Phosphoenolpyruvate carboxylase in Arabidopsis leaves balances carbon and nitrogen metabolism.
Abstract
Phosphoenolpyruvate carboxylase (PEPC) is a crucial enzyme that catalyzes an irreversible primary metabolic reaction in plants. Previous studies have used transgenic plants expressing ectopic PEPC forms with diminished feedback inhibition to examine the role of PEPC in carbon and nitrogen metabolism. To date, the in vivo role of PEPC in carbon and nitrogen metabolism has not been analyzed in plants. In this study, we examined the role of PEPC in plants, demonstrating that PPC1 and PPC2 were highly expressed genes encoding PEPC in Arabidopsis (Arabidopsis thaliana) leaves and that PPC1 and PPC2 accounted for approximately 93% of total PEPC activity in the leaves. A double mutant, ppc1/ppc2, was constructed that exhibited a severe growth-arrest phenotype. The ppc1/ppc2 mutant accumulated more starch and sucrose than wild-type plants when seedlings were grown under normal conditions. Physiological and metabolic analysis revealed that decreased PEPC activity in the ppc1/ppc2 mutant greatly reduced the synthesis of malate and citrate and severely suppressed ammonium assimilation. Furthermore, nitrate levels in the ppc1/ppc2 mutant were significantly lower than those in wild-type plants due to the suppression of ammonium assimilation. Interestingly, starch and sucrose accumulation could be prevented and nitrate levels could be maintained by supplying the ppc1/ppc2 mutant with exogenous malate and glutamate, suggesting that low nitrogen status resulted in the alteration of carbon metabolism and prompted the accumulation of starch and sucrose in the ppc1/ppc2 mutant. Our results demonstrate that PEPC in leaves plays a crucial role in modulating the balance of carbon and nitrogen metabolism in Arabidopsis.
Phosphoenolpyruvate carboxylase (PEPC; EC 4.1.1.31) is a crucial enzyme that functions in primary metabolism by irreversibly catalyzing the conversion of phosphoenolpyruvate (PEP) and HCO3− to oxaloacetate (OAA) and inorganic phosphate. PEPC is found in all plants, green algae, and cyanobacteria, and in most archaea and nonphotosynthetic bacteria, but not in animals or fungi (Chollet et al., 1996; O’Leary et al., 2011a). Several isoforms of PEPC are present in plants, including plant-type PEPCs and one bacterium-type PEPC (Sánchez and Cejudo, 2003; Sullivan et al., 2004; Mamedov et al., 2005; Gennidakis et al., 2007; Igawa et al., 2010). Arabidopsis (Arabidopsis thaliana) possesses three plant-type PEPC genes, AtPPC1, AtPPC2, and AtPPC3, and one bacterium-type PEPC gene, AtPPC4. Unlike plant-type PEPCs, bacterium-type PEPCs lack a seryl-phosphorylation domain near the N terminus, a typical domain conserved in plant-type PEPCs (Sánchez and Cejudo, 2003). Plant-type PEPCs form class 1 PEPCs, which exist as homotetramers. Recently, bacterium-type PEPCs have been reported to interact with plant-type PEPCs to form heterooctameric class 2 PEPCs in several species, including unicellular green algae (Selenastrum minutum), lily (Lilium longiflorum), and castor bean (Ricinus communis; O’Leary et al., 2011a).
Because of the irreversible nature of the enzymatic reactions catalyzed by PEPC isoforms, they are strictly regulated by a variety of mechanisms. PEPC is an allosteric enzyme and is activated by its positive effector, Glc-6-P, and inhibited by its negative effectors, malate, Asp, and Glu (O’Leary et al., 2011a). Control by reversible phosphorylation is another important mechanism that regulates the activity of PEPC. In this reaction, phosphorylation catalyzed by PEPC kinase changes the sensitivity of PEPC to its allosteric effectors (Nimmo, 2003). In addition, monoubiquitination may also regulate plant-type PEPC activity (Uhrig et al., 2008). Recent research in castor oil seeds suggests that bacterium-type PEPC is a catalytic and regulatory subunit of class 2 PEPCs, as class 1 and class 2 PEPCs show significant differences in their sensitivity to allosteric inhibitors (O’Leary et al., 2009, 2011b).
A number of studies on PEPC function have been performed in a variety of organisms (O’Leary et al., 2011a). The best described function of PEPC is in fixing photosynthetic CO2 during C4 and Crassulacean acid metabolism photosynthesis. However, in most nonphotosynthetic tissues and the photosynthetic tissues of C3 plants, the fundamental function of PEPC is to anaplerotically replenish tricarboxylic acid cycle intermediates (Chollet et al., 1996). PEPC also functions in malate production in guard cells and legume root nodules (Chollet et al., 1996). A chloroplast-located PEPC isoform in rice (Oryza sativa) was recently found to be crucial for ammonium assimilation (Masumoto et al., 2010). In addition, previous work in Arabidopsis suggested that AtPPC4 might play a role in drought tolerance (Sánchez et al., 2006).
Transgenic plants expressing ectopic PEPC forms with diminished feedback inhibition showed an increase in overall organic nitrogen content at the expense of starch and soluble sugars (Rademacher et al., 2002; Chen et al., 2004; Rolletschek et al., 2004). However, the in vivo function of PEPC in carbon and nitrogen metabolism has not been reported previously.
To further investigate the function of PEPC in higher plants, we isolated and characterized mutants of Arabidopsis deficient in the expression of the PEPC-encoding genes PPC1 and PPC2. We demonstrated that PPC1 and PPC2 were the most highly expressed PEPC genes in the leaves. To further define their role, we produced a double mutant (ppc1/ppc2) deficient in the expression of the PPC1 and PPC2 genes. We then conducted a detailed molecular, biochemical, and physiological characterization of this double mutant.
RESULTS
Mutation of PPC1 and PPC2 Causes Growth Arrest in Arabidopsis
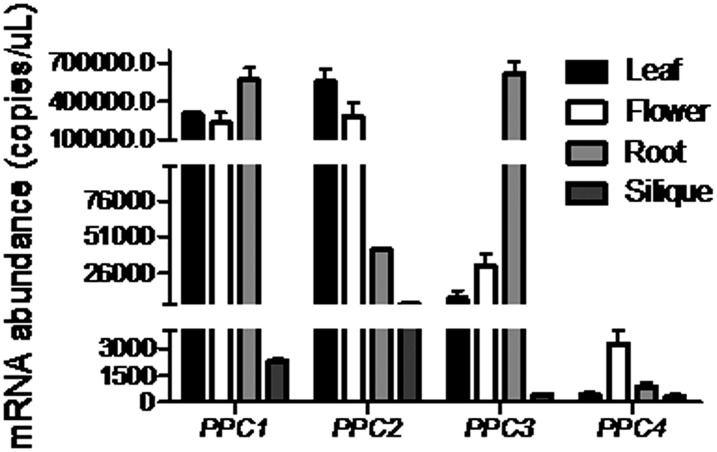
The Arabidopsis genome contains four PEPC genes, PPC1 (AT1G53310), PPC2 (AT2G42600), PPC3 (AT3G14940), and PPC4 (AT1G68750). To investigate the spatial expression patterns and abundance of PEPC genes in Arabidopsis, absolute quantitative real-time PCR was performed in flowers, siliques, and 14-d-old leaves and roots (Fig. 1). All four PPC genes were expressed in all organs tested, with great variance in relative abundance. PPC1 was expressed in flowers, leaves, and roots at high levels (more than 105 copies μL−1) and had low abundance in siliques (103–104 copies μL−1); PPC2 was highly abundant in flowers and leaves (more than 105 copies μL−1) and moderately abundant in roots and siliques (104–105 copies μL−1); PPC3 was expressed in roots at high levels (more than 105 copies μL−1), in leaves and flowers at moderate levels (104–105 copies μL−1), and in siliques at extremely low levels (102–103 copies μL−1); PPC4 was expressed in flowers at low levels (103–104 copies μL−1) and had very low expression levels in leaves, roots, and siliques (102–103 copies μL−1). Thus, our results indicated that PPC1 and PPC2 were the most highly expressed PEPC genes in the leaves. These results confirmed the findings of several earlier Arabidopsis PEPC studies (Gousset-Dupont et al., 2005; Gregory et al., 2009).
Figure 1.
Tissue-specific expression of PPC genes determined by absolute quantitative real-time PCR analysis. Wild-type plants were grown on one-half-strength Murashige and Skoog (1/2 MS) medium for 2 weeks and then transferred to soil for further growth. Fourteen-day-old leaves and roots, flowers, and siliques were harvested and subjected to gene expression analysis. PPC genes were expressed in flowers, siliques, and 14-d-old leaves and roots. However, they showed great variance in abundance.
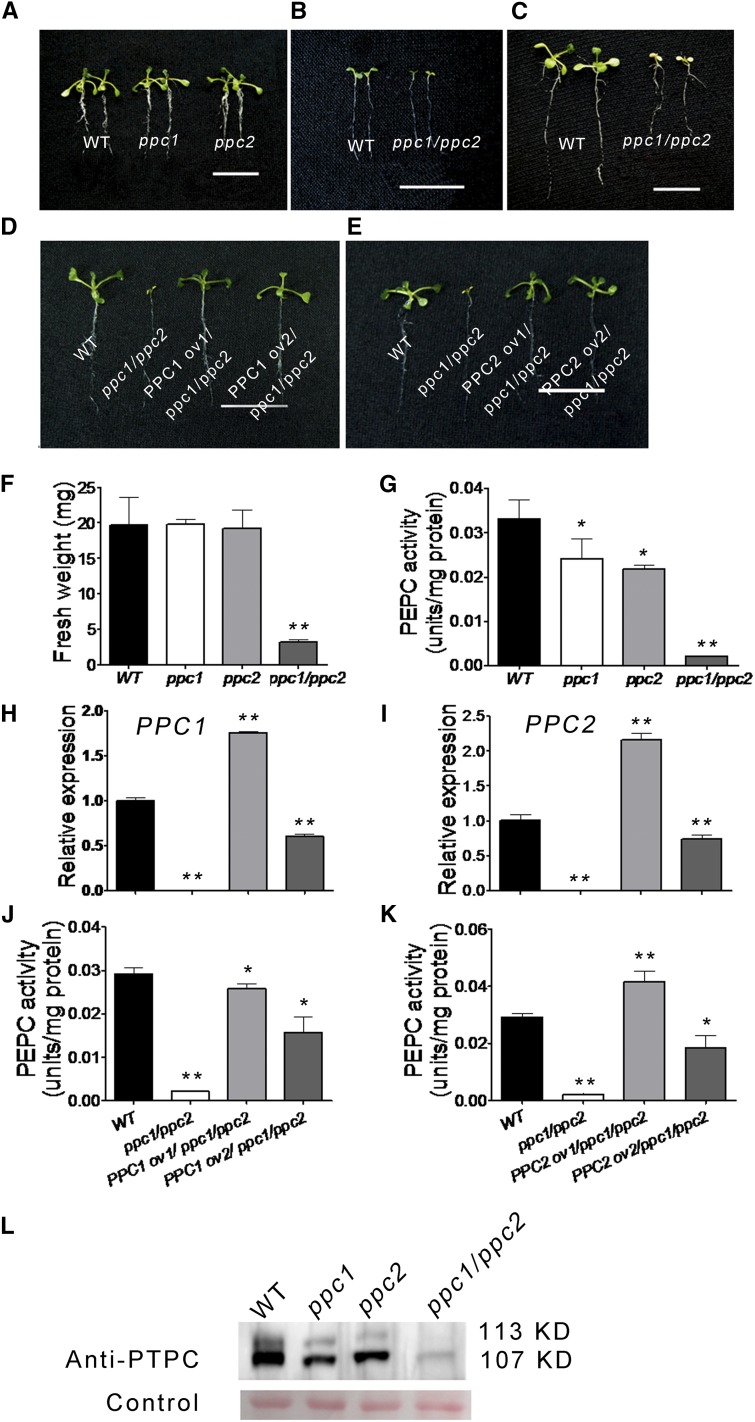
PEPC is an important enzyme in the glycolytic pathway and links with photosynthesis and respiration in the plant. So we hypothesize that PPC1 and PPC2 may play important roles in the growth and development of the aerial part. To test our hypothesis, two independent transfer DNA (T-DNA) insertion lines (PPC1 [Salk_088836] and PPC2 [Salk_025005]) from the Salk insertion mutant collection were selected and characterized. The two Salk lines each harbors an insertion in an intron (Supplemental Fig. S1A). No full-length transcripts were detected in any of the two homozygous mutants (Supplemental Fig. S1C). The single mutant plants did not exhibit any obvious morphological phenotypes (Fig. 2A) and did not show significant differences in fresh weight compared with the wild-type plants (Fig. 2F). To address whether this was because of the redundant functions of the two PEPC isoforms, crosses between the two lines were made to generate plants homozygous for the two T-DNA insertions. All possible genotypes were produced in the F2 population, as determined by PCR (Supplemental Table S1). The results showed that the ppc1/ppc2 double mutant displayed a severe growth-arrest phenotype (Fig. 2, B and C).
Figure 2.
Mutation of PPC1 and PPC2 leads to a growth-arrest phenotype in Arabidopsis. The wild type (WT), ppc mutants, and ppc1/ppc2 transgenic lines were grown on 1/2 MS medium. A, Phenotypes of 10-d-old single ppc mutants. B and C, Phenotypes of the ppc1/ppc2 double mutant at 5 d old (B) and 10 d old (C). D and E, Arabidopsis seedlings of the wild type, the ppc1/ppc2 mutant, and two ppc1/ppc2 mutant lines complemented by the expression of PPC1 (D) or PPC2 (E) cDNA. F, Biomass of 10-d-old seedlings (entire plants) of the wild type and ppc mutants. Values are means ± se (n = 30). G, Leaf PEPC activity of 10-d-old wild-type and ppc mutant seedlings. Data are given as means ± se (n = 5). H and I, Quantitative real-time PCR results of PPC1 and PPC2 in leaves of the wild type, ppc1/ppc2, and two ppc1/ppc2 transgenic lines expressing PPC1 (H) or PPC2 (I) cDNA. Relative expression levels are given after being normalized to the expression level of ACTIN and compared with the expression level of the wild type (represented as 1-fold). Data are given as means ± se (n = 3). J and K, Leaf PEPC activity analyzed in 10-d-old wild-type seedlings, ppc1/ppc2, and two ppc1/ppc2 transgenic lines expressing PPC1 (J) or PPC2 (K) cDNA. Data are given as means ± se (n = 5). L, The anti-PEPC antibody recognizes both a monoubiquitinated Arabidopsis plant-type PEPC (AtPTPC) band (approximately 113 kD) and an AtPTPC band (approximately 107 kD) in leaves of the wild type and ppc mutants. The protein levels of both bands show great variance in wild-type and ppc mutant leaves. Asterisks indicate significant differences between genotypes (**P < 0.01 and *P < 0.05 by Student’s t test). Bars = 1 cm in B and C and 2 cm in A, D, and E.
As shown in Figure 2F, the growth of the ppc1/ppc2 mutant was severely inhibited: the fresh weight of 10-d-old ppc1/ppc2 seedlings grown on 1/2 MS medium was approximately 16% of that of wild-type seedlings. In addition, small aerial part size and pale yellow cotyledons were evident, and growth of the ppc1/ppc2 mutant was arrested at the seedling stage (Fig. 2, B and C). The ppc1/ppc2 mutant could neither flower nor make seeds and, thus, could not complete its life cycle. Next, PEPC activity was examined in the leaves of 10-d-old wild-type, ppc1, ppc2, and ppc1/ppc2 mutant seedlings grown on 1/2 MS medium. PEPC activity levels in ppc1 and ppc2 plants were approximately 73% and 66% of those of the wild type, respectively, and the PEPC activity levels in ppc1/ppc2 plants were significantly lower than in the ppc1 and ppc2 single mutant plants, reaching only 6.5% of those of the wild-type plants (Fig. 2G). Protein gel-blot analysis revealed that the levels of plant-type PEPC (approximately 107 kD) were decreased to some extent in the leaves of the ppc1 and ppc2 mutants compared with the wild type and that the expression of PTPC was reduced to extremely low levels in leaves of the ppc1/ppc2 mutant by comparison with the wild type (Fig. 2L), which was in agreement with the results of the PEPC activity assays.
Confirmation that the disruption of PPC1 and PPC2 in the ppc1/ppc2 mutant was responsible for the mutant phenotype was achieved by genetic complementation. Wild-type complementary DNAs (cDNAs) of PPC1 (coding sequence [CDS] + 137 bp downstream termination codon TAG) and PPC2 (CDS) were inserted into 35S-pCAMBIA1301 plasmids (Zhou et al., 2008). The resulting constructs were used to introduce the complete PPC1 or PPC2 gene into the genome of the ppc1/ppc1 homozygous PPC2/ppc2 heterozygous mutant plants via Agrobacterium tumefaciens-mediated transformation (Clough and Bent, 1998). The insertions were confirmed by hygromycin resistance selection and PCR analysis. Our results showed that the transgenic mutant lines expressing PPC1 or PPC2 completely rescued the ppc1/ppc2 mutant phenotype (Fig. 2, D and E). Expression analysis of PPC1 and PPC2 in rescued lines was performed using quantitative real-time PCR (Fig. 2, H and I). The rescued lines showed significantly increased expression of PPC1 or PPC2 relative to the ppc1/ppc2 mutant. However, individual lines showed increased or lowered expression of PPC1 or PPC2 compared with the wild type. In addition, levels of PEPC activity were examined in rescued lines (Fig. 2, J and K). The rescued lines showed significantly increased PEPC activity levels relative to the ppc1/ppc2 mutant; however, individual lines showed increased or lowered PEPC activity compared with the wild type.
Significant Carbohydrate Accumulation in the ppc1/ppc2 Mutant
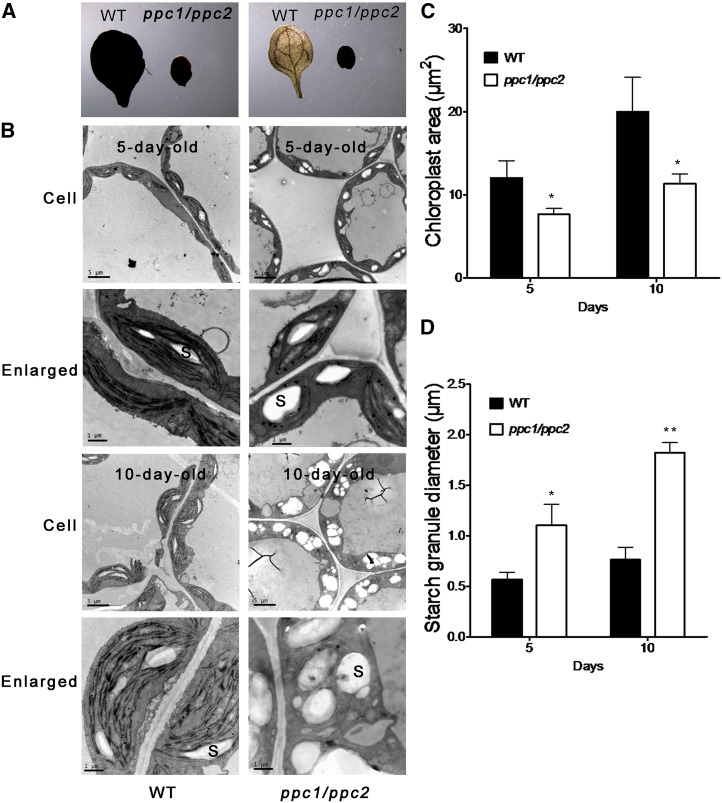
In order to assess whether knockout of PPC1 and PPC2 affects starch biosynthesis, we compared the starch content in ppc1/ppc2 mutants and wild-type plants by Lugol staining. As shown in Figure 3A (left), starch was accumulated at similar levels in ppc1/ppc2 and wild-type plants at the end of the illumination period, indicating normal starch biosynthesis in the ppc1/ppc2 mutant. At the end of the dark period, all starch had been consumed in wild-type plants, but starch levels were still high in the ppc1/ppc2 mutant plants (Fig. 3A, right). These results suggest that the ppc1/ppc2 mutant has a defect in carbohydrate metabolism. Transmission electron microscopy was performed to assess any ultrastructural perturbations in the cotyledons of the ppc1/ppc2 mutant. The cross-section areas of chloroplasts in the mesophyll cells of 5-d-old ppc1/ppc2 mutants showed a 37% reduction compared with those in the wild type, while the diameters of starch granules in the ppc1/ppc2 chloroplasts were nearly double (190%) those of the wild type (Fig. 3, C and D). Thus, the chloroplasts of 5-d-old ppc1/ppc2 mutant plants were much smaller and simpler than those of the wild type but exhibited accumulation of much larger starch granules (Fig. 3B). The cross-section areas of chloroplasts in the mesophyll cells showed a 43% reduction in 10-d-old ppc1/ppc2 mutants compared with those in the wild type, whereas the diameters of starch granules in chloroplasts were 2.4-fold larger than those in the wild type (Fig. 3, C and D). Ten-day-old ppc1/ppc2 mutant plants showed severely impaired thylakoid membrane organization but still exhibited overaccumulation of starch grains in the chloroplasts (Fig. 3B). Next, the amounts of Suc, Glc, Fru, and starch were quantified in 10-d-old ppc mutants and wild-type seedlings grown on 1/2 MS medium. Suc levels in the ppc1/ppc2 mutant were approximately 2.3 times higher than in wild-type plants, while similar Suc levels were observed in the ppc1, ppc2, and wild-type plants (Fig. 4). The levels of Glc and Fru in all four plant genotypes were almost equal (data not shown). Starch levels in the ppc1/ppc2 mutant were approximately 3.3-fold higher than in the wild type, while ppc1, ppc2, and wild-type plants all showed similar levels of starch (Fig. 4).
Figure 3.
Disruption of PPC1 and PPC2 results in starch accumulation in the ppc1/ppc2 mutant. The wild type (WT) and ppc1/ppc2 mutants were grown on 1/2 MS medium. A, Starch content in 10-d-old wild-type and ppc1/ppc2 mutant cotyledons, as revealed by Lugol staining. Left, Samples taken at the end of the illumination period; right, samples taken at the end of the dark period. B, Ultrastructures of chloroplasts in the mesophyll cells of mutant seedlings. All transmission electron microscopy samples were obtained from the cotyledons of 5- or 10-d-old ppc1/ppc2 and wild-type seedlings at midday. S, Starch granule. Images in the bottom row are the enlarged plastids shown in the top row. Bars = 5 μm in the top row and 1 μm in the bottom row. C and D, Comparison of chloroplast (C) and starch granule (D) sizes in wild-type and ppc1/ppc2 mutant plants. Quantification was performed on 30 chloroplasts in mesophyll cells from each cotyledon. Three individual cotyledons of each genotype were quantified. Asterisks indicate significant differences between genotypes (**P < 0.01 and *P < 0.05 by Student’s t test).
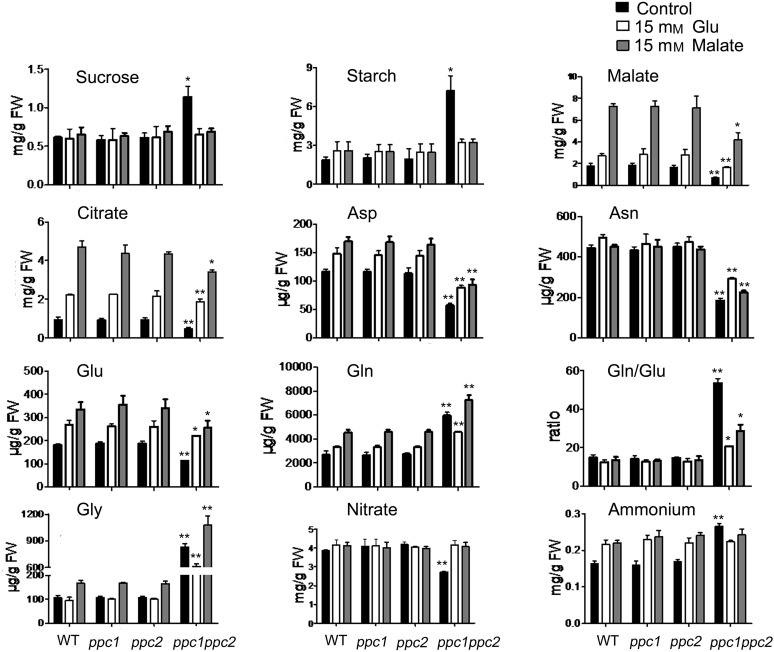
Figure 4.
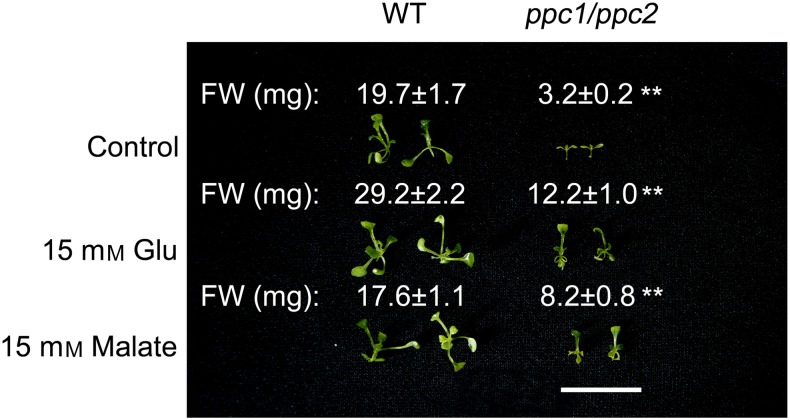
Metabolite changes in the ppc1/ppc2 mutants, which were partially complemented by the supply of 15 mm malate or Glu. Wild-type and ppc mutant plants were grown on 1/2 MS medium containing 15 mm malate or Glu for 10 d. Samples (entire plants) were taken at midday. Values are means ± se (n = 5). Asterisks indicate significant differences between genotypes (**P < 0.01 and *P < 0.05 by Student’s t test). FW, Fresh weight.
Metabolomics Analysis of the ppc1/ppc2 Double Knockout
To characterize the effect of the disruption of PPC1 and PPC2 on metabolite levels, seedlings from 10-d-old plants grown on 1/2 MS medium were harvested at midday and subjected to metabolite analysis. A number of amino acids and some organic acids were measured. The levels of some metabolites showed significant differences between the ppc1/ppc2 mutant and wild-type, ppc1, and ppc2 plants (P < 0.05; Fig. 4; Supplemental Table S2).
The levels of malate were markedly decreased in the ppc1/ppc2 mutant (Fig. 4). This likely reflects a decreased level of OAA, a product of PEPC that is readily converted into malate by malate dehydrogenase. Additionally, citrate levels were decreased by 49% in the ppc1/ppc2 mutant (Fig. 4). The levels of 2-oxoglutarate and succinate in all four plant genotypes remained almost equal (Supplemental Table S2).
Tyr levels in ppc1/ppc2 plants were almost 3.3-fold higher than in wild-type plants (Supplemental Table S2), which likely reflects an increased level of shikimate, the precursor of Tyr. Val and Leu levels were massively increased in the ppc1/ppc2 mutant (Supplemental Table S2), indicating an increased level of pyruvate, the precursor of Val and Leu. In addition, Gly and Ser levels were increased in ppc1/ppc2 mutant plants (Fig. 4; Supplemental Table S2), which may indicate the accumulation of photorespiratory intermediates.
The levels of Gln were increased by almost 2.2-fold in ppc1/ppc2 mutant plants, whereas the Glu levels were decreased by approximately 38% (Fig. 4). In addition, Asp and Asn levels were decreased by 51% and 58%, respectively, in the ppc1/ppc2 mutant (Fig. 4).
Changes of Nitrate and Ammonium Content in the ppc1/ppc2 Mutant
To determine whether the disruption of PPC1 and PPC2 affects nitrate assimilation, we measured the amounts of nitrate and ammonium in 10-d-old wild-type and ppc mutant seedlings grown on 1/2 MS medium. The results showed that nitrate levels in the ppc1/ppc2 mutant were decreased by approximately 37% compared with wild-type plants (Fig. 4); accordingly, the ammonium content in ppc1/ppc2 plants was approximately 1.5-fold higher than that of wild-type plants (Fig. 4). Nitrate and ammonium levels were almost equal in ppc1, ppc2, and wild-type seedlings (Fig. 4).
Metabolic Complementation of Growth Arrest in ppc1/ppc2 Mutants by Exogenous Malate or Glu
To assess whether decreased malate and Glu levels may be linked to the growth-arrest phenotype, ppc1/ppc2 mutants were grown on 1/2 MS medium containing 15 mm malate or Glu for 10 d. As shown in Figure 5, exogenously supplied malate or Glu rescued the growth-arrest phenotype of the ppc1/ppc2 mutants to some extent. When seedlings supplied with malate or Glu were grown for 10 d, the fresh weight of the wild-type seedlings was similar to or slightly higher than that of wild-type seedlings grown on 1/2 MS (Fig. 5). Conversely, the fresh weight of the malate- or Glu-supplemented ppc1/ppc2 seedlings was largely increased compared with that of ppc1/ppc2 seedlings grown on 1/2 MS (Fig. 5).
Figure 5.
Supplementation with 15 mm malate or Glu partially rescued the phenotype of ppc1/ppc2 seedlings. Phenotypes and biomass of ppc1/ppc2 mutants grown on 1/2 MS medium plus 15 mm Glu or malate for 10 d are shown. FW, Fresh weight; WT, wild type. Values are means ± se (n = 10). Asterisks indicate significant differences between genotypes (**P < 0.01 by Student’s t test). Bar = 2 cm.
To identify the metabolic changes that could account for complementation of the growth defect, metabolite analysis was performed in ppc mutants and wild-type seedlings supplied with malate or Glu. When the seedlings were treated with malate or Glu, starch and Suc were at similar levels in all four plant genotypes, although starch and Suc levels in ppc1/ppc2 mutants were decreased compared with ppc1/ppc2 seedlings grown on 1/2 MS medium (Fig. 4). Malate and citrate levels in malate- or Glu-supplemented wild-type plants were increased compared with those in untreated wild-type plants, while the levels of malate and citrate in the supplied ppc1/ppc2 mutants were considerably higher than those in untreated ppc1/ppc2 seedlings (Fig. 4). Levels of 2-oxoglutarate were increased, although only to levels close to those in ppc mutants and wild-type plants; succinate levels were similar in ppc mutants and wild-type seedlings (Supplemental Tables S2–S4).
When seedlings were supplied with malate or Glu for 10 d, the levels of Asp and Glu in wild-type plants were higher than those of untreated wild-type plants to some extent, while Asp and Glu levels in the supplied ppc1/ppc2 mutants were significantly higher than those in untreated ppc1/ppc2 seedlings (Fig. 4). Asn levels in malate- or Glu-supplied wild-type plants were similar to those in untreated wild-type plants, while the levels of Asn in the treated ppc1/ppc2 mutants were higher than those in untreated ppc1/ppc2 seedlings (Fig. 4). The levels of Gln and Gly in Glu- or malate-treated wild-type seedlings were similar to or higher than those of untreated wild-type seedlings, respectively, while Gln and Gly levels in the treated ppc1/ppc2 mutants were significantly lower and higher than those in untreated ppc1/ppc2 seedlings, respectively (Fig. 4). The ratio of Gln to Glu in malate- or Glu-supplied wild-type plants was similar to that of untreated wild-type plants, while Gln-Glu ratio in the supplied ppc1/ppc2 seedlings was much lower than that in untreated ppc1/ppc2 seedlings (Fig. 4).
The levels of nitrate and ammonium were similar in all four plant genotypes supplied with malate or Glu (Fig. 4). Compared with ppc1/ppc2 seedlings grown on 1/2 MS medium, ppc1/ppc2 mutants supplied with malate or Glu had increased nitrate and decreased ammonium (Fig. 4).
DISCUSSION
PEPC is an essential enzyme that catalyzes the biosynthesis of OAA from PEP and is crucial for primary metabolism in all plants. Here, we provide several lines of evidence showing that mutations in PPC1 and PPC2 lead to a severe growth-arrest phenotype (Fig. 2). PPC1 and PPC2 were the most highly expressed PEPC genes in the leaves, accounting for approximately 93% of total PEPC activity in the leaves (Figs. 1 and 2G). We also found that ppc1/ppc2 mutants accumulated starch and Suc (Figs. 3 and 4) and that disruption of PPC1 and PPC2 leads to the suppression of nitrogen assimilation (Fig. 4). Although quantitative real-time PCR analysis showed that PPC3 and PPC4 were expressed to some extent in leaves (Fig. 1), our results demonstrate that PPC1 and PPC2 are crucial for proper metabolism and growth in the aerial part of Arabidopsis.
The function of PPC1 and PPC2 was investigated by comparing the ppc1/ppc2 mutant with the wild type and ppc single mutants. Leaf metabolite analysis (Fig. 4) showed that knockout of PPC1 and PPC2 greatly decreased the levels of malate and citrate. Although 2-oxoglutarate levels in the knockouts did not show a significant difference when compared with wild-type plants (Supplemental Table S2), our results suggest that the anaplerotic flux from PEP to malate and hence 2-oxoglutarate is largely suppressed by the disruption of PPC1 and PPC2. In addition, the ppc1/ppc2 mutant had increased Gln levels and decreased levels of Glu, Asp, and Asn (Fig. 4). As the substrate and product of glutamate synthase (GOGAT), increased Gln and decreased Glu levels, respectively, suggest severe suppression of the GOGAT reaction in the ppc1/ppc2 mutant. Because Asp is synthesized by Asp aminotransferase using Glu and OAA as substrates, and Asn is synthesized by Asn aminotransferase using Asp and Gln as substrates, decreased Asp and Asn levels suggest that the activities of Asp aminotransferase and Asn aminotransferase are severely suppressed in the ppc1/ppc2 mutant. The fact that the ppc1/ppc2 mutants exogenously supplied with malate or Glu had a decreased Gln-Glu ratio (Fig. 4) may provide evidence that suppression of the Gln synthetase/GOGAT cycle was alleviated to some extent. Taken together, we suggest that knockout of PPC1 and PPC2 largely reduces the anaplerotic flux from PEP to malate and hence 2-oxoglutarate and, thereby, suppresses the Gln synthetase/GOGAT cycle and subsequent NH4+ assimilation.
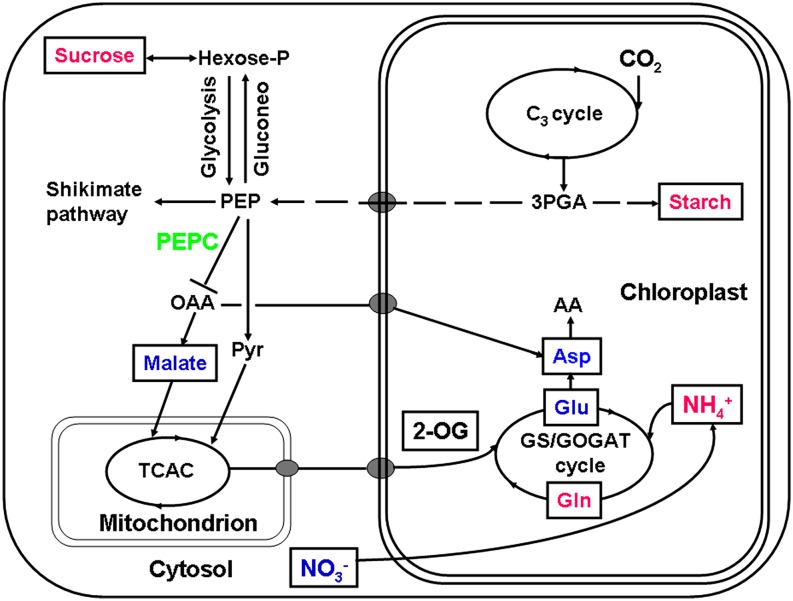
An interesting observation in this work is that starch accumulated in the ppc1/ppc2 mutant plants (Figs. 3B and 4). Previous reports studying the effects of a reduction of glycolytic enzymes by antisense RNA showed that the starch content in leaves was not increased but was significantly decreased in both transgenic tobacco (Nicotiana tabacum) with reduced enolase levels (Voll et al., 2009) and transgenic potato (Solanum tuberosum) plants with reduced expression of phosphoglycerate mutase (Westrama et al., 2002). In this study, we suggest that NH4+ assimilation in the ppc1/ppc2 mutant is suppressed by the Gln synthetase/GOGAT cycle (Fig. 4). However, previous studies demonstrated that decreased nitrogen fixation by reduced expression of nitrate reductase (Scheible et al., 1997) or 2-oxoglutarate/malate translocator (Schneidereit et al., 2006) did not enhance the accumulation of starch. Previous metabolic engineering studies expressed various forms of PEPC with diminished feedback inhibition in transgenic potato, Arabidopsis, and Vicia narbonensis seeds (Rademacher et al., 2002; Chen et al., 2004; Rolletschek et al., 2004). These alterations increased carbon flux into organic acids and amino acids and boosted the overall organic nitrogen content at the expense of starch and soluble sugars, revealing a coordinated balance in carbon/nitrogen metabolism in plants. Furthermore, a previous report studying the carbon/nitrogen balance showed that low nitrogen levels could stimulate the accumulation of starch (Scheible et al., 1997). Our results did show that the nitrate content in ppc1/ppc2 seedlings was notably decreased compared with that of the wild type (Fig. 4). Exogenous application of malate and Glu to ppc1/ppc2 mutants alleviated ammonium assimilation suppression, maintained nitrate level, and therefore prevented the accumulation of starch and Suc (Fig. 4). Taken together, we suggest that low nitrogen levels lead to starch and Suc accumulation in the ppc1/ppc2 mutants. Figure 6 shows a simplified schematic representation of the metabolic changes that occur in the ppc1/ppc2 mutants.
Figure 6.
Simplified schematic summary of metabolic changes in the ppc1/ppc2 mutant. Wild-type and ppc mutant plants were grown on 1/2 MS medium for 10 d. Samples were taken at midday and subjected to metabolite analysis (Fig. 4; Supplemental Table S2). Metabolites quantified are framed, and those whose levels showed significant differences (P < 0.05 by Student’s t test) in ppc1/ppc2 mutants are shown in colored letters. The activity of PEPC (in green) is decreased due to the disruption of PPC1 and PPC2, resulting in the accumulation of metabolites such as starch, Suc, Gln, and NH4+ (in magenta) and decreased levels of metabolites such as malate, Asp, Glu, and NO3− (in blue). AA, Amino acid; C3 cycle, Benson-Calvin cycle; Gluconeo, gluconeogenesis; GS/GOGAT, glutamine synthetase (GS)/glutamate synthase (GOGAT); 2-OG, 2-oxoglutarate; 3PGA, glycerate-3-phosphate; Pyr, pyruvate; TCAC, tricarboxylic acid cycle.
Glu occupies a central position in amino acid metabolism in plants and plays a central signaling and metabolic role at the interface of the carbon and nitrogen assimilatory pathways (Forde and Lea, 2007). The Glu receptor (AtGLR1.1) has been demonstrated to function as a regulator of carbon and nitrogen metabolism in Arabidopsis (Kang and Turano, 2003). In this study, Glu levels were notably decreased due to the disruption of PPC1 and PPC2 (Fig. 4). The decreased Glu status caused a series of alterations in carbon and nitrogen metabolism (starch and Suc accumulation and nitrogen assimilation suppression). Exogenous supply of Glu alleviated ammonium assimilation suppression, maintained the nitrate levels, and prevented starch and Suc accumulation in the ppc1/ppc2 mutants (Fig. 4). These results suggest that PPC1 and PPC2 are involved in the maintenance of steady-state Glu level in the plant and that the changed Glu status may act as a signal to regulate carbon and nitrogen metabolism in the ppc1/ppc2 mutants.
Ammonium assimilation is an important step in nitrogen metabolism and links with the nitrate assimilation and subsequent amino synthesis in the plant (Stitt et al., 2002). A recent study demonstrated that ammonium assimilation suppression due to decreased PEPC activity in rice resulted in a stunted phenotype at the vegetative stage but did not affect the completion of the life cycle (Masumoto et al., 2010). In the ppc1/ppc2 mutant, the growth was severely arrested at the seedling stage and the life cycle could not be completed (Fig. 2, B and C). Although ammonium assimilation suppression was found in the ppc1/ppc2 mutant, imbalance of carbon and nitrogen metabolism appeared to be the essential reason for the double mutant phenotype. The fact that ammonium assimilation suppression was still observed in the ppc1/ppc2 mutants supplied with exogenous malate and Glu may provide evidence to verify our hypothesis.
In many cases, reductions in enzyme levels do not lead to significant metabolic perturbations, probably due to the induction of compensatory pathways (Hodges, 2002). A previous study demonstrated that transgenic tobacco plants lacking the expression of cytosolic pyruvate kinase showed basically normal growth (Gottlob-McHugh et al., 1992). However, in a later report, inhibition of root growth and a decrease in the export of fixed carbon from the leaves to other organs were observed when these transgenic tobacco plants were cultivated under low light intensities (Knowles et al., 1998). In this study, the malate level in ppc1/ppc2 mutant plants was decreased by approximately 61% (Fig. 4). If malate translocation in different organs were considered, malate synthesis in leaves of the ppc1/ppc2 mutant would be suppressed more severely. Thus, it is evident that the anaplerotic metabolic pathway of PPC1 and PPC2 plays an important role in malate synthesis in Arabidopsis.
In the ppc1/ppc2 mutant, Gly levels were approximately 7.8-fold higher than in the wild type (Fig. 4). In photorespiration, Gly is synthesized by two different enzymes: one is Glu:glyoxylate aminotransferase, using Glu and glyoxylate as substrates, and the other is Ser:glyoxylate aminotransferase, using Ser and glyoxylate as substrates. In this study, Ser levels were greatly increased, while Glu levels were decreased to some extent, in the ppc1/ppc2 mutants (Fig. 4; Supplemental Table S2). It is likely that increased Ser and decreased Glu levels switch the balance to the Ser:glyoxylate aminotransferase reaction in the photorespiration reactions and, therefore, result in Gly accumulation.
In summary, the molecular genetic and metabolic analyses performed in this work revealed that disruption of PPC1 and PPC2 results in profound changes in carbon/nitrogen metabolism and plant growth in Arabidopsis. Decreased PEPC activity and its consequences (decreased malate synthesis, accumulated starch and Suc, suppressed ammonium assimilation, decreased nitrate levels, and arrested growth) are important metabolic events in the carbon/nitrogen balance of Arabidopsis. Although the responses of higher plants to carbon/nitrogen balance are complex and multiple mechanisms may be involved (Scheible et al., 1997), the insights gained in this work may offer clues for future attempts to understand more completely the molecular genetic and metabolic bases underlying the variations of carbon/nitrogen balance among different plant families and species.
MATERIALS AND METHODS
Plant Material and Growth Conditions
All of the Arabidopsis (Arabidopsis thaliana) alleles used in this study were in the Columbia background. Seeds of ppc1 and ppc2 mutant plants, corresponding to Salk_088836 and Salk_025005, respectively, were obtained from the Arabidopsis Biological Resource Center (http://abrc.osu.edu). The two Salk lines were characterized by the primers listed in Supplemental Table S5.
To create the ppc1/ppc2 double mutant, crosses between the two single mutants were made to generate plants homozygous for the two T-DNA insertions. All possible genotypes of progeny seedlings were determined in the F2 population by PCR-based assays. Among them, the ppc1/ppc2 double mutant displayed a severe growth-arrest phenotype and could neither flower nor make seeds. To obtain the ppc1/ppc2 double mutant seedlings, the ppc1/ppc1 homozygous PPC2/ppc2 heterozygous and the ppc2/ppc2 homozygous PPC1/ppc1 heterozygous mutant plants were screened in the F2 population, as determined by reverse transcription (RT)-PCR. The ppc1/ppc2 mutant seedlings were screened by phenotypic observation and RT-PCR assays on progeny from ppc2/ppc2 homozygous PPC1/ppc1 heterozygous or ppc1/ppc1 homozygous PPC2/ppc2 heterozygous parent lines.
All seeds were surface sterilized by incubation with 10% (v/v) bleach diluted with ethanol, washed three times with sterile distilled water, and then plated on 1/2 MS medium, pH 5.7, solidified with 0.7% (w/v) agar and supplemented with 1.5% (w/v) Suc. The seeds were kept at 4°C for 2 d and then germinated in incubators with a photon flux density of 60 to 80 μmol m−2 s−1 and a 16-h-light/8-h-dark photoperiod at 22°C. If needed, the seedlings were transferred from Murashige and Skoog medium to soil 10 d after germination. In the following experiments, the samples were harvested at midday and used for various metabolic and physiological measurements.
T-DNA Mutant Isolation and Characterization
Salk T-DNA insertion lines were obtained from the T-DNA insertion mutant population (Alonso et al., 2003). To confirm the mutants at the DNA level, PCR primers specific for the PPC gene sequences and the T-DNA border were used. Insertion sites were confirmed by sequencing the PCR product of the left border primer and a gene-specific primer. Expression of the target gene was analyzed using RT-PCR. RNA was extracted and cDNA was synthesized from 1 μg of RNA as described previously (Andre et al., 2007). RT-PCR was subsequently performed to quantify the cDNA. ACTIN (AT5G09810.1) was used as an internal control (Cao et al., 2008). Supplemental Table S5 lists the primers used in this study to genotype the T-DNA-tagged mutants.
Quantitative Real-Time PCR Analysis
Total RNA and cDNA were prepared from seedlings of the wild type and mutants as described above. For the analysis of tissue-specific expression, absolute quantitative real-time PCR was used to quantify the levels of mRNA encoding PPC genes. The primers used for the expression analysis of PPC1 (AT1G53310), PPC2 (AT2G42600), PPC3 (AT3G14940), and PPC4 (AT1G68750) were described previously (Sánchez and Cejudo, 2003). Absolute quantitative real-time PCR analysis was performed as described previously (Ismail et al., 2003; Vijgen et al., 2005).
Transmission Electron Microscopy
To investigate the status of chloroplast development, cotyledons of 5- and 10-d-old ppc1/ppc2 mutants and wild-type seedlings were prepared for transmission electron microscopy. Samples were cut into small pieces, fixed in 2.5% (w/v) glutaraldehyde in phosphate buffer (0.1 m; pH 7.2) for 4 h at 4°C, rinsed, and incubated in 1% (w/v) OsO4 in 0.1 m sodium phosphate buffer (pH 7.2) overnight at 4°C. The samples were then rinsed again in phosphate buffer (0.1 m; pH 7.2), dehydrated in an ethanol series, infiltrated with a graded series of Spurr’s resin in acetone, and then embedded in Spurr’s resin. Thin sections were obtained using a diamond knife and a Reichert OM2 ultramicrotome and then stained in 2% (w/v) uranyl acetate (pH 5) followed by 10 mm lead citrate (pH 12) and viewed with a transmission electron microscope (JEM-1230; JEOL). For measurements of chloroplasts and starch granules in fixed cotyledons, the area of chloroplasts and the diameter of starch granules from the cross sections were measured using ImageJ software (version 1.32; National Institutes of Health). The quantification was performed on 30 chloroplasts in the mesophyll cells from each of three cotyledons.
PEPC Activity Assays
Approximately 100 mg of leaves of 10-d-old wild-type and ppc mutant seedlings was ground in liquid nitrogen using an ice-cold mortar and a pestle with 1 mL of extraction buffer (200 mm HEPES-NaOH [pH 7], 10 mm MgCl2, 5 mm dithiothreitol, and 2% [w/v] polyvinylpyrrolidone-40; Li et al., 2010). The crude extracts were then transferred to prechilled Eppendorf tubes and centrifuged at 12,000g for 5 min. The supernatant was immediately used for the PEPC activity assay as described previously (Gregory et al., 2009). Reaction conditions were as follows: 50 mm HEPES-KOH (pH 8) containing 15% (v/v) glycerol, 2 mm PEP, 2 mm KHCO3, 5 mm MgCl2, 2 mm dithiothreitol, 0.15 mm NADH, and 5 units mL−1 malate dehydrogenase in a volume of 1 mL. One unit of enzyme activity was defined as 1 μmol of NADH oxidation per min at 25°C. The Bradford assay (Bradford, 1976) was used for protein quantification using bovine serum albumin as a standard.
Immunoblot Analysis
Total proteins were prepared from leaves of 10-d-old wild-type and ppc mutant seedlings as described (Deng et al., 2007). The extracted proteins were separated by SDS-PAGE and transferred to a nitrocellulose membrane (Millipore). The membrane was first probed with the anti-PEPC polyclonal antibody (AS09 458; Agrisera; 1:1,000 dilution) and then with an anti-rabbit alkaline phosphatase-conjugated secondary antibody (1:3,000). The antibody conjugates were detected in the reaction buffer (100 mm NaCl, 5 mm MgCl2, and 100 mm Tris-Cl, pH 9.5) containing 1 mg mL−1 nitroblue tetrazolium and 0.5 mg mL−1 5-bromo-4-chloro-3-indolyl phosphate, and the reaction was stopped with 1 mm EDTA.
Complementation Test
For the ppc1/ppc2 double mutant complementation assay, PPC1 (CDS + 137 bp downstream termination codon TAG; Supplemental Table S5) or PPC2 (CDS; Supplemental Table S5) cDNA was inserted into 35S-pCAMBIA1301 (Zhou et al., 2008) using the KpnI and XbaI or the KpnI and SalI restriction sites, respectively. The 35S promoter-driven PPC1 or PPC2 cDNA was then transformed into ppc1/ppc1 homozygous PPC2/ppc2 heterozygous mutant plants by the floral dip method using Agrobacterium tumefaciens EHA105 (Clough and Bent, 1998). Transformants were selected for resistance to hygromycin. DNA was extracted from selected plants and used for PCR analysis. The process was repeated to obtain ppc1/ppc2 homozygous double mutant lines containing the PPC1 or PPC2 transgene. Nine independent ppc1/ppc2 homozygous lines containing the PPC2 transgene and seven independent ppc1/ppc2 homozygous lines containing the PPC1 transgene were developed from 40 and 32 transgenic lines, respectively. Analysis of gene expression of PPC1 and PPC2 in rescued lines was done using quantitative real-time PCR. Total RNA and cDNA of the leaves were prepared as described above. The primers used for expression analysis of PPC1 and PPC2 are listed in Supplemental Table S5. In addition, the PEPC activity of rescued transgenic lines was determined as described above.
Carbohydrate Quantification
Ten-day-old wild-type and ppc mutant seedlings grown on 1/2 MS medium with or without 15 mm malate or Glu were collected at midday. Soluble sugars were extracted from samples using 80% (v/v) ethanol for 1 h at 80°C and quantified by high-performance anion-exchange chromatography with pulsed amperometric detection in an ICS-3000 system (Dionex) fitted with a CarboPac PA1 column. NaOH (15–300 mm; Fluka) in Millipore water (VWR) was used as the eluent. Quantitative calculation of sugars was performed using Chromeleon software 6.7 (Dionex; Poschet et al., 2011).
For starch measurement, 10-d-old wild-type and ppc mutant seedlings grown on 1/2 MS medium with or without 15 mm malate or Glu were harvested at midday, ground in liquid N2, and extracted in ice-cold 0.7 m perchloric acid. The samples were washed three times with 80% (v/v) ethanol to remove residual soluble glucans and pigments and then resuspended in water at 90°C for 20 min. The starch in the insoluble fraction was digested with α-amylase and amyloglucosidase (Megazyme), and the released Glc was measured spectrophotometrically, as described previously (Smith and Zeeman, 2006).
Determination of Metabolite Levels by Gas Chromatography-Mass Spectrometry
Metabolite analysis by gas chromatography-mass spectrometry was carried out essentially as described previously (Roessner et al., 2000, 2001). A total of 100 mg of 10-d-old wild-type and ppc mutant seedlings grown on 1/2 MS medium with or without 15 mm malate or Glu was collected at midday, frozen immediately in liquid nitrogen, and stored at −70°C until further analysis. The seedling powder was extracted with 1,400 μL of 100% methanol. An internal standard (50 μL of a 2 mg mL−1 ribitol solution) was added for quantification. The mixture was extracted for 15 min at 70°C, mixed vigorously with 1 volume of water, and centrifuged at 2,200g. Aliquots of the methanol/water supernatant were dried under a vacuum for 6 to 16 h. The dried residue was dissolved and derivatized for 90 min at 30°C in 80 μL of 20 mg mL−1 methoxyamine hydrochloride in pyridine followed by a 30-min treatment with 80 μL of N-methyl-N-(trimethylsilyl)trifluoroacetamide at 37°C. Standard substances used for peak identification were dissolved in water at 10 mg mL−1. A standard solution (5 μL) was derivatized as described above. An aliquot of the derivative was injected into a gas chromatography-mass spectrometry system composed of an AS7683 autosampler, a GC6890N gas chromatograph, and an MS5973N mass spectrophotometer (Agilent). Signals were normalized to an internal standard molecule introduced into the samples (ribitol), thus enabling the relative quantification of metabolites. The amounts of absolute metabolite were determined using calibration curves obtained with standard compound mixtures.
Amino Acid Quantification
Approximately 100 mg of 10-d-old wild-type and ppc mutant seedlings grown on 1/2 MS medium with or without 15 mm malate or Glu was collected at midday. Free amino acids were extracted as described previously (Toka et al., 2010). Tissues were ground in liquid nitrogen, extracted in 8% (w/v) 5-sulfosalicylic acid for 1 h, and centrifuged for 10 min at 12,000g. The supernatants were filtered through 0.22-μm filters. Free amino acids were measured with an automated amino acid analyzer (Hitachi L-8900).
Determination of Nitrate and Ammonium Content
Ten-day-old wild-type and ppc mutant seedlings grown on 1/2 MS medium with or without 15 mm malate or Glu were collected at midday. Tissues were ground in liquid nitrogen, extracted in distilled water, and centrifuged for 10 min at 12,000g. The supernatants were filtered through 0.22-μm filters. Nitrate and ammonium were quantified by high-performance anion-exchange chromatography with pulsed amperometric detection in an ICS-3000 system (Dionex).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Identification of SALK T-DNA mutants in PPC genes.
Supplemental Table S1. Segregation analysis of T-DNA-tagged alleles.
Supplemental Table S2. Metabolite changes in ppc1/ppc2 mutants grown on 1/2 MS medium.
Supplemental Table S3. Metabolite changes in ppc1/ppc2 mutants grown on 1/2 MS medium with 15 mm glutamate.
Supplemental Table S4. Metabolite changes in ppc1/ppc2 mutants grown on 1/2 MS medium with 15 mm malate.
Supplemental Table S5. List of primers used in this study.
Supplementary Material
Acknowledgments
We thank Alison Smith (John Innes Centre) and Zhengkai Xu and Wei Zhang (both Shanghai University) for critical reading and valuable suggestions, Dr. Zhiping Deng and Qinghuan Liang (both Zhejiang Academy of Agricultural Sciences) for assistance in immunoblot analysis, Dean Jiang (Zhejiang University) for assistance in PEPC activity assay, Xijiao Song (Zhejiang Academy of Agricultural Sciences) for assistance in transmission electron microscopy, and Dr. Rebecca Horn (scientificproofreading.co.uk) for assistance in editing the article.
Glossary
- PEPC
phosphoenolpyruvate carboxylase
- PEP
phosphoenolpyruvate
- OAA
oxaloacetate
- T-DNA
transfer DNA
- 1/2 MS
one-half-strength Murashige and Skoog
- cDNA
complementary DNA
- GOGAT
glutamate synthase
- RT
reverse transcription
Footnotes
This work was supported by the National Natural Science Foundation of China (grant nos. 30430450 and 31272114), the Innovation and Improving Fund from the Zhejiang Academy of Agricultural Sciences (grant nos. 2010R21Y03E02 and 2014CX016), and the Open Fund of the State Key Laboratory Breeding Base for Zhejiang Sustainable Pest and Disease Control (grant no. 2010DS700124–KF1402).
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Andre C, Froehlich JE, Moll MR, Benning C (2007) A heteromeric plastidic pyruvate kinase complex involved in seed oil biosynthesis in Arabidopsis. Plant Cell 19: 2006–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Cao Y, Dai Y, Cui S, Ma L (2008) Histone H2B monoubiquitination in the chromatin of FLOWERING LOCUS C regulates flowering time in Arabidopsis. Plant Cell 20: 2586–2602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LM, Li KZ, Miwa T, Izui K (2004) Overexpression of a cyanobacterial phosphoenolpyruvate carboxylase with diminished sensitivity to feedback inhibition in Arabidopsis changes amino acid metabolism. Planta 219: 440–449 [DOI] [PubMed] [Google Scholar]
- Chollet R, Vidal J, O’Leary MH (1996) Phosphoenolpyruvate carboxylase: a ubiquitous, highly regulated enzyme in plants. Annu Rev Plant Physiol Plant Mol Biol 47: 273–298 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Deng Z, Zhang X, Tang W, Oses-Prieto JA, Suzuki N, Gendron JM, Chen H, Guan S, Chalkley RJ, Peterman TK, et al. (2007) A proteomics study of brassinosteroid response in Arabidopsis. Mol Cell Proteomics 6: 2058–2071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forde BG, Lea PJ (2007) Glutamate in plants: metabolism, regulation, and signalling. J Exp Bot 58: 2339–2358 [DOI] [PubMed] [Google Scholar]
- Gennidakis S, Rao S, Greenham K, Uhrig RG, O’Leary B, Snedden WA, Lu C, Plaxton WC (2007) Bacterial- and plant-type phosphoenolpyruvate carboxylase polypeptides interact in the hetero-oligomeric class-2 PEPC complex of developing castor oil seeds. Plant J 52: 839–849 [DOI] [PubMed] [Google Scholar]
- Gottlob-McHugh SG, Sangwan RS, Blakeley SD, Vanlerberghe GC, Ko K, Turpin DH, Plaxton WC, Miki BL, Dennis DT (1992) Normal growth of transgenic tobacco plants in the absence of cytosolic pyruvate kinase. Plant Physiol 100: 820–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gousset-Dupont A, Lebouteiller B, Monreal J, Echevarria C, Pierre JN, Hodges M, Vidal JN, Hodges M, Vidal J (2005) Metabolite and post-translational control of phosphoenolpyruvate carboxylase from leaves and mesophyll cell protoplasts of Arabidopsis thaliana. Plant Sci 169: 1096–1101 [Google Scholar]
- Gregory AL, Hurley BA, Tran HT, Valentine AJ, She YM, Knowles VL, Plaxton WC (2009) In vivo regulatory phosphorylation of the phosphoenolpyruvate carboxylase AtPPC1 in phosphate-starved Arabidopsis thaliana. Biochem J 420: 57–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges M. (2002) Enzyme redundancy and the importance of 2-oxoglutarate in plant ammonium assimilation. J Exp Bot 53: 905–916 [DOI] [PubMed] [Google Scholar]
- Igawa T, Fujiwara M, Tanaka I, Fukao Y, Yanagawa Y (2010) Characterization of bacterial-type phosphoenolpyruvate carboxylase expressed in male gametophyte of higher plants. BMC Plant Biol 10: 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail MS, Wynendaele W, Aerts JL, Paridaens R, Van Mellaert L, Anné J, Gaafar R, Shakankiry N, Khaled HM, Christiaens MR, et al. (2003) Real-time quantitative RT-PCR and detection of tumour cell dissemination in breast cancer patients: plasmid versus cell line dilutions. Ann Oncol 14: 1241–1245 [DOI] [PubMed] [Google Scholar]
- Kang J, Turano FJ (2003) The putative glutamate receptor 1.1 (AtGLR1.1) functions as a regulator of carbon and nitrogen metabolism in Arabidopsis thaliana. Proc Natl Acad Sci USA 100: 6872–6877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles VL, McHugh SG, Hu Z, Dennis DT, Miki BL, Plaxton WC (1998) Altered growth of transgenic tobacco lacking leaf cytosolic pyruvate kinase. Plant Physiol 116: 45–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XR, Wang L, Ruan YL (2010) Developmental and molecular physiological evidence for the role of phosphoenolpyruvate carboxylase in rapid cotton fibre elongation. J Exp Bot 61: 287–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamedov TG, Moellering ER, Chollet R (2005) Identification and expression analysis of two inorganic C- and N-responsive genes encoding novel and distinct molecular forms of eukaryotic phosphoenolpyruvate carboxylase in the green microalga Chlamydomonas reinhardtii. Plant J 42: 832–843 [DOI] [PubMed] [Google Scholar]
- Masumoto C, Miyazawa S, Ohkawa H, Fukuda T, Taniguchi Y, Murayama S, Kusano M, Saito K, Fukayama H, Miyao M (2010) Phosphoenolpyruvate carboxylase intrinsically located in the chloroplast of rice plays a crucial role in ammonium assimilation. Proc Natl Acad Sci USA 107: 5226–5231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmo HG. (2003) Control of the phosphorylation of phosphoenolpyruvate carboxylase in higher plants. Arch Biochem Biophys 414: 189–196 [DOI] [PubMed] [Google Scholar]
- O’Leary B, Park J, Plaxton WC (2011a) The remarkable diversity of plant PEPC (phosphoenolpyruvate carboxylase): recent insights into the physiological functions and post-translational controls of non-photosynthetic PEPCs. Biochem J 436: 15–34 [DOI] [PubMed] [Google Scholar]
- O’Leary B, Rao SK, Kim J, Plaxton WC (2009) Bacterial-type phosphoenolpyruvate carboxylase (PEPC) functions as a catalytic and regulatory subunit of the novel class-2 PEPC complex of vascular plants. J Biol Chem 284: 24797–24805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Leary B, Rao SK, Plaxton WC (2011b) Phosphorylation of bacterial-type phosphoenolpyruvate carboxylase at Ser425 provides a further tier of enzyme control in developing castor oil seeds. Biochem J 433: 65–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poschet G, Hannich B, Raab S, Jungkunz I, Klemens PA, Krueger S, Wic S, Neuhaus HE, Büttner M (2011) A novel Arabidopsis vacuolar glucose exporter is involved in cellular sugar homeostasis and affects the composition of seed storage compounds. Plant Physiol 157: 1664–1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademacher T, Häusler RE, Hirsch HJ, Zhang L, Lipka V, Weier D, Kreuzaler F, Peterhänsel C (2002) An engineered phosphoenolpyruvate carboxylase redirects carbon and nitrogen flow in transgenic potato plants. Plant J 32: 25–39 [DOI] [PubMed] [Google Scholar]
- Roessner U, Luedemann A, Brust D, Fiehn O, Linke T, Willmitzer L, Fernie A (2001) Metabolic profiling allows comprehensive phenotyping of genetically or environmentally modified plant systems. Plant Cell 13: 11–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner U, Wagner C, Kopka J, Trethewey RN, Willmitzer L (2000) Technical advance: simultaneous analysis of metabolites in potato tuber by gas chromatography-mass spectrometry. Plant J 23: 131–142 [DOI] [PubMed] [Google Scholar]
- Rolletschek H, Borisjuk L, Radchuk R, Miranda M, Heim U, Wobus U, Weber H (2004) Seed-specific expression of a bacterial phosphoenolpyruvate carboxylase in Vicia narbonensis increases protein content and improves carbon economy. Plant Biotechnol J 2: 211–219 [DOI] [PubMed] [Google Scholar]
- Sánchez R, Cejudo FJ (2003) Identification and expression analysis of a gene encoding a bacterial-type phosphoenolpyruvate carboxylase from Arabidopsis and rice. Plant Physiol 132: 949–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez R, Flores A, Cejudo FJ (2006) Arabidopsis phosphoenolpyruvate carboxylase genes encode immunologically unrelated polypeptides and are differentially expressed in response to drought and salt stress. Planta 223: 901–909 [DOI] [PubMed] [Google Scholar]
- Scheible WR, Gonzalez-Fontes A, Lauerer M, Muller-Rober B, Caboche M, Stitt M (1997) Nitrate acts as a signal to induce organic acid metabolism and repress starch metabolism in tobacco. Plant Cell 9: 783–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneidereit J, Häusler RE, Fiene G, Kaiser WM, Weber AP (2006) Antisense repression reveals a crucial role of the plastidic 2-oxoglutarate/malate translocator DiT1 at the interface between carbon and nitrogen metabolism. Plant J 45: 206–224 [DOI] [PubMed] [Google Scholar]
- Smith AM, Zeeman SC (2006) Quantification of starch in plant tissues. Nat Protoc 1: 1342–1345 [DOI] [PubMed] [Google Scholar]
- Stitt M, Müller C, Matt P, Gibon Y, Carillo P, Morcuende R, Scheible WR, Krapp A (2002) Steps towards an integrated view of nitrogen metabolism. J Exp Bot 53: 959–970 [DOI] [PubMed] [Google Scholar]
- Sullivan S, Jenkins GI, Nimmo HG (2004) Roots, cycles and leaves: expression of the phosphoenolpyruvate carboxylase kinase gene family in soybean. Plant Physiol 135: 2078–2087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toka I, Planchais S, Cabassa C, Justin AM, De Vos D, Richard L, Savouré A, Carol P (2010) Mutations in the hyperosmotic stress-responsive mitochondrial BASIC AMINO ACID CARRIER2 enhance proline accumulation in Arabidopsis. Plant Physiol 152: 1851–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhrig RG, She YM, Leach CA, Plaxton WC (2008) Regulatory monoubiquitination of phosphoenolpyruvate carboxylase in germinating castor oil seeds. J Biol Chem 283: 29650–29657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijgen L, Keyaerts E, Moës E, Maes P, Duson G, Van Ranst M (2005) Development of one-step, real-time, quantitative reverse transcriptase PCR assays for absolute quantitation of human coronaviruses OC43 and 229E. J Clin Microbiol 43: 5452–5456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voll LM, Hajirezaei MR, Czogalla-Peter C, Lein W, Stitt M, Sonnewald U, Börnke F (2009) Antisense inhibition of enolase strongly limits the metabolism of aromatic amino acids, but has only minor effects on respiration in leaves of transgenic tobacco plants. New Phytol 184: 607–618 [DOI] [PubMed] [Google Scholar]
- Westrama A, Lloyd JR, Roessner U, Riesmeier JW, Kossmann J (2002) Increases of 3-phosphoglyceric acid in potato plants through antisense reduction of cytoplasmic phosphoglycerate mutase impairs photosynthesis and growth, but does not increase starch contents. Plant Cell Environ 25: 1133–1143 [Google Scholar]
- Zhou J, Jiao F, Wu Z, Li Y, Wang X, He X, Zhong W, Wu P (2008) OsPHR2 is involved in phosphate-starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiol 146: 1673–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.