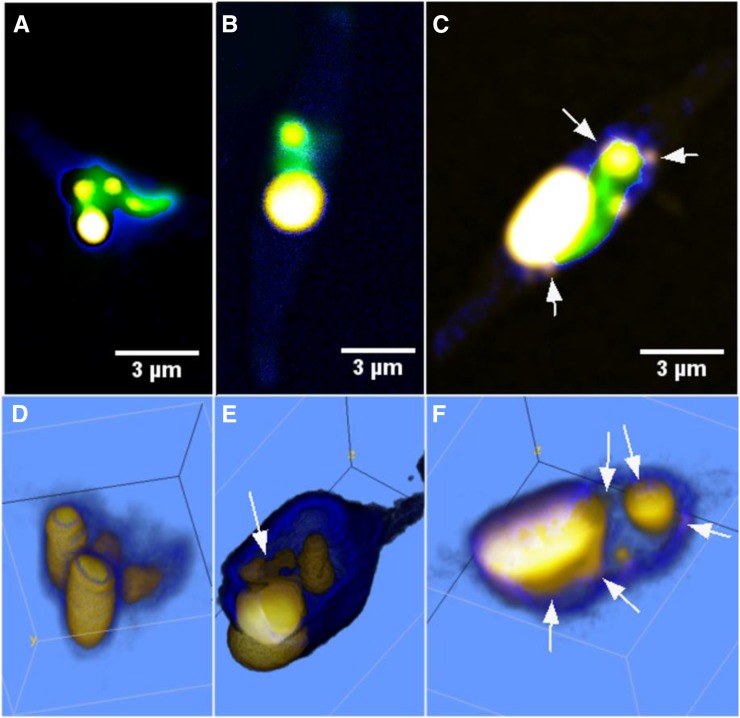
Lipids are accumulated as giant droplets alongside coalescing emerging droplets under excessive lipid storage, in contrast to the multiple micron-sized droplets formed at normal conditions.
Abstract
Microalgae have great prospects as a sustainable resource of lipids for refinement into nutraceuticals and biodiesel, which increases the need for detailed insights into their intracellular lipid synthesis/storage mechanisms. As an alternative strategy to solvent- and label-based lipid quantification techniques, we introduce time-gated coherent anti-Stokes Raman scattering (CARS) microscopy for monitoring lipid contents in living algae, despite strong autofluorescence from the chloroplasts, at approximately picogram and subcellular levels by probing inherent molecular vibrations. Intracellular lipid droplet synthesis was followed in Phaeodactylum tricornutum algae grown under (1) light/nutrient-replete (control [Ctrl]), (2) light-limited (LL), and (3) nitrogen-starved (NS) conditions. Good correlation (r2 = 0.924) was found between lipid volume data yielded by CARS microscopy and total fatty acid content obtained from gas chromatography-mass spectrometry analysis. In Ctrl and LL cells, micron-sized lipid droplets were found to increase in number throughout the growth phases, particularly in the stationary phase. During more excessive lipid accumulation, as observed in NS cells, promising commercial harvest as biofuels and nutritional lipids, several micron-sized droplets were present already initially during cultivation, which then fused into a single giant droplet toward stationary phase alongside with new droplets emerging. CARS microspectroscopy further indicated lower lipid fluidity in NS cells than in Ctrl and LL cells, potentially due to higher fatty acid saturation. This agreed with the fatty acid profiles gathered by gas chromatography-mass spectrometry. CARS microscopy could thus provide quantitative and semiqualitative data at the single-cell level along with important insights into lipid-accumulating mechanisms, here revealing two different modes for normal and excessive lipid accumulation.
The accumulation of lipids in microalgae is currently a field of intense research: with their high photosynthetic efficiency and rapid growth rates, these organisms hold great potential both for sustainable production of biofuels (Chisti, 2007) and as a nutrition source (de Jesus Raposo et al., 2013). As in all living cells, lipids in microalgae are present in membranes, such as the plasma and organelle membranes. Some microalgae also accumulate lipids, mainly triacylglycerols, in intracellular droplets (De Martino et al., 2011; White et al., 2012). One such microalga is Phaeodactylum tricornutum, a unicellular photoautotrophic diatom and a well-studied model organism. It has a sequenced genome and is known to produce long-chain n-3 polyunsaturated fatty acids (PUFAs), eicosapentaenoic acid (EPA), and small amounts of docosahexaenoic acid (DHA; Alonso et al., 1998). The long-chain n-3 PUFA-producing properties has made it particularly interesting within the areas of functional food and nutraceuticals. Under conditions of nitrogen starvation, P. tricornutum accumulates larger amounts of fatty acids, albeit of the more saturated nature (Yongmanitchai and Ward, 1991). However, in response to low irradiance, P. tricornutum has been reported to increase its content of PUFAs, especially EPA (Thompson et al., 1990). In order to optimize strain selection and algal cultivation conditions in relation to lipid accumulation/lipid profile, accurate tools to quantify total lipids as well as the degree of unsaturation are required.
Solvent extractions followed by methylation, gas chromatography coupled to flame ionization detection, and gas chromatography-mass spectrometry (GC-MS) detection belong to the standard techniques used for algal lipid analysis. However, these are cumbersome and require relatively large amounts of solvents and sample. The resulting quantitative information on lipid amounts is related to total cell mass, which may introduce artifacts, as the cell mass is also influenced by other metabolic parameters. Furthermore, these bioanalytical techniques provide rough population averages without information on the intracellular location or distribution. As an alternative, microscopy techniques are increasingly being used, with the benefit that lipid droplets can be evaluated directly in single cells with high precision. Fluorescence microscopy is the most widespread technique, relying on lipid-specific fluorescent markers: Nile Red is commonly used, but its poor permeability through the cell walls causes difficulties, as does its nonspecific binding (Chen et al., 2009). Other fluorophores like BODIPY 505/515 also have been suggested for live-cell studies (Cooper et al., 2010; Wong and Franz, 2013). Still, invasive techniques are needed, requiring solvents to facilitate the transport of the labeling molecules into the cell, potentially inducing stress responses that may affect cell metabolism. Furthermore, there is little knowledge available on how the accumulation of lipophilic dyes in lipid droplets and the fluorescence emission are influenced by environmental conditions such as temperature, pH, and deposited light doses. It has also been shown that the fluorescence intensity emitted from the dyes cannot be related directly to the local lipid concentration, excluding quantitative measurements (De la Hoz Siegler et al., 2012). In algae/plant cell biology, the applicability of fluorescence microscopy is also limited due to the fact that algae/plant cells generate strong autofluorescence, potentially interfering with the lipid/fatty acid signals. In order to take algal lipid quantification one step further, microscopy techniques, not being dependent on exogenous fluorophores and allowing efficient separation of the lipid/fatty acid signal from the autofluorescence, are desirable.
In label-free coherent anti-Stokes Raman scattering (CARS) microscopy, images are formed by probing intrinsic molecular vibrations through a nonlinear four-wave-mixing process (Cheng and Xie, 2004). Briefly, the frequency difference of two coherent near-infrared excitation beams (the pump beam at shorter wavelengths and the Stokes beam at longer wavelengths) are tuned to match the frequency of the target molecular vibration. As a result, resonant oscillators are coherently driven in the sample focal volume and an enhanced blue-shifted CARS signal is generated. Due to the nonlinear nature of the CARS process, emission is generated only in the high-intensity region of focused laser beams, allowing optical sectioning of the specimen. By tuning the frequency difference of the excitation beams to match the resonance frequency of carbon-hydrogen (C-H) vibrations, especially abundant in lipids, three-dimensional images of lipids can be recorded without any staining (Enejder et al., 2010). As the CARS emission scales with the square of the concentration of C-H bonds, quantitative data on intracellular amounts of lipids can be extracted (Cheng and Xie, 2004). However, cells with chromophores, such as algae and plant cells, generate exceptionally strong two-photon fluorescence, the spectral tails of which tend to bleed through the most efficient optical filters typically used for separation of the CARS signal. As an alternative approach, we have incorporated a time-correlated single-photon counting system (Enejder et al., 2010), allowing us to distinguish the long-decay fluorescence component from the instant CARS signal by time gating.
The capability of conventional CARS imaging for microalgae was recently demonstrated with proof-of-principle data, showing that individual, larger lipid droplets can be resolved visually, in contrast to conventional Raman microscopy (He et al., 2012). In this study, we introduce CARS microscopy with time-gated detection, also enabling the identification of subpicogram lipid-rich regions in the vicinity of strongly autofluorescent chloroplasts. This is particularly important because cellular storage lipids in algae are primarily synthesized in the chloroplasts and then budded off from the envelope membranes as nascent lipid droplets (Fan et al., 2011). Hence, time-gated CARS microscopy paves the way for high-precision quantification of the complete intracellular distribution of lipid stores, including the emerging droplets within and adjacent to chloroplasts. We show the feasibility of the approach for quantitative lipid analysis of large populations of living P. tricornutum cultivated under three different growth conditions: a control (Ctrl) group, a light-limited (LL) group to increase the EPA level, and a nitrogen-starved (NS) group to increase total lipid accumulation. We studied the lipid metabolism in approximately 100 cells per category over time and report detailed visual information on the dynamics of lipid droplet formation throughout a life cycle from budding droplets to, in some cases, giant lipid stores. We show that biologically relevant quantitative and qualitative data on intracellular lipid stores can be extracted at a precision of less than 1 pg cell−1 despite adjacent chromophores. Data extracted from the CARS images are further compared with solvent extraction and quantification of fatty acids using GC-MS. We further illustrate the potential to assess whether the different growth conditions promote the synthesis of more PUFAs by detecting shifts in lipid fluidity and saturation per individual lipid droplet from CARS C-H vibration ratio images.
RESULTS
Algae Growth and Viability during CARS Microscopy
In the cultures monitored by CARS microscopy, the algae grew exponentially for the first few days, with maximal specific growth rates of 0.003 to 0.005 h−1, similar for all types of cells. The final cell concentration was 2.3 × 107 cells mL−1 for Ctrl cultivation, 1.9 × 107 cells mL−1 for LL cultivation, and 1.5 × 107 cells mL−1 for NS cultivation at the last CARS measurement day when the cultures were in stationary phase.
The viability of the cells during CARS imaging was assessed by TO-PRO-1-iodide (Life Technologies Europe). It fluoresces at 531 nm when it penetrates dead cells and interacts with DNA. No increase in fluorescence was observed in cells imaged by CARS microscopy and treated with TO-PRO-1-iodide, indicating that the excitation powers used (approximately 60 mW in total before the objective and approximately 6 mW at the sample) allow studies of living cells. However, the light-harvesting system of the algae was affected by the excitation light, as assessed by collecting the two-photon-excited (TPE) autofluorescence (red and green) emitted by the chloroplasts. A gradual increase in the green autofluorescence was observed during image acquisition, together with a decrease in the red autofluorescence (data not shown), indicating a functional but degrading chloroplast protection system usually activated at high light intensities (Frank et al., 1994). No effects on lipid droplet morphology or content were observed during the image acquisition time (approximately 30–50 min per three-dimensional [3D] stack, consisting of approximately 30–50 two-dimensional images).
CARS Microscopy of Intracellular Lipid Droplet Distributions
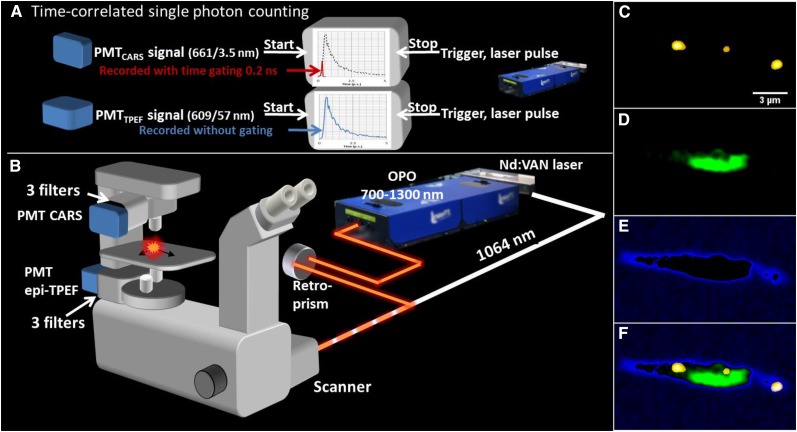
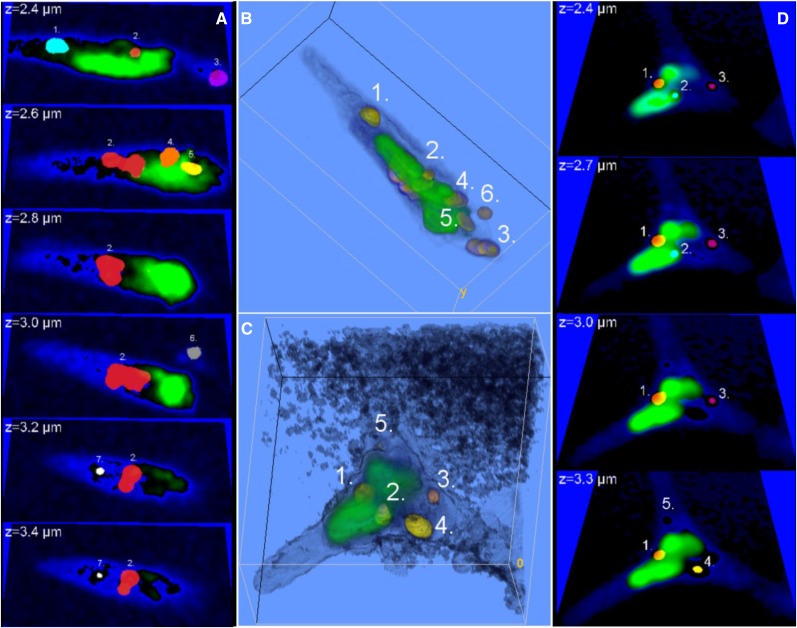
By complementing the spectral filtering with a short gating time (0.2 ns) for the recording of the CARS signal (for a schematic drawing of the time-gated CARS microscope, see Fig. 1, A and B), efficient separation was achieved also in regions with a strong contribution of the autofluorescence from the chloroplasts. By tuning to the symmetric C-H vibration at the frequency 2,845 cm−1, an enhanced CARS signal was collected from the lipid stores in the alga P. tricornutum (for a fusiform cell grown under Ctrl conditions, see the droplets in the gated CARS image in Fig. 1C). In order to investigate the position of the lipid droplets relative to the chloroplast(s), simultaneous TPE fluorescence images were collected pinpointing the chloroplast (color coded in green in Fig. 1D). As a guide for the eye, the outline of the cell, including the cell membrane, was deduced by low-intensity thresholding the CARS image, here color coded in blue (Fig. 1E). The overlay of the three images is shown in Figure 1F, illustrating the successful identification of lipids, including a small, central lipid droplet in the immediate vicinity of the chloroplast. However, this image merely illustrates the lipid distribution in a two-dimensional section of the cell. To get the full picture of the intracellular content and the distribution of lipid stores, 3D images are vital. The corresponding 3D distribution in the cell shown in Figure 1 is shown as a series of CARS/TPE fluorescence overlay images in Figure 2A at depths from 2.4 µm (the same plane as in Fig. 1) to 3.4 µm, with a separation of 0.2 µm. The ImageJ 3D object counter was used to link regions in each image plane that belong to the same lipid particle, identified by unique numbers and color codes. For instance, it can be noted that the lipid droplet that appeared smallest in Figure 1 (lipid droplet 2 in Fig. 2A, color coded in red) rather is the peripheral cross section of a large lipid droplet, spanning almost the full depth of the cell. Using the ImageJ 3D particle counter, the total lipid volume in the cell was 2.7 µm3 (2.4 pg; calculated using the density of 0.9 g mL−1 for vegetable oil) distributed over seven lipid droplets with individual volumes ranging from 0.04 to 1.8 µm3 (0.03–1.6 pg). A 3D rendering is shown in Figure 2B, illustrating that most lipid stores are located within or in close proximity to the chloroplast. While the number of droplets at the start was unusually high for the Ctrl cell category, the total volume of lipids was representative (for Ctrl) after approximately 1 week of culture. In Figure 2, C and D, the corresponding distributions of lipid stores and chloroplasts in one of the few triradiate cells are shown (Ctrl growth conditions). Five lipid droplets (each identified by a number) can be distinguished, all of which are found in close proximity to two chloroplast regions located in the central part of the cell, in contrast to the spatially elongated distribution observed in the fusiform cell.
Figure 1.
A, Schematic illustration of the time-gated CARS microscope allowing efficient separation of TPE fluorescence from chloroplasts in algae/plant cells using time-correlated single-photon counting. A neodymium vanadate pump laser generates the Stokes beam (1,064 nm) and seeds the optical parametric oscillator (OPO). The Stokes and pump/probe (typically at 816.9 nm) pulse trains are overlapped in time and space and coupled into the inverted microscope through a mirror scanner. Signals are recorded by single-photon counting photomultiplier tubes (PMTs) after spectral filtering (CARS in transmission mode, TPE fluorescence in epi mode) and processed by time-correlated single-photon counting. The instantly emitted CARS photons (spectrally filtered at 665/7.5 nm) are recorded during a gating time of 200 ps (red), whereas the full time decay is recorded for the TPE fluorescence (filtered at 684/24 nm; blue). The decay curve is symbolic. C to F, Spectrally filtered and gated images of algae grown for 159 h under Ctrl conditions. Shown are a CARS microscopy image (C-H vibration of 2,845 cm−1) of intracellular lipid droplets (yellow; C), TPE fluorescence of chloroplast (green; D), thresholded low-intensity CARS signal from the cell soma illustrating the outline of the cell (blue; E), and the overlay (F) of images C to F illustrating the location of the lipid droplets throughout the cell soma and in relation to the chloroplast.
Figure 2.
3D information on the lipid droplet distribution in fusiform and triradiate cells grown under Ctrl conditions for 159 h. Lipid droplets are identified and counted by the ImageJ 3D object counter. Object count maps (A and D; each lipid droplet is color coded in cyan, red, magenta, orange, or yellow and numbered throughout the z-stack) are overlaid on the cell soma (blue) and chromophore (green) images at depths spanning throughout the cells. The images span areas of 15.6 × 9.1 µm2 (A) and 12.3 × 12.9 µm2 (D). The volumes of the individual lipid droplets range between 0.01 and 1.8 µm3 (approximately 0.005–1.6 pg), and the total lipid volumes are 2.6 µm3 in A and 1.2 µm3 in D (approximately 2.3 and 1.1 pg). The corresponding 3D renderings are shown in B and C. They reveal that most lipid droplets are located in the vicinity of the chloroplasts. The patchy pattern in C originates from the nonresonant CARS signal from the medium, as also can be noted in D.
Lipid Droplet Accumulation and Morphology versus Culture Time
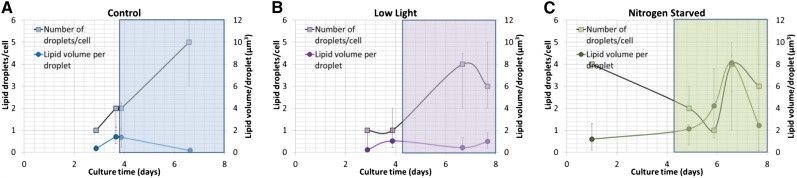
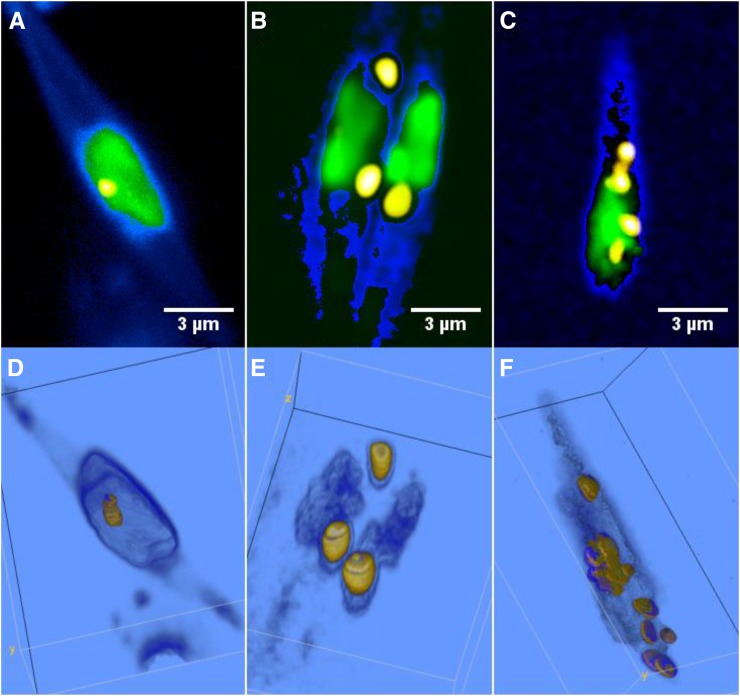
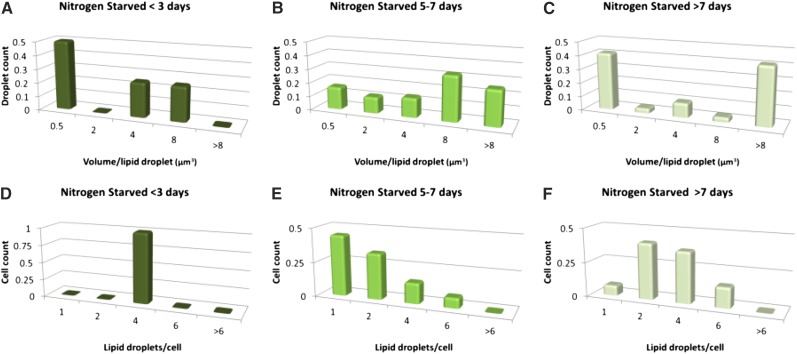
Lipid droplet formation in algae was monitored by CARS/TPE fluorescence microscopy during approximately 1 week under Ctrl, LL, and NS culture conditions for 98 (Ctrl), 102 (LL), and 60 (NS) cells. Under Ctrl and LL conditions, an increase in the number of lipid droplets (Fig. 3, A and B, squares) from a single lipid droplet per cell to three to nine droplets per cell was observed after 1 week of culture; most of these droplets were formed when entering the stationary phase after approximately day 2 to 3 (Ctrl) or day 4 (LL). This increase in the number of lipid droplets (and decrease in median volume due to budding droplets) was preceded by a slight increase in volume per lipid droplet throughout the exponential phase (Fig. 3, A and B, circles) from 0.34 to 1.4 µm3 per droplet for Ctrl and from 0.2 to 1 µm3 per droplet for LL. Representative CARS/TPE fluorescence images and volume renderings (here shown for the Ctrl category) illustrate this behavior in Figure 4, starting with a single, small lipid droplet per cell (Fig. 4, A and D), which increases slightly in volume during the exponential phase (Fig. 4, B and E; two cells with few lipid droplets) and then buds off to multiple, smaller lipid droplets (Fig. 4, C and F). The NS category showed a significantly different behavior in that cells exhibited several lipid droplets already after 1 d of growth while still being in the exponential phase (Fig. 3C, squares). The number of lipid droplets then decreased to a median value of one per cell at day 6 when entering the stationary phase, although accompanied by a significant increase in the volume per lipid droplet (median value of 4.2 µm3 at day 6 compared with 1.2 µm3 at day 1, and the quartiles of the two populations are separated; Fig. 3C). This indicates that multiple, micron-sized lipid droplets here coalesce to larger lipid droplets in the stationary phase. After 1 week of growth, the NS cells seemed to contain multiple (approximately four per cell), large (approximately 8 µm3) lipid droplets. However, the large 75% quartile values motivated a closer look at the data (Fig. 5). Figure 5, A, B, D, and E, confirms that multiple, normal-sized lipid droplets tend to coalesce during the stationary phase for NS conditions; however, rather than into multiple medium-sized lipid droplets, they seem to coalesce into a single giant lipid droplet with a volume from 8 to 47 µm3. This is followed by the appearance of new budding lipid droplets, confirmed in the histograms (Fig. 5, C and F) by a dual population, one of multiple, micron-sized droplets together with a single giant lipid droplet.
Figure 3.
Number of lipid droplets per cell (squares) and volume per lipid droplet (circles) versus culturing time for Ctrl (A), LL (B), and NS (C) conditions compiled from CARS microscopy images of 260 algae. Data show median and 25%/75% quartile values. Ctrl and LL cells host no more than one to two smaller (median values of 0.2–1.4 µm3) lipid droplets per cell during the exponential phase, followed by a gradual accumulation of up to nine similar-sized droplets per cell during the stationary phase (shaded regions in graphs). The NS cells accumulate multiple small-sized lipid droplets already during the exponential phase, which then coalesce during the stationary phase (decrease in number of droplets accompanied by increase in droplet volume). Smaller lipid droplets continue to form, and in the later stationary phase cells typically contain one giant lipid droplet surrounded by multiple submicron-sized droplets (low median lipid volume at approximately day 8 due to the budding lipid droplets, although with a 75% quartile of 15 µm3 due to the giant lipid droplet).
Figure 4.
Lipid accumulation and characteristic lipid droplet morphology of algae grown under Ctrl conditions for 3 d (A; exponential phase), 4 d (B; late exponential phase, two cells), and 7 d (C; stationary phase). CARS (lipid droplets: high C-H signal, yellow; cell soma: low C-H signal, blue) and TPE fluorescence (green) results are shown. In the early exponential phase, most cells have a single, small lipid droplet (0.27 µm3 in A), which with increasing culturing time increases slightly in volume (1.6–3.4 µm3 in B). During the stationary phase, cells instead show multiple, smaller lipid droplets (seven droplets in the range 0.036–1.8 µm3 in C). The corresponding volume renderings are shown in D to F. A similar phenomenon is observed for LL culture conditions. In B, the nonresonant CARS signal from the medium is thresholded in order to distinguish the cells better.
Figure 5.
Histograms showing the lipid droplet volume distributions (A–C) and droplet count distributions (D–F) for NS algae during exponential (A and D), early stationary (B and E), and later stationary (C and F) phases. They illustrate the shift from many submicron-sized droplets per cell (A and D) to a few larger droplets (B and E) when entering the stationary phase as a result of droplet fusion. Small lipid droplets continue to form, observed as a dual droplet population in C.
In Figure 6, representative CARS/TPE fluorescence images and volume renderings of NS cells are shown, illustrating the phenomenon underlying the dual population of lipid droplets shown in the histograms in Figure 5 (i.e. lipid droplet fusion). Already after 24 h of culturing, cells exhibit multiple micron-sized lipid droplets, as shown in Figure 6, A and D. After 6 d of culturing, there are clear signs of lipid droplet fusion, in particular in the volume renderings (Fig. 6, B and E). After 8 d of NS culturing conditions, the cells typically host one huge lipid droplet surrounded or docked by several smaller lipid droplets (Fig. 6, C and F). This wide range of lipid droplet volumes underlies the large quartile values in Figure 3C. The volume of the large droplets is in the range 8 to 47 µm3, while the satellite droplets are of more normal size, typically below 1 µm3.
Figure 6.
CARS (lipid droplets: high-density C-H signal, yellow; cell soma: low-density C-H signal, blue) and TPE fluorescence (green) overlay microscopy images of the lipid accumulation and morphology of NS cells after 1 d (A; exponential phase), 6 d (B; stationary phase), and 8 d (C; late stationary phase) of culturing. Corresponding volume renderings are shown in D to F. Images illustrate the phenomena described in Figures 3C and 5: many, smaller lipid droplets already present in the exponential phase (four lipid droplets in the range 0.07–4.1 µm3) fusing into fewer and larger lipid droplets in the stationary phase (two droplets of 1.3 and 7.8 µm3). Note the lipid droplets docking to the already oversized droplet in E. The volumes of these docking droplets are included in the volume count of the main droplet, as they start to coalesce and could not be reliably segmented. After 1 week during the late stationary phase, the cells typically host one giant lipid droplet surrounded by docking and satellite lipid droplets, as indicated by the arrows in C and F. The volume of the giant lipid droplet here is 20.1 µm3. The color coding of the CARS signal was optimized to distinguish the weaker emission from the smaller, docking lipid droplets. The color-code transfer function of the low-thresholded CARS signal in the volume rendering in E was set to 3D fill mode in order to highlight the weak cell soma signal in contrast to the two-dimensional gradient mode used in the other images.
Median and quartile values of lipid droplet sizes and numbers of lipid droplets per cell are summarized for each category (Ctrl, LL, and NS) in Table I. The median droplet sizes and counts were used to calculate the total lipid content per cell for the Ctrl and LL categories. This is a fair estimate due to the small cell-to-cell variation within each category and phase. For the NS category, the total lipid content had to be summed up for each single cell, and a median value was formed. This was necessary because the NS category consisted of a dual population in the stationary phase: several smaller lipid droplets and one giant droplet per cell. Volumes were converted into masses using the density of vegetable oil. The Ctrl category showed an increase in lipid weight from 0.31 (exponential phase) to 2 (stationary phase) pg cell−1 (i.e. by a factor of 6.5), the LL category showed a similar increase from 0.2 to 1.6 pg cell−1, whereas the primary lipid accumulation in the NS category seemed to take place already during the first culturing day followed by a minor accumulation from 10.4 to 11.9 pg cell−1 when entering the stationary phase. Compared with the Ctrl condition, nitrogen starvation induced an approximately 30-fold lipid accumulation during the early exponential phase.
Table I. Summary of CARS volume measurements.
The weight of lipid per cell was calculated on the assumption that the density of the lipid droplets is 0.9 g mL−1 (density of vegetable oil). Median and interquartile range (Q25%–Q75%) are shown.
| Growth Condition | Lipid Volume per Droplet |
Lipid Droplet Count per Cell |
Lipid Weight per Cell |
|||||
|---|---|---|---|---|---|---|---|---|
| Median (Q25%–Q75%; Days 1–3) | Median (Q25%–Q75%; Days 3–7) | No Droplet (Days 1–7) | Median (Q25%–Q75%; Days 1–3) | Median (Q25%–Q75%; Days 3–7) | No Cells (Days 1–7) | Median (Days 1–3) Calculated | Median (Days 3–7) Calculated | |
| µm3 | pg | |||||||
| Ctrl | 0.34 (0.17–0.63) | 1.1 (0.9–1.9) | 115 | 1 (1–1) | 2 (1–3) | 98 | 0.31 | 2.0 |
| LL | 0.22 (0.12–0.44) | 0.87 (0.59–1.7) | 108 | 1 (1–1) | 2 (1–4) | 102 | 0.20 | 1.6 |
| NS | 1.2 (1.0–2.6) | 3.3 (2.8–8.5) | 87 | 4 (4–4) | 2 (1–3) | 60 | 10.4 | 11.9 |
Total Content of Fatty Acids as Analyzed by GC-MS
The mean content of total identified fatty acids (95% or greater of the total peak area) in cells from the various growth conditions was calculated to be 82 mg g−1 dry weight in Ctrl cells, 74 mg g−1 dry weight in LL cells, and 114 mg g−1 dry weight in NS cells based on GC-MS measurements (Table II).
Table II. Major fatty acids analyzed by GC-MS in P. tricornutum under three different growth conditions at the end of the CARS experiment, culture: Ctrl (at 93 h), LL (at 93 h), and NS (at 141 h).
Data are from analyses of algae obtained during the last of the three culturing occasions, and the second column shows sd values of extraction replicates (n = 3).
| Fatty Acid | Ctrl | sd | LL | sd | NS | sd |
|---|---|---|---|---|---|---|
| mg g−1 dry algae | ||||||
| Total detected | 85.3 | 0.50 | 77.4 | 4.91 | 116.8 | 1.02 |
| Sum identified | 81.7 | 0.26 | 74.1 | 4.35 | 113.7 | 0.55 |
| C14:0 | 7.6 | 0.07 | 6.5 | 0.26 | 9.4 | 0.23 |
| C15:0 | 0.3 | 0.01 | 0.3 | 0.02 | 0.5 | 0.00 |
| C16:0 | 15.2 | 0.08 | 12.4 | 0.13 | 28.6 | 0.45 |
| C16:1 n-7 | 21.1 | 0.06 | 18.3 | 0.32 | 39.2 | 0.64 |
| C16:2 n-4 | 2.1 | 0.02 | 2.4 | 0.02 | 1.8 | 0.03 |
| C16:3 n-4 | 4.3 | 0.04 | 4.8 | 0.05 | 2.5 | 0.05 |
| C18:0 | 0.7 | 0.01 | 0.4 | 0.01 | 1.1 | 0.02 |
| C18:1 n-9 | 2.3 | 0.02 | 2.1 | 0.01 | 3.8 | 0.06 |
| C18:1 n-7 | 0.6 | 0.01 | 0.4 | 0.02 | 0.6 | 0.02 |
| C18:2 n-6 | 1.6 | 0.03 | 1.3 | 0.03 | 1.7 | 0.05 |
| C18:3 n-6 | 0.5 | 0.02 | 0.5 | 0.01 | 0.6 | 0.03 |
| C18:3 n-3 | 0.2 | 0.00 | 0.1 | 0.01 | 0.2 | 0.00 |
| C18:4 n-3 | 0.8 | 0.04 | 0.7 | 0.06 | 0.9 | 0.03 |
| C20:4 n-6 | 0.5 | 0.01 | 0.7 | 0.05 | 0.5 | 0.01 |
| C20:5 n-3 | 21.1 | 0.10 | 20.1 | 0.30 | 19.6 | 0.22 |
| C22:6 n-3 | 1.6 | 0.01 | 1.5 | 0.04 | 1.4 | 0.03 |
| C24:0 | 1.5 | 0.01 | 1.6 | 0.07 | 1.3 | 0.04 |
| PUFA-total fatty ratio | 0.40 | 0.43 | 0.26 | |||
The three major fatty acids in P. tricornutum were determined to be EPA (C20:5 n-3), palmitic acid (C16:0), and palmitoleic acid (C16:1 n-7). EPA was present in algae from all growth conditions at concentrations of 20 to 21 mg g−1. In the NS samples, palmitic (C16:0) and palmitoleic (C16:1) acid contents was 27 and 39 mg g−1, respectively, compared with approximately 14 and 20 mg g−1 in the Ctrl and LL groups. In the NS group, fewer PUFAs were present relative to the fatty acids in total, resulting in a lower PUFA:total fatty acid ratio (0.26) compared with 0.40 and 0.43 for Ctrl and LL, respectively (Table II). There was no significant difference in the PUFA-total fatty acid ratio between the LL and Ctrl groups.
Correlation between CARS and GC-MS Lipid Analysis
A simple model was formulated in order to relate CARS and GC-MS data; the GC-MS analysis provides the total amount of fatty acids per 1 g of dry cell content, while CARS microscopy provides the volume of lipid droplets contained in one cell. The following assumptions were made: (1) the average dry content (my) of the cell, excluding lipids, is constant for the three categories; (2) the dry content of lipids located outside lipid droplets (ml) varies insignificantly with culture conditions (constant); (3) CARS microscopy provides data on the average content of lipids in the droplets only (mld = ρVld, where Vld is the average total volume of lipid droplets in a cell and ρ is the lipid density); and (4) GC-MS provides data on the ratio (rl) of the dry mass of all fatty acids (mld + ml) to the overall dry mass of the cell (mld + ml + my):
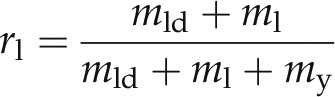 |
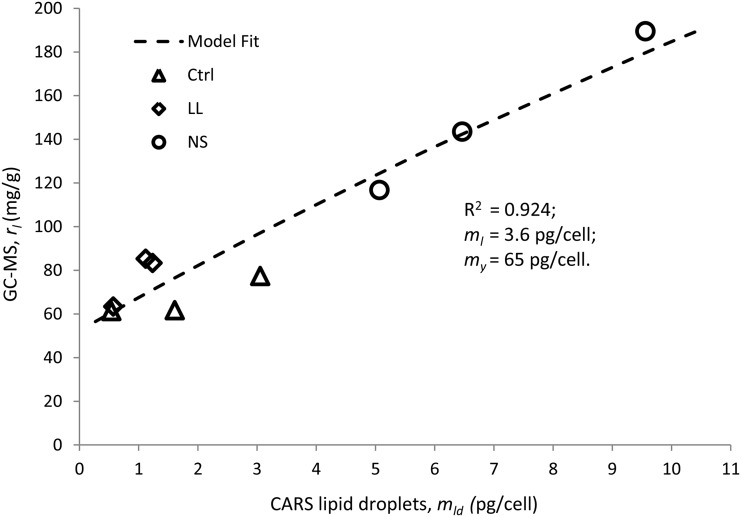
Model fit of the data using the Microsoft Office Excel Solver add-in (generalized reduced gradient nonlinear solving method) resulted in good correlation with parameter values ml = 3.6 pg cell−1 and my = 65 pg cell−1 (r2 = 0.924). Experimental data (symbols) and model fit (dashed line) are shown in Figure 7.
Figure 7.
Comparison of lipid amounts obtained from C-H CARS microscopy and GC-MS data. The good correlation indicates that the lipid droplets are responsible for the primary change in intracellular lipid content. Symbols represent experimental data of three repeated experiments for each growth condition: Ctrl, triangles; LL, diamonds; and NS, circles. The weight of lipids per cell was calculated on the assumption that the density of the lipid droplets is 0.9 g mL−1. The model (dashed line) assumes constant dry content per cell (my; excluding lipids) and dry content of nondroplet lipids (ml).
CARS Microspectroscopy of Lipid Fluidity/Unsaturation
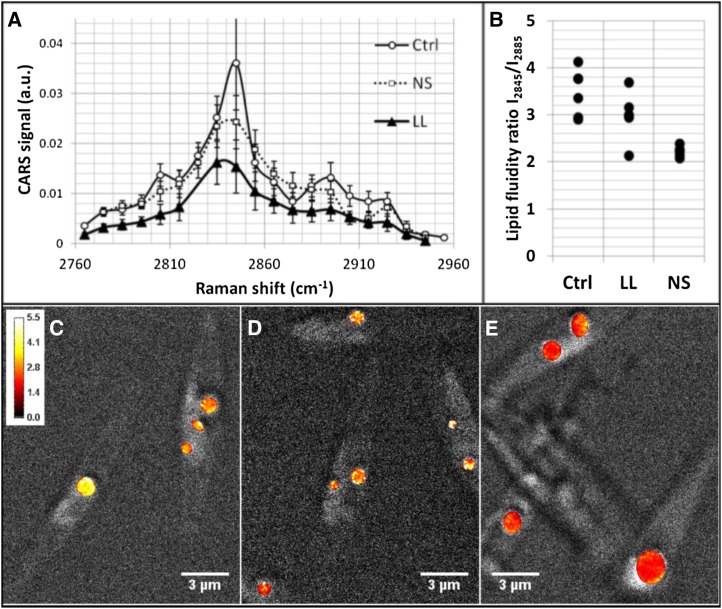
The single-vibration CARS images were complemented by spectral scans of CARS images containing information on all C-H vibrations with the purpose to evaluate differences in lipid fluidity and, hence, saturation between the categories by relating the symmetric (2,845 cm−1) to the asymmetric (approximately 2,880–2,895 cm−1) C-H vibrations of CH2 group. The ratio has been shown to decrease with decreasing fluidity, hence with increasing lipid saturation (Rinia et al., 2008). Spectra were collected from four to six lipid droplets per growth condition (Fig. 8A). Significantly lower lipid asymmetry ratios (2.2 ± 0.1) were obtained for the NS group compared with 3.4 ± 0.5 for the Ctrl group, and populations were separated (Fig. 8B). The LL category showed an intermediate state and a ratio of 3 ± 0.5. Ratio images were computed and overlaid the gray-scaled CARS microscopy in Figure 7, C to E, clearly showing lower values for the NS cells. Altogether, these findings indicate lower lipid fluidity for droplets in NS cells and, hence, larger amounts of saturated lipids.
Figure 8.
CARS microspectroscopy. A full spectral series of CARS microscopy images covering the C-H vibrational region 2,760 to 2,960 cm−1 was scanned for Ctrl, LL, and NS cells. Normalized and power-compensated CARS signals averaged over lipid droplets are shown as spectra in A. a.u., Arbitrary units. Average lipid fluidity ratios (I2845/I2885) for lipid droplets in the different cell categories are plotted in B, illustrating a significantly lower value for NS compared with Ctrl. This is further confirmed in the fluidity ratio images in C to E, where the lipid droplets in Ctrl (C) and LL (D) appear in yellow in contrast to the lipid droplets for NS in E, which all are color coded in red (see intensity bar in C). All measurements were conducted at room temperature. Lower lipid fluidity is reported for saturated lipids at constant temperatures. This indicates that NS cells contain larger amounts of saturated lipids.
DISCUSSION
In the attempt to identify algal strains and environmental conditions that promote the accumulation of large amounts of intracellular lipids to be harvested as biofuels and nutraceuticals, a growing body of studies involving batch (Breuer et al., 2012; Griffiths et al., 2012) or high-throughput (Lee et al., 2013) lipid screening techniques has appeared. For instance, nitrogen-limited conditions have repeatedly been shown to promote significant accumulation of neutral lipids for a wide collection of algal strains (Breuer et al., 2012; Griffiths et al., 2012; Erickson and Jimenez, 2013), and elevated PUFA levels have been reported at low light exposure (Thompson et al., 1990). This raises questions on the underlying cellular mechanisms, knowledge that could guide scientists to identify growth conditions and protocols that more efficiently target the intracellular pathways involved in lipid accumulation. Hence, an increasing interest in emerging single-cell imaging technologies has been raised within the algae research community. However, the strong pigmentation of algae makes it a challenging object for optical microscopy; the probe light is readily absorbed, potentially activating/damaging the cells, and the strong autofluorescence makes it difficult to reliably distinguish lipid stores from the pigmented organelles. Here, we introduce time-gated CARS microscopy for quantitative and semiqualitative analysis of lipid stores based on inherent C-H vibrations, requiring no exogenous reporter molecules, in algae. In this approach, a tight time gating (approximately 200 ps) together with narrow band-pass filters facilitate the separation of the TPE autofluorescence from the CARS signal and, hence, imaging of lipid droplets also in the vicinity of the chloroplasts. This study was designed to investigate the power of time-gated CARS microscopy as a tool to provide quantitative information on algae lipid levels, information on lipid store morphology and the underlying kinetics, as well as qualitative measures of lipid fluidity as an indirect measure of the degree of unsaturation. Therefore, an important part of the study was to obtain algae hosting lipid stores differing both qualitatively (unsaturation degree) and quantitatively. For this purpose, P. tricornutum was cultivated under Ctrl, LL, and NS conditions and subjected to GC-MS as well as time-gated C-H CARS analyses after harvest.
Fatty Acid Profiles as Determined by GC-MS
From the GC-MS analyses, we conclude that the lipid profiles of the LL and Ctrl categories were comparatively similar, while the lipid composition of the NS category was significantly different (Table I). The intention with the group of algae grown under LL conditions was to include a study object with elevated PUFA levels. This was done partly based on the research by Thompson et al. (1990), where most of the eight marine phytoplankton species investigated, including P. tricornutum, had their greatest proportion of EPA at low levels of irradiance. Despite previous findings, our GC-MS analyses could not discern any increase in EPA (all approximately 20 mg g−1) or in DHA (all approximately 1.5 mg g−1) at harvest of P. tricornutum grown under LL conditions compared with the Ctrl and NS categories (Table I). A likely explanation could be a low average light dose per cell for all three algae categories in our study compared with Thompson et al. (1990) due to the high cell density obtained (approximately 107 cells mL−1). This is supported by the low specific growth rate, here 0.003 to 0.005 h−1 for all three cultivation conditions, which was 1 order of magnitude lower compared with Thompson et al. (1990). It is likely that inoculating cultures with fewer cells per mL, as well as bubbling air, rather than shaking the cultures, for efficient mass transfer of carbon dioxide, would have resulted in detectable differences in long-chain PUFA content between the three growth conditions.
When comparing the fatty acid patterns of the Ctrl and NS cells in Table I, we conclude that our results agree with previous observations that nitrogen starvation induces an increase in total amounts of fatty acids, in particular saturated and monounsaturated species (C16:0 and C16:1), in P. tricornutum (Yongmanitchai and Ward, 1991; Tonon et al., 2002).
In general, the total amounts of fatty acids detected in P. tricornutum with GC-MS under the different growth conditions (83.3, 77.4, and 116.8 mg g−1 dry algae in Ctrl, LL, and NS conditions, respectively) were in the lower range of what has been reported by others (Fernández-Reiriz et al., 1989; Gatenby et al., 2003). This could partly be explained by the general light and carbon limitations in this study. Another explanation could be that several earlier studies are reporting total lipids in algae based on gravimetric lipid determination, a technique that covers lipids in a broader sense than GC-MS analyses of fatty acid methyl esters (including hydrophobic compounds dissolved in organic solvents, the glycerol backbone of triacylglycerols and phospholipids, as well as the sugar unit of glycolipids). For a comparison with CARS microscopy, primarily probing CH2 vibrations in acyl chains, GC-MS was judged to provide more relevant data.
Altogether, the GC-MS analyses indicate that the lipid profile of the NS category should be significantly distinguished from those of the Ctrl and LL categories in the CARS microscopy measurements, both quantitatively and qualitatively, whereas no major differences can be expected between the Ctrl and LL categories.
Quantitative Algal Lipid Analysis: Comparison of C-H CARS Microscopy and GC-MS
In order to evaluate the precision and sensitivity of lipid quantification using time-gated C-H CARS microscopy, measurements were conducted along with GC-MS analyses on all algae cultures. It was found that a strong correlation existed between lipid volumes in a single cell determined by C-H CARS microscopy and total fatty acids as determined by GC-MS. The total amount of fatty acids per cell, as calculated from the number of cells per mL at harvest, falls into the range of pg per cell. This is a reasonable amount (White et al., 2012) and compares favorably with the lipid volumes obtained by CARS microscopy on the final day of measurement (in the range of 1–5 µm3 cell−1), assuming the density of algal oil to be 0.9 g mL−1. Also, the more thorough comparison of lipid amounts, using a simple model, provided a good fit (r2 = 0.924), despite the simplifying assumptions (e.g. that the primary variations could be attributed to the lipid droplet pool of a single algae and not to variations in the dry mass or the membrane lipid mass). However, model parameter values were higher quantitatively; the total dry mass of a single cell was 65 pg cell−1, in contrast to the measured approximately 20 to 30 pg cell−1. Potential explanations could be losses of lipids and/or overall mass during the extraction for GC-MS analysis, lipid density variations within droplets, as well as the limited spatial resolution of the microscope. Further investigations are needed in order to improve this model.
Lipid Droplet Morphology and Kinetics
While neutral lipid metabolism has been studied extensively in a range of cells and organisms (Murphy, 2012), comparatively little information is available regarding the mechanisms and dynamics of the biosynthesis of lipids in microalgae. Fan et al. (2011) recently showed that microalgae employ a unique pathway for the synthesis of storage lipids: neutral lipids originate primarily from the chloroplasts rather than from the endoplasmic reticulum, as in plant and mammalian cells. By means of electron microscopy, they showed that lipid droplets are found not only in the cytosol but also in the chloroplasts themselves. This observation is confirmed by the three-dimensional colocalization CARS/two-photon fluorescence images, as shown in Figures 2, 4, and 6, revealing that most lipid droplets can be found within or in close proximity to the chloroplasts. This further highlights the importance of a technology capable of visualizing lipid stores in a strongly fluorescent environment for studies of the lipid metabolism in microalgae.
The lipid accumulation in Ctrl and LL groups could temporally be characterized by three phases (Fig. 3, A and B). In the first phase, during exponential growth, regular cell division resulted in a constant number of lipid droplets per cell (i.e. approximately one submicron-sized droplet per cell). At the end of the first 4 d, phase 2 (early stationary phase), lipid accumulation occurred as a result of a slight budding of more lipid droplets and increased lipid droplet volumes, in agreement with Fernández-Reiriz et al. (1989). This is also in agreement with Wong and Franz (2013), who used BODIPY 505/515 combined with laser scanning confocal microscopy. They found that P. tricornutum stores intracellular lipids in one or two large subcellular compartments, which then continuously increase in size when maintaining the cells in the early stationary phase under Ctrl conditions long term. When entering the late stationary phase, the third phase (days 4–6 in this study), the average droplet volumes of Ctrl and LL cells turned smaller again, indicating the budding of new, smaller droplets and/or lipid metabolism, a phenomenon that is characteristic for the stationary phase (Fernández-Reiriz et al., 1989).
The NS category showed a significantly different behavior in that these algae exhibited several lipid droplets already after 1 d of growth. This is in agreement with studies on the oil-producing green alga Chlorella sorokiniana C3, where lipid droplets started to form following 0.5 to 2 d of nitrogen starvation (Zhang et al., 2013). The increased lipid accumulation has been shown to be triggered by the oxidative stress that is built up during approximately 0.5 d of nitrogen starvation, when there are insufficient amounts of nitrogen for protein synthesis and growth (Zhang et al., 2013). Cells acclimate by reducing the photosynthetic rate and instead ramping up the cyclic electron transportation to produce more ATP for triacylglycerol synthesis (Zhang et al., 2013). We further noted that the number of lipid droplets subsequently decreased to a median value of one per cell at day 6, although of a significantly larger volume than droplets at day 1 (Fig. 3C). These data, together with the visual evidence in Figure 6, prove that the accumulation of lipid stores in conjunction with nutritional stress takes place by the coalescence of emerging lipid droplets with the main lipid store, forming a giant lipid droplet, rather than by a gradual accretion. This is in agreement with Wong and Franz (2013), who reported that the number of lipid bodies per cell ranged from one to six in P. tricornutum throughout cultivation, with volumes of individual lipid bodies in a broad range up to 38 µm3 (comparable to 47 µm3 observed in this study). They also described a nonspherical lipid body size, indicating that lipid storage proceeds by merging newly synthesized lipids with an existing lipid body in the cytoplasm. In contrast, the cytosolic lipid droplets, stained by BODIPY PC, in the green alga Chara corallina were not shown to form by fusion (Foissner, 2009). It could be seen that lipid droplets that moved together with, or along, the cortical endoplasmic reticulum cisternae in a cytoskeleton-independent manner collided, but never fused. These findings suggest that the growth of cytosolic lipid droplets in C. corallina occurs by continuous incorporation of neutral lipids. However, the impact of the lipid marker molecule on lipid droplet formation also may play a role. In our study, lipid droplet fusion in P. tricornutum was illustrated without labeling, which ascertains that the natural fusion mechanisms are not affected by labeling molecules. This is crucial, considering the significant impact that the physiochemical environment has on the organization of lipids.
A number of recent proteomic studies of lipid droplets from different animal tissues (Cermelli et al., 2006; Walther and Farese, 2009), Saccharomyces cerevisiae (Athenstaedt et al., 1999), and plants (Jolivet et al., 2004; Katavic et al., 2006) have revealed that droplet fusion as well as lipolytic activity at the lipid droplet surface are regulated by distinct proteins (e.g. perilipins in animals and fungi and oleosin in plants) associated with the lipid droplets. In algae, neither perilipins nor oleosin are found, but other proteins (nonhomologous to perelipin or oleosin) provide similar function, such as Major Lipid Droplet Protein (MLDP; Moellering and Benning, 2010; Davidi et al., 2012) and Lipid Droplet Surface Protein (LDSP; Vieler et al., 2012). These studies show that increased expression of MLDP or LDSP is correlated with triacylglycerol accumulation. However, their role in lipid droplet fusion needs further investigation, where CARS microscopy could provide important insights.
CARS Microspectroscopy of Lipid Fluidity/Unsaturation
Complementing single-vibration CARS images by spectral scans of CARS images revealed lower lipid fluidity for droplets in NS cells, indicating larger amounts of saturated lipids. It compared well with the mass spectrometry data. This finding opens up the possibility of using CARS microspectroscopy to screen lipids with respect to their saturation degree in order to identify local pools of unsaturated lipids at the single cell level or to identify optimal algae harvest time points when producing long-chain n-3 PUFA-rich oils. Tonon et al. (2002) reported how partitioning of DHA and EPA into the triacylglycerol fraction varies throughout the growth cycle. The interpretation of scanned CARS spectra to indirectly assess unsaturation through the analysis of lipid packing, however, requires reliable monitoring of the excitation power. A more efficient approach than scanning each individual vibration would be to collect a full CARS spectrum using a white-light excitation source (Heinrich et al., 2008; Pohling et al., 2014). Rinia et al. (2008) have exploited this possibility for 3T3-L1 cells (i.e. cancer cells that accumulate large amounts of lipids) and have shown that not only lipid fluidity and unsaturation degree varies between lipid droplets in a single cell with access to different fatty acids but also that there are variations within individual lipid droplets.
Limitations with C-H CARS Microscopy
Although proven to be a highly promising tool in algae lipid droplet analysis, a few precautions have to be taken when implementing CARS for specific purposes. It was observed that the light-harvesting system of the algae was affected by the excitation light during CARS measurements, as assessed by collecting the TPE autofluorescence (red and green) emitted by the chloroplasts. This acute stress was of limited importance in our study, as the impact on the lipid metabolism is significantly slower than the image acquisition time. However, for long-term time-lapse CARS measurements on living algae, such stress must be considered, and sufficient time between data acquisition must be planned in order to ascertain the recovery of the light-harvesting systems.
When using CARS microscopy to detect lipid droplets on a single cell, thresholding by visual inspection is used, which can result in the smallest lipid droplets not being detected. Furthermore, a certain amount of C-H vibration is needed to generate a CARS signal, which can result in missing budding lipid droplets and potentially the periphery of droplets. It is possible that these features contributed to the fact that lipid amounts in P. tricornutum grown under Ctrl conditions determined by CARS microscopy were smaller than those determined in previous studies (Fernández-Reiriz et al., 1989).
It also must be stressed that the CARS microscope is a highly sophisticated piece of equipment; thus, despite the promising data shown in this study, high investment costs prevent a full replacement of classic solvent extraction and chromatographic methods for routine total lipid analyses. However, turn-key systems are under development.
CONCLUSION
In conclusion, our results show that time-gated CARS microscopy is a promising technique to study lipid synthesis in microalgae, using P. tricornutum as a model organism. Average changes observed at large population levels by biochemical analyses can be detected with exceptional picogram precision at the subcellular level by merely investigating 20 to 60 cells at minimal sample preparation. CARS microscopy shows that lipid storage in microalgae takes place as an increasing number of isolated micron-sized lipid droplets with culturing time. However, in growth conditions promoting excessive lipid accumulation toward commercial harvest as biofuels and nutritional lipids, lipid droplets coalesce and form a single giant lipid store surrounded by docking, emerging lipid microvesicles. Besides high-precision quantitative and mechanistic information, CARS microspectroscopy provides estimates of lipid fluidity, an important qualitative evaluation parameter closely related to the degree of unsaturation. We envision that time-gated CARS microscopy may significantly contribute to the understanding of lipid synthesis not only in microalgae but in plant cells in general, being a challenge for other microscopy techniques due to their strongly emitting chromophores.
MATERIALS AND METHODS
Cultivation of Microalgae
Phaeodactylum tricornutum GUMACC 2 (elsewhere deposited as UTEX 642) was obtained from the Gothenburg University Microalgae Algal Culture Collection. Conditions for all types of cultivation were kept constant with 12-h-light/12-h-dark cycles of illumination by cool-white fluorescent tubes, gentle shaking of growth flasks (80 rpm), and temperature at 17°C ± 1°C. Cells were initially maintained in f/2 medium augmented with 30 mg L−1 sodium metasilicate (f/2 + Si medium), based on filtered natural seawater (Andersen, 2005), with illumination at 20 to 80 µE m−2 s−1. Light intensities (lux) were measured at the culture’s surface with LT Lutron Lx-101 (Elfa Distrelec) and recalculated to photosynthetic photon flux with a converter (International Light Technologies). One week before starting the main CARS cultivation, single precultures with 200 mL of each growth medium (Ctrl, LL, and NS) in 250-mL tissue culture flasks (with ventilated cap; Sarstedt) were initiated and placed inclined under the light. The Ctrl condition consisted of f/2 + Si medium at 120 to 160 µE m−2 s−1 irradiance, the LL condition consisted of f/2 + Si medium at 15 to 74 µE m−2 s−1 irradiance, and the NS condition consisted of f/2 + Si without the addition of nitrate at 120 to 160 µE m−2 s−1 irradiance. To start the main CARS cultures, cells from precultures were harvested by centrifugation (2,000g for 3 min), transferred into fresh medium (Ctrl, LL, or NS) at high cell density (107 cells mL−1), and incubated at similar conditions as for the respective preculture. Cell growth was monitored for up to 8 d by optical density measurements at 750 nm with a CaryUV 50 Bio spectrophotometer (Varian). After 1 to 6 d, cell samples were taken for CARS microscopy measurement (5-µL culture broth, withdrawn within the first 3 h of the 12-h light cycle). On the last day of CARS microscopy measurements, cells were sampled for biochemical fatty acid analysis by centrifugation (5,000g for 10 min). Between the sampling and gas chromatography analysis, cell pellets were stored at −80°C. Immediately prior to the sampling, the final cell concentration (cells per mL) was determined by counting cells in a disposable C-Chip hemocytometer (Digital Bio). The full experiments (algae cultivation, CARS microscopy, and fatty acid analysis) were repeated three times on separate occasions with minor variations in growth conditions.
The CARS Microscope
An in-house-built nonlinear microscopy setup was used, composed of a picosecond laser source, a laser scanning microscope, and a single-photon-counting detection system. The fundamental (1,064 nm) output of a neodymium vanadate laser (HighQ, picoTRAIN; 7 ps, 76 MHz) served as a Stokes beam. A frequency-doubled (532 nm) output from the laser seeded the optical parametric oscillator (APE Levante Emerald), providing a tunable-wavelength (690–990 nm) pump beam, covering a 700 to 5,000 cm−1 Raman shift region. The pump and Stokes beams were overlapped spatially, using a dichroic mirror (Thorlabs; DMSP1000R), and temporally, by an adjustable delay line in the pump beam path. After the overlap, both beams propagated in a collinear manner and were directed to the mirror-scanning unit (Nikon C1) of the confocal microscope (Nikon Eclipse TE2000-E). The average power of each beam was controlled by a half-wave plate and polarizing beam splitter cube combination. A high-numerical aperture (NA) objective (Nikon; Plan Fluor 40×/NA 1.30, oil) was used to focus the excitation light on the sample and to collect nonlinear emission in the epi-direction. Forward-propagating nonlinear signals were collected by an aspheric lens (Thorlabs; AL2520-A; NA 0.5). Emission signals were separated from excitation light by using long-pass dichroic mirrors (FF775-Di01-25x36), a pair of short-pass filters (FF01-790/SP-25), and single band-pass filters. Epi-propagating emission was split into two detection channels by a long-pass dichroic mirror (Di02-R635-25x36). Three channels were acquired simultaneously: two epifluorescence channels, red (FF02-684/24-25) and green (FF01-514/30-25), and forward-propagating CARS (FF01-665/7.5 for imaging at 2,845 cm−1 and FF01-661/20-25 for microspectral measurements). Emission light was detected using single-photon-counting hybrid photomultiplier tubes (Becker & Hickl; two HPM-100-40 and one HPM-100-50) operated by three parallel time-correlated single-photon-counting modules (Becker & Hickl; SPC-150).
In addition to spectral filtering, we time gated the detection of the CARS signal to suppress the influence of the TPE fluorescence from chloroplasts, emitted with a maximum at 678 nm. Even a narrow band (7.5 nm width) filter centered at 665 nm lets through substantial amounts of TPE fluorescence into the CARS channel without gating. To discriminate between the two signals, we exploited the fact that CARS emission is instantaneous with the laser excitation, while chloroplast TPE fluorescence has a decay time on the order of 1 ns. Hence, most of the TPE fluorescence signal was discarded by the use of the shortest possible gating time, corresponding to approximately 200 ps (limited by instrument response function), still allowing the recording of the full CARS signal.
Microscopy Measurements
For microscopy, coverslips were coated with 0.01% (w/v) poly-l-Lys solution (Sigma) at 37°C for 1 h and dried before applying approximately 5 µL of microalgal culture. Samples were used for no more than 4 h, since poly-l-Lys immobilization started to fail after this length of time. Three main measurements were performed on algae from the three growth conditions: (1) measurements of lipid droplet volume probing the C-H vibration at 2,845 cm−1, for subsequent comparisons with GC-MS results; (2) spectral CARS measurements covering the entire C-H vibrational region from 2,775 to 2,945 cm−1; and (3) TPE fluorescence microscopy of the chloroplasts analyzed in the spectral region 500 to 530 nm. The origin of all fluorescence data reported is native (i.e. autofluorescence) apart from that by TO-PRO-1-iodide (Life Technologies Europe).
A relatively large field of view (50 µm) was used in order to capture eight to 12 algae per stack for the CARS measurements of lipid droplets and the simultaneous TPE fluorescence from the chloroplasts. Frames of 1,024 × 1,024 pixels were recorded at a pixel dwell time of 50.16 µs. A stack typically consisted of 40 slices, 0.2 µm apart, which took 48 min to scan. For the Ctrl, LL, and NS groups, 98, 102, and 60 cells were imaged, respectively, containing a total of 115, 108, and 87 lipid droplets in total and one to two fluorescent regions per cell.
For spectral analysis, single images of microalgae were recorded in a smaller field of view (24.8 µm) at 512 × 512 pixels per frame. For Ctrl and NS, CARS spectra of four lipid droplets were analyzed, and for LL, five droplets were analyzed.
To determine if cells survived the imaging process, TO-PRO-1-iodide was carefully applied to imaged samples before reimaging. TO-PRO-1-iodide stains dead cells and was used as described previously (Gorokhova et al., 2012).
CARS Image Analysis
For image analysis, ImageJ (National Institutes of Health; versions 1.47a–1.47k) was used. Volumes of lipid droplets were estimated by applying the 3D object counter (ImageJ plugin) to stacks recorded in the CARS detection channel. Lipid droplets were segmented from the rest of the image by manual thresholding. These high-density CH2 CARS signals were color coded in yellow in Figures 1, 2, 5, and 6. Corresponding low-density CH2 CARS signals, potentially originating from organic matter in the cell soma, were thresholded by visual inspection and color coded in blue. The TPE fluorescence signal was color coded in green and overlaid the CARS images. In order to facilitate the 3D rendering, the CARS images of the lipid droplets were processed with Gaussian blur (ImageJ). 3D renderings were formed of the overlay stacks using the ImageJ plugin Volume Viewer using the projection mode and a two-dimensional gradient color-code transfer function.
CARS Microspectroscopy Analysis
CARS spectra were recorded in the C-H vibrational region between 2,775 and 2,945 cm−1 in 10-cm−1 increments for all three growth conditions (Ctrl, LL, and NS) by scanning the optical parametric oscillator output (pump beam) in the wavelength range of 821.5 to 810.1 nm in 0.66-nm steps. Recorded spectra were corrected for excitation power fluctuations during the wavelength scan. Spectra were calculated as averages of the signal emitted by all (four to six) lipid droplets in a field of view using ImageJ. The ratio of CARS signals at 2,845 cm−1 relative to 2,885 cm−1 (the acyl chain CH2 symmetric and asymmetric stretch modes) was computed as an indicator of the acyl chain ordering and, hence, lipid fluidity (Rinia et al., 2008). Higher amounts of saturated lipids result in lower fluidity and, hence, lower ratios. Ratio images were formed, color coded, and overlaid over the gray-coded CARS images.
Analysis of Total Fatty Acids
Extraction and analysis of fatty acids was done according to Cavonius et al. (2014). In brief, freeze-dried sample was spiked with C17:0 internal standard (Larodan) and subjected to a digestion with KOH (pro analysi; Riedel-de Haën) at 70°C for 120 min. After extracting lipids, these were transesterified by 10% (v/v) acetyl chloride in methanol. After extracting the fatty acid methyl esters, these were separated by GC-MS. The limit of detection was estimated as 0.1 μg mL−1 under the described running conditions (100 pg injected on column). Identification of fatty acids was carried out by comparison of retention time with known standards (GLC 463 [Nu-Chek Prep] and PUFA-3 [Supelco; a kind gift from Mats Andersson]). Quantification was carried out by integration of peak areas and comparison with the internal standard (C17:0).
Acknowledgments
We thank Angela Wulff (Gothenburg University) for granting access to P. tricornutum GUMACC 2, Mats Andersson (Gothenburg University) for the kind gift of PUFA standard, Henning Hagman (Chalmers University of Technology) for support and suggestions related to microscopy measurements, and Thomas Andlid (Chalmers University of Technology) for microbiology support.
Glossary
- PUFA
polyunsaturated fatty acid
- EPA
eicosapentaenoic acid
- DHA
docosahexaenoic acid
- GC-MS
gas chromatography-mass spectrometry
- CARS
coherent anti-Stokes Raman scattering
- C-H
carbon-hydrogen
- Ctrl
control
- LL
light-limited
- NS
nitrogen-starved
- TPE
two-photon-excited
- 3D
three-dimensional
- NA
numerical aperture
Footnotes
This work was supported by Chalmers University of Technology, Department of Chemical and Biological Engineering; the Kristina Stenborg Foundation; the Wallenberg Foundation; and the Swedish Research Council.
Articles can be viewed without a subscription.
References
- Alonso DL, Belarbi EL, Rodriguez-Ruiz J, Segura CI, Gimenez A (1998) Acyl lipids of three microalgae. Phytochemistry 47: 1473–1481 [Google Scholar]
- Andersen RA, editor (2005) Algal Culturing Techniques. Elsevier, Hong Kong, China [Google Scholar]
- Athenstaedt K, Zweytick D, Jandrositz A, Kohlwein SD, Daum G (1999) Identification and characterization of major lipid particle proteins of the yeast Saccharomyces cerevisiae. J Bacteriol 181: 6441–6448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuer G, Lamers PP, Martens DE, Draaisma RB, Wijffels RH (2012) The impact of nitrogen starvation on the dynamics of triacylglycerol accumulation in nine microalgae strains. Bioresour Technol 124: 217–226 [DOI] [PubMed] [Google Scholar]
- Cavonius LR, Carlsson NG, Undeland I (2014) Quantification of total fatty acids in microalgae: comparison of extraction and transesterification methods. Anal Bioanal Chem 406: 7313–7322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermelli S, Guo Y, Gross SP, Welte MA (2006) The lipid-droplet proteome reveals that droplets are a protein-storage depot. Curr Biol 16: 1783–1795 [DOI] [PubMed] [Google Scholar]
- Chen W, Zhang C, Song L, Sommerfeld M, Hu Q (2009) A high throughput Nile red method for quantitative measurement of neutral lipids in microalgae. J Microbiol Methods 77: 41–47 [DOI] [PubMed] [Google Scholar]
- Cheng JX, Xie XS (2004) Coherent anti-Stokes Raman scattering microscopy: instrumentation, theory, and applications. J Phys Chem B 108: 827–840 [Google Scholar]
- Chisti Y. (2007) Biodiesel from microalgae. Biotechnol Adv 25: 294–306 [DOI] [PubMed] [Google Scholar]
- Cooper MS, Hardin WR, Petersen TW, Cattolico RA (2010) Visualizing “green oil” in live algal cells. J Biosci Bioeng 109: 198–201 [DOI] [PubMed] [Google Scholar]
- Davidi L, Katz A, Pick U (2012) Characterization of major lipid droplet proteins from Dunaliella. Planta 236: 19–33 [DOI] [PubMed] [Google Scholar]
- de Jesus Raposo MF, de Morais RM, de Morais AM (2013) Health applications of bioactive compounds from marine microalgae. Life Sci 93: 479–486 [DOI] [PubMed] [Google Scholar]
- De la Hoz Siegler H, Ayidzoe W, Ben-Zvi A, Burrell RE, McCaffrey WC (2012) Improving the reliability of fluorescence-based neutral lipid content measurements in microalgal cultures. Algal Research 1: 176–184 [Google Scholar]
- De Martino A, Bartual A, Willis A, Meichenin A, Villazán B, Maheswari U, Bowler C (2011) Physiological and molecular evidence that environmental changes elicit morphological interconversion in the model diatom Phaeodactylum tricornutum. Protist 162: 462–481 [DOI] [PubMed] [Google Scholar]
- Enejder A, Brackmann C, Svedberg F (2010) Coherent anti-Stokes Raman scattering microscopy of cellular lipid storage. IEEE J Sel Top Quantum Electron 16: 506–515 [Google Scholar]
- Erickson RA, Jimenez R (2013) Microfluidic cytometer for high-throughput measurement of photosynthetic characteristics and lipid accumulation in individual algal cells. Lab Chip 13: 2893–2901 [DOI] [PubMed] [Google Scholar]
- Fan J, Andre C, Xu C (2011) A chloroplast pathway for the de novo biosynthesis of triacylglycerol in Chlamydomonas reinhardtii. FEBS Lett 585: 1985–1991 [DOI] [PubMed] [Google Scholar]
- Fernández-Reiriz MJ, Perez-Camacho A, Ferreiro MJ, Blanco J, Planas M, Campos MJ, Labarta U (1989) Biomass production and variation in the biochemical profile (total protein, carbohydrates, RNA, lipids and fatty acids) of seven species of marine microalgae. Aquaculture 83: 17–37 [Google Scholar]
- Foissner I. (2009) Fluorescent phosphocholine: a specific marker for the endoplasmic reticulum and for lipid droplets in Chara internodal cells. Protoplasma 238: 47–58 [DOI] [PubMed] [Google Scholar]
- Frank HA, Cua A, Chynwat V, Young A, Gosztola D, Wasielewski MR (1994) Photophysics of the carotenoids associated with the xanthophyll cycle in photosynthesis. Photosynth Res 41: 389–395 [DOI] [PubMed] [Google Scholar]
- Gatenby CM, Orcutt DM, Kreeger DA, Parker BC, Jones VA, Neves RJ (2003) Biochemical composition of three algal species proposed as food for captive freshwater mussels. J Appl Phycol 15: 1–11 [Google Scholar]
- Gorokhova E, Mattsson L, Sundström AM (2012) A comparison of TO-PRO-1 iodide and 5-CFDA-AM staining methods for assessing viability of planktonic algae with epifluorescence microscopy. J Microbiol Methods 89: 216–221 [DOI] [PubMed] [Google Scholar]
- Griffiths MJ, van Hille RP, Harrison EH (2012) Lipid productivity, settling potential and fatty acid profile of 11 microalgal species grown under nitrogen replete and limited conditions. J Appl Phycol 24: 989–1001 [Google Scholar]
- He XN, Allen J, Black PN, Baldacchini T, Huang X, Huang H, Jiang L, Lu YF (2012) Coherent anti-Stokes Raman scattering and spontaneous Raman spectroscopy and microscopy of microalgae with nitrogen depletion. Biomed Opt Express 3: 2896–2906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich C, Hofer A, Ritsch A, Ciardi C, Bernet S, Ritsch-Marte M (2008) Selective imaging of saturated and unsaturated lipids by wide-field CARS-microscopy. Opt Express 16: 2699–2708 [DOI] [PubMed] [Google Scholar]
- Jolivet P, Roux E, D’Andrea S, Davanture M, Negroni L, Zivy M, Chardot T (2004) Protein composition of oil bodies in Arabidopsis thaliana ecotype WS. Plant Physiol Biochem 42: 501–509 [DOI] [PubMed] [Google Scholar]
- Katavic V, Agrawal GK, Hajduch M, Harris SL, Thelen JJ (2006) Protein and lipid composition analysis of oil bodies from two Brassica napus cultivars. Proteomics 6: 4586–4598 [DOI] [PubMed] [Google Scholar]
- Lee TH, Chang JS, Wang HY (2013) Current developments in high-throughput analysis for microalgae cellular contents. Biotechnol J 8: 1301–1314 [DOI] [PubMed] [Google Scholar]
- Moellering ER, Benning C (2010) RNA interference silencing of a major lipid droplet protein affects lipid droplet size in Chlamydomonas reinhardtii. Eukaryot Cell 9: 97–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy DJ. (2012) The dynamic roles of intracellular lipid droplets: from archaea to mammals. Protoplasma 249: 541–585 [DOI] [PubMed] [Google Scholar]
- Pohling C, Brackmann C, Duarte A, Buckup T, Enejder A, Motzkus M (2014) Chemical imaging of lignocellulosic biomass by CARS microscopy. J Biophotonics 7: 126–134 [DOI] [PubMed] [Google Scholar]
- Rinia HA, Burger KNJ, Bonn M, Müller M (2008) Quantitative label-free imaging of lipid composition and packing of individual cellular lipid droplets using multiplex CARS microscopy. Biophys J 95: 4908–4914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PA, Harrison PJ, Whyte JNC (1990) Influence of irradiance on the fatty-acid composition of phytoplankton. J Phycol 26: 278–288 [Google Scholar]
- Tonon T, Harvey D, Larson TR, Graham IA (2002) Long chain polyunsaturated fatty acid production and partitioning to triacylglycerols in four microalgae. Phytochemistry 61: 15–24 [DOI] [PubMed] [Google Scholar]
- Vieler A, Brubaker SB, Vick B, Benning C (2012) A lipid droplet protein of Nannochloropsis with functions partially analogous to plant oleosins. Plant Physiol 158: 1562–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther TC, Farese RV Jr (2009) The life of lipid droplets. Biochim Biophys Acta 1791: 459–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White DA, Pagarette A, Rooks P, Ali ST (2012) The effect of sodium bicarbonate supplementation on growth and biochemical composition of marine microalgae cultures. J Appl Phycol 25: 153–165 [Google Scholar]
- Wong DM, Franz AK (2013) A comparison of lipid storage in Phaeodactylum tricornutum and Tetraselmis suecica using laser scanning confocal microscopy. J Microbiol Methods 95: 122–128 [DOI] [PubMed] [Google Scholar]
- Yongmanitchai W, Ward OP (1991) Growth of and omega-3 fatty acid production by Phaeodactylum tricornutum under different culture conditions. Appl Environ Microbiol 57: 419–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YM, Chen H, He CL, Wang Q (2013) Nitrogen starvation induced oxidative stress in an oil-producing green alga Chlorella sorokiniana C3. PLoS ONE 8: e69225. [DOI] [PMC free article] [PubMed] [Google Scholar]