A KNOTTED1-LIKE HOMEOBOX protein regulates abscission through modulating auxin concentration and transport.
Abstract
A gene encoding a KNOTTED1-LIKE HOMEOBOX PROTEIN1 (KD1) is highly expressed in both leaf and flower abscission zones. Reducing the abundance of transcripts of this gene in tomato (Solanum lycopersicum) by both virus-induced gene silencing and stable transformation with a silencing construct driven by an abscission-specific promoter resulted in a striking retardation of pedicel and petiole abscission. In contrast, Petroselinum, a semidominant KD1 mutant, showed accelerated pedicel and petiole abscission. Complementary DNA microarray and quantitative reverse transcription-polymerase chain reaction analysis indicated that regulation of abscission by KD1 was associated with changed abundance of genes related to auxin transporters and signaling components. Measurement of auxin content and activity of a DR5::β-glucuronidase auxin reporter assay showed that changes in KD1 expression modulated the auxin concentration and response gradient in the abscission zone.
Abscission is a highly programmed process and plays a critical role in the physiology and survival of plants, allowing plants to shed nonfunctional, stressed, or infected organs and disperse progeny (Bleecker and Patterson, 1997; Roberts et al., 2002). Shedding of plant organs occurs at predetermined positions called abscission zones (AZs). The AZ tissue is composed of small isodiametric cells with dense cytoplasm, and it is anatomically distinct long before the initiation of abscission (Roberts and González-Carranza, 2007). In tomato (Solanum lycopersicum), genetic analysis has revealed that several genes are involved in AZ differentiation. Three members of the MADS-box family, JOINTLESS, MACROCALYX (MC), and SEPALLATA MADS-BOX PROTEIN21, play a key role in differentiation of the pedicel AZ in tomato (Szymkowiak and Irish, 1999; Nakano et al., 2012; Liu et al., 2014). Studies of the tomato mutants lateral suppressor and blind have shown that GRAS (for Gibberellic Acid-Insensitive [GAI], Repressor of GAI, and Scarecrow) and MYELOBLASTOSIS family transcription factors are also involved in the formation of the pedicel AZ (Schumacher et al., 1999; Szymkowiak and Irish, 1999; Mao et al., 2000; Ampomah-Dwamena et al., 2002).
After the AZ is formed, it remains in a quiescent state from days to months until receiving the signals that initiate abscission (Roberts and González-Carranza, 2007; Nakano et al., 2013). Abscission initiation is triggered by developmental and environmental cues (Taylor and Whitelaw, 2001) and mediated by the interaction of two hormones, auxin and ethylene (Roberts et al., 2002; Estornell et al., 2013). Abscission cannot occur while there is a continuous polar flow of auxin passing through the AZ. Auxin depletion results in the initiation of abscission by making the AZ sensitive to ethylene (Abeles and Rubinstein, 1964; Addicott, 1982; Taylor and Whitelaw, 2001; Roberts et al., 2002; Meir et al., 2006). In tomato, elimination or reduction in auxin flow by removal of the subtending organ (leaf or flower) or application of polar auxin transport inhibitors initiates abscission (Meir et al., 2010). Reduced auxin flow changes the transcript abundance of many genes involved in auxin biosynthesis, transport, and signal transduction. In Arabidopsis (Arabidopsis thaliana), it has been reported that indole-3-acetic acid (IAA) signaling in the AZ is essential for organ shedding (Basu et al., 2013). Functional studies of Auxin Response Factor1 (ARF1), ARF2, ARF7, and ARF19 show that these transcriptional regulators have functions in floral organ abscission (Ellis et al., 2005; Okushima et al., 2005). However, it is still unclear what mechanism modulates auxin level and auxin signaling in the AZ during the onset of abscission.
A putative peptide ligand-receptor signal transduction pathway plays a role in the control of the onset of abscission in Arabidopsis (Cho et al., 2008; Stenvik et al., 2008). Analysis of inflorescence deficient in abscission (ida), haesa (hae) and haesa-like2 (hsl2) mutants in Arabidopsis indicated that IDA, a putative ligand, interacts with the receptor-like kinases HAE and HSL2 in regulating flower organ abscission (Jinn et al., 2000; Cho et al., 2008; Stenvik et al., 2008). In addition, several genes that may affect Arabidopsis floral organ abscission through interaction with the IDA-signaling pathway have been identified, including an ADP-ribosylation factor-GTPase-activating gene NEVERSHED (Liljegren et al., 2009; Liu et al., 2013) and three receptor-like kinases SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1 (Lewis et al., 2010), EVERSHED (Leslie et al., 2010), and CAST AWAY (Burr et al., 2011). In Arabidopsis, evidence suggests that IDA signaling represses the KNOTTED-LIKE HOMEOBOX (KNOX) family member KNOTTED-LIKE FROM ARABIDOPSIS THALIANA1 (KNAT1), which in turn, induces KNAT2 and KNAT6 to activate abscission (Shi et al., 2011).
KNOX proteins comprise a small family of 3-amino acid loop extension homeobox proteins, which fall into three subclasses: classes I, II, and M (Hay and Tsiantis, 2010). In our previous microarray studies, we found that three KNOX genes, TOMATO KNOTTED3 (TKN3), TNK4, and KNOTTED1-LIKE HOMEOBOX PROTEIN1 (KD1), were highly expressed in the pedicel AZ of tomato (Meir et al., 2010). TKN3 and TKN4 are class I KNOX genes with Arabidopsis homologs KNAT6 and KNAT2 that have been shown to be involved in the IDA abscission signaling pathway (Shi et al., 2011). Apart from our demonstration of high KD1 transcript abundance in the tomato AZ, there has been no suggestion of a role for class M KNOX proteins in abscission. Rather, these proteins have been associated with formation of compound leaves (Hay and Tsiantis, 2010). For example, Petroselinum (Pts), a semidominant KD1 mutant in tomato with multiply compound leaves, results from a single-nucleotide deletion in the promoter region of the KD1 gene that results in increased abundance of KD1 transcripts in the leaves (Kimura et al., 2008). Koenig et al. (2009) showed that auxin gradients play a primary role in controlling morphogenesis in compound leaves. Given the primary importance of an auxin gradient in determining the onset of abscission, we hypothesized that the KD1 protein might also play a role in the initiation of abscission. We report here the results of experiments designed to test that hypothesis. We examined the AZ transcriptome and physiology resulting from reduction in KD1 expression by virus-induced gene silencing (VIGS) or silencing in stably transformed plants under the control of an abscission-specific promoter. In addition, we examined the effect on abscission of up-regulation of KD1 in the semidominant Pts mutant.
RESULTS
KD1 Is Specifically Expressed in AZs
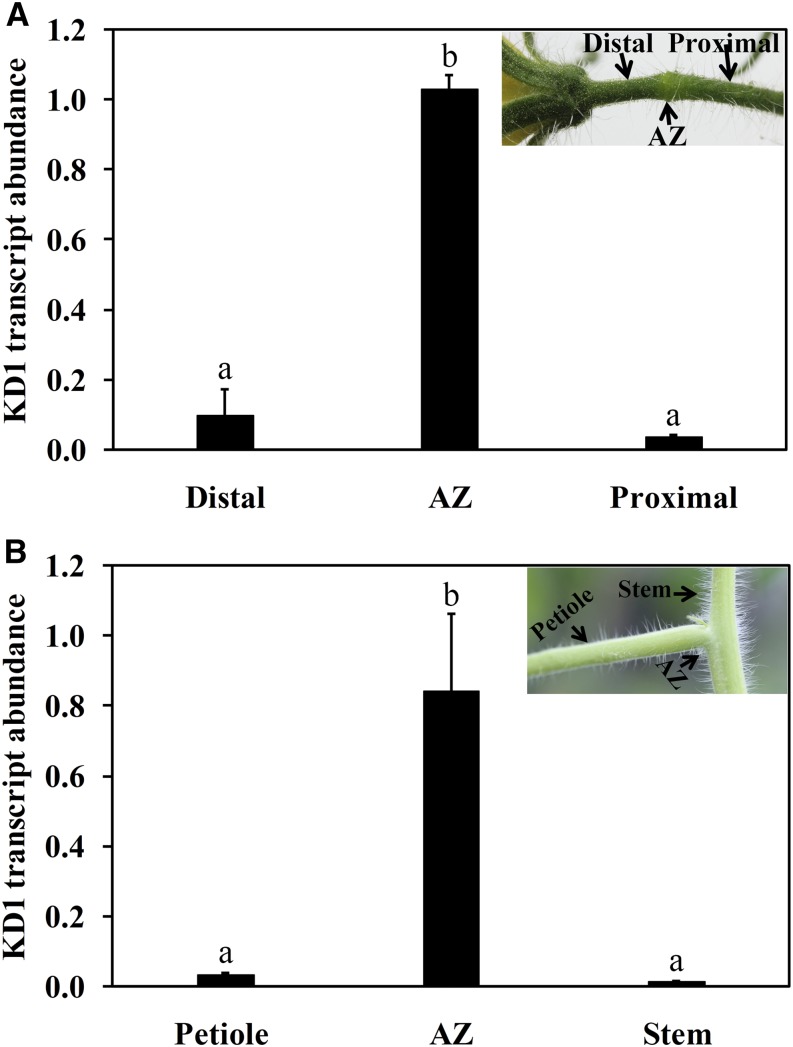
In previous microarray studies, we found, in tomato, that KD1 is predominately expressed in the pedicel AZ (Meir et al., 2010). To clarify the involvement of KD1 in tomato abscission, we first assayed KD1 transcripts in the pedicel and petiole AZs by quantitative reverse transcription (qRT)-PCR. We divided the pedicel into three parts: AZ, distal region (between the flower and the AZ), and proximal region (between the AZ and the peduncle). Transcripts of KD1 were detected in the AZ but not in the distal region or the proximal region (Fig. 1A). We then analyzed the transcript abundance of KD1 in the petiole AZ and adjacent regions, the petiole and the main axis. Transcripts of KD1 were much more abundant in the AZ than in the adjacent tissues (Fig. 1B). This AZ-specific expression pattern suggests that KD1 could be involved in the abscission process.
Figure 1.
Abundance of KD1 transcripts in pedicel (A) and petiole (B) AZs. A, Abundance of KD1 transcripts in AZ, distal, and proximal tissues of the pedicel. B, Abundance of KD1 transcripts in the petiole AZ, petiole, and main stem tissues. The abundance of KD1 transcripts was determined by qRT-PCR. Abundance of tomato 26S rRNA was used as an internal control. The abundance of KD1 transcripts was normalized against the abundance of 26S rRNA. Different letters indicate significant differences among tissues (Student’s t test, P < 0.05). The results are the means of three biological replicates ± sd.
Silencing KD1 Delays Abscission
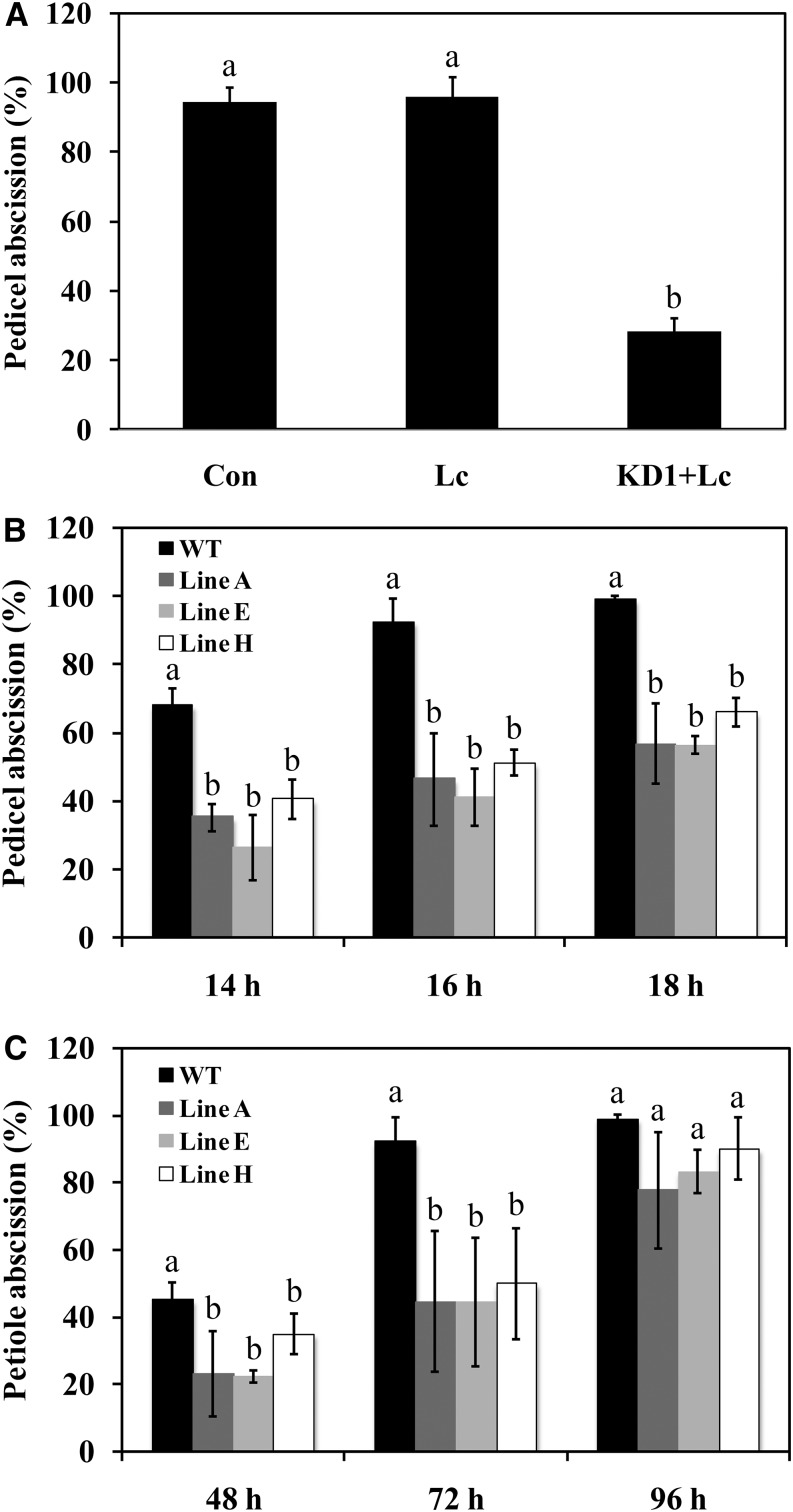
To understand the function of KD1 in abscission, we first examined the effect of silencing KD1 on abscission using VIGS. A purple transgenic tomato ‘New Yorker’ overexpressing a maize (Zea mays) anthocyanin gene Leaf color (Lc; Goldsbrough et al., 1996) was used as a silencing reporter. Silencing the Lc gene reversed the color of the transgenic plants from purple to green, providing a visual reporter of silenced tissue (Jiang et al., 2008). In this study, we used a silencing construct designed to silence both KD1 and Lc in the purple tomato line. Apart from the color change, there was no visible effect of silencing KD1 on the plants. Only 23% of the pedicels had abscised 16 h after flower removal in pedicels of green plants where both Lc and KD1 genes were silenced (Fig. 2A). In contrast, by that time, almost all pedicels had abscised in the controls for both the purple pedicels of the Lc plants and the green pedicels of plants where only Lc was silenced (Fig. 2A).
Figure 2.
Effects of silencing KD1 on the kinetics of abscission. A, VIGS of KD1. The effect of flower removal on pedicel abscission after 16 h was determined in pedicels of control (Con) Lc-overexpressed plants and silenced pedicels of Lc-overexpressed plants infected with TRV vectors containing fragments of the Lc gene alone (Lc) or combined with a fragment of KD1 (KD1+Lc). Different letters indicate significant differences among Con, Lc, and KD1+Lc (Student’s t test, P < 0.05). Pedicel (B) and petiole (C) abscission in wild-type (cv New Yorker) and TAPG4::antisense KD1 transgenic plants (lines A, E, and H). The percentages of pedicel and petiole abscission were determined at intervals after flower removal or during ethylene treatment (3 μL L−1), respectively. Different letters at each time point indicate significant differences between wild-type and transgenic lines (Student’s t test, P < 0.05). The results are the means of three replicates ± sd, with at least 15 samples per replicate. WT, Wild type.
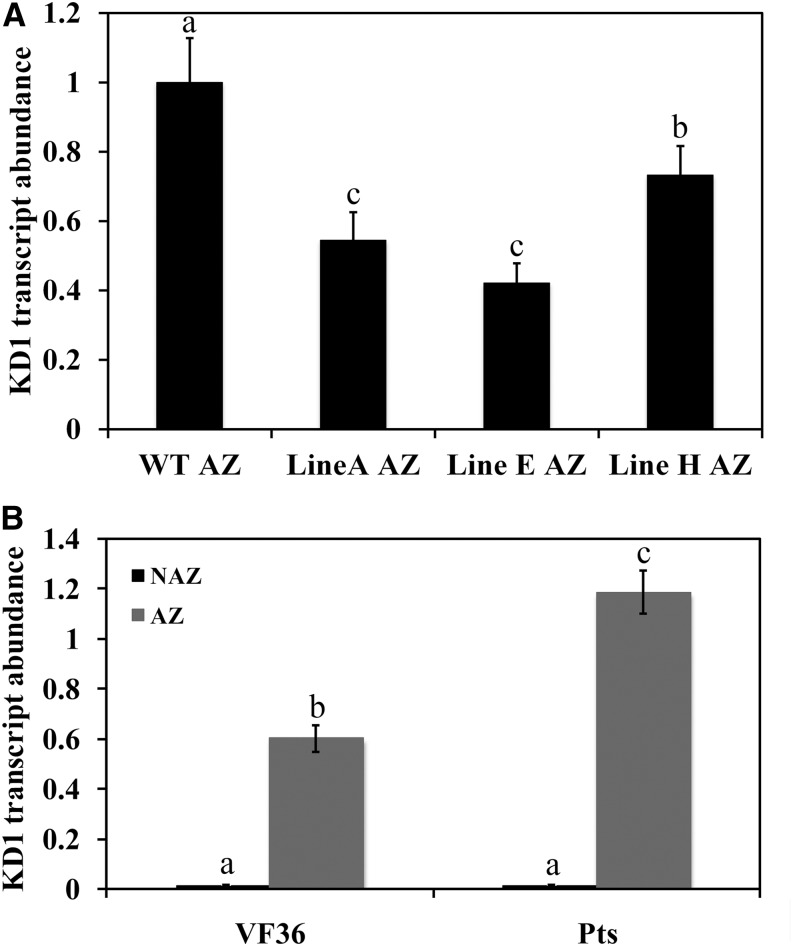
We further generated transgenic tomato ‘New Yorker’ plants, in which the KD1 antisense construct was driven by an abscission-specific promoter TOMATO POLYGALACTURONASE4 (TAPG4) (Kalaitzis et al., 1997; Hong et al., 2000). Ten independent transgenic lines were generated, and three representative transgenic lines (lines A, E, and H) were selected for additional analysis. The transformed plants were visually indistinguishable from wild-type (cv New Yorker) controls. KD1 transcript abundance measured in pedicel AZs 4 h after flower removal was significantly less in TAPG4::antisense KD1 transgenic lines than in wild-type plants (Fig. 3A). Among them, line E showed the lowest KD1 transcript abundance (Fig. 3A). Delayed abscission was seen in both pedicel and petiole abscission of the TAPG4::antisense KD1 transgenic plants (Fig. 2, B and C). At 18 h after flower removal, all of the pedicels of wild-type plants had abscised, whereas only 56% of pedicels had abscised in lines A and E, and 66% of pedicels had abscised in line H (Fig. 2B). Most (92%) of the petioles in wild-type plants had abscised after 72 h of ethylene treatment, whereas only 44% of both lines A and E petioles were abscised, and 50% of pedicels had abscised in line H (Fig. 2C).
Figure 3.
Abundance of KD1 transcripts in TAPG4::antisense KD1 transgenic plants and the Pts mutant. The abundance of KD1 transcripts was measured by qRT-PCR in AZ tissues of wild-type (cv New Yorker) and TAPG4::antisense KD1 transgenic lines (lines A, E, and H) 4 h after flower removal (A). Different letters indicate significant differences between wild-type and transgenic lines (Student’s t test, P < 0.05). In freshly harvested AZ tissues of wild-type (cv VF36) and Pts mutant plants, RNA was extracted from pedicel AZ or nonabscission zone (NAZ) pedicel tissues (B). Abundance of tomato 26S rRNA was used as an internal control. Different letters indicate significant differences among tissues (Student’s t test, P < 0.05). Results are the means of three biological replicates ± sd. WT, Wild type.
Overexpression of KD1 Promotes Abscission
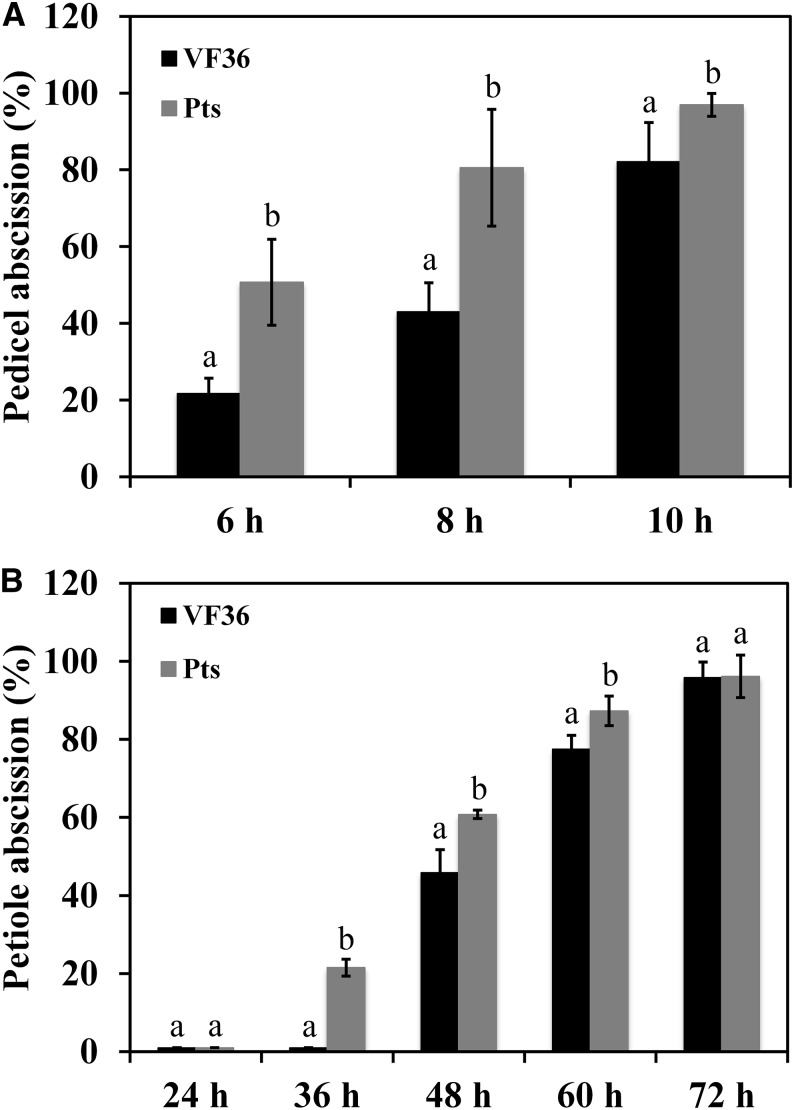
To further test the function of KD1 in abscission, we examined the abscission phenotype of the semidominant KD1 mutant Pts (Kimura et al., 2008). The transcript abundance of KD1 in the AZ of the Pts mutant plants was higher than that in wild-type (cv VF36) plants (Fig. 3B). KD1 transcripts were undetectable in non-AZ pedicel tissues of both Pts and wild-type plants (Fig. 3B). In the Pts mutant plants, 81% of the pedicels had abscised 8 h after flower removal, but only 43% of the pedicels of wild-type plants had abscised (Fig. 4A). In the petiole abscission assay, 22% of the petioles of Pts plants had abscised after 36 h of ethylene treatment, but few of those in the wild-type plants had (Fig. 4B).
Figure 4.
Kinetics of pedicel and petiole abscission in the KD1 semidominant mutant Pts. Pedicel (A) or petiole (B) abscission assays compared wild-type (cv VF36) plants with plants of the Pts mutant. The percentages of pedicel and petiole abscission were determined at intervals after flower removal or during ethylene treatment (3 μL L−1), respectively. Different letters indicate significant differences (Student’s t test, P < 0.05). Results are the means of three replicates ± sd, with at least 15 samples per replicate.
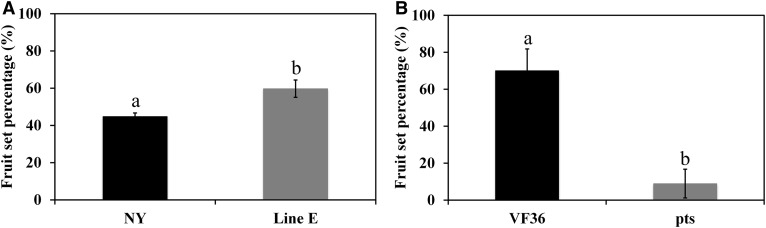
KD1 Expression Affects Fruit Set
Fruit set percentage was higher in TAPG4::antisense KD1 transgenic plants than in wild-type plants (cv New Yorker). Under our greenhouse conditions, 59.7% of flower buds set fruits in TAPG4::antisense KD1 transgenic plants, whereas 44.8% of wild-type flower buds set fruits (Fig. 5A). Moreover, fruit set percentage was significantly lower in the Pts mutant than in wild-type plants (cv VF36); 70% of flower buds set fruits in the wild-type plants, but only 9% of Pts flower buds set fruits (Fig. 5B).
Figure 5.
Fruit set in the TAPG4::antisense KD1 transgenic and Pts mutant. A, The percentage of accumulated fruit set was monitored in wild-type (cv New Yorker [NY]) and TAPG4::antisense KD1 transgenic plants (line E) in the greenhouse. B, The percentage of accumulated fruit set was monitored in wild-type (cv VF36) and Pts mutant plants in the greenhouse. Letters indicate significant differences (Student’s t test, P < 0.05). The results are the means of three replicates ± sd.
Gene Expression Analysis Identifies Genes Regulated by KD1 in the AZ
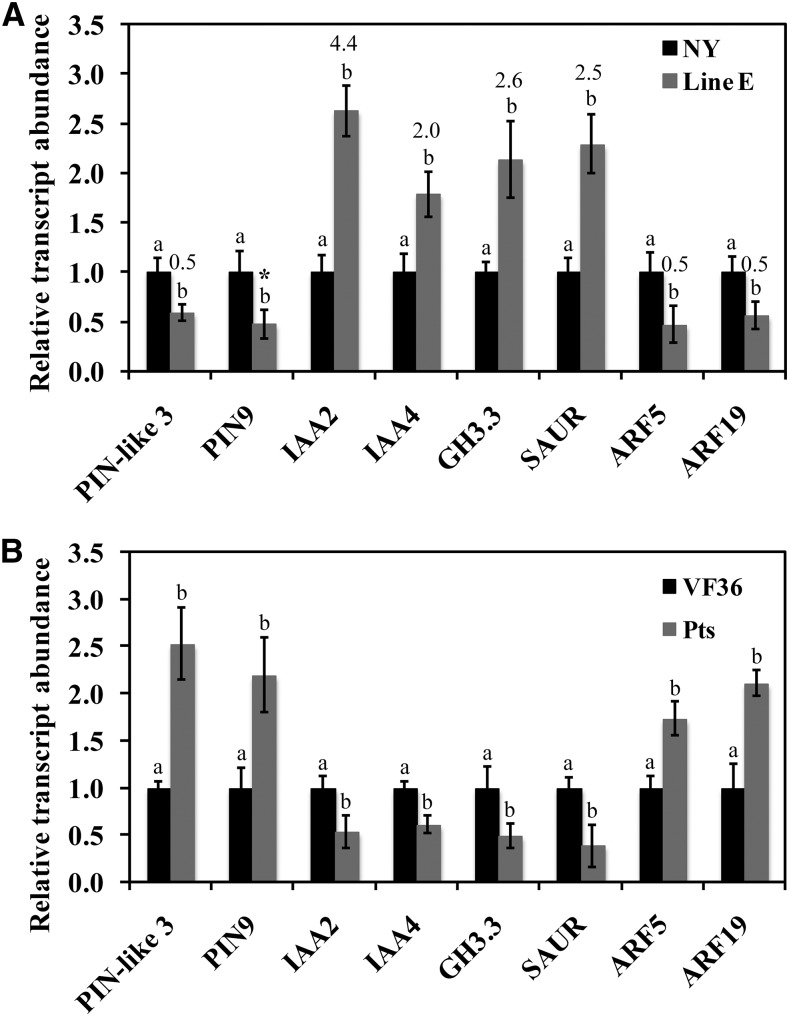
To investigate the transcriptional mechanisms underlying the delay of pedicel abscission in TAPG4::antisense KD1 plants, we compared gene expression in the AZ 4 h after flower removal in wild-type and TAPG4::antisense KD1 plants using a custom microarray. In TAPG4::antisense KD1 AZs, we identified 555 up-regulated and 593 down-regulated genes with known function (Supplemental Tables S1 and S2). Among these, 19 auxin-related genes were up-regulated, including AUXIN (Aux)/IAA2 (SL_NP000208), Aux/IAA4 (SL_TC198378), GRETCHEN HAGEN3.3 (GH3.3; SL_TC192282), and two SMALL AUXIN-UP RNA (SAUR) family genes (SL_TC202903 and SL_TC206415; Supplemental Table S3), and 11 auxin-related genes were down-regulated, including PIN-FORMED (PIN)-like3 (SL_TC197872), ARF5 (SL_TZXJ369TF), ARF8 (SL_BF097763), ARF19 (NM_001247676), and two IAA-amino acid hydrolase ILR1 precursors (SL_AW929186 and SL_AI781477; Supplemental Table S4). In addition, two KNOX family members, KNAT3 and SHOOT MERISTEMLESS, were up-regulated and down-regulated, respectively, in the AZs of the TAPG4::antisense KD1 line (Supplemental Tables S1 and S2).
As confirmation, we measured transcript abundance of selected genes in AZs of TAPG4::antisense KD1 plants using qRT-PCR (Fig. 6A). The results were consistent with the data obtained from the microarray (Fig. 6A) showing decreased abundance of auxin-efflux transporter (PIN) family and ARF genes and increased abundance of Aux/IAA, GH3.3, and SAUR genes compared with wild-type (cv New Yorker) control plants. We also examined transcript abundance of the same genes in AZs of the Pts mutant (Fig. 6B) and found that it was consistently the inverse of the pattern from the antisense plants, with higher abundance of transcripts of efflux transporter and ARF genes and reduced abundance of Aux/IAA, GH3.3, and SAUR genes compared with the cv VF36 control plants.
Figure 6.
Abundance of transcripts of KD1 downstream genes. Total RNA isolated from pedicel AZs of wild-type (cv New Yorker [NY]) and TAPG4::antisense KD1 transgenic (line E) plants 4 h after flower removal (A), and freshly harvested wild-type (cv VF36) and Pts mutant (B) plants were used to determine abundance of transcripts of each gene by qRT-PCR. Abundance of tomato 26S rRNA was used as an internal control. The expression ratio of each gene from microarray analysis is shown above the corresponding qRT-PCR column. Different letters indicate significant differences between NY and line E or between cv VF36 and Pts at each time point (Student’s t test, P < 0.05). Results are the means of three biological replicates ± sd. *, Data representing PIN9 were absent in the microarray.
KD1 Regulates the Auxin Content of the AZ
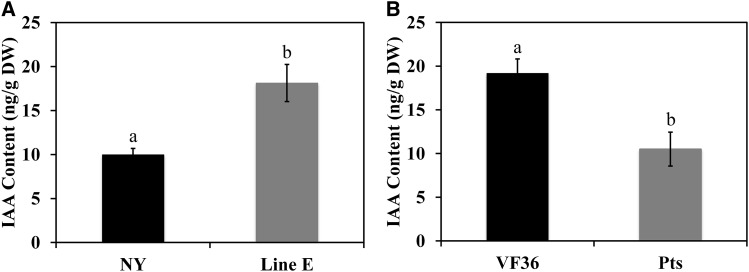
To test whether the observed differential gene expression related to auxin transport affects the auxin content, we examined IAA concentrations in tomato pedicel AZs by liquid chromatography-tandem mass spectrometry (LC-MS/MS). IAA concentrations in the petiole AZs of TAPG4::antisense KD1 plants were approximately 80% greater than those in the AZs of wild-type (cv New Yorker) plants (Fig. 7A). In contrast, IAA concentrations in the AZs of Pts mutant plants were 45% less than those in the AZs of wild-type (cv VF36) plants (Fig. 7B).
Figure 7.
Auxin concentrations in pedicel AZs. Tissues isolated from pedicel AZs were used to analyze the free IAA content of petiole AZs. A, AZs from wild-type (cv New Yorker [NY]) and TAPG4::antisense KD1 transgenic (line E) plants. B, AZs from wild-type (cv VF36) and Pts mutant plants. Different letters indicate significant differences (Student’s t test, P < 0.05). Values are the means of five replicates ± sd. DW, Dry weight.
KD1 Regulates Auxin Response Gradient in the AZ
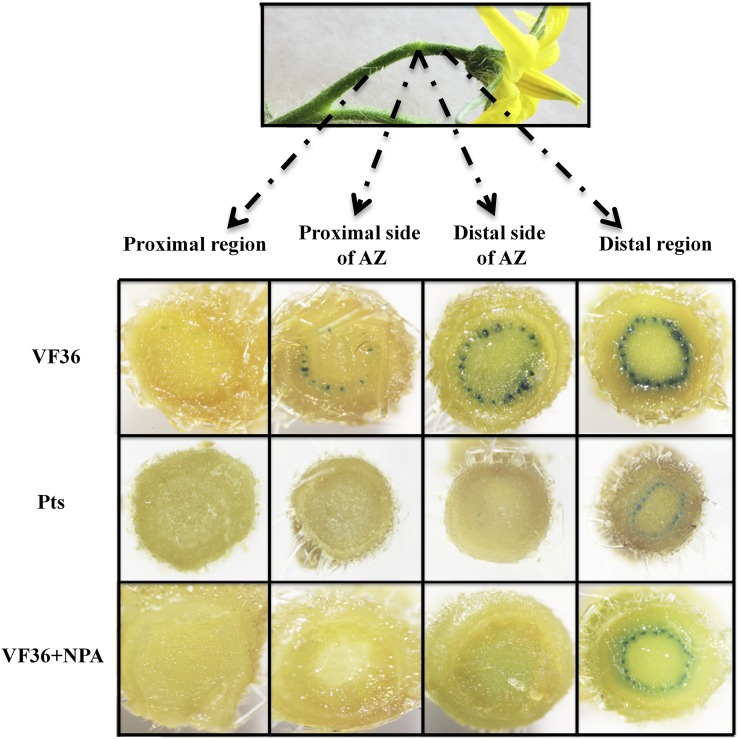
We introgressed the DR5::GUS auxin reporter transgene into cv VF36 and the Pts mutant background and compared GUS staining in pedicels of inflorescences from Pts DR5::GUS and cv VF36 DR5::GUS plants with staining in pedicels from cv VF36 DR5::GUS inflorescences treated with the auxin transport inhibitor N-1-napthylphthalamic acid (NPA; Fig. 8). The presence of GUS indicates activity of the auxin response pathway (Ulmasov et al., 1997; Koenig et al., 2009). In the distal region (between the flower and the AZ), GUS staining was similar in the cv VF36 and Pts mutant plants (Fig. 8). However, in the pedicel AZ itself, there was a clear gradient in GUS staining between the distal and proximal sides of the pedicel AZ in the cv VF36 plants but no detectable staining in the AZs from the Pts mutant (Fig. 8).
Figure 8.
Expression of DR5::GUS in AZ tissues of Pts mutant and NPA-treated wild-type plants. DR5::GUS expression in wild-type (cv VF36) and Pts mutant plants and from flower inflorescences of tomato (cv VF36) placed in a 25 μm NPA solution for 8 h (VF36+NPA). Transverse sections were taken from the proximal region (between the AZ and the peduncle), the proximal side of the AZ, the distal side of the AZ, and the distal region (between the flower and the AZ).
We also examined GUS staining in cv VF36 DR5::GUS plants treated with NPA, which accelerates pedicel abscission in intact inflorescences (Fig. 9). Application of NPA resulted in a GUS staining pattern similar to that seen in the Pts DR5::GUS plants, with staining in the distal region but none in the AZ or the proximal region (Fig. 8).
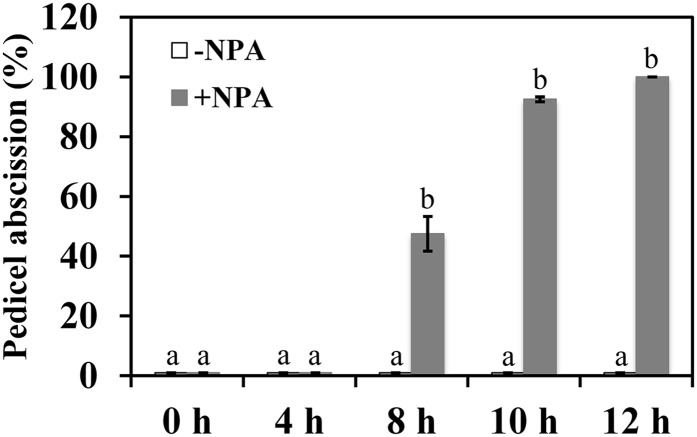
Figure 9.
Effects of NPA treatment on the kinetics of pedicel abscission. The percentage of pedicel abscission was determined at intervals from inflorescences placed in 25 μm NPA solution (+) or control (−). Different letters indicate significant differences between treatments at each time point (Student’s t test, P < 0.05). The results are means of three biological replicates ± sd, with at least 15 samples per replicate.
DISCUSSION
Auxin is known to be a key hormone in the initiation of abscission; the conventional model suggests that a change in the transport of auxin through the AZ results in sensitization of the AZ to ethylene, which then induces the hydrolytic enzymes and other components of the separation process (Abeles and Rubinstein, 1964; Addicott, 1982; Taylor and Whitelaw, 2001; Roberts et al., 2002; Meir et al., 2006). Our data are consistent with this model: treatment of tomato inflorescences with the auxin transport inhibitor NPA not only dramatically changed the activity of the auxin response pathway across the AZ but also stimulated pedicel abscission (Figs. 8 and 9).
Our data support the hypothesis that, in addition to its well-documented role in leaf development, KD1 plays a role in abscission. KD1 is known to modulate boundary separation and proximal-distal axis development in leaves (Kimura et al., 2008; Magnani and Hake, 2008; Peng et al., 2011), and our previous microarray analysis suggested a role for KD1 in leaf and flower abscission (Meir et al., 2010). Manipulating expression of KD1 by silencing or overexpression affects various aspects of plant development. In Arabidopsis, transgenic lines carrying KNATM (a KD1 ortholog) under control of the 35S promoter showed a series of leaf development defects (Magnani and Hake, 2008). However, Magnani and Hake (2008) reported difficulty in reducing expression of KNATM by RNA interference or artificial microRNA constructs. To overcome this problem and test the function of KD1 in abscission, we silenced KD1 in tomato using VIGS. VIGS analysis avoids potential lethality, because we inoculate plants that have already reached the seedling stage. Our results show that silencing KD1 expression significantly delays abscission (Fig. 2A). For additional analysis, we created KD1 antisense plants driven by an abscission-specific promoter TAPG4 (Hong et al., 2000; Meir et al., 2010). Abscission was significantly delayed in the TAPG4::antisense KD1 transgenic plants (Fig. 2, B and C), which otherwise were normal in their growth and development. Although reducing KD1 transcript abundance delayed abscission, it did not prevent it, suggesting that KD1 is part of a complex system controlling the abscission process. In our experiments, reduction in KD1 transcript abundance delayed abscission of both pedicels and petioles. In some tomato mutants, the abscission of pedicels and petioles is uncoupled. For example, in the Jointless1 and mc mutants, pedicels do not abscise, but petiole abscission is normal (Szymkowiak and Irish, 1999; Nakano et al., 2012). These MADS-box mutants seem to affect early events in the establishment of the AZ, whereas KD1 seems to be involved in the later physiological events leading to organ separation. It would be interesting to determine the genetic interaction among the Pts, Jointless1, and mc mutants.
Plants bearing the semidominant KD1 mutation Pts, which has a single-nucleotide deletion in the promoter region of the KD1 gene, have high KD1 transcript levels in their leaves (Kimura et al., 2008). We found that KD1 is also up-regulated in the AZ of the mutant plants, but the tissue specificity seen in wild-type plants remains—transcript abundance is much lower in the non-AZ pedicel tissues (Fig. 3B). The importance of KD1 in the abscission process is also suggested by the fact that pedicel abscission was accelerated in the Pts plants (Fig. 4). The possibility that KD1 is involved in auxin responses is suggested by the phenotype of the mutant plants, where growth alterations include not only the reported changes in leaf morphogenesis (which also involves auxin) but also, greatly reduced apical dominance (Supplemental Fig. S1). The Pts mutant was introgressed from the wild species (Solanum galapagense) into cv VF36 with only two backcrosses, and therefore, its genetic background is only 75% cv VF36. This means that we need to be cautious in interpreting results that might reflect the genome of the source species rather than the effects of the mutation. However, apart from KD1 and its downstream genes, transcripts of genes that we tested in the AZ showed similar abundance in the Pts mutant and cv VF36 (data not shown).
Altered auxin contents (Fig. 7) were observed in petiole AZs from plants with enhanced (Pts mutant) or reduced (TAPG4::antisense KD1) KD1 expression, suggesting that KD1 may play a role in modulating auxin levels. We detected no differential expression of auxin biosynthesis genes in KD1 antisense plants, and three genes encoding for IAA-amino acid conjugate hydrolases ILR were found to be down-regulated (Supplemental Table S4); therefore, neither increased synthesis nor increased release of auxin from conjugates (Woodward and Bartel, 2005) can explain the increased concentration of auxin in the antisense plants. Our microarray analysis showed that an auxin efflux transporter PIN-like3 (SL_TC197872; Fig. 6) was down-regulated in KD1 antisense plants. Recently, 10 members of the auxin efflux transporter gene family were identified in tomato (Pattison and Catalá, 2012). Because few of these genes were included in our custom microarray, we examined the transcript levels of other PIN genes in tomato plant AZs by qRT-PCR. In addition to PIN-like3, one other auxin efflux transporter, PIN9, was down-regulated in KD1 antisense plants (Fig. 6). The observed effect of manipulation of KD1 expression on the transcript abundance of these two auxin efflux transporters in the AZ is consistent with our hypothesis that KD1 plays a role in modulating auxin levels in the AZ, perhaps by controlling auxin efflux transport. In Arabidopsis, reduced auxin concentrations and reduced response to auxin are required for fruit valve margin separation. Reduced auxin concentrations in that system are also attributed to an increase in auxin efflux transporters (Sorefan et al., 2009).
Our microarray results show that transcript abundance of four auxin response genes ARFs was down-regulated in KD1 antisense transgenic plants (Supplemental Table S2). In Arabidopsis, genetic evidence suggests that ARFs play important roles in modulating the abscission process. ARF1, ARF2, ARF7, and ARF19 were identified as being involved in abscission (Ellis et al., 2005). Knockout or silencing of ARF2 in Arabidopsis delays flower senescence and organ shedding, and the delay is enhanced by suppressing the activity of ARF1, ARF7, or ARF19. The tomato homolog of the Arabidopsis ARF19 was also down-regulated in our TAPG4::antisense KD1 transgenic plants (Supplemental Table S4). Interestingly, transcript abundance of the tomato ARF2 homolog increased in KD1 antisense plants (Supplemental Table S3), implying a complex interplay between different components of the auxin response pathway during abscission.
Two AUX/IAA gene family members and two SAUR gene family members were also up-regulated in the AZs of TAPG4::antisense KD1 plants (Supplemental Table S3). This is consistent with our previous microarray analysis showing that seven Aux/IAA genes were down-regulated during tomato pedicel abscission (Meir et al., 2010). It seems highly likely that KD1 modulates the abscission process through the auxin-signaling pathway, perhaps by controlling the auxin response gradient through the AZ.
KNOX-type proteins are known to be involved in other auxin responses; in maize, the KNOX protein Knotted1 directly controls the auxin pathway at all levels, including auxin synthesis, transport, and signaling (Bolduc et al., 2012). Koenig et al. (2009) suggested that an auxin gradient plays fundamental roles in controlling morphogenesis in the compound leaves of tomato (Koenig et al., 2009). The fact that KD1 has been shown to be a genetic determinant of compound leaves leads us to postulate a role for KD1 in auxin gradient responses.
A transcript profiling study on tomato pedicels showed a gradient in auxin-induced gene expression in the pedicel, which may maintain the AZ in its quiescent state (Nakano et al., 2013). Our results indicate that enhanced expression of KD1 in the Pts mutant reduces auxin level and inhibits the auxin response gradient in the AZ. Our DR5::GUS auxin reporter assay (Fig. 8) revealed that the distal-to-proximal auxin response gradient was greatly reduced in the Pts mutant, especially in the proximal region, suggesting that KD1 modulates abscission by controlling the flow of auxin through the AZ. We hypothesize that enhanced expression of KD1 in the Pts mutant modulates an auxin flow gate in the AZ, thereby reducing the auxin gradient in the pedicel. Intriguingly, the elimination of the DR5::GUS response in the Pts mutant occurred, although there was only a 45% reduction in gross IAA content of the AZ (Fig. 7). It is known that the auxin response gradient across AZ is more important in the initiation of abscission than the auxin concentration itself (Nakano et al., 2013). Consistent with this result, NPA treatment, which disrupts the auxin polar transport, also eliminated the DR5:GUS auxin response signals in the AZ (Fig. 8). It seems, therefore, that disruption of the auxin response gradient is a key to the onset of abscission.
Canonical KNOX proteins contain an MEINOX domain at the N terminus and a homeodomain at the C terminus (Hake et al., 2004). The KD1 protein lacks a DNA-binding homeodomain, which suggests that the function of KD1 might depend on other proteins that bind to DNA. This possibility is indicated by presence of the MEINOX domain, which is known to mediate interaction with transcription factors (for example, the (BEL-LIKE homeodomain) transcription factor BIPINNATA; Kimura et al., 2008; Magnani and Hake, 2008). Additional studies are required to determine whether BIPINNATA or similar proteins have a function in abscission. In addition, it will be interesting to determine whether homologs of KNOX proteins known to have a function downstream from KD1 in Arabidopsis, such as KNAT1, KNAT2, and KNAT6 (Shi et al., 2011), play a role in organ abscission in tomato.
MATERIALS AND METHODS
Plant Materials and Treatments
Tomato (Solanum lycopersicum) germplasm ‘New Yorker’ (LA2009), Pts (LA2532), and ‘VF36’ (LA0490) plants were provided by the Tomato Genetics Resource Center, University of California. Pedicel and petiole abscission assays were performed as previously described (Jiang et al., 2008; Meir et al., 2010).
For the pedicel abscission assay, tomato inflorescences were harvested at 10 am from plants grown in the greenhouse. Inflorescences with at least two newly opened flowers were placed in a vial containing 10 mL of water and held in a high humidity chamber. Flowers were removed with a sharp razor blade, and abscission of the remaining pedicel from the peduncle was monitored at intervals.
To assay petiole abscission, the middle (second or third) petioles from young plants with four or five expanded leaves were used to prepare explants. Explants comprised a petiole and its subtending internode. The leaf and lateral shoots at the node were removed using a sharp razor blade. The explants were placed in a vial containing 10 mL of water and then exposed continuously to 3 μL L−1 of ethylene. Petiole abscission was monitored at intervals.
For NPA treatment, NPA was prepared as a 100 mm stock solution in dimethyl sulfoxide and diluted with water to 25 μm NPA. Tomato inflorescences with at least two newly opened flowers were placed in a vial containing 10 mL of NPA solution. Control inflorescences were placed in a vial containing a solution of the equivalent concentration of dimethyl sulfoxide.
Determination of Transcript Abundance
For examining transcript abundance in the pedicel AZs, 20 segments containing the AZ (less than 2 mm in length) were excised from pedicel AZs for each time point. Twenty segments of similar size were also excised from the distal and proximal regions of the pedicel. For examining transcript abundance in the petiole AZs, similar segments were excised from the petiole AZ, the distal region of the petiole, and the main stem axis.
Total RNA was extracted from the tissue samples using TRIzol reagent (Invitrogen) and treated with RNase-free DNase I (Promega) to remove any contaminating genomic DNA. First-strand complementary DNA (cDNA) was synthesized using 2 μg of total RNA, oligo d(T) primers, random hexamers, and Superscript Reverse Transcriptase (Invitrogen).
Quantitative real-time PCR was performed with the 7300 Real-Time PCR System (Applied Biosystems) using SYBR Green Real-Time PCR Master Mix (Applied Biosystems). To normalize sample variance, 26S ribosomal RNA (rRNA) was used as the internal control. Relative quantification of the transcript abundance of each gene was performed using the 2−∆∆CT method (Livak and Schmittgen, 2001). Primers used for determining transcript abundance are listed in Supplemental Table S5.
VIGS
A 203-bp fragment of the Lc gene was PCR amplified from tomato cDNA generated from Lc-overexpressed transgenic tomato lines using primers 5′-AGCGACGAGAGAAGCTCAAC-3′ and 5′-GGAGGGCCTTGTTATTAGCC-3′. The resulting product was cloned into tobacco rattle virus construct pTRV2 to form the pTRV2-Lc construct. A 227-bp fragment of the KD1 gene was PCR amplified from tomato cDNA using the primers 5′-TCTCAGCTCAGTGAACTCATGG-3′ and 5′-TTGTGGCAATCTAGCCATACAT-3′ and then subcloned into pTRV2-Lc to generate the pTRV2-KD1+Lc construct. Purple seedlings of transgenic tomato plants overexpressing Lc were infected with a mixed culture of Agrobacterium tumefaciens containing the pTRV1 vector and the pTRV2-KD1+Lc or pTRV2-Lc vector. The infection method has previously been described in detail (Jiang et al., 2008).
Vector Construction and Plant Transformation
To generate TAPG4::antisense KD1 transgenic plants, a 2,379-bp fragment of the TAPG4 abscission-specific promoter and the same fragment of the KD1 gene used in VIGS were amplified and then subcloned into the binary vector GSA1285, which carried the NEOMYCIN PHOSPHOTRANSFERASE II gene conferring kanamycin resistance to positive transformants. The plasmid construct used for transformation was introduced into Agrobacterium spp. LBA4404 by electroporation. Tomato ‘New Yorker’ was transformed by the tissue-culture method (Fillatti et al., 1987).
Microarray
Custom oligonucleotide microarrays were fabricated by NimbleGen Systems, Inc. using photolithography directed by the Maskless Array Synthesizer (Singh-Gasson et al., 1999). The custom oligonucleotide microarray contained probe sets for a total of 46,024 gene models derived from annotation of S. lycopersicum (The Dana Farber Cancer Institute Tomato Plant Gene Index; ftp://occams.dfci.harvard.edu/pub/bio/tgi/software/). Oligonucleotide probes (60-mer) were designed based on the NimbleGen standard procedure that optimizes the uniqueness of the targeted region and gas chromatography content, while minimizing self-complementarity and homopolymer runs. The highest ranking six probes (probe set) were selected to represent each gene model, with optimal probe spacing leading to uniformly distributed, nonoverlapping coverage. The 4-plex models used were based on the NimbleGen 385K design format (NimbleGen Systems, Inc.).
Ten micrograms of total RNA from mixed samples of three biological replicates was used for microarray hybridization. Concentration and purity of the RNA were evaluated using the 2100 Bioanalyzer (Agilent). Double-stranded cDNA was synthesized using the Invitrogen SuperScript Double-Stranded cDNA Synthesis Kit (Invitrogen). The cDNA samples were labeled with one-color cyanine3 random nonamers using the NimbleGen One-Color DNA Labeling Kit (Roche) followed by hybridizing to NimbleGen customized array slides according to the manufacturer’s instructions. Array slides were scanned by an MS 200 Microarray Scanner, and data were collected and analyzed using the MS 200 Data Collection and NimbleScan Software (Roche). Probe signal summarization, normalization, and background subtraction were performed using Robust Multichip Analysis (Irizarry et al., 2003) using the default parameters. Genes with an expression ratio of more than 2 or less than 0.5 between TAPG4::antisense KD1 and wild-type tissues were identified as up-regulated and down-regulated, respectively.
IAA Quantification
IAA content was measured by an LC-MS/MS system (LC20AD-MS/MS 8030 Plus; Shimadzu). Thirty pedicel AZ segments were frozen in liquid nitrogen and ground with a mortar and pestle; then, they were freeze dried in a low-temperature vacuum oven; 10 mg of each sample was extracted in 1 mL of 80% (v/v) methanol overnight at 4°C. Supernatants were collected after centrifugation at 15,200g for 10 min and then dried using a Jouan RCT-60 Concentrator. The dried extract was dissolved in 200 µL of sodium phosphate solution (0.1 mol L−1, pH 7.8) and passed through a Sep-Pak C18 cartridge (Waters). The cartridge was eluted with 1.5 mL of 50% (v/v) methanol, and the eluent was collected and vacuum dried again. The samples were redissolved in 80 µL of 10% (v/v) methanol; 10 µL of this solution was loaded onto the LC20AD-MS/MS 8030 Plus System. LC was performed using a 2.0- × 75-mm Shim-Pack XR-ODS II Column (2.2 μm; Shimadzu) with a column temperature of 40°C. The mobile phase comprising solvent A (0.05% [v/v] aqueous acetic acid) and solvent B (100% [v/v] methanol) was used in a gradient mode (time:concentration of A:concentration of B for 0:80:20, 6.0:35:65, 7.0:0:100, and 7.01:80:20) at an eluant flow rate of 0.3 mL min−1. MS/MS was performed under previously optimized conditions. The mass system was set to multiple reaction monitoring mode using electrospray ionization in the positive ion mode. Operating conditions were a nebulizing gas flow of 3 L min−1, drying gas flow of 15 L min−1, desolvation temperature of 180°C, and a heating block temperature of 480°. For IAA, a quadrupole 1 pre-bias of −18 eV, a quadrupole 3 pre-bias of −24 V, a collision energy of −16 eV, and mass:charge of 176:130 were used, whereas for deuterium-labeled IAA, a quadrupole 1 pre-bias of −19 eV, a quadrupole 3 pre-bias of −24 eV, a collision energy of −18 eV, and mass:charge of 181:134 were used. The IAA contents of the samples were calculated using a calibration curve established by using an internal 2H5-IAA standard (Olchemim).
GUS Staining
Tomato inflorescences were fixed in 90% (v/v) acetone for 20 min and then placed into GUS staining buffer of 0.5 mm 5-bromo-4-chloro-3-indolyl-β-glucuronic acid, 0.15 m NaH2PO4 (pH 7), 2 mm K3Fe(CN)6, 2 mm K4Fe(CN)6, and 0.05% (v/v) Triton X-100. The inflorescences were infiltrated in a capped 60-mL syringe by depressing the plunger for 3 min and then vacuuming for 1 h. After incubation in the dark at 37°C for 16 h, GUS-stained tissues were cleared and stored in 70% (v/v) ethanol. Images are representative of >20 observed samples stained in three independent experiments.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Phenotypes of the KD1 semidominant mutant Pts.
Supplemental Table S1. List of genes that are up-regulated in TAPG4::antisense KD1 plants.
Supplemental Table S2. List of genes that are down-regulated in TAPG4::antisense KD1 plants.
Supplemental Table S3. List of auxin-related genes that are up-regulated in TAPG4::antisense KD1 plants.
Supplemental Table S4. List of auxin-related genes that are down-regulated in TAPG4::antisense KD1 plants.
Supplemental Table S5. Primers used for quantitative PCR analysis.
Supplementary Material
Acknowledgments
We thank Dr. Andrew Macnish for initial characterization of KD1 using VIGS, Dr. Junping Gao for consultation on Auxin measurement, Dr. John Yoder for transgenic purple tomatoes expressing Lc, Dr. Neelima Sinha for the DR5::GUS tomato line, the Tomato Genetics Resource Center of the University of California for providing tomato seeds, and the Ralph M. Parsons Foundation Plant Transformation Facility of the University of California for conducting tomato transformations.
Glossary
- AZ
abscission zone
- cDNA
complementary DNA
- LC
liquid chromatography
- MS
mass spectrometry
- NPA
N-1-napthylphthalamic acid
- qRT
quantitative reverse transcription
- VIGS
virus-induced gene silencing
Footnotes
This work was supported by the U.S.-Israel Binational Agricultural Research and Development Fund (grant no. IS–4073–08C) and the National Natural Science Foundation of China (grant no. 91317312).
Articles can be viewed without a subscription.
References
- Abeles FB, Rubinstein B (1964) Regulation of ethylene evolution and leaf abscission by auxin. Plant Physiol 39: 963–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addicott FT. (1982) Abscission. University of California Press, Berkeley, CA [Google Scholar]
- Ampomah-Dwamena C, Morris BA, Sutherland P, Veit B, Yao JL (2002) Down-regulation of TM29, a tomato SEPALLATA homolog, causes parthenocarpic fruit development and floral reversion. Plant Physiol 130: 605–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu MM, González-Carranza ZH, Azam-Ali S, Tang S, Shahid AA, Roberts JA (2013) The manipulation of auxin in the abscission zone cells of Arabidopsis flowers reveals that indoleacetic acid signaling is a prerequisite for organ shedding. Plant Physiol 162: 96–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleecker AB, Patterson SE (1997) Last exit: senescence, abscission, and meristem arrest in Arabidopsis. Plant Cell 9: 1169–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolduc N, Yilmaz A, Mejia-Guerra MK, Morohashi K, O’Connor D, Grotewold E, Hake S (2012) Unraveling the KNOTTED1 regulatory network in maize meristems. Genes Dev 26: 1685–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burr CA, Leslie ME, Orlowski SK, Chen I, Wright CE, Daniels MJ, Liljegren SJ (2011) CAST AWAY, a membrane-associated receptor-like kinase, inhibits organ abscission in Arabidopsis. Plant Physiol 156: 1837–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SK, Larue CT, Chevalier D, Wang H, Jinn TL, Zhang S, Walker JC (2008) Regulation of floral organ abscission in Arabidopsis thaliana. Proc Natl Acad Sci USA 105: 15629–15634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis CM, Nagpal P, Young JC, Hagen G, Guilfoyle TJ, Reed JW (2005) AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana. Development 132: 4563–4574 [DOI] [PubMed] [Google Scholar]
- Estornell LH, Agustí J, Merelo P, Talón M, Tadeo FR (2013) Elucidating mechanisms underlying organ abscission. Plant Sci 199-200: 48–60 [DOI] [PubMed] [Google Scholar]
- Fillatti JJ, Kiser J, Rose R, Comai L (1987) Efficient transfer of a glyphosate tolerance gene into tomato using a binary Agrobacterium tumefaciens vector. Bio/Technology 5: 726–730 [Google Scholar]
- Goldsbrough AP, Tong Y, Yoder JI (1996) Lc as a non-destructive visual reporter and transposition excision marker gene for tomato. Plant J 9: 927–933 [Google Scholar]
- Hake S, Smith HM, Holtan H, Magnani E, Mele G, Ramirez J (2004) The role of knox genes in plant development. Annu Rev Cell Dev Biol 20: 125–151 [DOI] [PubMed] [Google Scholar]
- Hay A, Tsiantis M (2010) KNOX genes: versatile regulators of plant development and diversity. Development 137: 3153–3165 [DOI] [PubMed] [Google Scholar]
- Hong SB, Sexton R, Tucker ML (2000) Analysis of gene promoters for two tomato polygalacturonases expressed in abscission zones and the stigma. Plant Physiol 123: 869–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Jiang CZ, Lu F, Imsabai W, Meir S, Reid MS (2008) Silencing polygalacturonase expression inhibits tomato petiole abscission. J Exp Bot 59: 973–979 [DOI] [PubMed] [Google Scholar]
- Jinn TL, Stone JM, Walker JC (2000) HAESA, an Arabidopsis leucine-rich repeat receptor kinase, controls floral organ abscission. Genes Dev 14: 108–117 [PMC free article] [PubMed] [Google Scholar]
- Kalaitzis P, Solomos T, Tucker ML (1997) Three different polygalacturonases are expressed in tomato leaf and flower abscission, each with a different temporal expression pattern. Plant Physiol 113: 1303–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Koenig D, Kang J, Yoong FY, Sinha N (2008) Natural variation in leaf morphology results from mutation of a novel KNOX gene. Curr Biol 18: 672–677 [DOI] [PubMed] [Google Scholar]
- Koenig D, Bayer E, Kang J, Kuhlemeier C, Sinha N (2009) Auxin patterns Solanum lycopersicum leaf morphogenesis. Development 136: 2997–3006 [DOI] [PubMed] [Google Scholar]
- Leslie ME, Lewis MW, Youn JY, Daniels MJ, Liljegren SJ (2010) The EVERSHED receptor-like kinase modulates floral organ shedding in Arabidopsis. Development 137: 467–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis MW, Leslie ME, Fulcher EH, Darnielle L, Healy PN, Youn JY, Liljegren SJ (2010) The SERK1 receptor-like kinase regulates organ separation in Arabidopsis flowers. Plant J 62: 817–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljegren SJ, Leslie ME, Darnielle L, Lewis MW, Taylor SM, Luo R, Geldner N, Chory J, Randazzo PA, Yanofsky MF, et al. (2009) Regulation of membrane trafficking and organ separation by the NEVERSHED ARF-GAP protein. Development 136: 1909–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Butenko MA, Shi CL, Bolivar JL, Winge P, Stenvik GE, Vie AK, Leslie ME, Brembu T, Kristiansen W, et al. (2013) NEVERSHED and INFLORESCENCE DEFICIENT IN ABSCISSION are differentially required for cell expansion and cell separation during floral organ abscission in Arabidopsis thaliana. J Exp Bot 64: 5345–5357 [DOI] [PubMed] [Google Scholar]
- Liu D, Wang D, Qin Z, Zhang D, Yin L, Wu L, Colasanti J, Li A, Mao L (2014) The SEPALLATA MADS-box protein SLMBP21 forms protein complexes with JOINTLESS and MACROCALYX as a transcription activator for development of the tomato flower abscission zone. Plant J 77: 284–296 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆CT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Magnani E, Hake S (2008) KNOX lost the OX: the Arabidopsis KNATM gene defines a novel class of KNOX transcriptional regulators missing the homeodomain. Plant Cell 20: 875–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao L, Begum D, Chuang HW, Budiman MA, Szymkowiak EJ, Irish EE, Wing RA (2000) JOINTLESS is a MADS-box gene controlling tomato flower abscission zone development. Nature 406: 910–913 [DOI] [PubMed] [Google Scholar]
- Meir S, Hunter DA, Chen JC, Halaly V, Reid MS (2006) Molecular changes occurring during acquisition of abscission competence following auxin depletion in Mirabilis jalapa. Plant Physiol 141: 1604–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meir S, Philosoph-Hadas S, Sundaresan S, Selvaraj KSV, Burd S, Ophir R, Kochanek B, Reid MS, Jiang CZ, Lers A (2010) Microarray analysis of the abscission-related transcriptome in the tomato flower abscission zone in response to auxin depletion. Plant Physiol 154: 1929–1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T, Fujisawa M, Shima Y, Ito Y (2013) Expression profiling of tomato pre-abscission pedicels provides insights into abscission zone properties including competence to respond to abscission signals. BMC Plant Biol 13: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T, Kimbara J, Fujisawa M, Kitagawa M, Ihashi N, Maeda H, Kasumi T, Ito Y (2012) MACROCALYX and JOINTLESS interact in the transcriptional regulation of tomato fruit abscission zone development. Plant Physiol 158: 439–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y, Mitina I, Quach HL, Theologis A (2005) AUXIN RESPONSE FACTOR 2 (ARF2): a pleiotropic developmental regulator. Plant J 43: 29–46 [DOI] [PubMed] [Google Scholar]
- Pattison RJ, Catalá C (2012) Evaluating auxin distribution in tomato (Solanum lycopersicum) through an analysis of the PIN and AUX/LAX gene families. Plant J 70: 585–598 [DOI] [PubMed] [Google Scholar]
- Peng J, Yu J, Wang H, Guo Y, Li G, Bai G, Chen R (2011) Regulation of compound leaf development in Medicago truncatula by Fused Compound Leaf1, a class M KNOX gene. Plant Cell 23: 3929–3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts JA, Elliott KA, González-Carranza ZH (2002) Abscission, dehiscence, and other cell separation processes. Annu Rev Plant Biol 53: 131–158 [DOI] [PubMed] [Google Scholar]
- Roberts JA, González-Carranza ZH (2007) Abscission. InEncyclopedia of Life Sciences. John Wiley & Sons Ltd, Chichester, UK, pp 1–8 [Google Scholar]
- Schumacher K, Schmitt T, Rossberg M, Schmitz G, Theres K (1999) The Lateral suppressor (Ls) gene of tomato encodes a new member of the VHIID protein family. Proc Natl Acad Sci USA 96: 290–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi CL, Stenvik GE, Vie AK, Bones AM, Pautot V, Proveniers M, Aalen RB, Butenko MA (2011) Arabidopsis class I KNOTTED-like homeobox proteins act downstream in the IDA-HAE/HSL2 floral abscission signaling pathway. Plant Cell 23: 2553–2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh-Gasson S, Green RD, Yue Y, Nelson C, Blattner F, Sussman MR, Cerrina F (1999) Maskless fabrication of light-directed oligonucleotide microarrays using a digital micromirror array. Nat Biotechnol 17: 974–978 [DOI] [PubMed] [Google Scholar]
- Sorefan K, Girin T, Liljegren SJ, Ljung K, Robles P, Galván-Ampudia CS, Offringa R, Friml J, Yanofsky MF, Østergaard L (2009) A regulated auxin minimum is required for seed dispersal in Arabidopsis. Nature 459: 583–586 [DOI] [PubMed] [Google Scholar]
- Stenvik GE, Tandstad NM, Guo Y, Shi CL, Kristiansen W, Holmgren A, Clark SE, Aalen RB, Butenko MA (2008) The EPIP peptide of INFLORESCENCE DEFICIENT IN ABSCISSION is sufficient to induce abscission in Arabidopsis through the receptor-like kinases HAESA and HAESA-LIKE2. Plant Cell 20: 1805–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymkowiak EJ, Irish EE (1999) Interactions between jointless and wild-type tomato tissues during development of the pedicel abscission zone and the inflorescence meristem. Plant Cell 11: 159–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JE, Whitelaw CA (2001) Signals in abscission. New Phytol 151: 323–339 [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward AW, Bartel B (2005) Auxin: regulation, action, and interaction. Ann Bot (Lond) 95: 707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.