Arabidopsis accessions show different phenotypes in response to mild drought, yet a robust transcriptome response is conserved between the accessions.
Abstract
Although the response of plants exposed to severe drought stress has been studied extensively, little is known about how plants adapt their growth under mild drought stress conditions. Here, we analyzed the leaf and rosette growth response of six Arabidopsis (Arabidopsis thaliana) accessions originating from different geographic regions when exposed to mild drought stress. The automated phenotyping platform WIWAM was used to impose stress early during leaf development, when the third leaf emerges from the shoot apical meristem. Analysis of growth-related phenotypes showed differences in leaf development between the accessions. In all six accessions, mild drought stress reduced both leaf pavement cell area and number without affecting the stomatal index. Genome-wide transcriptome analysis (using RNA sequencing) of early developing leaf tissue identified 354 genes differentially expressed under mild drought stress in the six accessions. Our results indicate the existence of a robust response over different genetic backgrounds to mild drought stress in developing leaves. The processes involved in the overall mild drought stress response comprised abscisic acid signaling, proline metabolism, and cell wall adjustments. In addition to these known severe drought-related responses, 87 genes were found to be specific for the response of young developing leaves to mild drought stress.
Since plants started colonizing land, they had to adapt to fluctuations and restrictions in water availability. Morphological adaptations, such as the development of a waxy cuticle, vasculature, stomata, more complex root systems, and a diverse set of molecular mechanisms, allow plants to live in sometimes extreme conditions. Numerous studies have elucidated the adaptations of plants to severe drought conditions (McDowell et al., 2008; Akhtar et al., 2012; Golldack et al., 2014), often by withholding water until wilting or by cutting leaves and letting them dry to impose severe water deficits (Iuchi et al., 2001; Llorente et al., 2002; Taji et al., 2002; Cheong et al., 2003, 2007; Tran et al., 2004; Umezawa et al., 2004, 2006; Cominelli et al., 2005; Chen et al., 2006; Nelson et al., 2007; Park et al., 2007; Zhu et al., 2007; Zhang et al., 2008). However, the sudden infliction of such severe drought is unlikely to reflect what naturally happens in the field. In actual field conditions, plants have to adapt continuously to fluctuating environmental parameters, and only rarely will water be present in extreme excess (e.g. flooding) or, conversely, be so low in abundance that it actually threatens plant survival. Frequently, plants experience mild drought stress that, depending on the developmental stage, causes yield losses to various degrees. Despite its potential importance for agriculture, the response of plants to mild drought stress is poorly understood compared with severe dehydration stress (Aguirrezabal et al., 2006; Bouchabke et al., 2008; Harb et al., 2010; Baerenfaller et al., 2012; Des Marais et al., 2012).
Imposing mild drought stress requires a precise and well-monitored experimental setup, including a tight control of the soil water content and defining the precise timing of the drought onset, since the response to stress depends on the developmental stage of the plant (Skirycz et al., 2010; Verelst et al., 2010). Leaf development involves two main processes: cell proliferation and cell expansion. During cell proliferation, corresponding to the first phase of leaf development, all cells are dividing. This phase is followed by cell expansion, starting at the tip of the leaf and then moving as a front toward the leaf base. The so-called transition phase marks the developmental stage in which fully proliferating leaves convert to leaves with mainly expanding cells (Donnelly et al., 1999; Andriankaja et al., 2012; Gonzalez et al., 2012). Although it is well documented that both cell division and cell expansion are affected by drought stress (Aguirrezabal et al., 2006; Tardieu et al., 2010; Baerenfaller et al., 2012), little is known about the mechanisms involved in the response of the earliest phases of leaf development to mild drought stress.
Arabidopsis (Arabidopsis thaliana) is found all around the northern hemisphere, with some small patches along the African coast (Hoffmann, 2002; Koornneef et al., 2004), comprising many different habitats, each with specific environmental conditions, and therefore leading to differences in evolutionary pressure. Since the different populations are genetically isolated due to selfing, it is expected that different Arabidopsis accessions are evolutionarily highly adapted to their local environments. Experiments in laboratory conditions revealed that there is a high plasticity in drought tolerance (Bouchabke et al., 2008), nutrient uptake (Chardon et al., 2010), and salt tolerance (Katori et al., 2010) between different Arabidopsis accessions. With recent advances in sequencing technologies, it is feasible to study the intraspecies variations at the genome and transcriptome level that have risen through the adaptations of the different Arabidopsis accessions to their specific habitats, and also to find what has been evolutionarily conserved.
In order to identify the mechanisms that are active in young, growing leaves exposed to mild drought stress, out of a set of 24 accessions, six accessions capturing most of the variation in drought stress responses were selected (Bouchabke et al., 2008) and subjected to mild drought stress using the automated phenotyping platform WIWAM (Skirycz et al., 2011b), measuring growth-related phenotypes such as rosette area, leaf area, leaf epidermal pavement cell area, cell number, and the stomatal index. It was found that drought affected leaf growth throughout the entire course of development by the interplay of both reduced cell division and expansion and that the accessions behaved differently to mild drought stress.
We also harvested early developing leaf tissue for RNA sequencing and identified a list of 354 genes with common differential expression patterns under mild drought stress in all six accessions. These genes are involved in abscisic acid (ABA) signaling, Pro metabolism, and cell wall adjustments. In addition to these known drought-related genes, 87 genes were found to be specific for the response of young developing leaves to mild drought stress. Integration of coexpression and regulatory interaction information showed that the differentially expressed genes are highly connected. Moreover, a few genes were identified as hubs and thus are potential important players in the mild drought stress response in early developing leaf tissue. However, not all differentially expressed genes had a known function or were annotated to a specific process involved in stress responses. Therefore, those genes are interesting candidates to further unravel their role in the mild drought response.
RESULTS
Growth Measurements of Six Arabidopsis Accessions under Mild Drought
To study the effect of mild drought stress on leaf growth, a protocol was established in which stress is applied to young seedlings using the automated phenotyping platform WIWAM (Skirycz et al., 2011b). This platform allows for the automated weighing, watering, and imaging of the plants and, therefore, strictly controlling the applied watering regime. Because mild drought stress has been shown previously to have a profound effect on the cell number (Aguirrezabal et al., 2006; Pereyra-Irujo et al., 2008; Tardieu et al., 2010; Baerenfaller et al., 2012), drought treatment was started when the third leaf emerges from the shoot apical meristem, at 4 d after stratification (DAS), since at this point during development all cells of the third leaf are dividing. To enable the stress treatment at this early time point, plants were germinated in wet soil and transferred at 4 DAS to mild drought conditions on the WIWAM (Fig. 1; see “Materials and Methods”).
Figure 1.
Transfer protocol of seedlings to the WIWAM. A, Seeds were germinated on soil-filled 96-well plates. Average-sized seedlings (54; red circles) were selected using an in-house-developed image analysis algorithm. The seedling selection was done only for phenotyping experiments. B, At 4 DAS, seedlings were manually transferred from 96-well plates to the pretreated soil (drought or control). C, Pots were placed on the WIWAM for automatic phenotyping. D, Control soil water content was maintained at a constant value of 2.2 g water g−1 dry soil (solid line) during the entire experiment. For the mild drought condition (dashed line), the soil water content started at 1.2 g water g−1 dry soil after transfer to pots at 4 DAS. The stress level increased from 11 DAS onward until it reached 0.7 g water g−1 dry soil.
Multiple growth parameters were measured (Supplemental Fig. S1): rosette growth was followed over time by calculating the projected rosette area (PRA) daily; the area of the third leaf was measured at the transition from cell proliferation to cell expansion (10–11 DAS) and at maturity (22 DAS); the mean cell number and cell size of the third leaf were determined at maturity together with the stomatal index. Next to these leaf size measurements and cellular analyses, genome-wide transcriptome profiling using RNA sequencing was performed.
To study the effect of genetic variability in the growth response under mild drought stress applied during the leaf cell proliferation phase, six Arabidopsis accessions (Antwerp-1 [An-1], Bulhary-1 [Blh-1], Columbia-0 [Col-0], Cape Verdi Islands-0 [Cvi-0], Oystese-0 [Oy-0], and Shahdara [Sha]), representing different geographic regions (Supplemental Table S1) and shown to capture most of the variation in drought responses present in a set of 24 accessions (Bouchabke et al., 2008), were used.
Rosette Growth in Response to Mild Drought
The dynamic nature of rosette growth was grasped by following the PRA over time with daily intervals for each individual plant grown on the automated imaging platform WIWAM. To identify significant differences in growth between the accessions, a mixed model was used to analyze measurements of PRA over time (see “Materials and Methods”). This statistical model, using time, genotype, and treatment as fixed factors, showed that there was a significant difference in PRA between the accessions’ responses to mild drought stress over time (time × genotype × treatment interaction; P = 0.01).
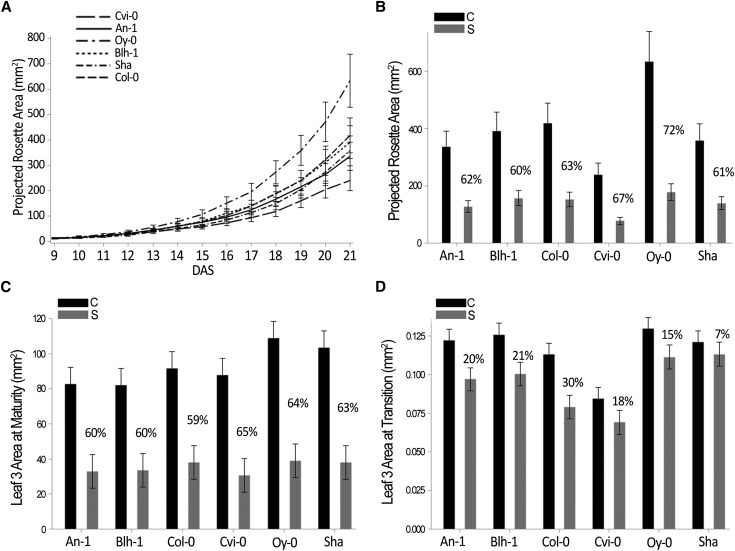
Next, we determined which accessions were significantly different by performing a pairwise comparison. This comparison was done for each time point: for the PRA in control conditions, for the PRA under mild drought stress, and for the relative difference of PRA under mild drought compared with control conditions. Results of the pairwise comparisons for all phenotypes can be found in Supplemental Table S2. Under control conditions, over time, Oy-0 was the largest and Cvi-0 was the smallest of the six accessions (P < 0.05; Fig. 2A). At maturity (21 DAS), Oy-0 reached a PRA of 633 mm2, while Cvi-0 measured 239 mm2 (Fig. 2, A and B), both differing significantly (P < 0.05) from the PRAs of An-1, Blh-1, Col-0, and Sha, which were not significantly different from each other.
Figure 2.
Measurements of rosette area and leaf size. A, Rosette areas at maturity. B, Leaf 3 areas at maturity. Control conditions (C) are indicated in black, and mild drought conditions (S) are indicated in gray. C, Leaf 3 areas at the transition from proliferation to expansion. D, Projected rosette area over time in control conditions for the six accessions. All values are least-square means ± se estimated from the mixed model, and percentages represent reductions under mild drought relative to the control.
After exposure to mild drought conditions, from 14 DAS onward, significant reductions in PRA were detected (Fig. 3). Therefore, the response over time to the imposed mild drought stress was analyzed from that time point onward by performing pairwise comparisons, showing that Oy-0 and Cvi-0 differed significantly from Blh-1 and Sha in their growth response over time to mild drought stress (P < 0.05; Fig. 3). At maturity (21 DAS), Oy-0 and Cvi-0 were 72% and 67% smaller, respectively, under mild drought compared with control conditions (Fig. 2B). Whereas the response of the PRA over time was different for Cvi-0 (Fig. 3), at the final time point the reduction in PRA of Cvi-0 did not differ from An-1, Blh-1, Col-0, and Sha, in contrast to that of Oy-0, which was significantly different from all other accessions except Cvi-0 (P < 0.05).
Figure 3.
Projected rosette area over time under control (C) and mild drought (S) conditions for the six accessions. Accession names indicated in the figure in black are significantly different from the reference accession in the time × genotype × treatment interaction at a P value cutoff of 0.05, and those indicated in gray are not significant. Values are least-square means ± se estimated from the mixed model.
In conclusion, our results demonstrate that mild drought has different effects on leaf growth in different accessions.
Leaf Growth in Response to Mild Drought
In order to analyze the cellular nature of the differences in growth response to mild drought stress, we studied in detail the third leaf harvested from the six accessions grown under control and stress conditions and quantified the leaf area, cell number, cell size, and stomatal index of the abaxial epidermis.
Under control conditions, the mature third leaf area ranged from 82 mm2 in Blh-1 to 109 mm2 in Oy-0 (Fig. 2C). The two accessions with the largest third leaf, Oy-0 and Sha (103 mm2), did not differ significantly from each other, but both had a significantly larger third leaf than the other four accessions (P < 0.05). The leaf area decreased for all six accessions when exposed to mild drought stress, ranging from 31 mm2 in Cvi-0 to 39 mm2 in Oy-0 (P < 0.05). Upon mild drought treatment, no significant differences in leaf area were detected between the accessions. The accessions showing the highest reduction at maturity were Cvi-0 (65%), Oy-0 (64%), and Sha (63%; Fig. 2C).
In order to know to what extent the drought treatment affected the third leaf early during development, leaf size was measured at the transition from proliferation to expansion. Upon microscopic investigation for the presence of puzzle-shaped pavement cells near the leaf tip, we concluded that for Cvi-0 the transition started at 11 DAS, whereas for all other accessions this was at 10 DAS. At this transition, the areas of leaves grown in control conditions ranged from 0.084 mm2 in Cvi-0 to 0.130 mm2 in Oy-0 (Fig. 2D). After mild drought treatment, Oy-0 and Sha (0.111 and 0.113 mm2, respectively) showed the largest third leaf, whereas that of Cvi-0 measured 0.069 mm2 and therefore was the smallest of the six accessions (Fig. 2D). The reductions in size under mild drought stress relative to the control condition varied from 7% in Sha to 30% in Col-0 (Fig. 2D). Whereas the 7% size reduction in Sha was not found to be significant, the leaf area of the other accessions was reduced significantly under mild drought stress at this early developmental time point.
In order to elucidate the cellular characteristics of the changes in leaf size, cellular drawings were made of the epidermis of the third leaf at maturity. These drawings were analyzed using an in-house-developed algorithm (Andriankaja et al., 2012) to obtain the cell number, cell area, and stomatal index. Under mild drought stress, we found that both pavement cell area and number were significantly reduced in all six accessions (P < 0.05). The reduction for pavement cell area (Fig. 4A) was quite similar for all accessions, ranging from 43% (Col-0) to 54% (Sha and Oy-0), in contrast to the number of pavement cells, which differed significantly more between accessions and varied from 18% (Blh-1 and Oy-0) up to 41% (An-1; Fig. 4B). The reduction in cell size and leaf size led to a higher cell density (Fig. 4C); however, the stomatal index was not significantly different between control and mild drought conditions (Fig. 4D).
Figure 4.
Measurements of cellular parameters of the mature third leaf of six accessions in control and mild drought conditions. A, Averaged cell areas of the pavement cells (µm2). B, Total number of pavement cells in the leaf. C, Pavement cell density (cells mm−2). D, Stomatal index as a percentage of stomata on total cell number. Control conditions (C) are indicated in black, and mild drought conditions (S) are indicated in gray. All values are means ± se, and percentages represent reductions under mild drought relative to the control.
In conclusion, the size of the third leaf was reduced in all six accessions after mild drought treatment, although to a different extent, due to decreases in cell number and average cell size.
Transcriptome Analysis
Previous work has shown that the transition from cell proliferation to cell expansion is of pivotal importance in determining the final leaf size (for review, see Gonzalez et al., 2012). This study aims to further unravel the effect of mild drought stress on this size-determining phase of leaf development. In our experiments, the third leaf was harvested at the beginning of the transition from cell proliferation to cell expansion (10–11 DAS), when the first expanding cells are discerned at the tip of the leaf. RNA was extracted and gene expression was determined through RNA sequencing.
Gene Expression under Mild Drought Stress
Common and accession-specific differentially expressed genes upon mild drought treatment were prioritized using a stage-wise statistical analysis (for details, see “Materials and Methods”; Supplemental Fig. S2). In the first stage, the differential expression was assessed for each gene. This stage consisted of two tests: stage I.a (for accession) checked for differential expression in at least one of the six accessions, resulting in 265 genes; and stage I.c (for common) checked for differential expression on average over the six accessions, giving 359 genes. The union of both tests delivered 439 genes that were differentially expressed during mild drought (Supplemental Table S3).
In order to find genes with an accession-specific response to mild drought stress, stage II tested the genes with significant differential expression in at least one accession (from stage I.a) for accession specificity (i.e. accession × treatment interaction). The stage II test identified 60 (Supplemental Table S3) out of the 265 stage I.a genes to show accession-specific differential expression, referred to as accession-specific genes. However, further analysis with pairwise tests assessing for differences in mild drought stress induced by differential expression between accessions in stage III (see “Materials and Methods”) unveiled that none of the 60 genes showed a response to the mild drought stress in one accession that was significantly different from the response in all five remaining accessions. The expression profiles of the 60 accession-specific genes in the six accessions for the three biological repeats are shown in Supplemental Figure S3.
In the next step, the 359 genes from the stage I.c analysis were analyzed further. For statistical reasons (detailed in “Materials and Methods” and Supplemental Fig. S2), five genes were removed. The remaining 354 genes are referred to as common drought genes (Supplemental Table S3). The expression profiles of all 354 genes in the six accessions for the three biological repeats are shown in Supplemental Figure S4.
A Gene Ontology (GO) enrichment analysis was performed to gain insight into the functional categories of the 354 common drought genes (Maere et al., 2005; Table I; Supplemental Table S4). The top enriched categories involve various abiotic stress responses, such as ABA signaling, osmotic stress, reactive oxygen species, and salt stress. Besides stress-related GO categories, cell wall modification- and cell growth-related genes also were clearly enriched among the common drought genes.
Table I. Main enriched GO categories in the 354 common drought genes.
The full list of enriched GO categories can be found in Supplemental Table S4. The Q value is the P value of the enrichment corrected with the Bonferroni method for multiple testing.
| Description | Q Value | No. of Genes |
|---|---|---|
| Cellular response to hormone stimulus | 1.39E-05 | 15 |
| Plant-type cell wall loosening | 1.39E-05 | 7 |
| ABA-mediated signaling pathway | 1.55E-05 | 8 |
| Cell wall modification | 3.32E-05 | 11 |
| Hormone-mediated signaling pathway | 4.65E-05 | 14 |
| Cell wall organization | 5.35E-05 | 12 |
| Plant-type cell wall modification | 6.25E-05 | 7 |
| Response to osmotic stress | 9.38E-05 | 18 |
| Plant-type cell wall organization | 1.89E-04 | 8 |
| Response to biotic stimulus | 7.16E-04 | 20 |
| Cell wall organization or biogenesis | 7.67E-04 | 13 |
| Regulation of transcription, DNA dependent | 5.77E-03 | 23 |
| Response to oxidative stress | 6.14E-03 | 11 |
| Response to salt stress | 1.18E-02 | 13 |
| Cellular response to reactive oxygen species | 1.22E-02 | 3 |
| Regulation of gene-specific transcription | 1.27E-02 | 3 |
| Cell wall thickening | 1.54E-02 | 3 |
| Developmental growth involved in morphogenesis | 1.76E-02 | 8 |
| Regulation of primary metabolic process | 1.88E-02 | 34 |
| Ethylene-mediated signaling pathway | 2.11E-02 | 4 |
| Cellular response to ethylene stimulus | 2.25E-02 | 4 |
| Glucan metabolic process | 2.25E-02 | 6 |
| Cell growth | 2.70E-02 | 9 |
| Response to cold | 2.75E-02 | 9 |
| Developmental growth | 3.05E-02 | 8 |
| Cell wall modification involved in multidimensional cell growth | 3.43E-02 | 3 |
| Response to ethylene stimulus | 3.43E-02 | 6 |
| Pro metabolic process | 3.44E-02 | 2 |
| Response to temperature stimulus | 3.92E-02 | 11 |
Hormone Signaling
The imposed mild drought stress had a major impact on ABA signaling in young developing leaves. Six of the 14 members of the PYRABACTIN RESISTANCE (PYR)/ PYRABACTIN RESISTANCE1-LIKE (PYL)/REGULATORY COMPONENTS OF ABA RECEPTORS (RCAR) ABA receptor protein family (PYR1, PYL1, PYL4, PYL5, PYL6, and PYL8) were down-regulated, whereas six of the nine protein phosphatase 2Cs (ABSCISIC ACID INSENSITIVE1 [ABI1], ABI2, HIGHLY ABSCISIC ACID-INDUCED PP2C GENE1 [HAI1], HAI2, PROTEIN PHOSPHATASE 2CA [AHG3], and HYPERSENSITIVE TO ABSCISIC ACID1 [HAB1]), two of the nine ABA-responsive element-binding factors (ABFs; ABF2/ABSCISIC ACID-RESPONSIVE ELEMENT-BINDING PROTEIN1 [AREB1] and ABF3), and six of the ABF target genes (ABSCISIC ACID-INSENSITIVE FIVE BINDING PROTEIN1 [AFP1], AHG3, HAI1, 3-KETOACYL-COA SYNTHASE2 [KCS2], RESPONSIVE TO DESSICATION20 [RD20], and AT5G53390; Yoshida et al., 2010, 2014) were up-regulated. In addition, 37 genes previously associated with ABA (Nemhauser et al., 2006; The Arabidopsis Information Resource; Supplemental Table S5) were found to be differentially expressed. For example, Δ1-pyrroline-5-carboxylate synthase (P5CS1), a rate-limiting enzyme in the Pro biosynthesis pathway (Strizhov et al., 1997), was up-regulated, whereas the gene encoding Pro dehydrogenase (PROLINE DEHYDROGENASE1 [PDH1]/EARLY RESPONSIVE TO DEHYDRATION5 [ERD5]/PROLINE OXIDASE [POX]) was down-regulated. Taken together, 57 of the 354 differentially expressed genes were ABA related.
Ethylene was previously associated with the regulation of leaf growth in vitro under mannitol-mediated growth reduction (Skirycz et al., 2011a; Dubois et al., 2013). Under mild drought conditions in soil, two 1-aminocyclopropane-1-carboxylic acid (ACC) oxidases (ACO4 and ACO2) were down-regulated as well as four ethylene response factors (ERFs; ERF2, ERF15, AT5G07580, and AT5G61590).
DELLAs are known negative regulators of growth under various stresses (Achard et al., 2006, 2008; Magome et al., 2008; Navarro et al., 2008; Claeys et al., 2012). Therefore, we compared the common differentially expressed genes upon mild stress with a list of putative DELLA targets (Claeys and Inzé, 2013). Of the 354 common drought genes, 43 were putative DELLA targets (Supplemental Table S6), which was a highly significant enrichment (P = 2.67E-45).
Cell Wall Modifications
At least 21 common drought genes were involved in cell wall modifications. The cell wall-loosening expansins (EXPA1, EXPA3, EXPA4, EXPA15, EXPB1, and EXPB3), pectin lyases (AT1G10640, AT1G60590, AT1G67750, AT3G61490, AT4G13710, and AT4G24780; Cosgrove, 2005), and pectin methylesterase inhibitors (AT1G23205 and AT2G26440) were up-regulated. On the other hand, genes encoding cell wall-strengthening enzymes, xyloglucan endotransglucosylases/hydrolases (XTHs) and fasciclin-like arabinogalactans (FLAs; Cosgrove, 2005; MacMillan et al., 2010), were consistently down-regulated (FLA2, FLA9, MERISTEM5 [MERI5B], XTH6, XTH9, XTH15, and XTH16).
Coexpression Network and Regulatory Interactions
The mild drought stress imposed on the six accessions clearly provoked a differential regulation of genes that were previously associated with drought stress, such as ABA signaling genes. However, of the 354 common differentially expressed genes, 216 were previously not associated with drought-responsive mechanisms (based on drought-related GO terms, relation to ABA, and involvement in cell wall modifications; Supplemental Table S7), of which 37 have yet unknown functions.
Next, we used the online tool CORNET (De Bodt et al., 2010, 2012) to perform coexpression and regulatory interaction network analyses. All 354 common differentially expressed genes were tested against different predefined microarray data sets in CORNET (see “Materials and Methods”) and resulted in a network of 202 coexpressed genes. In addition, a regulatory network analysis was performed in CORNET to get a view on the regulatory interactions between the coexpressed genes. This analysis used the confirmed regulatory interactions from the AGRIS (Davuluri et al., 2003; Palaniswamy et al., 2006; Yilmaz et al., 2011) and microarray gene-target relations databases. In addition, the text-mining database EVEX (Van Landeghem et al., 2013) was used to find extra regulatory interactions described in the literature. Interactions with neighbor genes were included for the regulatory interactions, adding 144 genes to the network, bringing the total to 346 genes. The coexpression and regulatory interaction analyses showed high connectivity between the differentially expressed genes (Fig. 5).
Figure 5.
Coexpression and regulatory interaction network of common differentially expressed genes in the six accessions. Edges colored blue connect coexpressed genes, and thickness increases with rising coexpression coefficients. Red, green, and black lines represent regulatory interactions, which can be direct (solid lines) or indirect (dashed lines). Regulatory interactions can be activating (green), repressing (red), or unknown (black). The top 20 most interacting genes in the network are indicated in yellow and can be found in Supplemental Table S8. Squares and diamonds are query genes, and circles are neighbor genes. Visualization is based on the coexpression analysis done in CORNET with a coexpression coefficient of 0.7 and confirmed regulatory interactions from AGRIS, microarray gene-target relations, and EVEX.
The 20 genes with the highest number of interactions (both coexpression and regulatory) are given in Supplemental Table S8. The genes with the most interactions encode MITOGEN-ACTIVATED PROTEIN KINASE3 (MPK3; 37 interactions) and a protein of unknown function (AT4G36500; 28 interactions). Also, a jasmonic acid (JA)-synthesizing lipoxygenase (LOX2; 17 interactions) is among the 20 most interacting genes.
Comparison with Severe Drought Studies
In the past, a large number of studies focused on severe stress. In order to make a comparison with our mild drought study, three studies imposing severe progressive drought stress on soil-grown plants were selected (Huang et al., 2008; Matsui et al., 2008; Harb et al., 2010). Whereas Harb et al. (2010) used Affymetrix ATH1 micro arrays (approximately 21,000 genes), the two other studies applied custom-made arrays (approximately 30,000 genes) with probes for all 354 common drought genes except two. A total of 188 of the common drought genes were differentially expressed in the same direction in at least one of the three studies compared (Supplemental Fig. S5). Forty-two genes were part of the core ABA signaling machinery (12 genes) or responded to ABA (30 genes), including the Pro-biosynthesizing P5CS1 and Pro dehydrogenase (PDH1/ERD5/POX). Eight from the 20 cell wall-modifying genes, including all four XTHs and the XTH-similar MERI5B, found in mild drought, behaved similarly in severe drought. Interestingly, four out of five differentially expressed expansins in mild drought showed opposing expression in severe drought. From the 43 putative DELLA targets differentially expressed in mild drought, 25 showed a similar behavior to severe drought.
In the end, 166 common drought genes were not transcriptionally modified in severe drought imposed on mature leaf tissue. Subsequently, we analyzed the behavior of the common drought genes in mature tissue exposed to mild drought stress.
Comparison with Mild Drought in Mature Tissue
Only three studies have addressed the transcriptomic response of soil-grown Arabidopsis plants to mild drought stress (Harb et al., 2010; Baerenfaller et al., 2012; Des Marais et al., 2012). In all cases, the transcriptional responses of mature tissues were analyzed, with the exception of the study by Baerenfaller et al. (2012), which pooled different developmental stages to detect mild drought stress-responsive genes. A comparison of the 354 common drought genes described in this study with the above-listed experiments showed that 216 of the 354 common drought genes in young developing tissue were expressed in the same direction in mature tissue under mild drought (Supplemental Fig. S6). Except for one protein phosphatase type 2C (PP2C; HAB1) and ABF2, all ABA signaling genes overlapped with the studies in mature tissue. Also, 21 ABA-responsive genes behaved similarly in mature and young developing leaf tissue. Furthermore, 15 of the 20 cell wall-modifying common drought genes and 12 of the 43 putative DELLA targets also played putative roles in mature leaves exposed to mild drought stress.
Genes Specifically Expressed in Young Developing Tissue Subjected to Mild Drought Stress
By comparing with the above-listed studies, 87 genes were identified to be differentially expressed specifically in young developing tissue under mild drought stress conditions (Fig. 6; Supplemental Table S9). Strikingly, none of these genes, with the exception of MYO-INOSITOL POLYPHOSPHATE 5-PHOSPHATASE2, COLD-REGULATED47, MPK3, NACL-INDUCIBLE GENE1, and AT5G53390, were previously found to be associated with ABA. Five of the 87 common drought genes were involved in cell wall modification: EXPA15, three genes encoding pectin lyases (AT1G60590, AT1G67750, and AT3G61490), and one pectin methylesterase inhibitor (AT1G23205).
Figure 6.

Venn diagram showing the overlap between genes involved in severe drought studies (Huang et al., 2008; Matsui et al., 2008; Harb et al., 2010), genes in mature tissue in mild drought studies (Harb et al., 2010; Baerenfaller et al., 2012; Des Marais et al., 2012), and the common drought genes.
Many of the 87 genes have not been associated previously with known growth responses in mild drought. However, some of the common drought genes might play a role in regulating growth in mild drought in young developing leaf tissue; interesting candidates are MPK3 and CYTOCHROME P450 78A7 (CYP78A7).
DISCUSSION
Mild Drought Scenarios, Their Effects and Robustness
Most of our knowledge about the responses of plants to drought stress is obtained by studying severe dehydration stress. The physiological relevance of this type of experiments compared with field conditions, however, has been questioned (Verslues et al., 2006; Claeys and Inzé, 2013; Lawlor, 2013). Moreover, increased tolerance to severe drought often comes with a yield penalty (Yang et al., 2010) and does not result in sustained growth under mild drought conditions (Skirycz et al., 2011b; Claeys and Inzé, 2013). The aim of this study was to analyze the effect of mild drought stress on young developing leaves. To this end, the automated phenotyping platform WIWAM was used to impose stress early during leaf development when the third leaf emerges from the shoot apical meristem. Daily imaging allowed for the analysis of stress effects in six different accessions on rosette growth over time. Although the six accessions analyzed responded differently to the imposed stress at the phenotypic level, the molecular response, investigated by RNA sequencing of developing leaves at the transition from proliferation to expansion, revealed that similar stress-related processes were differentially regulated in all accessions.
The protocol described here allows for an early onset of the mild drought stress when the third leaf emerges from the shoot apical meristem. In previous studies, stress onset started at the emergence of leaf 6 (Aguirrezabal et al., 2006; Bouchabke et al., 2008), explaining some major differences observed in the sensitivity of accessions to mild drought stress. For example, the effect of mild drought stress in An-1 previously was found to be rather limited (Aguirrezabal et al., 2006; Bouchabke et al., 2008), whereas in this study, a considerable size reduction of the third leaf and PRA was found. Differences in the developmental timing at which the drought stress was applied and in the drought scenario could lead to different outcomes in terms of drought tolerance or sensitivity (Tardieu, 2012). Therefore, it is important to screen different drought scenarios in order to come to a robust consensus response, which might be of great interest for agricultural applications. To increase the robustness of this consensus response, it is favorable to test plants with different genetic backgrounds.
Cellular Response to Mild Drought Stress
Mild drought stress in all six accessions caused reductions in cell area and cell number. Pavement cell area was reduced by 42% to 55%, depending on the accession, and cell number was decreased by 16% to 24%, except for An-1, having a 41% reduction. Reductions in cell number and cell size by drought have been reported previously (Aguirrezabal et al., 2006; Pereyra-Irujo et al., 2008; Baerenfaller et al., 2012), and quantitative trait loci linked to this reduction have been described (Tisné et al., 2008). In our experimental setup, cell expansion was relatively more affected by the mild drought stress than cell proliferation. Although the mild drought stress started and was effective during the proliferation phase, the water content was further lowered during the expansion phase from 11 DAS onward (Fig. 1D). This increase in drought severity during the transition phase from proliferation to expansion can explain the larger effect on cell expansion compared with that on cell proliferation. Alternatively, an initial reduction in cell division could have been compensated for during leaf development by meristemoid divisions, generating extra pavement cells while forming stomata (Geisler et al., 2000; Bergmann and Sack, 2007). Compensation by meristemoid division has been observed previously in the leaf growth response to mannitol (Skirycz et al., 2011a).
Transcriptome Response to Mild Drought in Six Accessions
In total, 354 genes were found to be differentially expressed similarly in all six accessions under mild drought. Despite the phenotypic differences between the accessions, which often can be explained by differences in regulation of the same genes (Chen et al., 2005; Delker et al., 2010), our analysis revealed few differences in the transcriptome response to the mild drought stress between the accessions. Only 60 genes, with functions in seemingly unrelated pathways, were found to have a drought response that differs between the accessions. None of these 60 genes showed a differential expression response that was specific for one accession only.
This small number of accession-specific genes was in contrast to a previous report (Des Marais et al., 2012) showing that, in mature leaves exposed to mild drought stress, there are many more genes accession-specifically expressed. A possible explanation of this discrepancy is that our experimental setup with RNA sequencing of microdissected young developing leaves introduced a certain amount of variation between biological repeats. This additional variation lowered the power for finding interactions and significant pairwise differences. Furthermore, it is also plausible that the transcriptional response to mild drought conditions in young developing leaves, containing mainly dividing cells, was more conserved than the response observed in mature leaves.
Therefore, we focused on the genes that were found to be common and differentially expressed under mild drought stress in the six accessions. These genes were involved in ABA signaling, Pro metabolism, and cell wall metabolism.
ABA Plays a Key Role in the Response of Soil-Grown Plants to Mild Drought Stress
The role of ABA in severe drought stress applied to mature plants is well known (Yamaguchi-Shinozaki and Shinozaki, 2006; Finkelstein, 2013). Also in the mild drought stress setup described here, numerous ABA signaling genes were found to be differentially expressed in early developing leaves exposed to mild drought stress. Among the ABA signaling genes were genes encoding ABA receptors (PYR/PYL/RCAR proteins), PP2Cs, and ABFs. The PYR/PYL/RCAR protein family contains 14 members that are all capable of activating signaling in response to ABA. Most members have been described as ABA receptors that, in the presence of ABA, inhibit PP2C through protein binding (for review, see Cutler et al., 2010; Umezawa et al., 2010). This, in turn, allows the accumulation of phosphorylated SUCROSE NONFERMENTING1-RELATED PROTEIN KINASE2s (SnRK2s) and subsequent phosphorylation of ABFs (Wang et al., 2013), which were shown to interact with DEHYDRATION-RESPONSIVE ELEMENT-BINDING PROTEIN (DREB)/C-REPEAT-BINDING PROTEINs such as DREB1A, DREB2A, and DREB2C, playing a central role in the drought response (Cutler et al., 2010; Lee et al., 2010). Mild drought stress imposed during early leaf development down-regulated the expression of six ABA receptor-encoding genes (PYR1, PYL1, PYL4, PYL5, PYL6, and PYL8). PYR1 and PYL1 are able to inhibit PP2Cs only in the presence of ABA, whereas PYL4, PYL5, PYL6, and PYL8 can inhibit PP2Cs in an ABA-independent way (Miyakawa et al., 2013). The absence of differentially expressed ABA biosynthesis genes such as 9-CIS-EPOXYCAROTENOID DIOXYGENASE (Tan et al., 2003), together with the down-regulation of the receptors and up-regulation of six PP2Cs (ABI1, ABI2, HAI1, HAI2, AHG3, and HAB1) as inhibitors of ABA signaling, might reflect a negative feedback on the ABA response. Yet, two of the four main ABFs (ABF2/AREB1 and ABF3; Yoshida et al., 2014) and six of the ABF target genes (AFP1, AHG3, HAI1, KCS2, RD20, and AT5G53390; Yoshida et al., 2010, 2014) were up-regulated, suggesting that the ABA response is still active despite such a putative negative feedback. In addition to these six ABF target genes, 37 other ABA-induced genes were differentially expressed in our data set, confirming the importance of ABA in the transcriptome response to mild drought stress also in proliferating leaves. The SnRK2 genes were not found to be differentially expressed, which is not surprising, since their activity is regulated through phosphorylation (Finkelstein, 2013).
One of the biochemical pathways controlled by ABA is Pro metabolism. Pro acts as an osmolyte, but it also functions as osmoprotectant and reactive oxygen species detoxifier. In addition, Pro has been suggested to have a regulatory role in mitochondrial functioning, cell proliferation, cell death, and the expression of rehydration-inducible genes (for review, see Szabados and Savouré, 2010). In our data, we found that the Pro biosynthesis gene P5CS1 was up-regulated, whereas the catabolic Pro dehydrogenase-encoding gene (PDH1/ERD5/POX) was down-regulated. This is in agreement with previous observations of increased Pro levels in response to drought stress (Verslues and Sharma, 2010).
Ethylene is one of the hormones involved in the biotic stress response and was found to play a role in the growth inhibitory response to low concentrations of mannitol administered to in vitro-grown plants (Verelst et al., 2010; Dubois et al., 2013). This treatment was generally thought to trigger osmotic stress, but the recent identification of specific mannitol receptors changed this view. Mannitol is now thought to trigger a biotic stress response besides its function as osmoticum (Claeys et al., 2014b; Trontin et al., 2014). Nevertheless, after mannitol treatment, the transcription factor ERF6 triggers, in an ACC-dependent manner, the expression of GA2-OXIDASE6 (GA2-OX6). Increased expression of GA2-OX6 leads to GA inactivation and reduced GA levels, in turn stabilizing the DELLA proteins, which will inhibit leaf growth (Dubois et al., 2013). The transcriptome analysis of developing leaves exposed to mild drought stress provided no evidence for the up-regulation of the ethylene signaling pathway. On the contrary, down-regulation of the expression of the genes encoding ACC oxidases (ACO2 and ACO4), one putative ACC oxidase (AT2G25450), and four ERFs (ERF2, ERF15, AT5G07580, and AT5G61590) was observed. Similar results were found in leaf tissue by Baerenfaller and colleagues (2012), who found that genes encoding two ACC oxidases (ACO2 and ACO5) and two ACC synthases (ACS10 and ACS11) were down-regulated, whereas two other genes encoding ACC synthases (ACS8 and ACS12) were up-regulated upon drought stress. However, as in the mannitol response, there was a clear growth reduction. Thus, other growth-regulating mechanisms must be at work during mild drought stress.
The general down-regulation of ethylene biosynthesis and signaling, and the suggested active role of ABA in the mild drought response, are in agreement with a proposed antagonism between both hormones. ABA and ethylene show contrasting effects on a number of processes, such as stomatal aperture (Tanaka et al., 2005), hyponastic growth (Benschop et al., 2005), seed germination (Beaudoin et al., 2000; Ghassemian et al., 2000), and defense gene expression and disease resistance (Anderson et al., 2004; De Paepe et al., 2004). Both hormones are suggested to antagonistically regulate each other’s metabolism and signaling (Cheng et al., 2009). Our data suggest that this also might be the case in response to mild drought stress.
DELLA proteins are known to play a role in stress-induced mitotic exit and cell differentiation (for review, see Claeys et al., 2014a). DELLA proteins are degraded in the presence of GA and are important players in GA signaling. In total, 43 putative DELLA targets were differentially expressed in all six accessions exposed to mild drought stress. DELLAs thus seem to play a role in the growth response of transitioning tissue under mild drought. Besides ethylene, GA and ABA also are known antagonists in different processes (Gómez-Cadenas et al., 2001; Ye and Zhang, 2012; Lü et al., 2014). Under mild drought, ABA might inhibit the GA signaling pathway, leading to DELLA stabilization and thus a reduction in growth. Further research is required to elucidate the molecular mechanism of this antagonism under mild drought stress.
Cell Wall Adjustments
In drought conditions, cell walls of organs and tissues that need to maintain their growth are loosened to keep them in a growth-ready state, while other tissues stiffen their cell walls (Wu and Cosgrove, 2000; Moore et al., 2008). In total, 21 genes involved in cell wall modifications were differentially expressed in young proliferating leaves exposed to mild drought stress. The up-regulated genes encode the cell wall-loosening expansins, pectin lyases, and pectin methylesterase inhibitors. On the other hand, genes encoding XTHs and fasciclin-like arabinogalactans, both involved in cell wall strengthening (Cosgrove, 2005; MacMillan et al., 2010), were down-regulated. The finding that mild drought stress results in the up-regulation of genes involved in cell wall loosening and the down-regulation of genes encoding cell wall-strengthening enzymes is an indication that, in this condition, the growing leaf tissue is kept in a growth-ready state. Moreover, since the sampled tissue was transitioning from proliferation to expansion, it is likely that under mild drought the cells are preparing for expansion earlier in comparison with control conditions, resulting in a differential up-regulation of cell wall-loosening enzymes. The earlier preparation for expansion, and thus the earlier arrest of cell proliferation, might be the driving force for the reduction in cell number observed under mild drought. Up-regulation of cell wall-loosening enzymes under mild drought also allows for growth with less turgor pressure and might facilitate cell expansion after rewatering (Lechner et al., 2008).
Central Players in the Drought-Regulated Network and New Drought-Responsive Genes
Besides known stress-related genes, more than half of the differentially expressed genes have no previously described function in drought responses. However, coexpression analysis revealed that a majority of the genes were coexpressed in a large set of microarray data sets. A further examination of the coexpression network allowed for the identification of genes connected to up to 37 other genes either by coexpression or regulatory interactions.
The most connected gene or hub in the obtained network encoded MPK3 and could potentially be an important regulator in the mild drought response. MPK3 has been shown to be involved in the regulation of the biotic stress response (Nakagami et al., 2005; Pedley and Martin, 2005; Beckers et al., 2009). In both transcriptional (through WRKY33) and posttranscriptional ways, MPK3 is able to induce and maintain ethylene biosynthesis (Li et al., 2012). Furthermore, MPK3/MPK6 is capable of phosphorylating ERF6 (Meng et al., 2013), a central player in the ethylene-regulated growth response to stress (Dubois et al., 2013). Under mild drought, we found that the expression of MPK3 was down-regulated specifically in young developing tissues in all six accessions, in agreement with the observed down-regulation of ethylene biosynthesis and signaling. In addition, MPK3 has been suggested to be a mediator of the MITOGEN-ACTIVATED PROTEIN KINASE KINASE4-induced osmotic stress response (Kim et al., 2011). In conclusion, our data suggest that MPK3 could play a central role in the growth response to mild drought stress.
Another highly connected gene in the network was LOX2, which is required for JA synthesis (Bell et al., 1995), a hormone involved in pathogen defense, wounding response, and plant growth (Zhang and Turner, 2008). The reduction in plant growth upon JA treatment is caused by an arrest at the G2 phase of the cell cycle (Świątek et al., 2002, 2004; Pauwels et al., 2008). Recently, LOX2 was shown to be under antagonistic control of TEOSINTE BRANCHED1/CYCLOIDEA/PROLIFERATING CELL FACTOR4 (TCP4) and TCP20. TCP4, similar to the rest of the class II TCPs, is regulated by JAGGED AND WAVY (Palatnik et al., 2003) and is known to cause a decrease in cell proliferation resulting in smaller leaves when hyperactivated (Sarvepalli and Nath, 2011). TCP20 (a class I TCP), on the other hand, is suggested to stimulate the cell cycle and organ growth through binding to the regulatory sequences of CYCLIN B1;1, PROLIFERATING CELL NUCLEAR ANTIGEN, and ribosomal genes (Li et al., 2005). The class I and class II TCP transcription factors are proposed to regulate leaf development through JA as an intermediary hormonal signal. According to this model, JA levels would increase during leaf development and, as such, induce the transition from proliferation to expansion (Danisman et al., 2012). Thus, the up-regulation of LOX2 under mild drought might lead to increased JA concentrations and to an earlier transition to cell expansion compared with control conditions. This faster transition is in agreement with the response of the genes encoding cell wall modifiers. A faster transition might explain the reduced cell number and, thus, part of the size reduction induced by mild drought stress. The importance of JA for the growth reduction is strengthened by the observation that JA-insensitive mutants (coronatine-insensitive1 and jamsonate-insensitive1) lack growth reduction under mild drought (Harb et al., 2010).
Strikingly, the second and eighth most interacting genes in the coexpression and regulatory interaction network are of unknown function (AT4G36500 and AT5G44580) and are interesting candidates for further functional characterization.
The Specific Response of Young Developing Leaf Tissue to Mild Drought Stress
With a large number of studies focusing on severe drought and the awareness that tolerance to severe drought does not necessarily mean better growth under mild drought (Skirycz et al., 2011b), the question remains how specific the response of plants to these two different manifestations of water deficit is.
A bit more than half of the common drought genes found here to be transcriptionally altered by mild drought also have been reported previously to be affected by severe drought. Many of the genes affected by both mild and severe drought have a role in ABA perception and signaling. Also, Harb et al. (2010) found a strong overlap for ABA-related genes between mild and severe drought in mature leaves. Here, we show that ABA also plays a role in the response of young developing leaves to mild drought. A striking difference between the response of plants to mild or severe drought stress, however, is the behavior of genes encoding cell wall-modifying proteins. Whereas genes encoding XTHs showed the same expression behavior in severe and mild drought stress, genes encoding expansins were, opposite to mild drought stress, down-regulated in severe drought. A plausible explanation is that in severe drought, no growth can be allowed, whereas in mild drought conditions, the expansins allow cell growth under reduced turgor pressure.
Of particular interest are the 87 genes specific for young developing leaf tissue exposed to mild drought. Twelve of the 87 genes have no known function and are interesting candidates for further functional analysis. Another 10 genes encode proteins that are involved in cell wall synthesis, loosening, or remodeling: four pectin lyases (AT1G60590, AT1G67750, AT3G61490, and AT4G13710), two cellulose synthase-like proteins (CSLB03 and CSLC6), one expansin (EXPA15), one extensin (EXT3), one arabinogalactan protein (AGP9), and one pectin methylesterase inhibitor (AT1G23205). All these genes are up-regulated specifically in young developing tissue under mild drought. Although growth is reduced, loosening the cell wall allows a reduced growth under lower turgor evoked by the mild drought.
Three genes encoding transcription factors, a myb-like protein (AT5G56840), BASIC HELIX-LOOP-HELIX42, and TEMPRANILLO1 (TEM1), are found among the genes that are differently expressed in young developing leaves exposed to mild drought. TEM1 has been shown to directly repress GA3-OX1 and GA3-OX2 expression (Osnato et al., 2012), both involved in one of the main bioactive GA-forming steps. Here, a down-regulation of TEM1 was noticed, which would allow an increase in GA, which might antagonize the observed ABA response. Other potential regulators specific for developing leaves are MPK3 and CYP78A7. As detailed above, MPK3 is the most central player in the coexpression and regulatory interactions network of mild drought stress-responsive genes.
CYP78A7 and CYP78A5/KLUH encode cytochrome P450 proteins and have redundant roles in positively regulating the relative growth of the shoot apical meristem and developing leaves and flowers (Anastasiou et al., 2007; Wang et al., 2008; Eriksson et al., 2010). The observed up-regulation of CYP78A7 is seemingly in disagreement with the coinciding growth reduction. However, to maintain growth under mild drought at reduced levels, there probably is a complex network of both growth promoters and growth inhibitors forcing each other into a certain balance.
In conclusion, the use of different accessions allowed for the detection of a robust set of genes that play a role in the mild drought response in different genetic backgrounds. Whereas some of these genes also were found to be altered in response to severe drought stress or in the response to mild drought in mature tissue, a unique set of 87 genes was found to specifically play a role in the response to mild drought in young developing leaves.
MATERIALS AND METHODS
Plant Growth and Experimental Setup
Six Arabidopsis (Arabidopsis thaliana) accessions (An-1, Blh-1, Col-0, Cvi-0, Oy-0, and Sha) were grown in a growth chamber under controlled environmental conditions (21°C, 55% relative humidity, 16-h day/8-h night, and 110–120 μmol m−2 s−1 light intensity).
Seeds of Blh-1 were provided by Olivier Loudet (Institut National de la Recherche Agronomique). Other accessions were obtained from the Nottingham Arabidopsis Stock Centre (N22660). The decision to screen these six accessions was based on an article by Bouchabke et al. (2008), where these six accessions covered a large part of the variation in response to mild drought present in their collection of 24 accessions mainly for total leaf area, but electrolyte leakage, relative water content, and cut rosette water loss also were taken into account.
All accessions were bulked at the same time. The geographical origin of the six accessions is provided in Supplemental Table S1. Plants were germinated on soil-filled 96-well plates after 4 d of stratification at 4°C in the dark. The timing of transition was checked for each accession; therefore, Cvi-0 was put to germinate 1 d earlier, as it reached the transition phase of the third leaf 1 d later compared with the other accessions. Seedlings were transferred to pots at 4 DAS. Fifty-four average-sized seedlings were selected (Fig. 1A) using an in-house-developed image algorithm in order to reduce seedling size effects at the beginning of the experiment. Before transfer, the relative water content of the pots was set at 1.2 g water g−1 dry soil for the mild drought treatment; the control condition was set at 2.2 g water g−1 dry soil (Fig. 1D). Once the seedlings were transferred to pots (Fig. 1B), they were placed on the automated phenotyping platform WIWAM (Skirycz et al., 2011b). The water content of the soil was kept constant until 10 DAS, after which it was lowered daily to target 0.7 g water g−1 dry soil for the mild-drought-treated plants (Fig. 1D). Images of the rosette of each plant were taken daily until 21 DAS and analyzed for the PRA (Skirycz et al., 2011b).
Size measurements of the third leaf were done at the transition from cell proliferation to cell expansion (10–11 DAS) and at maturity (22 DAS). For practical reasons, the mature third leaf was harvested 1 d later than the last PRA measurement at 21 DAS. To this end, the leaves were cut from the rosette, cleared in ethanol, and transferred to lactic acid before mounting on microscope slides. Measurements based on microscope images were done using ImageJ (http://imagej.nih.gov/ij/), and analysis of drawings made from the abaxial epidermis allowed for quantification of the cell area, cell number, and stomatal index (Andriankaja et al., 2012).
Statistical Analysis of Phenotypic Measurements
All statistical analyses were performed with SAS/STAT software, version 9.4 of the SAS System for Windows 7 64bit. (SAS Institute; http://www.sas.com).
A mixed model was fitted to the leaf 3 area data, both at transition and maturity. The fixed effects part of the model contained the main effects genotype and treatment and their interaction. The replication factor experiment was included in the model as a random factor to account for the correlation between observations belonging to the same experiment. Type 3 F tests were performed to estimate the pairwise comparisons between the ecotypes at control and stress levels as well as the pairwise comparisons between the ecotypes of the decrease in leaf 3 area due to stress. P values were adjusted for multiple comparisons with the false discovery rate (FDR) implemented in the SAS multtest procedure. Effects were considered significant at an FDR threshold of 0.05.
Cellular measurements were analyzed like the leaf 3 area data. In the case of a significant interaction (at the 0.05 significance level) between genotype and treatment, simple F tests of effects were performed to estimate differences between the stress and control treatments for each genotype. In the absence of a significant interaction but the presence of significant main effects, all pairwise comparisons were calculated between the genotypes and between the treatments. P values were adjusted with Tukey’s method as implemented in SAS.
A longitudinal data analysis was conducted in SAS on the daily measured PRA. A mixed model was used including the main effects of genotype, treatment, time, and all two-way and three-way interaction terms. The PRA was log transformed to stabilize the variance. Various covariance structures were tested to model the correlations between measurements done on the same plant. The Toeplitz covariance structure was selected based on Akaike’s information criterion. The replication factor experiment was included in the model as a random factor. The Kenward-Rogers approximation for computing the denominator degrees of freedom for the tests of fixed effects was used (Schaalje et al., 2001). The appropriate contrasts were defined in the estimate statement to conduct pairwise comparisons for the different factors and final time point.
Type 3 F tests were used to test significant differences. P values were adjusted for multiple comparisons with the FDR. An effect was considered significant at an FDR threshold of 0.05. Residual diagnostics were carefully examined.
Transcriptome Analysis
Sampling
To ensure sufficient material for transcriptome analysis, 60 seedlings were grown per accession per treatment. Plants were harvested at 10 DAS expect for Cvi-0, which was harvested at 11 DAS. Plants were flash frozen in liquid nitrogen immediately upon harvest. RNAlater-ICE (Ambion), which prevents RNA from degrading, was cooled at −70°C, added to the samples, and allowed to penetrate the tissue at −20°C for 5 d. The third leaf was collected by microdissection with a microscope. Samples were microdissected in a petri dish on dry ice to keep the samples below room temperature. Dissected leaves were ground with a Retsch machine and 3-mm metal balls. Samples were obtained from three independent biological repeats.
RNA Extraction
RNA was extracted with Trizol (Invitrogen) according to the manufacturer’s instructions. RNA samples were subjected to DNA digestion with the RNase-free DNase I kit (Qiagen), and, subsequently, impurities were removed with the RNeasy mini kit (Qiagen).
RNA Sequencing
Library preparation was done using the TruSeq RNA Sample Preparation Kit version 2 (Illumina). In brief, poly(A)-containing mRNA molecules are reverse transcribed, double-stranded complementary DNA is generated, and adapters are ligated. After quality control using the 2100 Bioanalyzer (Agilent), clusters are generated through amplification using the TruSeq PE Cluster Kit v3-cBot-HS (Illumina) followed by sequencing on the Illumina HiSeq2000 with the TruSeq SBS Kit v3-HS (Illumina). Sequencing was performed in paired-end mode with a read length of 100 bp.
The quality of the raw data was verified with FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/; version 0.9.1). Next, quality filtering was performed using the FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/; version 0.0.13): reads were globally filtered in which, for at least 75% of the reads, the quality exceeds Q20, and 3′ trimming was performed to remove bases with a quality below Q10, ensuring a minimum length of 90 bp remaining. Repair was performed using a custom Perl script. Reads were subsequently mapped to the Arabidopsis reference genome (The Arabidopsis Information Resource 10) using GSNAP version 2012-07-20 (Wu and Nacu, 2010), allowing maximally five mismatches. The concordantly paired reads that uniquely map to the genome were used for quantification at the gene level with htseq-count from the HTSeq.py python package (Anders et al., 2014). Data were normalized using TMM, implemented in the edgeR package (version 3.5.27) in R (version 3.1.0; Robinson et al., 2010; McCarthy et al., 2012).
Differential Expression Analysis
The samples were harvested in four different batches. The transcriptomes for both control and mild drought stress were observed for each accession within a batch. Gene-wise RNA sequencing counts were analyzed using a negative binomial model with batch, accession, and treatment main effects, an accession × treatment interaction, and a tag-wise overdispersion parameter for each gene.
We aimed to discover genes for which the drought stress response is accession specific as well as genes with a common drought-induced differential expression pattern in the different accessions. The former genes can be identified by testing for changes in stress-induced differential expression between accessions. Our design, however, implies 15 pairwise differential expression comparisons for each gene. It is well known that high-throughput experiments with a huge number of simultaneous hypothesis tests typically have a low power for detecting significant multiple treatment effects (Jiang and Doerge, 2006). Similar to Jiang and Doerge (2006), we propose a stepwise procedure for increasing the power to detect accession-specific differentially expressed genes between control and mild drought. In stage I, the null hypothesis of no differential expression between control and mild drought is tested against the alternative hypothesis that there is differential expression in at least one accession (test I.a). The list with significant I.a genes will be enriched for genes with a drought stress-induced expression response that is accession specific. Significant I.a genes are tested for a drought stress × accession interaction in stage II. Finally, stage II genes with a significant interaction are further dissected in stage III by assessing all multiple pairwise comparisons of mild drought stress-induced differential expression between the different accessions. Genes with a common differential expression between mild drought and control can be prioritized by assessing the average differential expression between mild drought stress and control over all accessions. This involves only one contrast for each gene and thus can be assessed using a single-stage hypothesis test, which is referred to as test I.c and will complement test I.a in stage I. Five of the genes detected in test I.c also appeared in the stage II list of genes with accession × treatment interaction and were removed from the downstream analysis.
We follow the same rationale as Jiang and Doerge (2006) to control the joint FDR over all three stages of the proposed test procedure. They showed that the overall FDR in a stepwise multiple comparison procedure can be controlled below the prespecified FDR significance level α when the sum of the FDR significance levels used in each of the different stages is below α. They also showed that it is advantageous to choose α1 in the first stage larger than the remaining stages, so that more genes can enter stage II. Here, the overall FDR over tests I.a and I.c in the first stage is controlled at 4%, while the FDR in stage II and III is set at 0.5%. All analyses were conducted with the edgeR package (version 3.5.27) in R (version 3.1.0; Robinson et al., 2010; McCarthy et al., 2012).
GO Enrichment
GO enrichment analyses were conducted with BiNGO (Maere et al., 2005).
Coexpression Analysis
The online tool CORNET (https://cornet.psb.ugent.be/; De Bodt et al., 2010) was used to perform coexpression analysis. Four sets of microarray experiments were selected: Compendium 2 (a global collection of various microarray experiments without bias to specific conditions or tissues), Leaf (microarray experiments of leaf tissue), Development (microarray experiments from different tissues, developmental stages, and developmental mutants), and Stress (microarray experiments under various biotic and abiotic stresses). A Pearson correlation cutoff of 0.7 was used for the coexpression analysis. Only coexpression between the 354 common drought genes was allowed. The four obtained coexpression networks were merged and visualized in Cytoscape (Cline et al., 2007).
Regulatory interactions were retrieved with the TF tool in CORNET. Interactions between query genes and neighbors were included. From the AGRIS database, only confirmed interactions were used. Computationally inferred regulatory interactions from microarray data were added to the network together with regulatory interactions retrieved from the literature using the EVEX text-mining tool (Van Landeghem et al., 2012). The network of regulatory interactions was merged with the coexpression network using Cytoscape.
Transcriptome (RNA-Sequencing) data from this article can be found in the ArrayExpress data libraries under accession number E-MTAB-3279.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Overview of the experimental setup.
Supplemental Figure S2. Schematic overview of the three-stage statistical analysis.
Supplemental Figure S3. Expression profiles of the 60 accession-specific genes.
Supplemental Figure S4. Expression profiles of the 354 common drought genes.
Supplemental Figure S5. Venn diagram showing overlap between common drought genes and the selected severe drought studies.
Supplemental Figure S6. Venn diagram showing overlap between common drought genes and the selected mild drought stress studies in mature tissue.
Supplemental Table S1. Geographic origins of the six Arabidopsis accessions.
Supplemental Table S2. Pairwise comparisons of projected rosette area and leaf 3 size measurements.
Supplemental Table S3. List of differential expressed genes.
Supplemental Table S4. List of enriched GO categories.
Supplemental Table S5. Common drought genes previously associated with ABA.
Supplemental Table S6. Putative DELLA targets among the common drought genes.
Supplemental Table S7. Common drought genes not previously annotated to be drought responsive.
Supplemental Table S8. Top 20 interactors from the coexpression and transcriptional regulation network.
Supplemental Table S9. Eighty-seven genes specific for young developing tissue exposed to mild drought.
Supplementary Material
Acknowledgments
We thank the entire Systems Biology of Yield group for creating a stimulating scientific environment with many fruitful discussions. We also thank Dr. Annick Bleys for help in improving the article and Marieke Dubois for fruitful discussions.
Glossary
- ABA
abscisic acid
- DAS
days after stratification
- PRA
projected rosette area
- Col-0
Columbia-0
- GO
Gene Ontology
- ACC
1-aminocyclopropane-1-carboxylic acid
- XTH
xyloglucan endotransglucosylase/hydrolase
- ABF
abscisic acid-responsive element-binding factor
- PP2C
type 2C protein phosphatase
- JA
jasmonic acid
- FDR
false discovery rate
Footnotes
This work was supported by the European Research Council under the European Union’s Seventh Framework Programme (grant no. FP7/2007-2013, European Research Council grant agreement no. [339341-AMAIZE]11), by Ghent University (Bijzonder Onderzoeksfonds Methusalem grant no. BOF08/01M00408 and Multidisciplinary Research Partnership Biotechnology for a Sustainable Economy grant no. 01MRB510W), by the Interuniversity Attraction Poles Programme (grant no. IUAP P7/29 “MARS”) initiated by the Belgian Science Policy Office, by the Agency for Innovation by Science and Technology in Flanders (predoctoral fellowship to P.C.), by the Multidisciplinary Research Partnership Bioinformatics: From Nucleotide to Networks (grant no. 01MR0310W from Ghent University to L.C.), and by the Research Foundation Flanders (postdoctoral fellowship to S.D.).
Articles can be viewed without a subscription.
References
- Achard P, Cheng H, De Grauwe L, Decat J, Schoutteten H, Moritz T, Van Der Straeten D, Peng J, Harberd NP (2006) Integration of plant responses to environmentally activated phytohormonal signals. Science 311: 91–94 [DOI] [PubMed] [Google Scholar]
- Achard P, Gong F, Cheminant S, Alioua M, Hedden P, Genschik P (2008) The cold-inducible CBF1 factor-dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell 20: 2117–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguirrezabal L, Bouchier-Combaud S, Radziejwoski A, Dauzat M, Cookson SJ, Granier C (2006) Plasticity to soil water deficit in Arabidopsis thaliana: dissection of leaf development into underlying growth dynamic and cellular variables reveals invisible phenotypes. Plant Cell Environ 29: 2216–2227 [DOI] [PubMed] [Google Scholar]
- Akhtar M, Jaiswal A, Taj G, Jaiswal JP, Qureshi MI, Singh NK (2012) DREB1/CBF transcription factors: their structure, function and role in abiotic stress tolerance in plants. J Genet 91: 385–395 [DOI] [PubMed] [Google Scholar]
- Anastasiou E, Kenz S, Gerstung M, MacLean D, Timmer J, Fleck C, Lenhard M (2007) Control of plant organ size by KLUH/CYP78A5-dependent intercellular signaling. Dev Cell 13: 843–856 [DOI] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W (2015) HTSeq: a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JP, Badruzsaufari E, Schenk PM, Manners JM, Desmond OJ, Ehlert C, Maclean DJ, Ebert PR, Kazan K (2004) Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell 16: 3460–3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andriankaja M, Dhondt S, De Bodt S, Vanhaeren H, Coppens F, De Milde L, Mühlenbock P, Skirycz A, Gonzalez N, Beemster GTS, et al. (2012) Exit from proliferation during leaf development in Arabidopsis thaliana: a not-so-gradual process. Dev Cell 22: 64–78 [DOI] [PubMed] [Google Scholar]
- Baerenfaller K, Massonnet C, Walsh S, Baginsky S, Bühlmann P, Hennig L, Hirsch-Hoffmann M, Howell KA, Kahlau S, Radziejwoski A, et al. (2012) Systems-based analysis of Arabidopsis leaf growth reveals adaptation to water deficit. Mol Syst Biol 8: 606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin N, Serizet C, Gosti F, Giraudat J (2000) Interactions between abscisic acid and ethylene signaling cascades. Plant Cell 12: 1103–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckers GJM, Jaskiewicz M, Liu Y, Underwood WR, He SY, Zhang S, Conrath U (2009) Mitogen-activated protein kinases 3 and 6 are required for full priming of stress responses in Arabidopsis thaliana. Plant Cell 21: 944–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell E, Creelman RA, Mullet JE (1995) A chloroplast lipoxygenase is required for wound-induced jasmonic acid accumulation in Arabidopsis. Proc Natl Acad Sci USA 92: 8675–8679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benschop JJ, Jackson MB, Gühl K, Vreeburg RAM, Croker SJ, Peeters AJM, Voesenek LACJ (2005) Contrasting interactions between ethylene and abscisic acid in Rumex species differing in submergence tolerance. Plant J 44: 756–768 [DOI] [PubMed] [Google Scholar]
- Bergmann DC, Sack FD (2007) Stomatal development. Annu Rev Plant Biol 58: 163–181 [DOI] [PubMed] [Google Scholar]
- Bouchabke O, Chang F, Simon M, Voisin R, Pelletier G, Durand-Tardif M (2008) Natural variation in Arabidopsis thaliana as a tool for highlighting differential drought responses. PLoS ONE 3: e1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chardon F, Barthélémy J, Daniel-Vedele F, Masclaux-Daubresse C (2010) Natural variation of nitrate uptake and nitrogen use efficiency in Arabidopsis thaliana cultivated with limiting and ample nitrogen supply. J Exp Bot 61: 2293–2302 [DOI] [PubMed] [Google Scholar]
- Chen WJ, Chang SH, Hudson ME, Kwan WK, Li J, Estes B, Knoll D, Shi L, Zhu T (2005) Contribution of transcriptional regulation to natural variations in Arabidopsis. Genome Biol 6: R32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Zhang H, Jablonowski D, Zhou X, Ren X, Hong X, Schaffrath R, Zhu JK, Gong Z (2006) Mutations in ABO1/ELO2, a subunit of holo-Elongator, increase abscisic acid sensitivity and drought tolerance in Arabidopsis thaliana. Mol Cell Biol 26: 6902–6912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng WH, Chiang MH, Hwang SG, Lin PC (2009) Antagonism between abscisic acid and ethylene in Arabidopsis acts in parallel with the reciprocal regulation of their metabolism and signaling pathways. Plant Mol Biol 71: 61–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong YH, Kim KN, Pandey GK, Gupta R, Grant JJ, Luan S (2003) CBL1, a calcium sensor that differentially regulates salt, drought, and cold responses in Arabidopsis. Plant Cell 15: 1833–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong YH, Pandey GK, Grant JJ, Batistic O, Li L, Kim BG, Lee SC, Kudla J, Luan S (2007) Two calcineurin B-like calcium sensors, interacting with protein kinase CIPK23, regulate leaf transpiration and root potassium uptake in Arabidopsis. Plant J 52: 223–239 [DOI] [PubMed] [Google Scholar]
- Claeys H, De Bodt S, Inzé D (2014a) Gibberellins and DELLAs: central nodes in growth regulatory networks. Trends Plant Sci 19: 231–239 [DOI] [PubMed] [Google Scholar]
- Claeys H, Inzé D (2013) The agony of choice: how plants balance growth and survival under water-limiting conditions. Plant Physiol 162: 1768–1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claeys H, Skirycz A, Maleux K, Inzé D (2012) DELLA signaling mediates stress-induced cell differentiation in Arabidopsis leaves through modulation of anaphase-promoting complex/cyclosome activity. Plant Physiol 159: 739–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claeys H, Van Landeghem S, Dubois M, Maleux K, Inzé D (2014b) What is stress? Dose-response effects in commonly used in vitro stress assays. Plant Physiol 165: 519–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B, et al. (2007) Integration of biological networks and gene expression data using Cytoscape. Nat Protoc 2: 2366–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cominelli E, Galbiati M, Vavasseur A, Conti L, Sala T, Vuylsteke M, Leonhardt N, Dellaporta SL, Tonelli C (2005) A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Curr Biol 15: 1196–1200 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. (2005) Growth of the plant cell wall. Nat Rev Mol Cell Biol 6: 850–861 [DOI] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR (2010) Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol 61: 651–679 [DOI] [PubMed] [Google Scholar]
- Danisman S, van der Wal F, Dhondt S, Waites R, de Folter S, Bimbo A, van Dijk ADJ, Muino JM, Cutri L, Dornelas MC, et al. (2012) Arabidopsis class I and class II TCP transcription factors regulate jasmonic acid metabolism and leaf development antagonistically. Plant Physiol 159: 1511–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davuluri RV, Sun H, Palaniswamy SK, Matthews N, Molina C, Kurtz M, Grotewold E (2003) AGRIS: Arabidopsis Gene Regulatory Information Server, an information resource of Arabidopsis cis-regulatory elements and transcription factors. BMC Bioinformatics 4: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bodt S, Carvajal D, Hollunder J, Van den Cruyce J, Movahedi S, Inzé D (2010) CORNET: a user-friendly tool for data mining and integration. Plant Physiol 152: 1167–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bodt S, Hollunder J, Nelissen H, Meulemeester N, Inzé D (2012) CORNET 2.0: integrating plant coexpression, protein-protein interactions, regulatory interactions, gene associations and functional annotations. New Phytol 195: 707–720 [DOI] [PubMed] [Google Scholar]
- Delker C, Pöschl Y, Raschke A, Ullrich K, Ettingshausen S, Hauptmann V, Grosse I, Quint M (2010) Natural variation of transcriptional auxin response networks in Arabidopsis thaliana. Plant Cell 22: 2184–2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Paepe A, Vuylsteke M, Van Hummelen P, Zabeau M, Van Der Straeten D (2004) Transcriptional profiling by cDNA-AFLP and microarray analysis reveals novel insights into the early response to ethylene in Arabidopsis. Plant J 39: 537–559 [DOI] [PubMed] [Google Scholar]
- Des Marais DL, McKay JK, Richards JH, Sen S, Wayne T, Juenger TE (2012) Physiological genomics of response to soil drying in diverse Arabidopsis accessions. Plant Cell 24: 893–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly PM, Bonetta D, Tsukaya H, Dengler RE, Dengler NG (1999) Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev Biol 215: 407–419 [DOI] [PubMed] [Google Scholar]
- Dubois M, Skirycz A, Claeys H, Maleux K, Dhondt S, De Bodt S, Vanden Bossche R, De Milde L, Yoshizumi T, Matsui M, et al. (2013) Ethylene Response Factor6 acts as a central regulator of leaf growth under water-limiting conditions in Arabidopsis. Plant Physiol 162: 319–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson S, Stransfeld L, Adamski NM, Breuninger H, Lenhard M (2010) KLUH/CYP78A5-dependent growth signaling coordinates floral organ growth in Arabidopsis. Curr Biol 20: 527–532 [DOI] [PubMed] [Google Scholar]
- Finkelstein R. (2013) Abscisic acid synthesis and response. The Arabidopsis Book 11: e0166, doi/10.1199/tab.0166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler M, Nadeau J, Sack FD (2000) Oriented asymmetric divisions that generate the stomatal spacing pattern in Arabidopsis are disrupted by the too many mouths mutation. Plant Cell 12: 2075–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghassemian M, Nambara E, Cutler S, Kawaide H, Kamiya Y, McCourt P (2000) Regulation of abscisic acid signaling by the ethylene response pathway in Arabidopsis. Plant Cell 12: 1117–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golldack D, Li C, Mohan H, Probst N (2014) Tolerance to drought and salt stress in plants: unraveling the signaling networks. Front Plant Sci 5: 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Cadenas A, Zentella R, Walker-Simmons MK, Ho THD (2001) Gibberellin/abscisic acid antagonism in barley aleurone cells: site of action of the protein kinase PKABA1 in relation to gibberellin signaling molecules. Plant Cell 13: 667–679 [PMC free article] [PubMed] [Google Scholar]
- Gonzalez N, Vanhaeren H, Inzé D (2012) Leaf size control: complex coordination of cell division and expansion. Trends Plant Sci 17: 332–340 [DOI] [PubMed] [Google Scholar]
- Harb A, Krishnan A, Ambavaram MMR, Pereira A (2010) Molecular and physiological analysis of drought stress in Arabidopsis reveals early responses leading to acclimation in plant growth. Plant Physiol 154: 1254–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann MH. (2002) Biogeography of Arabidopsis thaliana (L.) Heynh. (Brassicaceae). J Biogeogr 29: 125–134 [Google Scholar]
- Huang D, Wu W, Abrams SR, Cutler AJ (2008) The relationship of drought-related gene expression in Arabidopsis thaliana to hormonal and environmental factors. J Exp Bot 59: 2991–3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S, Kobayashi M, Taji T, Naramoto M, Seki M, Kato T, Tabata S, Kakubari Y, Yamaguchi-Shinozaki K, Shinozaki K (2001) Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J 27: 325–333 [DOI] [PubMed] [Google Scholar]
- Jiang H, Doerge RW (2006) A two-step multiple comparison procedure for a large number of tests and multiple treatments. Stat Appl Genet Mol Biol 5: e28. [DOI] [PubMed] [Google Scholar]
- Katori T, Ikeda A, Iuchi S, Kobayashi M, Shinozaki K, Maehashi K, Sakata Y, Tanaka S, Taji T (2010) Dissecting the genetic control of natural variation in salt tolerance of Arabidopsis thaliana accessions. J Exp Bot 61: 1125–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Woo DH, Kim JM, Lee SY, Chung WS, Moon YH (2011) Arabidopsis MKK4 mediates osmotic-stress response via its regulation of MPK3 activity. Biochem Biophys Res Commun 412: 150–154 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Alonso-Blanco C, Vreugdenhil D (2004) Naturally occurring genetic variation in Arabidopsis thaliana. Annu Rev Plant Biol 55: 141–172 [DOI] [PubMed] [Google Scholar]
- Lawlor DW. (2013) Genetic engineering to improve plant performance under drought: physiological evaluation of achievements, limitations, and possibilities. J Exp Bot 64: 83–108 [DOI] [PubMed] [Google Scholar]
- Lechner L, Pereyra-Irujo GA, Granier C, Aguirrezábal LAN (2008) Rewatering plants after a long water-deficit treatment reveals that leaf epidermal cells retain their ability to expand after the leaf has apparently reached its final size. Ann Bot (Lond) 101: 1007–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Kang JY, Park HJ, Kim MD, Bae MS, Choi HI, Kim SY (2010) DREB2C interacts with ABF2, a bZIP protein regulating abscisic acid-responsive gene expression, and its overexpression affects abscisic acid sensitivity. Plant Physiol 153: 716–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Potuschak T, Colón-Carmona A, Gutiérrez RA, Doerner P (2005) Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proc Natl Acad Sci USA 102: 12978–12983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Meng X, Wang R, Mao G, Han L, Liu Y, Zhang S (2012) Dual-level regulation of ACC synthase activity by MPK3/MPK6 cascade and its downstream WRKY transcription factor during ethylene induction in Arabidopsis. PLoS Genet 8: e1002767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorente F, López-Cobollo RM, Catalá R, Martínez-Zapater JM, Salinas J (2002) A novel cold-inducible gene from Arabidopsis, RCI3, encodes a peroxidase that constitutes a component for stress tolerance. Plant J 32: 13–24 [DOI] [PubMed] [Google Scholar]
- Lü P, Zhang C, Liu J, Liu X, Jiang G, Jiang X, Khan MA, Wang L, Hong B, Gao J (2014) RhHB1 mediates the antagonism of gibberellins to ABA and ethylene during rose (Rosa hybrida) petal senescence. Plant J 78: 578–590 [DOI] [PubMed] [Google Scholar]
- MacMillan CP, Mansfield SD, Stachurski ZH, Evans R, Southerton SG (2010) Fasciclin-like arabinogalactan proteins: specialization for stem biomechanics and cell wall architecture in Arabidopsis and Eucalyptus. Plant J 62: 689–703 [DOI] [PubMed] [Google Scholar]
- Maere S, Heymans K, Kuiper M (2005) BiNGO: a Cytoscape plugin to assess overrepresentation of Gene Ontology categories in biological networks. Bioinformatics 21: 3448–3449 [DOI] [PubMed] [Google Scholar]
- Magome H, Yamaguchi S, Hanada A, Kamiya Y, Oda K (2008) The DDF1 transcriptional activator upregulates expression of a gibberellin-deactivating gene, GA2ox7, under high-salinity stress in Arabidopsis. Plant J 56: 613–626 [DOI] [PubMed] [Google Scholar]
- Matsui A, Ishida J, Morosawa T, Mochizuki Y, Kaminuma E, Endo TA, Okamoto M, Nambara E, Nakajima M, Kawashima M, et al. (2008) Arabidopsis transcriptome analysis under drought, cold, high-salinity and ABA treatment conditions using a tiling array. Plant Cell Physiol 49: 1135–1149 [DOI] [PubMed] [Google Scholar]
- McCarthy DJ, Chen Y, Smyth GK (2012) Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 40: 4288–4297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell N, Pockman WT, Allen CD, Breshears DD, Cobb N, Kolb T, Plaut J, Sperry J, West A, Williams DG, et al. (2008) Mechanisms of plant survival and mortality during drought: why do some plants survive while others succumb to drought? New Phytol 178: 719–739 [DOI] [PubMed] [Google Scholar]
- Meng X, Xu J, He Y, Yang KY, Mordorski B, Liu Y, Zhang S (2013) Phosphorylation of an ERF transcription factor by Arabidopsis MPK3/MPK6 regulates plant defense gene induction and fungal resistance. Plant Cell 25: 1126–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyakawa T, Fujita Y, Yamaguchi-Shinozaki K, Tanokura M (2013) Structure and function of abscisic acid receptors. Trends Plant Sci 18: 259–266 [DOI] [PubMed] [Google Scholar]
- Moore JP, Vicré-Gibouin M, Farrant JM, Driouich A (2008) Adaptations of higher plant cell walls to water loss: drought vs desiccation. Physiol Plant 134: 237–245 [DOI] [PubMed] [Google Scholar]
- Nakagami H, Pitzschke A, Hirt H (2005) Emerging MAP kinase pathways in plant stress signalling. Trends Plant Sci 10: 339–346 [DOI] [PubMed] [Google Scholar]
- Navarro L, Bari R, Achard P, Lisón P, Nemri A, Harberd NP, Jones JDG (2008) DELLAs control plant immune responses by modulating the balance of jasmonic acid and salicylic acid signaling. Curr Biol 18: 650–655 [DOI] [PubMed] [Google Scholar]
- Nelson DE, Repetti PP, Adams TR, Creelman RA, Wu J, Warner DC, Anstrom DC, Bensen RJ, Castiglioni PP, Donnarummo MG, et al. (2007) Plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proc Natl Acad Sci USA 104: 16450–16455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser JL, Hong F, Chory J (2006) Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126: 467–475 [DOI] [PubMed] [Google Scholar]
- Osnato M, Castillejo C, Matías-Hernández L, Pelaz S (2012) TEMPRANILLO genes link photoperiod and gibberellin pathways to control flowering in Arabidopsis. Nat Commun 3: 808. [DOI] [PubMed] [Google Scholar]
- Palaniswamy SK, James S, Sun H, Lamb RS, Davuluri RV, Grotewold E (2006) AGRIS and AtRegNet: a platform to link cis-regulatory elements and transcription factors into regulatory networks. Plant Physiol 140: 818–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D (2003) Control of leaf morphogenesis by microRNAs. Nature 425: 257–263 [DOI] [PubMed] [Google Scholar]
- Park JE, Park JY, Kim YS, Staswick PE, Jeon J, Yun J, Kim SY, Kim J, Lee YH, Park CM (2007) GH3-mediated auxin homeostasis links growth regulation with stress adaptation response in Arabidopsis. J Biol Chem 282: 10036–10046 [DOI] [PubMed] [Google Scholar]
- Pauwels L, Morreel K, De Witte E, Lammertyn F, Van Montagu M, Boerjan W, Inzé D, Goossens A (2008) Mapping methyl jasmonate-mediated transcriptional reprogramming of metabolism and cell cycle progression in cultured Arabidopsis cells. Proc Natl Acad Sci USA 105: 1380–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedley KF, Martin GB (2005) Role of mitogen-activated protein kinases in plant immunity. Curr Opin Plant Biol 8: 541–547 [DOI] [PubMed] [Google Scholar]
- Pereyra-Irujo GA, Velázquez L, Lechner L, Aguirrezábal LAN (2008) Genetic variability for leaf growth rate and duration under water deficit in sunflower: analysis of responses at cell, organ, and plant level. J Exp Bot 59: 2221–2232 [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarvepalli K, Nath U (2011) Hyper-activation of the TCP4 transcription factor in Arabidopsis thaliana accelerates multiple aspects of plant maturation. Plant J 67: 595–607 [DOI] [PubMed] [Google Scholar]
- Schaalje GB, McBride JB, Fellingham GW (2001) Approximations to distributions of test statistics in complex mixed linear models using SAS Proc MIXED. In Proceedings of SAS User Group International 26th Annual Conference, Long Beach, CA, April 22–25 2001, paper 262-26. SAS Institute Inc., Cary, NC [Google Scholar]
- Skirycz A, Claeys H, De Bodt S, Oikawa A, Shinoda S, Andriankaja M, Maleux K, Eloy NB, Coppens F, Yoo SD, et al. (2011a) Pause-and-stop: the effects of osmotic stress on cell proliferation during early leaf development in Arabidopsis and a role for ethylene signaling in cell cycle arrest. Plant Cell 23: 1876–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skirycz A, De Bodt S, Obata T, De Clercq I, Claeys H, De Rycke R, Andriankaja M, Van Aken O, Van Breusegem F, Fernie AR, et al. (2010) Developmental stage specificity and the role of mitochondrial metabolism in the response of Arabidopsis leaves to prolonged mild osmotic stress. Plant Physiol 152: 226–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skirycz A, Vandenbroucke K, Clauw P, Maleux K, De Meyer B, Dhondt S, Pucci A, Gonzalez N, Hoeberichts F, Tognetti VB, et al. (2011b) Survival and growth of Arabidopsis plants given limited water are not equal. Nat Biotechnol 29: 212–214 [DOI] [PubMed] [Google Scholar]
- Strizhov N, Abrahám E, Okrész L, Blickling S, Zilberstein A, Schell J, Koncz C, Szabados L (1997) Differential expression of two P5CS genes controlling proline accumulation during salt-stress requires ABA and is regulated by ABA1, ABI1 and AXR2 in Arabidopsis. Plant J 12: 557–569 [DOI] [PubMed] [Google Scholar]
- Świątek A, Azmi A, Stals H, Inzé D, Van Onckelen H (2004) Jasmonic acid prevents the accumulation of cyclin B1;1 and CDK-B in synchronized tobacco BY-2 cells. FEBS Lett 572: 118–122 [DOI] [PubMed] [Google Scholar]
- Świątek A, Lenjou M, Van Bockstaele D, Inzé D, Van Onckelen H (2002) Differential effect of jasmonic acid and abscisic acid on cell cycle progression in tobacco BY-2 cells. Plant Physiol 128: 201–211 [PMC free article] [PubMed] [Google Scholar]
- Szabados L, Savouré A (2010) Proline: a multifunctional amino acid. Trends Plant Sci 15: 89–97 [DOI] [PubMed] [Google Scholar]
- Taji T, Ohsumi C, Iuchi S, Seki M, Kasuga M, Kobayashi M, Yamaguchi-Shinozaki K, Shinozaki K (2002) Important roles of drought- and cold-inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana. Plant J 29: 417–426 [DOI] [PubMed] [Google Scholar]
- Tan BC, Joseph LM, Deng WT, Liu L, Li QB, Cline K, McCarty DR (2003) Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J 35: 44–56 [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Sano T, Tamaoki M, Nakajima N, Kondo N, Hasezawa S (2005) Ethylene inhibits abscisic acid-induced stomatal closure in Arabidopsis. Plant Physiol 138: 2337–2343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardieu F. (2012) Any trait or trait-related allele can confer drought tolerance: just design the right drought scenario. J Exp Bot 63: 25–31 [DOI] [PubMed] [Google Scholar]
- Tardieu F, Parent B, Simonneau T (2010) Control of leaf growth by abscisic acid: hydraulic or non-hydraulic processes? Plant Cell Environ 33: 636–647 [DOI] [PubMed] [Google Scholar]
- Tisné S, Reymond M, Vile D, Fabre J, Dauzat M, Koornneef M, Granier C (2008) Combined genetic and modeling approaches reveal that epidermal cell area and number in leaves are controlled by leaf and plant developmental processes in Arabidopsis. Plant Physiol 148: 1117–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran LSP, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2004) Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16: 2481–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trontin C, Kiani S, Corwin JA, Hématy K, Yansouni J, Kliebenstein DJ, Loudet O (2014) A pair of receptor-like kinases is responsible for natural variation in shoot growth response to mannitol treatment in Arabidopsis thaliana. Plant J 78: 121–133 [DOI] [PubMed] [Google Scholar]
- Umezawa T, Nakashima K, Miyakawa T, Kuromori T, Tanokura M, Shinozaki K, Yamaguchi-Shinozaki K (2010) Molecular basis of the core regulatory network in ABA responses: sensing, signaling and transport. Plant Cell Physiol 51: 1821–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T, Okamoto M, Kushiro T, Nambara E, Oono Y, Seki M, Kobayashi M, Koshiba T, Kamiya Y, Shinozaki K (2006) CYP707A3, a major ABA 8′-hydroxylase involved in dehydration and rehydration response in Arabidopsis thaliana. Plant J 46: 171–182 [DOI] [PubMed] [Google Scholar]
- Umezawa T, Yoshida R, Maruyama K, Yamaguchi-Shinozaki K, Shinozaki K (2004) SRK2C, a SNF1-related protein kinase 2, improves drought tolerance by controlling stress-responsive gene expression in Arabidopsis thaliana. Proc Natl Acad Sci USA 101: 17306–17311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Landeghem S, De Bodt S, Drebert ZJ, Inzé D, Van de Peer Y (2013) The potential of text mining in data integration and network biology for plant research: a case study on Arabidopsis. Plant Cell 25: 794–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Landeghem S, Hakala K, Rönnqvist S, Salakoski T, Van de Peer Y, Ginter F (2012) Exploring biomolecular literature with EVEX: connecting genes through events, homology, and indirect associations. Adv Bioinformatics 2012: 582765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verelst W, Skirycz A, Inzé D (2010) Abscisic acid, ethylene and gibberellic acid act at different developmental stages to instruct the adaptation of young leaves to stress. Plant Signal Behav 5: 473–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verslues PE, Agarwal M, Katiyar-Agarwal S, Zhu J, Zhu JK (2006) Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J 45: 523–539 [DOI] [PubMed] [Google Scholar]
- Verslues PE, Sharma S (2010) Proline metabolism and its implications for plant-environment interaction. The Arabidopsis Book 8: e0140, doi/10.1199/tab.0140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JW, Schwab R, Czech B, Mica E, Weigel D (2008) Dual effects of miR156-targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. Plant Cell 20: 1231–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Xue L, Batelli G, Lee S, Hou YJ, Van Oosten MJ, Zhang H, Tao WA, Zhu JK (2013) Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effectors of abscisic acid action. Proc Natl Acad Sci USA 110: 11205–11210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TD, Nacu S (2010) Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26: 873–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Cosgrove DJ (2000) Adaptation of roots to low water potentials by changes in cell wall extensibility and cell wall proteins. J Exp Bot 51: 1543–1553 [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57: 781–803 [DOI] [PubMed] [Google Scholar]
- Yang S, Vanderbeld B, Wan J, Huang Y (2010) Narrowing down the targets: towards successful genetic engineering of drought-tolerant crops. Mol Plant 3: 469–490 [DOI] [PubMed] [Google Scholar]
- Ye N, Zhang J (2012) Antagonism between abscisic acid and gibberellins is partially mediated by ascorbic acid during seed germination in rice. Plant Signal Behav 7: 563–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz A, Mejia-Guerra MK, Kurz K, Liang X, Welch L, Grotewold E (2011) AGRIS: the Arabidopsis Gene Regulatory Information Server, an update. Nucleic Acids Res 39: D1118–D1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Fujita Y, Maruyama K, Mogami J, Todaka D, Shinozaki K, Yamaguchi-Shinozaki K (2014) Four Arabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic acid signalling in response to osmotic stress. Plant Cell Environ 38: 35–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Fujita Y, Sayama H, Kidokoro S, Maruyama K, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2010) AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J 61: 672–685 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Turner JG (2008) Wound-induced endogenous jasmonates stunt plant growth by inhibiting mitosis. PLoS ONE 3: e3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YY, Li Y, Gao T, Zhu H, Wang DJ, Zhang HW, Ning YS, Liu LJ, Wu YR, Chu CC, et al. (2008) Arabidopsis SDIR1 enhances drought tolerance in crop plants. Biosci Biotechnol Biochem 72: 2251–2254 [DOI] [PubMed] [Google Scholar]
- Zhu SY, Yu XC, Wang XJ, Zhao R, Li Y, Fan RC, Shang Y, Du SY, Wang XF, Wu FQ, et al. (2007) Two calcium-dependent protein kinases, CPK4 and CPK11, regulate abscisic acid signal transduction in Arabidopsis. Plant Cell 19: 3019–3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.