Transcription factors in seedling cotyledons are quantified using novel silicon photonic crystal protein arrays and their levels are correlated with transcript abundances.
Abstract
Transcription factors control important gene networks, altering the expression of a wide variety of genes, including those of agronomic importance, despite often being expressed at low levels. Detecting transcription factor proteins is difficult, because current high-throughput methods may not be sensitive enough. One-dimensional, silicon-substrate photonic crystal (PC) arrays provide an alternative substrate for printing multiplexed protein microarrays that have greater sensitivity through an increased signal-to-noise ratio of the fluorescent signal compared with performing the same assay upon a traditional aminosilanized glass surface. As a model system to test proof of concept of the silicon-substrate PC arrays to directly detect rare proteins in crude plant extracts, we selected representatives of four different transcription factor families (zinc finger GATA, basic helix-loop-helix, BTF3/NAC [for basic transcription factor of the NAC family], and YABBY) that have increasing transcript levels during the stages of seedling cotyledon development. Antibodies to synthetic peptides representing the transcription factors were printed on both glass slides and silicon-substrate PC slides along with antibodies to abundant cotyledon proteins, seed lectin, and Kunitz trypsin inhibitor. The silicon-substrate PC arrays proved more sensitive than those performed on glass slides, detecting rare proteins that were below background on the glass slides. The zinc finger transcription factor was detected on the PC arrays in crude extracts of all stages of the seedling cotyledons, whereas YABBY seemed to be at the lower limit of their sensitivity. Interestingly, the basic helix-loop-helix and NAC proteins showed developmental profiles consistent with their transcript patterns, indicating proof of concept for detecting these low-abundance proteins in crude extracts.
Transcription factors act as master regulators controlling the expression of suites of multiple genes. Despite this important function, they are often expressed at very low levels, both as RNA and as protein, because only a small amount may be necessary to activate a cascade of other genes. This makes transcription factors difficult to study, especially at the protein level, where fewer sensitive, high-throughput tools are currently available.
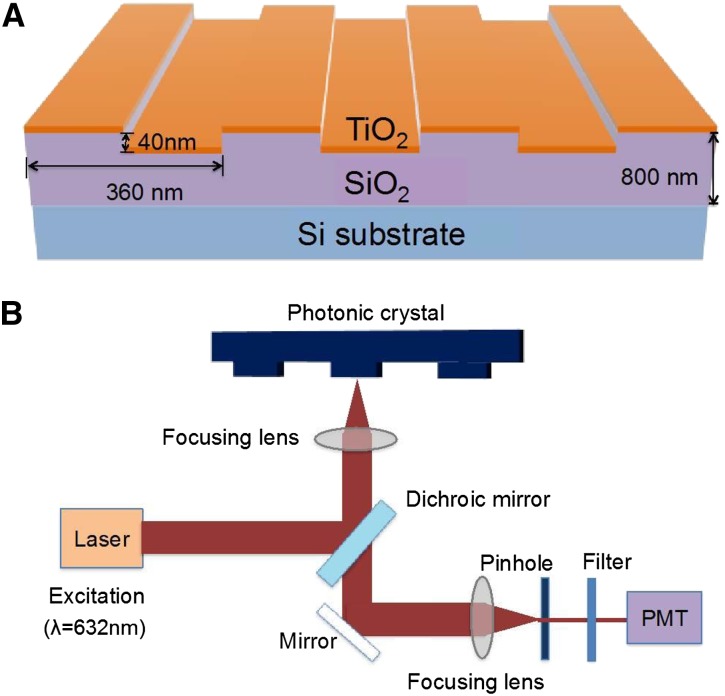
One-dimensional photonic crystals (PCs) have been developed as an alternative surface to the aminosilanized glass slides that have been successfully used for high-throughput study of gene expression by complementary DNA (cDNA) microarrays (Jones et al., 2010). As shown in Figure 1, a precise, nanoscale grating of silicon dioxide topped with layers of highly refractive materials, such as titanium dioxide, allows the structure to be tuned to provide two resonance wavelengths—one at the excitation wavelength of a desired fluorescent reporter molecule and the other at the fluorophore’s emission wavelength (for review, see Cunningham and Zangar, 2012; Chaudhery et al., 2013). Referred to as photonic crystal enhanced fluorescence (PCEF), this dual-resonance property increases the signal-to-noise ratio for fluorescent tags that are captured on the PC surface, allowing fluorescent molecules from the sample that attach to capture spots on the surface to be more readily distinguished from the background than they are on a regular aminosilanized glass slide. The sensitivity of the PC structure was further enhanced by the use of low-autofluorescent silicon as the bottommost layer. In this report, we refer to arrays printed on these PC devices as silicon-substrate PCs arrays, whereas those printed on aminosilanized GAPSII (Corning) glass slides are referred to, for simplicity, as glass slides.
Figure 1.
Schematics of the PC structure and detection instrument. The PC device (A) is comprised of a periodic surface structure fabricated in a low-refractive index SiO2 layer on a silicon substrate and then overcoated with a thin film of high-refractive index TiO2. The device is further coated with a layer of epoxysilane, and the protein array is printed on top. The detection instrument (B) is a Tecan LS Reloaded Confocal Laser Microarray Scanner fitted with a 632.8-nm, 5-mW laser for Cyanine5 (Cy5) excitation and a Cy5 emission filter (band pass, 670–715 nm). See “Materials and Methods” for more details. PMT, Photomultiplier tube.
Work with a cDNA array printed on a PC surface (Mathias et al., 2010) showed that the PC surface doubled or tripled the number of genes that could be detected above background compared with traditional aminosilanized glass slides. Furthermore, studies in which PC surfaces (Huang et al., 2011; George et al., 2013) were printed with antibodies related to a set of cancer biomarkers showed detection of proteins at concentrations in the range of 0.3 pg mL−1 to 10 ng mL−1.
Other types of PC structures that do not use fluorescence include biosensors, in which detection is performed by measuring shifts in the PC resonant wavelength for various analytes that bind to the surface (Pal et al., 2011; Scullion et al., 2011; Chakravarty et al., 2013; Zou et al., 2014). One recent report illustrates use of the microcavity biosensors to detect a lung cancer antigen in lysates of a recombinant lung cancer cell line, in which expression of the antigen is induced (Chakravarty et al., 2013). However, neither PCEF arrays nor PC biosensors have been tested for performance to detect proteins in plant systems. Using antibodies to well-known seed proteins as controls alongside antibodies generated to synthetic peptides representing transcription factors, we tested the silicon-substrate PCs for biological performance with plant crude extracts to detect changes in these low-abundance transcription factors in cotyledons during early seedling growth.
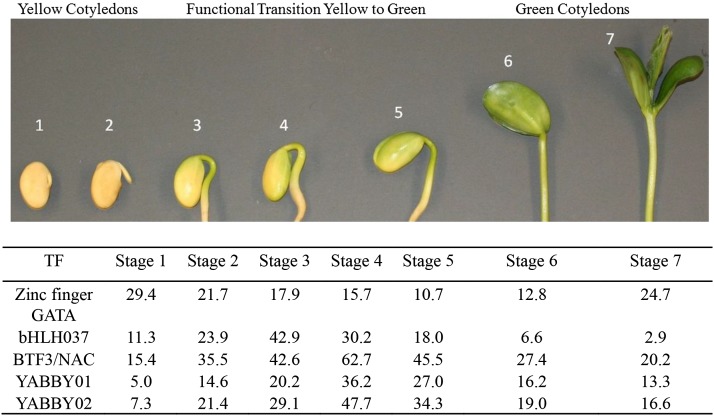
The antibodies were printed as arrays on silicon-substrate PCs in parallel during the same printing run with traditional aminosilanized glass substrate slides to examine the sensitivity of each to detect changes in abundances of six proteins over seven stages of cotyledons dissected from germinating seedlings. During development of the immature seed, the cotyledons are the storage organs of the seed, filling with a large number of transcripts and proteins, some highly abundant. The seeds dehydrate as they mature and may be quiescent for months or years before imbibition of water causes germination to begin (Bewley, 1997). The previously stored products in the cotyledons are used as food for the seedling in its early growth, but around stage 3 (approximately 3 d after germination), the cotyledons begin to turn from yellow to green and become photosynthesizing organs that produce nutrients for the seedlings, in essence serving as the first leaves. This is a complete transformation in function that occurs in just a few days.
After approximately a week, the first true leaves become functional, and the cotyledons eventually senesce. This complex developmental process is certainly accompanied by many changes in gene regulation, because the cellular machinery is repurposed. Global views of the gene expression changes over these seven stages of seedling cotyledon development in soybean (Glycine max) have been performed using both microarrays (Gonzalez and Vodkin, 2007) and high-throughput RNA sequencing (RNA-Seq; Shamimuzzaman and Vodkin, 2014). Here, we examine six proteins—including four transcription factors—at these same stages, pairing the protein abundance data from silicon PC slides with gene expression data starting from the very early stages of immature seed development. We show proof of concept with this sensitive technology by using it to gain a more complete understanding of the changes in low-abundance transcription factors occurring during this dynamic sequence of seedling cotyledon development.
RESULTS
Selection of Four Transcription Factor Genes That Increase in Expression in the Cotyledons during Early Seedling Growth
Cotyledons were taken from seven stages of early seedling development of soybean beginning with dry mature seeds that had been soaked in water for approximately 24 h (stage 1) and ending with seedlings approximately 6- to 7-cm tall above ground and fully green, with the first true leaves prominent (stage 7) as shown in Figure 2. Unless otherwise stated, all biological samples and data were from cv Williams. Seedling cotyledons from the same seven stages of this cultivar had been used to produce high-throughput next generation transcriptome sequencing data using Illumina technology to yield 46 to 75 million RNA-Seq reads per stage, which are sufficient to detect low-abundance transcripts representing transcription factors (Shamimuzzaman and Vodkin, 2014). The soybean genome contains approximately 5,600 gene models that are annotated as transcription factors (Jones and Vodkin, 2013). In each of the seven stages of seedling cotyledons, approximately 1,000 to 1,700 transcription factors were expressed with normalized reads per kilobase of gene model per million mapped reads (RPKM) levels of ≥5, and 479 showed notable expression of at least 30 RPKM in at least one of seven seedling cotyledon stages (Shamimuzzaman and Vodkin, 2014). From that data set, we selected four transcription factors that increased in the cotyledons during early seedling growth, as shown in Figure 2 and Supplemental Table S1. The four transcription factors included members of the zinc finger, basic helix-loop-helix (bHLH), nascent polypeptide-associated complex (NAC), and YABBY families that have been investigated in other plant systems (Bailey et al., 2003; Manfield et al., 2007; Bartholmes et al., 2012; Kang et al., 2013) using genetic and other approaches. However, their detection at the protein level has rarely been described without the use of reporter fusions or overexpression in transgenic plants. Our goal here was to directly detect these low-abundance proteins from crude plant extracts using a potentially high-throughput, highly sensitive microarray format.
Figure 2.
Seven stages of soybean cotyledon development and transcription factor expression. Stage 1 is after 24 h of imbibition of dry mature seed. Stage 7 is approximately 7 d after planting. During stages 3 to 5, the cotyledons began to turn green and undergo a functional transition from a storage organ to a photosynthesizing organ. Stage numbers are indicated above each image. RNA-Seq data for transcription factor gene models at each stage are included at bottom. (YABBY is represented by two gene models.) For the full RNA-Seq data and gene model numbers, see Supplemental Table S1. TF, Transcription factor.
Polyclonal Antibodies Generated to Synthetic Peptides Representing Epitopes of the Four Selected Transcription Factor Proteins
The four selected transcription factor proteins (bHLH037, Basic Transcription Factor3 [BTF3]/NAC, YABBY, and zinc finger GATA) are presumably of very low abundance based on RNA-Seq data, and therefore, they are not amenable to direct purification. Thus, we chose to synthesize peptides of 15 amino acids to predicted antigenic regions for each transcription factor as shown in Table I (formore information, see Supplemental Table S2). The rabbit polyclonal antibodies were purified by affinity chromatography with the cognate peptide. For two of these transcription factors, BTF3/NAC and YABBY, the same antibodies have been shown to bind the transcription factors in chromatin immunoprecipitation experiments with the same tissue of seedling cotyledons coupled with high-throughput sequencing to reveal the suites of genes that they regulate (Shamimuzzaman and Vodkin, 2013).
Table I. Antibodies printed on arrays used to detect transcription factors and control seed proteins.
Antigen sequences, concentrations, number of spots printed per slide, and other information are shown. Soybean gene model names here are identical between genome versions Gm109 a1.v1.0 and Gm189 a1.v1.1 unless otherwise noted. The sequences from Gm189 a1.v1.1 were used to determine the amino acid position numbers. All matches between peptide sequence and gene model were 100%. All antibodies are polyclonal rabbit and affinity purified. N/A, Not applicable.
| Antibody | Peptide Sequence (Antigen) | Soybean Gene Model | Amino Acid Position | Concentration | Spots/Slide | Reference |
|---|---|---|---|---|---|---|
| mg mL−1 | ||||||
| Zinc finger GATAa | SQSQDRGEDDSNSNC | Glyma06g01110.1 | 73–86 | 0.404 | 4 | This work |
| bHLH037a | DPESIKPPKRRNVKC | Glyma17g19830.1 | 140–153 | 0.810 | 4 | This work |
| BTF3/NACa | VRTGGKGTMRRKKKC | Glyma01g18780.1 | 13–26 | 0.376 | 4 | Shamimuzzaman and Vodkin (2013) |
| YABBYa | CTEERVVNRPPEKRQ | Glyma13g22620.1 | 99–112 | 0.622 | 4 | Shamimuzzaman and Vodkin (2013) |
| Glyma17g12200.1 | 99–112 | |||||
| Lectin | Whole protein | Glyma02g01590.1 | N/A | 0.880 | 8 | Vodkin (1981) |
| Trypsin inhibitor | Whole protein | Glyma01g10900.1 | N/A | 0.340 | 4 | Vodkin (1981) |
| Glyma08g45530.1b |
From synthetic peptides produced by GenScript Corporation. A C residue in the first or last position (C terminus or N terminus) of the antigen sequence was added to facilitate conjugation.
Glyma08g45531.1 in a1.v1.1 of the soybean genome.
In addition, we used two polyclonal antibodies that had previously been produced against the native purified seed proteins, soybean lectin and soybean trypsin inhibitor, to serve as controls. The lectin and trypsin inhibitor antibodies were also affinity purified and have been used many times in conventional immunoprecipitation experiments and western blots (Vodkin, 1981; Jofuku and Goldberg, 1989; Lindstrom et al., 1990) showing their specificity.
Comparison of Conventional Glass Arrays with Silicon-Substrate PC Arrays
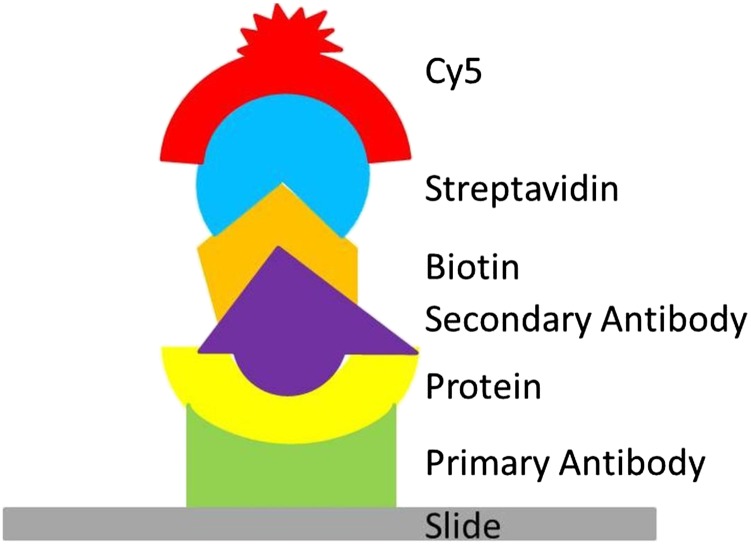
Primary antibodies for the six soybean proteins were printed on either glass slides or silicon-substrate PC devices attached to glass slides during the same printer run. Crude protein was extracted from the seedling cotyledon tissue at each stage and diluted to 25 μg mL−1 of total protein. A sandwich assay approach was used, with an equal number of glass and silicon-substrate PC slides. Figure 3 illustrates the assay, in which slides were incubated in first, the crude protein mixture, then, biotinylated secondary antibodies, and finally, streptavidin-Cy5, providing a reporter molecule visible when the slides were scanned. The slide images were then quantitated, yielding intensity values on a per-spot basis. A total of four slides across two biological replicates was used for each stage. Background and negative controls were subtracted from the average of all replicate spots for each protein.
Figure 3.
Illustration of the sandwich assay as described in the text. Primary antibodies were printed onto silicon-substrate PCs or glass slides. Slides were then sequentially incubated with crude protein extract, biotinylated secondary antibodies, and streptavidin-Cy5. Cy5 is the reporter molecule visible to laser scanners.
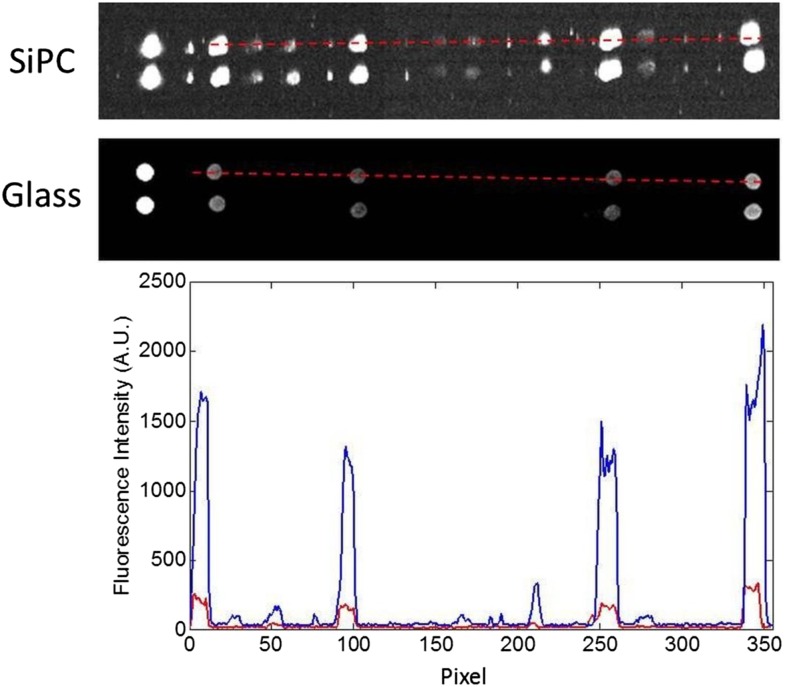
Spots printed on silicon-substrate PC slides consistently generated higher fluorescence intensity values than spots on glass slides under the same incubation and scanning conditions. Figure 4 shows fluorescence intensity of pixels on images of identical rows on both a silicon-substrate PC slide and a glass slide for the same stage. As can be seen both by eye and in the plot of values, the silicon-substrate PC slide produced a higher intensity for most protein spots, whether high or low abundance. The optical properties of the silicon-substrate PC slide allowed it to have a better signal-to-noise ratio than the ordinary glass slide, meaning that the intensity of Cy5-labeled pixels stood out better from the background of the slide. Thus, fainter spots were distinguished on the silicon-substrate PC that were not differentiated from background on the glass slide.
Figure 4.
Fluorescence intensity of spots on silicon-substrate PCs and glass slides. Top and middle, Identical rows from a silicon-substrate PC (SiPC) slide and a glass slide. The red dotted lines indicate spots with intensity plotted in the graph below. Spots to the left of red dotted lines are orientation spots and negative control spot (from left to right). Bottom, Silicon-substrate PC slide intensity values are in blue, and glass slide intensity values are in red. Slides were incubated with the same tissue extract and scanned under identical conditions.
Profiles of Transcription Factor Proteins Detected in Early Seedling Cotyledons on Silicon-Substrate PC versus Glass Arrays
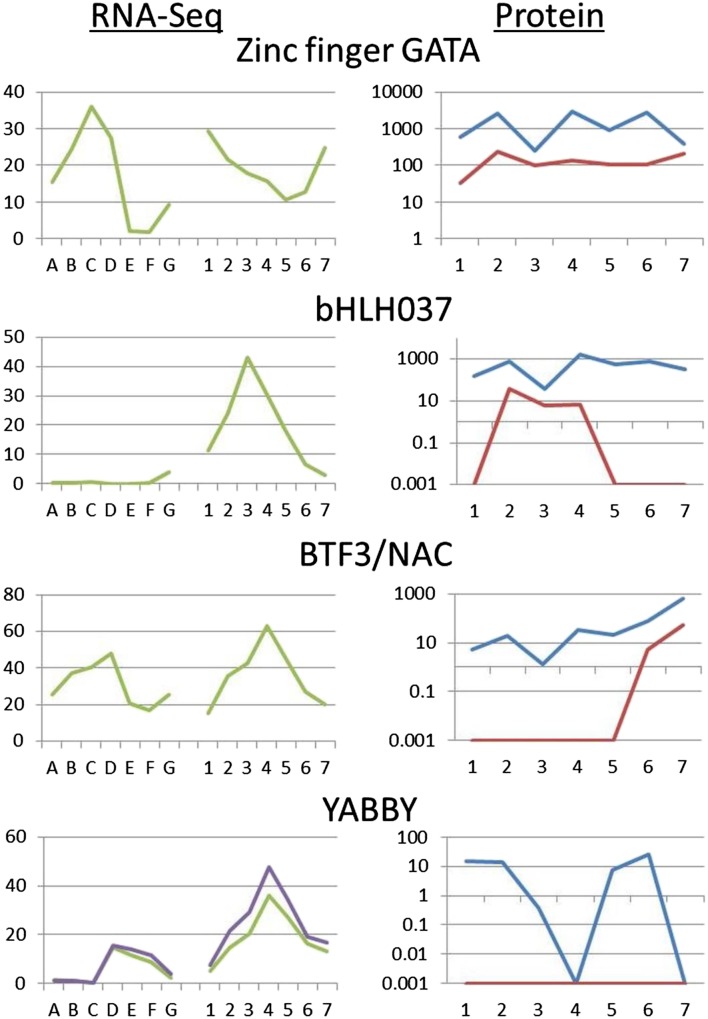
The protein expression patterns of four transcription factors are shown in Figure 5, and direct values are detailed in Table II for each of the seven stages of seedling cotyledon development. Each value represents 16 spots (32 for the control lectin protein) across four slides (including two biological replicates). The four transcription factor proteins showed differing patterns across seedling cotyledon development as well as differing results from the two types of slides. Two of the transcription factors, zinc finger GATA and bHLH037, had high values at most stages on the silicon-substrate PCs, although bHLH037 had lower numbers than zinc finger GATA at every stage. However, on the glass slides, the two proteins showed different profiles. Zinc finger GATA was found at every stage, but bHLH037 was not found above background at four of seven stages on the glass slides.
Figure 5.
Developmental gene expression and protein abundance data for four transcription factors. RNA-Seq data (left) in RPKMs on the y axis show gene expression at seven stages of immature seeds (stages A–G) and seven stages of seedling cotyledons (stages 1–7). Multiple lines indicate data from multiple gene models. Protein array data (right) show protein abundance at seven stages of seedling cotyledons for both silicon-substrate PC slides (blue) and glass slides (red). A data point of 0.001 indicates that the protein was undetected at that stage. All protein data are shown on a log scale. Numbers 1 to 7 are seedling cotyledon stages as described in Figure 2 and the text. Stages on the x axis are defined as A to G. A, 4 DAF; B, 12 to 14 DAF; C, 22 to 24 DAF; D, 5- to 6-mg whole seeds; E, 100- to 200-mg cotyledon; F, 400- to 500-mg cotyledon; G, dry mature seeds.
Table II. Protein array data over seven stages of seedling cotyledon development on glass and silicon-substrate PC slides.
Average intensity of 16 measurements (32 measurements for lectin) per protein from four slides minus background and negative controls. For details of calculations, see “Materials and Methods.” Negative values are transformed to 0.001.
| Antibody | Slide Type | Stage 1 | Stage 2 | Stage 3 | Stage 4 | Stage 5 | Stage 6 | Stage 7 |
|---|---|---|---|---|---|---|---|---|
| Zinc finger GATA | PC | 583.3 | 2,555.1 | 250.4 | 2,878.5 | 891.9 | 2,821.5 | 396.2 |
| bHLH037 | PC | 146.8 | 747.7 | 37.1 | 1,589.5 | 568.6 | 791.2 | 337.1 |
| BTF3/NAC | PC | 5.3 | 19.9 | 1.3 | 34.3 | 20.5 | 79.1 | 647.5 |
| YABBY | PC | 15.2 | 13.6 | 0.4 | 0.001 | 7.6 | 25.6 | 0.001 |
| Lectin | PC | 232.2 | 762.4 | 151.1 | 782.5 | 931.6 | 1,580.3 | 643.8 |
| Trypsin inhibitor | PC | 288.6 | 2,345.6 | 346.0 | 1,598.4 | 665.4 | 267.0 | 866.7 |
| Zinc finger GATA | Glass | 32.9 | 237.8 | 97.4 | 138.9 | 103.1 | 104.2 | 214.9 |
| bHLH037 | Glass | 0.001 | 36.6 | 6.1 | 6.7 | 0.001 | 0.001 | 0.001 |
| BTF3/NAC | Glass | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 5.2 | 53.9 |
| YABBY | Glass | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 |
| Lectin | Glass | 87.7 | 99.9 | 33.7 | 203.1 | 418.1 | 294.1 | 144.5 |
| Trypsin inhibitor | Glass | 139.1 | 181.4 | 18.5 | 300.8 | 234.7 | 339.6 | 1,622.8 |
The other two transcription factors in this study, BTF3/NAC and YABBY, showed much lower protein abundance throughout the seedling cotyledon stages. BTF3/NAC displayed a dramatic rise in protein abundance on the silicon-substrate PCs, increasing over 500 times between stages 3 and 7. On the glass slides, BTF3/NAC was not detected above background until the final two stages of highest abundance. In contrast, the YABBY protein showed consistently low abundance on the silicon-substrate PCs, and at two stages, it was not detected above background; however, on the glass slides, it was not detected above background at any stage.
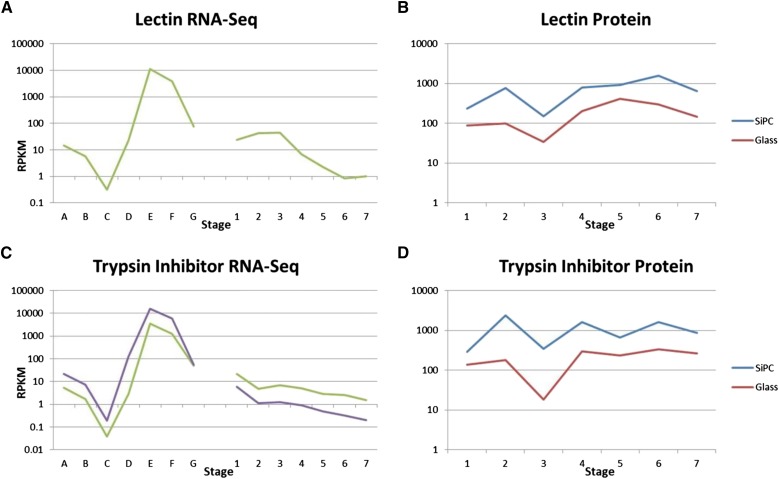
The protein abundance data for two seed storage proteins in the same tissues are shown in Table II and Figure 6. These proteins displayed very high abundance on both types of slides. On the silicon-substrate PCs, they both had values over a range of 8- to 10-fold. Thus, the abundance of these proteins was relatively stable compared with some of the transcription factors, such as BTF3/NAC, with its increase of 500-fold over time. On the glass slides, the seed proteins had similar abundance profiles, although always with smaller numbers, indicative of the higher sensitivity of the silicon-substrate PC slides. The most variation in values was introduced by the biological replicates as shown in Supplemental Figure S1; the se calculations for final values are shown in Supplemental Table S3.
Figure 6.
Developmental gene expression and protein abundance data for two storage proteins. All graphs are log scale. RNA-Seq data (A and C) in RPKMs on the y axis show gene expression at seven stages of immature seeds (A–G) and seven stages of seedling cotyledons (1–7). Multiple lines indicate data from multiple gene models. Protein array data (B and D) show protein abundance at seven stages of seedling cotyledons for both silicon-substrate PC slides (SiPCs; blue) and glass slides (red). A data point of 0.001 indicates that the protein was undetected at that stage. All protein data are shown on log scale. Numbers 1 to 7 are seedling cotyledon stages as described in Figure 2 and the text. Stages on the x axis are defined as A to G. A, 4 DAF; B, 12 to 14 DAF; C, 22 to 24 DAF; D, 5- to 6-mg whole seeds; E, 100- to 200-mg cotyledon; F, 400- to 500-mg cotyledon; G, dry mature seeds.
In summary, the transcription factors and seed proteins display a variety of expression profiles, including dramatic changes in abundance across the seven stages of development. The silicon-substrate PC slides also detect low-abundance transcription factors, which had values that fell below background on the glass slides.
Comparative Transcript Profiles of the Six Proteins throughout Immature Cotyledon and Seedling Cotyledon Development
As shown in Figures 5 and 6 and Supplemental Table S1, we charted the expression of the genes representing these six proteins from RNA-Seq data derived from immature seed cotyledons (Jones and Vodkin, 2013) as well as seedling cotyledons (Shamimuzzaman and Vodkin, 2014). The RPKM values for the transcription factors tended to be fairly low, with the highest value being around 60 for BTF3/NAC. Both BTF3/NAC and YABBY had similar gene expression profiles, which peaked at the 5- to 6-mg whole-seed stage (BTF3/NAC being about 2 times as high as YABBY). In the seedling cotyledon stages, both genes peaked sharply at stage 4, with YABBY having slightly lower expression overall.
In the immature seed stages, bHLH037 had extremely low gene expression, with RPKMs less than 1 at all stages until a slight increase at the dry seed stage, which seemed to be part of the buildup to the peak at seedling cotyledon stage 3. In contrast, zinc finger GATA peaked sharply at 22 to 24 days after flowering (DAF) whole seed, although it also started to increase again at the dry whole-seed stage, leading to a relatively high value at stage 1 in the seedling cotyledons. After dropping to a low at stage 5, its gene expression began to increase again.
The two storage protein genes had very similar expression profiles to each other as expected. Both had tremendously high expression in the 100- to 200-mg cotyledons, with RPKMs around 10,000, but then, declined rapidly. During the seedling cotyledon stages, gene expression for these gene models was quite low, with RPKMs less than 40 at all stages.
Use of a Lectin-Negative Isoline Shows Lack of Nonspecific Cross Reaction in Cotyledon Extracts Using the Silicon-Substrate PC Arrays
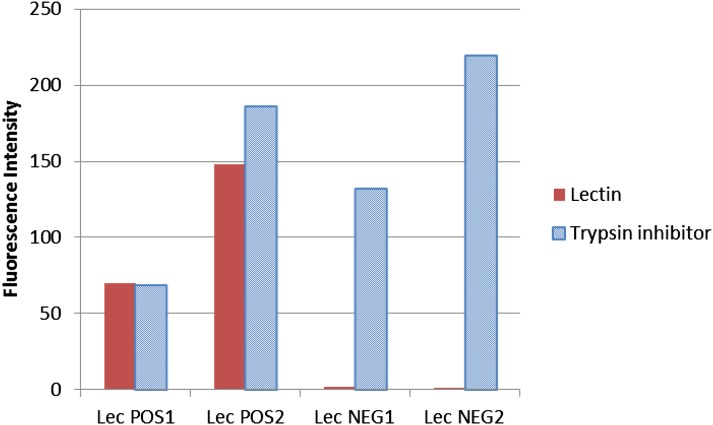
A naturally occurring transposon insertion that abolishes lectin mRNA and protein production has previously been thoroughly described at the molecular level (Pull et al., 1978; Vodkin, 1981; Vodkin et al., 1983; Goldberg et al., 1983). This mutation has been backcrossed into the genetic background of cv Williams to create genetic isolines that are either lectin positive or lectin negative. Crude extracts from either normal or lectin-negative soybean cotyledons were incubated with the silicon-substrate PC arrays. As shown in Figure 7, lectin is detected comparably with trypsin inhibitor in the normal lectin-positive line. However, in the lectin-negative soybean line, trypsin inhibitor levels are unchanged, but lectin is undetectable, indicating that there was no spurious attachment of other proteins to the lectin antibody and also, showing the low autofluorescence of the silicon-substrate PC arrays.
Figure 7.
Differential detection on silicon-substrate PC slides of trypsin inhibitor and lectin proteins in lectin-positive and -negative isolines. Histogram showing relative fluorescence levels representing either lectin (red) or trypsin inhibitor (blue) in dry seed from soybean lines with normal lectin levels (Lec POS) or no lectin (Lec NEG). Two slides (technical replicates) are shown for each soybean line. Lectin is the average of eight spots per slide, and trypsin inhibitor is the average of four spots per slide; average slide background and negative controls were subtracted out, and zero and negative values were transformed to 0.001.
DISCUSSION
Abundant Proteins in Crude Extracts Saturate the Sensitive Silicon-Substrate PC Arrays
The carbohydrate-binding lectin and the protease inhibitor Kunitz trypsin inhibitor are both well-characterized proteins in soybean that may have roles in nutrient storage and defense (Vodkin, 1981; Jofuku and Goldberg, 1989; Van Damme et al., 2008; De Hoff et al., 2009). They are both highly abundant proteins, especially in seedling cotyledons, where previous studies have found lectin and trypsin inhibitor at approximately 2 and 3 mg, respectively, per 1 g of fresh weight (Pueppke and Bauer, 1978; Tan-Wilson et al., 1982) during the early stages of germination. This amount does not markedly decrease until approximately 8 d after germination (Pueppke and Bauer, 1978; Wilson et al., 1988), which is beyond the timeframe of our study. This pattern is consistent with the results that we found of high, relatively steady levels of protein throughout all stages examined. These high protein abundances in the seedling cotyledons result from high levels of gene expression in the immature cotyledons and storage of the proteins in specialized storage vacuoles, the protein bodies.
The consistent, relatively narrow range of protein levels displayed by lectin and trypsin inhibitor may be caused by a saturation effect on both types of slides, both glass and silicon-substrate PC, whereby the proteins were so abundant that neither technique could accurately quantify them past a certain point. A previous report with PCEF slides has suggested that their range is from 0.3 pg mL−1 to 10 ng mL−1 (Huang et al., 2011; Cunningham and Zangar, 2012). With total seed protein at 25 μg mL−1 in our assays and lectin representing approximately 1% of the total extractable seed protein (Pull et al., 1978), the estimated amount of lectin in the solution is about 250 ng mL−1, well over the amount needed to saturate both the glass and the silicon-substrate PC arrays. Because our goal was to detect low-abundance transcription factors as shown below, the higher abundance proteins in the same crude extracts will necessarily be out of range.
Silicon-Substrate PC Arrays Outperform Glass to Directly Detect Scarce Proteins, Such as Transcription Factors, in Crude Plant Extracts
Silicon-substrate PC slides distinctly outperformed glass slides by detecting in measurable amounts the proteins that otherwise fell below background on the glass slides. For example, bHLH037 had a protein abundance pattern on the silicon-substrate PC slides similar to that of zinc finger GATA, being relatively high; however, on the glass slides, it was undetectable above background at four stages. Likewise, BTF3/NAC, which increased in abundance in the seedling cotyledons during development as determined by the silicon-substrate PC arrays, was not detected at all during the first five stages on the glass slides. YABBY was not detected on glass at all; it had such low abundance that even the silicon-substrate PCs did not detect it at two stages.
These results highlight the strength of the silicon-substrate PC arrays in detecting low but critical proteins, such as transcription factors, which act as master regulators controlling the expression of suites of other genes. Such proteins are found in too low abundance to be purified from tissue by conventional techniques, but the timing of their appearance can have major downstream effects. A technique that allows a large number of such low-abundance proteins to be measured in crude extracts of many stages and tissues would be very useful for elucidating large-scale protein changes and effects. Our results also show the practicality of using antibodies generated to synthetic peptides representing regions of the proteins.
Biological Context of the Developmental Changes in Levels of the Four Transcription Factor Proteins
Because we had both protein abundance data and gene expression data from the same seedling cotyledon stages for these proteins as well as gene expression data from the immature seed stages immediately preceding the seedling cotyledon stages, we could examine how changes in gene expression levels might affect changes in protein levels. For many transcription factors, there are few data measuring their levels directly over developmental time. It is likely that they have high turnover, because their protein causes changes in the expression of other genes, which is a tightly regulated part of the developmental program. For the transcription factors shown in Figure 5, three had single, sharp peaks in gene expression during the seedling cotyledon stages, which could be driving a sudden increase in protein abundance a stage or two later. Notably, these peaks occurred in stages 3 or 4, when the seedling cotyledon undergoes a functional transition from yellow storage organ to green photosynthesizing tissue. The zinc finger GATA protein had a more sustained presence at both the transcript and protein levels.
After Its Transcript Pattern, bHLH037 Protein Levels Increase in Seedling Cotyledons
In the case of bHLH037, its gene expression peaked at stage 3. This stage is the point of minimum protein abundance according to the silicon-substrate PCs; however, protein abundance peaked at the next stage, stage 4, before decreasing. It is possible that this delay indicates the time needed for the transcripts to be translated and accumulate. The timing of gene expression seemed to be sharply defined, whereas the protein abundance persisted at measureable levels on the silicon-substrate PCs throughout the rest of the stages studied.
The bHLH037 factor is part of the basic-helix-loop-helix family of transcription factors, which is found in a wide variety of eukaryotes, including humans, plants, and yeast (Saccharomyces cerevisiae; Heim et al., 2003). The bHLH037 protein examined in this study corresponds to the soybean homolog of the HECATE family, which was first identified in Arabidopsis (Gremski et al., 2007). This family has mainly been studied in the context of floral development in Arabidopsis (Gremski et al., 2007) as well as in shoot apical meristem tissue under short-day treatment in soybean (Wong et al., 2013). However, little additional function beyond their important role in female reproductive tissue development has been found for the HECATE genes (Feller et al., 2011). With the peak in gene expression in the seedling cotyledons and notable protein abundance throughout seedling cotyledon development, there may be additional unknown functions for this gene family.
The Zinc Finger GATA Protein Is Found at Appreciable Levels in All Seven Seedling Cotyledon Stages
In contrast to bHLH037, zinc finger GATA had a peak of gene expression during the immature cotyledon stage. Its expression increased again during very late seed development, carrying over into relatively high gene expression during stage 1 of the seedling cotyledons (which are separated from the dry mature seeds only by 24-h imbibition in water). After this, the zinc finger GATA protein was detectable at appreciable levels in all seven stages, although there was about 10-fold variation in the levels (Table II). It is possible that these results indicate that the zinc finger GATA protein is not turned over rapidly during seedling development and that it initially accumulates to a higher level, perhaps enough to cause a saturation effect with the sensitive silicon-substrate PC slides. Some of the highest average values recorded from the silicon-substrate PC slides were found with this zinc finger GATA protein during the seedling cotyledon stages.
The zinc finger GATA protein used in this study corresponds to the GATA transcription factor5-like (GATA5) protein from soybean. GATA5 has been specifically noted to have low but constant gene expression in many parts of the adult Arabidopsis plant, such as roots, stems, and leaves (Manfield et al., 2007). Another GATA gene in the same subfamily, GATA8 or Blue Micropylar End3, has been implicated in the initial process of seed germination (Liu et al., 2005), which would be consistent with the high values for gene expression and protein abundance that we see in the early stages of the seedling cotyledons. However, the GATA family, like bHLH, is found in a number of eukaryotes, and the plant members are associated with a wide variety of processes, such as light and circadian responsiveness, sugar and nitrogen metabolism, and flower and shoot apical meristem development (Manfield et al., 2007). The relatively high abundance of GATA5 protein in the seedling cotyledons suggests that it may have an important role to play in early seedling development.
The BTF3/NAC Protein Accumulates Significantly in Stages 6 and 7 after Peak Transcript Abundance
Another transcription factor, BTF3/NAC, showed the dynamic range of the silicon-substrate PC slides, with its protein abundance increasing dramatically from values around 1 at stage 3 to about 650 at stage 7. The RNA-Seq data peaked sharply at stage 4, whereas the protein abundance began to rise noticeably in stage 6. Similar to bHLH037, the delay may indicate the time needed to translate the gene transcripts and accumulate the protein, which likely peaks later in development than stage 7. It is notable that the glass slides only detected this protein during the last two stages, when its abundance was highest according to the silicon-substrate PCs, showing the threshold below which the glass slides are not as sensitive.
BTF3 is part of the NAC transcription factor family, with members found in plants, animals, and yeast (Kang et al., 2013). It may be involved in directing newly synthesized peptides to organelles, such as chloroplasts and mitochondria; silencing has been found to result in yellowed leaves, because chloroplasts become smaller with less chlorophyll and reduced gene expression (Yang et al., 2007). This finding meshes well with the results from our study, in that this gene’s expression peaked at stage 4 (in the middle of the seedling cotyledon’s functional transition from yellow storage organ to green photosynthesizing organ), with the protein increasing dramatically in abundance coincident with the greening of the seedling cotyledon. Other studies have suggested that BTF3 has roles in biotic and abiotic stress response and pollen development (Huh et al., 2012; Wang et al., 2012; Kang et al., 2013).
Recently, we elucidated the binding sites for the same members of the NAC and YABBY transcription factor families as well as identified some of the genes that they may regulate in seedling cotyledon development using chromatin immunoprecipitation coupled with high-throughput sequencing (ChIP-Seq; Shamimuzzaman and Vodkin, 2013). BTF3/NAC was found to have up to 72 targets that it may regulate, including lipoxygenase and pectin methyl esterase inhibitor.
The YABBY Protein Seems to Be Turned Over Rapidly in the Seedling Cotyledons
The YABBY protein is an interesting case, being of such low abundance that even the silicon-substrate PCs could not detect its protein above background at two stages. However, it was not detected at all on the glass slides, and thus, the silicon-substrate PCs provided information that would, otherwise, have been unattainable. As with BTF3/NAC, the gene expression of the two YABBY models peaked at stage 4 (before the peak in protein abundance at stage 6). It would seem that the YABBY protein is turned over quickly by the seedling cotyledons, because its abundance dropped to undetectable at the next stage (stage 7).
In contrast to other transcription factors studied here, the YABBY family is found only in flowering plants (Bartholmes et al., 2012). YABBY5, the protein used here, is part of the vegetative YABBY branch, which is mainly involved in various aspects of leaf development (Juarez et al., 2004; Liang et al., 2009; Bartholmes et al., 2012). It is possible that YABBY or some of the other transcription factors discussed here are found only in sublayers of the seedling cotyledons and not throughout the whole organ, which would dilute their concentrations in the crude extracts. Another hypothesis for the low protein abundance of YABBY is that its transcripts are not as efficiently translated as, for example, bHLH037, given that their gene expression levels are comparable but that bHLH037 has much higher protein abundance. It is also possible that YABBY protein is turned over at a much higher rate than bHLH037.
Alternatively, the antibodies to the YABBY proteins may not be as effective in recognizing the protein as the antibodies to bHLH037, zinc finger GATA, and BTF3/NAC. However, this seems unlikely, because the same antibodies to the YABBY protein have been used in ChIP-Seq experiments with stages 4 and 5 seedling cotyledons, identifying 96 genes potentially regulated by the YABBY transcription factor, including an APETALA2 transcription factor, fatty acid desaturase, and a WRKY transcription factor (Shamimuzzaman and Vodkin, 2013). Thus, the antibodies seem to be functional by two different experimental techniques, ChIP-Seq and direct detection of the YABBY protein on the silicon-substrate PC arrays, albeit at a level near the lower limit of the arrays.
CONCLUSION
Our results show the power of the silicon-substrate PC arrays to directly detect low-abundance, critical proteins, such as transcription factors, in crude plant extracts. Direct detection of transcription factors has been very limited; generally, the presence of the protein is inferred indirectly from transcript data or detection in transgenic plants after the protein is fused to fluorescent reporter molecules. Alternatively, the binding sites of the transcription factor may be explored by ChIP-Seq. As a model system to test proof of concept of the silicon-substrate PC arrays to directly detect rare proteins, we selected representatives of four different transcription factor families that showed increasing transcript levels during seedling cotyledon development. Our results also show the practicality of using antibodies generated to synthetic peptides representing regions of the low-abundance proteins. For antibodies to both NAC and YABBY proteins, the antibodies were confirmed as functional by a second experimental technique, ChIP-Seq (Shamimuzzaman and Vodkin, 2013). Here, the zinc finger GATA transcription factor was detected at high and possibly, saturating levels on the silicon-substrate PC arrays. Most interestingly, the bHLH037 and BTF3/NAC proteins showed developmental profiles consistent with their transcript patterns, indicating proof of concept for detecting these low-abundance proteins in crude extracts.
MATERIALS AND METHODS
Silicon-Substrate PC Fabrication
The silicon-substrate PC is composed of a periodic surface structure fabricated in a low-refractive index SiO2 layer on a silicon substrate (Fig. 1). The grating structure is coated with a high-refractive index TiO2 thin film with a thickness of 130 nm. The PC has a period of 360 nm, a duty cycle of 36%, and a grating depth of 40 nm. A commercial vendor (Novati Technologies Inc.) was contracted for performing photolithography and reactive ion etching of the SiO2 grating structure over 8-in-diameter wafers, whereas TiO2 thin films were deposited upon whole wafers at a second vendor (Intlvac Inc.). After lithography, etching, and TiO2 deposition, the wafers were diced into 1.0- × 0.5-in2 pieces. These pieces were affixed to standard microscope slides with optically transparent double-sided adhesives (3 M).
Plant Material and Protein Extraction
Soybean (Glycine max ‘Williams’; maturity group III) seeds were used. Separate lots of seed, which had been grown and harvested in different years, were used for the two biological replicates to account for natural variation in protein abundance. Dry mature seeds were soaked in water for approximately 24 h to obtain stage 1 cotyledons, from which the seed coats and radicles were removed. For stages 2 to 7, seed were grown in a standard soil mix in the greenhouse for approximately 2 to 7 d, and cotyledons were harvested at the appropriate seedling developmental stage as described in Shamimuzzaman and Vodkin (2013). Tissue was fresh frozen in liquid nitrogen for 10 min and then stored at −80°C. Total protein was extracted from ground tissue with 0.01 m phosphate-buffered saline (PBS), pH 7.0 (Sigma-Aldrich). Extractions generally represented two to three cotyledons from different plants. Seed of cv Williams (Plant Introduction no. 548631) or cv Williams lectin-negative isoline L90-8047 (Plant Introduction no. 591534) can be requested from the U.S. Department of Agriculture at the Germplasm Resources Information Network (http://www.ars-grin.gov).
Origin of Primary and Secondary Antibodies
Primary antibodies for the four transcription factor proteins (bHLH037, BTF3/NAC, YABBY, and zinc finger GATA) were produced by the GenScript Corporation. Using the sequences indicated by the GenBank accession numbers in Supplemental Table S2, GenScript Corporation designed synthetic peptides for the production of polyclonal antibodies. These antibodies were purified using affinity chromatography to the synthetic peptides. For the two seed proteins, lectin and trypsin inhibitor, affinity-purified primary antibodies were produced to the native whole protein as described in Vodkin, 1981.
To create secondary antibodies, primary antibodies were biotinylated using the EasyLink Biotin (Type A) Conjugation Kit (Abcam).
Printing of Arrays
Arrays were printed using a QArray2 Robot (Genetix/Molecular Devices) on either aminosilanized GAPSII glass slides (Corning) or epoxysilanized silicon-substrate PC devices mounted on glass slides. Printing was performed at 50% to 55% humidity and ambient temperature in subdued light.
The array consisted of four identical rows, each containing 20 spots. Of 20 spots per row, 10 represented antibodies (0.24–0.973 mg mL−1; includes some proteins not discussed here). Each type of antibody was printed one time per row, except for lectin, which was printed two times per row. Thus, each antibody had four replicate spots per slide, except for lectin, which had eight replicate spots per slide (Table I). Other spots in the array included an orientation spot (0.1 mg mL−1; Alexa-633-labeled donkey anti-sheep IgG antibody; Molecular Probes/Life Technologies) and PBS spots (0.01 m), which functioned as negative controls.
Incubations
The incubation protocol was adapted from Huang et al. (2011). Arrays were generally incubated within 1 week of being printed. First, slides were blocked for 1 h in a solution of 1% (v/v) casein in PBS (Bio-Rad). Second, slides were incubated overnight at room temperature in the dark with gentle shaking (50 rpm) in the protein extract, which was diluted to 25 μg mL−1 of total protein with 0.1% (v/v) casein in PBS. Third, slides were incubated overnight under the same conditions in the secondary antibody mixture, which was diluted to about 0.025 μg mL−1 for each antibody using phosphate-buffered saline with 0.05% (v/v) Tween (PBS-T; Sigma-Aldrich). Fourth, slides were incubated for 30 min under the same conditions with streptavidin-Cy5 (GE Healthcare/Amersham) at 1 μg mL−1 in PBS-T. Washes between steps were performed in PBS-T.
Slide Scanning
Both silicon-substrate PCs and glass slides were scanned from the top in the same manner with a commercially available confocal laser microarray scanner (Tecan LS Reloaded; Fig. 1). This scanner was fitted with a 632.8-nm, 5-mW laser for Cy5 excitation and a Cy5 emission filter (band pass, 670–715 nm). The incident light was transverse magnetic polarized and made incident on the substrates at an angle of 5° so that maximum laser coupling efficiency could be achieved. Scans were obtained at a resolution of 10 μm, and the photomultiplier tube gain was adjusted to 80 so that the largest fluorescence intensities did not saturate the photomultiplier tube.
Data Analysis
GenePix Pro 6.1 software (Molecular Devices) was used to find the spots and quantitate their fluorescence intensity levels. The final value for each protein represents 16 spots across four slides (32 spots for lectin), with slide background and negative control values removed, and includes two biological replicates. More specifically, the F635 median values of replicate spots on one slide were averaged; then, the average background (B635) of all 80 spots on the slide was subtracted from this number. The average F635 median value of all 36 PBS spots on a slide (negative controls minus background) was also subtracted from the protein spot average. The values for each protein from all replicate slides (four glass or four silicon-substrate PC) were then averaged together to give the final values. Negative values (less than background and negative controls) were transformed to 0.001. The largest variation in values was introduced by the biological replicates (Supplemental Fig. S1). se calculations for final values are shown in Supplemental Table S3.
RNA-Seq Data and Gene Models
For additional details about the seedling cotyledon or immature seed RNA-Seq data, see Shamimuzzaman and Vodkin (2014) and Jones and Vodkin (2013), respectively. Briefly, total RNA was sequenced using the Illumina method by the Keck Center, and the sequence data were processed through the standard Illumina pipeline. The reads were then aligned to 78,773 Glyma1 cDNA soybean gene models (a1.v1.0) determined by the Soybean Genome Project, Department of Energy, Joint Genome Institute (Schmutz et al., 2010) using the alignment program Bowtie (Langmead et al., 2009). Data are given in RPKMs (Mortazavi et al., 2008).
Seedling cotyledon RNA-Seq data represent the average of two biological replicates across the same seven stages as were used for the protein extractions described above. Immature seed RNA-Seq data represent the average of two biological replicates across seven stages of either whole seeds (4, 12–14, or 22–24 d after flowering; 5–6 mg fresh weight; dry mature seeds) or just cotyledons (fresh weight of whole green seeds; either 100–200 or 400–500 mg). RNA-Seq data derived from Jones and Vodkin (2013) and Shamimuzzaman and Vodkin (2014) are shown in Supplemental Table S1.
To connect protein sequences to soybean gene models, the GenBank protein sequences listed in Supplemental Table S2 were used to search the soybean gene models at Phytozome (www.phytozome.net) using BLASTP with default parameters. The gene models with the best match (e value and percentage identity) to the proteins were selected, with a minimum of 90% identity (for more information, see Supplemental Table S2). For the BTF3/NAC, three other gene models have high matches at the antigenic region (Shamimuzzaman and Vodkin, 2013), but the entire protein is only an 85% match; therefore, these gene models did not meet the criteria used.
Sequence data used in this paper were derived from previously entered RNA-Seq data sets GSE42550 (Shamimuzzaman and Vodkin, 2014) for seedling cotyledons and GSE42871 (Jones and Vodkin, 2013) for developing seed and cotyledons and are found in the Gene Expression Omnibus database at the National Center for Biotechnology Information.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Intensity values for six proteins from four silicon-substrate PC slides using stage 1 or 5 seedling cotyledon tissue.
Supplemental Table S1. RNA-Seq data for gene models linked to six proteins.
Supplemental Table S2. Six soybean proteins linked to gene models.
Supplemental Table S3. se of intensity values for six proteins.
Supplementary Material
Glossary
- cDNA
complementary DNA
- ChIP-Seq
chromatin immunoprecipitation coupled with high-throughput sequencing
- DAF
days after flowering
- PBS
phosphate-buffered saline
- PBS-T
phosphate-buffered saline with 0.05% Tween
- PC
photonic crystal
- PCEF
photonic crystal-enhanced fluorescence
- RPKM
reads per kilobase of gene model per million mapped reads
Footnotes
This work was supported by the National Institutes of Health (grant no. R01 GM086382 to B.T.C and L.V.) and the Illinois Soybean Association and United Soybean Board through the National Science Foundation Center for Innovative Instrumentation Technology.
Articles can be viewed without a subscription.
References
- Bailey PC, Martin C, Toledo-Ortiz G, Quail PH, Huq E, Heim MA, Jakoby M, Werber M, Weisshaar B (2003) Update on the basic helix-loop-helix transcription factor gene family in Arabidopsis thaliana. Plant Cell 15: 2497–2502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholmes C, Hidalgo O, Gleissberg S (2012) Evolution of the YABBY gene family with emphasis on the basal eudicot Eschscholzia californica (Papaveraceae). Plant Biol (Stuttg) 14: 11–23 [DOI] [PubMed] [Google Scholar]
- Bewley JD. (1997) Seed germination and dormancy. Plant Cell 9: 1055–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarty S, Lai WC, Zou Y, Drabkin HA, Gemmill RM, Simon GR, Chin SH, Chen RT (2013) Multiplexed specific label-free detection of NCI-H358 lung cancer cell line lysates with silicon based photonic crystal microcavity biosensors. Biosens Bioelectron 43: 50–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhery V, George S, Lu M, Pokhriyal A, Cunningham BT (2013) Nanostructured surfaces and detection instrumentation for photonic crystal enhanced fluorescence. Sensors (Basel) 13: 5561–5584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham BT, Zangar RC (2012) Photonic crystal enhanced fluorescence for early breast cancer biomarker detection. J Biophotonics 5: 617–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Hoff PL, Brill LM, Hirsch AM (2009) Plant lectins: the ties that bind in root symbiosis and plant defense. Mol Genet Genomics 282: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feller A, Machemer K, Braun EL, Grotewold E (2011) Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J 66: 94–116 [DOI] [PubMed] [Google Scholar]
- George S, Chaudhery V, Lu M, Takagi M, Amro N, Pokhriyal A, Tan Y, Ferreira P, Cunningham BT (2013) Sensitive detection of protein and miRNA cancer biomarkers using silicon-based photonic crystals and a resonance coupling laser scanning platform. Lab Chip 13: 4053–4064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg RB, Hoschek G, Vodkin LO (1983) An insertion sequence blocks the expression of a soybean lectin gene. Cell 33: 465–475 [DOI] [PubMed] [Google Scholar]
- Gonzalez DO, Vodkin LO (2007) Specific elements of the glyoxylate pathway play a significant role in the functional transition of the soybean cotyledon during seedling development. BMC Genomics 8: 468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gremski K, Ditta G, Yanofsky MF (2007) The HECATE genes regulate female reproductive tract development in Arabidopsis thaliana. Development 134: 3593–3601 [DOI] [PubMed] [Google Scholar]
- Heim MA, Jakoby M, Werber M, Martin C, Weisshaar B, Bailey PC (2003) The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol Biol Evol 20: 735–747 [DOI] [PubMed] [Google Scholar]
- Huang CS, George S, Lu M, Chaudhery V, Tan R, Zangar RC, Cunningham BT (2011) Application of photonic crystal enhanced fluorescence to cancer biomarker microarrays. Anal Chem 83: 1425–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh SU, Kim KJ, Paek KH (2012) Capsicum annuum basic transcription factor 3 (CaBtf3) regulates transcription of pathogenesis-related genes during hypersensitive response upon Tobacco mosaic virus infection. Biochem Biophys Res Commun 417: 910–917 [DOI] [PubMed] [Google Scholar]
- Jofuku KD, Goldberg RB (1989) Kunitz trypsin inhibitor genes are differentially expressed during the soybean life cycle and in transformed tobacco plants. Plant Cell 1: 1079–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SI, Gonzalez DO, Vodkin LO (2010) Flux of transcript patterns during soybean seed development. BMC Genomics 11: 136–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SI, Vodkin LO (2013) Using RNA-Seq to profile soybean seed development from fertilization to maturity. PLoS ONE 8: e59270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juarez MT, Twigg RW, Timmermans MC (2004) Specification of adaxial cell fate during maize leaf development. Development 131: 4533–4544 [DOI] [PubMed] [Google Scholar]
- Kang G, Ma H, Liu G, Han Q, Li C, Guo T (2013) Silencing of TaBTF3 gene impairs tolerance to freezing and drought stresses in wheat. Mol Genet Genomics 288: 591–599 [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang D, Wong CE, Singh MB, Beveridge CA, Phipson B, Smyth GK, Bhalla PL (2009) Molecular dissection of the pea shoot apical meristem. J Exp Bot 60: 4201–4213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom JT, Vodkin LO, Harding RW, Goeken RM (1990) Expression of soybean lectin gene deletions in tobacco. Dev Genet 11: 160–167 [DOI] [PubMed] [Google Scholar]
- Liu PP, Koizuka N, Martin RC, Nonogaki H (2005) The BME3 (Blue Micropylar End 3) GATA zinc finger transcription factor is a positive regulator of Arabidopsis seed germination. Plant J 44: 960–971 [DOI] [PubMed] [Google Scholar]
- Manfield IW, Devlin PF, Jen CH, Westhead DR, Gilmartin PM (2007) Conservation, convergence, and divergence of light-responsive, circadian-regulated, and tissue-specific expression patterns during evolution of the Arabidopsis GATA gene family. Plant Physiol 143: 941–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathias PC, Jones SI, Wu HY, Yang F, Ganesh N, Gonzalez DO, Bollero G, Vodkin LO, Cunningham BT (2010) Improved sensitivity of DNA microarrays using photonic crystal enhanced fluorescence. Anal Chem 82: 6854–6861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Pal S, Guillermain E, Sriram R, Miller BL, Fauchet PM (2011) Silicon photonic crystal nanocavity-coupled waveguides for error-corrected optical biosensing. Biosens Bioelectron 26: 4024–4031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pull SP, Pueppke SG, Hymowitz T, Orf JH (1978) Soybean lines lacking the 120,000-dalton seed lectin. Science 200: 1277–1279 [DOI] [PubMed] [Google Scholar]
- Pueppke SG, Bauer WD (1978) Role of lectins in plant-microorganism interactions: II. Distribution of soybean lectin in tissues of Glycine max (L.) Merr. Plant Physiol 61: 779–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al. (2010) Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183 [DOI] [PubMed] [Google Scholar]
- Scullion MG, Di Falco A, Krauss TF (2011) Slotted photonic crystal cavities with integrated microfluidics for biosensing applications. Biosens Bioelectron 27: 101–105 [DOI] [PubMed] [Google Scholar]
- Shamimuzzaman M, Vodkin L (2013) Genome-wide identification of binding sites for NAC and YABBY transcription factors and co-regulated genes during soybean seedling development by ChIP-Seq and RNA-Seq. BMC Genomics 14: 477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamimuzzaman M, Vodkin L (2014) Transcription factors and glyoxylate cycle genes prominent in the transition of soybean cotyledons to the first functional leaves of the seedling. Funct Integr Genomics 14: 683–696 [DOI] [PubMed] [Google Scholar]
- Tan-Wilson AL, Rightmire BR, Wilson KA (1982) Different rates of metabolism of soybean proteinase inhibitors during germination. Plant Physiol 70: 493–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme EJM, Lannoo N, Peumans WJ (2008) Plant lectins. InKader JC, Delseny M, eds, Advances in Botanical Research, Vol 3 Elsevier, Waltham, MA, pp 107–209 [Google Scholar]
- Vodkin LO. (1981) Isolation and characterization of messenger RNAs for seed lectin and Kunitz trypsin inhibitor in soybeans. Plant Physiol 68: 766–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vodkin LO, Rhodes PR, Goldberg RB (1983) cA lectin gene insertion has the structural features of a transposable element. Cell 34: 1023–1031 [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhang X, Lu S, Wang M, Wang L, Wang W, Cao F, Chen H, Wang J, Zhang J, et al. (2012) Inhibition of a basal transcription factor 3-like gene Osj10gBTF3 in rice results in significant plant miniaturization and typical pollen abortion. Plant Cell Physiol 53: 2073–2089 [DOI] [PubMed] [Google Scholar]
- Wilson KA, Papastoitsis G, Hartl P, Tan-Wilson AL (1988) Survey of the proteolytic activities degrading the Kunitz trypsin inhibitor and glycinin in germinating soybeans (Glycine max). Plant Physiol 88: 355–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CE, Singh MB, Bhalla PL (2013) The dynamics of soybean leaf and shoot apical meristem transcriptome undergoing floral initiation process. PLoS ONE 8: e65319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang KS, Kim HS, Jin UH, Lee SS, Park JA, Lim YP, Pai HS (2007) Silencing of NbBTF3 results in developmental defects and disturbed gene expression in chloroplasts and mitochondria of higher plants. Planta 225: 1459–1469 [DOI] [PubMed] [Google Scholar]
- Zou Y, Chakravarty S, Kwong DN, Lai WC, Xu X, Lin X, Hosseini A, Chen RT (2014) Cavity-waveguide coupling engineered high sensitivity silicon photonic crystal microcavity biosensors with high yield. IEEE J Sel Top Quantum Electron 20: 6900710 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.