A network of transcription factors regulates arbuscular mycorrhizal symbiosis in Lotus.
Abstract
Arbuscular mycorrhizal (AM) fungi, in symbiosis with plants, facilitate acquisition of nutrients from the soil to their host. After penetration, intracellular hyphae form fine-branched structures in cortical cells termed arbuscules, representing the major site where bidirectional nutrient exchange takes place between the host plant and fungus. Transcriptional mechanisms underlying this cellular reprogramming are still poorly understood. GRAS proteins are an important family of transcriptional regulators in plants, named after the first three members: GIBBERELLIC ACID-INSENSITIVE, REPRESSOR of GAI, and SCARECROW. Here, we show that among 45 transcription factors up-regulated in mycorrhizal roots of the legume Lotus japonicus, expression of a unique GRAS protein particularly increases in arbuscule-containing cells under low phosphate conditions and displays a phylogenetic pattern characteristic of symbiotic genes. Allelic rad1 mutants display a strongly reduced number of arbuscules, which undergo accelerated degeneration. In further studies, two RAD1-interacting proteins were identified. One of them is the closest homolog of Medicago truncatula, REDUCED ARBUSCULAR MYCORRHIZATION1 (RAM1), which was reported to regulate a glycerol-3-phosphate acyl transferase that promotes cutin biosynthesis to enhance hyphopodia formation. As in M. truncatula, the L. japonicus ram1 mutant lines show compromised AM colonization and stunted arbuscules. Our findings provide, to our knowledge, new insight into the transcriptional program underlying the host’s response to AM colonization and propose a function of GRAS transcription factors including RAD1 and RAM1 during arbuscule development.
Plant roots absorb inorganic phosphorus and most other nutrients from the soil to maintain normal growth. Phosphorus depletion in the rhizosphere favored the evolution of adaptive strategies in response to low phosphate (Pi) conditions, such as the formation of symbioses with arbuscular mycorrhizal fungi (AMF), which improves Pi uptake at the cost of photosynthesis-derived carbon. About 80% of terrestrial plants are colonized by AMF.
Under conditions of Pi starvation, plant roots exude strigolactones into the rhizosphere, which induce the germination of arbuscular mycorrhizal (AM) fungal spores and subsequent branching of fungal hyphae (Akiyama et al., 2005; Besserer et al., 2006). Strigolactones are short-lived molecules because of an ester bond that readily hydrolyzes in water. As a consequence, it is presumed that germinated external hyphae grow following the concentration gradient of the strigolactones toward the roots. Growing hyphae then contact the root surface and form hyphopodia, preparing for penetration. AM fungal hyphae secrete short-chain lipochitin oligosaccharides (LCOs) and short-chain chitin oligomers to provoke plant roots into an early invasion response (Maillet et al., 2011; Genre et al., 2013). Subsequently, in root epidermal and subjacent cortex cells, nuclei move toward the hyphal penetration sites and direct the formation of the prepenetration apparatus (PPA), lined with microtubules and endoplasmic reticulum, which guide the hyphae into the root endosphere (Genre et al., 2005, 2008). Once the hyphae grow into the cortical cells, they form highly branched structures, the arbuscules (Latin for little tree). Arbuscules are separated from the cytoplasm by the periarbuscular membrane, which is the continuous plant plasma membrane surrounding the arbuscule. The periarbuscular membrane contains a repertoire of proteins that facilitate its distinct functions, such as mycorrhiza-specific Pi transporters (Rausch et al., 2001; Harrison et al., 2002; Paszkowski et al., 2002; Karandashov et al., 2004; Kobae and Hata, 2010; Pumplin et al., 2012) and half-size ATP-binding cassette transporters stunted arbuscule (STR)/STR2 (Zhang et al., 2010). Arbuscules have a short life span of approximately 4 to 15 d with mature arbuscules subsequently undergoing collapse concomitant with the appearance of septa before disappearing, leaving the cells for recolonization (Sanders et al., 1977).
During evolution, AM symbiosis (AMS) emerged about 400 million years earlier than the rhizobium-legume symbiosis (RLS; Redecker et al., 2000; Parniske, 2008) and is thought to have played a critical role during land colonization by plants (Humphreys et al., 2010). Genetic evidence shows that both symbioses share a common symbiosis signaling pathway (CSSP), which is composed of Leu-rich repeat receptor-like kinase (SYMBIOSIS RECEPTOR-LIKE KINASE [SYMRK] in Lotus japonicus/DOES NOT MAKE INFECTIONS2 [DMI2] in Medicago truncatula), cation channels (CASTOR and POLLUX in L. japonicus/DMI1 in M. truncatula), lectin nucleotide phosphohydrolase, calcium/calmodulin-dependent protein kinase (CCAMK; DMI3), an interacting protein of DMI3 (CYCLOPS in L. japonicus/INTERACTING PROTEIN OF DOES NOT MAKE INFECTIONS3 [IPD3] in M. truncatula), components of the nuclear pore complex (NUP85, NUP133, and NENA), and the GRAS (for GIBBERELLIC ACID-INSENSITIVE, REPRESSOR of GAI, and SCARECROW [SCR]) transcription factor NODULATION SIGNALING PATHWAY2 (NSP2) (Catoira et al., 2000; Endre et al., 2002; Stracke et al., 2002; Ané et al., 2004; Lévy et al., 2004; Imaizumi-Anraku et al., 2005; Kistner et al., 2005; Kanamori et al., 2006; Messinese et al., 2007; Saito et al., 2007; Gutjahr et al., 2008; Groth et al., 2010; Maillet et al., 2011; Roberts et al., 2013). SYMRK, nucleoporins, and cation channels are genetically upstream of calcium spiking, which is subsequently decoded by CCaMK (Saito et al., 2007; Charpentier et al., 2008; Kosuta et al., 2008).
Plant-specific GRAS transcription factors play an important regulatory role in root and shoot development, the GA3 and phytochrome A signaling pathways, abiotic stress, and symbiosis (Di Laurenzio et al., 1996; Peng et al., 1997; Pysh et al., 1999; Bolle et al., 2000; Greb et al., 2003; Kaló et al., 2005; Smit et al., 2005; Fode et al., 2008). The GRAS protein family was divided into DELLA, HAIRY MERISTEM, LILIUM LONGIFLORUM SCARECROW-LIKE, PHYTOCHROME A SIGNAL TRANSDUCTION1, LATERAL SUPPRESSOR, SCR, SHORT-ROOT, and SCARECROW-LIKE3 subfamilies, each with distinct conserved domains and functions (Tian et al., 2004). In symbiosis, the CSSP is divergent from GRAS complexes. NSP2 forms a DNA-binding complex with another GRAS transcription factor, NODULATION SIGNALING PATHWAY1 (NSP1), to elevate early nodulin gene expression during rhizobial infection (Hirsch et al., 2009). NSP2 interacts with GRAS transcription factor REDUCED ARBUSCULAR MYCORRHIZATION1 (RAM1) to induce RAM2, encoding a glycerol-3-phosphate acyl transferase (GPAT), to facilitate hyphopodia formation on the root surface during mycorrhizal colonization (Gobbato et al., 2012). GPAT initiates de novo glycerolipid synthesis and participates in the biosynthesis of cutin and suberin, which are the polymer matrices of the cell wall (Beisson et al., 2007; Li et al., 2007). Phenotypically, RAM1, but not NSP1, is required for mycorrhizal factor-induced lateral root growth (Maillet et al., 2011; Gobbato et al., 2012). Moreover, DELLA proteins and GA3 were reported to antagonistically regulate arbuscule development in pea (Pisum sativum), M. truncatula, and rice (Oryza sativa; Floss et al., 2013; Foo et al., 2013; Yu et al., 2014). A GA3-deficient mutant showed enhanced mycorrhization in pea and exogenous GA3 repressed mycorrhization in three species. Meanwhile, severely impaired arbuscules were observed in mutants of DELLA orthologs. DELLA interacting protein (DIP1) was identified in rice, and RAM1 was shown to interact with DIP1 (Yu et al., 2014). This suggests that formation of multicomponent GRAS transcription factor complexes is a prerequisite for elicitation of nodulation or mycorrhization (Oldroyd, 2013).
Arbuscule development occurs in genetically separable stages, including (1) PPA formation, (2) fungal cell entry, (3) birdsfoot formation (lowly branched arbuscule resembling a bird’s foot), (4) highly branched hyphae forming a mature arbuscule, and (5) arbuscule collapse including formation of septa to disconnect old arbuscules from the hyphal network (Gutjahr and Parniske, 2013). Defective CCaMK and CYCLOPS resulted in the arrest of PPA formation. The Vapyrin gene from M. truncatula and its ortholog PENETRATION AND ARBUSCULE MORPHOGENESIS1 (PAM1) from Petunia hybrida are required for both AMS and RLS (Feddermann et al., 2010; Pumplin et al., 2010). Down-regulation of Vapyrin by RNA interference (RNAi) displayed a significant reduction in hyphopodium penetration into the epidermal cell, and hyphae failed to enter the cortical cells (Pumplin et al., 2010). In pam1 mutants, although few arbuscules were observed in the cortical cells, their growth was arrested at the early point of branching (Feddermann et al., 2010; Pumplin et al., 2010). Half-size ATP-binding cassette transporters STR/STR2 were shown to be essential for mature arbuscule formation. In M. truncatula and rice, loss of function mutations in STR genes resulted in stunted arbuscules (Zhang et al., 2010; Gutjahr et al., 2012), suggesting a role of STR proteins in the transport of metabolites that are essential for a functional root-AMF interaction. Mycorrhiza-specific Pi transporters, such as M. truncatula PHOSPHATE TRANSPORTER4 (MtPT4), Rice PHOSPHATE TRANSPORTER11 (OsPT11), and maize (Zea mays) PHOSPHATE TRANSPORTER1;6 (Pht1;6), localize to arbuscule-containing cells and are indispensable for maintenance of arbuscule function (Javot et al., 2007; Yang et al., 2012; Willmann et al., 2013). Loss-of-function mutants and RNAi lines of MtPT4, OsPT11, and maize pht1;6 mutant led to reduced root colonization with premature degeneration of the arbuscules. In addition, RNAi knockdown of VESICLE ASSOCIATED MEMBRANE PROTEIN72 in M. truncatula or SOLUBLE N-ETHYLMALEIMIDE-SENSITIVE FACTOR ATTACHMENT PROTEIN RECEPTOR family member L. japonicus VESICLE TRANSPORT THROUGH T-SNARE INTERACTION12 in L. japonicus led to distorted arbuscular development, which provided evidence for a role of these proteins in vesicle trafficking during arbuscule formation (Ivanov et al., 2012; Lota et al., 2013). Moreover, two L. japonicus mutants were reported to be affected in distinct steps of arbuscule development but not during RLS (Groth et al., 2013).
While transcriptional changes in arbuscule-containing cortical cells were recently reported (Hogekamp et al., 2011), the sophisticated regulatory mechanisms underlying these changes are still poorly understood. Here we report that a unique GRAS protein, REQUIRED FOR ARBUSCULE DEVELOPMENT1 (RAD1), regulates arbuscule development. RAD1 interacts with RAM1 and LjNSP2, respectively, providing support for the existence of a GRAS protein-protein interaction network in arbuscule development.
RESULTS
Gene Expression Profiling Reveals Transcription Factors Related to AM Colonization in L. japonicus
To investigate transcriptional regulation of arbuscule development and to screen for candidate transcription factor genes, roots of L. japonicus Gifu-129, colonized for 8 weeks by Rhizophagus irregularis and resupplied or not with Pi, were subjected to deep sequencing analysis of messenger RNA (RNAseq). Two independent biological replicates exhibiting similar mycorrhization rates and induction of arbuscular marker gene expression (LjPT4) were used. Application of a minimum 2-fold induction at a significance level of P < 0.01 was used as a cutoff for significantly up-regulated genes. As a result, 45 genes encoding transcription factors or transcriptional regulators were significantly up-regulated by R. irregularis compared with nonmycorrhizal roots grown in a similar low-Pi environment (Fig. 1A; Supplemental Table S1). These genes encoded GRAS (18 members), APETALA2 (AP2)/ETHYLENE RESPONSE FACTOR (ERF; eight), NO APICAL MERISTEM, ARABIDOPSIS TRANSCRIPTION ACTIVATION FACTOR and CUP-SHAPED COTYLEDON (NAC)-domain family members (four), MYELOBLASTOSIS (MYB; three), C2H2 zinc finger (five), MADS DNA-binding domain (MADS [from MINICHROMOSOME MAINTENANCE1, AGAMOUS, DEFICIENS and SERUM RESPONSE FACTOR]; two), BASIC LEUCINE ZIPPER (bZIP; one), AUXIN RESPONSE FACTOR (ARF; one), WRKY DNA binding domain (one), NODULE INCEPTION (NIN)-LIKE protein (one), and LATERAL ORGAN BOUNDARIES (LOB) domain (one) transcription factors, indicating the complexity of transcriptional regulation of AMS. They are distinguishable in four general expression patterns: genes that were overall highly expressed (11), genes that were only responsive to AM under low-phosphorus conditions (23), genes with high expression under low Pi regardless of AM status (nine), and the rest of intermediate status (i.e. with no exact classification; two; Supplemental Table S2). Although many of these transcription factors were not reported previously, some of them or their homologs have been shown to be up-regulated in AM (Guether et al., 2009; Hogekamp et al., 2011; Gaude et al., 2012; Gobbato et al., 2012; Devers et al., 2013; Volpe et al., 2013).
Figure 1.
Global analysis of mycorrhiza up-regulated transcription factors. A, Hierarchical clustering of mycorrhizal (+M) and nonmycorrhizal wild-type Gifu-129 expression profiles based on transcript abundance of the genome-wide 45 AM up-regulated transcription factors identified by RNAseq (adjusted P < 0.01, fold change > 2). Gifu-129 plants were grown for 8 weeks at low Pi (5.5 μm; low P) or high Pi (7.5 mm; high P) conditions, respectively, or they were grown at low Pi for 6 weeks with a subsequent shift from low Pi to high Pi for 2 weeks (shift). Significantly up-regulated transcription factors are shown in a heat map drawn by using R. Log10 values of normalized counts per transcript plus one were used. Transcription factor families are labeled above the genes. The closest homologs and RAD1 were labeled below the plot. Asterisks indicate the genes whose expression was confirmed by qRT-PCR in Supplemental Figure S1. B, The 43 AM up-regulated transcription factors were clustered into three groups in AM host and nonhost species. Amino acid sequences of these L. japonicus AM up-regulated transcription factors were utilized for reciprocal blast in AM nonhost Brassicaceae (Capsella rubella, Thellungiella halophila, Brassica rapa, Arabidopsis lyrata, and Arabidopsis [Arabidopsis thaliana]), Lupinus angustifolius, and AM hosts (M. truncatula, Populus trichocarpa, Carica papaya, Gossypium raimondii, and Vitis vinifera). Symbols indicate the presence (+) or absence (−) of the capacity for symbiosis. Detected homologs are labeled with dark blue. Lack of homolog detection was labeled with white.
To investigate the distribution of these genes in plant species, a reciprocal blast survey in AM nonhosts (L. angustifolius, C. rubella, T. halophila, B. rapa, A. lyrata, and Arabidopsis) and AM hosts (M. truncatula, P. trichocarpa, C. papaya, G. raimondii, and V. vinifera) was carried out. Except for the partial sequences of chr1.CM1413.480.r2.d and L. japonicus SELECTED GENOME SHOTGUN ASSEMBLY (LjSGA)_055804.0.1, the remaining 43 transcription factor coding sequences were clustered into three groups: conserved genes that occur in host and nonhost species (8), host-specific or correlated (16+4), and potential L. japonicus-specific genes (15), respectively (Fig. 1B). Interestingly, out of the 16 AM host-specific genes, 10 were absent in L. angustifolius (Fig. 1B), which is a known AM nonhost but still retains the ability to associate with nitrogen-fixing rhizobia (Oba et al., 2001; Schulze et al., 2006). This suggested the important role of these 10 transcription factors in AMS. To further determine the absence of the AM host-specific transcription factors in the Brassicaceae, microsynteny analysis was performed. We compared genomic blocks encompassing these genes in M. truncatula and P. trichocarpa and the corresponding blocks in Arabidopsis. Indeed, this synteny analysis supports the orthology between both host species for most of the putative host-specific genes (15 of 16; Supplemental Text S1). By contrast, none of the syntenic regions in Arabidopsis contains homologs of these genes, supporting their absence in this well-characterized nonhost species (Supplemental Text S1).
To verify AM-dependent expression of the previously described genes identified by RNAseq, quantitative reverse transcription (qRT)-PCR was performed. We tested eight genes, and all of them showed a similar expression pattern compared to the RNAseq results, although the fold induction rates were slightly different (Supplemental Fig. S1). Mycorrhization and AM marker gene expression are generally suppressed under high-Pi conditions, which was recently suggested to be mediated by GA3 signaling and low levels of DELLA proteins (Nagy et al., 2009; Breuillin et al., 2010; Chen et al., 2011; Floss et al., 2013; Lota et al., 2013). Consistently, the expression of all eight tested mycorrhiza up-regulated transcription factors was repressed after a 2-week high-Pi treatment (Supplemental Fig. S1).
LORE1a Insertion Mutant rad1 Displays Impaired Mycorrhization
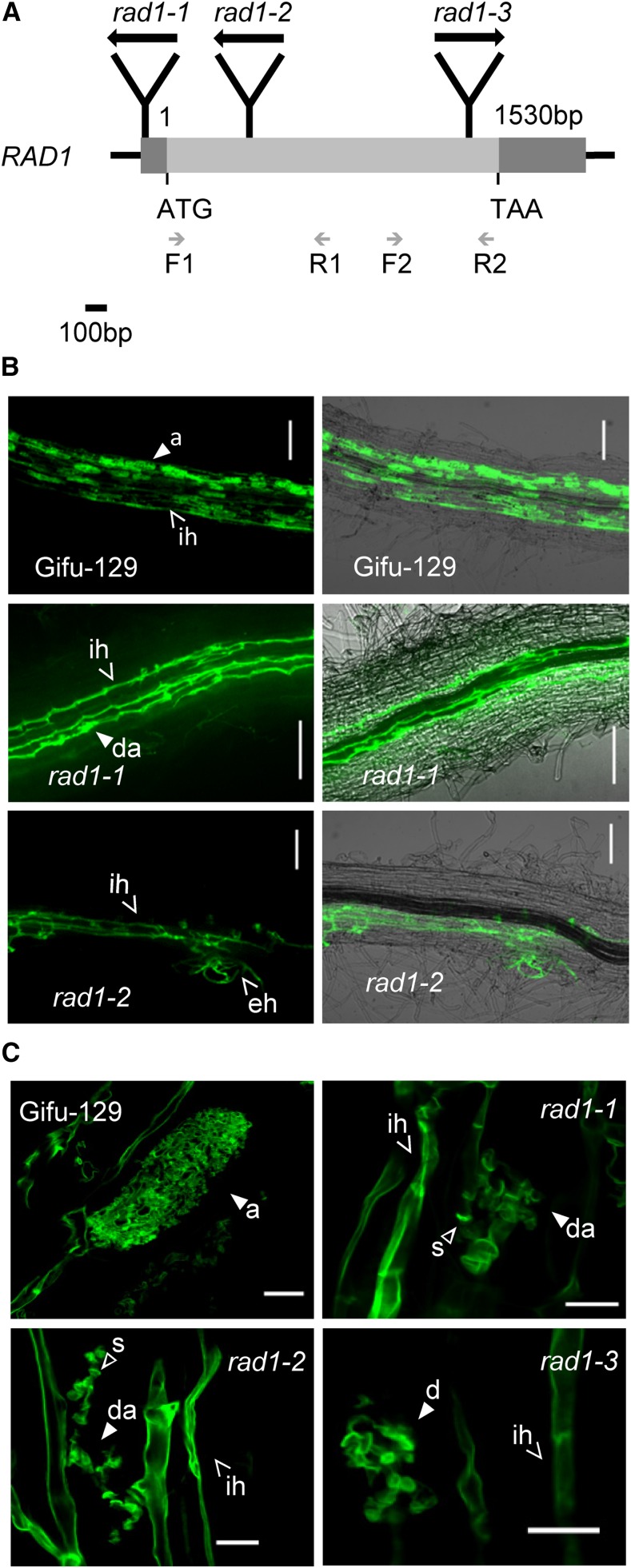
To identify important transcriptional regulators in AMS, mutants of these candidate transcription factor genes, containing LOTUS RETROTRANSPOSON 1A (LORE1a) transposon insertions (Fukai et al., 2012; Urbański et al., 2012), were screened for potential defects in AMS with R. irregularis. Subsequently, mycorrhization rate and arbuscule morphology were surveyed at 8 weeks postinoculation (wpi). Mycorrhizal roots of a mutant with a LORE1a insertion in the 5′ untranslated region (UTR) of a GRAS protein gene were found to contain a high number of stunted arbuscules (Supplemental Fig. S2A). We therefore named the gene RAD1 (chr4.CM1864.540.r2.m) and the corresponding mutant as rad1-1. The full-length RAD1 complementary DNA (cDNA) sequence corresponding to the intronless gene is 1530 bp long, and the predicted protein size is 56.7 kD. To further confirm the function of RAD1 in AM development, two other alleles, rad1-2 and rad1-3, were identified with LORE1a insertions in the RAD1 coding region (Fig. 2A). The RAD1 transcripts were detected in three allelic rad1 mutants (Supplemental Fig. S2B). Both the 5′ and 3′ partial transcripts of RAD1 were significantly reduced in rad1-1, which indicated that the insertion in the 5′ UTR leads to a knockdown of RAD1 gene expression. In rad1-2, the 5′ partial transcript of RAD1 was barely detectable, whereas the 3′ transcript could be identified at a very low level. In rad1-3, the 5′ transcript level was comparable to that in Gifu-129, whereas the 3′ transcript was not detectable.
Figure 2.
Mycorrhizal phenotype of rad1 allelic mutants. A, RAD1 (chr4.CM1864.540.r2.m) gene structure is shown at scale, and LORE1a insertion sites are illustrated in mutants rad1-1 (−71 bp upstream of the translation start codon [ATG]), rad1-2 (359 bp downstream of start ATG), and rad1-3 (1,379 bp downstream of start ATG). TAA indicates the translation stop codon. Black arrows indicate the 5′ to 3′ direction of the LORE1a transposon sequence. Gray arrows indicate the specific primers used for RT-PCR. Light-gray bar represents the coding region, and dark-gray bars represent the 5′ UTR and 3′ UTR. B, Confocal microscopy images of R. irregularis in the roots of Gifu-129 and the rad1 allelic homozygous mutants. Images at right show the overlay of fluorescence and bright-field images. Scale bars = 100 µm. C, Defective arbuscules in rad1 allelic homozygous mutants. Fungal structures were stained with WGA-Alexa Fluor 488. White arrowheads indicate characteristic mycorrhizal structures: a, arbuscule; ih, intraradical hyphae; eh, extraradical hyphae; da, degenerated arbuscule; and s, septa. Scale bars = 10 µm.
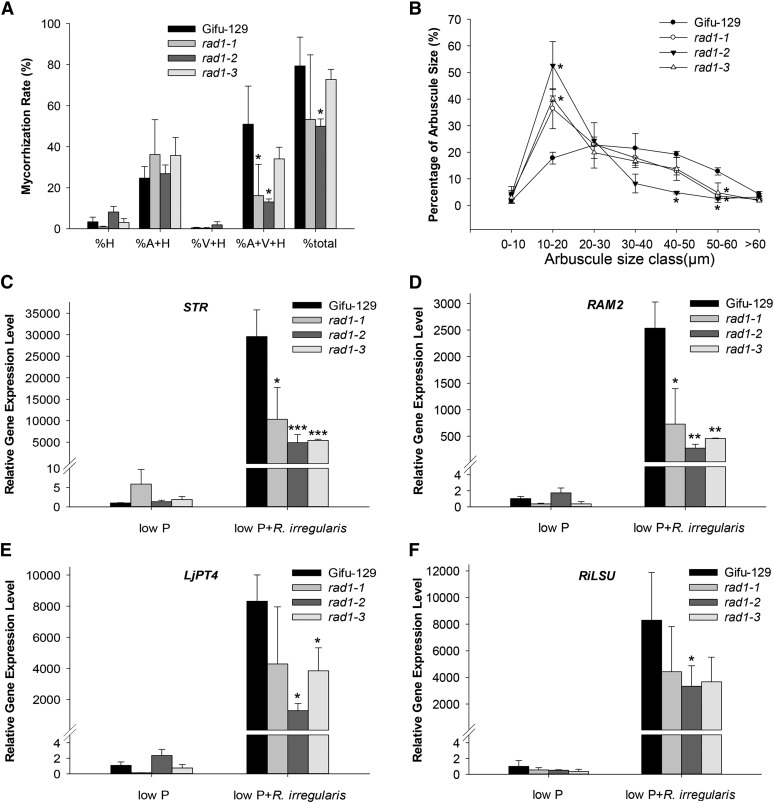
Although the total mycorrhization rates in both rad1-1 and rad1-3 were indistinguishable from that in the wild type at 6 wpi, a significantly reduced degree of mycorrhization was observed in rad1-2, especially concerning the percentage of sectors with arbuscules, vesicles, and hyphae (Fig. 3A). In both rad1-2 and rad1-3, arbuscule number and size were significantly less than in wild-type Gifu-129, accompanied by accelerated arbuscule degeneration (Figs. 2B and 3B). In rad1-2 and rad1-3, 52% and 40%, respectively, of arbuscules ranged from 10 to 20 μm in size, whereas 18% in the wild type were of this diameter (Fig. 3B). Likewise, 36% of arbuscules in rad1-1 ranged from 10 to 20 μm in size, which was an insignificant difference from the control due to relatively large variation between biological replicates. In addition, the percentage of arbuscules ranging from 50 to 60 μm was much less in all three mutant alleles. In particular, arbuscules larger than 40 μm were rarely observed in rad1-2. The arbuscule phenotype was more severe in rad1-2 than that in rad1-1 and rad1-3, which was consistent with the different RAD1 transcript levels in the three allelic mutants. Intriguingly, the arbuscules in the mutants collapsed prematurely with increased numbers of septa compared with Gifu-129 (Fig. 2C). This indicated that the GRAS transcription factor RAD1 is essential for regulating arbuscule morphology and senescence.
Figure 3.
Mycorrhizal phenotype and marker gene expression in allelic rad1 mutants. A, Mycorrhization rate of Gifu-129 and three allelic rad1 mutant lines in the presence of R. irregularis at 6 wpi. Four plants per pot served as one biological replicate. Three biological replicates were used. Asterisks indicate significant differences between the wild type and rad1 mutant using Student’s t test (*P < 0.05). Error bars represent sd (n = 3). At least four independent experiments were performed with similar results. Percent total colonization and percentage of different fungal structures in colonized roots as indicated on x axis were counted. H, Hyphae; V, vesicle; A, arbuscule. B, Frequency of arbuscule size classes in the wild-type Gifu-129 and three allelic rad1 mutant lines. Mean value of frequency was from three biological replicates with approximately 90 root segments per genotype. Approximately 400 arbuscules were measured per genotype. Error bars represent sd (n = 3). Student’s t test was utilized between Gifu-129 and rad1 mutant at each size cluster. Asterisks indicate significant differences between frequency values of Gifu-129 and genotypes corresponding to the respective symbol closest to the asterisk (P < 0.05). C to F, Marker gene expression of STR, RAM2, LjPT4, and R. irregularis large subunit (RiLSU) in the presence or absence of R. irregularis under low-Pi conditions. Four plants per pot served as a biological replicate. Mean values of three biological replicates are presented. Error bars represent sd. Asterisks indicate significant differences (Student’s t test, *P ≤ 0.05, **P < 0.01, and ***P < 0.001).
To reveal transcriptional changes resulting from defective RAD1, expression of AM marker genes correlating to mature arbuscule formation was studied in rad alleles relative to Ubiquitin. Six weeks postinoculation, expression of marker genes of arbuscule development stage IV, such as STR and RAM2 as well as LjPT4, was significantly reduced in rad1 allelic mutants (Fig. 3, C–E). Primers specific for R. irregularis large subunit ribosomal RNA were used to demonstrate fungal RNA levels (van Tuinen et al., 1998; Fig. 3F). Expression of AM-inducible marker genes was also compromised in rad1-2 and rad1-3 in a time course experiment (Supplemental Fig. S3). These data indicated that impairment of RAD1 inhibits the formation of highly branched arbuscules and the corresponding marker gene expression. Additionally, constitutive overexpression of RAD1 in L. japonicus hairy roots resulted in a significantly increased mycorrhization rate and number of sectors with arbuscules (Supplemental Fig. S4A). Exemplarily, three root samples were analyzed for RAD1 overexpression, exhibiting on average a 4-fold higher RAD1 expression in the Cauliflower mosaic virus (CaMV) 35S promoter 35Spro:RAD1 transgenic roots compared with hairy roots harboring the empty vector (Supplemental Fig. S4B). Moreover, expression levels of LjPT4 were also increased in these roots (Supplemental Fig. S4C). This result further confirmed RAD1 function in arbuscule development.
Expression of RAD1 Is Root Specific and Enhanced in Arbuscule-Containing Cells
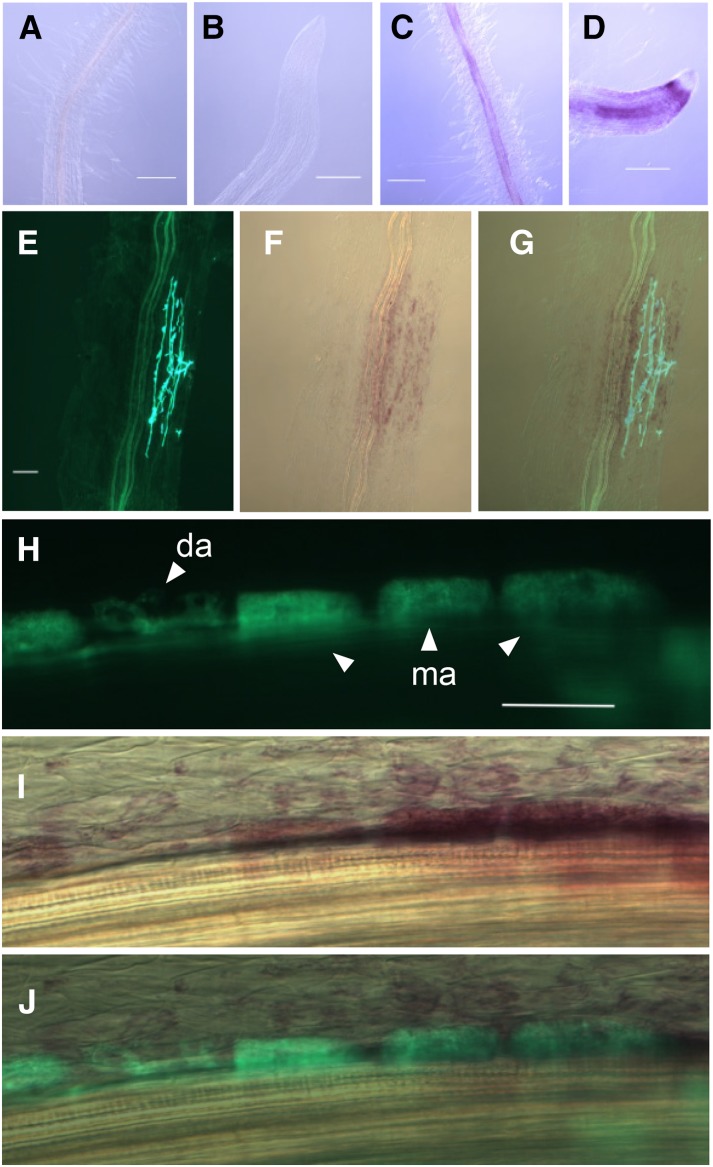
The gene expression of RAD1 was detectable in root tissue by reverse transcription (RT)-PCR in colonized and noncolonized plants under low-Pi conditions (Supplemental Fig. S5A). To elucidate the spatial pattern of RAD1 gene expression, a 1.9-kb promoter region of RAD1 was fused to GUS generating the chimeric gene RAD1pro:GUS for Agrobacterium rhizogenes-mediated transformation of hairy roots. Discosoma spp. RED FLUORESCENT PROTEIN1 encoded by the pRedroot vector was used for the selection of successful genetic transformation. Consistent with the qRT-PCR results (Supplemental Fig. S1), GUS was not detected under high-Pi conditions (Fig. 4, A and B). RAD1 could be induced by low Pi, and moderate GUS staining was detected in the cells closest to the vascular cylinder (Fig. 4, C and D). After colonization with R. irregularis, RAD1pro:GUS expression was observed in the presence of intraradical hyphae in epidermal cells and in the outer cortical cells (Fig. 4, E–G). Interestingly, as the development of arbuscules in the cortical cells progressed, the highest GUS activity was observed in the well-developed arbuscule-containing cells (Fig. 4, H–J). GUS activity progressively decreased in cells harboring aging and senescent arbuscules. The activity of RAD1 promoter was induced as early as internal hyphae appeared in the root tissue, which suggests that RAD1 could influence the growth of intracellular hyphae upon induction of the gene by inter- and/or intracellular hyphal growth. Overall, the temporal pattern of RAD1pro:GUS expression during arbuscule development supports the role of RAD1 in arbuscule formation.
Figure 4.
Histochemical analysis of proRAD1:GUS expression in mycorrhizal and nonmycorrhizal hairy roots of L. japonicus. A and B, Absence of GUS activity in nonmycorrhizal roots and root tips under high-Pi conditions. Scale bars = 100 µm. C and D, GUS activity in nonmycorrhizal roots and root tips under low-Pi conditions. Scale bars = 100 µm. E to G, GUS activity was observed in arbuscule-containing and adjacent cells. E, Fluorescence image of WGA-Alexa Fluor 488-stained intraradical fungal structures. F, Bright-field image of magenta GUS-stained root sector shown in E. G, Alexa Fluor 488 and bright-field image were merged to show colocalization of intraradical AM fungal hyphae with GUS activity. Scale bar = 10 µm. Fluorescence of Alexa Fluor 488 (H), bright-field image of magenta GUS-stained arbusculated inner cortical cells (I), and merged images (J). Scale bar = 50 µm. da, Developing arbuscule; ma, mature arbuscule.
Symbiotic Function of RAD1 Is Characteristic for AMS
AMS and RLS share common symbiosis regulatory pathways at the early stages of microbial root colonization. To check whether RAD1 is in line with or divergent from the CSSP, transcription of RAD1 was studied in roots forming nodules. The nodulation marker gene L. japonicus NODULE INCEPTION was induced significantly, but the expression levels of AM marker genes LjPT4 and SUBTILASE M1, as well as RAD1 remained unchanged in roots carrying nodules (Supplemental Fig. S6A). Accordingly, visual inspection revealed that normal pink nodules were formed in all three genotypes, and there was no significant difference in the number of nodules between Gifu-129, rad1-1, and rad1-3 plants (Supplemental Fig. S6B). These observations led us to conclude that RAD1 function is essential for normal mycorrhization, rather than a common prerequisite for both root symbioses.
RAD1 Displays an Evolutionary Pattern Characteristic of Phylogenetically AMS-Related Genes
To better characterize RAD1 and determine its specificity among GRAS proteins, all 18 mycorrhiza-up-regulated L. japonicus GRAS protein family members, 33 GRAS family members from Arabidopsis, as well as known GRAS transcription factors involved in symbiosis such as LjNSP1, LjNSP2, MtNSP1, MtNSP2, OsDIP1, MtDELLA1/2, and MtRAM1, were subject to phylogenetic tree analysis (Fig. 5). RAD1 and the closely related protein LjSGA_122341.1 clustered separately from the Arabidopsis GRAS members. Such an absence in Arabidopsis is a common pattern of genes involved in AMS (Delaux et al., 2014). We further investigated the phylogenetic distribution of RAD1 in land plants by looking in genomes and transcriptomes of 18 other hosts and nine additional nonhost species (Supplemental Text S2). Putative orthologs of RAD1 were selected based on e values <10−90 in BLAST and reciprocal analyses, and were found in all the angiosperms tested except in nonhost species. Even L. angustifolius, which forms RLS but not AMS, seems to lack RAD1, confirming its specific function for AMS. A tree was then generated to better characterize the evolutionary pattern of RAD1 (Fig. 5). Clear orthologs were found for most dicot species clustered in clade I. Interestingly, an additional member, likely resulting from ancient gene duplication, was found for some of the species clustered in clade II. Whether the additional copy was lost in species with a single RAD1 sequence, such as M. truncatula and L. japonicus, or this duplication took place only in specific lineages would require a much deeper sampling within these clades. In addition, putative orthologs of RAD1 were also identified in monocots, lycophytes, and liverworts, suggesting an ancient origin of this transcription factor (clade III). To further test the orthology of these potential homologs, we conducted a synteny analysis on a subset of host species and Arabidopsis as a nonhost representative. This analysis supported the orthology of all the host RAD1, including those from monocots and Selaginella moellendorffii (Supplemental Fig. S7).
Figure 5.
Phylogenetic analysis of GRAS proteins. An unrooted phylogenetic tree was generated based on amino acid alignment including 18 mycorrhiza-inducible GRAS proteins identified in this work (blue dots); 33 GRAS family members from Arabidopsis, symbiotic GRAS MtRAM1, MtDELLA1/MtDELLA2, OsDIP1, MtNSP1, MtNSP2, LjNSP1, LjNSP2 (pink dots); and the RAD1 homologs from dicots (green dots), monocots (light-green dots), lycophytes (orange dot), and liverworts (cyan dots). The alignment of protein sequences was performed using ClustalX, and the phylogenetic tree was constructed by MEGA5 using the Neighbor-Joining algorithm. Bootstrap values were labeled at each node. The RAD1 orthologs (clade I), paralogs (clade II), and RAD1 orthologs in monocots, lycophytes, and liverworts (clade III) with color background were selected based on e values <10−90 in BLAST using reciprocal analysis. Red star indicates the putative genome duplication event.
GRAS Proteins Interact with RAD1 in Vitro and in Vivo
As GRAS transcription factors have been reported to form dimers with GRAS family members (Itoh et al., 2002; Hirsch et al., 2009; Gobbato et al., 2012), RAD1 was used as bait and seven GRAS proteins representative of the different clades in the phylogenetic tree were used as prey in yeast two-hybrid (Y2H) assays to identify potential interacting proteins that may facilitate RAD1 function. Our data showed that RAD1 failed to form a homodimer in the yeast system. Two specific interactions were demonstrated in vitro (Supplemental Fig. S5B). One of them was the protein encoded by chr1.cm1852.30.r2.m, which shares 80% sequence identity with MtRAM1, suggesting it is the orthologous RAM1 in L. japonicus. Interestingly, RAD1 also directly interacted with LjNSP2 (Supplemental Fig. S5B), a protein with dual regulatory functions in root symbiosis signaling and appropriate establishment of RLS and AMS (Kaló et al., 2005; Hirsch et al., 2009; Liu et al., 2011; Maillet et al., 2011; Lauressergues et al., 2012).
GRAS domains are composed of five highly conserved sequential motifs: Leu heptad repeat I (LHRI), VHIID, Leu heptad repeat II, PFYRE, and SAW (Bolle, 2004). The LHRI motif and the C-terminal PFYRE and SAW motifs are required for GRAS transcription factor interaction (Hirsch et al., 2009). To study the specificity of interaction between RAD1 and RAM1, truncated versions of RAD1 were generated. As RAM1 exhibited strong autoactivation, it was used as prey in our Y2H assay. As shown in Figure 6A, deletion of the N terminus including the LHRI motif or specifically the SAW motif from RAD1 abolished the interaction between RAD1 and RAM1. None of the RAD1 versions interacted with the GRAS control protein, indicating the specificity of the interaction between RAD1 and RAM1 (Fig. 6A).
Figure 6.
RAD1 interacts with RAM1 in vitro and in vivo. A, Interaction between truncated versions of RAD1 and full-length RAM1. Conserved domains in GRAS proteins are color coded. BD, GAL4 DNA-binding domain; SD, synthetic minimal media based upon yeast (Saccharomyces cerevisiae) nitrogen base supplemented with Glc and amino acids (aa) except those indicated (L, W, H); 3AT, 3-amino-1,2,4-triazole. cGRAS encoding GRAS protein fused to the GAL4 activation domain (AD) was used as a negative control. B, In vitro pull-down assay. GST fusion proteins RAM1 (GST-RAM1) and NSP2 (GST-NSP2) or GST alone (control) were used to precipitate His-tag fused RAD1 (His-RAD1). + indicates the presence of the recombinant protein. WB: anti-His, Western blotting with anti-His antibody detecting His-bait fusion proteins. Purified GST fusion protein through glutathione-Sepharose beads was analyzed by SDS-PAGE and Coomassie Brilliant Blue in-gel protein staining (CBB staining). Arrows at left demonstrate presence of GST fusion proteins as designated. C, In vivo Co-IP of HA-RAD1 by GFP-RAM1. In vivo Co-IP was performed using N. benthamiana leaves infiltrated with A. rhizogenes GV3101 strain harboring the desired interacting pair (HA:RAD1/GFP:RAM1). Leaf extracts were subjected to immunoprecipitation (IP) using anti-GFP antibody. Anti-HA and anti-GFP antibodies were used for western blotting of Co-IP sample. Symbols represent the presence (+) or absence (–) of the various proteins. D, Interaction between RAM1 and RAD1 by BiFC assay in N. benthamiana. Leaves were infiltrated with mixtures of A. tumefaciens strain GV3101 to coexpress RAD1 and RAM1 fused to one-half of split YFP, respectively, as indicated at left. Three days after infiltration, images were captured from five different leaves for each plasmid combination. Scale bars = 50 µm.
An in vitro glutathione S-transferase (GST) pull-down assay was subsequently performed to verify the direct interaction between RAD1, LjNSP2, and RAM1, respectively. Our results showed that recombinant RAM1 and LjNSP2 proteins purified from Escherichia coli have the ability to precipitate RAD1 in vitro, thus confirming the Y2H results (Fig. 6B).
To further support the RAD1-RAM1 interaction, the subcellular localization of both proteins was investigated using GFP-protein fusions. Particle bombardment of onion (Allium cepa) epidermal cells demonstrated GFP-RAD1 localization in the nucleus (Supplemental Fig. S8A). A. rhizogenes-mediated transformation of L. japonicus hairy roots confirmed the nuclear localization of GFP-RAD1 (Supplemental Fig. S8B). The RAD1-RAM1 and RAD1-LjNSP2 interaction was confirmed in the nucleus by bimolecular fluorescence complementation (BiFC) assay (Fig. 6D; Supplemental Fig. S9). Moreover, RAD1 did not form homodimers in vivo, whereas RAM1 homodimerized in the nucleus and cytoplasm. RAM1, RAD1, and LjNSP2 did not interact with the split C terminus of Yellow Fluorescent Protein (YFP) or the N terminus of Yellow Fluorescent Protein alone, which suggested the specific interaction between them. Coimmunoprecipitation (Co-IP) was also used to confirm the interaction between RAD1 and RAM1 in vivo. To this end, hemagglutinin (HA)-tagged RAD1 and GFP-tagged RAM1 were transiently expressed in Nicotiana benthamiana leaves. Indeed, RAM1 was able to capture RAD1 in vivo when both proteins were transiently and simultaneously expressed (Fig. 6C). In sum, these data support our view that RAD1 forms multicomponent GRAS transcription factor complexes including RAM1 and/or NSP2 involved in late fungal colonization of host roots in AMS.
L. japonicus ram1 Mutant Fails to Establish Mycorrhizae
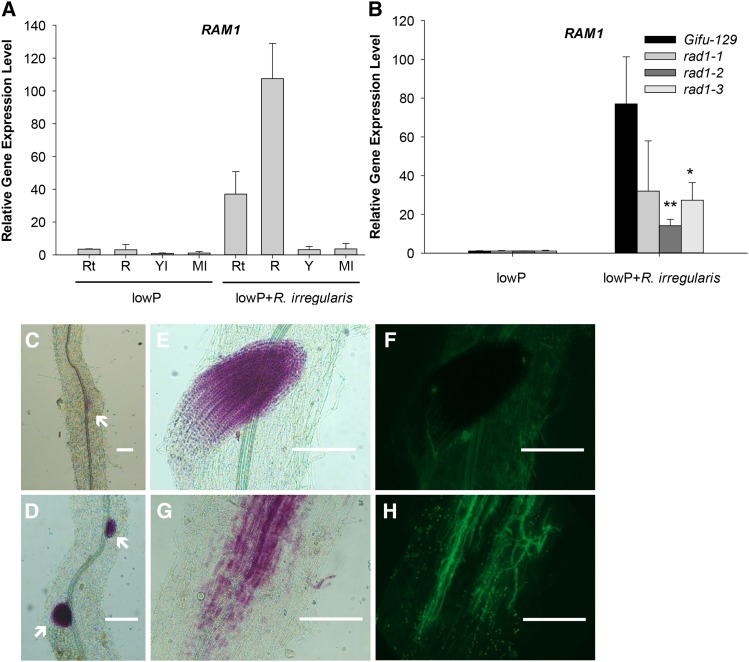
To elucidate the significance of RAM1 in L. japonicus-R. irregularis association, two ram1 insertion alleles were identified. The ram1-1 contains a LORE1a insertion in the second exon of the RAM1 gene, and the second allele ram1-2 has a LORE1a insertion in the first exon (Supplemental Fig. S10A). Homozygosity of ram1-1 and ram1-2 plants was confirmed by genotyping with sequence-specific primers (Supplemental Fig. S10B). When inoculated with R. irregularis, Gifu-129 established a normal mycorrhiza. In contrast, in ram1-1, not only was the total mycorrhization rate lower compared with the wild type, but intracellular branched hyphae instead of arbuscules appeared in the cortex (Fig. 7, A and B). In ram1-2, although the total mycorrhization rate was similar to the wild type, the arbuscules were stunted and arrested at birdsfoot stage III. This indicates that RAM1 is required for arbuscule development, which was consistent with RAM1 function in M. truncatula (Gobbato et al., 2012, 2013).
Figure 7.
Mycorrhizal phenotype in two allelic RAM1 defective mutants. A, Gifu-129, ram1-1, and ram1-2 were inoculated with R. irregularis for 6 weeks. Scale bars = 50 µm. Fungal structures were stained with Alexa Fluor 488. White arrows indicate arbuscules. B, Mycorrhization rate in Gifu-129 and ram1 alleles. Four biological replicates were used. Error bars represent SD (n = 4). Student’s t test was used between Gifu-129 and the two ram1 alleles. Asterisks indicate P < 0.05. C to F, Mycorrhizal marker gene expression in wild-type and ram1 alleles. Mean values (±sd) of four biological replicates are presented. Student’s t test was used. Asterisks indicate significant difference relative to wild-type Gifu-129. *P < 0.05; **P < 0.01.
Subsequently, we studied the expression pattern of L. japonicus RAM1. Our qRT-PCR result showed that transcript levels of RAM1 were detected in all tissues tested and were strongly enhanced in mycorrhizal roots and root tips (Fig. 8A). Induction of RAM1 gene expression cohered with the colonization of R. irregularis, which was suppressed in rad1 alleles (Fig. 8B). The 1.8-kb promoter region of RAM1 was fused to GUS to test the tissue-specific expression pattern of RAM1. Under low-Pi conditions, GUS activity could be detected weakly in root primordia, where lateral roots emerge from the primary root (Fig. 8C). In the presence of R. irregularis, RAM1 was induced in the cortical cells (Fig. 8, G and H). Interestingly, GUS activity could also be detected in the root tips where intercellular hyphae barely reach (Fig. 8, E and F). Likewise, RAM1 promoter activity could also be detected in lateral root primordia (Fig. 8D). This indicates that RAM1 is strongly expressed in arbuscule-containing cells and root meristems, suggesting a dual role in arbuscule development and in root development.
Figure 8.
Transcriptional expression pattern of RAM1 in L. japonicus. A, RT-PCR analysis of RAM1 transcript levels in different tissues of the wild type in the presence or absence of R. irregularis. Rt, Root tip; R, root; Yl, young leaves; Ml, mature leaves. Mean values from three biological replicates are shown (±sd, n = 3). B, RAM1 transcript levels in three rad1 alleles in the presence or absence of R. irregularis. Mean values from three biological replicates are shown (±sd, n = 3). Student’s t test was used. *P < 0.05; **P < 0.01. C to H, Histochemical analysis of proRAM1:GUS expression in the absence of R. irregularis (C) and in the presence of R. irregularis (D–H) in hairy roots of L. japonicus. Arrows indicate lateral root primordia. Scale bars = 200 µm.
DISCUSSION
Here we demonstrated that the GRAS protein encoding gene RAD1, the expression of which was increased in arbuscule-containing cells, was required for arbuscule development. RAD1 was shown to be specific for AMS, rather than for RLS (Supplemental Fig. S6). This phenotype is supported by a phylogenetic pattern specific to AM-related genes (Figs. 1B and 5). Premature collapse of arbuscules was observed in three rad1 allelic mutants, and the temporal profile of marker gene expressions was recorded, supporting the role of RAD1 in arbuscule development as well as in overall fungal colonization of the cortex. RAD1-RAM1 interaction could be demonstrated both in vitro and in vivo (Fig. 6; Supplemental Fig. S5B). Our results support a molecular role of RAD1 oligomerization with at least two GRAS-type transcriptional regulators, RAM1 and LjNSP2, in Pi-dependent arbuscule development.
Transcriptional Network Underlying Cellular Reprogramming during Late Colonization in AMS
Microbial reprogramming of root cellular development in the AMS is under intense study and is accompanied by remarkable progress in understanding the molecular mechanisms underlying mainly the precontact and early colonization phase of AM development (Oldroyd, 2013). Only a few cis-regulatory elements were reported to be essential for AM-specific gene expression in arbuscule-containing cells, whereas little is known about components of the corresponding transcriptional machinery (Chen et al., 2011; Lota et al., 2013; Xie et al., 2013). Overall, we found 45 transcriptional regulators that were significantly induced during mycorrhiza formation and the expression of which was under the control of Pi (Fig. 1; Supplemental Fig. S1; Supplemental Table S1). This common expression pattern of multiple regulatory proteins strongly suggested simultaneous and multilevel regulation of developmental and functional processes during late colonization in AMS, which remain to be mechanistically dissected. It has previously been reported that several transcription factor genes were induced after a 6-h treatment by AM fungal Myc-LCO signals in M. truncatula (Czaja et al., 2012). Comparing the encoded proteins of these early induced M. truncatula genes with the 45 mycorrhiza-regulated transcription factors from L. japonicus did not result in the identification of orthologous pairs, which suggested that expression of transcription factors during arbuscule development is likely to be triggered in a Myc-LCO-independent way.
Only a few of these transcription factor genes were identified as mycorrhiza regulated in earlier works (Guether et al., 2009; Hogekamp et al., 2011; Gaude et al., 2012; Gobbato et al., 2012; Volpe et al., 2013). According to the Plant Transcription Factor Database, there are 46 GRAS family members in L. japonicus (Guo et al., 2008). Eighteen of these were significantly induced in mycorrhizal roots compared with nonmycorrhizal roots, 10 of which appear in the AM host and are not detected by an extensive BLAST survey in the nonhost species (Fig. 1B; Supplemental Text S1). In sum, we have expanded the number of mycorrhiza-regulated GRAS proteins and members of other transcription factor families, providing a global view of transcriptional regulators in mycorrhizal L. japonicus roots. It was previously postulated that GRAS protein complex formation is crucial in the activation of either RLS or AMS (Gobbato et al., 2012). Our work will serve as a basis for more detailed protein interaction network and coexpression analysis, respectively, in the context of arbuscule development.
Regulatory Network of GRAS Proteins in Arbuscular Mycorrhizae
To date, several GRAS transcription factors involved in mycorrhizal signaling or mycorrhizal colonization have been identified, namely NSP1, NSP2, RAM1, DELLA, and DELLA-interacting proteins (Liu et al., 2011; Maillet et al., 2011; Gobbato et al., 2012; Delaux et al., 2013; Floss et al., 2013; Yu et al., 2014). In M. truncatula, NSP1 and NSP2 are essential for Nod factor-induced nodulation (Kaló et al., 2005; Smit et al., 2005). Their homologs LjNSP1 and LjNSP2 in L. japonicus were also reported to be required for nodulation (Heckmann et al., 2006). On the other hand, NSP1 and NSP2 were reported to regulate strigolactone biosynthesis both in M. truncatula and rice, but nsp1nsp2 double mutants exhibited well-developed and more arbuscules compared with wild-type roots, while the colonization was significantly reduced (Liu et al., 2011). In contrast, nsp2-2 mutant exhibited abolished Myc-LCO-triggered root branching, and quantitative reassessment of the mycorrhizal phenotype revealed a significantly reduced degree of colonization in nsp2-2 and nsp1 mutants when compared with wild-type plants, showing that NSP2 and NSP1 are involved in mycorrhizal signaling (Maillet et al., 2011; Delaux et al., 2013). In M. truncatula, the GRAS transcription factor RAM1 was reported to be required for AMS and to regulate the expression of RAM2 encoding a GPAT of the cutin biosynthesis pathway, which facilitates hyphopodia formation (Gobbato et al., 2012; Wang et al., 2012). Further research showed that RAM1 and RAM2 were also involved in arbuscule development (Gobbato et al., 2013). It remains to be investigated whether cooperative activity between RAD1 and RAM1 directs RAM2 expression in roots undergoing mycorrhization. Gobbato et al. (2012) also reported on protein-protein interaction between RAM1 and NSP2 in support of a role for NSP2 in AMS. Our work showed that RAD1 interacted with LjNSP2 in vitro and in vivo (Figure 6B; Supplemental Figs. S5B and S9). Recent work demonstrated that della1/della2 double mutant exhibited severely impaired development of arbuscules, and dominant DELLA protein lacking the DELLA domain restored the arbuscule formation in the presence of GA3, which indicated the positive regulatory role of DELLA in arbuscule development (Floss et al., 2013). In rice, DIP1 was identified as a protein interacting with DELLA, and the knockdown of DIP1 led to less mycorrhizal colonization (Yu et al., 2014). Interestingly, Yu et al. also found that DIP1 interacted with RAM1, although RAM1 did not interact with DELLA directly. Further genetic studies will likely reveal common functions of RAD1 and its interaction partners in AMS.
Proposed RAD1 Mode of Action
The mycorrhizal colonization in the rad1-2 mutant line was reduced, and all three rad1 alleles showed accumulation of prematurely stunted arbuscules. Likewise, the ram1-1 mutant exhibited compromised mycorrhization, and both ram1 mutant alleles showed arrested arbuscules (Fig. 7, A and B). Promoter-GUS activity strongly suggested the concurrence of RAD1 and RAM1 expression in the cortex (Figs. 4 and 8). Like RAM1, gene expression analysis revealed a broad effect of RAD1 on AM-related genes, including RAM2 (Figs. 3D and 7E). Like NSP2, RAD1 has no predicted DNA-binding domains, whereas predicted DNA-binding residues are enriched in the N terminus of RAM1 when the BindN+ web-based tool was used for prediction (Hirsch et al., 2009; Wang et al., 2010). Considering the different proteins interacting with RAD1 and their proposed roles throughout AM development, we propose that RAD1 regulates subsets of mycorrhiza-inducible genes in cooperation with RAM1, which is likely to have DNA-binding activity (Oldroyd, 2013). However, to date we cannot exclude that the reduced induction of these genes may be a secondary effect of abnormal arbuscules. Nevertheless, we speculate that the decision to activate either one of these subsets of genes is defined by the formation of specific heterocomplexes of RAD1 with other regulatory proteins, promoting mycorrhiza-specific responses. The mycorrhiza-signaling pathway(s), which promotes RAD1 protein complex formation, and the mechanisms that dictate complex formation remain to be defined.
Many open questions still offer food for thought concerning AM research. The gene Medtr8g109760.1 (M. truncatula genome version 3.0) or Medtr4g104060 (M. truncatula genome version 3.5), the closest homolog of RAD1 in M. truncatula sharing 74% identity at its corresponding amino acid sequence level, was reported to be expressed in arbuscule-containing cells and in adjacent cells as was shown by laser-capture microdissection of mycorrhizal roots followed by microarray hybridization of labeled cDNA (Gaude et al., 2012). This suggested that RAD1 shares similar functions across different plant species. This hypothesis is also supported by the conservation of RAD1 in mycorrhizal host plants, including basal land plants (Fig. 5). Interestingly, microRNA miR5204* has the ability to cleave the messenger RNA derived from Medtr8g109760.1 (Devers et al., 2011). As the expression of miR5204* was correlated with the plant Pi status and up-regulated in mycorrhizae, it was proposed that miR5204* spatially regulates GRAS gene expression during AMS. Future work will likely shed more light on posttranscriptional regulation of the RAD1-dependent regulatory network.
MATERIALS AND METHODS
Plant Materials, Growth Conditions, and Transformation
LORE1a insertion lines used in this article were kindly provided by Dr. Stig U. Andersen and Jens Stougaard (http://users-mb.au.dk/pmgrp/) under the following accession numbers: rad1-1 (30001039-2), rad1-2 (30030576), rad1-3 (30052260), ram1-1 (30002740), and ram1-2 (30082472). Lotus japonicus Gifu-129 was used in all experiments, and all LORE1a insertion mutants that we used were in the Gifu-129 background. Gifu-129, rad1-1, rad1-2, rad1-3, ram1-1, and ram1-2 were grown in the greenhouse at 22°C and 16-h-day/8-h-night periods. For phenotypic and gene expression analysis, plants were grown on 0.8% (w/v) agar for 5 d in the dark and then transplanted to a soil:sand mixture (1:9 [w/w]) with fungal inoculum (Rhizophagus irregularis, BEG75). Plants were grown in one-half-strength Hoagland solution with 5 μM NH4H2PO4 for 6 to 8 weeks. Agrobacterium rhizogenes-mediated hairy root transformation of L. japonicus was conducted as described previously (Lota et al., 2013). Transgenic roots were selected based on Discosoma spp. RED FLUORESCENT PROTEIN1 (pRedRoot)- or GFP-derived fluorescence (pH7WG2D). Hairy root transformation was mediated by A. rhizogenes strain 15384 as described.
Analysis of Mycorrhization and Measurement of Arbuscule Size
The procedures for mycorrhizal colonization analysis were based on the magnified intersection method from McGonigle et al. (1990) with some modification. For the mycorrhization rate, 15 root fragments, each about 1 cm long, were stained with trypan blue or wheat germ agglutinin (WGA) Alexa Fluor 488, and placed onto microscopic slides. Using 20× object magnification, over 150 views per slide were surveyed and classified into five groups: no colonization, only hyphae (H), hyphae with arbuscules (H+A), hyphae with vesicles (V+H), and hyphae with arbuscules and vesicles (A+V+H). The percentages of each group were calculated by the number of each sector divided by total views. For arbuscule size measurement, after WGA-488 staining, about 30 images were taken from each slide and viewed using 40× magnification of Leica CLSM5500. Each ecotype had three slides from three biological replicates. For the wild type, the images were taken close to the end of the infection units, as described previously (Javot et al., 2011). For rad1 mutants, images were randomly taken in the colonized region. Each arbuscule was measured along the longitudinal direction of the root. Then, the sizes of arbuscules from each slide were classified into seven clusters from 0 to 10 µm to >60 µm. The numbers in different clusters were divided by the number of all measured arbuscules from each slide to get the percentage of arbuscule size classes. Mean values of the respective percentages from three biological replica were used for the diagram shown in Figure 3B.
Constructs and Plasmids
For tissue-specific RAD1 gene expression analysis, a 1.9-kb promoter region was amplified by primers 5′-ACTggtaccTGCTTTCTTACTACTGTGTTC-3′ and 5′-AGGcccgggAACTCAAGATTATGCAAACTC-3′ from transformation competent artificial chromosome clone TM2604, with lower case letters indicating suitable restriction sites. ggtacc and cccggg are the Kpn1 and Sma1 enzyme digest sites, respectively. The PCR fragment was digested by Kpn1 and Sma1 and inserted into the binary vector pRedRoot-GUS to generate pRedRoot-RAD1pro-GUS. The 1.8-kb promoter region of RAM1 was amplified by primers 5′-ACTggtaccGCTTTTAGTGACTCATATG-3′ and 5′-gggACACCTAGTTGGCTAGCT-3′ to generate the pRedRoot-RAM1pro-GUS. For the overexpression assay of RAD1, the cDNA sequence of RAD1 was amplified by PCR. Then the PCR product with attB sequence ends was used for a Gateway recombination reaction to generate pH7WG2D-35Spro-RAD1. For subcellular localization of RAD1 protein in onion (Allium cepa) epidermal cells, the pGWB6-RAD1 plasmid encoding an N-terminal GFP fusion to RAD1 was generated by a Gateway recombination reaction and delivered into onion cells by particle bombardment. Cells expressing the 35S CaMV promoter-driven GFP gene were used as a control.
Gateway vectors pAS/pACT were used for Y2H and Gateway vectors RfA-sYFPn-pBatTL-B and RfA-sYFPc-pBatTL-B (from Dr. Fransicka Turk, MPI) were used for BiFC assay. The primers are described in Supplemental Table S3.
RNA Transcript Profiling
RNAseq was performed as described previously (Willmann et al., 2013). In brief, >1 µg of molecular grade DNA-free RNA from each sample was used for transcriptome analysis. Library construction was performed according to the manufacturer´s protocol (Illumina, TruSeq). RNAseq was performed on Illumina HiSeq2000 at the Cologne Center for Genomics (http://portal.ccg.uni-koeln.de/ccg/). Paired-end reads of 100 bp each were generated, and six samples were loaded onto one flow cell lane using multiplexing. Barcodes of sequences were removed, and the quality control of reads was done with the Galaxy FASTQ tool (Giardine et al., 2005; Blankenberg et al., 2010; Goecks et al., 2010). Short reads were mapped to the L. japonicus transcriptome (Sato et al., 2008) using Burrows-Wheeler Alignment tool with default parameters and an insert size of 200 (Li and Durbin, 2010). Hits per gene were counted using HTseq-count (developed by Simon Anders from EMBL Heidelberg). Statistical analysis of differentially expressed genes was performed with DESeq (Anders and Huber, 2010). Genes with fold change ≥ 2 and P value ≤ 0.05 were identified as being differentially expressed. Gene annotation was performed using Blast2GO (Conesa et al., 2005).
Quantitative Real-Time PCR Analysis
Total RNA was extracted using the NucleoSpin RNA Plant extraction kit (Macherey-Nagel) according to the manufacturer’s instructions. Total RNA of 0.5 to 1 μg was used to synthesize first stand cDNA using Thermo Scientific RevertAid H Minus Reverse Transcriptase. Gene expression was determined by qRT-PCR (ABI 7500) using the SYBR green PCR master mix (Applied Biosystems). Relative changes in gene expression from real-time quantitative PCR experiments were analyzed with the 2−ΔΔCT method, which was normalized to the endogenous reference Ubiquitin gene and relative to the reference sample (Livak and Schmittgen, 2001). Gifu-129 grown under low-Pi conditions was used as a reference sample without annotation. The gene-specific primers used in this article can be found in Supplemental Table S3.
Trypan Blue Staining and WGA-Alexa Fluor 488 Staining
For mycorrhizal structure visualization, trypan blue staining was performed as described (Brundrett et al., 1984). For WGA-Alexa Fluor 488 staining, roots were first washed with distilled water and then soaked in 50% (v/v) ethanol overnight. Roots were then incubated in a 20% (w/v) KOH solution for 3 d at room temperature. After rinsing with water, the roots were incubated in 0.1 m HCl for 2 h. After rinsing with water and subsequently with 1× phosphate-buffered saline, roots were stained in 1× phosphate-buffered saline buffer containing 0.2 μg mL−1 WGA-Alexa Fluor 488 overnight in the dark. For long-term storage, roots were stored at 4°C in the staining solution. Zeiss confocal microscopy (LSM 700) was used to detect the WGA-Alexa Fluor 488 (excitation/emission maxima at approximately 495/519 nm) signal.
Sequence Collection and Phylogenetic Analysis
To collect sequences corresponding to potential RAD1 orthologs, RAD1 amino acid sequence was searched in publicly available genomes using BLASTp. The list of species and the sequences used are indicated in Supplemental Table S3. Hits with an e value of less than 10−90 were selected for the phylogenetic analysis. In addition, for nonhost species, the best hits were reciprocally blasted on the Medicago truncatula genome to confirm the absence of orthologs. Collected sequences were aligned using MAFFT (http://mafft.cbrc.jp/alignment/server/) and the alignment manually curated with Bioedit. Phylogenetic trees were generated by both the Neighbor-Joining and maximum-likelihood algorithms in MEGA5. Partial gap deletion (90%) was used together with the JTT substitution model (gamma 2 parameter rate). Bootstrap values were calculated using 100 replicates.
Reciprocal Blast Analysis
Reciprocal blasts were carried out using amino acid sequences of the 43 transcription factors (TFs) (Supplemental Table S1). First, each TF was used as a query in a BLASTp search on genomes of host and nonhost species (Supplemental Table S3). Best equal hits were collected for each species and used as a query in reciprocal BLASTp on the L. japonicus genome (http://www.kazusa.or.jp/lotus/). A reciprocal blast was considered positive if the initial TF was among the equal best hits.
Synteny Analysis
As L. japonicus is not available in GEvo (https://genomevolution.org/CoGe/GEvo.pl), we used M. truncatula. Given the extremely high identity between L. japonicus and M. truncatula genes, we assumed that M. truncatula was a good proxy. An approximately 200-kb-sized region in the M. truncatula genome containing transcription factors identified by RNAseq was compared to the syntenic region in Arabidopsis (Arabidopsis thaliana; Columbia-0) and Populus trichocarpa using CoGe:GEvo (https://genomevolution.org/CoGe/GEvo.pl) as described in Delaux et al. (2014).
For RAD1, in addition to the previously mentioned genome, we included rice (Oryza sativa), Vitis vinifera, Sorghum bicolor, maize (Zea mays), and Selaginella moellendorffii.
GST Pull-Down Assay
The pull-down assay was processed as previously described, with modifications (Cui et al., 2010). pGEX-LjNSP2, pGEX-RAM1, and pET-RAD1 were generated by Gateway recombination reactions, and GST was used as a negative control. In brief, 50 mL of bacterial cell culture expressing GST-tagged proteins was resuspended with 15 mL of lysis buffer (25 mm Tris-HCl, pH 7.5, 150 mm NaCl, 3 mm dithiothreitol [DTT], and 1× complete mini-protease inhibitor cocktail [Roche]) after centrifugation at 5,000 rpm for 10 min. The sample was sonicated (2 min twice on ice at 40% power using Sonics vibra-cell ultrasonic processor VCX500) and centrifuged at 15,000 rpm for 20 min; then, the supernatant was incubated with 100 µL of glutathione agarose beads for 30 min with rotation at 4°C and washed three times with wash buffer (25 mm Tris-HCl, pH 7.5, 150 mm NaCl, 3 mm DTT, 0.2% [v/v] Triton X-100). Then, the beads containing purified GST protein, GST-LjNSP2, and GST-RAM1 were incubated with soluble bacterial lysate (20 mg) containing His-RAD1 for 20 min at 4°C, respectively. After washing four times with wash buffer, the proteins were eluted with 100 µL of 15 mm reduced glutathione (25 mm Tris-HCl, pH 9.0). Western blotting using approximately 1% of input and 6.7% of eluted protein was performed for protein detection.
Co-IP and BiFC
The Co-IP assay was performed as described previously (Cui et al., 2010). For transient expression of proteins in Nicotiana benthamiana, leaves were infiltrated with mixed Agrobacterium tumefaciens GV3101 pMP90 strains (final optical density at 600 nm of 0.5) harboring a vector that generates HA-RAD1 or GFP-RAM1 protein under the control of the CaMV 35S promoter, respectively, in combination with GV3101 strain expressing RNA silencing suppressor p19 protein. One gram of leaf tissue was harvested 3 d after infiltration and homogenized in 1.5 mL of IP buffer (50 mM Tris-Cl, pH 7.5, 150 mm NaCl, 5 mm EDTA, 2 mm DTT, 0.1% [v/v] Triton X-100, and 1× complete mini-protease inhibitor cocktail [Roche]). After two times of centrifugation at 20,000g for 10 min, the supernatants were incubated with 50 µL of HA agarose beads for 1 h at 4°C and then washed six times with IP buffer. The beads were eluted with 60 μL of SDS loading buffer (100 mm Tris-Cl, pH 6.8, 4% [w/v] SDS, 0.2% [w/v] bromophenol blue, 20% [v/v] glycerol, and 200 mm DTT), and then 20 μL of each elution was analyzed by western immunoblotting.
For the BiFC assay, leaves of N. benthamiana were infiltrated with A. tumefaciens GV3101 pMP90RG strains harboring either pBatTL-RAM1-YFPn or pBatTL-RAD1-YFPn, in combination with pBatTL-RAM1-YFPc, pBatTL-RAD1-YFPc, or empty vector, as described by Hu et al. (2014). Cells were kept in infiltration buffer (10 mM MgCl2, 10 mM MES, pH 5.6, 0.5 mM acetosyringone) for 2 to 3 h and mixed with p19 before infiltration. Images were taken by Leica CLSM DM5500 3 d after infiltration.
Subcellular Protein Localization
pGWB6-RAD1 was used for subcellular localization in onion epidermal cells by particle bombardment, as described by Folkers et al.( 2002). In addition, transgenic hairy roots containing pGWB6-RAD1, generated by inoculation of L. japonicus seeding stems with transformed A. rhizogenes, containing pGWB6-RAD1 from 1-month-old wild-type L. japonicus plants, were used for RAD1 subcellular localization. GFP fluorescence was detected via Zeiss LSM700 confocal microscopy.
Histochemical GUS Analysis
pRedRoot-RAD1pro:GUS was introduced into hairy roots of Gifu-129 using A. rhizogenes-mediated transformation. Transformed hairy roots were harvested 6 and 8 weeks after colonization by R. irregularis. Magenta GUS staining was used to detect GUS activity, and WGA-Alexa Fluor 488 staining was used to detect the presence of R. irregularis in root tissues. Fluorescence imaging was performed with a Zeiss fluorescent microscope (Microscope Axio Imager.D2).
Y2H Assay
Using pAS-RAD1 as bait, cDNA sequences of eight GRAS family members were amplified by PCR and subcloned into the pACT2 vector to be used as prey. Ten microliters of each 1, 0.1, 0.01, and 0.001 optical density at 600 nm dilutions of yeast (Saccharomyces cerevisiae) cell suspension was dropped on synthetic minimal media plates based upon yeast nitrogen base supplemented with Glc and amino acids without Leu and Trp or without Leu, Trp, and His. An AP2 transcription factor encoded by the chr2.CM0608.1100.r2.m gene was used as a negative control. To determine the domains required for RAD1 and RAM1 interaction, the DNA fragments encoding full-length and truncations of RAD1ΔN (121–509 amino acids), ΔNL (200–509 amino acids), ΔS (1–420 amino acids), and ΔPS (1–340 amino acids) were cloned into pAS bait vector by using the Gateway system. cGRAS, encoding mycorrhiza-induced GRAS gene chr3.CM0106.470.r2.d, was used as a negative control. One millimolar 3-amino-1,2,4-triazole was applied to suppress autoactivation.
Nodulation Assay
To generate nodulated roots, plants were inoculated with Mesorhizobium loti strain as described in Liu et al. (2003). One week after germination, seedlings were transferred to the sand and soil mixture as described previously. Ten days later, seedlings were inoculated with an M. loti cell suspension of 0.1 OD. Three weeks postinoculation, plants were harvested, and nodules were counted and examined by microscopy.
Statistical analysis was performed using Microsoft Excel and SigmaPlot (Systat Software).
Sequence data from this article can be found in the Lotus database (http://www.kazusa.or.jp/lotus/) or GenBank under the following accession numbers: RAD1, chr4.CM1864.540.r2.m; RAM1, chr1.CM1852.30.r2.m; LjNSP2, chr1.CM1976.90.r2.m; STR, chr4.CM0042.2570.r2.d; RAM2, CM0905.50.r2.d and CM0905.160.r2.d; SbtM1, chr2.CM0021.2780.r2.m; LjPT4, chr1.CM2121.10.r2.a; cGRAS, chr3.CM0106.470.r2.d; and Ubiquitin, DQ249171.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. AM-inducible transcription factor genes repressed by high-Pi conditions.
Supplemental Figure S2. Mycorrhiza morphology in rad1-1 and RAD1 transcript analysis in rad1 LORE1a insertion lines.
Supplemental Figure S3. Mycorrhization rate and AM-inducible marker gene expression in rad1-2 and rad1-3 at 5 and 7 wpi.
Supplemental Figure S4. Mycorrhizal structures and RAD1 gene expression in transgenic hairy roots overexpressing chimeric 35Spro:RAD1.
Supplemental Figure S5. RAD1 gene expression profile in mycorrhizal and nonmycorrhizal L. japonicus and GRAS proteins that interact with RAD1.
Supplemental Figure S6. RAD1 gene expression in nodulated roots and nodulation in rad1-1 and rad1-3.
Supplemental Figure S7. Synteny analysis of RAD1 on AM host species and nonhost Arabidopsis.
Supplemental Figure S8. Nuclear localization of RAD1.
Supplemental Figure S9. Interaction among RAM1, RAD1, and LjNSP2 by BiFC assay in N. benthamiana.
Supplemental Figure S10. Gene structure of RAM1 in wild-type L. japonicus and ram1 mutant.
Supplemental Table S1: List of mycorrhiza up-regulated transcription factors generated by DESeq.
Supplemental Table S2: Forty-five mycorrhiza up-regulated transcription factors under different treatments.
Supplemental Table S3: Sequences of oligonucleotide primer pairs used.
Supplemental Text S1: Microsynteny analysis of potential symbiotic transcription factors.
Supplemental Text S2: Sequences used for phylogenetic tree.
Supplementary Material
Acknowledgments
We thank Jens Stougaard and Stig U. Andersen (Department of Molecular Biology and Genetics, Aarhus University) for providing the LORE1a mutants studied in this work; Shusei Sato (Kazusa DNA Research Institute) for transformation competent artificial chromosome clones and genome sequence information; Franziska Turck (Max Planck Institute for Plant Breeding Research) for RfA-pBatTL plasmids for BiFC; Jane Parker (Max Planck Institute for Plant Breeding Research) for supporting parts of this work; Christian Becker, Janine Altmüller, and Peter Nürnberg (Cologne Center for Genomics at University of Cologne) for expert support in RNAseq analysis; Karl Pioch (Cologne Biocenter, University of Cologne) for correcting the article; Jan Grossbach and Juliana Almario (Cologne Biocenter, University of Cologne) for drawing the heat map; and Dilyana Filipova (Cologne Biocenter, University of Cologne) for assistance with plant genotyping.
Glossary
- AM
arbuscular mycorrhizal
- Pi
phosphate
- LCO
lipochitin oligosaccharide
- PPA
prepenetration apparatus
- AMS
arbuscular mycorrhizal symbiosis
- RLS
rhizobium-legume symbiosis
- CSSP
common symbiosis signaling pathway
- GPAT
glycerol-3-phosphate acyl transferase
- RNAi
RNA interference
- AMF
arbuscular mycorrhizal fungi
- RNAseq
deep sequencing analysis of mRNA
- qRT
quantitative reverse transcription
- wpi
weeks postinoculation
- UTR
untranslated region
- cDNA
complementary DNA
- Y2H
yeast two-hybrid
- LHRI
Leu heptad repeat I
- GST
glutathione S-transferase
- BiFC
bimolecular fluorescence complementation
- Co-IP
coimmunoprecipitation
- HA
hemagglutinin
- DTT
dithiothreitolRT: reverse transcription
- RT
reverse transcription
Footnotes
This work was supported by the Alexander von Humboldt Foundation (to L.X.), by the Priority Program 1212 of the German Research Foundation (grant no. BU 2250/1 to M.B.), and by the Center of Excellence CEPLAS (Cluster of Excellence on Plant Sciences), University of Cologne.
Articles can be viewed without a subscription.
References
- Akiyama K, Matsuzaki K, Hayashi H (2005) Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435: 824–827 [DOI] [PubMed] [Google Scholar]
- Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ané JM, Kiss GB, Riely BK, Penmetsa RV, Oldroyd GE, Ayax C, Lévy J, Debellé F, Baek JM, Kalo P, et al. (2004) Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science 303: 1364–1367 [DOI] [PubMed] [Google Scholar]
- Beisson F, Li Y, Bonaventure G, Pollard M, Ohlrogge JB (2007) The acyltransferase GPAT5 is required for the synthesis of suberin in seed coat and root of Arabidopsis. Plant Cell 19: 351–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besserer A, Puech-Pagès V, Kiefer P, Gomez-Roldan V, Jauneau A, Roy S, Portais JC, Roux C, Bécard G, Séjalon-Delmas N (2006) Strigolactones stimulate arbuscular mycorrhizal fungi by activating mitochondria. PLoS Biol 4: e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenberg D, Von Kuster G, Coraor N, Ananda G, Lazarus R, Mangan M, Nekrutenko A, Taylor J (2010) Galaxy: a web-based genome analysis tool for experimentalists. Curr Protoc Mol Biol Chapter 19: Unit 19 10 11-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolle C. (2004) The role of GRAS proteins in plant signal transduction and development. Planta 218: 683–692 [DOI] [PubMed] [Google Scholar]
- Bolle C, Koncz C, Chua NH (2000) PAT1, a new member of the GRAS family, is involved in phytochrome A signal transduction. Genes Dev 14: 1269–1278 [PMC free article] [PubMed] [Google Scholar]
- Breuillin F, Schramm J, Hajirezaei M, Ahkami A, Favre P, Druege U, Hause B, Bucher M, Kretzschmar T, Bossolini E, et al. (2010) Phosphate systemically inhibits development of arbuscular mycorrhiza in Petunia hybrida and represses genes involved in mycorrhizal functioning. Plant J 64: 1002–1017 [DOI] [PubMed] [Google Scholar]
- Brundrett MC, Piche Y, Peterson RL (1984) A new method for observing the morphology of vesicular-arbuscular mycorrhizae. Can J Bot 62: 2128–2134 [Google Scholar]
- Catoira R, Galera C, de Billy F, Penmetsa RV, Journet EP, Maillet F, Rosenberg C, Cook D, Gough C, Dénarié J (2000) Four genes of Medicago truncatula controlling components of a nod factor transduction pathway. Plant Cell 12: 1647–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier M, Bredemeier R, Wanner G, Takeda N, Schleiff E, Parniske M (2008) Lotus japonicus CASTOR and POLLUX are ion channels essential for perinuclear calcium spiking in legume root endosymbiosis. Plant Cell 20: 3467–3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A, Gu M, Sun S, Zhu L, Hong S, Xu G (2011) Identification of two conserved cis-acting elements, MYCS and P1BS, involved in the regulation of mycorrhiza-activated phosphate transporters in eudicot species. New Phytol 189: 1157–1169 [DOI] [PubMed] [Google Scholar]
- Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676 [DOI] [PubMed] [Google Scholar]
- Cui H, Wang Y, Xue L, Chu J, Yan C, Fu J, Chen M, Innes RW, Zhou JM (2010) Pseudomonas syringae effector protein AvrB perturbs Arabidopsis hormone signaling by activating MAP kinase 4. Cell Host Microbe 7: 164–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaja LF, Hogekamp C, Lamm P, Maillet F, Martinez EA, Samain E, Dénarié J, Küster H, Hohnjec N (2012) Transcriptional responses toward diffusible signals from symbiotic microbes reveal MtNFP- and MtDMI3-dependent reprogramming of host gene expression by arbuscular mycorrhizal fungal lipochitooligosaccharides. Plant Physiol 159: 1671–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaux PM, Bécard G, Combier JP (2013) NSP1 is a component of the Myc signaling pathway. New Phytol 199: 59–65 [DOI] [PubMed] [Google Scholar]
- Delaux PM, Varala K, Edger PP, Coruzzi GM, Pires JC, Ané JM (2014) Comparative phylogenomics uncovers the impact of symbiotic associations on host genome evolution. PLoS Genet 10: e1004487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devers EA, Branscheid A, May P, Krajinski F (2011) Stars and symbiosis: microRNA- and microRNA*-mediated transcript cleavage involved in arbuscular mycorrhizal symbiosis. Plant Physiol 156: 1990–2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devers EA, Teply J, Reinert A, Gaude N, Krajinski F (2013) An endogenous artificial microRNA system for unraveling the function of root endosymbioses related genes in Medicago truncatula. BMC Plant Biol 13: 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Laurenzio L, Wysocka-Diller J, Malamy JE, Pysh L, Helariutta Y, Freshour G, Hahn MG, Feldmann KA, Benfey PN (1996) The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86: 423–433 [DOI] [PubMed] [Google Scholar]
- Endre G, Kereszt A, Kevei Z, Mihacea S, Kaló P, Kiss GB (2002) A receptor kinase gene regulating symbiotic nodule development. Nature 417: 962–966 [DOI] [PubMed] [Google Scholar]
- Feddermann N, Muni RR, Zeier T, Stuurman J, Ercolin F, Schorderet M, Reinhardt D (2010) The PAM1 gene of petunia, required for intracellular accommodation and morphogenesis of arbuscular mycorrhizal fungi, encodes a homologue of VAPYRIN. Plant J 64: 470–481 [DOI] [PubMed] [Google Scholar]
- Floss DS, Levy JG, Lévesque-Tremblay V, Pumplin N, Harrison MJ (2013) DELLA proteins regulate arbuscule formation in arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci USA 110: E5025–E5034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fode B, Siemsen T, Thurow C, Weigel R, Gatz C (2008) The Arabidopsis GRAS protein SCL14 interacts with class II TGA transcription factors and is essential for the activation of stress-inducible promoters. Plant Cell 20: 3122–3135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkers U, Kirik V, Schöbinger U, Falk S, Krishnakumar S, Pollock MA, Oppenheimer DG, Day I, Reddy AS, Jürgens G, et al. (2002) The cell morphogenesis gene ANGUSTIFOLIA encodes a CtBP/BARS-like protein and is involved in the control of the microtubule cytoskeleton. EMBO J 21: 1280–1288; erratum Folkers U, Kirik V, Schöbinger U, Falk S, Krishnakumar S, Pollock MA, Oppenheimer DG, Day I, Reddy AR, Jürgens G, et al (2002) EMBO J 21: 2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo E, Ross JJ, Jones WT, Reid JB (2013) Plant hormones in arbuscular mycorrhizal symbioses: an emerging role for gibberellins. Ann Bot (Lond) 111: 769–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukai E, Soyano T, Umehara Y, Nakayama S, Hirakawa H, Tabata S, Sato S, Hayashi M (2012) Establishment of a Lotus japonicus gene tagging population using the exon-targeting endogenous retrotransposon LORE1. Plant J 69: 720–730 [DOI] [PubMed] [Google Scholar]
- Gaude N, Bortfeld S, Duensing N, Lohse M, Krajinski F (2012) Arbuscule-containing and non-colonized cortical cells of mycorrhizal roots undergo extensive and specific reprogramming during arbuscular mycorrhizal development. Plant J 69: 510–528 [DOI] [PubMed] [Google Scholar]
- Genre A, Chabaud M, Balzergue C, Puech-Pagès V, Novero M, Rey T, Fournier J, Rochange S, Bécard G, Bonfante P, et al. (2013) Short-chain chitin oligomers from arbuscular mycorrhizal fungi trigger nuclear Ca2+ spiking in Medicago truncatula roots and their production is enhanced by strigolactone. New Phytol 198: 190–202 [DOI] [PubMed] [Google Scholar]
- Genre A, Chabaud M, Faccio A, Barker DG, Bonfante P (2008) Prepenetration apparatus assembly precedes and predicts the colonization patterns of arbuscular mycorrhizal fungi within the root cortex of both Medicago truncatula and Daucus carota. Plant Cell 20: 1407–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genre A, Chabaud M, Timmers T, Bonfante P, Barker DG (2005) Arbuscular mycorrhizal fungi elicit a novel intracellular apparatus in Medicago truncatula root epidermal cells before infection. Plant Cell 17: 3489–3499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, Zhang Y, Blankenberg D, Albert I, Taylor J, et al. (2005) Galaxy: a platform for interactive large-scale genome analysis. Genome Res 15: 1451–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobbato E, Marsh JF, Vernié T, Wang E, Maillet F, Kim J, Miller JB, Sun J, Bano SA, Ratet P, et al. (2012) A GRAS-type transcription factor with a specific function in mycorrhizal signaling. Curr Biol 22: 2236–2241 [DOI] [PubMed] [Google Scholar]
- Gobbato E, Wang E, Higgins G, Bano SA, Henry C, Schultze M, Oldroyd GE (2013) RAM1 and RAM2 function and expression during arbuscular mycorrhizal symbiosis and Aphanomyces euteiches colonization. Plant Signal Behav 8: e26049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goecks J, Nekrutenko A, Taylor J, Galaxy Team (2010) Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol 11: R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greb T, Clarenz O, Schafer E, Muller D, Herrero R, Schmitz G, Theres K (2003) Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes Dev 17: 1175–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth M, Kosuta S, Gutjahr C, Haage K, Hardel SL, Schaub M, Brachmann A, Sato S, Tabata S, Findlay K, et al. (2013) Two Lotus japonicus symbiosis mutants impaired at distinct steps of arbuscule development. Plant J 75: 117–129 [DOI] [PubMed] [Google Scholar]
- Groth M, Takeda N, Perry J, Uchida H, Dräxl S, Brachmann A, Sato S, Tabata S, Kawaguchi M, Wang TL, et al. (2010) NENA, a Lotus japonicus homolog of Sec13, is required for rhizodermal infection by arbuscular mycorrhiza fungi and rhizobia but dispensable for cortical endosymbiotic development. Plant Cell 22: 2509–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guether M, Balestrini R, Hannah M, He J, Udvardi MK, Bonfante P (2009) Genome-wide reprogramming of regulatory networks, transport, cell wall and membrane biogenesis during arbuscular mycorrhizal symbiosis in Lotus japonicus. New Phytol 182: 200–212 [DOI] [PubMed] [Google Scholar]
- Guo AY, Chen X, Gao G, Zhang H, Zhu QH, Liu XC, Zhong YF, Gu X, He K, Luo J (2008) PlantTFDB: a comprehensive plant transcription factor database. Nucleic Acids Res 36: D966–D969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutjahr C, Banba M, Croset V, An K, Miyao A, An G, Hirochika H, Imaizumi-Anraku H, Paszkowski U (2008) Arbuscular mycorrhiza-specific signaling in rice transcends the common symbiosis signaling pathway. Plant Cell 20: 2989–3005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutjahr C, Parniske M (2013) Cell and developmental biology of arbuscular mycorrhiza symbiosis. Annu Rev Cell Dev Biol 29: 593–617 [DOI] [PubMed] [Google Scholar]
- Gutjahr C, Radovanovic D, Geoffroy J, Zhang Q, Siegler H, Chiapello M, Casieri L, An K, An G, Guiderdoni E, et al. (2012) The half-size ABC transporters STR1 and STR2 are indispensable for mycorrhizal arbuscule formation in rice. Plant J 69: 906–920 [DOI] [PubMed] [Google Scholar]
- Harrison MJ, Dewbre GR, Liu J (2002) A phosphate transporter from Medicago truncatula involved in the acquisition of phosphate released by arbuscular mycorrhizal fungi. Plant Cell 14: 2413–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckmann AB, Lombardo F, Miwa H, Perry JA, Bunnewell S, Parniske M, Wang TL, Downie JA (2006) Lotus japonicus nodulation requires two GRAS domain regulators, one of which is functionally conserved in a non-legume. Plant Physiol 142: 1739–1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch S, Kim J, Muñoz A, Heckmann AB, Downie JA, Oldroyd GE (2009) GRAS proteins form a DNA binding complex to induce gene expression during nodulation signaling in Medicago truncatula. Plant Cell 21: 545–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogekamp C, Arndt D, Pereira PA, Becker JD, Hohnjec N, Küster H (2011) Laser microdissection unravels cell-type-specific transcription in arbuscular mycorrhizal roots, including CAAT-box transcription factor gene expression correlating with fungal contact and spread. Plant Physiol 157: 2023–2043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JY, Zhou Y, He F, Dong X, Liu LY, Coupland G, Turck F, de Meaux J (2014) miR824-Regulated AGAMOUS-LIKE16 Contributes to Flowering Time Repression in Arabidopsis. Plant Cell 26: 2024–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys CP, Franks PJ, Rees M, Bidartondo MI, Leake JR, Beerling DJ (2010) Mutualistic mycorrhiza-like symbiosis in the most ancient group of land plants. Nat Commun 1: 103. [DOI] [PubMed] [Google Scholar]
- Imaizumi-Anraku H, Takeda N, Charpentier M, Perry J, Miwa H, Umehara Y, Kouchi H, Murakami Y, Mulder L, Vickers K, et al. (2005) Plastid proteins crucial for symbiotic fungal and bacterial entry into plant roots. Nature 433: 527–531 [DOI] [PubMed] [Google Scholar]
- Itoh H, Ueguchi-Tanaka M, Sato Y, Ashikari M, Matsuoka M (2002) The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell 14: 57–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov S, Fedorova EE, Limpens E, De Mita S, Genre A, Bonfante P, Bisseling T (2012) Rhizobium-legume symbiosis shares an exocytotic pathway required for arbuscule formation. Proc Natl Acad Sci USA 109: 8316–8321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javot H, Penmetsa RV, Breuillin F, Bhattarai KK, Noar RD, Gomez SK, Zhang Q, Cook DR, Harrison MJ (2011) Medicago truncatula mtpt4 mutants reveal a role for nitrogen in the regulation of arbuscule degeneration in arbuscular mycorrhizal symbiosis. Plant J 68: 954–965 [DOI] [PubMed] [Google Scholar]
- Javot H, Penmetsa RV, Terzaghi N, Cook DR, Harrison MJ (2007) A Medicago truncatula phosphate transporter indispensable for the arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci USA 104: 1720–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaló P, Gleason C, Edwards A, Marsh J, Mitra RM, Hirsch S, Jakab J, Sims S, Long SR, Rogers J, et al. (2005) Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308: 1786–1789 [DOI] [PubMed] [Google Scholar]
- Kanamori N, Madsen LH, Radutoiu S, Frantescu M, Quistgaard EM, Miwa H, Downie JA, James EK, Felle HH, Haaning LL, et al. (2006) A nucleoporin is required for induction of Ca2+ spiking in legume nodule development and essential for rhizobial and fungal symbiosis. Proc Natl Acad Sci USA 103: 359–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karandashov V, Nagy R, Wegmüller S, Amrhein N, Bucher M (2004) Evolutionary conservation of a phosphate transporter in the arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci USA 101: 6285–6290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistner C, Winzer T, Pitzschke A, Mulder L, Sato S, Kaneko T, Tabata S, Sandal N, Stougaard J, Webb KJ, et al. (2005) Seven Lotus japonicus genes required for transcriptional reprogramming of the root during fungal and bacterial symbiosis. Plant Cell 17: 2217–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobae Y, Hata S (2010) Dynamics of periarbuscular membranes visualized with a fluorescent phosphate transporter in arbuscular mycorrhizal roots of rice. Plant Cell Physiol 51: 341–353 [DOI] [PubMed] [Google Scholar]
- Kosuta S, Hazledine S, Sun J, Miwa H, Morris RJ, Downie JA, Oldroyd GE (2008) Differential and chaotic calcium signatures in the symbiosis signaling pathway of legumes. Proc Natl Acad Sci USA 105: 9823–9828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauressergues D, Delaux PM, Formey D, Lelandais-Brière C, Fort S, Cottaz S, Bécard G, Niebel A, Roux C, Combier JP (2012) The microRNA miR171h modulates arbuscular mycorrhizal colonization of Medicago truncatula by targeting NSP2. Plant J 72: 512–522 [DOI] [PubMed] [Google Scholar]
- Lévy J, Bres C, Geurts R, Chalhoub B, Kulikova O, Duc G, Journet EP, Ané JM, Lauber E, Bisseling T, et al. (2004) A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science 303: 1361–1364 [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R (2010) Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26: 589–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Beisson F, Koo AJ, Molina I, Pollard M, Ohlrogge J (2007) Identification of acyltransferases required for cutin biosynthesis and production of cutin with suberin-like monomers. Proc Natl Acad Sci USA 104: 18339–18344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Blaylock LA, Endre G, Cho J, Town CD, VandenBosch KA, Harrison MJ (2003) Transcript profiling coupled with spatial expression analyses reveals genes involved in distinct developmental stages of an arbuscular mycorrhizal symbiosis. Plant Cell 15: 2106–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Kohlen W, Lillo A, Op den Camp R, Ivanov S, Hartog M, Limpens E, Jamil M, Smaczniak C, Kaufmann K, et al. (2011) Strigolactone biosynthesis in Medicago truncatula and rice requires the symbiotic GRAS-type transcription factors NSP1 and NSP2. Plant Cell 23: 3853–3865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lota F, Wegmüller S, Buer B, Sato S, Bräutigam A, Hanf B, Bucher M (2013) The cis-acting CTTC-P1BS module is indicative for gene function of LjVTI12, a Qb-SNARE protein gene that is required for arbuscule formation in Lotus japonicus. Plant J 74: 280–293 [DOI] [PubMed] [Google Scholar]
- Maillet F, Poinsot V, André O, Puech-Pagès V, Haouy A, Gueunier M, Cromer L, Giraudet D, Formey D, Niebel A, et al. (2011) Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature 469: 58–63 [DOI] [PubMed] [Google Scholar]
- McGonigle T, Miller M, Evans D, Fairchild G, Swan J (1990) A new method which gives an objective measure of colonization of roots by vesicular-arbuscular mycorrhizal fungi. New Phytol 115: 495–501 [DOI] [PubMed] [Google Scholar]
- Messinese E, Mun JH, Yeun LH, Jayaraman D, Rougé P, Barre A, Lougnon G, Schornack S, Bono JJ, Cook DR, et al. (2007) A novel nuclear protein interacts with the symbiotic DMI3 calcium- and calmodulin-dependent protein kinase of Medicago truncatula. Mol Plant Microbe Interact 20: 912–921 [DOI] [PubMed] [Google Scholar]
- Nagy R, Drissner D, Amrhein N, Jakobsen I, Bucher M (2009) Mycorrhizal phosphate uptake pathway in tomato is phosphorus-repressible and transcriptionally regulated. New Phytol 181: 950–959 [DOI] [PubMed] [Google Scholar]
- Oba H, Tawaray K, Wagatsuma T (2001) Arbuscular mycorrhizal colonization in Lupinus and related genera. Soil Sci Plant Nutr 47: 685–694 [Google Scholar]
- Oldroyd GE. (2013) Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat Rev Microbiol 11: 252–263 [DOI] [PubMed] [Google Scholar]
- Parniske M. (2008) Arbuscular mycorrhiza: the mother of plant root endosymbioses. Nat Rev Microbiol 6: 763–775 [DOI] [PubMed] [Google Scholar]
- Paszkowski U, Kroken S, Roux C, Briggs SP (2002) Rice phosphate transporters include an evolutionarily divergent gene specifically activated in arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci USA 99: 13324–13329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Carol P, Richards DE, King KE, Cowling RJ, Murphy GP, Harberd NP (1997) The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev 11: 3194–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pumplin N, Mondo SJ, Topp S, Starker CG, Gantt JS, Harrison MJ (2010) Medicago truncatula Vapyrin is a novel protein required for arbuscular mycorrhizal symbiosis. Plant J 61: 482–494 [DOI] [PubMed] [Google Scholar]
- Pumplin N, Zhang X, Noar RD, Harrison MJ (2012) Polar localization of a symbiosis-specific phosphate transporter is mediated by a transient reorientation of secretion. Proc Natl Acad Sci USA 109: E665–E672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pysh LD, Wysocka-Diller JW, Camilleri C, Bouchez D, Benfey PN (1999) The GRAS gene family in Arabidopsis: sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J 18: 111–119 [DOI] [PubMed] [Google Scholar]
- Rausch C, Daram P, Brunner S, Jansa J, Laloi M, Leggewie G, Amrhein N, Bucher M (2001) A phosphate transporter expressed in arbuscule-containing cells in potato. Nature 414: 462–470 [DOI] [PubMed] [Google Scholar]
- Redecker D, Kodner R, Graham LE (2000) Glomalean fungi from the Ordovician. Science 289: 1920–1921 [DOI] [PubMed] [Google Scholar]
- Roberts NJ, Morieri G, Kalsi G, Rose A, Stiller J, Edwards A, Xie F, Gresshoff PM, Oldroyd GE, Downie JA, et al. (2013) Rhizobial and mycorrhizal symbioses in Lotus japonicus require lectin nucleotide phosphohydrolase, which acts upstream of calcium signaling. Plant Physiol 161: 556–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Yoshikawa M, Yano K, Miwa H, Uchida H, Asamizu E, Sato S, Tabata S, Imaizumi-Anraku H, Umehara Y, et al. (2007) NUCLEOPORIN85 is required for calcium spiking, fungal and bacterial symbioses, and seed production in Lotus japonicus. Plant Cell 19: 610–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders FE, Tinker PB, Black RLB, Palmerley SM (1977) The development of endomycorrhizal root systems: I. spread of infection and growth-promoting effects with four species of vesicular-arbuscular endophyte. New Phytol 78: 257–268 [Google Scholar]
- Sato S, Nakamura Y, Kaneko T, Asamizu E, Kato T, Nakao M, Sasamoto S, Watanabe A, Ono A, Kawashima K, et al. (2008) Genome structure of the legume, Lotus japonicus. DNA Res 15: 227–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze J, Temple G, Temple SJ, Beschow H, Vance CP (2006) Nitrogen fixation by white lupin under phosphorus deficiency. Ann Bot (Lond) 98: 731–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit P, Raedts J, Portyanko V, Debellé F, Gough C, Bisseling T, Geurts R (2005) NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308: 1789–1791 [DOI] [PubMed] [Google Scholar]
- Stracke S, Kistner C, Yoshida S, Mulder L, Sato S, Kaneko T, Tabata S, Sandal N, Stougaard J, Szczyglowski K, et al. (2002) A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 417: 959–962 [DOI] [PubMed] [Google Scholar]
- Tian C, Wan P, Sun S, Li J, Chen M (2004) Genome-wide analysis of the GRAS gene family in rice and Arabidopsis. Plant Mol Biol 54: 519–532 [DOI] [PubMed] [Google Scholar]
- Urbański DF, Małolepszy A, Stougaard J, Andersen SU (2012) Genome-wide LORE1 retrotransposon mutagenesis and high-throughput insertion detection in Lotus japonicus. Plant J 69: 731–741 [DOI] [PubMed] [Google Scholar]
- van Tuinen D, Jacquot E, Zhao B, Gollotte A, Gianinazzi-Pearson V (1998) Characterization of root colonization profiles by a microcosm community of arbuscular mycorrhizal fungi using 25S rDNA-targeted nested PCR. Mol Ecol 7: 879–887 [DOI] [PubMed] [Google Scholar]
- Volpe V, Dell’Aglio E, Giovannetti M, Ruberti C, Costa A, Genre A, Guether M, Bonfante P (2013) An AM-induced, MYB-family gene of Lotus japonicus (LjMAMI) affects root growth in an AM-independent manner. Plant J 73: 442–455 [DOI] [PubMed] [Google Scholar]
- Wang E, Schornack S, Marsh JF, Gobbato E, Schwessinger B, Eastmond P, Schultze M, Kamoun S, Oldroyd GE (2012) A common signaling process that promotes mycorrhizal and oomycete colonization of plants. Curr Biol 22: 2242–2246 [DOI] [PubMed] [Google Scholar]
- Wang L, Huang C, Yang MQ, Yang JY (2010) BindN+ for accurate prediction of DNA and RNA-binding residues from protein sequence features. BMC Syst Biol (Suppl 1) 4: S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmann M, Gerlach N, Buer B, Polatajko A, Nagy R, Koebke E, Jansa J, Flisch R, Bucher M (2013) Mycorrhizal phosphate uptake pathway in maize: vital for growth and cob development on nutrient poor agricultural and greenhouse soils. Front Plant Sci 4: 533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Huang W, Liu F, Tang N, Liu Y, Lin H, Zhao B (2013) Functional analysis of the novel mycorrhiza-specific phosphate transporter AsPT1 and PHT1 family from Astragalus sinicus during the arbuscular mycorrhizal symbiosis. New Phytol 198: 836–852 [DOI] [PubMed] [Google Scholar]
- Yang SY, Grønlund M, Jakobsen I, Grotemeyer MS, Rentsch D, Miyao A, Hirochika H, Kumar CS, Sundaresan V, Salamin N, et al. (2012) Nonredundant regulation of rice arbuscular mycorrhizal symbiosis by two members of the PHOSPHATE TRANSPORTER1 gene family. Plant Cell 24: 4236–4251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu N, Luo D, Zhang X, Liu J, Wang W, Jin Y, Dong W, Liu J, Liu H, Yang W, et al. (2014) A DELLA protein complex controls the arbuscular mycorrhizal symbiosis in plants. Cell Res 24: 130–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Blaylock LA, Harrison MJ (2010) Two Medicago truncatula half-ABC transporters are essential for arbuscule development in arbuscular mycorrhizal symbiosis. Plant Cell 22: 1483–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.