A membrane-bound prenyltransferase complex is responsible for all prenylations in the bitter acid pathway of hop glandular trichomes.
Abstract
Bitter acids (α and β types) account for more than 30% of the fresh weight of hop (Humulus lupulus) glandular trichomes and are well known for their contribution to the bitter taste of beer. These multiprenylated chemicals also show diverse biological activities, some of which have potential benefits to human health. The bitter acid biosynthetic pathway has been investigated extensively, and the genes for the early steps of bitter acid synthesis have been cloned and functionally characterized. However, little is known about the enzyme(s) that catalyze three sequential prenylation steps in the β-bitter acid pathway. Here, we employed a yeast (Saccharomyces cerevisiae) system for the functional identification of aromatic prenyltransferase (PT) genes. Two PT genes (HlPT1L and HlPT2) obtained from a hop trichome-specific complementary DNA library were functionally characterized using this yeast system. Coexpression of codon-optimized PT1L and PT2 in yeast, together with upstream genes, led to the production of bitter acids, but no bitter acids were detected when either of the PT genes was expressed by itself. Stepwise mutation of the aspartate-rich motifs in PT1L and PT2 further revealed the prenylation sequence of these two enzymes in β-bitter acid biosynthesis: PT1L catalyzed only the first prenylation step, and PT2 catalyzed the two subsequent prenylation steps. A metabolon formed through interactions between PT1L and PT2 was demonstrated using a yeast two-hybrid system, reciprocal coimmunoprecipitation, and in vitro biochemical assays. These results provide direct evidence of the involvement of a functional metabolon of membrane-bound prenyltransferases in bitter acid biosynthesis in hop.
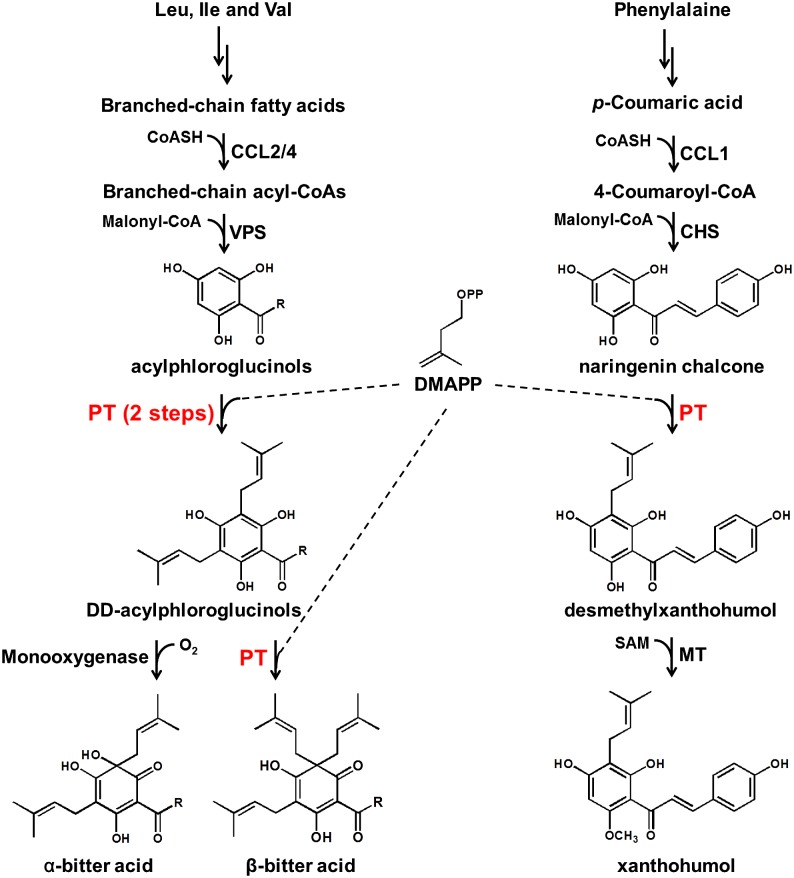
Hop (Humulus lupulus; Cannabaceae) is an important commodity in the beer-brewing industry, mostly due to its high content of bitter acids (a mixture of humulone and lupulone derivatives) and prenylated flavonoids in the glandular trichomes of female flowers (Stevens et al., 1998; Stevens and Page, 2004; Wang et al., 2008; Van Cleemput et al., 2009). Recent studies have demonstrated that hop terpenophenolics (a term for both bitter acids and prenylated flavonoids) exhibit diverse biological activities and have a high potential for use in drug development (Goto et al., 2005; Zanoli and Zavatti, 2008; Saugspier et al., 2012). For these reasons, a combination of large-scale trichome-targeted EST sequencing (thus far, there are more than 22,000 ESTs deposited in the TrichOME database [http://www.planttrichome.org], and a large amount of RNA sequencing data from different hop tissues [and cultivars] has been deposited in the National Center for Biotechnology Information [http://www.ncbi.nlm.nih.gov/sra?term=humulus%20lupulus]), reverse genetics, and in vitro biochemical assays have been employed in the study of terpenophenolic biosynthesis; several key enzymes have been identified and functionally characterized in recent years (Okada et al., 2004; Nagel et al., 2008; Wang et al., 2008; Wang and Dixon, 2009; Dai et al., 2010; Tsurumaru et al., 2012; Xu et al., 2013). Thus far, we know that the precursors for terpenophenolic biosynthesis are derived from amino acid catabolism (Leu, Ile, and Val for bitter acids and Phe for xanthohumol [XN]) and the dimethylallyl diphosphate (DMAPP) generated from the plastid-localized 2-C-methyl-d-erythritol 4-phosphate (Fig. 1). The prenylation steps are critical for final product formation in hop trichomes and include one prenylation for XN, two prenylations for α-acids, and three prenylations for β-acids. It is known that all of these prenylations use DMAPP as the prenyl donor (Fig. 1). The efficiency of prenyltransferase enzymes in hop trichomes is assumed to be very high, as intermediate compounds (e.g. acylphloroglucinol derivatives and naringenin chalcone [NC]) are barely detectable as compared with the very high concentrations of the prenylated products (Nagel et al., 2008; Clark et al., 2013; Xu et al., 2013). In addition to the aforementioned major prenylated chemicals, geranylated chemicals also have been detected in hop tissues, although these occur at lower concentrations (Stevens et al., 2000).
Figure 1.
The proposed biosynthetic pathway for bitter acids and XN in hop glandular trichomes. The major prenylation steps in the pathway are highlighted in red, and the possible R group in acylphloroglucinols, DD-acylphloroglucinols, and bitter acids are isobutyryl, isopropyl, and butan-2-yl groups. CCL, Carboxyl CoA ligase; CHS, chalcone synthase; CoASH, CoA; MT, methyltransferase.
In plants, the prenylation reactions are catalyzed by aromatic prenyltransferases, which contribute an additional facet to the metabolic diversity in plant-specialized metabolism. Unlike the soluble aromatic prenyltransferases from fungi and bacteria, all aromatic prenyltransferases isolated from plants thus far are membrane-bound proteins (Yazaki et al., 2009). In addition to being membrane proteins, these plant aromatic prenyltransferases also share other common characteristics, including plastid localization, divalent ion dependency, and relatively narrow substrate specificities. The first flavonoid prenyltransferase gene, which catalyzes the prenylation of naringenin at position 8, was recently identified from Sophora flavescens root (designated as SfN8DT-1; Sasaki et al., 2008). Several plant aromatic prenyltransferase genes were cloned from various plant species using homologous gene-cloning strategies (Akashi et al., 2009; Shen et al., 2012; Karamat et al., 2014; Munakata et al., 2014). Likewise, one membrane-bound prenyltransferase gene (HlPT-1) was recently cloned from hop (Tsurumaru et al., 2012). Biochemical results showed that HlPT-1 recognized three acylphloroglucinols (phlorisovalerophenone [PIVP], phlorisobutyrophenone [PIBP], and phlormethylbutanophenone [PMBP]) and NC as substrates. However, HlPT-1 transferred only one DMAPP to these prenyl acceptors, and the enzymatic conversion rate was relatively low (688 ± 29.7 pmol mg−1 microsomal protein h−1; Tsurumaru et al., 2012). This result suggests that some unknown factors are required to complete bitter acid biosynthesis in hops, particularly the second and third prenylation steps in the pathway. There has been extensive interest in the large-scale production of these value-added prenylated chemicals, due to their diverse biological activities and use in the brewing industry. However, simply overexpressing known aromatic prenyltransferase genes, either in plant or in microbial hosts, has not yet generated satisfactory results (Sasaki et al., 2009; Koeduka et al., 2011; Sugiyama et al., 2011). Additionally, no PT that catalyzes more than one prenylation reaction has been reported, although multiprenylated polyketides occur widely in plants (Singh and Bharate, 2006).
Previously, we functionally characterized two branched short-chain fatty acid CoA ligase genes (HlCCL2 and HlCCL4). We further successfully reconstructed the upstream steps of the bitter acid pathway by cointroducing HlCCL2, HlCCL4, and HlVPS (for valerophenone synthase) in a yeast (Saccharomyces cerevisiae) system to produce PIVP, PIBP, and PMBP. This yeast system now serves as a bioplatform that can be used to characterize the remaining enzymatic steps in the bitter acid biosynthesis pathway (Xu et al., 2013). Here, we used an optimized yeast system, combined with site-directed mutagenesis, split-ubiquitin membrane yeast two-hybrid (MbY2H), reciprocal coimmunoprecipitation, and in vitro biochemical assays to characterize two candidate aromatic prenyltransferase genes (HlPT1L and HlPT2) from hop trichomes. Our results showed that PT2 interacted physically with PT1L to form a functional heterocomplex that catalyzed the major prenylations in the bitter acid pathway with high efficiency. PT1L catalyzed the first prenylation step and PT2 catalyzed the subsequent two prenylation steps. Identification of a heteromeric membrane-bound prenyltransferase complex in this study provides tools for the metabolic engineering of high-value prenylated natural products using the strategies of synthetic biology.
RESULTS AND DISCUSSION
Yeast Strain Selection for Membrane-Bound Aromatic Prenyltransferases
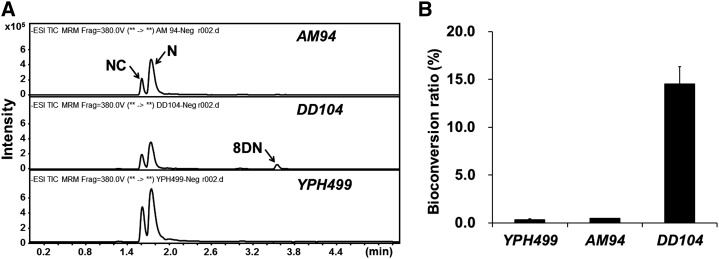
We selected a yeast system to characterize the genes involved in the bitter acid pathway due to the lack of a hop transformation system, including the transient gene-silencing technique (Xu et al., 2013). To test whether yeast was a suitable host for plant aromatic prenyltransferase, we coexpressed HlCCL1/HlCCL8 (the coumarate CoA ligase and malonyl CoA ligase that we cloned previously from hop [GenBank accession nos. JQ740203 and JQ740210]; Xu et al., 2013), HlCHS (for chalcone synthase), and SfN8DT-1 (the first characterized flavonoid prenyltransferase from plants; Sasaki et al., 2008) together in a wild-type yeast strain (YPH499) fed with 1 mm coumarate. The chemical analysis of the resulting culture medium by liquid chromatography-mass spectrometry (LC-MS) showed that only tiny amounts of 8-dimethylallyl naringenin (8DN) could be detected (the conversion ratio to 8DN from the sum of naringenin and NC was 0.33% ± 0.099%; n = 3). This suggested that the availability of DMAPP in YPH499 may be a limiting factor for the prenylation of naringenin by SfN8DT-1. In yeast cells, the DMAPP and isopentenyl diphosphate generated from the mevalonate pathway are used in the biosynthesis of isoprenoids such as sterols and ubiquinone. To obtain a yeast strain with a larger pool of DMAPP than that of YPH499, two engineered yeast strains were tested: AM94 (this strain produces more farnesyl diphosphate for sesquiterpene production; Ignea et al., 2011) and DD104 (down-regulation of endogenous farnesyl pyrophosphate synthase by site mutation of K197G, which was originally designed for monoterpene production; Fischer et al., 2011; Supplemental Fig. S1). When coexpressing the same set of genes that had been introduced into YPH499, the bioconversion of 8DN from naringenin and NC in DD104 increased 44-fold compared with YPH499 (14.52% ± 1.79% versus 0.33% ± 0.099%; n = 3). No significant difference in 8DN production was observed between AM94 and YPH499 (0.46% ± 0.003% versus 0.33% ± 0.099%; n = 3; Fig. 2). A previous study had shown that the bioconversion from naringenin to 8DN by SfN8DT-1 ranged from 0.5% to 2% in the W303-1A-Δcoq2 yeast strain (for complete details of the genetic background, see Supplemental Table S1 and Supplemental Fig. S1D; Sasaki et al., 2009). We thus selected the DD104 strain for use in our hop prenyltransferase study.
Figure 2.
In vivo production of 8DN in various yeast strains. A, Chemical profiling of different yeast strains by liquid chromatography-electrospray ionization-mass spectrometry. For 8DN, retention time = 3.54 min and the MRM transition is mass-to-charge ratio (m/z) 339→119; for naringenin (N), retention time = 1.73 min and the MRM mode is m/z 271→119; for NC, retention time = 1.60 min and the multireaction monitoring (MRM) mode is m/z 271→151. B, Bioconversion ratio of 8DN from the sum of naringenin and NC in different yeast strains. All experiments were performed in triplicate.
Identification of Aromatic Prenyltransferase Candidates from a Hop Trichome-Specific Database
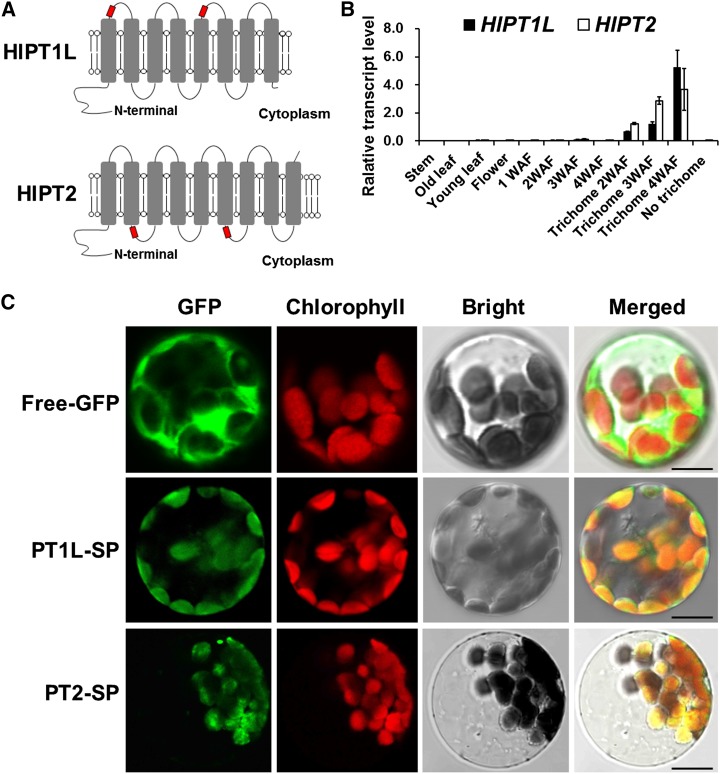
As mentioned previously, bitter acid and XN biosynthesis occurs predominantly in the glandular trichomes of female hop flowers (Nagel et al., 2008; Xu et al., 2013). Two putative aromatic prenyltransferase genes were identified with BLAST against the hop trichome-specific library in the TrichOME Web site using SfN8DT-1 as a query, and the full-length complementary DNA (cDNA) of these two putative aromatic prenyltransferase genes were obtained with RACE. One candidate gene (encoding a peptide of 414 amino acids, TCHL40971, in the TrichOME database) was found to be 98.5% identical to the prenyltransferase gene (GenBank accession no. AB543053; designated as HlPT-1) cloned by Yazaki’s group from hop ‘Kirin II’ (Tsurumaru et al., 2010). The sequence difference between these two proteins was probably due to the different hop cultivar (‘Nugget’) we used for cDNA library construction; we thus designated this HlPT-1 allele as HlPT1-like (HlPT1L). Another candidate gene, designated as HlPT2, encodes a polypeptide of 408 amino acids (GD252109 and GD245380 in the TrichOME database). HlPT1L and HlPT2 share 47% identity to each other at the protein level and share 24.2% and 22.5% identity to SfN8DT-1, respectively (Supplemental Fig. S2). Both HlPT1L and HlPT2 contain two conserved Asp-rich motifs and plastid signal peptides at the N terminus predicted by TargetP (www.cbs.dtu.dk/services/TargetP/), which we confirmed with GFP experiments (Fig. 3C; Supplemental Fig. S2). Analysis of the peptide sequences using the TMHMM Server version 2.0 indicated that HlPT1L and HlPT2 have eight and nine predicted transmembrane domains, respectively (Fig. 3A). The trichome-specific expression pattern of HlPT1L and HlPT2, together with their localization in plastids, suggested that both genes are likely involved in bitter acid and XN biosynthesis (Fig. 3, B and C). Phylogenetic analysis showed that both HlPT1L and HlPT2 shared a common ancestor with the VTE2-2 (for vitamin E 2-2; HlPT1L and HlPT2 share 43.2% and 40.2% identity, respectively, with VTE2-2 at the protein level) enzymes, which are involved in plastoquinone biosynthesis in Arabidopsis (Arabidopsis thaliana; Tian et al., 2007; Supplemental Fig. S3).
Figure 3.
Characterization of HlPT1L and HlPT2 in hop. A, Membrane topology diagram of HlPT1L and HlPT2. The transmembrane domains and the Asp-rich motifs are highlighted in gray and red, respectively. B, Quantitative reverse transcription-PCR analysis of two PT genes in different tissues and different development stages of cones and trichomes. Transcript levels are expressed relative to the Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) gene (n = 3). WAF, Weeks after flowering. C, Subcellular localizations of HlPT1L (the first 86 amino acids at the N terminus) and HlPT2 (the first 83 amino acids at the N terminus) in Arabidopsis leaf mesophyll protoplasts as revealed by laser confocal microscopy. Chloroplasts are revealed by red chlorophyll autofluorescence. SP, Signal peptide. Bars = 10 μm.
Coexpression of Codon-Optimized PT1L and PT2 Led to Bitter Acid Production in Yeast
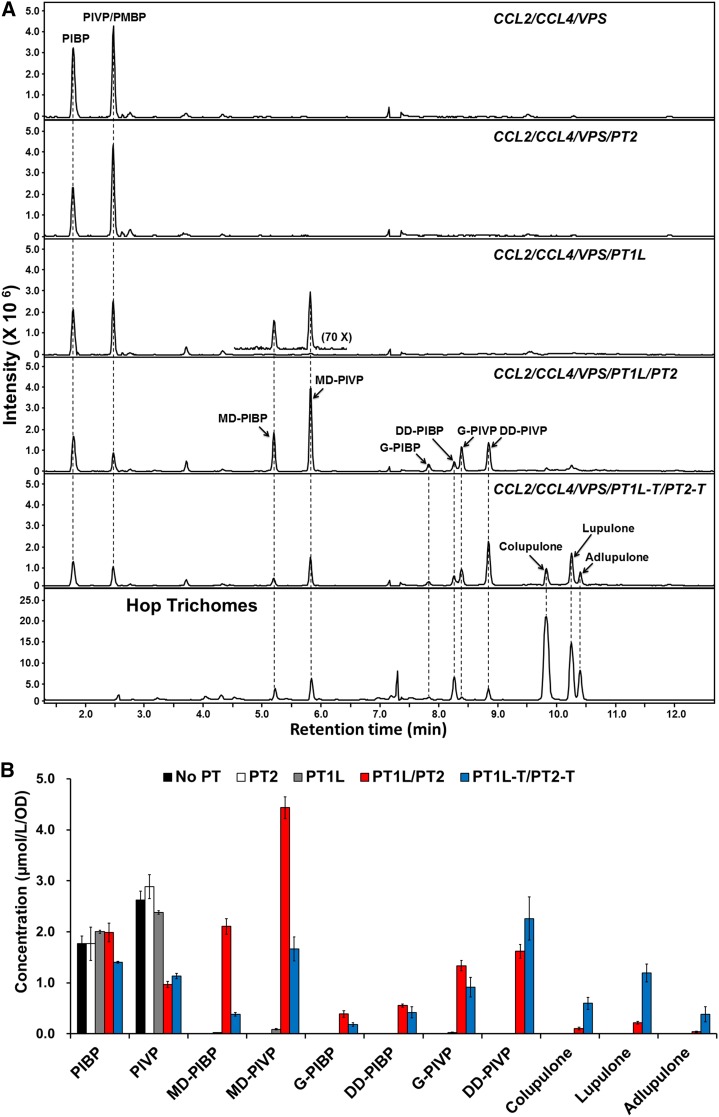
We initially tested whether the coexpression of HlPT1L and HlPT2 in the DD104 yeast strain harboring CCL2/CCL4/VPS genes led to the production of prenylated acylphloroglucinols. Under flask-shake culture conditions, the engineered yeast strain produced monoprenylated acylphloroglucinol (including monodimethylallylated [MD] and geranylated [G]) acylphloroglucinols (total, 1.2 μmol L−1 OD−1, where OD means the optical density of yeast cell culture measured at 600 nm) and a trace amount of didimethylallylated (DD) acylphloroglucinol (total, 0.2 μmol L−1 OD−1), but no bitter acids (Supplemental Fig. S4). We further tested whether codon optimization of hop PT genes would increase the production of prenylated acylphloroglucinol. The yeast and Arabidopsis (designed for functional identification of PT genes in planta) codon-optimized versions of these two hop prenyltransferase genes were synthesized (for detailed sequences, see Supplemental Figs. S5 and S6). Unexpectedly, the yeast strain harboring the yeast-optimized PT1L/PT2 (designated as PT1L-Sc and PT2-Sc, respectively; Supplemental Fig. S6) produced even less prenylated acylphloroglucinols than did the yeast strain harboring HlPT1L/HlPT2 of hop origin (Supplemental Fig. S4). However, the yeast strain harboring the Arabidopsis codon-optimized PT1L/PT2 (designated as PT1L-At and PT2-At, respectively; Supplemental Fig. S5) produced the highest amount of prenylated acylphloroglucinol (total, 10.8 μmol L−1 OD−1), including 6.54 μmol L−1 OD−1 of MD-acylphloroglucinol, 1.72 μmol L−1 OD−1 of G-acylphloroglucinol, 2.17 μmol L−1 OD−1 of DD-acylphloroglucinol, and 0.37 μmol L−1 OD−1 of total β-acids, after Gal induction for 48 h (Fig. 4; Supplemental Fig. S4). Based on these results, we used Arabidopsis codon-optimized PT1L and PT2 for further studies (designated as PT1L-At and PT2-At, respectively). It is noteworthy that our LC-MS method did not distinguish PIVP and PMBP (which are isomeric) and their corresponding derivatives, with the exception of lupulone (a PIVP derivative) and adlupulone (a PMBP derivative). We thus use the term PIVP to represent the mixture of PIVP and PMBP for convenience. We further tested the effect of PT1L-At alone or PT2-At alone on the production of prenylated acylphloroglucinols. The PT1L-At alone strain produced 4.38 μmol L−1 OD−1 of acylphloroglucinol, 0.11 μmol L−1 OD−1 of MD-acylphloroglucinol, and 0.026 μmol L−1 OD−1 of G-acylphloroglucinol. No prenylated compounds were produced by the strain harboring only PT2-At or the control strain lacking the PT genes (Fig. 4). The retention times and mass spectra of PIBP, PIVP, MD-PIBP, and PIVP were identical to those of authentic standards (here, we used the chemical extract of hop trichomes as standards for cohumulone, humulone, and adhumulone); the chemical structures of G-PIBP, G-PIVP, DD-PIVP (also known as 4-deoxyhumulone), and DD-PIBP (also known as 4-deoxycohumulone) were tentatively elucidated from their mass spectra (the mass spectra of standards and other compounds are shown in Supplemental Fig. S7).
Figure 4.
In vivo production of prenylated chemicals in yeast cells coexpressing HlCCL2, HlCCL4, HlVPS, and different hop aromatic prenyltransferase genes. A, Chromatogram of selected ions of m/z 197.0808 for PIBP, m/z 211.0965 for PIVP, m/z 265.1434 for MD-PIBP, m/z 279.1591 for MD-PIVP, m/z 333.206 for DD-PIBP and G-PIBP, m/z 347.2217 for DD-PIVP and G-PIVP, m/z 401.2715 for colupulone, and m/z 415.2863 for lupulone and adlupulone, all with a mass accuracy in the 20 parts per million range. Some regions of these chromatograms have been rescaled to allow easier visualization. B, Production of different chemicals by yeast strains harboring no PT gene (control), PT1L, PT2, PT1L/PT2, or PT1L-T/PT2-T (the combination of truncated HlPT1L and HlPT2). Data are means ± sd for at least three independent clones. All PT genes used in this experiment were Arabidopsis codon-optimized sequences.
It has been demonstrated that the N-terminal signal peptide somehow inhibits the enzymatic activity of isoflavonoid-specific prenyltransferase (LaPT1) from white lupin (Lupinus albus; Shen et al., 2012). We thus coexpressed truncated PT1L-At (with an 86-amino acid deletion from the start codon, based on the signal peptide prediction by TargetP; truncated PT1L-At was renamed as PT1L-T) and truncated PT2-At (with an 83-amino acid deletion from the start codon; truncated PT2-At was renamed as PT2-T) to test the effect of these signal peptides on the activity of hop prenyltransferase proteins. The resulting yeast produced much higher amounts of bitter acids than did the respective yeast strains harboring full-length PT1L-At and PT2-At (0.37 μmol L−1 OD−1 of total β-acids): 2.18 μmol L−1 OD of total β-acids after Gal induction for 48 h (Fig. 4). Unlike the acylphloroglucinols, most of which were secreted into culture medium, greater than 70% of the prenylated acylphloroglucinols remained inside the yeast cells (Supplemental Fig. S8). No α-acids could be detected in any of the tested yeast strains, indicating that there is an additional enzymatic reaction that is required for the final oxidation step that converts DD-acylphloroglucinols into α-acids.
Different Prenylation Steps Catalyzed by PT1L and PT2 in the Bitter Acid Pathway
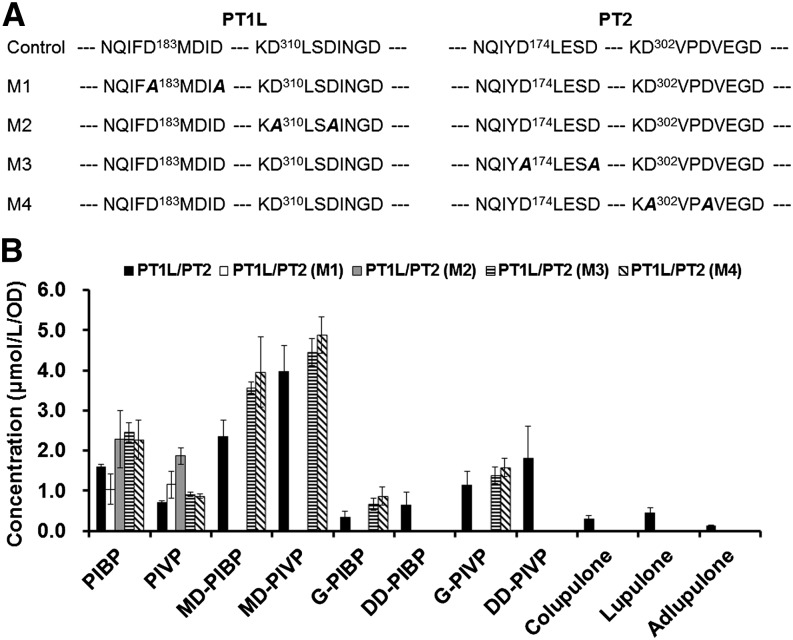
Although bitter acids were produced when PT1L-At and PT2-At were coexpressed together, the roles of PT1L and PT2 in the bitter acid pathway have not been clearly defined. To address this question, we mutated the Asp in Asp-rich motifs (DXXXD or DXXD; where X is any amino acid) in PT1L and PT2 to Ala (AXXXA or AXXA; Fig. 5A) and coexpressed one normal PT gene and a mutated PT gene in DD104 yeast harboring CCL2/CCL4/VPS. Previous studies had shown the critical role of the Asp-rich motif for aromatic prenyltransferase activity. Thus, mutation in the Asp-rich motif would very likely lead to inactive aromatic prenyltransferases (Ohara et al., 2009; Cheng and Li, 2014). The yeast strain harboring mutated PT1L-At and wild-type PT2-At did not produce any prenylated products, which revealed that PT1L is responsible for at least the first prenylation step in the bitter acid pathway (this result is consistent with a previous report; Tsurumaru et al., 2012). Wild-type PT2 did not recognize PIVP/PIBP as a substrate. Interestingly, the strain harboring wild-type PT1L-At and mutated PT2-At produced higher amounts of monoprenylated compounds than did the strain harboring PT1L-At alone, implying that the inactive PT2 protein, like its wild-type isoform, increases the prenylation efficiency of PT1L (Fig. 5). Meanwhile, no diprenylated or triprenylated compounds were produced by the strain harboring wild-type PT1L-At and mutated PT2-At, suggesting that PT2 is a functional protein that is responsible for the second and third prenylation steps in the β-acid pathway.
Figure 5.
Effects of the Asp-rich motif on bitter acid production in engineered yeast strains. A, Sequence comparison of wild-type (control) and mutant PT1L/PT2. Only the Asp-rich motifs are shown, and the mutated amino acids are in boldface and italics. B, Production of different compounds by yeast strains harboring wild-type and mutated PT1L/PT2 pairs. Data are means ± sd for at least three independent clones. All PT genes used in this experiment were Arabidopsis codon-optimized sequences.
HlPT1L Interacts Physically with HlPT2
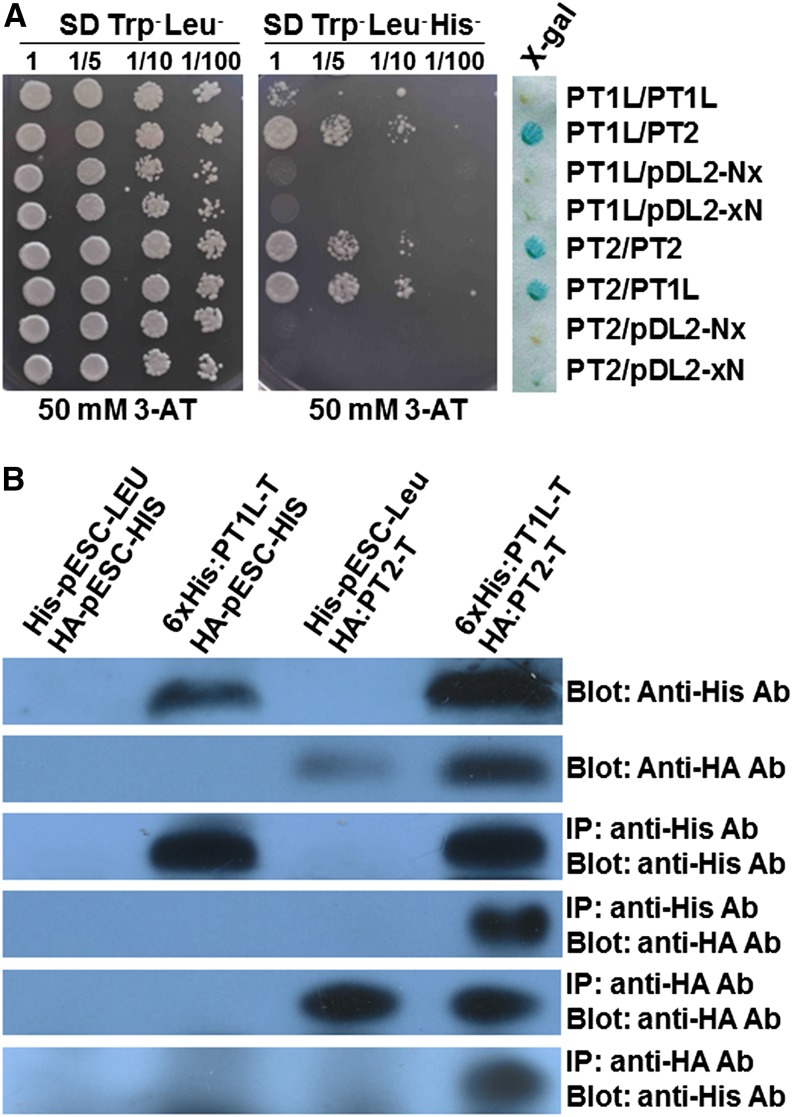
Higher levels of monoprenylated compounds were observed when PT1L-At and PTL2-At were coexpressed, no matter whether they were wild-type or mutated forms, than when expressing PT1L-At alone (Figs. 4 and 5). This finding drove us to hypothesize that PT2 may interact with PT1L to form a complex that enables efficient prenyltransferase activity. We thus used a split-ubiquitin MbY2H to test this hypothesis. The results showed that PT2 was able to form a homomer with itself or form a heteromer with PT1L; this result was further confirmed with positive β-galactosidase assays (Fig. 6A). The lack of growth in the negative controls (pDL2-Nx and pDL2-xN are empty vectors) indicated that the detected interactions between PT1L and PT2 were not false positives (Fig. 6A). To further confirm the interaction between PT1L and PT2, we performed coimmunoprecipitation experiments. N-terminal His-tagged PT1L-T and N-terminal hemagglutinin (HA)-tagged PT2-T were coexpressed in yeast. The results showed that PT2 recombinant protein could be coimmunoprecipitated when the lysates of transformed cells were incubated with an anti-His antibody. The anti-HA antibody also could be coimmunoprecipitated with His-tagged PT1L recombinant protein. However, no interactions between His-tagged PT1L-T and HA tag (as a negative control) or HA-tagged PT2-T and His tag (as a negative control) could be detected (Fig. 6B). Together, these results support the hypothesis that HlPT1L/HlPT2 forms and functions as a complex in yeast membranes.
Figure 6.
Direct interaction between PT1L and PT2 in yeast membranes. A, Split-ubiquitin assays for PT1L and PT2 protein interactions. The different transformed yeast cells were serially diluted and spotted onto synthetic dextrose growth medium (SD, Trp− Leu−) and synthetic dextrose selection medium (SD, Trp− Leu− His−), both supplemented with 50 mm 3-amino-1,2,4-triazole (3-AT). The positive interaction between PT1L and PT2 was confirmed by filter assay for the detection of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside acid (X-gal) activity. B, Reciprocal coimmunoprecipitation of His-tagged truncated PT1L and HA-tagged truncated PT2 in yeast cells using anti-HA and anti-His antibodies. Total protein extracts from transformed yeast strains harboring truncated PT1L-At and PT2-At alone or together were incubated with anti-His and anti-HA antibodies, and the immunoprecipitates (IP) were separated by SDS-PAGE and subjected to immunoblot analysis using anti-HA and anti-His antibodies. His-pESC-LEU and HA-pESC-HIS (producing the His-tag or the HA-tag peptide only in yeast cells) were used as negative controls. Ab, Antibody.
Biochemical Characterization of the Heteromeric Prenyltransferase Complex
To better understand the effect of the heteromeric prenyltransferase complex on the efficiency of prenylation, kinetic analyses of the activities of the prenyltransferase complex were carried out. Consistent with the data in yeast, microsomes containing full-length PT1L had very weak activity for PIBP and PIVP in vitro (too low to be suitable for Km determination; Supplemental Fig. S9). The truncated PT1L showed much higher activity to PIBP and PIVP compared with that of the full-length PT1L protein (Table I). Microsomes containing full-length PT1L and mutated full-length PT2 had 7- to 10-fold higher prenyltransferase activity toward PIVP, PIBP, DMAPP, and geranyl diphosphate (GPP) than did truncated PT1L alone, although these had similar Km values (Table I). These results revealed that physical interaction with PT2 increased the turnover rate of PT1L without changing the affinity of PT1L to substrates. However, the much higher efficiency of PT1L toward GPP than to DMAPP is not consistent with the fact that the dimethylallylated chemicals accumulate to much higher levels than the geranylated chemicals in hop trichomes (Fig. 4; Stevens and Page, 2004; Xu et al., 2013). Likely, most of the GPP was consumed by myrcene synthase for monoterpenoid biosynthesis (Wang et al., 2008) and not by the prenyltransferase complex for bitter acid production in hop trichomes. It is noteworthy that, in our test conditions, truncated PT1L (Arabidopsis codon optimized) also exhibited activity toward NC to form desmethylxanthohumol, albeit at a much lower level (3.4% ± 0.35%; n = 3) compared with that of PIVP (Supplemental Fig. S10). This activity also was detected in a previous report (Tsurumaru et al., 2012).
Table I. Kinetic parameters for PT1L, PT1L-T, and PT1L/PT2 (M3).
All PT genes used in this experiment were Arabidopsis codon-optimized sequences. The data are presented as means ± sd (n = 3). N.D., Not determined due to the low PT activity.
| Enzyme | Substrate | Km | Vmax |
|---|---|---|---|
| μm | μmol mg−1 microsomal protein min−1 | ||
| PT1L | PIVPa | N.D. | N.D. |
| PIBPa | N.D. | N.D. | |
| DMAPPb | N.D. | N.D. | |
| GPPb | N.D. | N.D. | |
| PT1L-T | PIVPa | 1.36 ± 0.43 | 1.08 ± 0.19 |
| PIBPa | 5.1 ± 0.81 | 0.41 ± 0.03 | |
| DMAPPb | 43.52 ± 3.7 | 0.47 ± 0.04 | |
| GPPb | 5.70 ± 1.07 | 0.69 ± 0.02 | |
| PT1L/PT2 (M3) | PIVPa | 3.39 ± 0.73 | 7.96 ± 1.77 |
| PIBPa | 6.68 ± 1.08 | 3.06 ± 0.38 | |
| DMAPPb | 43.64 ± 23.35 | 4.75 ± 1.94 | |
| GPPb | 6.18 ± 1.37 | 5.36 ± 0.82 |
As mentioned above, PT2 catalyzed the last two prenylation steps in the β-bitter acid pathway in yeast cells. The yeast microsomes containing the combination of truncated HlPT1L and truncated HlPT2 (PT1L-T/PT2-T) proteins catalyzed PIVP and DMAPP to form DD-PIVP in vitro, and no lupulone could be found (Supplemental Fig. S11H). However, prepared yeast microsomes containing the PT2 protein did not further prenylate MD-PIVP in vitro, regardless of whether PT2 was present singly or together with PT1L or if PT2 was full length or truncated (Supplemental Fig. S11, B–J). One plausible explanation for this is that a metabolon is formed by PT1L and PT2 and this complex is essential for PT2 enzymatic function. In this scenario, exogenously supplied intermediate (MD-acylphloroglucinol in this case) may not be able to access the enzyme active sites in the metabolon (Winkel, 2004).
CONCLUSION
Two major conclusions can be drawn from our study. First, we functionally characterized a complex of two aromatic prenyltransferases that catalyzed three sequential prenylation steps in the bitter acid pathway. Biochemical data, together with the results from the yeast two-hybrid and the reciprocal coimmunoprecipitation experiments, further supported the hypothesis that a metabolon was formed by HlPT1 and HlPT2 in hop glandular trichomes. The aromatic prenyltransferase metabolon demonstrated here, together with the high trichome-specific expression level of the genes involved in the bitter acid pathway (HlCCL2/4, HlVPS, and HlPT1L/2), are responsible for the high efficiency of bitter acid biosynthesis in hop trichomes (Okada et al., 2004; Xu et al., 2013). Second, we successfully reconstructed the entire β-bitter acid pathway in a yeast system. Under flask-shake conditions, the engineered yeast produced up to 2.18 μmol L−1 OD of total β-acids, and it should be possible to further increase bitter acid production by using a combination of strategies from synthetic biology and optimized fermentation conditions (Keasling, 2010; Zhou et al., 2014). The engineered strains in this study also provide a starting point for the functional identification of other components involved in the bitter acid pathway. Among these, the elucidation of the enzyme responsible for the last oxidation step in the α-bitter acid pathway has the high priority due to its critical role in flavoring beer (Caballero et al., 2012). Several P450s and monooxygenase genes have been identified from the TrichOME database, and together with the fact that DD-acylphloroglucinol (the proposed precursor for the reaction) is produced in our engineered yeast, these hold promise for the completion of the whole jigsaw puzzle of the complete bitter acid pathway.
MATERIALS AND METHODS
Plant Materials, RNA Analysis, and Chemicals
The growth of hop (Humulus lupulus ‘Nugget’), RNA isolation and cDNA preparation from hop tissues, and quantitative real-time PCR were performed as described previously (Wang et al., 2008). The codon-optimized sequences of PT1L and PT2 were generated using the JCat tool (http://www.jcat.de/; Grote et al., 2005) and synthesized by TaKaRa Bio.
All chemicals used in this study were purchased from Sigma-Aldrich except PIBP/PIVP and their monoprenylated derivatives, which were synthesized as described previously (George et al., 2010). The purity and concentration of these chemicals were determined using ultra-HPLC-mass spectrometry and NMR (Supplemental Table S2).
Hop Prenyltransferase Gene Isolation and in Vitro Mutagenesis
To obtain the full-length sequences of PT1L and PT2 from hop trichomes, 3′ RACE was performed with the SMART RACE cDNA Amplification Kit following the manufacturer’s instructions. The open reading frames of PT1L and PT2 obtained by reverse transcription-PCR were subcloned into the pGEM-T Easy vector (Promega) and verified by sequencing of at least five independent clones (for primer information, see Supplemental Table S3). Nucleotide sequences of HlPT1L and HlPT2 can be found in the GenBank database under accession numbers KM222441 and KM222442, respectively.
The Asp in the conserved Asp-rich motifs in PT1L (NQIYD183LESD187 and KD310VPD313VEGD; D in bold indicates the Asp to be mutated) and PT2 (NQIFD174MDID178 and KD302LSD305INGD; D in bold indicates the Asp to be mutated) were mutated to Ala using a previously described PCR-mediated method (Ho et al., 1989). The primers used are detailed in Supplemental Table S3.
Reconstitution of the Bitter Acid Pathway in Yeast and Analysis of Products
HlVPS (GenBank accession no. AB015430) was subcloned into the pESC-HIS vector (Agilent Technologies). The two HlCCL genes were subcloned into the pESC-LEU vector (CCl2/4 for the bitter acid pathway; GenBank accession nos. JQ740204 and JQ740206), and PT1L and PT2 (either wild-type or mutated copy) were inserted into the pESC-URA vector singly or together (for primer information, see Supplemental Table S3). Different combinations of three constructs (either empty vector or the construct with inserts) were cotransformed into the appropriate yeast (Saccharomyces cerevisiae) strain using a high-efficiency lithium acetate transformation protocol (Gietz and Schiestl, 2007). The resulting positive yeast clones, after being verified by PCR, were cultured in 30 mL of synthetic dextrose dropout medium (−Leu, −His, −uracil), with d-Glc as the carbon source, for 2 d. Then, the yeast cells were harvested and resuspended into synthetic galactose dropout medium (−Leu, −His, −uracil) with 2% (w/v) Gal. After induction for an appropriate period, 5-mL aliquots of the cultures were lysed by sonication after measuring cell density and subsequently extracted with 5 mL of ethyl acetate (10 μm naringenin was added as an internal standard). Extracts were dried under a stream of nitrogen and subsequently dissolved in 60% (v/v) methanol for LC-MS analysis (1290 liquid chromatograph coupled to a 6550 quadrupole time-of-flight mass spectrometer; Agilent Technologies). Two microliters of sample was loaded onto an HPLC column (ZORBAX Extend-C18, 2.1 × 50 mm, 1.8 μm; Agilent) run at a flow rate of 0.4 mL min−1. The gradient (solvent A, 0.05% (v/v) formic acid and 5 mm ammonium formate in 10% (v/v) acetonitrile; solvent B, 0.05% (v/v) formic acid and 5 mm ammonium formate in 90% (v/v) acetonitrile) program was set as follows at a flow rate of 0.4 mL min−1: 0 to 10 min, a linear gradient from 20% to 90% (v/v) B; and 10 to 12 min, 90% B. Mass spectra were acquired in positive-ion mode. The operating parameters were set as follows: capillary voltage, 4,000 V; nebulizer pressure, 20 pounds per square inch gauge; drying gas flow rate, 13 L min−1; gas temperature, 225°C; sheath gas flow rate, 12 L min−1; sheath gas temperature, 350°C; and fragmentor, 365 V. The content of prenylated compounds in the samples was quantified based on a standard curve prepared with PIVP or PIBP.
Split-Ubiquitin MbY2H Assays
The MbY2H assays were performed with a DUAL membrane kit following the manufacturer’s instructions (Dualsystems Biotech). The primers used are listed in Supplemental Table S3.
Coimmunoprecipitation Assays of HlPT1L and HlPT2
N-terminal His-tagged PT1L-T (+1 to +258 bp from ATG was removed; designated as 6xHis:HlPT1L-T) and N-terminal HA-tagged PT2-T (+1 to +249 bp from ATG was removed; designated as HA:PT2-T) were subcloned into the pESC-LEU and pESC-HIS vectors, respectively. The correct plasmids, confirmed by sequencing, were cotransformed into DSY-1 yeast cells. The pESC-LEU and pESC-HIS vectors also were modified using a PCR-mediated method for expressing His tag or HA tag in yeast cells, respectively (for primer information, see Supplemental Table S3). The resulting plasmids, renamed as His-pESC-LEU or HA-pESC-HIS, were used as negative controls. The transformed yeast cells were precultured in synthetic dropout medium containing 2% (w/v) Glc to an optical density at 600 nm (OD600) value greater than 2, diluted to OD600 = 0.4, and cultivated with 2% (w/v) Gal until the OD600 value reached 1. Cultures (400 mL) of yeast cells were harvested and disrupted with glass beads with the Mini-Bead-Beater (Biospec Products) in an extraction buffer (2 mL g−1 cells) containing 50 mm Tris-HCl, pH 7.5, 1 mm EDTA, 1% Triton X-100, 1 mm phenylmethylsulfonyl fluoride, 1 μg mL−1 leupeptin, and 1 μg mL−1 pepstatin. Glass beads and cell debris were removed by two rounds of centrifugation at 12,000g for 15 min. The supernatant was incubated with the first antibody (anti-His or anti-HA) at 4°C overnight with shaking, followed by incubation with protein G agarose beads for an additional 4 h. The beads were washed twice with extraction buffer supplemented with 150 mm NaCl and then resuspended in SDS-PAGE sample buffer. The immunoprecipitated proteins were separated by 15% SDS-PAGE, transferred to a polyvinylidene difluoride membrane, and blotted with appropriate antibodies (anti-His or anti-HA).
In Vitro Prenyltransferase Assay
Yeast microsome preparations were carried out as described by Pompon et al. (1996). The microsomal proteins were used directly for prenyltransferase assays after quantification by Bradford assay. The standard prenyltransferase assay reaction contained 1 mm dithiothreitol, 10 mm Tris-HCl (pH 7), 10 mm MgCl2, 30 mm NaF as a phosphatase inhibitor, 100 µm prenyl donor chemicals, and 100 µm DMAPP. The reaction mixtures were incubated in a total volume of 250 µL at 30°C. The enzymatic reaction mixtures were extracted twice with 250 µL of ethyl acetate. The organic phases of the extracts were combined and dried under nitrogen gas. Extracts were then dissolved in 250 µL of 40% (w/v) methanol, and 1 µL of the resulting solution was analyzed with liquid chromatography-quadrupole time-of-flight-mass spectrometry. For the determination of the apparent Km values of different substrates, 100 µm prenyl donor was added in the standard PT reactions (200 µL volume) containing a series of concentrations of prenyl acceptors, and vice versa. The enzymatic products were extracted and quantified using the aforementioned protocol. The apparent Km data were calculated by using Hanes plots (Hyper32, version 1.0.0).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers KM222441 and KM222442.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. The prenyl diphosphate pathway in the different yeast strains YPH499, DD104, AM94, and W303-1A-Δcoq2 used in this study.
Supplemental Figure S2. Alignment of HlPT1L and HlPT2 with SfN8DT-1 (cloned from S. flavescens; GenBank accession no. AB325579).
Supplemental Figure S3. Phylogenetic analyses of HlPT1L, HlPT2, and their homologs from other plant lineages.
Supplemental Figure S4. Production of acylphloroglucinols and prenylated acylphloroglucinols in yeast strains harboring different codon-optimized PT1L/PT2 pairs.
Supplemental Figure S5. Nucleotide alignment of HlPT2, PT2-At, and PT2-Sc.
Supplemental Figure S6. Nucleotide alignment of HlPT1L, PT1L-At, and PT1L-Sc.
Supplemental Figure S7. Electrospray ionization-mass spectrometry (in positive mode) spectra of the compounds produced in an engineered yeast strain harboring CCL2/CCL4/VPS/HlPT1L/HlPT2 or hop glandular trichomes.
Supplemental Figure S8. Distribution of acylphloroglucinols and prenylated acylphloroglucinols produced by a yeast strain harboring CCL2/CCL4/VPS/PT1L-T/PT2-T in cells and in the culture medium.
Supplemental Figure S9. Liquid chromatography-quadrupole time-of-flight-mass spectrometry analysis of products generated by yeast microsomes containing different prenyltransferase proteins, using PIVP or PIBP as substrate.
Supplemental Figure S10. Substrate specificity of yeast microsome harboring truncated HlPT1L.
Supplemental Figure S11. Liquid chromatography-quadrupole time-of-flight-mass spectrometry analysis of enzymatic products generated by yeast microsomes containing different prenyltransferase proteins, using monoprenylated PIVP as a substrate.
Supplemental Table S1. Yeast strains and plasmids used in this study.
Supplemental Table S2. 1H-NMR and 13C-NMR signal assignment for PIBP, PIVP, MD-PIBP, and MD-PIVP.
Supplemental Table S3. Primers used in the present study.
Supplementary Material
Acknowledgments
We thank Dr. Francis Karst (University of Strasbourg) for providing the DD104 yeast strain, Dr. Antonios Makris (Mediterranean Agronomic Institute of Chania) for providing the AM94 yeast strain, Dr. Kazufumi Yazaki (Kyoto University) for providing the W303-1A-Δcoq2 yeast strain and the SfN8DT clone (in pDR196 vector), and Dr. Ben Moulton (York University) for help with the chemical synthesis of PIBP and PIVP.
Glossary
- cDNA
complementary DNA
- XN
xanthohumol
- DMAPP
dimethylallyl diphosphate
- NC
naringenin chalcone
- PIVP
phlorisovalerophenone
- PIBP
phlorisobutyrophenone
- PMBP
phlormethylbutanophenone
- MbY2H
membrane yeast two-hybrid
- 8DN
8-dimethylallyl naringenin
- LC-MS
liquid chromatography-mass spectrometry
- MD
monodimethylallylated
- G
geranylated
- DD
didimethylallylated
- HA
hemagglutinin
- OD600
optical density at 600 nm
- m/z
mass-to-charge ratio
Footnotes
This work was supported by the National Program on Key Basic Research Projects (973 Program grant no. 2012CB113900 to G.W.), the National Natural Sciences Foundation of China (grant no. 31470387 to G.W.), and the State Key Laboratory of Plant Genomics of China (grant nos. SKLPG2011B0104 and SKLPG2011A0104 to G.W).
Articles can be viewed without a subscription.
References
- Akashi T, Sasaki K, Aoki T, Ayabe S, Yazaki K (2009) Molecular cloning and characterization of a cDNA for pterocarpan 4-dimethylallyltransferase catalyzing the key prenylation step in the biosynthesis of glyceollin, a soybean phytoalexin. Plant Physiol 149: 683–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero I, Blanco CA, Porras M (2012) Iso-alpha-acids, bitterness and loss of beer quality during storage. Trends Food Sci Technol 26: 21–30 [Google Scholar]
- Cheng W, Li W (2014) Structural insights into ubiquinone biosynthesis in membranes. Science 343: 878–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SM, Vaitheeswaran V, Ambrose SJ, Purves RW, Page JE (2013) Transcriptome analysis of bitter acid biosynthesis and precursor pathways in hop (Humulus lupulus). BMC Plant Biol 13: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Wang G, Yang DS, Tang Y, Broun P, Marks MD, Sumner LW, Dixon RA, Zhao PX (2010) TrichOME: a comparative omics database for plant trichomes. Plant Physiol 152: 44–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer MJ, Meyer S, Claudel P, Bergdoll M, Karst F (2011) Metabolic engineering of monoterpene synthesis in yeast. Biotechnol Bioeng 108: 1883–1892 [DOI] [PubMed] [Google Scholar]
- George JH, Hesse MD, Baldwin JE, Adlington RM (2010) Biomimetic synthesis of polycyclic polyprenylated acylphloroglucinol natural products isolated from Hypericum papuanum. Org Lett 12: 3532–3535 [DOI] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH (2007) High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc 2: 31–34 [DOI] [PubMed] [Google Scholar]
- Goto K, Asai T, Hara S, Namatame I, Tomoda H, Ikemoto M, Oku N (2005) Enhanced antitumor activity of xanthohumol, a diacylglycerol acyltransferase inhibitor, under hypoxia. Cancer Lett 219: 215–222 [DOI] [PubMed] [Google Scholar]
- Grote A, Hiller K, Scheer M, Münch R, Nörtemann B, Hempel DC, Jahn D (2005) JCat: a novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res 33: W526–W531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77: 51–59 [DOI] [PubMed] [Google Scholar]
- Ignea C, Cvetkovic I, Loupassaki S, Kefalas P, Johnson CB, Kampranis SC, Makris AM (2011) Improving yeast strains using recyclable integration cassettes, for the production of plant terpenoids. Microb Cell Fact 10: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karamat F, Olry A, Munakata R, Koeduka T, Sugiyama A, Paris C, Hehn A, Bourgaud F, Yazaki K (2014) A coumarin-specific prenyltransferase catalyzes the crucial biosynthetic reaction for furanocoumarin formation in parsley. Plant J 77: 627–638 [DOI] [PubMed] [Google Scholar]
- Keasling JD. (2010) Manufacturing molecules through metabolic engineering. Science 330: 1355–1358 [DOI] [PubMed] [Google Scholar]
- Koeduka T, Shitan N, Kumano T, Sasaki K, Sugiyama A, Linley P, Kawasaki T, Ezura H, Kuzuyama T, Yazaki K (2011) Production of prenylated flavonoids in tomato fruits expressing a prenyltransferase gene from Streptomyces coelicolor A3(2). Plant Biol (Stuttg) 13: 411–415 [DOI] [PubMed] [Google Scholar]
- Munakata R, Inoue T, Koeduka T, Karamat F, Olry A, Sugiyama A, Takanashi K, Dugrand A, Froelicher Y, Tanaka R, et al. (2014) Molecular cloning and characterization of a geranyl diphosphate-specific aromatic prenyltransferase from lemon. Plant Physiol 166: 80–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel J, Culley LK, Lu Y, Liu E, Matthews PD, Stevens JF, Page JE (2008) EST analysis of hop glandular trichomes identifies an O-methyltransferase that catalyzes the biosynthesis of xanthohumol. Plant Cell 20: 186–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohara K, Muroya A, Fukushima N, Yazaki K (2009) Functional characterization of LePGT1, a membrane-bound prenyltransferase involved in the geranylation of p-hydroxybenzoic acid. Biochem J 421: 231–241 [DOI] [PubMed] [Google Scholar]
- Okada Y, Sano Y, Kaneko T, Abe I, Noguchi H, Ito K (2004) Enzymatic reactions by five chalcone synthase homologs from hop (Humulus lupulus L.). Biosci Biotechnol Biochem 68: 1142–1145 [DOI] [PubMed] [Google Scholar]
- Pompon D, Louerat B, Bronine A, Urban P (1996) Yeast expression of animal and plant P450s in optimized redox environments. Methods Enzymol 272: 51–64 [DOI] [PubMed] [Google Scholar]
- Sasaki K, Mito K, Ohara K, Yamamoto H, Yazaki K (2008) Cloning and characterization of naringenin 8-prenyltransferase, a flavonoid-specific prenyltransferase of Sophora flavescens. Plant Physiol 146: 1075–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K, Tsurumaru Y, Yazaki K (2009) Prenylation of flavonoids by biotransformation of yeast expressing plant membrane-bound prenyltransferase SfN8DT-1. Biosci Biotechnol Biochem 73: 759–761 [DOI] [PubMed] [Google Scholar]
- Saugspier M, Dorn C, Thasler WE, Gehrig M, Heilmann J, Hellerbrand C (2012) Hop bitter acids exhibit anti-fibrogenic effects on hepatic stellate cells in vitro. Exp Mol Pathol 92: 222–228 [DOI] [PubMed] [Google Scholar]
- Shen G, Huhman D, Lei Z, Snyder J, Sumner LW, Dixon RA (2012) Characterization of an isoflavonoid-specific prenyltransferase from Lupinus albus. Plant Physiol 159: 70–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh IP, Bharate SB (2006) Phloroglucinol compounds of natural origin. Nat Prod Rep 23: 558–591 [DOI] [PubMed] [Google Scholar]
- Stevens JF, Miranda CL, Buhler DR, Deinzer ML (1998) Chemistry and biology of hop flavonoids. J Am Soc Brew Chem 56: 136–145 [Google Scholar]
- Stevens JF, Page JE (2004) Xanthohumol and related prenylflavonoids from hops and beer: to your good health! Phytochemistry 65: 1317–1330 [DOI] [PubMed] [Google Scholar]
- Stevens JF, Taylor AW, Nickerson GB, Ivancic M, Henning J, Haunold A, Deinzer ML (2000) Prenylflavonoid variation in Humulus lupulus: distribution and taxonomic significance of xanthogalenol and 4′-O-methylxanthohumol. Phytochemistry 53: 759–775 [DOI] [PubMed] [Google Scholar]
- Sugiyama A, Linley PJ, Sasaki K, Kumano T, Yamamoto H, Shitan N, Ohara K, Takanashi K, Harada E, Hasegawa H, et al. (2011) Metabolic engineering for the production of prenylated polyphenols in transgenic legume plants using bacterial and plant prenyltransferases. Metab Eng 13: 629–637 [DOI] [PubMed] [Google Scholar]
- Tian L, DellaPenna D, Dixon RA (2007) The pds2 mutation is a lesion in the Arabidopsis homogentisate solanesyltransferase gene involved in plastoquinone biosynthesis. Planta 226: 1067–1073 [DOI] [PubMed] [Google Scholar]
- Tsurumaru Y, Sasaki K, Miyawaki T, Momma T, Umemoto N, Yazaki K (2010) An aromatic prenyltransferase-like gene HlPT-1 preferentially expressed in lupulin glands of hop. Plant Biotechnol 27: 199–204 [Google Scholar]
- Tsurumaru Y, Sasaki K, Miyawaki T, Uto Y, Momma T, Umemoto N, Momose M, Yazaki K (2012) HlPT-1, a membrane-bound prenyltransferase responsible for the biosynthesis of bitter acids in hops. Biochem Biophys Res Commun 417: 393–398 [DOI] [PubMed] [Google Scholar]
- Van Cleemput M, Cattoor K, De Bosscher K, Haegeman G, De Keukeleire D, Heyerick A (2009) Hop (Humulus lupulus)-derived bitter acids as multipotent bioactive compounds. J Nat Prod 72: 1220–1230 [DOI] [PubMed] [Google Scholar]
- Wang G, Dixon RA (2009) Heterodimeric geranyl(geranyl)diphosphate synthase from hop (Humulus lupulus) and the evolution of monoterpene biosynthesis. Proc Natl Acad Sci USA 106: 9914–9919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Tian L, Aziz N, Broun P, Dai X, He J, King A, Zhao PX, Dixon RA (2008) Terpene biosynthesis in glandular trichomes of hop. Plant Physiol 148: 1254–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel BSJ. (2004) Metabolic channeling in plants. Annu Rev Plant Biol 55: 85–107 [DOI] [PubMed] [Google Scholar]
- Xu H, Zhang F, Liu B, Huhman DV, Sumner LW, Dixon RA, Wang G (2013) Characterization of the formation of branched short-chain fatty acid:CoAs for bitter acid biosynthesis in hop glandular trichomes. Mol Plant 6: 1301–1317 [DOI] [PubMed] [Google Scholar]
- Yazaki K, Sasaki K, Tsurumaru Y (2009) Prenylation of aromatic compounds, a key diversification of plant secondary metabolites. Phytochemistry 70: 1739–1745 [DOI] [PubMed] [Google Scholar]
- Zanoli P, Zavatti M (2008) Pharmacognostic and pharmacological profile of Humulus lupulus L. J Ethnopharmacol 116: 383–396 [DOI] [PubMed] [Google Scholar]
- Zhou J, Du G, Chen J (2014) Novel fermentation processes for manufacturing plant natural products. Curr Opin Biotechnol 25: 17–23 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.