A poplar repressor protein negatively regulates flavonoid and proanthocyanidin pathway genes by interacting with other transcription factors.
Abstract
Trees in the genus Populus (poplar) contain phenolic secondary metabolites including the proanthocyanidins (PAs), which help to adapt these widespread trees to diverse environments. The transcriptional activation of PA biosynthesis in response to herbivory and ultraviolet light stress has been documented in poplar leaves, and a regulator of this process, the R2R3-MYB transcription factor MYB134, has been identified. MYB134-overexpressing transgenic plants show a strong high-PA phenotype. Analysis of these transgenic plants suggested the involvement of additional MYB transcription factors, including repressor-like MYB factors. Here, MYB182, a subgroup 4 MYB factor, was found to act as a negative regulator of the flavonoid pathway. Overexpression of MYB182 in hairy root culture and whole poplar plants led to reduced PA and anthocyanin levels as well as a reduction in the expression of key flavonoid genes. Similarly, a reduced accumulation of transcripts of a MYB PA activator and a basic helix-loop-helix cofactor was observed in MYB182-overexpressing hairy roots. Transient promoter activation assays in poplar cell culture demonstrated that MYB182 can disrupt transcriptional activation by MYB134 and that the basic helix-loop-helix-binding motif of MYB182 was essential for repression. Microarray analysis of transgenic plants demonstrated that down-regulated targets of MYB182 also include shikimate pathway genes. This work shows that MYB182 plays an important role in the fine-tuning of MYB134-mediated flavonoid metabolism.
Flavonoids are widely distributed secondary plant metabolites that include the anthocyanins, flavonols, flavones, and proanthocyanidins (PAs; Koes et al., 2005). Collectively, these compounds play many roles in the interaction of plants with their environment, such as protection against UV and visible light stress, resistance to herbivores and pathogens, attraction of pollinators and seed dispersers, and modulation of physiological and developmental signals (Mol et al., 1998). The PAs, also known as condensed tannins, are polymers of flavan-3-ols, typically consisting of two to 50 subunits. PAs are present in many plants but are especially abundant in trees, where they are found in vegetative tissues such as leaves, bark, and roots (Barbehenn and Constabel, 2011). They are also present in seeds and many types of fruit, where they are thought to discourage premature consumption by frugivores or prevent spoilage by fungi (Cipollini and Stiles, 1993).
In humans, a diet high in PAs has been linked to reduced risk of chronic cardiovascular diseases (Rasmussen et al., 2005; Scalbert et al., 2005). PAs are thought to prevent atherosclerosis by inhibiting the oxidation of low-density lipoproteins, based on their electron-donating properties and ability to scavenge reactive oxygen species (Scalbert et al., 2005). Berry fruits, whole grains, wine, and beer are considered good sources of PAs (Prior and Gu, 2005). In the diets of ruminants such as sheep and cattle, PAs can be important modulators of excess rumen bacterial activity, preventing bloat and improving protein use efficiency (Min et al., 2003). This effect makes these compounds important components of silage and forage crops and targets for manipulation via agricultural biotechnology.
In nature, the PAs are associated with diverse ecological functions. They typically possess broad antimicrobial properties, and by inhibiting bacterial activity in soils they affect nutrient cycling. As a result, the PA content of litter from keystone trees species such as poplar (genus Populus) correlates strongly with community structure and ecosystem dynamics (Schweitzer et al., 2008). Many studies have attempted to show that PAs are important in defense against insect herbivores. The results of such experiments are mixed, however, as PAs do not consistently show strong effects against major tree pests (Barbehenn and Constabel, 2011; Boeckler et al., 2014). By contrast, they can adversely affect performance and reproductive success in mammals due to their ability to bind dietary protein in mammalian digestive systems (Barbehenn and Constabel, 2011). In planta, PAs also may function to inhibit pathogenic microorganisms (Constabel et al., 2014). Tannins also are often found in the roots of trees, although their roles there are poorly defined.
The flavonoids, including PAs, are derived from phenylpropanoids and malonyl-CoA, which serve as substrates for chalcone synthase (CHS), the first enzyme in the flavonoid biosynthetic pathway. Following isomerization and hydroxylation by flavanone 3-hydroxylase (F3H), intermediates are reduced by dihydroflavonol 4-reductase (DFR; Marles et al., 2003). Anthocyanin synthase (ANS) then catalyzes the last common step in the biosynthesis of anthocyanins and PAs. Anthocyanidin reductase (ANR) and leucoanthocyanidin reductase are specific to the PA branch of the pathway and produce flavan-3-ols, typically epicatechin and catechin, respectively (Abrahams et al., 2003; Xie et al., 2003). These are polymerized via an unknown mechanism and stored in the vacuole. Genetic studies in Arabidopsis (Arabidopsis thaliana) revealed that multidrug and toxic compound extrusion transporter protein (TRANSPARENT TESTA12 [TT12]), glutathione S-transferase (TT19), and autoinhibited H1-ATPase isoform 10 are required for the transport of PAs to the vacuole.
The biosynthesis of anthocyanins and PAs is regulated by three types of transcription factors: R2R3-MYB factors, basic helix-loop-helix (bHLH) proteins, and conserved WD40 repeat (WDR) proteins (Koes et al., 2005; Ramsay and Glover, 2005). The MYB, bHLH, and WDR proteins interact physically to form the MBW complex that activates transcription, which has best been described using genetic tools in Arabidopsis. The MBW complex interacts directly with promoter DNA via multiple cis-elements. Known target sequences include the Myb-response element and AC elements for R2R3-MYB proteins and the E-box or bHLH-binding motif, which is bound by the bHLH proteins (Feller et al., 2011; Lai et al., 2013; Xu et al., 2014). In contrast, WDR proteins have not been shown to bind DNA but are thought to function by interacting with MYB and bHLH proteins (Baudry et al., 2004). It is the specific combination of interacting R2R3-MYB, bHLH, and WDR factors within the complex that determines which target genes are expressed in a given cell (Baudry et al., 2004; Broun, 2005; Koes et al., 2005; Ramsay and Glover 2005).
The bHLH and WDR cofactors are adaptable and can be involved in multiple processes. For example, the bHLH factor TT8 is required for PA synthesis in Arabidopsis when it interacts with the PA-specific R2R3-MYB TT2, but it is also involved in the synthesis of anthocyanins and seed coat mucilage when combined with other cofactors (Nesi et al., 2000). A second Arabidopsis bHLH protein, GLABROUS3 (GL3), belongs to a distinct bHLH subgroup and has several nonoverlapping functions in determining epidermal cell fate (Zhang et al., 2003). TT8 and GL3 are partially redundant, however, and can compensate for each other in knockout mutants (Broun, 2005). In petunia (Petunia hybrida), the two subgroups of bHLH cofactors are represented by ANTHOCYANIN1 (AN1) and JAF13, respectively (Koes et al., 2005). Both TT8 and GL3 have been shown to interact directly with the WDR protein TRANSPARENT TESTA GLABRA1 (TTG1; Zhang et al., 2003). TTG1 has roles in the regulation of trichome and root hair formation in addition to its function in the synthesis of Arabidopsis seed coat PAs (Ramsay and Glover, 2005). By contrast, the MYBs are functionally divergent and appear to be the major determinants of whether a gene or pathway is expressed in a given cell type (Koes et al., 2005). In Arabidopsis, the R2R3-MYB factor WEREWOLF functions exclusively in root hair patterning (Lee and Schiefelbein, 1999), while TT2 is specific to PA biosynthesis and PRODUCTION ANTHOCYANIN PIGMENT1 (PAP1) regulates only anthocyanin biosynthesis (Nesi et al., 2001; Teng et al., 2005). Nevertheless, all these MYBs can interact with the same bHLH protein such as TT8 to then activate distinct target promoters (Nesi et al., 2000; Gonzalez et al., 2008). In addition, a given MYB can interact with several different bHLHs, providing multiple functional combinations (Zimmermann et al., 2004).
Homologs of Arabidopsis TT2 and PAP1 are found in many species of plants, where they have conserved roles as PA and anthocyanin regulators, respectively. TT2-type PA regulators have been characterized in fruit, including DkMYB2 from persimmon (Diospyros kaki), VvMYBPA2 from grapevine (Vitis vinifera), and FaMYB9 in strawberry (Fragaria × ananassa; Terrier et al., 2009; Akagi et al., 2010; Schaart et al., 2013). MYBs from the TT2 group also are active in regulating PAs in vegetative tissues, such as LjTT2 from Japanese lotus (Lotus japonicus), TaMYB14 from clover (Trifolium arvense), and MYB134 from poplar (Mellway et al., 2009; Yoshida et al., 2010; Hancock et al., 2012). These MYB transcription factors act as part of the MBW complex that promotes flavonoid gene expression. For MYB134, we demonstrated direct binding to AC-like elements that are found in phenylpropanoid gene promoters (Mellway et al., 2009). A second type of PA MYB regulator was defined by the discovery of VvMYBPA1 from grapevine (Bogs et al., 2007). Orthologs of this MYB were subsequently described in other species (Akagi et al., 2009). These PA MYBs appear to act in parallel as well as in tandem with the TT2-type PA MYBs; in grapevine, both PA MYBs regulate flavonoid promoters directly, but VvMYBPA2 overexpression also induces transcripts of VvMYBPA1 (Terrier et al., 2009). Most species examined have both types of PA MYBs; Arabidopsis is a notable exception and has only the TT2 type. A further subgroup of MYB factors are the PAP1-like anthocyanin regulators. In Arabidopsis, PAP1 regulates stress-induced anthocyanins (Teng et al., 2005), and PAP1 orthologs are responsible for coloration in a variety of fruit (Lin-Wang et al., 2010). The floral color regulators AN2 from petunia and ROSEA1/2 from snapdragon (Antirrhinum majus) also are part of this MYB clade (Koes et al., 2005; Ramsay and Glover, 2005). In poplar, Wilkins et al. (2009) identified a group of PAP1-like MYBs that are highly expressed in pigmented tissues, suggesting conserved functions in anthocyanin synthesis, but none have been characterized to date.
Control of gene expression by the MBW complex can be further modulated by the involvement of repressor MYB proteins. The first such negative MYB regulators to be characterized include Antirrhinum spp. MYB308 and MYB330, which negatively affect phenolic acid and lignin biosynthesis (Tamagnone et al., 1998), and Arabidopsis MYB4 and MYB32, which regulate sinapate esters and lignin biosynthesis (Jin et al., 2000; Preston et al., 2004). These MYB repressors defined subgroup 4 within the MYB gene family, which to date includes only negative regulators. Recently, Arabidopsis MYB7, the downstream target of MYB4, was identified as a repressor of flavonol biosynthesis and demonstrated to directly target DFR and UDP-Glc-dependent glucosyltransferase (UGT) genes (Fornalé et al., 2014). Chrysanthemum spp. MYB1, the repressor of lignin biosynthesis, also down-regulates the flavonoid pathway (Zhu et al., 2013).
Repressor MYBs of the anthocyanin pathway have also been identified. These include strawberry FaMYB1 and FcMYB1 (for Fragaria chiloensis) and petunia PhMYB27 (Aharoni et al., 2001; Salvatierra et al., 2013; Albert et al., 2014). Overexpression of FaMYB1 causes the suppression of anthocyanin synthesis in tobacco (Nicotiana tabacum), and suppression of FcMYB1 by transient RNA interference in strawberry fruit increased the accumulation of anthocyanin. Likewise, RNA interference suppression of PhMYB27 increases the accumulation of anthocyanin in petunia flowers and vegetative tissues (Albert et al., 2014). In phylogenies, these flavonoid R2R3 repressor MYBs all cluster together within MYB subgroup 4 (Stracke et al., 2001), and all share the C2 motif (pfLNLD/ELxiG/S) with a core consensus sequence of LxLxL or DLNxxP (Aharoni et al., 2001; Dubos et al., 2010). These consensus sequences are the signature patterns of the EAR (for ethylene-responsive factor [ERF]-associated amphiphilic repression) motif, the predominant form of transcriptional repressor motif identified in plants (Kagale and Rozwadowski, 2011). Elegant work with PhMYB27 has begun to illuminate the mechanism of action of the subgroup 4 R2R3 repressor MYBs. In yeast two-hybrid assays, PhMYB27 interacts directly with bHLHs of both the GL3 and TT8 subgroups, although with different affinities (Albert et al., 2014). Furthermore, deletion of the DLNxxP-type EAR motif reduced PhMYB27 repressor activity; the function of this motif was not defined but may involve chromatin remodeling (Kagale and Rozwadowski, 2011). Recently, the first suppressor of the PA pathway, VvMYBC2-L1, was identified in grapevine as a new locus colocated with expression quantitative trait loci for PA-related genes (Huang et al., 2014). This subgroup 4 repressor MYB down-regulates PAs and flavonoid biosynthetic genes when overexpressed in hairy roots. Little is known about its mechanism of action, but it contains a partial LxLxL-type EAR motif.
In addition to subgroup 4 R2R3-MYB repressors, the Arabidopsis single-repeat R3-MYB repressors CAPRICE and ENHANCER OF TRYPTOCHON AND CAPRICE (ETC1) were shown to be involved in the down-regulation of anthocyanin biosynthesis (Zhu et al., 2009; Nemie-Feyissa et al., 2014). In petunia, the R3-MYB factor MYBx acts as a negative regulator of anthocyanin accumulation in parallel to PhMYB27 (Albert et al., 2014). These R3-MYB proteins do not contain the repressor motif but act as passive repressors by binding bHLH proteins required for the formation of MBW complexes (Albert et al., 2014). A distinct type of repressor from Arabidopsis, AtMYBL2, also contains only a single repeat but otherwise is more closely related to the R2R3-MYBs than other characterized R3-MYBs (Dubos et al., 2008; Matsui et al., 2008). This protein has a unique C-terminal TLLLFR motif that is required for repressor activity, and so the mechanism appears to be distinct from the small R3 MYB repressors.
In general, the flavonoid MYB repressors are associated with specific flavonoids, but they also may exert effects on related pathways. For example, FaMYB1 suppresses anthocyanin biosynthetic genes when overexpressed in tobacco, including ANS and UGT (Aharoni et al., 2001), and MYBL2 targets CHS, chalcone isomerase (CHI), F3H, DFR, ANS, and TT8 (Dubos et al., 2008; Matsui et al., 2008). However, when overexpressed in Lotus corniculatus, FaMYB1 also reduces PA biosynthesis (Paolocci et al., 2011), and AtMYBL2 suppresses ANR expression in the seed coat (Dubos et al., 2008). Likewise, the VvMYBC2-L1-overexpressing grapevine hairy roots have a reduction in stilbene content in addition to reduced PAs and PA precursors (Huang et al., 2014). Therefore, the determinants of target specificity and details about the mechanism of repression still remain to be elucidated, especially in perennial plants that accumulate significant concentrations of PAs.
The genus Populus consists of widespread trees of the northern hemisphere that are commonly known as poplars, aspens, and cottonwoods. Populus spp. typically contain substantial amounts of phenolic metabolites, including hydroxycinnamate esters, salicinoids, and PAs, which in Populus tremuloides can accumulate to 25% dry weight of leaves (Donaldson et al., 2006). Furthermore, in Populus spp., PA biosynthesis and accumulation can be rapidly induced by stresses including wounding, herbivore damage, pathogen attack, nitrogen deficiency, and UV light (Osier and Lindroth, 2004, Miranda et al., 2007; Mellway et al., 2009), suggesting a complex system of regulators. Stress induction of PAs in poplar involves the up-regulation of flavonoid biosynthesis genes. This response is mediated by MYB134, a TT2-type R2R3-MYB transcription factor (Mellway et al., 2009).
In the poplar genome, 192 genes encode R2R3-MYB transcription factors (Wilkins et al., 2009), but very few poplar MYBs with roles in flavonoid regulation are functionally characterized. MYB134 is an activator of PA biosynthesis and stimulates the poplar ANR1 promoter in transient expression assays in Arabidopsis (Zifkin et al., 2012; Gesell et al., 2014). Transgenic poplar plants overexpressing MYB134 accumulate up to 50 times more PAs in their leaves, yet they have normal anthocyanin levels and only slightly elevated flavonol contents, suggesting that MYB134 regulates PAs specifically (Mellway et al., 2009). Concurrently, microarray analysis revealed that all known early and late structural genes for PA biosynthesis were up-regulated in these plants (Mellway, 2009). In addition, several MYB genes predicted to encode both positive and negative regulators were expressed at elevated levels in the MYB134 overexpressors. One such regulator is MYB115, which belongs to the MYBPA1 subgroup of R2R3-MYB activators (Terrier et al., 2009). We also identified several genes encoding R2R3 repressor MYBs of subgroup 4 and one single-repeat R3 MYB. Here, we characterize the MYB182 R2R3-MYB repressor-like gene in detail. We demonstrate that, when MYB182 is overexpressed in poplar hairy roots and transgenic poplar plants, PA accumulation is reduced, as is the expression of flavonoid biosynthetic genes. Using transient expression assays, we further show that MYB182 represses gene expression induced by the activator MYBs and that this repression requires interaction with a bHLH cofactor. In addition, MYB182 may repress other regulatory genes as well as other enzyme-encoding genes important for phenolic metabolism in poplar.
RESULTS
The Poplar Repressor-Like MYB165, MYB182, and MYB194 Belong to a Separate Subclade within R2R3-MYB Subgroup 4
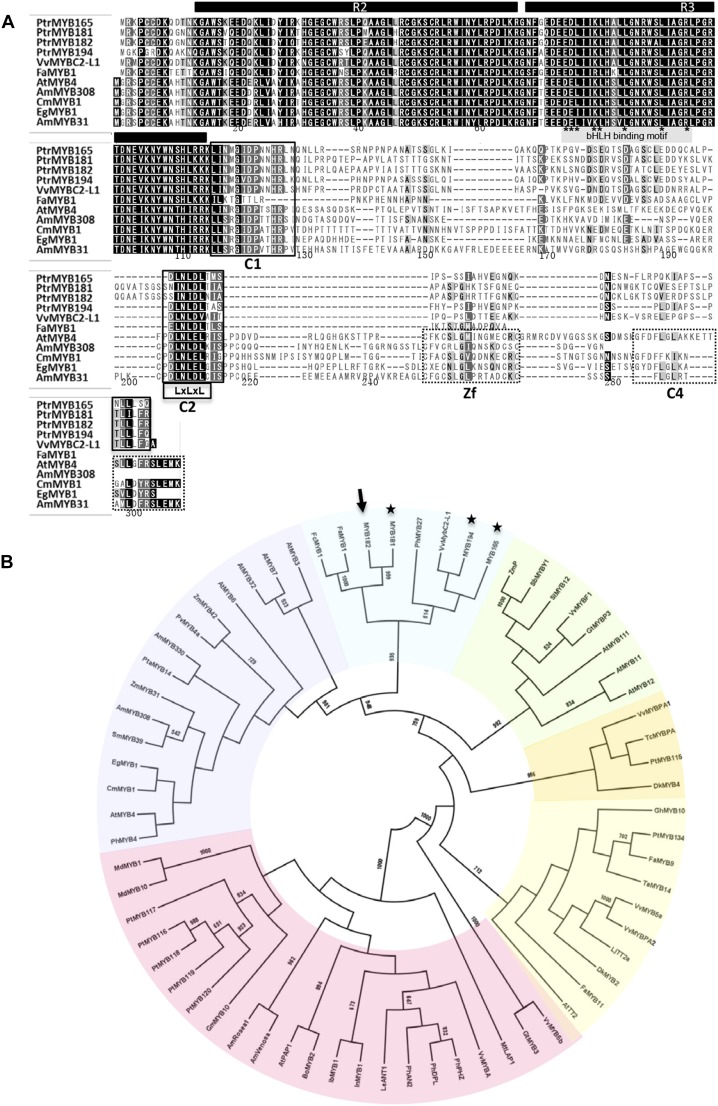
The protein sequences of the poplar repressor-like MYB genes MYB165, MYB182, and MYB194, which were identified by their enhanced expression in MYB134 overexpressor plants, were aligned and compared with characterized R2R3-MYBs from other species (Fig. 1; Supplemental Table S1). MYB181 was included in the phylogenic tree as it showed high identity (86%) to MYB182, although unlike the other three MYBs this, gene was not up-regulated in MYB134 overexpressors. The alignment shows that the N-terminal R2R3 domain is highly conserved and includes a bHLH-binding domain, while the C-terminal domain is very divergent (Fig. 1A). However, the latter region contains two conserved protein motifs, the C1 motif (LlsrGIDPxT/SHRxI/L; Shen et al., 2012) and the C2 motif (pdLNLD/ELxiG/S), which is diagnostic for subgroup 4 MYB proteins and contains the EAR repressor domain (Jin et al., 2000; Kagale and Rozwadowski, 2011). The new poplar repressor MYBs share the LxLxL-type EAR motif with the lignin/phenylpropanoid MYBs, although in MYB181 and MYB182 this motif contains conservative substitutions. Thus, whereas LNLDL is most common, MYB181 and MYB182 C2 domains contain INLDL and INIDL, respectively. The poplar repressors also diverge in the xiG/S portion of the C2 motif (Fig. 1A). In the lignin or phenylpropanoid repressor MYBs such as AtMYB4 and Eucalyptus gunnii EgMYB1 (Jin et al., 2000; Legay et al., 2010), a zinc-finger motif (CX1-2CX7-12CX2C) and a C4 motif (FLGLx4-7V⁄LLD⁄GF⁄YR⁄Sx1LEMK) were present. These motifs were not found in FaMYB1, VvMYBC2-L1, PhMYB27, and the new poplar MYBs. In its place, these MYB repressor proteins all contained a complete (MYB165 and MYB194) or partial (MYB181 and MYB182) TLLLFR repressor motif (Matsui et al., 2008).
Figure 1.
Sequence analysis of repressor-like R2R3-MYBs relevant to poplar PA regulation. A, ClustalW alignment of the amino acid sequences of poplar MYB165, MYB181, MYB182, MYB194, and other R2R3-MYB subgroup 4 proteins. The R2 and R3 MYB domains are indicated by black bars. The boxed sequences are potential functional motifs. White letters on a black background represent residues that are identical in the sequences aligned. White letters on a gray background indicate conservative changes. Boxes indicate conserved elements in the primary sequence: C1 motif, LLsrGIDPX(T/S)HRX(I/L); C2 motif, pdLNL(D/E)LXi(G/S); C4 motif, GYDFLG(L/M)X4-7LX(Y/F)(R/S)XLEMK; zinc-finger (Zf) motif, CX1-2CX7-12CX1-2C; and TLLLFR motif. The bHLH-binding region is also marked, with key residues indicated by asterisks. B, Phylogenetic tree of poplar MYB165, MYB181, MYB182, MYB194, and related functionally characterized R2R3-MYB transcription factors from other plants, constructed from the N-terminal DNA-binding domains using the maximum likelihood method with all bootstrap values over 500 shown (1,000 replicates). Stars indicate the poplar repressor-like MYBs discussed here, and the arrow marks MYB182. Clades are indicated in color as follows: light blue, flavonoid repressors; purple, lignin/phenylpropanoid repressors; green, flavonol activators; light yellow, TT2-type PA activators; dark yellow, MYBPA1-type PA activators; and pink, anthocyanidin activators. GenBank accession numbers for all protein sequences are listed in Supplemental Table S1.
To better define the structure of subgroup 4 and to identify closely related R2R3-MYB factors, a phylogenetic analysis of the repressor MYBs and known anthocyanidin and PA regulators was performed (Fig. 1B; Supplemental Fig. S1). This phylogeny resolved a number of clades according to function. As expected, clades for known anthocyanin, flavonol, and two types of PA activators (TT2 and MYBPA1 types) were clearly defined with strong bootstrap support. The new poplar repressor MYBs grouped together with the previously characterized anthocyanin repressors FaMYB1 and PhMYB27 from strawberry and petunia and the PA repressor VvMYBC2-L1 from grapevine (Aharoni et al., 2001; Albert et al., 2014; Huang et al., 2014). This subclade was separated from the second major group of R2R3 repressor MYBs that comprises the general phenylpropanoid and lignin repressors from a variety of plants, including monocots. Together with the presence of different motifs in the C terminus, this analysis confirms that the R2R3-MYB repressors of flavonoid metabolism are distinct from other types of repressors.
In silico expression profiles using the Populus trichocarpa expression database and the eFP browser were generated for the new repressor MYBs in different poplar tissues (Supplemental Fig. S2). For comparison, the genes encoding the poplar PA regulator MYB134 and ANR1, a key enzyme for PA biosynthesis, were also profiled. Both genes are strongly expressed in young leaves, roots, and seedlings, all tissues with high PA synthesis. In etiolated seedlings, transcripts of MYB134 are also slightly induced by light. Except for MYB182, all of the repressors are expressed mainly in young leaves. By contrast, the expression profile for MYB182 was the opposite of that for MYB134, the positive PA regulator. This is consistent with the idea that MYB182 could have a negative effect on PA accumulation. Such negative correlations in developmental expression have been observed with AtMYBL2, PhMYB27, and VvMYBC2-L1 repressors (Dubos et al., 2008; Albert et al., 2011; Huang et al., 2014). We next used reverse transcription-quantitative PCR (RT-qPCR) to confirm that MYB182 expression is up-regulated in MYB134-overexpressing poplars, as this is how it was first identified (Supplemental Fig. S3). The MYB182 repressor also was induced by wound stress, similar to MYB134 and the PA biosynthetic genes. Based on these results, we focused on MYB182 for more detailed analyses of the role of this gene in PA and flavonoid metabolism.
Poplar Hairy Roots Overexpressing MYB182 Show Reduced PA Levels and Flavonoid Gene Expression
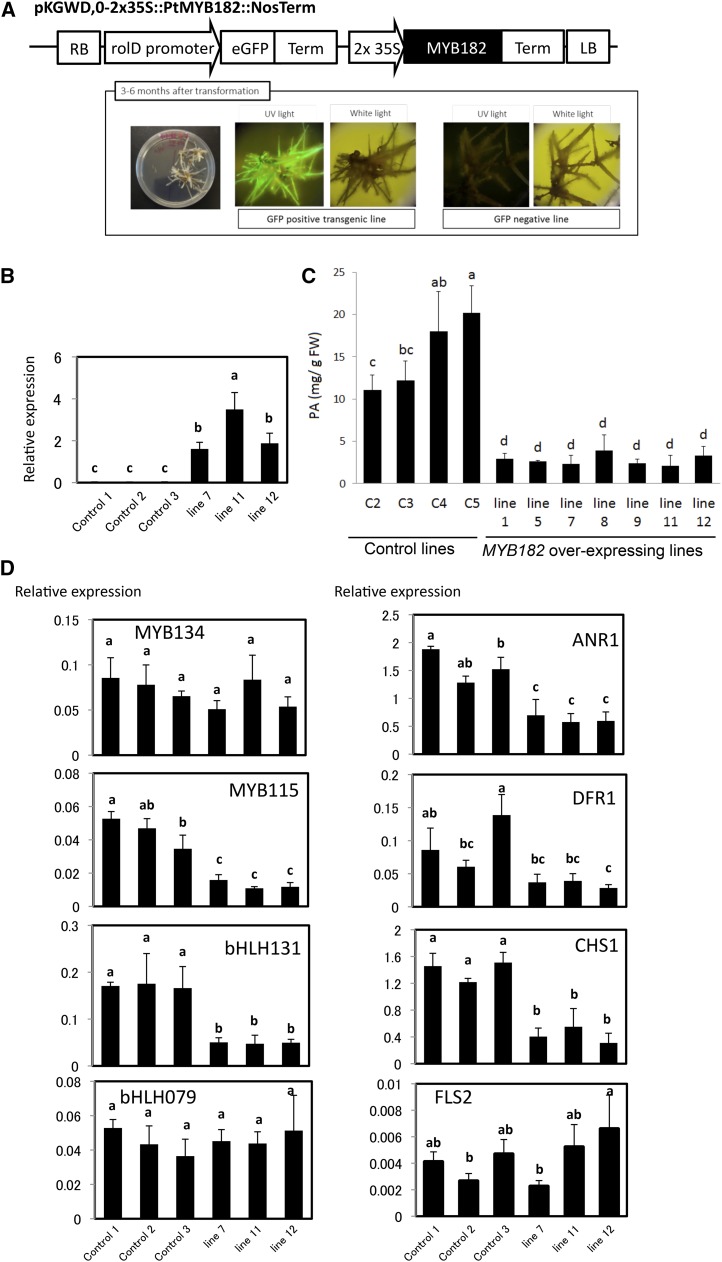
Since poplar roots accumulate high levels of PAs and express both positive and negative PA MYB regulators, we first used a hairy root expression system to directly test the function of MYB182. Induction of hairy roots by Agrobacterium rhizogenes is a relatively fast and efficient way of obtaining transgenic tissues (Giri and Narasu, 2000). Transgenic hairy roots have been used successfully for the analysis of transcriptional regulation of secondary metabolism in alfalfa (Medicago sativa), grape, and soybean (Glycine max; Terrier et al., 2009; Pang et al., 2013; Huang et al., 2014). The MYB182 coding sequence was cloned into a binary vector harboring the GFP marker gene under the control of a root-specific promoter. Two weeks following cocultivation with A. rhizogenes, leaf explants began to develop hairy roots. GFP-expressing roots were subcultured and analyzed further. Susceptibility to A. rhizogenes is known to vary among Populus spp. and genotypes; we obtained the highest frequency of hairy root formation with Populus tremula × Populus alba clone 717-1-B4 and the ARqua1 A. rhizogenes strain (Fig. 2A). Only GFP-expressing roots were analyzed further. All GFP-positive lines that expressed MYB182 showed dramatically reduced PA levels compared with empty vector control hairy roots (Fig. 2C). RT-qPCR confirmed the overexpression of MYB182 in all three lines examined (Fig. 2B).
Figure 2.
Ectopic overexpression of MYB182 in poplar hairy roots. A, Plasmid construct used for hairy root transformation of poplar. The GFP marker gene is driven by a rolD promoter and MYB182 by the double cauliflower mosaic virus 35S promoter (shown as 2x 35S). The images show GFP expression in hairy roots indicating the presence of the construct. LB, Left border; RB, right border. B, Expression of MYB182 in transgenic hairy root and control lines. C, PA content in independent MYB182 overexpression and empty vector control hairy root lines. FW, Fresh weight. D, Relative expression of PA-related genes assayed by RT-qPCR. bHLH131 and bHLH079 are poplar members of the TT8/AN1 and GL3/DEL/JAF13 subgroups, respectively (Supplemental Fig. S7). FLS2, FLAVONOL SYNTHASE2. MYB115 is an R2R3-MYB-positive PA regulator. Relative gene expression is normalized to the mean of elongation factor1B and ubiquitin/ribosomal protein27a expression. All data points are means of three biological replicates for each of three control and three transgenic lines, with error bars indicating sd. Labeled columns not connected by the same letter are significantly different at P < 0.05, based on a Tukey-Kramer honestly significant difference test.
To test the prediction that MYB182 overexpression leads to reduced PA levels by down-regulating the flavonoid pathway, the expression of flavonoid structural genes was examined by RT-qPCR. Reduced transcript levels were observed for the ANR1 and CHS1 genes (Fig. 2D). This suggested an effect on both early and late flavonoid biosynthesis genes. DFR1 showed a similar pattern, but its overall expression was too low to observe a consistent reduction in transcripts. By contrast, the FLAVONOL SYNTHASE2 gene, which does not participate in PA or anthocyanin synthesis, was not affected significantly by MYB182 overexpression (Fig. 2D). These experiments suggested that MYB182 specifically impacts PA and core flavonoid transcripts. We also assayed the expression of the positive PA regulator MYB134 and other regulatory genes that we previously observed to be up-regulated in MYB134-overexpressing poplar. MYB134 and a GL3-like bHLH cofactor (bHLH079; see below) were not affected by MYB182 overexpression. However, MYB115, the MYBPA1-type positive PA regulator, and bHLH131, a PA synthesis cofactor of the TT8/DEL/JAF13 group (Gesell et al., 2014), were affected in the MYB182-overexpressing hairy roots, and both showed reduced expression (Fig. 2D).
MYB182 Overexpression in Transgenic Poplar Plants Leads to Reduced Anthocyanin and PA Synthesis and Accumulation
To investigate the role of MYB182 in whole poplar plants, it was overexpressed in transgenic poplar plants under the control of the double cauliflower mosaic virus 35S promoter. Multiple independently transformed lines were generated, propagated, and grown in the greenhouse together with wild-type poplar plants (see “Materials and Methods”). Overexpression of MYB182 in poplar did not lead to any visible abnormalities when plants were grown under normal greenhouse conditions (Supplemental Fig. S4)
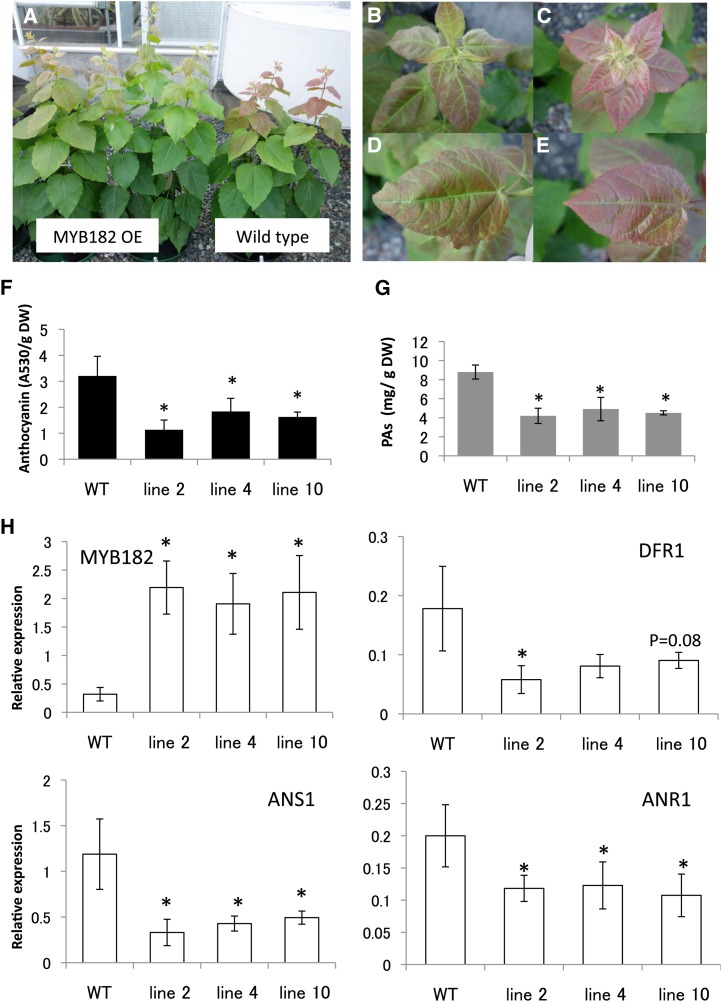
Previous work in our laboratory had shown that, in the greenhouse, poplar produces relatively low concentrations of PAs and other phenolics, but these accumulate to much higher levels when plants are exposed to direct sunlight (Mellway et al., 2009). Therefore, we exposed plants to natural summer sunlight (up to 2,080 µmol m−2 s−1) outside the greenhouse in July 2013. Within 1 week, the effect of the increased light exposure was apparent, observed in the reddening of the young leaves of both transgenic and control lines. However, this light-induced accumulation of red anthocyanin pigment was much less marked in the transgenic lines than in the controls (Fig. 3, A–E). Direct measurement of anthocyanins confirmed significant differences between transgenic and control leaf extracts (Fig. 3F). Likewise, the BuHCl assay revealed a significant decrease in PA concentration in leaves of the MYB182 overexpressors compared with control lines (Fig. 3G). The direct effect of MYB182 on the flavonoid pathway was confirmed by RT-qPCR analysis: while MYB182 was overexpressed in each transgenic line, the structural genes DFR1, ANS1, and ANR1 were repressed relative to controls (Fig. 3H). This down-regulation is consistent with the observed decrease in both anthocyanin and PA levels, as DFR and ANS are common to both flavonoid branches.
Figure 3.
Effect of MYB182 overexpression in transgenic poplars grown under natural sunlight. A, Visual comparison of wild-type control and MYB182-overexpressing (OE) poplar plants after 10 weeks of growth. B to E, Closeup images of young leaves of MYB182 OE (B and D) and control (C and E) plants. B and C show top views of the youngest leaves of MYB182 OE and control plants, and D and E show leaves at leaf plastochron index 5. F and G, Anthocyanin and PA contents of MYB182 OE lines and wild-type (WT) control lines. DW, Dry weight. H, Quantitative reverse transcription-PCR analysis of MYB182 and DFR1, ANS1, and ANR1 genes in the wild type and three independent MYB182 OE lines. Relative gene expression is normalized to the mean of elongation factor1B and ubiquitin/ribosomal protein27a expression. Data points shown are means from three biological replicates (individual plants), with error bars indicating sd. Asterisks above the bars indicate values determined by Student’s t test to be significantly different from control (P < 0.05).
HPLC analysis of methanolic extracts of MYB182 overexpressor and control leaves was carried out to determine if MYB182 expression altered the abundance of other phenolics in the transgenic plants. The HPLC method separates the major poplar phenolic constituents, including salicinoids, hydroxycinnamate esters, and flavonol glycosides (Supplemental Fig. S5). We could detect no significant differences between control and MYB182 overexpressor extracts, suggesting that the other phenolic pathways were not significantly affected by MYB182.
Transcriptome Analysis of MYB182 Overexpressor Plants Reveals Additional Down-Regulated Genes and Potential Targets of MYB182
To identify additional genes down-regulated by MYB182, the GeneChip Poplar Genome Array was used to compare leaf transcriptomes of three wild-type and three MYB182 overexpressor plants. In order to reduce environmentally induced variability in expression levels, the array was probed with complementary DNA isolated from leaves of greenhouse-grown plants. We defined a fold-change threshold of less than 0.5 and greater than 2, with P < 0.05 and Q < 0.1. Using these criteria, 318 and 153 probes were classified as down-regulated and up-regulated, respectively. This supports the idea that MYB182 has an overall repressive effect. To validate the microarray data, RT-qPCR for MYB182 and a set of down-regulated genes was carried out for the MYB182 overexpressors (Supplemental Fig. S6). A strong correlation of deregulated genes seen in both microarray and RT-qPCR confirmed the robustness of the array data.
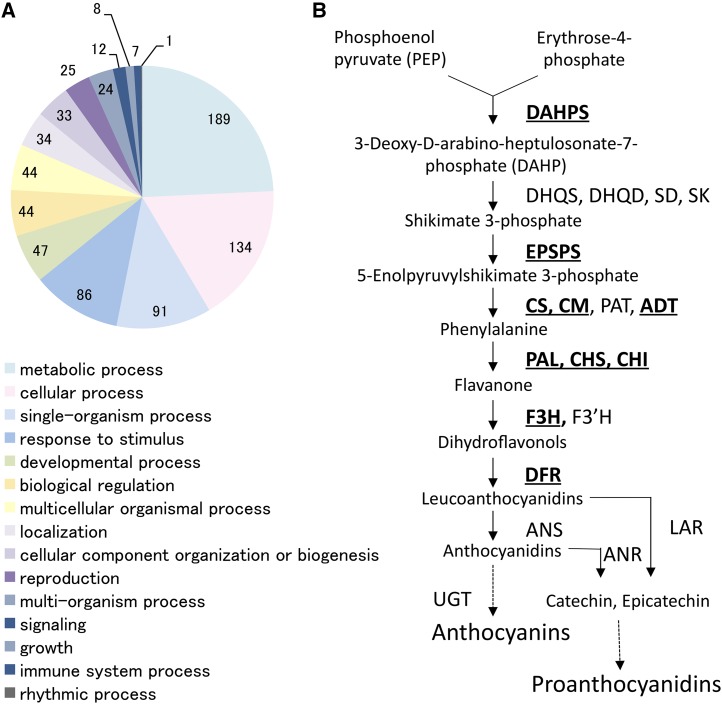
Significantly down-regulated genes were annotated by Blast2GO (Conesa et al., 2005) and categorized by predicted function. In total, 189 of the 318 down-regulated gene probes were classified by Gene Ontology identifier as being involved in metabolic process (Fig. 4A); this is consistent with a role of MYB182 in secondary metabolism (Fig. 4B). All annotations for genes with flavonoid or phenylpropanoid functions were verified by manual BLAST searches of the GenBank database. A number of core flavonoid enzymes were significantly down-regulated, including CHS2, F3H, DFR2, and CHI2 (Table I). In poplar, F3H and CHI are both single-copy genes, while DFR exists as two isoforms; these genes are all wound stress induced and contribute to PA synthesis (Tsai et al., 2006). While we did not detect significant down-regulation of all the isoforms for the flavonoid enzymes using our cutoffs, the analysis confirms that, when overexpressed, MYB182 has repressive effects on the flavonoid pathway.
Figure 4.
Analysis of target genes based on microarray profiles from MYB182-overexpressing poplar plants. A, Pie chart of Gene Ontology categories corresponding to the 381 down-regulated probes classified as biological process. Note that any one probe can belong to more than one Gene Ontology category. B, Summary of the shikimate, phenylpropanoid, and flavonoid pathways leading to PA and anthocyanin synthesis. Abbreviated enzyme names are indicated for each biosynthetic step. Boldface and underlined names represent genes significantly down-regulated in MYB182-overexpressing plants. DAHPS, 3-Deoxy-d-arabino-heptulosonate 7-phosphate synthase; DHQS, dehydroquinate synthase; DHQD, dehydroquinate dehydratase; SD, shikimate dehydrogenase; SK, shikimate kinase; EPSPS, 5-enolpyruvylshikimate-3-phosphate synthase; CM, chorismate mutase; ADT, arogenate dehydratase; PAL, Phe ammonia lyase; F3′H, flavonoid 3′-hydroxylase; LAR, leucoanthocyanidin reductase.
Table I. List of selected genes that were down-regulated in MYB182 overexpression plants.
Descriptions are based on Blast2GO annotations (Conesa et al., 2005), unless noted otherwise.
| Gene Model Identifier | Description | Fold Change | P |
|---|---|---|---|
| Phenylpropanoid and flavonoid biosynthesis | |||
| Potri.016G014500 | UDP-Glc flavonoid 3-O-glucosyltransferase-like | 0.14 | 2.33E-04 |
| Potri.003G066800 | Flavonoid 3′-monooxygenase-like | 0.20 | 2.42E-04 |
| Potri.016G091100 | Phe ammonia lyase (PAL3a) | 0.22 | 2.18E-04 |
| Potri.001G167900 | Flavonoid 3′-monooxygenase-like | 0.23 | 2.08E-04 |
| Potri.003G066400 | Flavonoid 3′-monooxygenase-like | 0.23 | 3.30E-04 |
| Potri.001G051600 | Chalcone synthase (CHS2b) | 0.27 | 4.55E-04 |
| Potri.018G051300 | Flavonoid 3′-monooxygenase-like | 0.28 | 1.39E-04 |
| Potri.006G141400 | Flavonoid 3′-monooxygenase-like | 0.32 | 5.71E-05 |
| Potri.006G171100 | Flavonoid 3-O-glucosyltransferase-like (UGT78M1c) | 0.32 | 3.61E-04 |
| Potri.005G113700 | Flavanone 3-hydroxylase (F3H) | 0.34 | 2.03E-03 |
| Potri.004G139700 | Flavonol synthase (FLS1b) | 0.34 | 5.71E-04 |
| Potri.T071600 | 4-Coumarate:ligase (4CL4) | 0.43 | 1.87E-05 |
| Potri.005G229500 | Dihydroflavonol 4-reductase (DFR2b) | 0.44 | 9.13E-04 |
| Potri.006G171200 | Flavonoid 3-O-glucosyltransferase-like (UGT78M1c) | 0.45 | 1.55E-05 |
| Potri.019G057800 | Chalcone isomerase (CHIL2b) | 0.48 | 1.60E-03 |
| Phe, Tyr, and Trp biosynthesis | |||
| Potri.005G073300 | Phospho-2-dehydro-3-deoxyheptonate aldolase chloroplastic-like (DAHPS) | 0.30 | 8.58E-05 |
| Potri.004G188100 | Arogenate dehydratase prephenate dehydratase chloroplastic-like (ADT1a) | 0.31 | 6.74E-06 |
| Potri.002G099200 | Phospho-2-dehydro-3-deoxyheptonate aldolase chloroplastic-like (DAHPS2a) | 0.37 | 1.08E-04 |
| Potri.005G162800 | Phospho-2-dehydro-3-deoxyheptonate aldolase chloroplastic-like (DAHPS1a) | 0.39 | 5.89E-04 |
| Potri.002G146400 | 5-Enolpyruvylshikimate-3-phosphate synthase (EPSPS1a) | 0.48 | 2.50E-04 |
| Potri.017G088700 | Chorismate mutase (CM1b) | 0.49 | 6.28E-05 |
| Potri.010G221600 | Chorismate synthase family protein (CS1a) | 0.49 | 1.19E-03 |
Described by Hamberger et al. (2007). bDescribed by Tsai et al. (2006). cDescribed by Veljanovski and Constabel (2013).
In addition to the core flavonoid genes, two genes annotated as UGT-flavonoid glycosyltransferases also were down-regulated, as well as two core phenylpropanoid genes: Phe ammonia lyase3 and 4-coumarate-CoA ligase4. The down-regulation of a suite of uncharacterized poplar genes annotated as flavonoid 3′-monooxygenase-like genes was intriguing; these are being investigated further, but their connection to flavonoid metabolism and their specific biochemical functions are unknown. Interestingly, transcripts encoding five shikimate pathway genes were identified among the significantly down-regulated genes. These include phospho-2-dehydro-3-deoxyheptonate aldolase/3-deoxy-d-arabino-heptulosonate 7-phosphate synthase, 5-enolpyruvylshikimate-3-phosphate synthase, chorismate mutase, and arogenate dehydratase-like genes (Table I). The shikimate pathway is well known and is required by plants to provide aromatic amino acids for proteins as well as phenylpropanoids. Thus, it is possible that MYB182 not only affects PA and anthocyanin metabolism but also has the potential to down-regulate the shikimic acid pathway at the level of gene expression (Fig. 4B).
MYB182 Inhibits the Transactivation of a PA Promoter by Positive MYB Regulators
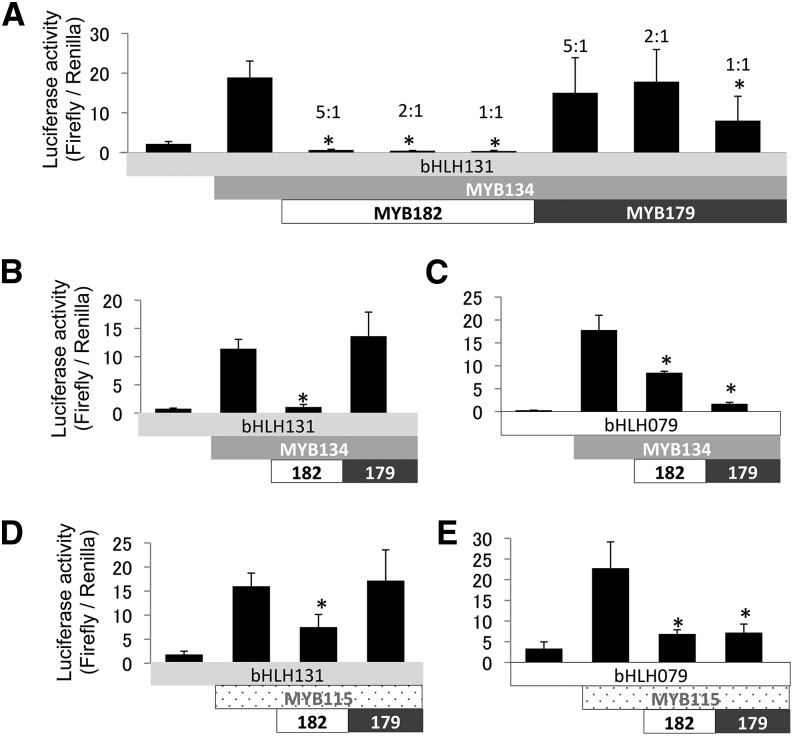
To determine if MYB182 can directly repress PA synthesis, we performed promoter activation assays in transiently transformed poplar suspension cells using promoter-luciferase fusion constructs. We had shown previously that the poplar ANR1 promoter is activated by MYB134 when cotransformed with the poplar bHLH131 cofactor into a transient expression system (Zifkin et al., 2012; Gesell et al., 2014). To show negative regulation by MYB182, a positive regulator such as MYB134 was used in combination with the MYB182 repressor construct, and the reduction in activation was measured. We first tested the activator and repressor constructs at different ratios, with up to a 5-fold excess of activator. Even at this reduced ratio (repressor:activator, 1:5), MYB182 was able to repress the activation of poplar ANR1 (Fig. 5A). Based on these results, for subsequent experiments a repressor:activator ratio of 1:4 was used. For comparison, we also tested constructs encoding the single-repeat R3 MYB, MYB179, since this gene also is up-regulated by MYB134 and is similar to CPC and MYBx, single-repeat R3 MYB repressors that down-regulate anthocyanin biosynthesis in Arabidopsis and petunia (Wester et al., 2009; Zhu et al., 2009; Albert et al., 2014). In our experiments, MYB179 only repressed ANR1 activation at a repressor:activator ratio of 1:1. When the relative amount of MYB179 was reduced, it was no longer active (Fig. 5A). This difference with MYB182 suggested potentially distinct mechanisms of action for MYB179 and MYB182.
Figure 5.
Transient expression assays to test the activation and repression of the poplar ANR1 promoter by poplar MYB and bHLH regulators in suspension-cultured poplar cells. MYB activators and repressor plasmids were tested in a ratio of 4:1 (250:62.5 ng of DNA, which gives a similar ratio of molecular weight) unless indicated otherwise. A, Transcriptional activation by MYB134 (R2R3 activator MYB) and poplar bHLH131 at different activator-to-repressor ratios. B and C, Effects of MYB182 (R2R3-MYB repressor) and MYB179 (R3-MYB) on the activation of the ANR1 promoter by MYB134 in the presence of bHLH131 (TT8/DEL/AN1 homolog) or poplar bHLH079 (GL3/JAF13 homolog). D and E, Transcriptional activation by MYB115 (R2R3 activator MYB) with bHLH131 or bHLH079 and repression by MYB182 and MYB179. All data points are means of three biological replicates, with error bars indicating sd. Asterisks indicate values determined by Student’s t test to be significantly different from control (P < 0.05).
The R2R3 repressor-like MYBs contain a bHLH-binding region (Fig. 1) and thus are predicted to interact directly with bHLH cofactors. To probe this interaction, we assayed repressor and activator constructs together with two poplar bHLHs belonging to different subclades: poplar bHLH131, which is part of the TT8 subclade (Gesell et al., 2014), and poplar bHLH079, which belongs to the JAF13/GL3 subclade (Supplemental Fig. S7). For these experiments, we also included poplar MYB179. In Arabidopsis, GL3 interacts with the PA or anthocyanin activator MYBs to form an MBW complex and activate the DFR promoter, while the single-repeat R3 MYB CPC disrupts this interaction (Zimmermann et al., 2004; Wester et al., 2009). Our luciferase assays indicated that MYB182 was less effective as a repressor when cobombarded with bHLH079 than with bHLH131 (Fig. 5B). For MYB179, however, we found the opposite pattern: greater repression with bHLH079 than with bHLHL131 (Fig. 5C). This dependency on the type of bHLH cofactor suggested that the bHLH interaction is important for the mechanism of action of MYB repressors. We also tested MYB182 and MYB179 repressor activity with the second PA MYB activator, MYB115; the pattern of repression here was generally similar to that seen using MYB134 as the activator (Fig. 5, D and E).
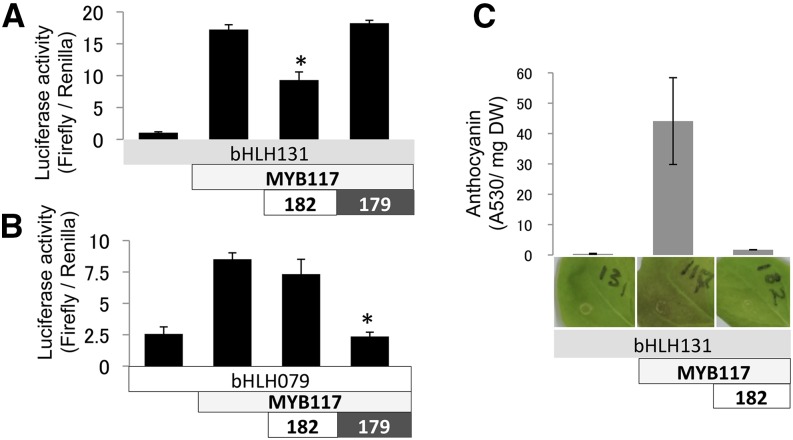
Since the transgenic poplar overexpressing MYB182 showed reduced anthocyanin accumulation (Fig. 3), we designed experiments to test whether MYB182 could be involved directly in the negative regulation of anthocyanin biosynthesis (Fig. 6). Based on the many characterized anthocyanin regulators of the PAP1 type (Fig. 1), we chose MYB117 as a likely ortholog for functional studies. In bombardment experiments carried out as previously, MYB117 activated the DFR promoter from Arabidopsis with both bHLH cofactors (Fig. 6). However, unlike the previous activation experiments using MYB134 or MYB115, MYB117 activation of DFR was repressed by MYB182 only when cotransformed with bHLH131. As before, MYB179 showed the opposite pattern, repressing MYB117 only if bHLH079 was used as a cofactor. The repression of anthocyanin biosynthesis was confirmed using transient transformation in agroinfiltrated Nicotiana benthamiana leaves. In this experiment, MYB117 induced visible anthocyanin accumulation when infiltrated with the bHLH131 cofactor (Fig. 6C). Coinfiltration of MYB182 with the above activators, however, prevented anthocyanin synthesis and red leaf coloration.
Figure 6.
Functional analysis and repression of the poplar anthocyanin regulator MYB117 in transiently transformed poplar suspension cells and agroinfiltrated N. benthamiana leaves. A and B, Activation of the Arabidopsis DFR promoter by anthocyanin regulators and effects of MYB repressors in poplar cell culture. Assays of MYB117 were carried out with the repressor MYB182 or MYB179 and using either bHLH131 (A) or bHLH079 (B) as a cofactor. C, Anthocyanin content and corresponding images of red leaf coloration at 8 d following agroinfiltration of leaves in the presence and absence of MYB182 and MYB117. The ratio of MYB activators to repressors was 4:1. All data points are means of three independent replicates, with error bars indicating sd. Asterisks above the bars indicate values determined by Student’s t test to be significantly different from the control (P < 0.05). DW, Dry weight.
Together, the transient expression experiments demonstrated that MYB182 repressed promoter activation driven by both anthocyanin and PA activator MYBs. With the PA regulators MYB134 and MYB115, the repressor activity of MYB182 was clear with either bHLH cofactor. With the MYB117 anthocyanin activator, however, repression by MYB182 was detected only in combination with bHLH131.
MYB182 Repressor Activity Requires Interaction with the bHLH Cofactor
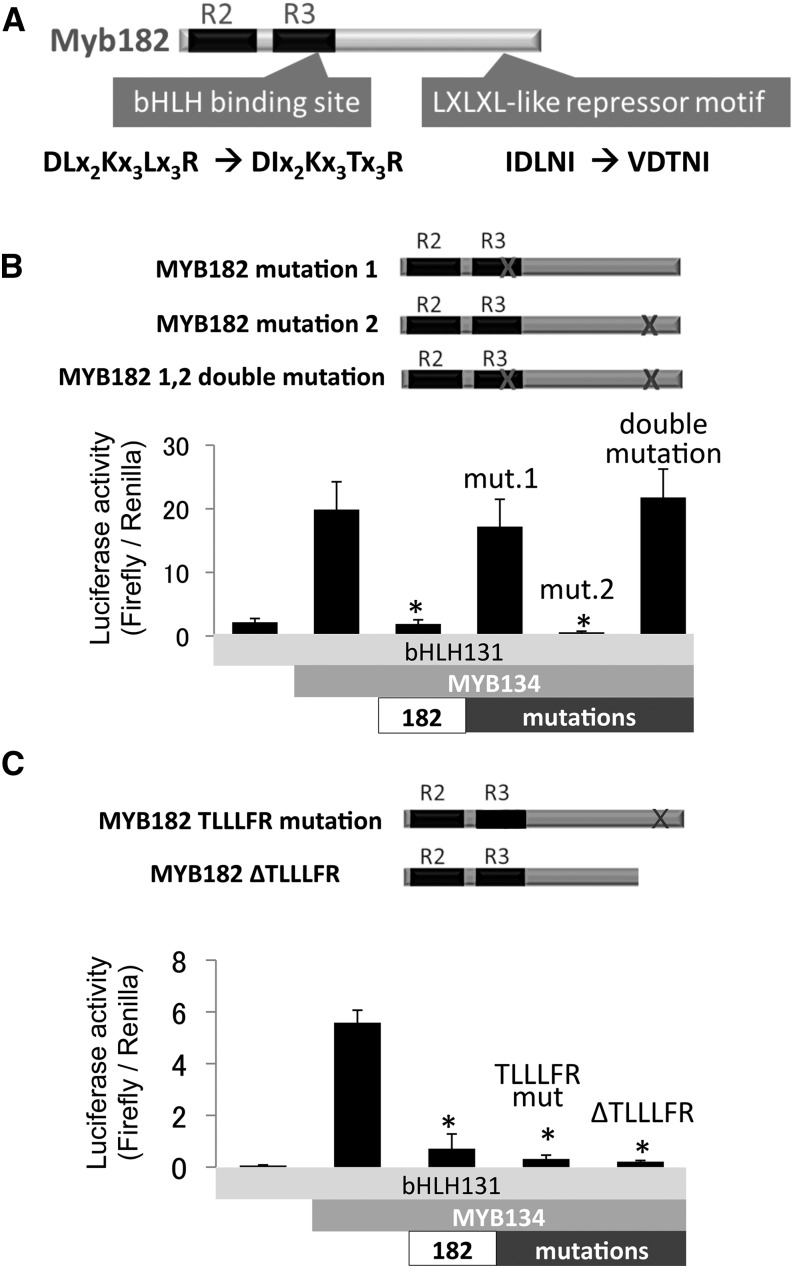
The alignment of MYB182 and other repressor-like MYBs had revealed the two common motifs found in all R2R3 repressor-like MYBs: the bHLH-binding motif in the R3 domain and a C2 region with a partial EAR motif on the C-terminal region (Figs. 1A and 7A). To probe the functions of these conserved regions, we created mutant versions of MYB182 for promoter activation and repression constructs, in which one or both of the motifs were altered. For the bHLH mutation, we altered the first and last L residues to I and T, respectively, whereas to generate a C2 mutant, we replaced IDLNI with VDTNI (Fig. 7A). MYB182 without a functional bHLH-binding site (mutant 1) lost its ability to repress MYB134 activation of the ANR promoter (Fig. 7B). In contrast, mutation of the C2 motif (mutant 2) did not result in a detectable reduction of repressor function. A construct with both motifs altered (double mutant) showed the same results as mutant 1. These results corroborate the idea that the primary mechanism of action for MYB182 involves an interaction with the bHLH cofactors. In addition, based on the presence of the TLLLFR motif in the C-terminal region, we created mutant constructs to test whether this motif might be required for the repressor activity of MYB182. Neither a complete deletion nor a partial modification of this sequence prevented MYB182 from repressing promoter activation (Fig. 7C).
Figure 7.
Functional analysis of conserved motifs in MYB182. A, Structure of MYB182 protein showing the locations of the conserved bHLH-binding domain and the C2 repressor motif. The design strategy for the mutation of amino acids in the conserved motifs is shown below each site in question. B, Locations of mutations within MYB182 mutant constructs used in the promoter activation/repression assay and corresponding results showing activation of the poplar ANR1 promoter by MYB134 with bHLH131 and repression by MYB182. mut.1 and mut.2 refer to mutants 1 and mutant 2 shown above. C, Schematic and results of TLLLFR mutant tests, where the motif was either mutated or deleted. Data points are means of three biological replicates, with error bars indicating sd. Asterisks above the bars indicate values that were determined by Student’s t test to be significantly different from the control (P < 0.05).
DISCUSSION
The PA pathway in poplar responds to multiple developmental and environmental signals, reflecting the action of both positive and negative regulators. Here, we characterize a novel poplar repressor-like R2R3-MYB, MYB182, and demonstrate that it can down-regulate PA as well as anthocyanin metabolism. MYB182 is the second MYB repressor of PA metabolism described to date. Our analysis shows that MYB182 acts on flavonoid gene promoters and may also affect other regulatory genes. Our data further indicate that MYB182 can repress activation by both anthocyanin- and PA-specific MYB activators and that interaction with a bHLH cofactor is required for this repression. These conclusions are based on two lines of evidence. First, transgenic poplars and hairy root cultures that overexpress MYB182 have diminished PA concentrations, and the transgenic leaves also show lower levels of anthocyanins. The reduction in PAs in transgenic tissues correlates with reduced transcript levels of key flavonoid pathway genes. Second, transient expression and promoter activation assays directly demonstrate the repressive effects of MYB182 on relevant promoters activated with both PA and anthocyanin activator MYBs.
MYB182 Is a Repressor That Reduces PA and Anthocyanin Accumulation and Represses Flavonoid Gene Expression When Overexpressed in Transgenic Poplar
Our data from MYB182-overexpressing hairy roots clearly indicate that PA accumulation is decreased and transcripts of relevant flavonoid genes are reduced in abundance in transgenic hairy roots (Fig. 2). Effects on anthocyanin accumulation could not be studied in this system, as hairy roots do not make these pigments. In parallel, our whole-plant transformations with MYB182 also showed reduced PA levels and decreased anthocyanin accumulation in leaves (Fig. 3). In contrast to plants overexpressing other repressor-like MYBs such as AmMYB308, AtMYB4, and PvMYB4 (Tamagnone et al., 1998; Jin et al., 2000; Shen et al., 2012), in MYB182 overexpressors no phenotypic abnormalities were detected.
The MYB182-overexpressing plants showed only mild effects of the transgene when grown in the greenhouse but developed a clearer phenotype when they were moved into natural sunlight to induce flavonoid biosynthesis (Fig. 3, A–E). While the light levels in such an experimental setup are difficult to control, the intensity of the light was not excessive, and we have used this treatment previously to reliably stimulate flavonoid metabolism (Mellway et al., 2009; Gesell et al., 2014). However, the transition from greenhouse conditions with no UV-B light penetration and lower intensity visible light to natural sunlight likely constitutes a mild stress treatment (see “Materials and Methods”). Nevertheless, the reduction of PA metabolites and gene expression in transgenic plants was less marked than that in the hairy root culture system. This was perhaps due to a difference between the relative expression of the PA pathway in leaves and roots. Whereas leaves accumulate PAs together with large amounts of other phenolics, including salicinoids, flavonols, and hydroxycinnamic acid esters (Constabel and Lindroth, 2010), poplar roots have a much simpler phenolic profile that is dominated by the PAs (C.P. Constabel, unpublished data). Hairy roots are thus more sensitive to manipulations of the PA pathway, which will make them an ideal system for testing PA repressor activity. In grapevine, hairy root transformation has been used to validate the function of the VvMYBC2-L1 gene (Huang et al., 2014).
Microarray analysis and RT-qPCR of the transgenic plants demonstrated the repressor function of MYB182, showing the down-regulation of genes encoding flavonoid enzymes (Fig. 3; Table I), and is similar to the RT-qPCR analysis of hairy root flavonoid gene expression. An effect of MYB182 on PA, anthocyanin, and core flavonoid genes is consistent with several features of the MYB182 protein sequence, which contains amino acid motifs shared by other flavonoid regulators. A recent study by Heppel et al. (2013) identified two regions of conserved amino acid residues in the R2 and R3 domains of activator MYB transcription factors and proposed that these residues are important for the specific regulation of the PA and anthocyanin pathway. The MYB182 sequence contains a Gly at position 49 in the R2 domain, a conserved feature in all PA regulators and in the other R2R3-MYB repressors (Fig. 1A). At positions 90 to 93 of the C-terminal end of the R3 repeat, a short stretch of amino acid residues was observed by Heppel et al. (2013) to be different in the PA MYBs (Asp-Asn-Glu-Ile) from the anthocyanin MYBs (Ala-Asn-Asp-Val). In MYB182, this region has residues characteristic of both the PA and anthocyanin regulators (Asp-Asn-Glu-Val), which could reflect MYB182’s capacity to repress both pathways. However, other regions also must be important, since the Asp-Asn-Glu-Ile motif is found in some subgroup 4 lignin and phenylpropanoid repressor MYBs as well.
In addition to the down-regulation of core flavonoid genes, in the MYB182 overexpressors we also noted the reduced transcript abundance of other genes with annotations suggesting peripheral roles in flavonoid synthesis. For example, two UGTs are repressed by MYB182, including UGT78M1, an enzyme that we had found to be induced in Melampsora medusae-infected poplar leaves (Miranda et al., 2007). However, the recombinant UGT78M1 protein was not active with any anthocyanin, flavonol, or flavan-3-ol substrate that we tested, so its biochemical function remains undefined (Veljanovski and Constabel, 2013). The functions of the other UGT are also unknown; however, it is expressed in herbivore-stressed poplars (Ralph et al., 2006). A conspicuous group of down-regulated genes is annotated as encoding flavonoid 3′-monooxygenase-like enzymes (i.e. flavonoid 3′-hydroxylase-like), which belong to the cytochrome P450 gene family. A phylogenetic analysis of the most related P450s indicated that the genes identified in our experiment are not highly similar to any characterized genes (Supplemental Fig. S8). They are closest to GhDDWF1 and NtCYP71D20 (approximately 69%–79% protein identity), which are involved in the biosynthesis of brassinosteroids and capsidiol, respectively (Kang et al., 2001; Ralston et al., 2001). Until other genes in this subgroup are functionally characterized, the significance of the down-regulation of the poplar flavonoid 3′-monooxygenase-like genes will remain unclear.
It must be stressed that the genes affected by MYB182 in overexpression experiments remain tentative targets of this repressor, as overexpression experiments in themselves are not sufficient to unequivocally demonstrate function. Analysis of transgenic and wild-type plants under stress conditions also may help illuminate the role of MYB182 in poplar adaptation. Ultimate proof of the function of MYB182 and validation of its targets will require the creation of MYB182 knockout plants.
A bHLH-Interacting Site in the R3 Domain of MYB182 Is Critical for Repression
Analysis of amino acid sequences revealed that the MYBs in the PA/anthocyanin clade of subgroup 4 repressors harbor neither the zinc-finger nor the C4 motif, but they do contain the conserved C1 and C2 motifs as well as the bHLH-binding region (Fig. 1). Previous work had demonstrated direct protein-protein interactions of bHLH proteins with MYB repressors, including FaMYB1, AtMYB4, and PhMYB27 (Aharoni et al., 2001; Zimmermann et al., 2004; Albert et al., 2014). In vitro mutagenesis of our MYB182 constructs allowed us to test the functions of the putative motifs directly. Disruption of the bHLH-binding site clearly abolished the repressor activity of MYB182 (Fig. 7), indicating that its activity depends on interaction with bHLH factors. This is consistent with our observation that the pattern of repression is altered depending on which type of bHLH factor is used as a cofactor. MYB182 showed repression with either bHLH079 (GL3-type) or bHLH131 (TT8-like) cofactor using the PA activators, although to varying degrees. When activation was driven by the PAP1-like anthocyanin regulator MYB117, however, MYB182 could repress promoter activation only if the bHLH079 was coexpressed (Fig. 6). These data corroborate the importance of the bHLH cofactor for repressor function, so that MYB182 could be acting in part by competing for bHLH with the activator MYBs. Furthermore, it may also act as part of a MBW complex, as suggested by experiments on PhMYB27 in petunia (Albert et al., 2014). Those authors suggest that, in this case, specificity may depend on the activator and that the R2R3 repressor MYBs switches the complex from activation to repression. In any event, the strength of the MYB repressor-bHLH interaction is likely to be a critical element. The correlation between the strength of protein-protein interactions and repressor function has not yet been examined for the R2R3 subgroup 4 MYBs. For the R3 single-repeat MYBs, including Arabidopsis CPC, gentian (Gentiana triflora) MYBR1, and petunia MYBx, protein-binding affinities do differ among these R3-MYB factors, and this can affect their function (Kirik et al., 2004; Wester et al., 2009; Nakatsuka et al., 2013). Quantitative analysis of protein-protein interactions may reveal the interaction and affinity of MYB182, MYB179, and MYB117 with bHLH131 and bHLH079.
A key question that still needs to be answered directly is if MYB182 itself can bind to flavonoid promoters and thus directly repress flavonoid gene expression. While we could not test this possibility, mutating the R3 domain of Arabidopsis AtMYB4 impeded its ability to bind DNA and abolished its ability to repress transcription. This would suggest that DNA binding is also essential for its repressor function (Jin et al., 2000). Other subgroup 4 MYB factors are reported to directly bind target gene promoters (Jin et al., 2000; Fornalé et al., 2010; Shen et al., 2012; Bomal et al., 2014). We cannot rule out that mutation of the MYB182 bHLH-binding site also might have affected its DNA-binding ability, since Baudry et al. (2004) had shown that the Arabidopsis MYB TT2 can only bind to its target gene promoter in yeast (Saccharomyces cerevisiae) in the presence of a bHLH or WDR factor. Currently, our data do not allow us to distinguish this possibility from a mechanism based on competition for bHLH cofactors, although we note that these are not mutually exclusive mechanisms. Protein-protein interaction experiments and yeast one-hybrid assays should be able to distinguish between these mechanisms.
Surprisingly, mutation of the C2 motif in MYB182 did not diminish its repressor activity, despite the documented role of this motif in MYB subgroup 4 lignin and phenylpropanoid repressors and the anthocyanin repressor PhMYB27. It is possible that the differences in the motif in MYB182, although conservative (INIDL versus LNLDL), reduce the motif’s functionality. However, even the previous studies showed that repressor activity could not be completely eliminated by disruption or removal of the C2 motif (Jin et al., 2000; Shen et al., 2012; Albert et al., 2014). This suggests that additional mechanisms are important. Nevertheless, the C2 region with the core EAR motif is considered to be the predominant transcriptional repression motif in plants (Ohta et al., 2001; Hiratsu et al., 2003; Tiwari et al., 2004). In tobacco ERF3 (NtERF3), where the EAR motif was first characterized, deletion mutations within the EAR motif eliminated its ability to repress transcription (Ohta et al., 2001). Although the mechanism of repression is still unclear, some reports show that the EAR motif is important for interaction with histone modification proteins (Kagale and Rozwadowski, 2011). In Arabidopsis, the INDOLE-3-ACETIC ACID12/BODENLOS auxin transcriptional repressor, and NOVEL INTERACTOR OF JAZ, which functions in transcriptional repression of JA signaling, have been shown to interact with the TOPLESS (TPL) corepressor (Szemenyei et al., 2008; Pauwels et al., 2010). The genetic interaction between TPL or TPL-RELATED1 and the histone deacetylase AtHDA19 supports a model for EAR motif-mediated repression via the recruitment and action of chromatin modifiers (Kagale and Rozwadowski, 2011). Such a model could explain why deletion of the C2 motif did not impact MYB182 in our transient expression assays, since the target promoter is transferred into the cell as a plasmid and is not present as chromatin. An alternative explanation would be that the effects of C2 motif deletion were obscured by the action of the bHLH-binding site, since binding to a bHLH cofactor seems to be sufficient to repress transcriptional activation (Fig. 7, mutant 1). This could explain why Ohta et al. (2001) saw effects of the EAR motifs in transient assays using plasmids while we did not, since the repressor used by these authors does not require such cofactors. Disruption of the TLLLFR motif likewise did not inhibit the repressor activity of MYB182, unlike what was reported for AtMYBL2 (Matsui et al., 2008). The discrepancy could be due to different assay systems, as Matsui et al. (2008) used the yeast Gal4-binding domain, or the dominant effect of the bHLH interaction. In order to better understand the mechanism(s) by which MYB182 acts, as well as the potential role of the different motifs within MYB182, the ratios of activator, cofactor, and mutant repressor will likely need to be examined carefully. However, overall, our experimental system indicates a greater importance of the bHLH-binding site of MYB182 relative to the conserved C2 motif, at least against the MYB134/bHLH131 activator complex.
MYB182 Is Part of a Complex Network of Positive and Negative Regulators for Flavonoid Metabolism in Poplar
MYB182 was first identified due to its enhanced expression in transgenic plants that overexpress MYB134 and was hypothesized to help modulate PA induction. Our data have corroborated this idea, and core flavonoid genes are indeed targets of the MYB182 repressor. However, our work has uncovered additional connections to other parts of the flavonoid and PA regulatory network in poplar. Poplar hairy roots overexpressing MYB182 showed reduced expression of MYB115 and bHLH131, both positive regulators which stimulate PA synthesis (Fig. 2; Gesell et al., 2014). An inhibitory effect of MYB182 on these and other positive transcriptional regulators still needs to be confirmed but would provide another level of control with additional feedback loops for down-regulating the PA pathway. Poplar MYB165 and MYB194 also need to be studied in more detail. Their roles are unknown to date, but they could provide additional features that will contribute to the development of a regulatory network model for stress-induced PA regulation in poplar.
The recent characterization of the anthocyanin repressor PhMYB27 in petunia provides a model for such multilevel regulation: This protein acts to repress the bHLH cofactor AN1, MYBx, and its own expression, in addition to down-regulating the structural genes (Albert et al., 2014). Such negative regulation via repression of the MYB PA activators was also suggested by work on the regulation of grapevine PA biosynthesis (Terrier et al., 2009; Huang et al., 2014), where expression levels of the positive regulators VvMYBPA1 and VvMYBPA2 in VvMYBC2-L1-overexpressing hairy roots were reduced. Work in these distinct experimental systems all suggest that there are common regulatory mechanisms among subgroup 4 R2R3-MYB factors regulating late flavonoid genes in higher plants.
Overall, our data suggest that MYB182 can inhibit flavonoid biosynthesis at three levels. First, it acts to repress the transcription of genes encoding flavonoid enzymes that are required for PA synthesis. Second, it counteracts the effects of the positive regulators MYB134 and MYB115 by repressing the expression of components of the PA-activating MBW complexes themselves. In their work on petunia PhMYB27 and anthocyanin regulation, Albert et al. (2014) refer to this two-level repression as a “double lock-down” mechanism: direct down-regulation of flavonoid structural genes and the parallel down-regulation of positive flavonoid regulators. Third, based on our microarray analysis of MYB182 overexpressor plants, we speculate that the availability of shikimate phenylpropanoid precursors also may be reduced, although this will need to be validated directly.
CONCLUSION
Our work has used both transient and stable transformation of poplar to illuminate the role of MYB182 as a repressor of the flavonoid pathway leading to anthocyanins and PAs. Its repressor activity in this pathway is consistent with its position in the phylogeny of subgroup 4 MYB factors, and it helps to define a subclade of flavonoid R2R3-MYB repressor proteins. While we could not demonstrate a role for the C2 motif or TLLLFR motif of MYB182, its bHLH-binding region is clearly essential for repression. The mechanism by which MYB182 down-regulates flavonoid genes appears to be similar to the recently characterized anthocyanin repressors in petunia. MYB182 is part of a complex network of positive and negative transcriptional regulators for PAs that now includes several activator MYBs and bHLHs as well as three additional MYB repressors. These remain to be characterized in future work.
MATERIALS AND METHODS
Plant Growth Conditions and Stress Treatments
Populus tremula × Populus tremuloides (clone INRA 353-38) and P. tremula × Populus alba (clone INRA 717-1-B4) were micropropagated in vitro on solid Lloyd & McCown’s Woody Plant Medium (WPM; Caisson Laboratories) supplemented with 0.5 µg mL−1 indole butyric acid in growth chambers under long-day conditions (16 h of light/8 h of dark, 25°C). These plants were the source of material for stable transformation experiments. For greenhouse experiments, rooted in vitro plantlets were transplanted to soil and acclimated in a mist chamber for 3 weeks prior to being moved into the greenhouse.
For natural sunlight exposure, 2-month-old potted poplar plants were moved from the greenhouse (mean maximum photosynthetically active radiation, 392 µmol s−1 m−2; UV-B irradiance, 0.003 mW cm−2; temperature, 17°C–28°C) to full natural sunlight during July in Victoria, British Columbia, Canada (mean maximum photosynthetically active radiation, 2,080 µmol s−1 m−2; UV-B irradiance, 0.2 mW cm−2; temperature, 12°C–32°C). For wounding experiments, leaves of greenhouse-grown, 3-month-old wild-type 353-38 plants (leaf plastochron index, 10–12) were crushed along their margins with pliers. Leaves were rewounded 1 h following initial wounding and harvested at 24 h.
Suspension cell cultures of Populus trichocarpa × Populus deltoides (H11-11) were maintained in 40 mL of Murashige and Skoog (MS) liquid medium (Sigma) supplemented with 3% (w/v) Suc and maintained in 250-mL flasks on a rotary shaker (130 rpm) in the dark at 22°C. Seven milliliters of suspension culture was subcultured into 40 mL of fresh MS medium every 7 d, and four individual 7-d-old cultures were used for each bombardment.
Nicotiana benthamiana plants for agroinfiltration were grown in a growth chamber under long-day conditions (16 h of light/8 h of dark, at 22°C) and used when 4 weeks old.
Phylogenetic and Sequence Analyses
Full-length amino acid sequences were retrieved from public databases. For phylogenetic tree construction of R2R3-MYB proteins, multiple sequence alignments were performed using Dialign (Al Ait et al., 2013; http://dialign.gobics.de). Only sequence regions with at least 10% diagonal similarity were retained, which restricted the alignment to the conserved DNA-binding N-terminal region. After first using ProtTest (Abascal et al., 2005; http://darwin.uvigo.es/software/prottest2_server.html), which determined that the most suitable model for maximum likelihood analysis was JTT+I+G, we generated the phylogenetic tree using a local PhyML (Guindon et al., 2010) via JTT model with manual modification of γ-distribution and proportion of invariable sites. Bootstrapping was carried out with 1,000 replicates using the same evolution model for the original tree using local PhyML. Trees were displayed using Figtree (http://tree.bio.ed.ac.uk/software/figtree/) and rooted at the midpoint. For phylogenetic tree construction of flavonoid 3′-monooxygenase-like and bHLH genes, multiple alignments of full-length protein sequences were performed with the ClustalW algorithm implemented in MEGA package version 5.2 (Tamura et al., 2011) with default parameters. The phylogenetic tree was constructed using the means of maximum likelihood method with bootstrapping (500 replicates).
Cloning of Poplar MYB Factors and Plasmid Construction
Total RNA was isolated from young leaves of P. tremula × P. tremuloides INRA 353-38 plants using the method outlined below. Reverse transcription was carried out using SuperScript II reverse transcriptase (Invitrogen) according to the manufacturer’s instructions. The coding regions of MYB117, MYB179, MYB182, and bHLH079 were amplified with Phusion High Fidelity DNA Polymerase (New England Biolabs) using the gene-specific primers designed based on the coding sequences for P. trichocarpa in the Phytozome database (http://www.phytozome.net/; Supplemental Table S2). Each PCR product was digested with SpeI and KpnI, then ligated behind the double 35S promoter of the pMDC32 overexpression vector (Curtis and Grossniklaus, 2003). For hairy root constructs, the region containing a double 35S promoter, the MYB182 coding sequence, and the nopaline synthase terminator was amplified with Phusion High Fidelity DNA Polymerase from pMDC32-MYB182 with the primers containing the attB site for Gateway cloning (Supplemental Table S2). This PCR product was transferred into the Gateway binary vector pKGWD,0 (Karimi et al., 2002) through LR recombination (LR Clonase II; Invitrogen) to yield the MYB182 overexpressor construct with a GFP marker. Mutations for pMDC32-MYB182 mutant plasmids were performed by site-directed mutagenesis via overlap extension using PCR with mutated primers (Supplemental Table S2). Activator and cofactor plasmids (pMDC32-bHLH131, pMDC32-MYB134, and pMDC32-MYB115) and reporter plasmid pGREEN800LUC-ANR1 promoter constructs were described previously (Zifkin et al., 2012; Gesell et al., 2014; Hellens et al., 2005). The Arabidopsis (Arabidopsis thaliana) DFR promoter (1-kb upstream region) was amplified from Arabidopsis (ecotype Columbia) leaf genomic DNA extracted using the DNeasy Plant Mini Kit (Qiagen). The PCR products were digested with SpeI and KpnI and then ligated upstream of the firefly luciferase gene in the same reporter vector.
Generation of Transgenic Poplar Hairy Roots and Whole Plants
For hairy root generation, leaves of in vitro-grown P. tremula × P. alba clone INRA 717-1-B4 were excised and placed on preculture solid medium (0.25 g of MES, 0.1 g of myo-inositol, 30 g of Suc, and 4.33 g of MS [Caisson] salts in 1 L, pH 5.7) for 16 h. These leaves were wounded by cutting the leaf veins gently before transformation with Agrobacterium rhizogenes. pKGWD,0-MYB182 and pKGWD,0 (empty vector) were moved into A. rhizogenes strain ARqua1 by electroporation. Transformed colonies were grown on MG/L (for 5 g L−1 tryptone, 2.5 g L−1 yeast extract, 5.2 g L−1 NaCl, 2.0 g L−1 glutamic acid, 10 g L−1 mannitol, 0.2 g L−1 MgSO4·7H2O, and 0.5 g L−1 K2HPO4)-agar medium at 28°C with spectinomycin and biotin for vector selection. Liquid cultures were grown in MG/L medium for 16 h before spinning down. The bacterial pellet was resuspended in induction broth (0.25 g of MES, 0.1 g of myo-inositol, 30 g of Suc, and 4.33 g of MS [Caisson] salts in 1 L) with 200 µm acetosyringone to an optical density of 0.6 to 0.8 and used to inoculate excised leaves. Hairy roots arising from transformed leaves were excised and maintained in the dark at 25°C in petri dishes on solid antibiotic-containing medium (preculture medium with carbenicillin, cefotaxime, and timentin). After hairy roots appeared on the leaves, infected leaves were moved to solid WPM supplemented with 0.25 mg L−1 indole-3-butyric acid and 0.25 mg L−1 1-naphthaleneacetic acid. Individual hairy root clones were subcultured every 21 d. Screening for positive transformants was performed by GFP detection under UV light using an Olympus SZX7 Zoom Stereomicroscope. Hairy roots were harvested and flash frozen in liquid nitrogen for nucleic acid extraction and PA analysis.
For generating transgenic plants, the binary vector pMDC32-MYB182 was transferred to the Agrobacterium tumefaciens strain GV3101::pMP90. P. tremula × P. tremuloides clone 353-38 leaf explants were transformed as described previously (Leplé et al., 1992; Mellway et al., 2009). Positive independently transformed lines were identified by selection of plantlets on hygromycin-containing shooting and rooting medium and subsequently confirmed by PCR. Transgenics were maintained and propagated on solid WPM as described above prior to acclimation to the greenhouse for further analysis.
RNA Extraction and Real-Time Quantitative PCR
Total RNA was extracted from frozen poplar hairy roots or ground leaf powder using the modified cetyl trimethyl ammonium bromide method for polyphenol-rich plant tissue described by Muoki et al. (2012). The RNA preparations were checked with agarose electrophoresis and analysis on a Nanodrop 2000 (Thermo Scientific). Prior to quantitative PCR analysis, total RNA (2.5 µg) was treated with amplification-grade DNase I (Invitrogen) according to the manufacturer’s instructions. DNase I-treated RNA (250 ng) was used for reverse transcription by SuperScript II reverse transcriptase (Invitrogen). For real-time quantitative PCR, two technical replicates were analyzed using the Stratagene Mx4000 thermal cycler. Each quantitative PCR (15-µL total volume) consisted of the QuantiTect SYBRGreen mix (Qiagen) with 0.67 µm gene-specific primers and 2 µL of complementary DNA template diluted 1:20 from original reverse transcription. The amplification protocol was 95°C for 3 min followed by 40 cycles of 94°C for 30 s, 58°C to 64°C for 30 s, and 72°C for 30 s. Annealing temperatures for each primer were determined based on tests to determine their efficiency (Supplemental Table S2). Gene expression levels were quantified by normalization to the mean of elongation factor1β (α-subunit) and ubiquitin/ribosomal protein27a expression (GenBank accession nos. XM_002299613 and XM_002320914.2) and calculated by the 2−ΔΔCT method (Livak and Schmittgen, 2001).
Microarray Analysis
DNase I-treated RNA from wild-type and MYB182 overexpressor transgenic plants (leaves 10–12) was further purified with the NucleoSpin RNA II clean-up kit (Clontech). These RNA preparations were used for Affymetrix GeneChip Poplar Genome Array microarray hybridizations conducted at the Genome Quebec Innovation Centre at McGill University. Data were normalized with FlexArray using a GC robust multiarray average normalization and an empirical Bayes (Rocke) significance test. Genes were considered differentially expressed in the transgenics if they showed a fold change of greater than 2 or less than 0.5 with P < 0.05 and Q < 0.1. The Q value was calculated using the R package QVALUE (Storey, 2002). Annotations of differentially expressed genes were performed with Blast2GO (Conesa et al., 2005; http://blast2go.com/b2ghome) and Phytozome (http://www.phytozome.net/) and were manually checked via BLAST database searches (National Center for Biotechnology Information) and literature searches. The categorization of gene ontology was performed with Blast2GO.
Transient Expression and Dual Luciferase Promoter Activation Assays
For each bombardment assay, aliquots of poplar suspension cells (P. trichocarpa × P. deltoides H11-11) were collected from four independently grown cultures and placed on four individual filter papers (7-cm diameter) divided into quarters. The filter papers plus cells were placed onto solid MS medium with 0.5 m mannitol in petri dishes and incubated for 1 h prior to bombardment. Aliquots (25 μL) of 0.7-µm-diameter tungsten beads (Bio-Rad; 60 mg mL−1) were coated with 250 ng of each plasmid (activator, cofactor, and reporter vectors) under constant vortexing, followed by the addition of 25 μL of 2.5 m CaCl2 and 10 μL of 0.1 m spermidine. Samples were vortexed for an additional 10 min at 4°C. The beads were washed first with 200 μL of 70% (v/v) ethanol, then with 200 μL of 100% ethanol, and resuspended in 20 μL of 100% ethanol. The beads were pipetted onto flying discs (Bio-Rad) and allowed to air dry. The bombardments were conducted with a PDS-1000/He Biolistic Particle Delivery System (Bio-Rad) under 900 p.s.i. Filter papers with cells were placed approximately 13 cm from the rupture disc. After bombardment, the cells were incubated in darkness for 48 h at room temperature prior to the luciferase assay.
Promoter activation was assayed using the Dual-Luciferase Reporter Assay System (Promega). The cells were scraped from the filter paper, homogenized with a micropestle, and mixed by vortexing in 150 μL of passive lysis buffer (Promega Dual-Luciferase Reporter Assay System). The extract was incubated on ice for 10 min and then centrifuged for 10 min at 13,000 rpm. Aliquots (10 μL) were assayed for firefly and Renilla luciferase luminescence, as described by the manufacturer (Promega).
N. benthamiana Agroinfiltration
The overexpression vectors pMDC32-bHLH131, pMDC32-MYB117, and pMDC32-MYB182 were transformed into A. tumefaciens strain GV3101:pMP90. A. tumefaciens cultures harboring these plasmids were grown in liquid Luria-Bertani medium at 28°C for 16 h. Bacteria were pelleted and resuspended in 10 mm MgCl2 solution to an optical density at 600 nm of 0.7. Equal amounts of A. tumefaciens suspensions harboring each construct were infiltrated into the abaxial surface of the oldest two leaves of 4-week-old N. benthamiana plants as described by Espley et al. (2007). Eight days after infiltration, photographs of infiltrated leaves were taken, and leaf samples were harvested, freeze dried, and analyzed for anthocyanins as described above.
Phytochemical Extraction and Assays
For PA analysis of hairy roots, 50 mg of fresh tissue was ground in liquid nitrogen and extracted for 16 h at room temperature in 5 mL of 100% (v/v) methanol. Extracts were centrifuged to remove solid debris, and an aliquot (0.5 mL) was assayed for PAs using the acid butanol assay as described (Porter et al., 1985) using purified P. tremuloides PAs as a standard. For analysis of other samples, 25 mg of ground freeze-dried powder was extracted into 1.5 mL of 100% methanol and sonicated for 10 min. Extracts were centrifuged and extracted two more times with 1.5 mL of 100% methanol and sonication. Following a final centrifugation step, the three extracts were pooled and assayed as above. To measure anthocyanins, approximately 50 mg of ground, freeze-dried leaf powder was extracted into 500 μL of 1% (v/v) HCl in methanol overnight at room temperature with shaking. One volume of distilled, deionized water was added to 500 μL of extract and mixed. The sample was extracted with 1 mL of chloroform and centrifuged at 13,000 rpm for 5 min (Nemie-Feyissa et al., 2014). Anthocyanins were measured in the aqueous upper phase at 530 nm (modified from Martin et al., 2002).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers KP723392 to KP723395.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Distance tree representation of the phylogenetic analysis in Figure 1.
Supplemental Figure S2. In silico tissue-specific gene expression for poplar MYB134 and repressor-like MYB genes.
Supplemental Figure S3. Up-regulation of MYB182 transcripts by MYB134 overexpression and stress treatment.
Supplemental Figure S4. Populus tremula × P. tremuloides wild-type and transgenic overexpressing MYB182.
Supplemental Figure S5. Sample HPLC chromatogram of phenolic profiles comparing the wild type and MYB182 overexpressor plants.
Supplemental Figure S6. Correlation of gene expression between microarray and qPCR data.
Supplemental Figure S7. Phylogenetic analysis of bHLH factors related to poplar bHLH079 and poplar bHLH131.
Supplemental Figure S8. Phylogenetic analysis of flavonoid 3'-monooxygenase-like genes.
Supplemental Table S1. GenBank accession numbers for proteins in Figure 1.
Supplemental Table S2. List of primers used.
Supplementary Material
Acknowledgments
We thank Mireille Chabaud and David Barker for A. rhizogenes strain ARqua1, Roger P. Hellens for providing the pGREENII0800 plasmid, Dr. Andreas Gesell for vector constructs for the transient assay, Brad Binges for help with plant care, Tieling Zhang for technical assistance, Dr. Jürgen Ehlting for help with phylogenetic analysis, Amy Franklin and Cuong Le for help with microarray data analysis, Dr. John Taylor for access to the stereomicroscope, and the Centre for Forest Biology for growth facilities.
Glossary
- PA
proanthocyanidin
- bHLH
basic helix-loop-helix
- WDR
WD40 repeat
- RT-qPCR
reverse transcription-quantitative PCR
- WPM
Lloyd & McCown’s Woody Plant Medium
- MS
Murashige and Skoog
Footnotes
This work was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC) via Discovery Grants and Accelerator Supplements as well as the NSERC CREATE Program in Forests and Climate Change, by the Japan Society for the Promotion of Science Institutional Program for Young Researcher Overseas Visits (to K.Y.), and by the University of Victoria’s Centre for Forest Biology.
Articles can be viewed without a subscription.
References
- Abascal F, Zardoya R, Posada D (2005) ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21: 2104–2105 [DOI] [PubMed] [Google Scholar]
- Abrahams S, Lee E, Walker AR, Tanner GJ, Larkin PJ, Ashton AR (2003) The Arabidopsis TDS4 gene encodes leucoanthocyanidin dioxygenase (LDOX) and is essential for proanthocyanidin synthesis and vacuole development. Plant J 35: 624–636 [DOI] [PubMed] [Google Scholar]
- Aharoni A, De Vos CHR, Wein M, Sun Z, Greco R, Kroon A, Mol JNM, O’Connell AP (2001) The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J 28: 319–332 [DOI] [PubMed] [Google Scholar]
- Akagi T, Ikegami A, Tsujimoto T, Kobayashi S, Sato A, Kono A, Yonemori K (2009) DkMyb4 is a Myb transcription factor involved in proanthocyanidin biosynthesis in persimmon fruit. Plant Physiol 151: 2028–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akagi T, Ikegami A, Yonemori K (2010) DkMyb2 wound-induced transcription factor of persimmon (Diospyros kaki Thunb.), contributes to proanthocyanidin regulation. Planta 232: 1045–1059 [DOI] [PubMed] [Google Scholar]
- Al Ait L, Yamak Z, Morgenstern B (2013) DIALIGN at GOBICS: multiple sequence alignment using various sources of external information. Nucleic Acids Res 41: W3–W7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert NW, Davies KM, Lewis DH, Zhang H, Montefiori M, Brendolise C, Boase MR, Ngo H, Jameson PE, Schwinn KE (2014) A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant Cell 26: 962–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert NW, Lewis DH, Zhang H, Schwinn KE, Jameson PE, Davies KM (2011) Members of an R2R3-MYB transcription factor family in Petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning. Plant J 65: 771–784 [DOI] [PubMed] [Google Scholar]
- Barbehenn RV, Constabel CP (2011) Tannins in plant-herbivore interactions. Phytochemistry 72: 1551–1565 [DOI] [PubMed] [Google Scholar]
- Baudry A, Heim MA, Dubreucq B, Caboche M, Weisshaar B, Lepiniec L (2004) TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J 39: 366–380 [DOI] [PubMed] [Google Scholar]
- Boeckler GA, Towns M, Unsicker SB, Mellway RD, Yip L, Hilke I, Gershenzon J, Constabel CP (2014) Transgenic upregulation of the condensed tannin pathway in poplar leads to a dramatic shift in leaf palatability for two tree-feeding Lepidoptera. J Chem Ecol 40: 150–158 [DOI] [PubMed] [Google Scholar]
- Bogs J, Jaffé FW, Takos AM, Walker AR, Robinson SP (2007) The grapevine transcription factor VvMYBPA1 regulates proanthocyanidin synthesis during fruit development. Plant Physiol 143: 1347–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomal C, Duval I, Giguère I, Fortin É, Caron S, Stewart D, Boyle B, Séguin A, MacKay JJ (2014) Opposite action of R2R3-MYBs from different subgroups on key genes of the shikimate and monolignol pathways in spruce. J Exp Bot 65: 495–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broun P. (2005) Transcriptional control of flavonoid biosynthesis: a complex network of conserved regulators involved in multiple aspects of differentiation in Arabidopsis. Curr Opin Plant Biol 8: 272–279 [DOI] [PubMed] [Google Scholar]
- Cipollini ML, Stiles EW (1993) Fruit rot, antifungal defense, and palatability of fleshy fruits for frugivorous birds. Ecology 74: 751–762 [Google Scholar]
- Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676 [DOI] [PubMed] [Google Scholar]
- Constabel CP, Lindroth R (2010) The impact of genomics on advances in herbivore defense and secondary metabolism in Populus. InJansson S, Bhalerao R, Groover A, eds, Genetics and Genomics of Populus, Vol 8 Springer, New York, pp 279–305 [Google Scholar]
- Constabel CP, Walker V, Yoshida K (2015) Diverse ecological roles of plant tannins: plant defense and beyond. InRomani A, Lattanzio V, Quideau S, eds, Recent Advances in Polyphenol Research, Vol 4 Wiley-Blackwell, Oxford, pp 115–142 [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson JR, Stevens MT, Barnhill HR, Lindroth RL (2006) Age-related shifts in leaf chemistry of clonal aspen (Populus tremuloides). J Chem Ecol 32: 1415–1429 [DOI] [PubMed] [Google Scholar]
- Dubos C, Le Gourrierec J, Baudry A, Huep G, Lanet E, Debeaujon I, Routaboul JM, Alboresi A, Weisshaar B, Lepiniec L (2008) MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J 55: 940–953 [DOI] [PubMed] [Google Scholar]
- Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L (2010) MYB transcription factors in Arabidopsis. Trends Plant Sci 15: 573–581 [DOI] [PubMed] [Google Scholar]
- Espley RV, Hellens RP, Putterill J, Stevenson DE, Kutty-Amma S, Allan AC (2007) Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J 49: 414–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feller A, Machemer K, Braun EL, Grotewold E (2011) Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J 66: 94–116 [DOI] [PubMed] [Google Scholar]
- Fornalé S, Lopez E, Salazar-Henao JE, Fernández-Nohales P, Rigau J, Caparros-Ruiz D (2014) AtMYB7, a new player in the regulation of UV-sunscreens in Arabidopsis thaliana. Plant Cell Physiol 55: 507–516 [DOI] [PubMed] [Google Scholar]
- Fornalé S, Shi X, Chai C, Encina A, Irar S, Capellades M, Fuguet E, Torres JL, Rovira P, Puigdomènech P, et al. (2010) ZmMYB31 directly represses maize lignin genes and redirects the phenylpropanoid metabolic flux. Plant J 64: 633–644 [DOI] [PubMed] [Google Scholar]
- Gesell A, Yoshida K, Tran LT, Constabel CP (2014) Characterization of an apple TT2-type R2R3 MYB transcription factor functionally similar to the poplar proanthocyanidin regulator PtMYB134. Planta 240: 497–511 [DOI] [PubMed] [Google Scholar]
- Giri A, Narasu ML (2000) Transgenic hairy roots: recent trends and applications. Biotechnol Adv 18: 1–22 [DOI] [PubMed] [Google Scholar]
- Gonzalez A, Zhao M, Leavitt JM, Lloyd AM (2008) Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J 53: 814–827 [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59: 307–321 [DOI] [PubMed] [Google Scholar]
- Hamberger B, Ellis M, Friedmann M, Souza CDA, Barbazuk B, Douglas CJ (2007) Genome-wide analyses of phenylpropanoid-related genes in Populus trichocarpa, Arabidopsis thaliana, and Oryza sativa: the Populus lignin toolbox and conservation and diversification of angiosperm gene families. Can J Bot 85: 1182–1201 [Google Scholar]
- Hancock KR, Collette V, Fraser K, Greig M, Xue H, Richardson K, Jones C, Rasmussen S (2012) Expression of the R2R3-MYB transcription factor TaMYB14 from Trifolium arvense activates proanthocyanidin biosynthesis in the legumes Trifolium repens and Medicago sativa. Plant Physiol 159: 1204–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA (2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heppel SC, Jaffé FW, Takos AM, Schellmann S, Rausch T, Walker AR, Bogs J (2013) Identification of key amino acids for the evolution of promoter target specificity of anthocyanin and proanthocyanidin regulating MYB factors. Plant Mol Biol 82: 457–471 [DOI] [PubMed] [Google Scholar]
- Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M (2003) Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J 34: 733–739 [DOI] [PubMed] [Google Scholar]
- Huang YF, Vialet S, Guiraud JL, Torregrosa L, Bertrand Y, Cheynier V, This P, Terrier N (2014) A negative MYB regulator of proanthocyanidin accumulation, identified through expression quantitative locus mapping in the grape berry. New Phytol 201: 795–809 [DOI] [PubMed] [Google Scholar]
- Jin H, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, Weisshaar B, Martin C (2000) Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J 19: 6150–6161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagale S, Rozwadowski K (2011) EAR motif-mediated transcriptional repression in plants: an underlying mechanism for epigenetic regulation of gene expression. Epigenetics 6: 141–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JG, Yun J, Kim DH, Chung KS, Fujioka S, Kim JI, Dae HW, Yoshida S, Takatsuto S, Song PS, et al. (2001) Light and brassinosteroid signals are integrated via a dark-induced small G protein in etiolated seedling growth. Cell 105: 625–636 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kirik V, Simon M, Huelskamp M, Schiefelbein J (2004) The ENHANCER OF TRY AND CPC1 gene acts redundantly with TRIPTYCHON and CAPRICE in trichome and root hair cell patterning in Arabidopsis. Dev Biol 268: 506–513 [DOI] [PubMed] [Google Scholar]
- Koes R, Verweij W, Quattrocchio F (2005) Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci 10: 236–242 [DOI] [PubMed] [Google Scholar]
- Lai Y, Li H, Yamagishi M (2013) A review of target gene specificity of flavonoid R2R3-MYB transcription factors and a discussion of factors contributing to the target gene selectivity. Front Biol 8: 577–598 [Google Scholar]
- Lee MM, Schiefelbein J (1999) WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell 99: 473–483 [DOI] [PubMed] [Google Scholar]
- Legay S, Sivadon P, Blervacq AS, Pavy N, Baghdady A, Tremblay L, Levasseur C, Ladouce N, Lapierre C, Séguin A, et al. (2010) EgMYB1, an R2R3 MYB transcription factor from eucalyptus negatively regulates secondary cell wall formation in Arabidopsis and poplar. New Phytol 188: 774–786 [DOI] [PubMed] [Google Scholar]
- Leple JC, Brasileiro AC, Michel MF, Delmotte F, Jouanin L (1992) Transgenic poplars: expression of chimeric genes using four different constructs. Plant Cell Rep 11: 137–141 [DOI] [PubMed] [Google Scholar]
- Lin-Wang K, Bolitho K, Grafton K, Kortstee A, Karunairetnam S, McGhie TK, Espley RV, Hellens RP, Allan AC (2010) An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol 10: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Marles MAS, Gruber MY, Scoles GJ, Muir AD (2003) Pigmentation in the developing seed coat and seedling leaves of Brassica carinata is controlled at the dihydroflavonol reductase locus. Phytochemistry 62: 663–672 [DOI] [PubMed] [Google Scholar]
- Martin T, Oswald O, Graham IA (2002) Arabidopsis seedling growth, storage lipid mobilization, and photosynthetic gene expression are regulated by carbon:nitrogen availability. Plant Physiol 128: 472–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui K, Umemura Y, Ohme-Takagi M (2008) AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J 55: 954–967 [DOI] [PubMed] [Google Scholar]
- Mellway RD (2009) The regulation of stress-induced proanthocyanidin metabolism in poplar. PhD thesis. University of Victoria, Victoria, Canada [Google Scholar]
- Mellway RD, Tran LT, Prouse MB, Campbell MM, Constabel CP (2009) The wound-, pathogen-, and ultraviolet B-responsive MYB134 gene encodes an R2R3 MYB transcription factor that regulates proanthocyanidin synthesis in poplar. Plant Physiol 150: 924–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min BR, Barry TN, Attwood GT, McNabb WC (2003) The effect of condensed tannins on the nutrition and health of ruminants fed fresh temperate forages: a review. Anim Feed Sci Technol 106: 3–19 [Google Scholar]
- Miranda M, Ralph SG, Mellway R, White R, Heath MC, Bohlmann J, Constabel CP (2007) The transcriptional response of hybrid poplar (Populus trichocarpa × P. deltoides) to infection by Melampsora medusae leaf rust involves induction of flavonoid pathway genes leading to the accumulation of proanthocyanidins. Mol Plant Microbe Interact 20: 816–831 [DOI] [PubMed] [Google Scholar]
- Mol J, Grotewold E, Koes R (1998) How genes paint flowers and seeds. Trends Plant Sci 3: 212–217 [Google Scholar]
- Muoki RC, Paul A, Kumari A, Singh K, Kumar S (2012) An improved protocol for the isolation of RNA from roots of tea (Camellia sinensis (L.) O. Kuntze). Mol Biotechnol 52: 82–88 [DOI] [PubMed] [Google Scholar]
- Nakatsuka T, Yamada E, Saito M, Fujita K, Nishihara M (2013) Heterologous expression of gentian MYB1R transcription factors suppresses anthocyanin pigmentation in tobacco flowers. Plant Cell Rep 32: 1925–1937 [DOI] [PubMed] [Google Scholar]
- Nemie-Feyissa D, Olafsdottir SM, Heidari B, Lillo C (2014) Nitrogen depletion and small R3-MYB transcription factors affecting anthocyanin accumulation in Arabidopsis leaves. Phytochemistry 98: 34–40 [DOI] [PubMed] [Google Scholar]
- Nesi N, Debeaujon I, Jond C, Pelletier G, Caboche M, Lepiniec L (2000) The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell 12: 1863–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesi N, Jond C, Debeaujon I, Caboche M, Lepiniec L (2001) The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell 13: 2099–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M (2001) Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13: 1959–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osier TL, Lindroth RL (2004) Long-term effects of defoliation on quaking aspen in relation to genotype and nutrient availability: plant growth, phytochemistry and insect performance. Oecologia 139: 55–65 [DOI] [PubMed] [Google Scholar]
- Pang YZ, Cheng XF, Huhman DV, Ma JY, Peel GJ, Yonekura-Sakakibara K, Saito K, Shen GA, Sumner LW, Tang YH, et al. (2013) Medicago glucosyltransferase UGT72L1: potential roles in proanthocyanidin biosynthesis. Planta 238: 139–154 [DOI] [PubMed] [Google Scholar]
- Paolocci F, Robbins MP, Passeri V, Hauck B, Morris P, Rubini A, Arcioni S, Damiani F (2011) The strawberry transcription factor FaMYB1 inhibits the biosynthesis of proanthocyanidins in Lotus corniculatus leaves. J Exp Bot 62: 1189–1200 [DOI] [PubMed] [Google Scholar]
- Pauwels L, Barbero GF, Geerinck J, Tilleman S, Grunewald W, Pérez AC, Chico JM, Bossche RV, Sewell J, Gil E, et al. (2010) NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464: 788–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter LJ, Hrstich LN, Chan BG (1985) The conversion of procyanidins and prodelphinidins to cyanidin and delphinidin. Phytochemistry 25: 223–230 [Google Scholar]
- Preston J, Wheeler J, Heazlewood J, Li SF, Parish RW (2004) AtMYB32 is required for normal pollen development in Arabidopsis thaliana. Plant J 40: 979–995 [DOI] [PubMed] [Google Scholar]
- Prior RL, Gu L (2005) Occurrence and biological significance of proanthocyanidins in the American diet. Phytochemistry 66: 2264–2280 [DOI] [PubMed] [Google Scholar]
- Ralph S, Oddy C, Cooper D, Yueh H, Jancsik S, Kolosova N, Philippe RN, Aeschliman D, White R, Huber D, et al. (2006) Genomics of hybrid poplar (Populus trichocarpa × deltoides) interacting with forest tent caterpillars (Malacosoma disstria): normalized and full-length cDNA libraries, expressed sequence tags, and a cDNA microarray for the study of insect-induced defences in poplar. Mol Ecol 15: 1275–1297 [DOI] [PubMed] [Google Scholar]
- Ralston L, Kwon ST, Schoenbeck M, Ralston J, Schenk DJ, Coates RM, Chappell J (2001) Cloning, heterologous expression, and functional characterization of 5-epi-aristolochene-1,3-dihydroxylase from tobacco (Nicotiana tabacum). Arch Biochem Biophys 393: 222–235 [DOI] [PubMed] [Google Scholar]
- Ramsay NA, Glover BJ (2005) MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci 10: 63–70 [DOI] [PubMed] [Google Scholar]
- Rasmussen SE, Frederiksen H, Struntze Krogholm K, Poulsen L (2005) Dietary proanthocyanidins: occurrence, dietary intake, bioavailability, and protection against cardiovascular disease. Mol Nutr Food Res 49: 159–174 [DOI] [PubMed] [Google Scholar]
- Salvatierra A, Pimentel P, Moya-León MA, Herrera R (2013) Increased accumulation of anthocyanins in Fragaria chiloensis fruits by transient suppression of FcMYB1 gene. Phytochemistry 90: 25–36 [DOI] [PubMed] [Google Scholar]
- Scalbert A, Manach C, Morand C, Rémésy C, Jiménez L (2005) Dietary polyphenols and the prevention of diseases. Crit Rev Food Sci Nutr 45: 287–306 [DOI] [PubMed] [Google Scholar]
- Schaart JG, Dubos C, Romero De La Fuente I, van Houwelingen AM, de Vos RC, Jonker HH, Xu W, Routaboul JM, Lepiniec L, Bovy AG (2013) Identification and characterization of MYB-bHLH-WD40 regulatory complexes controlling proanthocyanidin biosynthesis in strawberry (Fragaria × ananassa) fruits. New Phytol 197: 454–467 [DOI] [PubMed] [Google Scholar]
- Schweitzer J, Madritch M, Bailey J, LeRoy C, Fischer D, Rehill B, Lindroth R, Hagerman A, Wooley S, Hart S, et al. (2008) From genes to ecosystems: the genetic basis of condensed tannins and their role in nutrient regulation in a Populus model system. Ecosystems (N Y) 11: 1005–1020 [Google Scholar]
- Shen H, He X, Poovaiah CR, Wuddineh WA, Ma J, Mann DGJ, Wang H, Jackson L, Tang Y, Stewart CN Jr, et al. (2012) Functional characterization of the switchgrass (Panicum virgatum) R2R3-MYB transcription factor PvMYB4 for improvement of lignocellulosic feedstocks. New Phytol 193: 121–136 [DOI] [PubMed] [Google Scholar]
- Storey JD. (2002) A direct approach to false discovery rates. J R Stat Soc B 64: 479–498 [Google Scholar]
- Stracke R, Werber M, Weisshaar B (2001) The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 4: 447–456 [DOI] [PubMed] [Google Scholar]
- Szemenyei H, Hannon M, Long JA (2008) TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319: 1384–1386 [DOI] [PubMed] [Google Scholar]
- Tamagnone L, Merida A, Parr A, Mackay S, Culianez-Macia FA, Roberts K, Martin C (1998) The AmMYB308 and AmMYB330 transcription factors from Antirrhinum regulate phenylpropanoid and lignin biosynthesis in transgenic tobacco. Plant Cell 10: 135–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng S, Keurentjes J, Bentsink L, Koornneef M, Smeekens S (2005) Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiol 139: 1840–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrier N, Torregrosa L, Ageorges A, Vialet S, Verriès C, Cheynier V, Romieu C (2009) Ectopic expression of VvMybPA2 promotes proanthocyanidin biosynthesis in grapevine and suggests additional targets in the pathway. Plant Physiol 149: 1028–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle TJ (2004) Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 16: 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CJ, Harding SA, Tschaplinski TJ, Lindroth RL, Yuan Y (2006) Genome-wide analysis of the structural genes regulating defense phenylpropanoid metabolism in Populus. New Phytol 172: 47–62 [DOI] [PubMed] [Google Scholar]
- Veljanovski V, Constabel CP (2013) Molecular cloning and biochemical characterization of two UDP-glycosyltransferases from poplar. Phytochemistry 91: 148–157 [DOI] [PubMed] [Google Scholar]
- Wester K, Digiuni S, Geier F, Timmer J, Fleck C, Hülskamp M (2009) Functional diversity of R3 single-repeat genes in trichome development. Development 136: 1487–1496 [DOI] [PubMed] [Google Scholar]
- Wilkins O, Nahal H, Foong J, Provart NJ, Campbell MM (2009) Expansion and diversification of the Populus R2R3-MYB family of transcription factors. Plant Physiol 149: 981–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie DY, Sharma SB, Paiva NL, Ferreira D, Dixon RA (2003) Role of anthocyanidin reductase, encoded by BANYULS in plant flavonoid biosynthesis. Science 299: 396–399 [DOI] [PubMed] [Google Scholar]
- Xu W, Grain D, Bobet S, Le Gourrierec J, Thévenin J, Kelemen Z, Lepiniec L, Dubos C (2014) Complexity and robustness of the flavonoid transcriptional regulatory network revealed by comprehensive analyses of MYB-bHLH-WDR complexes and their targets in Arabidopsis seed. New Phytol 202: 132–144 [DOI] [PubMed] [Google Scholar]
- Yoshida K, Kume N, Nakaya Y, Yamagami A, Nakano T, Sakuta M (2010) Comparative analysis of the triplicate proathocyanidin regulators in Lotus japonicus. Plant Cell Physiol 51: 912–922 [DOI] [PubMed] [Google Scholar]
- Zhang F, Gonzalez A, Zhao M, Payne CT, Lloyd A (2003) A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130: 4859–4869 [DOI] [PubMed] [Google Scholar]
- Zhu HF, Fitzsimmons K, Khandelwal A, Kranz RG (2009) CPC, a single-repeat R3 MYB, is a negative regulator of anthocyanin biosynthesis in Arabidopsis. Mol Plant 2: 790–802 [DOI] [PubMed] [Google Scholar]
- Zhu L, Shan H, Chen S, Jiang J, Gu C, Zhou G, Chen Y, Song A, Chen F (2013) The heterologous expression of the Chrysanthemum R2R3-MYB transcription factor CmMYB1 alters lignin composition and represses flavonoid synthesis in Arabidopsis thaliana. PLoS ONE 8: e65680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zifkin M, Jin A, Ozga JA, Zaharia LI, Schernthaner JP, Gesell A, Abrams SR, Kennedy JA, Constabel CP (2012) Gene expression and metabolite profiling of developing highbush blueberry fruit indicates transcriptional regulation of flavonoid metabolism and activation of abscisic acid metabolism. Plant Physiol 158: 200–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann IM, Heim MA, Weisshaar B, Uhrig JF (2004) Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like bHLH proteins. Plant J 40: 22–34 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.