The virulence activity of an effector protein belonging to the widely conserved AvrE family is linked to its ability to cause system-wide reprogramming of phenylpropanoid metabolism in susceptible maize seedlings.
Abstract
AvrE family type III effector proteins share the ability to suppress host defenses, induce disease-associated cell death, and promote bacterial growth. However, despite widespread contributions to numerous bacterial diseases in agriculturally important plants, the mode of action of these effectors remains largely unknown. WtsE is an AvrE family member required for the ability of Pantoea stewartii ssp. stewartii (Pnss) to proliferate efficiently and cause wilt and leaf blight symptoms in maize (Zea mays) plants. Notably, when WtsE is delivered by a heterologous system into the leaf cells of susceptible maize seedlings, it alone produces water-soaked disease symptoms reminiscent of those produced by Pnss. Thus, WtsE is a pathogenicity and virulence factor in maize, and an Escherichia coli heterologous delivery system can be used to study the activity of WtsE in isolation from other factors produced by Pnss. Transcriptional profiling of maize revealed the effects of WtsE, including induction of genes involved in secondary metabolism and suppression of genes involved in photosynthesis. Targeted metabolite quantification revealed that WtsE perturbs maize metabolism, including the induction of coumaroyl tyramine. The ability of mutant WtsE derivatives to elicit transcriptional and metabolic changes in susceptible maize seedlings correlated with their ability to promote disease. Furthermore, chemical inhibitors that block metabolic flux into the phenylpropanoid pathways targeted by WtsE also disrupted the pathogenicity and virulence activity of WtsE. While numerous metabolites produced downstream of the shikimate pathway are known to promote plant defense, our results indicate that misregulated induction of phenylpropanoid metabolism also can be used to promote pathogen virulence.
Pantoea stewartii ssp. stewartii (Pnss) is a bacterial pathogen that causes Stewart’s wilt and leaf blight in maize (Zea mays). Wilt results when Pnss bacteria infect xylem vessels of seedlings and block water transport through the production of exopolysaccharide. In older plants, leaf blight develops as streaks along leaf veins and can result in the loss of leaves. Currently, the economic significance of Pnss-associated disease is limited to sweet maize production in the central and northeastern United States (White, 1999). While the economic impact of Pnss on maize has been reduced through the use of resistant cultivars, the tools available for both host and pathogen make the Pnss-maize interaction highly suitable for laboratory study. Maize is of major importance as a source of food, secondary products, and energy. As such, substantial tools have been developed for its study, including genome sequences for inbred lines B73 and Mo17 and the Mexican landrace Palomero Toluqueño as well as curated online tools and databases available to the maize research community (Brunner et al., 2005; Schnable et al., 2009; Vielle-Calzada et al., 2009; Schaeffer et al., 2011). For Pnss, targeted mutagenesis and transformation are relatively easy, and a large collection of genetically characterized mutants is available. Additionally, Pnss infection assays are quick and uniform. Maize seedlings are ready for inoculation as soon as 6 d after sowing, leaf cells can be uniformly targeted following vacuum infiltration, the appearance of water-soaking symptoms occurs in less than 1 d, and in planta bacterial growth can be measured over the course of several days. Thus, Pnss-maize is a highly tractable system for study of the interaction of a crop with a pathogenic bacterium.
Many bacterial pathogens of both plants and animals utilize type III secretion systems (T3SSs) to transport effector proteins (called type III effectors [T3Es]) directly into the cytoplasm of host cells. Remarkably, Pnss encodes two distinct T3SSs, one that is active in corn flea beetles, which harbor Pnss over the winter and transmit it to maize plants in the spring and late summer via feeding, and a second, hypersensitive response and pathogenicity (Hrp) T3SS that is active in maize tissues (Correa et al., 2012). The study of T3Es from plant pathogenic bacteria has been limited by the fact that deletion of an individual T3E often has little or no effect on virulence, due in part to functional redundancy within the suite of T3Es encoded by a pathogen. To study the action of T3Es in isolation, numerous studies have used transgenic expression of T3Es in planta, often under the control of chemically inducible promoters. Although this approach has been very useful, it includes uncertainties about the timing and level of T3Es inside plant cells compared with when the T3Es are naturally delivered. Thus, another advantage to studying Pnss is the key role of a single type III effector protein, WtsE, the deletion of which completely compromises the ability of Pnss to cause disease in maize (Frederick et al., 2001). Extensive mutagenesis revealed no other T3E that is needed for pathogenicity in maize (Frederick et al., 2001), although it cannot be ruled out that other, yet unknown, T3Es may have minor or redundant roles. Thus, due to the detectable contribution of WtsE to bacterial disease, the Pnss-maize pathosystem is attractive for the study of this T3E in particular.
WtsE is a member of the AvrE/HopR superfamily of T3Es that are widely distributed among plant-associated, gram-negative bacteria, including strains of Pseudomonas, Ralstonia, Erwinia, and Xanthomonas spp. (Kvitko et al., 2009). Members of this superfamily often have marked effects on pathogenicity. WtsE from Pnss is most closely related to AvrE family T3Es and in particular to DspA/E proteins from Erwinia spp. Just as a functional WtsE is required for Pnss pathogenicity (Frederick et al., 2001), DspA/E is required for the ability of Erwinia amylovora to elicit fire blight symptoms in apple (Malus domestica), Cotoneaster spp., and pear (Pyrus communis; Gaudriault et al., 1997; Bogdanove et al., 1998). Additionally, AvrE, along with the sequence-unrelated but functionally redundant T3E, HopM1, and the AvrE superfamily member HopR1 (Badel et al., 2006; Kvitko et al., 2009), is required for the virulence of Pseudomonas syringae pv tomato on tomato (Solanum lycopersicum), Arabidopsis (Arabidopsis thaliana), and Nicotiana benthamiana. Thus, members of this important superfamily of T3Es make key contributions to the pathogenicity of several genera of bacteria on plants ranging from grasses to fruit trees. Given the importance and widespread distribution of this superfamily, insights obtained from the study of WtsE are likely to be broadly relevant to plant-pathogen interactions.
Although the mechanisms by which AvrE/HopR T3Es promote virulence are not well understood, the consequences of their actions in plant cells have been examined. Many AvrE family T3Es, including WtsE, have been shown to elicit water-soaked disease symptoms in susceptible host plants and to promote the growth and survival of the bacteria in planta (Bogdanove et al., 1998; Badel et al., 2006; Boureau et al., 2006; Ham et al., 2009). Basal defense, as assessed by callose deposition, was suppressed by DspA/E of E. amylovora in apple and AvrE of P. syringae and WtsE of Pnss in Arabidopsis (DebRoy et al., 2004; Ham et al., 2008). In nonhost plants, both DspA/E of E. amylovora and WtsE of Pnss have been shown to increase bacterial growth before bacterial population decline begins, presumably due to the onset of host defenses (Oh et al., 2007; Ham et al., 2008). Our previous work indicates that there are at least two main functions for WtsE (Ham et al., 2006, 2008, 2009). First, it induces a water-soaking response in maize that may release water and nutrients to support initial and continued bacterial growth. Second, it suppresses plant defenses in Arabidopsis and likely does the same in maize.
A clue to the mechanism of action of AvrE family T3Es came from the identification of important protein motifs conserved within the family. Most AvrE family members contain a putative endoplasmic reticulum membrane retention signal (ERMRS) and one or two WxxxE motifs (Ham et al., 2009). The WxxxE motifs were first discovered in T3Es of animal pathogens (Alto et al., 2006). Derivatives of WtsE with either the putative ERMRS or both WxxxE motifs mutated (ΔFEMK and w12 mutants, respectively) failed to elicit disease symptoms in maize or to suppress defense responses in Arabidopsis (Ham et al., 2009). Interestingly, the growth of these site-directed wtsE mutants of Pnss in maize was only modestly reduced. However, given that many T3Es have multiple targets, and given the large size of AvrE/HopR T3Es, it seems likely that WtsE might have multiple functions and targets within plant cells. The WxxxE motifs of AvrE family members are hypothesized to form part of the fold that mimics the active site of a guanine-exchange factor (GEF) protein and thus to modulate the activity of host GTPase proteins, as has been demonstrated for Map/IpgB/Sif family members from bacterial pathogens of animals (Huang et al., 2009). Confirmation of this biochemical prediction, along with localization in plant cells, have proven difficult due to the cell toxicity of the AvrE family T3Es (Ham et al., 2006). Intriguingly, HopM1, which is functionally redundant to AvrE for promoting the full virulence of P. syringae pv tomato strain DC3000, targets for degradation an ADP ribosylation factor-GEF called Arabidopsis HopM-Interactor7 (AtMIN7; Nomura et al., 2006). AtMIN7 is required for pathogen-associated molecular pattern (PAMP)-triggered immunity and salicylic acid (SA)-regulated responses to pathogens, including callose deposition (Nomura et al., 2006). Because ADP ribosylation factor-GEFs are regulators of the plant cytoskeleton and vesicle traffic, it has been hypothesized that HopM1 and AvrE family effectors manipulate these pathways (Nomura et al., 2006; Ham et al., 2009).
The molecular targets of T3Es from plant pathogens are numerous and varied, including proteins involved in the recognition of PAMPs at the plant plasma membrane, defense-associated mitogen-activated protein kinase signaling cascades, the plant cytoskeleton, components of the chloroplast, isoflavanone biosynthesis, and proteins involved in vesicle trafficking (for review, see Block and Alfano, 2011; Deslandes and Rivas, 2012). While many, perhaps most, T3Es suppress plant defense responses, others may be involved in promoting bacterial growth by providing bacterial access to plant nutrients (Chen et al., 2010). In addition to their intended defense-suppressing function(s), T3Es also can elicit plant defenses via direct or indirect interaction with specific host resistance proteins. While the specific mechanisms by which WtsE and other AvrE family T3Es function remain largely a mystery, the data presented here indicate secondary metabolism, particularly the phenylpropanoid pathway, as a potential target.
Products of plant metabolism play important roles in plant-microbe interactions, including plant defense. For example, phytoalexin terpenoids, such as kauralexins and zealexins, accumulate in response to fungal attack (Ahuja et al., 2012). The shikimate pathway also gives rise to numerous products important to plant defense. Chorismate, the precursor to the synthesis of aromatic amino acids, is utilized for the production of the important pathogen defense-related plant hormone SA (Wildermuth et al., 2001). Trp and its indole precursor are converted to glucosinolates and benzoxazinoids in Arabidopsis and maize, respectively, with roles in defense signaling (Bednarek et al., 2009; Ahmad et al., 2011). Phe-derived compounds are a major class of defense-associated metabolites (Naoumkina et al., 2010). Preformed phenolics are involved in the lignification of cell walls associated with the hypersensitive response (Beckman, 2000). Hydroxycinnamic acid amides fortify the plant cell wall by reducing its digestibility to pathogens (Facchini et al., 2002). In addition to abiotic stress, flavonoids also contribute to plant defense against biotic stresses (Treutter, 2005). On the microbe side of the interactions, secondary metabolites derived from numerous plant-associated bacteria also can affect plant physiology and block host responses. For example, the coronatine toxin from P. syringae can reopen stomata and antagonize SA-dependent and SA-independent defense signaling (Melotto et al., 2006; Geng et al., 2012; Zheng et al., 2012).
Here, we show that WtsE modulates maize metabolism. The effect of this single effector is mediated, at least in part, through alteration of the expression of enzymes involved in phenylpropanoid metabolism. Introduction of WtsE into the cells of maize seedling leaves through an Escherichia coli delivery system (EcDS) elicited numerous plant responses, including genome-wide changes in transcription. Induced expression of genes involved in the phenylpropanoid pathway by WtsE was confirmed by quantitative reverse transcription (qRT)-PCR. WtsE was found to elicit the accumulation of coumaroyl tyramine (CouTyr), a hydroxycinammic acid amide typically associated with lignin in plant cell walls. The ability of derivatives of WtsE to elicit transcriptional and metabolic changes in susceptible maize seedlings was correlated with their ability to induce disease symptoms. Also, chemical inhibitors of the shikimate pathway or Phe ammonia lyase limited the ability of WtsE to induce CouTyr and to promote disease. These results indicate that the virulence activity of WtsE depends on the perturbation of phenylpropanoid metabolism.
RESULTS
Genome-Wide Transcriptional Response of Maize Seedlings to WtsE
To investigate the effect of WtsE on transcription in maize, we delivered WtsE into cells of maize seedling leaves using a heterologous EcDS in order to eliminate other Pnss-associated factors that might also affect maize transcription. This system utilized a strain of E. coli MC4100 carrying the entire Dickeya dadantii hrp gene cluster in plasmid pCPP2156 (Ham et al., 1998), which becomes a WtsE delivery system (EcWtsE) when it carries a second plasmid expressing wtsEF or derivatives thereof. Previous work has indicated that WtsE, when delivered by EcWtsE, induces disease-like water-soaking symptoms in leaves of susceptible maize seedlings (Ham et al., 2006, 2009). While validating the use of the EcDS to study the mechanism of action of WtsE, this study also highlighted the importance of selecting a time point that precedes potential secondary transcriptional changes associated with widespread cell death. Macroscopic investigation revealed that WtsE-induced symptoms first became apparent approximately 8 h after infiltration (hai; Supplemental Fig. S1). Using electrolyte leakage to measure the loss of cell integrity provided a more sensitive assay to assess WtsE-induced cell death (Supplemental Fig. S1). The conductivity of water-containing maize leaves remained consistently low until approximately 6 hai, at which time plants that received WtsE showed increasing signal relative to control plants. Thus, 6 hai was chosen as the time point for transcriptome analysis.
We used a microarray approach to examine the effect of WtsE on transcription in maize cells (http://www.maizegdb.org/microarray.php). Of the more than 43,000 maize oligonucleotides on the array, 1,047 oligonucleotides showed highly significant differences between treatment (EcWtsE) and control (EcDS) based on a minimum fold change of 2.83 and a P value of less than 0.02 (Supplemental Fig. S2). The oligonucleotides were annotated to genes from the maize B73 genome based on the work of Seifert et al. (2012). We found that 586 of the 1,047 oligonucleotides were annotated to single genes. Of these, 314 and 272 were up- or down-regulated by WtsE, corresponding to 290 and 249 unique genes, respectively (Supplemental Table S1). Another 285 of the 1,047 oligonucleotides remained unannotated, and 176 oligonucleotides could not be unambiguously annotated; they may have annealed to products from multiple genes during hybridization. Our results indicate that WtsE caused large-scale transcriptional reprogramming within maize seedling leaves within 6 hai of EcWtsE infiltration.
Differentially expressed probes were assigned Gene Ontology (GO) terms using the AgriGO platform (Du et al., 2010) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways using Kobas 2.0 (Xie et al., 2011). We assessed whether certain GO terms or KEGG pathways might be significantly enriched in our data sets (Table I). Photosynthesis was significantly represented among probes that were down-regulated in plants exposed to WtsE. Secondary metabolism, particularly the phenylpropanoid pathway, was highly represented in WtsE-induced probes. Additional probes up-regulated in response to WtsE included several that are associated with genes involved in the biosynthesis of aromatic amino acids that are precursors to phenylpropanoid metabolism (see below).
Table I. Significantly overrepresented GO biological process terms and KEGG pathways in annotations of up-regulated or down-regulated probes in WtsE-exposed versus control plants in microarray experiments.
| Group | Term | Probesa | Backgroundb | Pc | Genes (Minimum)d | Genes (Maximum)e |
|---|---|---|---|---|---|---|
| WtsE up-regulated (P < 0.02, fold change > 2.8) | Aromatic amino acid family metabolic process | 14 | 108 | 6.00E-12 | 10 | 24 |
| Cellular aromatic compound metabolic process | 17 | 332 | 7.20E-08 | 12 | 28 | |
| Cellular nitrogen compound metabolic process | 23 | 613 | 1.30E-07 | 18 | 35 | |
| Cellular carbohydrate metabolic process | 27 | 830 | 2.10E-07 | 16 | 33 | |
| Amine metabolic process | 25 | 904 | 9.10E-06 | 17 | 37 | |
| Carbohydrate metabolic process | 38 | 1,821 | 3.80E-05 | 23 | 44 | |
| Alcohol metabolic process | 17 | 681 | 0.00066 | 11 | 20 | |
| Cellular nitrogen compound biosynthetic process | 15 | 493 | 0.00018 | 11 | 22 | |
| Phe, Tyr, and Trp biosynthesisf | 6 | 41 | 0.00021 | 5 | 8 | |
| WtsE down-regulated (P < 0.02, fold change > 2.8) | Photosynthesis | 51 | 217 | 2.10E-52 | 27 | 72 |
| Generation of precursor metabolites and energy | 36 | 667 | 1.60E-15 | 22 | 52 | |
| Cellular homeostasis | 11 | 218 | 1.40E-05 | 6 | 12 | |
| Oxidation reduction | 28 | 1,184 | 4.00E-05 | 13 | 45 | |
| Carbohydrate metabolic process | 37 | 1,821 | 6.90E-05 | 25 | 46 | |
| Regulation of biological quality | 32 | 1,568 | 0.00018 | 19 | 40 | |
| Glc metabolic process | 11 | 336 | 0.00062 | 7 | 18 | |
| Photosynthesisf | 8 | 80 | 0.00021 | 5 | 16 |
The number of microarray probes matching each term is shown for each group (up- or down-regulated). The total numbers of probes with GO annotations in each group were 393 (up-regulated) and 390 (down-regulated). The numbers of probes with KEGG pathway annotations were 120 (up-regulated) and 105 (down-regulated). bThe number of microarray probes matching each term regardless of expression is shown. The total number of probes with GO annotations was 39,870, and the total number of probes with KEGG pathway annotations was 22,421. cP values correspond to hypergeometric distribution tests, where population is the microarray probes with each annotation and the samples were the up-regulated or down-regulated probes with each annotation. dDue to ambiguous annotations for some probes (matching more than one gene), two estimates of the amount of differentially expressed genes are shown; the minimum was calculated by leaving one gene per probe and collapsing probes matching the same gene. eMaximum number of genes corresponds to all genes matching the probes. fKEGG pathway terms are in boldface; all other terms are GO biological processes.
CouTyr Induction by WtsE in Maize
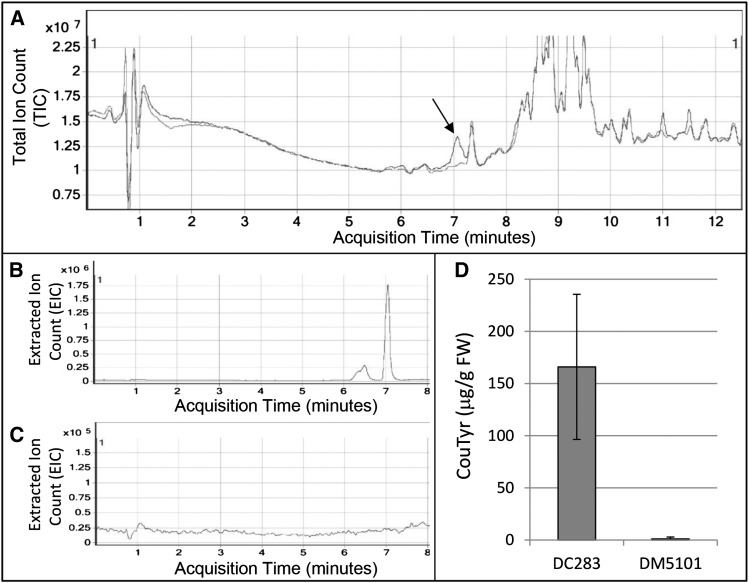
Given the significant induction of genes involved in the synthesis of phenolic and phenylpropanoid compounds, we sought to examine the effect of WtsE on the levels of these metabolites in maize seedlings. Seedling leaves infiltrated with either a wild-type Pnss strain (DC283) or a Pnss wtsE::miniTn5 mutant strain (DM5101) were harvested at 20 hai, which is approximately 2 h after the first macroscopic water-soaked symptoms produced by DC283 became apparent. Samples were extracted with 80% (v/v) methanol, diluted 100-fold in liquid chromatography-mass spectroscopy (LC-MS)-grade methanol and analyzed via LC-MS/MS (Fig. 1). Compounds were separated using an Agilent C18 Poroshell column, subjected to electrospray ionization, and monitored in positive ion mode. Initial precursor ion scans (which show the mass of the intact, nonfragmented parent molecule) showed several peaks eluting in the DC283-infiltrated leaves that were close to the limit of detection or absent in DM5101-infiltrated leaves (Fig. 1, A–C). The most prominent of these peaks exhibited a retention time of approximately 7.1 min and a mass-to-charge ratio (m/z) of 284 (Fig. 1A). Subsequent product ion scans indicated that this peak was likely a derivative of coumaric acid, and its mass (m/z = 284), combined with the function of genes induced by WtsE, led us to speculate that the peak at retention time = 7.1 min was CouTyr. Genuine CouTyr (para-N-transcoumaroyl tyramine) was synthesized (Pedersen et al., 2010) and used as an authentic standard to investigate the identity of the m/z = 284 peak. Comparison of retention time, precursor ion mass, and four precursor-product ion mass transitions (284→146.9, 284→118.8, 284→91.2, and 284→77.1; Supplemental Table S2) confirmed that the peak induced in DC283-induced leaves was CouTyr. Quantification based on standard curves generated using genuine CouTyr (Supplemental Fig. S3) revealed that infiltration with DC283 induced the accumulation of 160 μg g−1 fresh weight of CouTyr (Fig. 1D). Interestingly, while quantifying CouTyr, a peak with an identical precursor ion mass (m/z = 284) was observed to elute from the column approximately 0.5 min prior to CouTyr (retention time = 6.6 min). Further investigation revealed that this peak exhibited a fragmentation pattern identical to that observed with CouTyr, including the four mass transitions used to positively identify and quantify CouTyr by comparison with the genuine standard. Because of this, we hypothesize that the m/z = 284 peak eluting at retention time = 6.6 min is either a stereoisomer of CouTyr generated during the synthesis of this compound or an oxidative breakdown/catalysis product with an open-ring structure. While the structure of this putative stereoisomer and its potential role in WtsE virulence are currently unclear, since the CouTyr structural isomer most likely formed during the synthesis of this molecule (ortho-N-transcoumaroyl tyramine) exhibits a significant shift in both retention time and mass fragmentation patterns (data not shown), our current hypothesis favors the presence of an oxidized CouTyr isomer.
Figure 1.
WtsE induces CouTyr accumulation in sweet maize ‘Seneca Horizon’ seedlings. A, Liquid chromatography-tandem mass spectrometry (LC-MS/MS) chromatograms showing overlaid total ion counts (TIC) of precursor mass ion scans (positive ion mode) of extracts of cv Seneca Horizon seedlings from 20 h after infiltration with wild-type Pnss (DC283; black trace) and the wtsE null mutant (DM5101; gray trace). WtsE-dependent accumulation of CouTyr is apparent at an acquisition time of approximately 7.1 min (arrow). B and C, Extracted ion counts (EIC) of putative CouTyr species (m/z = 284) in extracts from plants infiltrated with DC283 (B) or DM5101 (C) as in A. D, Quantification of CouTyr in extracts of plants treated as in A. Shown are combined data and sd from six biological replicates (n = 12). FW, Fresh weight.
WtsE-Induced Expression of Genes Encoding Phenylpropanoid Biosynthetic Enzymes
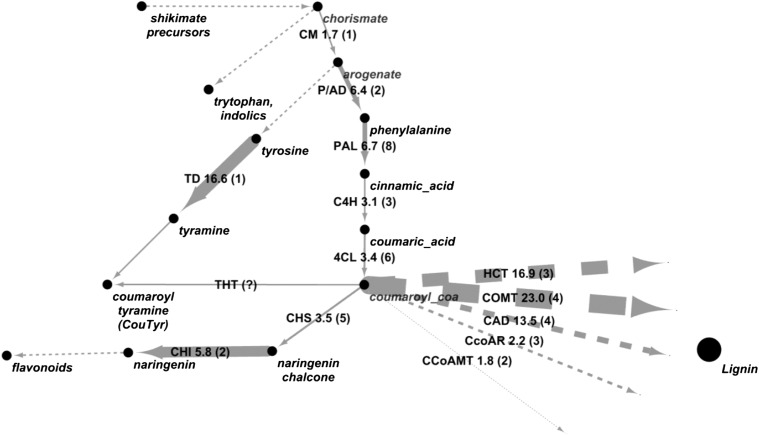
The observation that WtsE induces a high level of CouTyr accumulation led us to look more closely at the expression of genes encoding biosynthetic enzymes involved in the biosynthesis of coumaric acid and tyramine. Indeed, the microarray data showed that WtsE induces numerous genes that could account for the accumulation of these compounds as well as additional compounds generated from coumaroyl-CoA (Fig. 2).
Figure 2.
Generalized pathway of secondary metabolism from chorismate to aromatic amino acids and their secondary products. Enzyme abbreviations are shown over the step(s) they catalyze; numbers next to enzyme names indicate the maximum observed fold increases in expression in WtsE-exposed versus control plants in microarray experiments. Arrow thickness relates to this fold-change value. Numbers in parentheses indicate how many genes were found up-regulated with P < 0.05. Dashed arrows indicate steps not shown. CAD, Cinnamoyl alcohol dehydrogenase; CCoAMT, caffeoyl-CoA O-methyltransferase; CcoAR, cinnamoyl-CoA reductase; CHI, chalcone isomerase; CHS, chalcone synthase; CM, chorismate mutase; COMT, caffeic acid O-methyltransferase; HCT, hydroxycinnamoyl transferase; THT, tyramine N-hydroxycinnamoyltransferase.
Next, we used qRT-PCR to further investigate the expression of a subset of WtsE-induced genes. The results of these experiments served three purposes. The first was to validate the microarray data by independently confirming that these genes were WtsE induced. Second, the experiments provided additional information on the effects of the EcDS on the expression of these genes. The microarray only compared two treatments (EcWtsE versus the EcDS control) and thus only revealed changes in gene expression with or without WtsE over a possible background of responses to PAMPs from the EcDS. By including a buffer treatment, the qRT-PCR provided information about the effect of PAMPs on the expression of the WtsE-regulated genes. Third, the experiments provided information about how the expression of WtsE-induced genes changes over time in response to the different treatments.
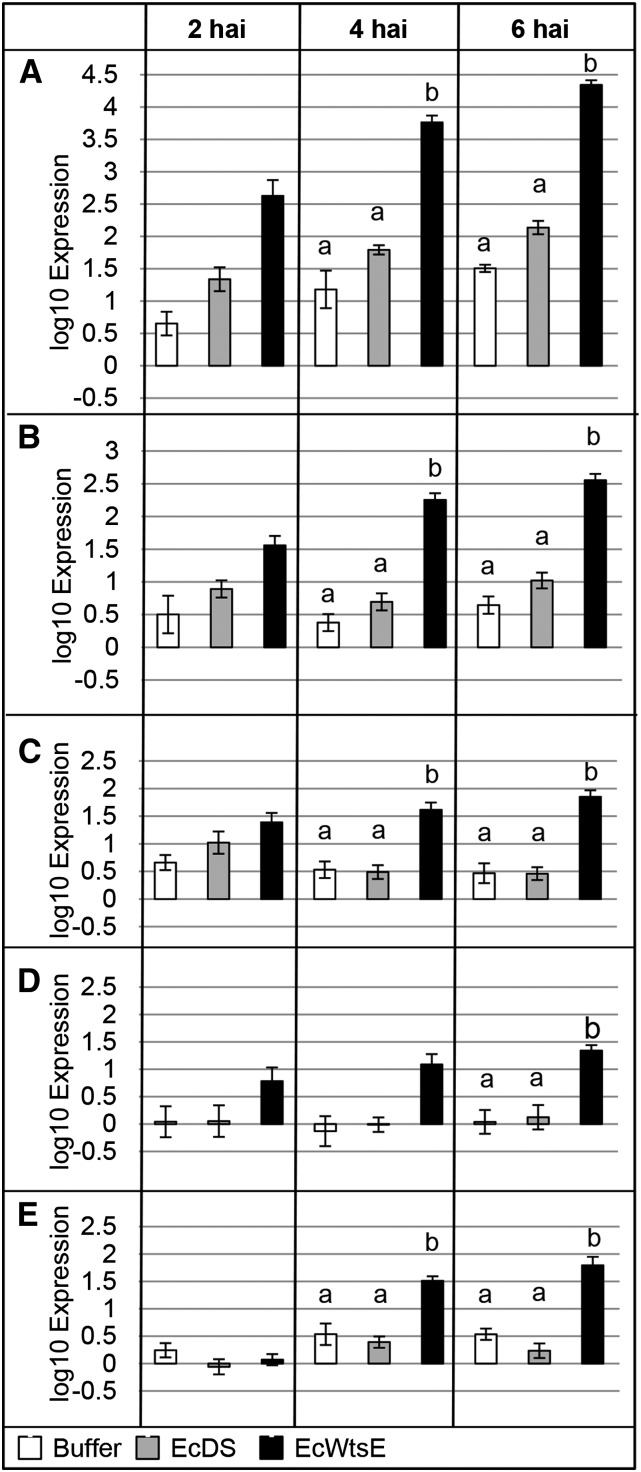
Maize seedlings were infiltrated with buffer, EcDS, or EcWtsE. The expression of WtsE-induced genes was analyzed by qRT-PCR at 2, 4, and 6 hai (Fig. 3). By 4 hai, plants infiltrated with EcWtsE contained significantly more transcripts for tyrosine decarboxylase (TD), 4-coumarate-coenzyme A ligase (4CL), cinnamate 4-hydroxylase (C4H), and phenylalanine ammonia lyase (PAL) than plants infiltrated with EcDS or buffer (Fig. 3, A–C and E, respectively). By 6 hai, prephanate/arogenate dehydratase (P/AD) also was more highly expressed in EcWtsE-infiltrated plants than those infiltrated with EcDS or buffer (Fig. 3D). These results confirm that the induction of these genes is dependent on WtsE. Furthermore, because buffer- and EcDS-infiltrated plants did not differ significantly for any gene at any of the times assessed, PAMPs associated with the EcDS did not induce the expression of any of these genes.
Figure 3.
Expression of five genes associated with phenolic metabolism in maize seedlings vacuum infiltrated with buffer, EcDS, or EcWtsE. Tissue was recovered at 2, 4, and 6 hai, and expression was determined for TD (A), 4CL (B), C4H (C), P/AD (D), and PAL (E) transcripts. Plants exposed to WtsE had greater mRNA accumulation of each of these genes than controls. Shown are average values and se of six qRT-PCRs (three reactions each for the complementary DNA from two biological replicates). Each biological replicate consisted of the first true leaves from two seedlings. Significantly different expression levels for a given gene and time point are indicated with letters (ANOVA and Fisher’s lsd, P < 0.05). No values at 2 hai, nor the 4-hai time point for P/AD, were found to significantly differ from controls.
The Functionality of WtsE Derivatives in Biological Assays Correlates with Transcriptional and Metabolic Perturbations
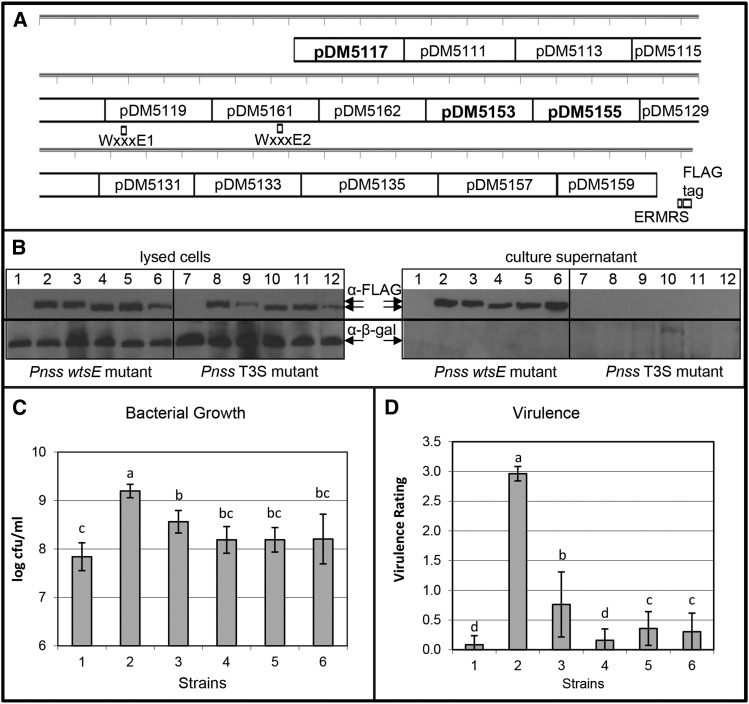
Fifteen partial deletions (approximately 300 bp each) distributed across the C terminally FLAG-tagged wtsE reading frame were constructed (Fig. 4A). Based on our previous finding that deletion of the C-terminal ERMRS abolished the virulence activity of WtsE, we suspected that placing a tag at the C terminus of WtsE might similarly disrupt the function of WtsE. However, to our surprise, wild-type WtsE with a C-terminal FLAG tag remained active; thus, this tagging strategy was employed for the other derivatives. One goal was to investigate which regions of the gene were dispensable for the plant cell death-inducing or bacterial growth-promoting functions of WtsE. A second goal was to use the derivatives to determine if the virulence activity of WtsE correlates with its ability to modulate gene expression and induce CouTyr synthesis.
Figure 4.
Pnss strains harboring mutated forms of wtsE were reduced in growth and virulence in maize seedlings. A, Map of WtsE showing the locations of the internal deletions in the listed plasmids and derivatives used for further analyses in boldface. Each tick represents 100 bp. B, Production and secretion of WtsE-FLAG and derivatives. Plasmid pJA017 encodes C terminally FLAG-tagged WtsE and the chaperone WtsF in vector pRK415. Pnss strains DM5101 (wtsE mutant) and DM711 (hrpJ type III secretion mutant) were transformed with pRK415, pJA017, or variants of pJA017 expressing wtsE derivatives. Proteins were visualized by anti-FLAG and anti-β-galactosidase (β-gal) immunoblotting from lysed cells (left) or from culture medium (right). Positions of WtsE derivatives and β-galactosidase, a cytoplasmic protein that serves as a loading control for cell lysis and a negative control for leakage of cell contents into the culture supernatant, are indicated by arrows. DM5101 (lanes 1–6) and DM711 (lanes 7–12) contained the following plasmids: lanes 1 and 7, pRK415; lanes 2 and 8, pJA017; lanes 3 and 9, pJA052 (wtsE w12 mutant, W694A, and W840A); lanes 4 and 10, pDM5117; lanes 5 and 11, pDM5153; and lanes 6 and 12, pDM5155. C and D, Average values and sd from three biological replicates measuring bacterial growth (C) and virulence (D) of DM5101 carrying the same plasmids as for lanes 1 to 6 in B. Both measurements were made 3 d after whorl inoculation with virulence rated on a scale from 0 to 3, with 3 being the most severe and 0 being symptomless. Different letters in C indicate significant differences in bacterial growth (ANOVA and Tukey’s honestly significant difference, P < 0.05). Different letters in D indicate significant differences in virulence (Kruskal-Wallis and Tukey’s mean rank tests, P < 0.05).
Each wtsE derivative was cloned into a broad-host-range plasmid and introduced into the wtsE null mutant strain DM5101. Western blotting of extracts from bacteria grown on an hrp-inducing medium demonstrated that 13 of the 15 WtsE derivatives were detectably produced inside the bacteria. However, of the 13 expressed deletion derivatives, only three (those produced from pDM5117 [wtsEΔ709-1017], pDM5153 [wtsEΔ2935-3234], and pDM5155 [wtsEΔ3235-3534]) were shown to be secreted in a type III-dependent manner as efficiently as wild-type WtsE (Fig. 4B). Each of these three expressed and secreted WtsE derivatives, along with the w12 derivative mutated in both WxxxE motifs, was significantly compromised, relative to wild-type WtsE, in its ability to complement either the growth or virulence defects of DM5101 (Fig. 4, C and D).
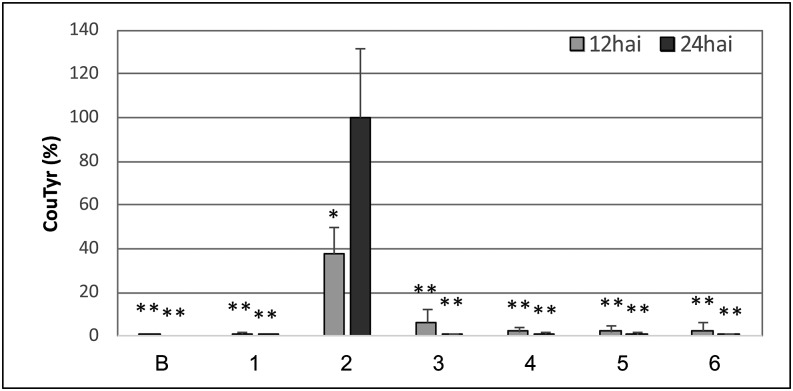
Next we used these four functionally compromised derivatives of WtsE (three internal deletions and the w12 mutant) to determine if virulence activity correlated with the perturbation of phenylpropanoid metabolism. First, the derivatives were tested for their ability to elicit transcripts of genes involved in the phenylpropanoid pathway (Fig. 5). We measured transcript levels of TD, 4CL, C4H, P/AD, and PAL at 4 hai, which was the earliest time that reliable transcriptional induction was observed using EcWtsE (Fig. 3). EcWtsE-FLAG (EcDS with pJA017) induced more of each transcript than EcDS, and this difference was significant for all but PAL. Relative to wild-type WtsE-FLAG, each of the derivatives elicited significantly less accumulation of TD, a subset of the derivatives induced significantly less accumulation of C4H and P/AD, and none of the derivatives differed significantly in the induction of PAL and 4CL. However, the clear trend among all derivatives was a reduction, relative to wild-type WtsE, in the expression of the tested genes. Similarly, we found that the WtsE derivatives each failed to induce CouTyr accumulation (Fig. 6). This was in contrast to EcWtsE, which induced 102 ± 16 and 285 ± 105 μg g−1 fresh weight of CouTyr at 12 and 24 hai, respectively. Collectively the results of Figures 4, 5, and 6 demonstrate that derivatives of WtsE that are unable to elicit transcriptional perturbations and CouTyr accumulation in maize seedlings also are unable to promote Pnss growth or cause disease symptoms.
Figure 5.
Effects of wild-type and mutated forms of WtsE on the expression of five genes associated with phenolic metabolism in maize seedlings. Six-day-old cv Seneca Horizon seedlings were vacuum infiltrated with EcDS (pRK415; 1), EcWtsE-FLAG (pJA017; 2), or the EcDS-expressing mutated forms of FLAG-tagged wtsE: w12 mutant (pJA052; 3), wtsEΔ709-1017 (pDM5109; 4), wtsEΔ2935-3234 (pDM5153; 5), or wtsEΔ3235-3534 (pDM5155; 6). Samples were collected at 4 hai, and the expression of TD (A), 4CL (B), C4H (C), P/AD (D), and PAL (E) was assessed. Shown are average values and se of qRT-PCRs done in triplicate for each of three biological replicates. Each biological replicate is two pooled first true leaves. Significantly different expression levels for a given gene and time point are indicated with letters (ANOVA and Fisher’s lsd, P < 0.05). For PAL, gene expression did not differ significantly among these treatments.
Figure 6.
Effects of wild-type and mutated forms of WtsE on the accumulation of CouTyr. B73 maize seedlings were infiltrated with buffer (B) or with the EcDS carrying empty vector (pLAFR3; 1) or plasmids encoding wild-type wtsE (pJA001; 2), w12 mutant (pDM5175; 3), wtsEΔ709-1017 (pDM5189; 4), wtsEΔ2935-3234 (pDM5184; 5), or wtsEΔ3235-3534 (pDM5182; 6). Samples were collected at 12 and 24 hai, and the accumulation of CouTyr was measured by LC-MS/MS. Data, from two samples each from three independent biological replicates (n = 6), are presented as percentages of CouTyr induced by EcDS carrying wild-type wtsE (pJA001) ± sd. Asterisks indicate statistically significant differences in CouTyr levels compared with pJA001, as determined by two-tailed Student’s t test (*P < 0.01, **P < 0.001).
WtsE Similarly Affects the Expression of Phenylpropanoid Metabolic Enzymes in the Context of Pnss Infection
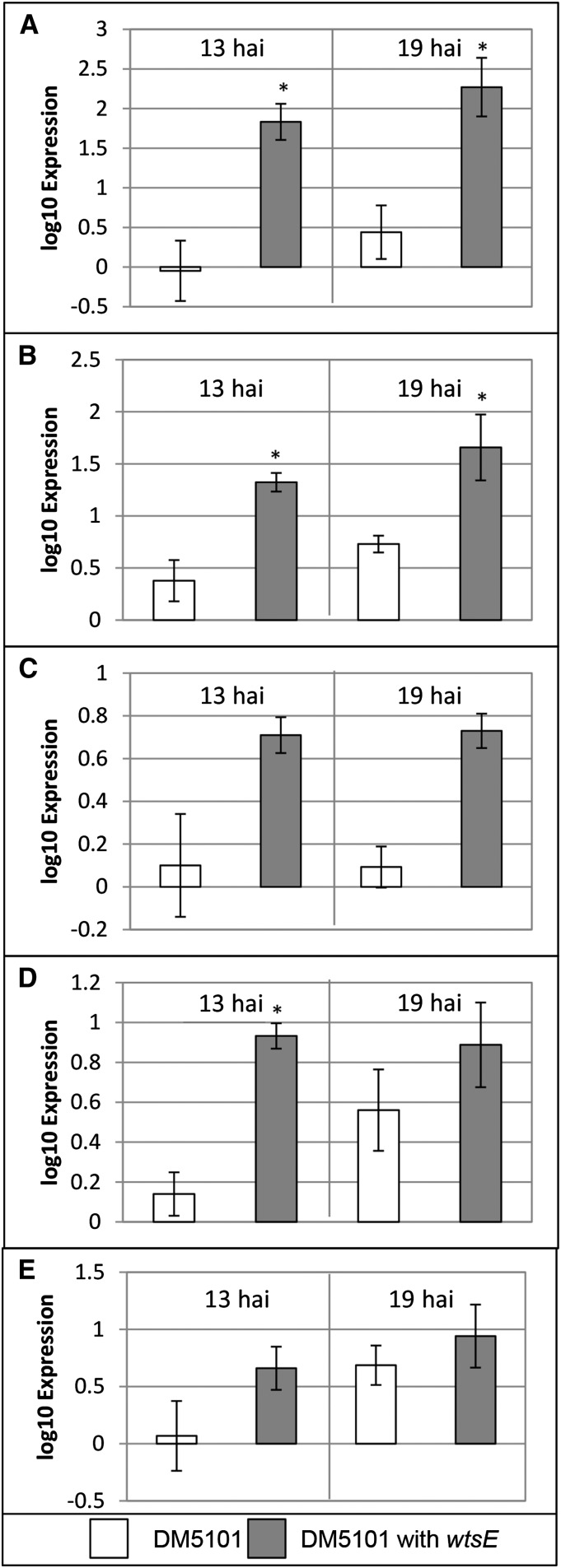
We sought to determine if the effects of WtsE on maize transcription observed when WtsE is delivered by the EcDS also occur during the interaction of Pnss with maize leaves. We compared wild-type strain DC283 with the wtsE mutant DM5101 (Supplemental Fig. S4) as well as strains of DM5101 carrying the empty vector (pRK415) and a complementing plasmid (pJA017; Fig. 7). The timing of measurements in these two comparisons preceded the appearance of macroscopic water-soaked symptoms induced by DC283 and DM5101 (pJA017) at approximately 18 hai and beyond 24 hai, respectively. The expression of TD, 4CL, C4H, P/AD, and PAL was measured by qRT-PCR analysis. For each of these genes, the trend was toward higher expression when the bacteria delivered WtsE, with the increase being statistically significant in a subset of comparisons and time points.
Figure 7.
Expression of five genes associated with phenolic metabolism in maize seedlings following infiltration with a Pnss wtsE null mutant and a complemented strain. Tissue from cv Seneca Horizon seedlings was harvested at 13 and 19 hai with a wtsE null mutant of Pnss (DM5101) carrying empty vector (pRK415; white bars) or plasmid-borne wtsE (pJA017; gray bars), and the accumulation of TD (A), 4CL (B), C4H (C), P/AD (D), and PAL (E) transcripts was assessed by quantitative PCR. Shown are average values and se of qRT-PCRs done in triplicate for each of three biological replicates. Asterisks indicate significantly different expression levels relative to the infiltration of DM5101 for a given gene at a single time point (two-tailed Student’s t test, P < 0.05).
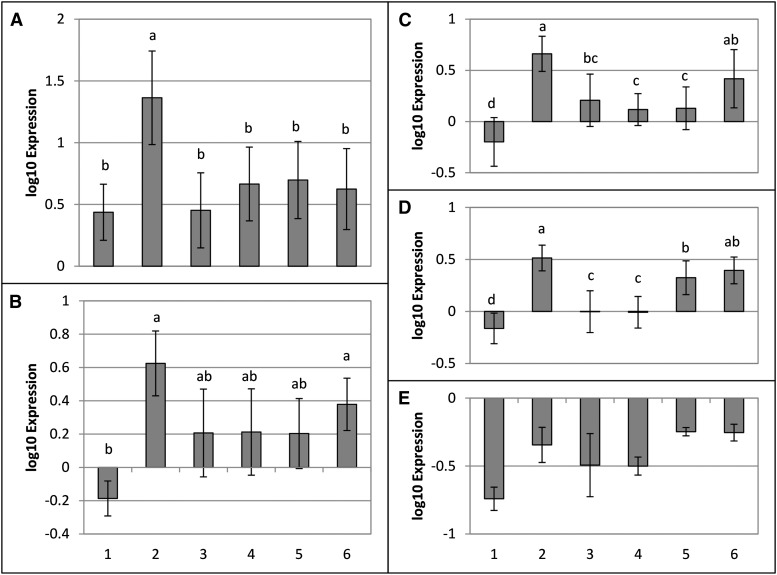
We also wished to determine if the ability of WtsE to induce the expression of genes involved in phenylpropanoid metabolism was conserved in a related AvrE family T3E. For this purpose, we used DspA/E from E. amylovora, which we have shown can complement the virulence defect of a Pnss wtsE mutant (Ham et al., 2006). DspA/E, when delivered by DM5101, as compared with DM5101 with empty vector, induced an increased expression of TD, 4CL, C4H, P/AD, and PAL at 13 and 19 hai; these differences were statistically significant at 13 hai for all but C4H (Supplemental Fig. S5). Collectively, these results indicate that WtsE and a closely related T3E from E. amylovora affect the transcription of common genes involved in phenylpropanoid metabolism in maize seedlings.
Metabolic Flux through the Shikimate Pathway and PAL Activity Are Required for Virulence Activities of WtsE
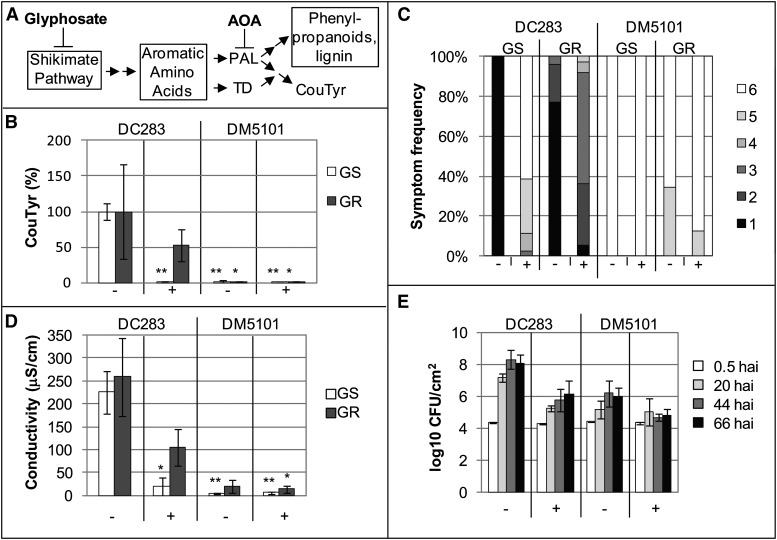
The data presented here indicate significant effects of WtsE on phenolic and phenylpropanoid metabolism and, for derivatives of WtsE, a correlation between these effects and virulence activity. To challenge the significance of the correlation, we tested the effect of metabolic inhibitors on WtsE activity. The herbicide glyphosate inhibits 5-enolpyruvoylshikimate-3-phosphate synthase (EPSPS; Steinrücken and Amrhein, 1980), an essential enzyme in the shikimate pathway, and thus reduces the production of chorismate, aromatic amino acids, and presumably downstream phenylpropanoid metabolites (Fig. 8A). Spraying maize seedlings with 0.5% (w/v) glyphosate caused no detectable symptoms for the first 2 d. Growth retardation relative to control plants was apparent by 3 to 4 d, and visible herbicidal symptoms appeared after 7 to 8 d (Supplemental Fig. S6). In the following experiments using glyphosate (Fig. 8), we examined the activity of WtsE during the first few hours up to a maximum of 3 d after glyphosate application.
Figure 8.
Inhibition of the shikimate pathway attenuates the virulence activity of WtsE. A, Schematic showing the point of action of glyphosate (and AOA). Boxes contain multiple enzymatic steps and/or processes. B to D, Glyphosate-susceptible cv Seneca Horizon (GS) or glyphosate-resistant Pioneer P1615XR (GR) maize seedlings were sprayed with buffer (−) or 0.5% (w/v) glyphosate (+). Six hours later, plants were infiltrated with wild-type Pnss strain DC283 or wtsE mutant strain DM5101. At 20 hai, assessments were made of CouTyr accumulation (B), symptom severity (C), and ion leakage (D). B, CouTyr level was measured in two samples each from three independent biological replicates (n = 6) by LC-MS/MS. Data are presented as percentages of the appropriate varietal treatment with DC283 in the absence of glyphosate ± sd. Asterisks indicate statistically significant differences in CouTyr levels compared with the DC283 treatment in the same variety and in the absence of glyphosate, as determined by two-tailed Student’s t tests (*P < 0.05 and **P < 0.001). C, Symptom development of inoculated plants was assessed on a scale from 1 to 6, with 1 representing plants with very severe symptoms, and 6 representing nonsymptomatic plants. Shown are combined data from three biological replicates composed of 30 or more observations per combination. D, Average values and sd from three biological replicates for cv Seneca Horizon and two biological replicates for Pioneer P1615XR. Asterisks indicate statistically significant differences compared with DC283 without glyphosate as determined by two-tailed Student’s t tests (*P < 0.05 and **P < 0.001). E, Plants of cv Seneca Horizon sprayed 6 h earlier with buffer (−) or 0.5% (w/v) glyphosate (+) were infiltrated with a low titer of DC283 or DM5101, and bacterial growth was measured over the following 66 h. Shown are average values and sd of pooled data from three independent biological replicates.
First, we determined that, as predicted, glyphosate inhibited the WtsE-induced accumulation of CouTyr (Fig. 8B). We next sought to determine whether glyphosate, by suppressing the production of CouTyr and other metabolites downstream from the shikimate pathway, could disrupt the virulence activity of WtsE. Indeed, the WtsE-dependent ability of wild-type Pnss strain DC283 to induce disease symptoms and ion leakage and to grow to high levels in maize seedlings was significantly impaired by glyphosate (Fig. 8, C–E).
Our results with glyphosate are consistent with metabolic flux through the shikimate pathway being required for the virulence activity of WtsE. However, we considered two alternative hypotheses. The first was that the inhibition of WtsE activity resulted from an adverse effect of glyphosate on the bacteria. For example, the observed ability of glyphosate to modestly compromise the in planta growth of the wtsE mutant strain (DM5101) in this experiment might have resulted from an adverse effect on the bacteria (Fig. 8E). To distinguish the effects of glyphosate on the bacteria from its effects on the plant, we utilized a Pnss-susceptible maize line (Pioneer P1615XR) that is resistant to glyphosate. Unlike some glyphosate-resistant plants, Pioneer P1615XR is not resistant due to altered uptake and subcellular partitioning of glyphosate, which could alter the exposure of the bacteria. Rather, these plants express a bacterial EPSPS enzyme that is glyphosate resistant and thus will more efficiently produce chorismate in the presence of glyphosate. Glyphosate did not significantly affect the ability of DC283 to induce CouTyr production in Pioneer P1615XR (Fig. 8B). Also, the effect of glyphosate on the ability of DC283 to cause disease symptoms and ion leakage in Pioneer P1615XR was less than that in cv Seneca Horizon (Fig. 8, C and D). Thus, while glyphosate may have a slight detrimental effect on the bacteria, this effect is insufficient to account for the observed inhibition of WtsE virulence activity inside plant cells.
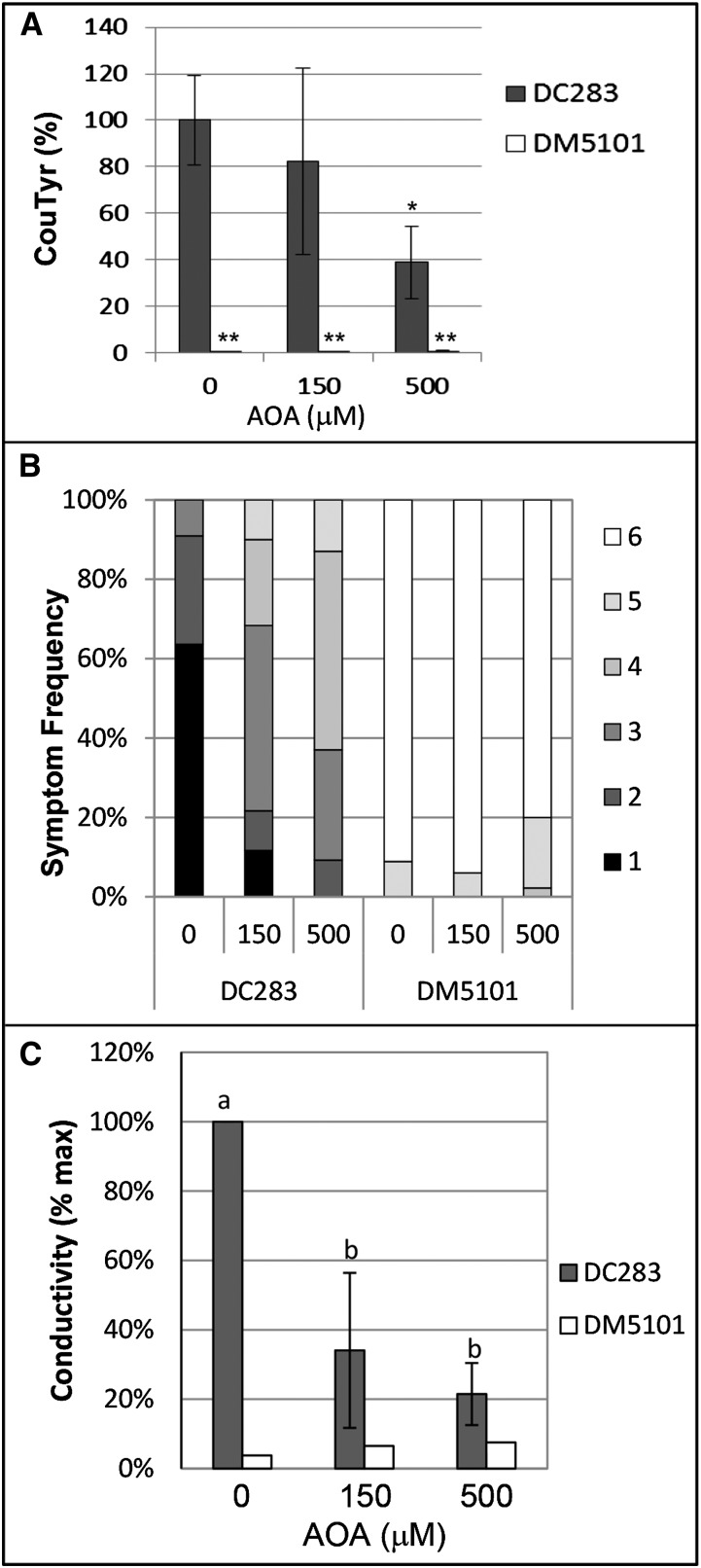
The second hypothesis we considered was that glyphosate inhibits WtsE activity by inhibiting amino acid production and thus protein synthesis. Our previous studies with cycloheximide (Ham et al., 2006, 2008) showed that inhibition of protein synthesis blocks the ability of WtsE to elicit hypersensitive response-like cell death in nonhost plants but has no effect on the ability of WtsE to induce disease-like symptoms in susceptible maize seedlings. These results argue against glyphosate inhibiting WtsE activity by inhibiting protein synthesis. As another approach to alleviate this concern, we tested the effect of (aminooxy)acetic acid (AOA), an inhibitor of PAL (Fig. 8A; Carver et al., 1991; Peiser et al., 1998). Our results indicate that, when coinfiltrated into maize seedling leaves along with Pnss strains, AOA could inhibit the ability of DC283 to induce the production of CouTyr (Fig. 9A). Furthermore, AOA attenuated WtsE-induced disease symptoms and ion leakage (Fig. 9, B and C). Thus, two independent inhibitors that block WtsE-induced CouTyr production each also blocks the virulence activity of WtsE.
Figure 9.
Inhibition of PAL attenuates the virulence activity of WtsE. Six-day-old maize seedlings were inoculated with DC283 or DM5101 along with no AOA (0) or the indicated concentrations of AOA (150 or 500 μm). A, CouTyr level was measured at 20 hai in two samples each from three independent biological replicates (n = 6) by LC-MS/MS. Within each biological replicate, the level of CouTyr induced by DC283 with no AOA was set to 100%. Error bars indicate sd, and asterisks indicate statistically significant differences in CouTyr levels compared with DC283 with no AOA (two-tailed Student’s t test, *P < 0.01 and **P < 0.001). B and C, Disease symptoms (B) and ion leakage (C) were assessed at 20 hai as in Figure 8, C and D. B, Combined data from three biological replicates composed of 45 or more observations per combination. C, Average values and sd of 10 technical replicates from three independent biological replicates, with letters indicating significant differences in CouTyr accumulation between DC283 treatments (ANOVA and Tukey’s honestly significant difference, P < 0.05).
Recent work with Ustilago maydis showed that the fungal effector Cmu1 is a chorismate mutase that attenuates SA production in infected maize (Djamei et al., 2011). We hypothesized that WtsE, through its perturbation of phenolic metabolism, might similarly suppress SA accumulation. To our surprise, Pnss, in a WtsE-dependent manner, induced strong accumulation of SA in maize seedling leaves (Supplemental Fig. S7). DC283-infiltrated leaves accumulated SA levels exceeding 2 ng g−1 fresh weight at 12 hai, prior to the onset of macroscopic disease symptoms, and more than 10 ng g−1 fresh weight at 24 hai, after the onset of water-soaked lesions. Relative to DC283, the wtsE mutant strain DM5101 induced significantly lower levels of SA at 12 and 24 h. Infiltration of maize leaves with buffer or DM711 (a type III secretion-deficient strain of Pnss) induced no detectable increase in SA above the background observed in untouched seedlings.
DISCUSSION
Despite the widespread distribution of AvrE family T3Es among important plant pathogenic bacteria and their significant contribution to the pathogenicity of these bacteria, the mode of action of these effector proteins and their effects on defense responses and pathogenesis are not well understood. Some T3Es have been shown to inhibit specific metabolic processes and thus alter the accumulation of specific plant hormones or metabolites. For example, Jelenska et al. (2007) showed that a P. syringae T3E, HopI1, inhibits SA production by targeting of a chloroplast-localized chaperone, and Zhou et al. (2011) showed that another P. syringae T3E, HopZ1b, suppresses the production of a phytoalexin precursor by targeting a soybean (Glycine max) 2-hydroxyisoflavanone dehydratase. We show here that WtsE alters the expression of an entire suite of metabolic enzymes, which results in the accumulation of CouTyr. We support the importance of these changes by showing that inactive derivatives of WtsE fail to induce the reprogramming and that chemical inhibitors specific to WtsE-induced pathways block the virulence activity of WtsE. Thus, by combining molecular and biochemical studies, we demonstrate that perturbation of phenylpropanoid metabolism is required for WtsE virulence activity. The generality of our findings to other AvrE family effectors awaits testing.
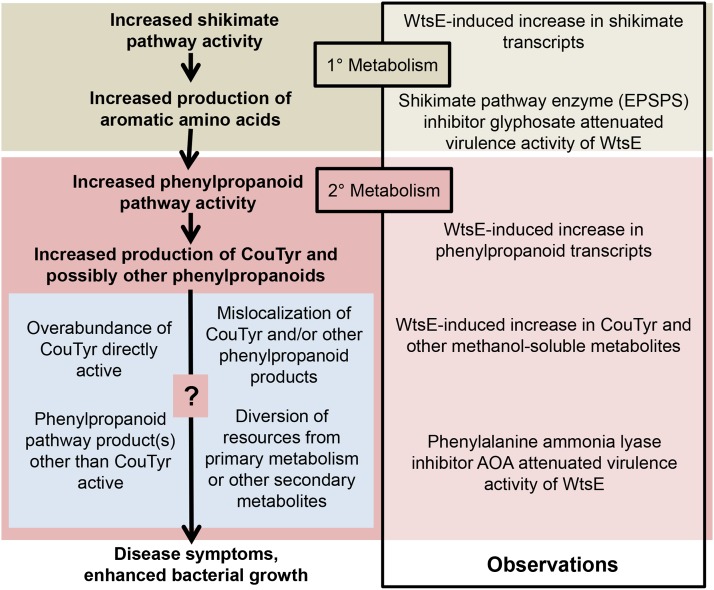
We considered that the observed metabolic effects of WtsE might be indirect. For example, by suppressing the defense response against possible elicitors associated with the EcDS or Pnss, WtsE could promote bacterial growth and/or prevent the normal feedback regulation of phenylpropanoid gene expression/metabolism, which in turn could cause exaggerated changes in gene expression and/or metabolic activity. We discount this hypothesis based on the observation that inhibitors of EPSPS and PAL block the ability of WtsE to carry out its virulence activity. We instead favor a model in which the virulence activity of WtsE is mediated through the perturbation of phenylpropanoid metabolism. Within this context, we put forth a model offering several nonexclusive possibilities for how metabolic changes might underlie the virulence activity of WtsE (Fig. 10).
Figure 10.
Model. WtsE induces phenylpropanoid metabolism to cause disease symptoms and promote bacterial growth in maize leaves. Observations made in this study are listed in the box to the right. Inferences consistent with these observations are listed in boldface type. Suggested modes of action of phenylpropanoid pathway products are listed in the blue box.
Our results reveal that WtsE affects numerous pathways localized to the chloroplast. WtsE suppressed the expression of numerous genes involved in photosynthesis, a common response in plant-microbe interactions. The perturbation of photosynthetic pathways can have profound effects on plant responses, but these are typically context specific, depending on both host and pathogen, and difficult to predict (Kangasjȁrvi et al., 2012). Conversely, WtsE induced the expression of genes in the shikimate and phenylpropanoid pathways. Chorismate, the end product of the shikimate pathway, is the precursor for the synthesis of several phytoalexins and defense-associated plant hormones as well as the aromatic amino acids Phe, Trp, and Tyr. Concomitant up-regulation of the shikimate and phenylpropanoid pathways could support the elevated production of phenolic and phenylpropanoid compounds, such as the observed WtsE-induced accumulation of SA and CouTyr. Our experiments with shikimate pathway and PAL inhibitors indicate that the virulence activity of WtsE requires flux through these pathways.
Phenylpropanoid metabolism produces an incredible variety of metabolites (Vogt, 2010) and appears to be a common target of phytopathogenic bacteria and other plant pathogens. P. syringae perturbed the expression of key enzymes of the phenylpropanoid pathway, as well as down-regulated photosynthesis, in Arabidopsis (Thilmony et al., 2006; Truman et al., 2006). In a compatible E. amylovora-apple pathosystem, the induced expression of phenylpropanoid pathway genes is delayed in a T3SS-dependent manner (Venisse et al., 2002). P. syringae type III effectors can alter the accumulation of chorismate- and Phe-derived metabolites (Jelenska et al., 2007; Zhou et al., 2011). U. maydis deploys effectors that suppress SA production (Djamei et al., 2011) and promote anthocyanin production (Tanaka et al., 2014).
The role of CouTyr during plant-microbe interactions is not clear. CouTyr was found to be bactericidal against Xanthomonas campestris (Newman et al., 2001) but not to P. syringae (Zacarés et al., 2007). In maize, CouTyr production has been demonstrated in leaf tissue in response to wounding (Ishihara et al., 2000), infection by the fungus Colletotrichum graminicola (Balmer et al., 2013), or herbivory by the caterpillar Spodoptera littoralis (Marti et al., 2013). However, in feeding experiments with S. littoralis, CouTyr did not demonstrate any biocidal activity (Marti et al., 2013). In potato (Solanum tuberosum) leaf and cell suspensions, the production of CouTyr has been shown to be induced in response to elicitors from both virulent and avirulent races of the oomycete Phytophthera infestans (Keller et al., 1996). CouTyr also is induced in the incompatible interaction of a resistant tomato cultivar with P. syringae (Zacarés et al., 2007) and in incompatible and nonhost interactions between pepper (Capsicum annuum) and X. campestris (Newman et al., 2001). Production of CouTyr by Pnss-challenged maize is an interesting case because it is dependent on a key virulence factor of a compatible pathogen. Thus, we hypothesize that WtsE-induced production of CouTyr is either directly beneficial to Pnss or an indirect consequence of other changes in phenylpropanoid metabolism that are directly beneficial to Pnss.
Efforts to identify a biological activity of CouTyr during the maize-Pnss interaction, including cell death induction and defense suppression, have been unsuccessful so far. These experiments are hindered by the poor solubility of CouTyr (maximum of 0.2 m in 2% [v/v] NaOH), and their interpretation is limited by uncertainty about whether vacuum-infiltrated CouTyr is delivered to the appropriate subcellular location in maize cells. The ultimate localization of CouTyr produced during Pnss infection is unknown. Deposition of phenolic compounds at the cell wall is considered a normal part of defense responses to fungal pathogens (Facchini et al., 2002). One possible function of CouTyr is to decrease the digestibility of cell walls, a function that would be beneficial when plants are attacked by pathogens expressing cell wall-degrading enzymes. However, Pnss is not known to produce any degradative enzymes in culture or to show evidence of cell wall degradation in planta. In potato cell suspensions treated with sufficient amounts of elicitor from P. infestans, CouTyr is secreted, and the soluble portion is almost 50 times greater than that in the cell wall or cytoplasm. An observation that is possibly related to the cell death induced by WtsE is that secretion of CouTyr by elicitor-treated cell suspensions is accompanied by their browning (Keller et al., 1996). The mechanism by which CouTyr moves out of the plant cell may involve transport carrier vesicles or transport carrier proteins. Given the potential for WtsE to affect vesicle traffic through the mimicry of GEFs, the manipulation of CouTyr localization may affect its accumulation and alter its fate in Pnss-infected maize and its role in Pnss pathogenesis.
Rather than possessing biological activity, CouTyr may indirectly affect the maize-Pnss interaction. The selective misregulation of genes involved in phenolic and phenylpropanoid metabolism could be a virulence strategy to promote or divert carbon flow into products beneficial or detrimental, respectively, to bacterial survival (Yao et al., 1995). For example, Tin2 from U. maydis is hypothesized to promote anthocyanin production to divert metabolites away from cell wall lignification that impedes the fungal infection (Tanaka et al., 2014). Similarly, WtsE-induced up-regulation of enzymes in the phenylpropanoid pathway, including those involved in the production of CouTyr and perhaps other hydroxycinnamic acid amides, could have the effect of directing carbon away from the biosynthesis of defense-associated metabolites.
The results of this work have revealed several previously unknown effects of WtsE on maize physiology. WtsE elicits major disruptions in several pathways, each focused in the chloroplast. These changes include the down-regulation of photosynthesis and the up-regulation of the shikimate and phenylpropanoid pathways. The latter causes a substantial increase in the production of the hydroxycinnamic acid amide CouTyr. Up-regulation of the shikimate pathway and phenylpropanoid pathways is directly linked by carbon flow. Although the proximity of photosynthesis enzymes to those involved in the shikimate and phenylpropanoid pathways is intriguing, it is unclear whether down-regulating photosynthesis has a direct impact on these or other pathways. These findings point to future study of WtsE, and perhaps other AvrE family T3Es, to examine how the accumulation and localization of CouTyr and other chorismate-derived metabolites affect maize and Pnss cells during their interaction.
MATERIALS AND METHODS
Bacterial Strains, Plasmids, and Growth Conditions
Strains and plasmids used in this study are listed in Table II. Bacteria were routinely grown in Luria-Bertani (LB) broth or agar (Sambrook and Russell, 2001) supplemented with the appropriate antibiotics and grown at 28°C for Pantoea stewartii ssp. stewartii or 37°C for Escherichia coli.
Table II. Strains and plasmids used in this study.
| Name | Description | Reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| MC4100 | araD139 Δ(argF-lac)205 λ− flbB5301 ptsF25 relA1 rpsL150 deoC1 | Casadaban (1976) |
| DH5α | mcrBC+ supE44 ΔlacU169 (f80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Grant et al. (1990) |
| Pnss | ||
| DC283 | A spontaneous nalidixic acid resistant (Nalr) derivative of the wild-type strain SS104 | Coplin et al. (1986) |
| DM711 | DC283 hrpJ79::Tn3HoHoI, ampicilln resistant (Apr) | Frederick et al. (2001) |
| DM5101 | A wtsE::miniTn5gus derivative of DC283, kanamycin resistant (Kmr) | Ham et al. (2006) |
| Plasmids | ||
| pCPP2156 | Plasmid encoding hrp type III secretion system gene cluster from D. dadantii, specinomycin resistant | Ham et al. (1998) |
| pCR4-TOPO | Vector for TOPO cloning PCR products, Apr, Kmr | Invitrogen |
| pDM5109 | pCR4-TOPO with wtsEΔ709-1017-FLAG and wtsF, Apr, Kmr | This study |
| pDM5117 | pRK415 with wtsEΔ709-1017-FLAG and wtsF, tetracycline resistant (Tcr) | This study |
| pDM5189 | pLAFR3 with wtsEΔ709-1017 and wtsF, Tcr | This study |
| pDM5141 | pCR4-TOPO with wtsEΔ2935-3234-FLAG and wtsF, Apr, Kmr | This study |
| pDM5153 | pRK415 with wtsEΔ2935-3234-FLAG and wtsF, Tcr | This study |
| pDM5184 | pLAFR3 with wtsEΔ2935-3234 and wtsF, Tcr | This study |
| pDM5143 | pCR4-TOPO with wtsEΔ3235-3534-FLAG and wtsF, Apr, Kmr | This study |
| pDM5155 | pRK415 with wtsEΔ3235-3534-FLAG and wtsF, Tcr | This study |
| pDM5182 | pLAFR3 with wtsEΔ3235-3534 and wtsF, Tcr | This study |
| pJA001 | pLAFR3 with wtsE-F operon and native promoter, Tcr | This study |
| pDM5175 | pLAFR3 with wtsE-w12 (W694A and W840A) and wtsF, Tcr | This study |
| pJA006 | Plasmid derived from pJH001 encoding WtsF and a C terminally FLAG-tagged WtsE driven by native promoter in pCR4-TOPO vector (Invitrogen), Apr, Kmr | This study |
| pJA017 | pRK415 with wtsE encoding a C-terminal FLAG tag driven by native promoter and wtsF, Tcr | This study |
| pJA045 | pCR4-TOPO with wtsE-w12 encoding a C-terminal FLAG tag and wtsF, Apr, Kmr | This study |
| pJA052 | pRK415 with wtsF and wtsE-w12 encoding a C-terminal FLAG tag, Tcr | This study |
| pJH001 | Plasmid containing wtsE-F operon with native promoter in pCR4-TOPO vector, Apr, Kmr | Ham et al. (2008) |
| pJH021 | Plasmid containing wtsE-F operon with native promoter in pBluescript SK(+) (Stratagene), Apr | Ham et al. (2008) |
| pJH055 | Plasmid containing wtsF and wtsE with w12 mutations in pBluescript SK(+), Apr | Ham et al. (2009) |
| pCPP1250 | Plasmid containing dspA/E-F from E. amylovora in pML123, gentamicin resistant | Bogdanove et al. (1998) |
| pLAFR3 | Incompatibility group P cosmid, αlacZ, Tcr | Staskawicz et al. (1987) |
| pRK415 | Incompatibility group P αlacZ, Tcr | Keen et al. (1988) |
For vacuum infiltrations and inoculations of maize (Zea mays) seedlings, overnight cultures of E. coli or Pnss were centrifuged at 2,500g for 10 min, and bacteria were resuspended in 10 mm potassium phosphate buffer, pH 7.2, containing 0.2% (v/v) Tween 40. For E. coli, the optical density at 540 nm was adjusted to 1. For Pnss, the optical density at 540 nm was adjusted to 0.57 (equivalent to 1 × 109 colony-forming units [cfu] mL−1). This 1 × 109 cfu mL−1 suspension was used for all vacuum infiltration-based experiments, with the exception of the growth curve in Figure 9E, for which the bacteria were diluted to 1 × 106 cfu mL−1 prior to infiltration. For whorl inoculations and associated growth and virulence measurements (Fig 4, C and D), the bacteria were diluted to 1 × 108 cfu mL−1 prior to inoculation.
Construction of Plasmids
pJA001 was created by digesting pJH021 DNA (Ham et al., 2009) with restriction enzymes EcoRI and BamHI (Invitrogen) and ligating the resulting 6.4-kb fragment (containing the wtsE-F operon and its native promoter from Pnss) into the vector pLAFR3 (Staskawicz et al., 1987), also cut with EcoRI and BamHI.
Plasmid pJA017 encodes WtsF and C terminally FLAG-tagged, but otherwise unmutated, WtsE (WtsE-FLAG) in the vector pRK415. In making pJA017, pJH021 was used as template in separate PCRs with primers 5′-ACAGCCTCAATGCTAAACTCACAGAAG-3′ (wtsE F1386A-F) with 5′-TCATTTGTCGTCATCGTCTTTGTAGTCGCTTTTCATTTCAAACCCTTC-3′ (wtsE-FLAG N R) and 5′-GACTACAAAGACGATGACGACAAATGAGGTTATGATTAATTCATC-3′ (wtsE-FLAG C F) with the M13 Reverse universal primer 5′-AGCGGATAACAATTTCACACAGG-3′. The products were used together as template for PCR with external primers wtsE F1386A-F and M13 Reverse. The newly created fragment was then digested with BamHI and SstI and ligated into pJH001, which harbors the same insert as pJH021 (Ham et al., 2008) cut with the same enzymes, thus creating the intermediate plasmid JA006 in the high copy number background of pCR4-TOPO (Invitrogen). The entire wtsE-F region was then excised from pJA006 using EcoRI and BamHI and ligated into pRK415 to create pJA017.
Plasmid pDM5153 contains wtsEΔ2935-3234-FLAG and wtsF. Two separate PCRs were performed using pJH021 as template and primers 5′-GCACTTCACAACCTTTCCGAT-3′ (wtsE outer 3-F) paired with 5′-GTTAAAGCTTTGTATTTGAAGATTGTATGCAATATTTTTCAG-3′ (wtsE d8-R) and 5′-GCATACAATCTTCAAATACAAAGCTTTAACGTTAACCG-3′ (wtsE d8-F) paired with 5′-ACGACCAAACGTGAGATTTATAAGC-3′ (wtsE outer 3-R). The resulting products were then used together as template in a PCR using wtsE outer 3-F and wtsE outer 3-R as primers. The PCR product was cut with PstI and KpnI and ligated into pJA006 cut with the same enzymes, creating the intermediate plasmid pDM5141. The insert from pDM5141 was excised using EcoRI and BamHI and cloned into pRK415, creating pDM5153.
Plasmid pDM5155 contains wtsEΔ3235-3534-FLAG and wtsF. Two separate PCR products were created using wtsE outer 3-F paired with 5′-ATGTAGCTGGGCTATCTGTGCCGCTTTACTGATCGG-3′ (wtsE d9-R) and 5′-ATCAGTAAAGCGGCACAGATAGCCCAGCTACAT-3′ (wtsE d9-F) with wtsE outer 3-R. The resulting products were then used together as template in PCR using wtsE outer 3-F and wtsE outer 3-R as primers. The resulting PCR product was cut with PstI and KpnI and ligated into pJA006 cut with the same enzymes, creating the intermediate plasmid pDM5143. The insert from pDM5143 was excised using EcoRI and BamHI and cloned into pRK415, creating pDM5155.
Plasmid pDM5117 contains wtsEΔ709-1017-FLAG and wtsF. To construct the partial deletion of wtsE, we took advantage of two naturally occurring restriction sites: ClaI (bases 502–507 of the wtsE-coding DNA sequence) and NdeI (bases 1,018–1,023). A fragment representing bases 315 to 708 of wtsE was amplified via PCR using the primers 5′-CAGTCGGATTCGGAGACCCAC-3′ (wtsE outer 1-F) and 5′-GCGCCATATGATCAAGATGCCGCTGCGGAAG-3′ (wtsE d1-R NdeI). This fragment was then digested with the enzymes ClaI and NdeI, creating a 206-bp fragment that was ligated into pJA006 cut with the same enzymes. This created the intermediate plasmid pDM5109. The insert from pDM5109 was then cut with EcoRI and BamHI and ligated into pRK415 to create pDM5117.
Plasmid pJA052 encodes wtsE-FLAG with the w12 mutations (i.e. W694A and W840A, mutations of both WxxxE motifs) and wtsF. In making pJA052, a 1,836-bp KpnI-to-PstI fragment containing wtsE w12 was cut from pJH055 (Ham et al., 2009), gel purified, and ligated into pJA006 cut with the same enzymes, creating the intermediate plasmid pJA045. Plasmid pJA045 was digested with EcoRI and BamHI and ligated into pRK415 cut with the same enzymes, creating pJA052.
To construct plasmid pDM5189 containing wtsEΔ709-1017 and wtsF, a fragment of WtsE (155–708 bp) was amplified from pJH021 using primers 5′-CCAGTATGCCCGCTATTCGTCAGC-3′ (wtsE outer F) and wtsE d1-R NdeI. The PCR product was digested with BspE and NdeI and ligated to pJH021 digested with the same enzymes to create an intermediate plasmid, which was then digested with EcoRI and BamHI and cloned into pLAFR3 to create pDM5189.
To construct plasmid pDM5184 containing wtsEΔ2935-3234 and wtsF, the same PCRs were performed as to construct plasmid pDM5153. The resulting PCR product was digested with PstI and KpnI, cloned into pJH021 digested by the same enzymes to create an intermediate plasmid, and the fragment created by digestion with EcoRI and BamHI was cloned into pLAFR3.
To construct plasmid pDM5182 containing wtsEΔ3235-3534 and wtsF, the same PCRs were performed as to construct plasmid pDM5155. The resulting PCR product was digested with PstI and KpnI, cloned into pJH021 to create an intermediate plasmid, and the fragment created by digestion with EcoRI and BamHI was cloned into pLAFR3.
To construct plasmid pDM5157, a fragment containing wtsE with the w12 mutations and wtsF was released from pJH055 by EcoRI and BamHI and ligated into pLAFR3.
PCR products to be used as template in further PCRs were resolved by agarose gel electrophoresis and excised from the gel prior to purification. PCR products were cleaned before digestion, and digested DNA was cleaned before ligation. DNA was cleaned using the QIAquick PCR purification kit (Qiagen). Digested vectors were routinely treated with calf intestinal alkaline phosphatase and cleaned prior to ligation. The sequences of DNA fragments cloned using PCR were confirmed by DNA sequencing at the Plant-Microbe Genomics Facility at Ohio State University. Manipulation of DNA was performed according to standard techniques (Ausubel et al., 1997).
Secretion Assays
Overnight cultures of Pnss in LB broth were pelleted by centrifugation, washed twice, and suspended in hrp-inducing medium (Ahmad et al., 2001) supplemented with 1 mm isopropylthio-β-galactoside and appropriate antibiotics. hrp-inducing medium cultures were grown overnight at 22°C and adjusted to equal optical densities before harvesting. Pellet and supernatant fractions were separated by centrifugation at 20,200g. To the supernatant fraction, phenylmethylsulfonyl fluoride was used at a concentration of 1 to 2 mm to prevent protein degradation. TCA was then added to a concentration of 10% (v/v), and the fraction was incubated on ice overnight to precipitate proteins. Proteins were pelleted by centrifugation at 20,200g for 30 min at 4°C and washed in acetone. Bacterial cell and TCA-precipitated protein pellets were resuspended in 1/50th and 1/350th of the original culture or supernatant volume, respectively, in 20 mm Tris, pH 7.5, supplemented with 1 mm phenylmethylsulfonyl fluoride.
Proteins were separated by SDS-PAGE using a 10% (w/v) resolving gel. Proteins were blotted onto Immobilon-P polyvinylidene difluoride (Millipore) using the Mini-Trans Blot Cell (Bio-Rad) according to the manufacturer’s instructions. Blots were developed with α-FLAG M2 and α-β-galactosidase primary antibodies and anti-mouse (Jackson ImmunoResearch) and anti-rabbit (Amersham, GE Healthcare) secondary antibodies, respectively.
Plant Growth, Infiltration, and Inoculation
Seeds of sweet maize (variety rugosa) ‘Seneca Horizon’ or maize lines Pioneer P1615XR (a glyphosate-resistant hybrid) and B73 were sown in 4.5-inch-diameter pots in Metro-Mix 360 (Sun Gro Horticulture), eight to 10 seeds per pot, and were watered daily for 5 d. Plants were grown in a growth chamber maintained at 30°C with a daily cycle of 18 h of daylight (85 μE m−2 s−1) and 6 h of darkness.
Six-day-old maize seedlings were vacuum infiltrated with bacteria as described above. A humidifier was placed in the growth chamber to increase the relative humidity to greater than 65%. For most infiltrations, the humidifier was placed in the chamber 2 d prior to infiltration. For plants sprayed with glyphosate, humidifiers were placed in the chamber the morning of infiltration. Plants lacking a visible second true leaf or that appeared unhealthy were culled. Soil was covered, and the pots were inverted over beakers containing inoculum or buffer so that the seedlings were submerged. Seedlings were then vacuum infiltrated three times, 5 min each time, at a minimum vacuum pressure of 200 mm Hg. First true leaves from infiltrated plants were examined against the light immediately following infiltration, and those appearing well infiltrated were labeled for harvesting and/or scoring at later times. WtsE-induced disease symptoms were scored on a scale from 1 to 6 (with 1 indicating most severe and 6 indicating no symptoms; Supplemental Fig. S8). At harvest, 3.5- to 5-cm lengths (including the tips) of the first true leaves were tested for ion leakage or immediately frozen in liquid nitrogen and stored at −80°C. The remaining, unharvested leaves were observed at 24 to 36 hai, and the experiment was carried forward only if WtsE-induced symptoms were robust.
For whorl inoculation of seedlings with Pnss strains (Fig. 4, C and D), plants were not watered for 2 d prior to inoculation. To inoculate the plants, 50 µL of inoculum was pipetted directly into the whorl of 6-d-old maize seedlings and allowed to dry. After drying, plants were watered and maintained in the growth chamber until growth measurement or symptom assessment was done 3 d following inoculation. Pnss-induced disease symptoms following whorl inoculation were rated on a scale from 0 to 3 (with 3 indicating most severe and 0 indicating no symptoms) as described previously (Ahmad et al., 2001; Ham et al., 2008).
Ion Leakage
Following bacterial infiltration, the tips of first true leaves were removed at the indicated times. Tissue alongside a ruler was optically scanned, and then two leaf tips per vial were placed in 30 mL of milli-Q purified water (Millipore) in glass screw-cap tubes. Because leaf pieces tended to float out of the water and cling to the sides of the tubes, plungers from 10-mL syringes were added to the tubes to raise the water level and gently force leaves to remain in the liquid. Leaf tissue was allowed to sit in water for approximately 30 min, vials were shaken to homogenize liquid, and then electrical conductivity was measured using a Cond 330i (WTW). Leaf area was determined using ImageJ (Schneider et al., 2012), and conductivity per cm2 of leaf area was calculated for each sample.
Chemical Inhibitors
For glyphosate treatment, 6-d-old cv Seneca Horizon maize seedlings were sprayed with 0% or 0.5% (w/v) glyphosate (Syngenta) in 0.2% (v/v) Tween 40. After 5 h, plants were inverted in water for 10 min to wash away glyphosate remaining on leaves. Plants then were infiltrated with Pnss strains, and tissue was collected, as described above.
For AOA treatment, 6-d-old cv Seneca Horizon maize seedlings were vacuum infiltrated with inoculum containing Pnss strains, as described above, and a final concentration of 0, 150, or 500 μm AOA (Sigma-Aldrich). A 25 mm stock solution of AOA in water was produced fresh on the day of the experiment and added to the inoculum.
Recovery and Enumeration of Bacteria from Leaf Tissue
To assess bacterial titer in plant tissues, maize seedlings were cut off at the soil line (Fig. 4C) and placed in 5 mL of 10 mm potassium phosphate buffer, pH 7.2, or leaf discs were harvested (Fig. 8E) and placed in 200 μL of the same buffer in a microcentrifuge tube. Maize tissue was thoroughly macerated using a Power Gen 700 homogenizer (Fisher Scientific) or microfuge grinding tip. Supernatant from the grinding was serially diluted, and the dilutions were spotted onto LB agar plates supplemented with appropriate antibiotics. The cfu per milliliter of supernatants was calculated and compared with the initial plated inoculum (Fig. 4C), or a measurement was made shortly after infiltration (Fig. 8E) to assess bacterial multiplication in planta.
Microarray Tissue Collection and Preparation of RNA
Seedlings of cv Seneca Horizon were vacuum infiltrated with EcWtsE (E. coli MC4100[pCPP2156, pJA001]) or EcDS with empty vector (E. coli MC4100[pCPP2156, pLAFR3]), and tissue was collected at 6 hai. Tissue for microarray experiments represented four biological replicates. Plants for one biological replicate were grown, infiltrated, and harvested before the seeds for the following biological replicate were sown. Tissue from the first true leaves of four separate plants that had been infiltrated and harvested on the same day was pooled to form each biological replicate. Tissue was ground with a mortar and pestle under liquid nitrogen, and RNA was isolated using the RNAqueous kit (Ambion) according to kit instructions. RNA yield and purity were estimated using a NanoDrop (Thermo Fisher Scientific) and visualized by agarose gel electrophoresis. RNA from the four separate biological replicates was used for hybridization of four separate two-channel microarrays, with RNA from treatment plants (those infiltrated with EcWtsE) being compared with that from control plants (those infiltrated with EcDS) within each array. RNA was sent to the Maize Oligonucleotide Array Project at the University of Arizona for RNA amplification, RNA labeling, hybridization, and reading of the arrays. Array protocols were done according to Galbraith et al. (2011). Arrays were scanned using a GenePix 4000AL microarray scanner, and raw data were extracted with the GenePix 6.0 program and analyzed according to Jandova et al. (2012).
Statistical analysis of microarrays also was performed at the Maize Oligonucleotide Array Project. Statistical analysis was performed in R (http://www.r-project.org/) using the Limma statistical package. Loess normalization was used to obtain the average intensity of each gene from the four arrays. Significant differences between treatment and control groups were assessed using Student’s t test (α = 0.02). Significant differences in dye effect were assessed using Student’s t test (α = 0.05).
Annotation of Microarray Results and Analysis of GO Terms and KEGG Pathways
GO terms were assigned to the differentially expressed probes using the AgriGO platform (Du et al., 2010); AgriGO was then used to do a hypergeometric distribution test with Yekutieli correction (false discovery rate under dependency) to identify overrepresented terms in the groups of repressed or induced probes when compared with GO terms assigned to all microarray probes. Probes also were assigned to maize KEGG genes and KEGG pathway terms using Kobas 2.0 (mapping e < 0.01; Xie et al., 2011). A hypergeometric distribution test with Yekutieli correction also was applied to identify overrepresented KEGG pathways.
Quantitative PCR
Genes involved in the phenylpropanoid pathway were chosen for examination by qRT-PCR. The genes chosen were TD (MaizeGDB gene identifier GRMZM2G093125), P/AD (GRMZM2G437912), 4CL (GRMZM2G174732), PAL (GRMZM2G170692), and cinnamic acid to 4-coumaric acid (GRMZM2G147245). Primers for actin (GRMZM2G126010) were used as a control for normalizing results. Because it was unclear from microarray results which possible splice variants were altered in expression between treatment and control plants, PCR primers were not designed to amplify products spanning introns (Supplemental Table S3).
First true leaves from bacteria-infiltrated plants were collected at 2, 4, 6, 13, or 19 hai. Two leaves were pooled for each biological replicate. RNA was isolated using the RNAqueous kit, and RNA quality was checked via Nanodrop. RNA preparations were treated with DNase I Amplification Grade (Invitrogen) according to the manufacturer’s instructions. Production of complementary DNA was done according to the Reverse Transcription System (Promega). Quantitative PCR assays were performed on two biological replicates, each in triplicate. Reactions were 25 µL in volume and run on 96-well plates in the iQ5 Multicolor Real-Time PCR Detection System (Bio-Rad) using the following program: 95°C for 3 min; 40 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s; followed by melting-curve analysis from 55°C to 95°C in 30-s cycles. Data were analyzed using the iQ5 Optical System Software.
Metabolite Extraction from Maize Seedling Leaves
Leaf tip samples were collected in 1.5-mL centrifuge tubes (DOT Scientific; part 609) and ground in liquid nitrogen using a plastic pestle. Approximately 22 mg of each sample was transferred to a second 1.5-mL centrifuge tube and ground a second time. One milliliter of 80% (v/v) methanol (LC-MS/MS grade; Fisher Scientific; part A456-1) was added into each sample, along with 10 μL of 50 ng μL−1 formononetin internal standard (Sigma-Aldrich) dissolved in 100% LC-MS/MS-grade methanol. Samples were vortexed, extracted at 4°C for 20 min on a nutator, and finally centrifuged at 12,000g for 15 min at 4°C. The resulting supernatants were then filtered through 0.2-μm nylon filters and diluted 100-fold with 100% methanol.
LC-MS/MS Analyses
During LC-MS/MS analyses, samples were stored at a temperature of 4°C in a chilled autosampler chamber. For quantification of CouTyr, 1 μL of each sample was injected into an Agilent 1260 Infinity LC system, and compounds were separated using an Agilent Poroshell 120 EC-C18 (3.5 × 50 mm, 2.7 μm) column at 30°C and an acidified water:methanol buffer system (buffer A, 0.1% [v/v] acetate and 5% [v/v] methanol in water; buffer B, 0.1% [v/v] acetate in water). Gradient conditions were as follows: 1-min hold at 20% B, 3 min at 20% to 80% B, 5 min at 80% to 100% B, 2-min hold at 100% B, and back to 20% B in 1.5 min. Eluted compounds were further separated and quantified using an Agilent G6460 triple quadrupole dual mass spectrometer equipped with an electrospray ionization source. LC-MS/MS settings were as follows: gas temperature of 250°C, gas flow rate of 10 L min−1, nebulizer pressure of 60 p.s.i., sheath gas flow of 12 L min−1, sheath gas temperature of 400°C, capillary voltage of 3,000 V, and nozzle voltage of 700 V. Compounds were quantified using positive ion mode, with four mass transitions (one quantification transition and three confirmatory qualification transitions) monitored for each compound of interest (Supplemental Table S2), and verified through comparison with authentic standards. The limit of quantitation for CouTyr for this method was calculated to be 51 ng g−1 tissue harvested.
Ultra High-Performance Liquid Chromatography Measurement of SA
Leaf tissues were weighed, frozen on liquid nitrogen, and ground with two 3.2-mm stainless steel beads in each tube using a Qiagen TissueLyser. SA was extracted twice with 500 μL per 100 mg fresh weight of 100% methanol containing 1 mm butylated hydroxyanisole (Sigma-Aldrich). Each extraction was for 24 h in the dark at 4°C, and the clear supernatant was removed after centrifugation at 13,000 rpm for 15 min in a tabletop microcentrifuge. The two pooled extractions were stored in darkness at −20°C. Ultra high-performance liquid chromatography analysis was done within 1 week of extraction. Extracts were run through a 0.2-μm, 2.1-mm guard filter (Waters) prior to chromatographic separation on a 2.1- × 100-mm, 1.7-μm ACQUITY UPLC RP18 BEH shield column (Waters) using the Acquity UPLC system (Waters). Column temperature was set to 30°C, and the sample manager was maintained at 4°C. HPLC-grade water with 2% (v/v) acetic acid (A) and HPLC-grade methanol with 2% (v/v) acetic acid (B) were used as solvents. An 11.33-min gradient program with a flow rate of 0.3 mL min−1 was used: 95% A and 5% B at 0 min, 90% A and 10% B at 0.67 min, 50% A and 50% B at 6.67 min, 20% A and 80% B at 10 min, 100% B at 10.67 min, and 95% A and 5% B at 11.33 min. SA was detected using fluorescence at emission wavelength of 400 nm. Peak areas of SA in standards and tissue samples were normalized to the butylated hydroxyanisole internal standard. Calibration curves were plotted by peak areas of a serial diluted standard of commercial SA (Sigma-Aldrich) with a correlation coefficient higher than 0.999.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Symptoms and ion leakage in maize leaves infiltrated with EcWtsE.
Supplemental Figure S2. Statistical significance versus fold change of maize transcripts with EcWtsE or EcDS.
Supplemental Figure S3. Standard curves for quantification of CouTyr.
Supplemental Figure S4. WtsE-induced phenolic metabolism gene expression in maize seedlings.
Supplemental Figure S5. DspA/E-induced phenolic metabolism gene expression in maize seedlings.
Supplemental Figure S6. Macroscopic effects of glyphosate on maize seedlings.
Supplemental Figure S7. Quantification of salicylic acid induction following infiltration of maize seedlings with Pnss.
Supplemental Figure S8. Leaves displaying virulence phenotypes as scored in Figures 8C and 9B.
Supplemental Table S1. WtsE elicited significantly altered expression of numerous genes from maize seedlings in microarray experiments.
Supplemental Table S2. Mass spectrometry settings for CouTyr and formononetin quantitations.
Supplemental Table S3. Primers used in quantitative PCR experiments.
Supplementary Material
Acknowledgments
We thank Terry Graham and Mike Kelly for help with LC-MS/MS, Enrico Bonello for help with ultra high-performance liquid chromatography, Katie Wilkins for help with data sorting, and Jason Parrish for assistance with glyphosate application and glyphosate-resistant maize.
Glossary
- Pnss
Pantoea stewartii subsp. stewartii
- T3SS
type III secretion system
- T3E
type III effector
- ERMRS
endoplasmic reticulum membrane retention signal
- GEF
guanine-exchange factor
- PAMP
pathogen-associated molecular pattern
- SA
salicylic acid
- qRT
quantitative reverse transcription
- CouTyr
coumaroyl tyramine
- EcDS
Escherichia coli delivery system
- EcWtsE
WtsE delivery system
- hai
hours after infiltration
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- LC-MS/MS
liquid chromatography and triple quadrupole dual mass-spectroscopy
- m/z
mass-to-charge ratio
- TD
tyrosine decarboxylase
- 4CL
4-coumarate-coenzyme A ligase
- C4H
cinnamate 4-hydroxylase
- PAL
phenylalanine ammonia lyase
- P/AD
prephanate/arogenate dehydratase
- EPSPS
5-enolpyruvoylshikimate-3-phosphate synthase
- AOA
(aminooxy)acetic acid
- LB
Luria-Bertani
- cfu
colony-forming units
Footnotes
This work was supported by the National Science Foundation (grant no. MCB–0718882), the U.S. Department of Agriculture (grant no. National Institute of Food and Agriculture 2008–35319–04506), the Korean Rural Development Administration Next-Generation BioGreen Program (System and Synthetic Agro-Biotech Center and grant nos. PJ009088 and PJ011091), and the Ohio Agricultural Research and Development Center of Ohio State University.
Articles can be viewed without a subscription.
References
- Ahmad M, Majerczak DR, Pike S, Hoyos ME, Novacky A, Coplin DL (2001) Biological activity of harpin produced by Pantoea stewartii subsp. stewartii. Mol Plant Microbe Interact 14: 1223–1234 [DOI] [PubMed] [Google Scholar]
- Ahmad S, Veyrat N, Gordon-Weeks R, Zhang Y, Martin J, Smart L, Glauser G, Erb M, Flors V, Frey M, et al. (2011) Benzoxazinoid metabolites regulate innate immunity against aphids and fungi in maize. Plant Physiol 157: 317–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahuja I, Kissen R, Bones AM (2012) Phytoalexins in defense against pathogens. Trends Plant Sci 17: 73–90 [DOI] [PubMed] [Google Scholar]
- Alto NM, Shao F, Lazar CS, Brost RL, Chua G, Mattoo S, McMahon SA, Ghosh P, Hughes TR, Boone C, et al. (2006) Identification of a bacterial type III effector family with G protein mimicry functions. Cell 124: 133–145 [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, More DD, Seidman JG, Smith JA, Struhl K (1997) Short Protocols in Molecular Biology: A Compendium of Methods from Current Protocols in Molecular Biology, Ed 3.John Wiley & Sons, New York [Google Scholar]
- Badel JL, Shimizu R, Oh HS, Collmer A (2006) A Pseudomonas syringae pv. tomato avrE1/hopM1 mutant is severely reduced in growth and lesion formation in tomato. Mol Plant Microbe Interact 19: 99–111 [DOI] [PubMed] [Google Scholar]
- Balmer D, de Papajewski DV, Planchamp C, Glauser G, Mauch-Mani B (2013) Induced resistance in maize is based on organ-specific defence responses. Plant J 74: 213–225 [DOI] [PubMed] [Google Scholar]
- Beckman CH. (2000) Phenolic-storing cells: keys to programmed cell death and periderm formation in wilt disease resistance and in general defence responses in plants? Physiol Mol Plant Pathol 57: 101–110 [Google Scholar]
- Bednarek P, Pislewska-Bednarek M, Svatos A, Schneider B, Doubsky J, Mansurova M, Humphry M, Consonni C, Panstruga R, Sanchez-Vallet A, et al. (2009) A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323: 101–106 [DOI] [PubMed] [Google Scholar]
- Block A, Alfano JR (2011) Plant targets for Pseudomonas syringae type III effectors: virulence targets or guarded decoys? Curr Opin Microbiol 14: 39–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanove AJ, Kim JF, Wei Z, Kolchinsky P, Charkowski AO, Conlin AK, Collmer A, Beer SV (1998) Homology and functional similarity of an hrp-linked pathogenicity locus, dspEF, of Erwinia amylovora and the avirulence locus avrE of Pseudomonas syringae pathovar tomato. Proc Natl Acad Sci USA 95: 1325–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boureau T, ElMaarouf-Bouteau H, Garnier A, Brisset MN, Perino C, Pucheu I, Barny MA (2006) DspA/E, a type III effector essential for Erwinia amylovora pathogenicity and growth in planta, induces cell death in host apple and nonhost tobacco plants. Mol Plant Microbe Interact 19: 16–24 [DOI] [PubMed] [Google Scholar]
- Brunner S, Fengler K, Morgante M, Tingey S, Rafalski A (2005) Evolution of DNA sequence nonhomologies among maize inbreds. Plant Cell 17: 343–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver TLW, Robbins MP, Zeyen RJ (1991) Effects of two PAL inhibitors on the susceptibility and localized autofluorescent host cell responses of oat leaves attacked by Erysiphe graminis DC. Physiol Mol Plant Pathol 39: 269–287 [Google Scholar]
- Casadaban MJ. (1976) Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J Mol Biol 104: 541–555 [DOI] [PubMed] [Google Scholar]
- Chen LQ, Hou BH, Lalonde S, Takanaga H, Hartung ML, Qu XQ, Guo WJ, Kim JG, Underwood W, Chaudhuri B, et al. (2010) Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 468: 527–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coplin DL, Frederick RD, Majerczak DR, Haas ES (1986) Molecular cloning of virulence genes from Erwinia stewartii. J Bacteriol 168: 619–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa VR, Majerczak DR, Ammar D, Merighi M, Pratt RC, Hogenhout SA, Coplin DL, Redinbaugh MG (2012) The bacterium Pantoea stewartii uses two different type III secretion systems to colonize its plant host and insect vector. Appl Environ Microbiol 78: 6327–6336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DebRoy S, Thilmony R, Kwack YB, Nomura K, He SY (2004) A family of conserved bacterial effectors inhibits salicylic acid-mediated basal immunity and promotes disease necrosis in plants. Proc Natl Acad Sci USA 101: 9927–9932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslandes L, Rivas S (2012) Catch me if you can: bacterial effectors and plant targets. Trends Plant Sci 17: 644–655 [DOI] [PubMed] [Google Scholar]
- Djamei A, Schipper K, Rabe F, Ghosh A, Vincon V, Kahnt J, Osorio S, Tohge T, Fernie AR, Feussner I, et al. (2011) Metabolic priming by a secreted fungal effector. Nature 478: 395–398 [DOI] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38: W64–W70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facchini PJ, Hagel J, Zulak KG (2002) Hydroxycinnamic acid amide metabolism: physiology and biochemistry. Can J Bot 80: 577–589 [Google Scholar]
- Frederick RD, Ahmad M, Majerczak DR, Arroyo-Rodríguez AS, Manulis S, Coplin DL (2001) Genetic organization of the Pantoea stewartii subsp. stewartii hrp gene cluster and sequence analysis of the hrpA, hrpC, hrpN, and wtsE operons. Mol Plant Microbe Interact 14: 1213–1222 [DOI] [PubMed] [Google Scholar]
- Galbraith DW, Janda J, Lambert GM (2011) Multiparametric analysis, sorting, and transcriptional profiling of plant protoplasts and nuclei according to cell type. Methods Mol Biol 699: 407–429 [DOI] [PubMed] [Google Scholar]
- Gaudriault S, Malandrin L, Paulin JP, Barny MA (1997) DspA, an essential pathogenicity factor of Erwinia amylovora showing homology with AvrE of Pseudomonas syringae, is secreted via the Hrp secretion pathway in a DspB-dependent way. Mol Microbiol 26: 1057–1069 [DOI] [PubMed] [Google Scholar]
- Geng X, Cheng J, Gangadharan A, Mackey D (2012) The coronatine toxin of Pseudomonas syringae is a multifunctional suppressor of Arabidopsis defense. Plant Cell 24: 4763–4774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant SG, Jessee J, Bloom FR, Hanahan D (1990) Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc Natl Acad Sci USA 87: 4645–4649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ham JH, Bauer DW, Fouts DE, Collmer A (1998) A cloned Erwinia chrysanthemi Hrp (type III protein secretion) system functions in Escherichia coli to deliver Pseudomonas syringae Avr signals to plant cells and to secrete Avr proteins in culture. Proc Natl Acad Sci USA 95: 10206–10211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ham JH, Majerczak D, Ewert S, Sreerekha MV, Mackey D, Coplin D (2008) WtsE, an AvrE-family type III effector protein of Pantoea stewartii subsp. stewartii, causes cell death in non-host plants. Mol Plant Pathol 9: 633–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ham JH, Majerczak DR, Arroyo-Rodriguez AS, Mackey DM, Coplin DL (2006) WtsE, an AvrE-family effector protein from Pantoea stewartii subsp. stewartii, causes disease-associated cell death in corn and requires a chaperone protein for stability. Mol Plant Microbe Interact 19: 1092–1102 [DOI] [PubMed] [Google Scholar]
- Ham JH, Majerczak DR, Nomura K, Mecey C, Uribe F, He SY, Mackey D, Coplin DL (2009) Multiple activities of the plant pathogen type III effector proteins WtsE and AvrE require WxxxE motifs. Mol Plant Microbe Interact 22: 703–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Sutton SE, Wallenfang AJ, Orchard RC, Wu X, Feng Y, Chai J, Alto NM (2009) Structural insights into host GTPase isoform selection by a family of bacterial GEF mimics. Nat Struct Mol Biol 16: 853–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara A, Kawata N, Matsukawa T, Iwamura H (2000) Induction of N-hydroxycinnamoyltyramine synthesis and tyramine N-hydroxycinnamoyltransferase (THT) activity by wounding in maize leaves. Biosci Biotechnol Biochem 64: 1025–1031 [DOI] [PubMed] [Google Scholar]
- Jandova J, Janda J, Sligh JE (2012) Changes in mitochondrial DNA alter expression of nuclear encoded genes associated with tumorigenesis. Exp Cell Res 318: 2215–2225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelenska J, Yao N, Vinatzer BA, Wright CM, Brodsky JL, Greenberg JT (2007) A J domain virulence effector of Pseudomonas syringae remodels host chloroplasts and suppresses defenses. Curr Biol 17: 499–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kangasjȁrvi S, Neukermans J, Li S, Aro EM, Noctor G (2012) Photosynthesis, photorespiration, and light signalling in defence responses. J Exp Bot 63: 1619–1636 [DOI] [PubMed] [Google Scholar]
- Keen NT, Tamaki S, Kobayashi D, Trollinger D (1988) Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70: 191–197 [DOI] [PubMed] [Google Scholar]
- Keller H, Hohlfeld H, Wray V, Hahlbrock K, Scheel D, Strack D (1996) Changes in the accumulation of soluble and cell wall-bound phenolics in elicitor-treated cell suspension cultures and fungus-infected leaves of Solanum tuberosum. Phytochemistry 42: 389–396 [Google Scholar]
- Kvitko BH, Park DH, Velásquez AC, Wei CF, Russell AB, Martin GB, Schneider DJ, Collmer A (2009) Deletions in the repertoire of Pseudomonas syringae pv. tomato DC3000 type III secretion effector genes reveal functional overlap among effectors. PLoS Pathog 5: e1000388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti G, Erb M, Boccard J, Glauser G, Doyen GR, Villard N, Robert CA, Turlings TC, Rudaz S, Wolfender JL (2013) Metabolomics reveals herbivore-induced metabolites of resistance and susceptibility in maize leaves and roots. Plant Cell Environ 36: 621–639 [DOI] [PubMed] [Google Scholar]
- Melotto M, Underwood W, Koczan J, Nomura K, He SY (2006) Plant stomata function in innate immunity against bacterial invasion. Cell 126: 969–980 [DOI] [PubMed] [Google Scholar]
- Naoumkina MA, Zhao Q, Gallego-Giraldo L, Dai X, Zhao PX, Dixon RA (2010) Genome-wide analysis of phenylpropanoid defence pathways. Mol Plant Pathol 11: 829–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MA, von Roepenack-Lahaye E, Parr A, Daniels MJ, Dow JM (2001) Induction of hydroxycinnamoyl-tyramine conjugates in pepper by Xanthomonas campestris, a plant defense response activated by hrp gene-dependent and hrp gene-independent mechanisms. Mol Plant Microbe Interact 14: 785–792 [DOI] [PubMed] [Google Scholar]
- Nomura K, Debroy S, Lee YH, Pumplin N, Jones J, He SY (2006) A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science 313: 220–223 [DOI] [PubMed] [Google Scholar]
- Oh CS, Martin GB, Beer SV (2007) DspA/E, a type III effector of Erwinia amylovora, is required for early rapid growth in Nicotiana benthamiana and causes NbSGT1-dependent cell death. Mol Plant Pathol 8: 255–265 [DOI] [PubMed] [Google Scholar]
- Pedersen HA, Steffensen SK, Christophersen C, Mortensen AG, Jørgensen LN, Niveyro S, de Troiani RM, Rodríguez-Enríquez RJ, Barba-de la Rosa AP, Fomsgaard IS (2010) Synthesis and quantitation of six phenolic amides in Amaranthus spp. J Agric Food Chem 58: 6306–6311 [DOI] [PubMed] [Google Scholar]
- Peiser G, López-Gálvez G, Cantwell M, Saltveit ME (1998) Phenylalanine ammonia lyase inhibitors control browning of cut lettuce. Postharvest Biol Technol 14: 171–177 [Google Scholar]
- Sambrook J, Russell DW (2001) Molecular Cloning: A Laboratory Manual, Ed 3.Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Schaeffer ML, Harper LC, Gardiner JM, Andorf CM, Campbell DA, Cannon EK, Sen TZ, Lawrence CJ (2011) MaizeGDB: curation and outreach go hand-in-hand. Database (Oxford) 2011: bar022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, et al. (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert F, Thiemann A, Pospisil H, Scholten S (2012) Re-annotation of the maize oligonucleotide array. Maydica 57: 49–55 [Google Scholar]
- Staskawicz B, Dahlbeck D, Keen N, Napoli C (1987) Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J Bacteriol 169: 5789–5794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinrücken HC, Amrhein N (1980) The herbicide glyphosate is a potent inhibitor of 5-enolpyruvyl-shikimic acid-3-phosphate synthase. Biochem Biophys Res Commun 94: 1207–1212 [DOI] [PubMed] [Google Scholar]
- Tanaka S, Brefort T, Neidig N, Djamei A, Kahnt J, Vermerris W, Koenig S, Feussner K, Feussner I, Kahmann R (2014) A secreted Ustilago maydis effector promotes virulence by targeting anthocyanin biosynthesis in maize. eLife 3: e01355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thilmony R, Underwood W, He SY (2006) Genome-wide transcriptional analysis of the Arabidopsis thaliana interaction with the plant pathogen Pseudomonas syringae pv. tomato DC3000 and the human pathogen Escherichia coli O157:H7. Plant J 46: 34–53 [DOI] [PubMed] [Google Scholar]
- Treutter D. (2005) Significance of flavonoids in plant resistance and enhancement of their biosynthesis. Plant Biol (Stuttg) 7: 581–591 [DOI] [PubMed] [Google Scholar]
- Truman W, de Zabala MT, Grant M (2006) Type III effectors orchestrate a complex interplay between transcriptional networks to modify basal defence responses during pathogenesis and resistance. Plant J 46: 14–33 [DOI] [PubMed] [Google Scholar]
- Venisse JS, Malnoy M, Faize M, Paulin JP, Brisset MN (2002) Modulation of defense responses of Malus spp. during compatible and incompatible interactions with Erwinia amylovora. Mol Plant Microbe Interact 15: 1204–1212 [DOI] [PubMed] [Google Scholar]
- Vielle-Calzada JP, Martínez de la Vega O, Hernández-Guzmán G, Ibarra-Laclette E, Alvarez-Mejía C, Vega-Arreguín JC, Jiménez-Moraila B, Fernández-Cortés A, Corona-Armenta G, Herrera-Estrella L, et al. (2009) The Palomero genome suggests metal effects on domestication. Science 326: 1078. [DOI] [PubMed] [Google Scholar]
- Vogt T. (2010) Phenylpropanoid biosynthesis. Mol Plant 3: 2–20 [DOI] [PubMed] [Google Scholar]
- White DG. (1999) Compendium of Corn Diseases, Ed 3.APS Press, St. Paul [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, Ausubel FM (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565 [DOI] [PubMed] [Google Scholar]
- Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, Kong L, Gao G, Li CY, Wei L (2011) KOBAS 2.0: a Web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res 39: W316–W322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao K, De Luca V, Brisson N (1995) Creation of a metabolic sink for tryptophan alters the phenylpropanoid pathway and the susceptibility of potato to Phytophthora infestans. Plant Cell 7: 1787–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacarés L, López-Gresa MP, Fayos J, Primo J, Bellés JM, Conejero V (2007) Induction of p-coumaroyldopamine and feruloyldopamine, two novel metabolites, in tomato by the bacterial pathogen Pseudomonas syringae. Mol Plant Microbe Interact 20: 1439–1448 [DOI] [PubMed] [Google Scholar]
- Zheng XY, Spivey NW, Zeng W, Liu PP, Fu ZQ, Klessig DF, He SY, Dong X (2012) Coronatine promotes Pseudomonas syringae virulence in plants by activating a signaling cascade that inhibits salicylic acid accumulation. Cell Host Microbe 11: 587–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Lin J, Johnson A, Morgan RL, Zhong W, Ma W (2011) Pseudomonas syringae type III effector HopZ1 targets a host enzyme to suppress isoflavone biosynthesis and promote infection in soybean. Cell Host Microbe 9: 177–186 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.