 wild type by whole-genome bisulfite sequencing (Zhao et al., 2014). Previous studies reported no significant difference in the whole-genome DNA methylation level between ros4 or ros5-1 and the wild type (Zhao et al., 2014). Similarly, the whole-genome DNA methylation level in mbd7 did not show too much difference when compared with that in the wild type (Supplemental Fig. S4A). However, by analyzing the windows with significant methylation differences (P < 0.01) between the C24 wild type and mbd7, we found that the methylation level of mbd7 was higher than that of C24 in these analyzed loci (Supplemental Fig. S4B), which indicates that MBD7 prevents hypermethylation at these genomic loci. The total methylation levels in mbd7 were 20.6% in CG, 6% in CHG, and 2.2% in CHH, which were a little higher than 20.2% (CG), 5.4% (CHG), and 2% (CHH) in the C24 wild type (Supplemental Fig. S4C). In total, we identified 2,664 differentially methylated regions (DMRs) with increased DNA methylation, including 497 hyper-DMRs in CG methylation, 192 hyper-DMRs in CHG methylation, and 2,041 hyper-DMRs in CHH methylation in mbd7 (Supplemental Table S1). The number of hyper-DMRs that are localized in gene bodies, transposon elements (TEs), upstream and downstream of gene bodies, and the intergenic regions are 701, 1,348, 241, 114, and 260 in mbd7, respectively (Supplemental Table S2). Among 2,041 CHH hyper-DMRs in mbd7, 1,235 are in TEs, 272 in gene bodies, 231 upstream of gene bodies, 93 downstream of gene bodies, and 210 in intergenic regions. Hyper-DMR distribution analysis on the whole genome indicated that hyper-DMRs in mbd7 were intensively localized around the chromocenters in five individual chromosomes (Fig. 4A). As more than 50% hyper-DMRs are TEs, we analyzed whether these TEs have any distinguishing features. The length of TEs that overlapped with hyper-DMRs in mbd7 was a little shorter than that of total TEs (Fig. 4B), suggesting that MBD7 has a bias to target short TEs. Comparing the ratio of individual TEs to total TEs in the whole genome with the ratio of MBD7-affected individual TEs to total MBD7-affected TEs, four categories of TEs, DNA/En-Spm, LINE/L1, LTR/Copia, and LTR/Gypsy, were more affected than other TEs by MBD7 (Fig. 4C). Among these four TEs, the Gypsy-type LTR retrotransposons (LTR/Gypsy) are most affected by MBD7 (Fig. 4C). The Gypsy-type LTR retrotransposons targeted by MBD7 were intensively distributed around the five chromocenters (Fig. 4A, blue line), which is consistent with the total Gypsy TE distribution in Arabidopsis (Zemach et al., 2013). These results suggest that MBD7 prefers to target genomic loci around chromocenters, and the Gypsy-type LTR retrotransposons are the most affected TEs.
Figure 4.
Hyper-DMRs in mbd7 are enriched around chromocenters. A, The distribution of hyper-DMRs in mbd7 on five chromosomes of Arabidopsis. The red lines in the diagrams represent the distribution of hyper-DMRs in mbd7, and the blue lines represent the distribution of Gypsy transposons that overlap with hyper-DMRs in mbd7. The red points on the bar under the diagrams represent the chromocenters of different chromosomes. B, Comparison of the average length of TEs overlapped with hyper-DMRs in mbd7 with that of total TEs in the whole genome. C, Comparison between the percentage of individual TEs to total TEs in the whole genome and the percentage of individual TEs to total TEs overlapping with hyper-DMRs in mbd7. Numbers above the bars are percentages. The classified information of TEs was downloaded from The Arabidopsis Information Resource (TAIR) 10 (ftp://ftp.arabidopsis.org/home/tair/Genes/TAIR10_genome_release/TAIR10_transposable_elements/TAIR10_Transposable_Elements.txt).
In order to compare with other mutants, we also included the DNA methylation profiles of ros1-1, ros4, and ros5-1 mutants (Zhao et al., 2014). The heat-map analysis of these DMRs indicated a high corelativity between mbd7 and ros1-1, ros4, or ros5-1 (Fig. 5A). The DMRs of mbd7 overlapped with those of each mutant with a different ratio (Fig. 5B). Among these DMRs in mbd7, about 34% in CG methylation, 29.1% in CHG methylation, and 2.6% in CHH methylation are found to be overlapped in all four mutants (mbd7, ros1-1, ros4, and ros5-1; Fig. 5B). The low overlapped ratio in CHH is likely due to the low methylation level, which reduces the number after quality control. We also found that 50.6% of the DMRs in mbd7 overlapped with TEs, which is higher than in ros1-1 (30.6%), ros4 (23.7%), or ros5-1 (16.3%). The CG, CHG, and CHH DMRs in ros1-1 were evenly distributed in different genomic regions, while CG DMRs in mbd7, ros4, and ros5-1 were localized mainly in genic regions and CHH DMRs were mainly in TEs (Fig. 5C). CHG DMRs in TEs were more abundant in mbd7 than in other mutants. These results suggest that although these proteins may work together to target some common genomic loci, each one has its own preference to some specific regions (Li et al., 2012; Qian et al., 2012, 2014; Zhao et al., 2014).
Figure 5.
Comparison of hyper-DMRs among different mutants. A, Heat map showing the hyper-DMRs in mbd7 overlapping with those regions in ros5-1, ros4, and ros1-1 in the CG, CHG, and CHH contexts. The color key is presented at right (light yellow indicates low methylation, and black indicates high methylation). B, The overlapping ratio of the hyper-DMRs (P < 0.01) in mbd7 with those regions in ros5-1, ros4, and ros1-1 mutants in the CG, CHG, and CHH contexts. C, Compositions of the genomic locations of hyper-DMRs in ros5-1, ros4, ros1-1, and mbd7 mutants in the CG, CHG, and CHH contexts. The regions upstream and downstream of a gene were 2 kb away from the transcription start site and the transcriptional end site.
MBD7 Binds Preferably to the High-Density DNA Methylation Regions
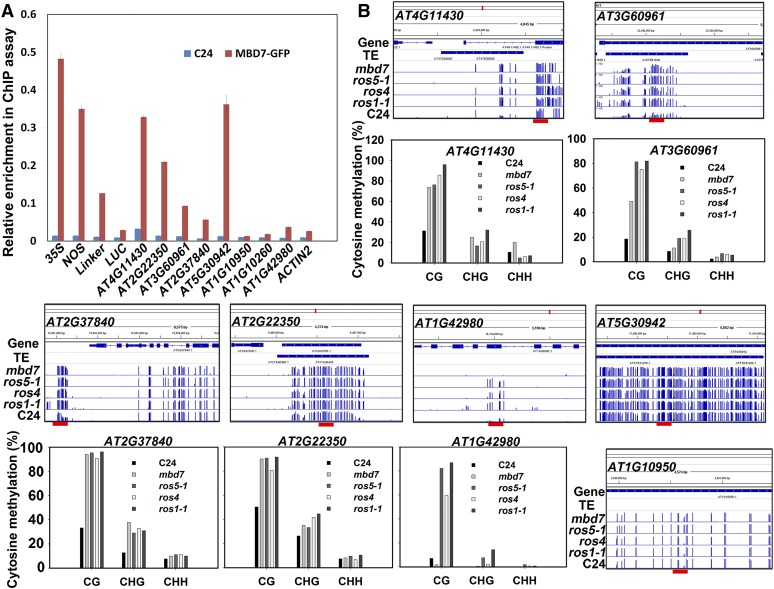
To determine which regions MBD7 binds to, we performed a chromatin immunoprecipitation (ChIP) assay using transgenic plants expressing Pro35S:MBD7-GFP. We selected several DNA regions that have higher DNA methylation in mbd7 than in the wild type, and we checked them by PCR after ChIP. In the T-DNA region of ProRD29A:LUC/Pro35S:NPTII transgenic plants, the affinity was highest near the 35S promoter region, followed by the 3′ NOS terminator and the linker region between 3′ NOS and the RD29A promoter (Fig. 6A; for the regions examined, see Fig. 2A). The affinity was lowest in the LUC coding region. These results indicate that MBD7 binds to most of the Pro35S:NTPII:NOS region with high affinity. We selected four hypermethylated loci (AT4G11430, AT3G60961, AT2G37840, and AT2G22350) that are regulated by MBD7, ROS1, ROS4, and ROS5, and we confirmed their DNA methylation levels by bisulfite sequencing in each mutant (Fig. 6B). AT3G60961 and AT2G22350 are TEs that are targeted by the RdDM pathway, and AT4G11430 is near transposons and not targeted by the RdDM pathway. As shown in Figure 6A, MBD7 bound to these four regions. MBD7 did not bind to AT1G42980 (a gene without nearby transposons, targeted by RdDM), whose methylation was increased by ros1-1, ros4, and ros5-1 mutations but not by the mbd7 mutation. The AT5G30942 locus is localized in the middle of a TE-rich region that has a high density of DNA methylation, that is in the chromocenter of chromosome V, and that is targeted by the DDM1 pathway; MBD7 bound to this locus (Fig. 6A), although DNA methylation in AT5G30942 was not changed by mbd7, ros1-1, ros4, or ros5-1 mutations (Fig. 6B). Perhaps MBD7 targets AT5G30942 but the DNA methylation level in this locus is already very high and thus cannot be enhanced by mbd7. In contrast to AT5G30942, AT1G10950 is localized in a gene region without TEs. AT1G10950 has hypermethylated DNA, but the DNA methylation density is lower than in AT5G30942, and its DNA methylation was not changed by the mbd7, ros1-1, ros4, and ros5-1 mutations (Fig. 6B). MBD7 did not bind to AT1G10950 (Fig. 6A). As negative controls, MBD7 did not bind to AT1G10260 or ACTIN2 loci that lacked DNA methylation. These results suggest that MBD7 binds to genomic regions with high densities of DNA methylation and might target loci in addition to those detected by the whole-genome bisulfite sequencing.
Figure 6.
MBD7 binds to DNA regions with high methylation density. A, ChIP assay indicates that MBD7 binds to different parts of the T-DNA region and to some DMRs. The C24 wild-type plants expressing MBD7-GFP were used for the ChIP assay with anti-GFP antibodies. The fragments used for PCR in the T-DNA region are indicated in Figure 2A with red bars and with the numbers 1 (35S), 2 (NOS), 3 (linker), and 4 (LUC). Other fragments for PCR in different DMRs are indicated by red bars in Figure 6B. B, DNA methylation in different DMRs as indicated by whole-genome bisulfite sequencing and individual bisulfite sequencing in mbd7, ros5-1, ros4, ros1-1, and the C24 wild type.
DISCUSSION
Active DNA demethylation is carried out by DNA demethylases in the ROS1 protein family through a base excision-repair pathway (Zhu, 2009; Gong and Zhu, 2011). However, how these DNA demethylases are recruited to the specific target regions for removal of DNA methylation is not known. Previous studies suggest that ROS4/IDM1 interacts with ROS5/IDM2, which regulates H3K18 acetylation, and creates a favorable chromatin environment for recruiting of ROS1 (Li et al., 2012; Qian et al., 2012, 2014; Zhao et al., 2014). By using the same genetic screen that was used to isolate the ros4 and ros5 mutants, we identified MBD7, a methyl-CpG-binding domain protein in this study. MBD7 interacts physically with ROS5/IDM2, and previous studies indicated that ROS5/IDM2 also interacts with ROS4/IDM1 (Qian et al., 2014; Zhao et al., 2014) and that ROS1 is in the same complex as ROS5/IDM2 (Zhao et al., 2014). These related proteins evidently target some common genomic sites for active DNA demethylation, because any mutation in these four genes increases the DNA methylation in some common sites and because the mbd7 ros5-1 double mutant does not have an additive effect on DNA methylation level relative to mbd7 and ros5-1 single mutants. Because ROS1, ROS4/IDM1, ROS5/IDM2, and MBD7 all belong to a small family of proteins, functional redundancy of the paralogous proteins may work at different loci for the regulation of active DNA demethylation. For example, for the AT1G42980 locus (Fig. 6), the DNA methylation in mbd7 is not changed and MBD7 does not bind to this locus, but DNA methylation is increased in ros5-1, ros4, and ros1-1 mutants. It is speculated that the MBD7 paralogs may function together with ROS4/IDM1, ROS5/IDM2, and ROS1 in this locus. The molecular mechanism for the different genomic regions preferably targeted by different proteins needs further exploring.
Our study indicates that MBD7 prefers to target the genomic regions with high density of DNA methylation around chromocenters, which is consistent with previous studies showing that MBD7 is intensively localized in the chromocenters (Zemach et al., 2008). The mutations in met1 and ddm1 abolish the recruitment of MBD7 to chromocenters (Zemach et al., 2005). MBD7 binds to CG methylation sites (Zemach and Grafi, 2003). These results suggest that the dense DNA methylation is required for MBD7 functions. Our study indicates that the hyper-DMRs in mbd7 are highly enriched with CHH methylation and that MBD7 prefers to target the short TEs, especially the Gypsy-type LTR retrotransposons. Given that CHH methylation is usually catalyzed by DRM2 in the RdDM pathway (Cao and Jacobsen, 2002; Cao et al., 2003) and by CMT2 in the DDM1-dependent pathway (Zemach et al., 2013), these results suggest that MBD7 is involved in active DNA demethylation at genomic sites targeted by both the RdDM pathway and the DDM1-dependent pathway, which is supported by our DNA methylation analyses on individual loci (Fig. 6).
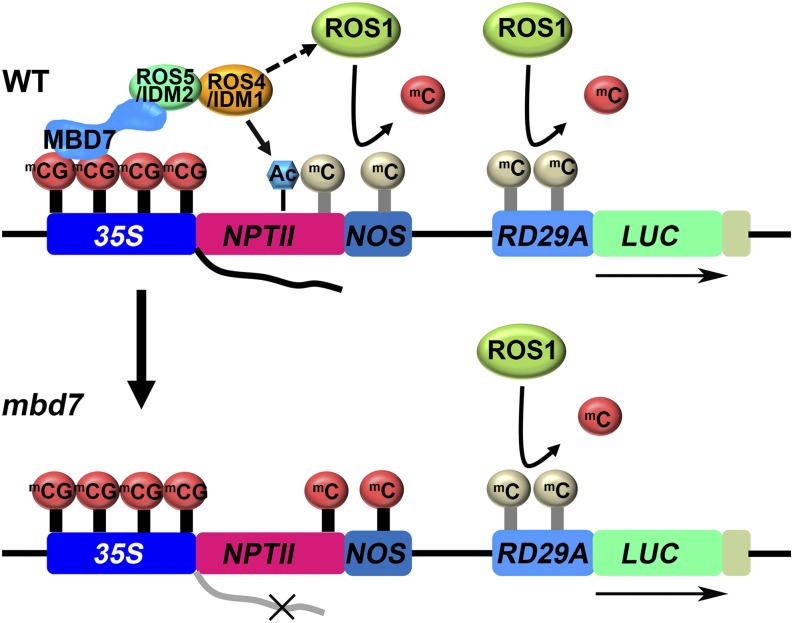
A previous study indicated that the MBD7 C-terminal domain has very strong chromatin-binding ability and that one of its MBD domains binds to chromosomal methylated DNA sites but does not affect MBD7 subnuclear localization (Zemach et al., 2009). In this study, we found that the MBD7 C-terminal domain, but not its MBD domains, interacts with ROS5/IDM2. ROS5/IDM2 is an α-crystallin domain-containing protein that also interacts with ROS4/IDM1, a histone acetyltransferase (Qian et al., 2014; Zhao et al., 2014). We speculate that MBD7 binds to methylated CG through one of its MBD domains in the N terminus and then the C-terminal domain recruits ROS5/IDM2, which further recruits ROS4/IDM1 and other proteins (Fig. 7; Zemach et al., 2005; Scebba et al., 2007). ROS4/IDM1 will acetylate histone H3K18 and H3K23 to create a chromatin environment that facilitates the recruitment of ROS1 protein for the removal of DNA methyl groups. Because MBD domain proteins are conserved in mammals, the results reported here also may apply to mammalian systems (Fournier et al., 2012).
Figure 7.
Proposed working model for MBD7. MBD7 binds to a genomic region with a high density of CG methylation, which recruits ROS5/IDM2 and then ROS4/IDM1. ROS4/IDM1 may acetylate H3K18 and H2K23 to create a chromatin environment to further facilitate the recruitment of ROS1 protein for the removal of DNA methyl groups. The proteins that directly recruit ROS1 need to be identified in a future study. RD29A is regulated by ROS1, which is independent of the MBD7-ROS5/IDM2-ROS4/IDM1 complex. Ac, H3K18 or H3K23 acetylation; mCG or mC, 5′ cytosine methylation; WT, wild type. The light color mC indicates dynamically removed mC.
MATERIALS AND METHODS
Plant Materials and Mutant Screening
All mutants used in this study are in the Arabidopsis (Arabidopsis thaliana) C24 background, which carries transgenes of ProRD29A:LUC and Pro35S:NPTII (which we refer to as the wild type in this study). The mutants ros1-1, ros4/idm1-3, ros5-1, nuclear RNA polymerase d2, and rfc1-1 were isolated previously in our laboratory. The mutant mbd7 was identified from an ethyl methanesulfonate-mutagenized population of the wild type as described previously (Li et al., 2012). In short, wild-type seedlings are resistant to 50 mg L−1 kanamycin. Individual mutants that were sensitive to 50 mg L−1 kanamycin were selected, and their kanamycin phenotype was confirmed in the next generation. The putative mutants were crossed with Columbia (the wild type without T-DNA insertion), and the F2 plants that were sensitive to a kanamycin-amended medium and that carried the NPTII gene (detected by PCR) were used for mapping. We obtained 582 F2 plants that contain NPTII but show kanamycin sensitivity for the map-based cloning.
For the complementation assay, the MBD7 genomic sequence (from −2,643 to +343 bp, which contains the 2,643-bp promoter, the coding region, and the 278-bp 3′ region) was cloned into pCAMBIA1300. Then, the construct in Agrobacterium tumefaciens GV3101 was transformed into the mbd7 mutant, and transgenic plants were selected and analyzed for their kanamycin phenotypes. All primers used in this study are listed in Supplemental Table S3.
Real-Time Reverse Transcription-PCR Assay
Real-time reverse transcription-PCR was used to assess the transcription levels of different genes. Total RNAs were extracted from 7-d-old seedlings by use of Trizol reagent (Invitrogen). The reverse-transcribed complementary DNAs (cDNAs) were used for real-time reverse transcription-PCR in SYBR Green Master Mix (Takara) on a Step One Plus machine (Applied Biosystems). Primers specific for each gene are listed in Supplemental Table S3. ACTIN was used as an internal control.
Histochemical GUS Staining
The MBD7 promoter region (from −2,643 to −1 bp) and the GUS coding region were cloned into the pCAMBIA1391 vector. Then, the recombinant plasmid was transformed into C24 wild-type plants with the help of A. tumefaciens GV3101. Histochemical GUS staining was performed on the T2 homozygous transgenic plants as described previously (Xia et al., 2006).
Bisulfite Sequencing
Genomic DNA was extracted from 7-d-old seedlings, and about 500 ng was treated with the EZ Methylation-Gold Kit (Zymo Research) following the protocol supplied by the manufacturer. About 80 ng of treated DNA was used in the PCR with the specific primers listed in Supplemental Table S3. The PCR products were cloned into the pMD18-T vector (Takara), and at least 15 independent clones of each sample were sequenced for each region.
Coimmunoprecipitation Assay
The full-length cDNA of MBD7 fused with the Flag coding sequence was cloned into the multiple cloning site of a modified pCAMBIA1300 vector. A vector expressing Myc-ROS5 was described previously (Zhao et al., 2014). The two purified plasmids were cotransformed into Arabidopsis protoplasts. After the protoplasts were cultured overnight in a light chamber, the proteins were extracted from protoplasts with the immunoprecipitation buffer (50 mm Tris-HCl, pH 7.6, 5 mm MgCl2, 150 mm NaCl, 10% [v/v] glycerol, 0.5% [v/v] Nonidet P-40, 0.5 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, and protease inhibitor cocktail [one tablet per 50 mL]; Roche). The protein solution was centrifuged, and the supernatant was added to 20 μL of Anti-C-Myc-Agarose Affinity Gel (Sigma-Aldrich). After incubation on a rotary shaker at 4°C for 2 h, the agarose beads were washed four times with the immunoprecipitation buffer in which the concentration of Nonidet P-40 was adjusted to 0.1% (v/v). The immunoprecipitated products were then detected by western blot.
Luciferase Complementation Imaging Assay
The full-length cDNA of MBD7 was fused with NLUC in the pCAMBIA vector. The vectors expressing CLUC-ROS1, CLUC-ROS4/IDM1, and CLUC-ROS5/IDM2 were constructed previously in our laboratory (Zhao et al., 2014). The FLS2-NLUC and CLUC-PUB13-ARM constructs were prepared as described previously (Lu et al., 2011). After the plasmid constructs were transformed into A. tumefaciens GV3101 and then into Nicotiana benthamiana, a fluorescence image was observed with a CCD camera (1300B; Roper).
Protein Purification and Pull-Down Assay
To prepare the purified GST-MBD7 protein, the GST in the pGEX4T-1 vector was fused with the full-length cDNA of MBD7; the construct was then transformed into Escherichia coli BL21 cells. Constructs of His-ROS5 and His-PYR1 in the pET30a vector were described previously (Zhao et al., 2014). A 2-μg quantity of GST-MBD7 and the same quantity of His-ROS5 or His-PYR1 protein were added in a binding buffer (50 mm Tris-HCl, pH 7.5, 250 mm NaCl, 0.1% [v/v] Nonidet P-40, and 1 mm phenylmethylsulfonyl fluoride). After incubating at 4°C overnight, each binding system was treated with 1 mL of binding buffer and 25 μL of nickel-nitrilotriacetic acid agarose beads and then was incubated at 4°C for another 2 h. The nickel-nitrilotriacetic acid agarose beads were then washed four times with the binding buffer and finally one time with the Nonidet P-40-free binding buffer. The proteins were eluted twice with 160 μL of elution buffer (20 mm Tris-HCl, pH 7.5, 300 mm NaCl, 5 mm MgCl2, and 250 mm imidazole). The pull-down products were detected by western blot using GST or His antibody.
Yeast Two-Hybrid Assay
We used the pGADT7 vector containing the GAL4 AD and pGBKT7 vector containing the GAL4 BD for the yeast (Saccharomyces cerevisiae) two-hybrid assay. BD-MBD7 was constructed by cloning full-length cDNA of MBD7 into the pGBKT7 vector. The plasmids of AD-ROS5, AD-ROS4, and AD-ROS1 were generated previously in our laboratory (Zhao et al., 2014). To determine which domain of MBD7 is required for interaction with ROS5, we generated two truncated MBD7 constructs: BD-MBD (1–231) and BD-CTD (for C-terminal domain; 232–306). About 400 ng of plasmid DNA of each construct was cotransformed into the yeast strain AH109, which was then cultured on two different synthetic dropout media that lacked either Trp and Leu or Trp, Leu, His, and adenine. After 3 d at 28°C, the growth of the strains on the medium lacking Trp and Leu indicates the transformation efficiency, and the fact that positive strains survived on the medium lacking Trp, Leu, His, and adenine indicates that two proteins interact with each other.
Whole-Genome Bisulfite Sequencing and Data Analysis
Genomic DNA was extracted from 7-d-old seedlings using the DNeasy Plant Mini Kit (Qiagen). We performed MethylC-seq with HiSeq 2000 (Illumina) after bisulfite treatment and Illumina library construction.
The reads obtained from sequencing were cleaned with SolexaQA software (Cox et al., 2010) and were mapped to the Arabidopsis reference sequence (TAIR 10) by use of Bismark (Krueger and Andrews, 2011). DMRs were determined as described previously (Zhao et al., 2014). Bins applied in this study were 100 bp from the reference genome. The DMR identification used the standard that the absolute methylation levels of mutants in CG, CHG, and CHH contexts were at least 0.4, 0.2, and 0.1, respectively.
The genome distribution of DMRs was annotated according to GFF files (including gene and TE) downloaded from TAIR 10. The Arabidopsis genome was divided into five parts: TE, gene, upstream, downstream, and intergenic regions. The regions upstream and downstream of a gene were 2 kb away from the transcription start site and the transcriptional end site.
ChIP Assay
Ten-day-old seedlings of wild-type C24 and its transgenic lines generated from the transformation of Pro35S:MBD7-GFP were used for ChIP assays as described previously (Saleh et al., 2008). Anti-GFP antibodies (AB290) were used. The ChIP products were dissolved in 100 μL of water, and 1.5 μL of the solution was used in each quantitative PCR with the specific primers listed in Supplemental Table S3.
Colocalization of MBD7 and ROS5
The full-length cDNA of MBD7 was fused with GFP under the control of the 35S promoter, and the construct was cloned into a modified pCAMBIA1300 vector. A vector expressing mCherry-ROS5 was described previously (Zhao et al., 2014). The procedures of transient expression in tobacco (Nicotiana tabacum) and image acquisition were also described previously (Liu et al., 2010).
The gene accession numbers used in this study are as follows: AT5G59800 (MBD7), AT2G36490 (ROS1), AT3G14980 (ROS4/IDM1), AT1G54840 (ROS5/IDM2), AT5G22010 (RFC1), AT5G46330 (FLS2), AT3G46510 (PUB13), AT4G17870 (PYR1), and AT3G18780 (ACTIN2). We used whole-genome bisulfite sequencing to analyze the methylome of mbd7 mutant plants. The original data set was deposited in the Gene Expression Omnibus database with the accession number SRP051989.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. GUS expression driven by the MBD7 promoter in seedlings growing on MS medium for 2.5, 3, 6, and 10 d.
Supplemental Figure S2. A DNA methylation inhibitor and a DNA replication mutation in rfc1-1 release Pro35S:NPTII gene silencing in mbd7.
Supplemental Figure S3. MBD7 does not interact with ROS4/IDM1 or ROS1.
Supplemental Figure S4. The whole-genome methylation difference between mbd7 and the wild type (C24).
Supplemental Table S1. Hyper-DMRs in mbd7.
Supplemental Table S2. The genomic locations of hyper-DMRs in mbd7.
Supplemental Table S3. Primers used in this study.
Supplementary Material
Glossary
- RdDM
RNA-directed DNA methylation
- BAC
bacterial artificial chromosome
- DMR
differentially methylated region
- TE
transposon element
- ChIP
chromatin immunoprecipitation
- cDNA
complementary DNA
- TAIR
The Arabidopsis Information Resource
- MS
Murashige and Skoog
- T-DNA
transfer DNA
Footnotes
This work was supported by the Natural Science Foundation of China (grant nos. 31330041 and 31421062).
Articles can be viewed without a subscription.
References
- Bartee L, Malagnac F, Bender J (2001) Arabidopsis cmt3 chromomethylase mutations block non-CG methylation and silencing of an endogenous gene. Genes Dev 15: 1753–1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender J. (2012) RNA-directed DNA methylation: getting a grip on mechanism. Curr Biol 22: R400–R401 [DOI] [PubMed] [Google Scholar]
- Cao X, Aufsatz W, Zilberman D, Mette MF, Huang MS, Matzke M, Jacobsen SE (2003) Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Curr Biol 13: 2212–2217 [DOI] [PubMed] [Google Scholar]
- Cao X, Jacobsen SE (2002) Role of the Arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr Biol 12: 1138–1144 [DOI] [PubMed] [Google Scholar]
- Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Tang X, Zhou JM (2008) Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol 146: 368–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox MP, Peterson DA, Biggs PJ (2010) SolexaQA: at-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinformatics 11: 485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier A, Sasai N, Nakao M, Defossez PA (2012) The role of methyl-binding proteins in chromatin organization and epigenome maintenance. Brief Funct Genomics 11: 251–264 [DOI] [PubMed] [Google Scholar]
- Gehring M, Huh JH, Hsieh TF, Penterman J, Choi Y, Harada JJ, Goldberg RB, Fischer RL (2006) DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell 124: 495–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z, Morales-Ruiz T, Ariza RR, Roldán-Arjona T, David L, Zhu JK (2002) ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell 111: 803–814 [DOI] [PubMed] [Google Scholar]
- Gong Z, Zhu JK (2011) Active DNA demethylation by oxidation and repair. Cell Res 21: 1649–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag JR, Pikaard CS (2011) Multisubunit RNA polymerases IV and V: purveyors of non-coding RNA for plant gene silencing. Nat Rev Mol Cell Biol 12: 483–492 [DOI] [PubMed] [Google Scholar]
- He XJ, Chen T, Zhu JK (2011) Regulation and function of DNA methylation in plants and animals. Cell Res 21: 442–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He XJ, Hsu YF, Pontes O, Zhu J, Lu J, Bressan RA, Pikaard C, Wang CS, Zhu JK (2009) NRPD4, a protein related to the RPB4 subunit of RNA polymerase II, is a component of RNA polymerases IV and V and is required for RNA-directed DNA methylation. Genes Dev 23: 318–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F, Andrews SR (2011) Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27: 1571–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE (2009) Molecular biology: dynamic DNA methylation. Science 323: 1568–1569 [DOI] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE (2010) Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 11: 204–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Qian W, Zhao Y, Wang C, Shen J, Zhu JK, Gong Z (2012) Antisilencing role of the RNA-directed DNA methylation pathway and a histone acetyltransferase in Arabidopsis. Proc Natl Acad Sci USA 109: 11425–11430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindroth AM, Cao X, Jackson JP, Zilberman D, McCallum CM, Henikoff S, Jacobsen SE (2001) Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science 292: 2077–2080 [DOI] [PubMed] [Google Scholar]
- Liu Q, Wang J, Miki D, Xia R, Yu W, He J, Zheng Z, Zhu JK, Gong Z (2010) DNA replication factor C1 mediates genomic stability and transcriptional gene silencing in Arabidopsis. Plant Cell 22: 2336–2352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D, Lin W, Gao X, Wu S, Cheng C, Avila J, Heese A, Devarenne TP, He P, Shan L (2011) Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 332: 1439–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke M, Aufsatz W, Kanno T, Daxinger L, Papp I, Mette MF, Matzke AJ (2004) Genetic analysis of RNA-mediated transcriptional gene silencing. Biochim Biophys Acta 1677: 129–141 [DOI] [PubMed] [Google Scholar]
- Matzke MA, Birchler JA (2005) RNAi-mediated pathways in the nucleus. Nat Rev Genet 6: 24–35 [DOI] [PubMed] [Google Scholar]
- Ortega-Galisteo AP, Morales-Ruiz T, Ariza RR, Roldán-Arjona T (2008) Arabidopsis DEMETER-LIKE proteins DML2 and DML3 are required for appropriate distribution of DNA methylation marks. Plant Mol Biol 67: 671–681 [DOI] [PubMed] [Google Scholar]
- Qian W, Miki D, Lei M, Zhu X, Zhang H, Liu Y, Li Y, Lang Z, Wang J, Tang K, et al. (2014) Regulation of active DNA demethylation by an α-crystallin domain protein in Arabidopsis. Mol Cell 55: 361–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian W, Miki D, Zhang H, Liu Y, Zhang X, Tang K, Kan Y, La H, Li X, Li S, et al. (2012) A histone acetyltransferase regulates active DNA demethylation in Arabidopsis. Science 336: 1445–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh A, Alvarez-Venegas R, Avramova Z (2008) An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat Protoc 3: 1018–1025 [DOI] [PubMed] [Google Scholar]
- Scebba F, De Bastiani M, Bernacchia G, Andreucci A, Galli A, Pitto L (2007) PRMT11: a new Arabidopsis MBD7 protein partner with arginine methyltransferase activity. Plant J 52: 210–222 [DOI] [PubMed] [Google Scholar]
- Stroud H, Do T, Du J, Zhong X, Feng S, Johnson L, Patel DJ, Jacobsen SE (2014) Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat Struct Mol Biol 21: 64–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia R, Wang J, Liu C, Wang Y, Wang Y, Zhai J, Liu J, Hong X, Cao X, Zhu JK, et al. (2006) ROR1/RPA2A, a putative replication protein A2, functions in epigenetic gene silencing and in regulation of meristem development in Arabidopsis. Plant Cell 18: 85–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, Gaspan O, Grafi G (2008) The three methyl-CpG-binding domains of AtMBD7 control its subnuclear localization and mobility. J Biol Chem 283: 8406–8411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, Grafi G (2003) Characterization of Arabidopsis thaliana methyl-CpG-binding domain (MBD) proteins. Plant J 34: 565–572 [DOI] [PubMed] [Google Scholar]
- Zemach A, Kim MY, Hsieh PH, Coleman-Derr D, Eshed-Williams L, Thao K, Harmer SL, Zilberman D (2013) The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 153: 193–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, Li Y, Wayburn B, Ben-Meir H, Kiss V, Avivi Y, Kalchenko V, Jacobsen SE, Grafi G (2005) DDM1 binds Arabidopsis methyl-CpG binding domain proteins and affects their subnuclear localization. Plant Cell 17: 1549–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, Paul LK, Stambolsky P, Efroni I, Rotter V, Grafi G (2009) The C-terminal domain of the Arabidopsis AtMBD7 protein confers strong chromatin binding activity. Exp Cell Res 315: 3554–3562 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Xie S, Li X, Wang C, Chen Z, Lai J, Gong Z (2014) REPRESSOR OF SILENCING5 encodes a member of the small heat shock protein family and is required for DNA demethylation in Arabidopsis. Plant Cell 26: 2660–2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Pontes O, Zhu J, Miki D, Zhang F, Li WX, Iida K, Kapoor A, Pikaard CS, Zhu JK (2008) ROS3 is an RNA-binding protein required for DNA demethylation in Arabidopsis. Nature 455: 1259–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. (2009) Active DNA demethylation mediated by DNA glycosylases. Annu Rev Genet 43: 143–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.