Arabidopsis immunity against multiple pathogens depends on unconventional G protein complexes.
Abstract
In animals, heterotrimeric G proteins, comprising Gα, Gβ, and Gγ subunits, are molecular switches whose function tightly depends on Gα and Gβγ interaction. Intriguingly, in Arabidopsis (Arabidopsis thaliana), multiple defense responses involve Gβγ, but not Gα. We report here that the Gβγ dimer directly partners with extra-large G proteins (XLGs) to mediate plant immunity. Arabidopsis mutants deficient in XLGs, Gβ, and Gγ are similarly compromised in several pathogen defense responses, including disease development and production of reactive oxygen species. Genetic analysis of double, triple, and quadruple mutants confirmed that XLGs and Gβγ functionally interact in the same defense signaling pathways. In addition, mutations in XLG2 suppressed the seedling lethal and cell death phenotypes of BRASSINOSTEROID INSENSITIVE1-associated receptor kinase1-interacting receptor-like kinase1 mutants in an identical way as reported for Arabidopsis Gβ-deficient mutants. Yeast (Saccharomyces cerevisiae) three-hybrid and bimolecular fluorescent complementation assays revealed that XLG2 physically interacts with all three possible Gβγ dimers at the plasma membrane. Phylogenetic analysis indicated a close relationship between XLGs and plant Gα subunits, placing the divergence point at the dawn of land plant evolution. Based on these findings, we conclude that XLGs form functional complexes with Gβγ dimers, although the mechanism of action of these complexes, including activation/deactivation, must be radically different form the one used by the canonical Gα subunit and are not likely to share the same receptors. Accordingly, XLGs expand the repertoire of heterotrimeric G proteins in plants and reveal a higher level of diversity in heterotrimeric G protein signaling.
Heterotrimeric GTP-binding proteins (G proteins), classically consisting of Gα, Gβ, and Gγ subunits, are essential signal transduction elements in most eukaryotes. In animals and fungi, ligand perception by G protein-coupled receptors leads to replacement of GDP with GTP in Gα, triggering activation of the heterotrimer (Li et al., 2007; Oldham and Hamm, 2008). Upon activation, GTP-bound Gα and Gβγ are released and interact with downstream effectors, thereby transmitting signals to multiple intracellular signaling cascades. Signaling terminates when the intrinsic GTPase activity of Gα hydrolyzes GTP to GDP and the inactive heterotrimer reforms at the receptor. The large diversity of mammalian Gα subunits confers specificity to the multiple signaling pathways mediated by G proteins (Wettschureck and Offermanns, 2005). Five distinct classes of Gα have been described in animals (Gαi, Gαq, Gαs, Gα12 and Gαv), with orthologs found in evolutionarily primitive organisms such as sponges (Oka et al., 2009). Humans possess four classes of Gα involving 23 functional isoforms encoded by 16 genes (McCudden et al., 2005), while only a single prototypical Gα is usually found per plant genome (Urano et al., 2013). Multiple copies of Gα are present in some species with recently duplicated genomes, such as soybean (Glycine max) with four Gα genes (Blanc and Wolfe, 2004; Bisht et al., 2011). In the model plant Arabidopsis (Arabidopsis thaliana), a prototypical Gα subunit (GPA1) is involved in a number of important processes, including cell proliferation (Ullah et al., 2001), inhibition of inward K+ channels and activation of anion channels in guard cells by mediating the abscisic acid pathway (Wang et al., 2001; Coursol et al., 2003), blue light responses (Warpeha et al., 2006, 2007), and germination and postgermination development (Chen et al., 2006; Pandey et al., 2006).
It is well established that heterotrimeric G proteins play a fundamental role in plant innate immunity. In Arabidopsis, two different Gβγ dimers (Gβγ1 and Gβγ2) are generally considered to be the predominant elements in G protein defense signaling against a variety of fungal pathogens (Llorente et al., 2005; Trusov et al., 2006, 2007, 2009; Delgado-Cerezo et al., 2012; Torres et al., 2013). By contrast, these studies attributed a small or no role to Gα, because mutants deficient in Gα displayed only slightly increased resistance against the fungal pathogens (Llorente et al., 2005; Trusov et al., 2006; Torres et al., 2013). The Gβγ-mediated signaling also contributes to defense against a model bacterial pathogen Pseudomonas syringae, by participating in programmed cell death (PCD) and inducing reactive oxygen species (ROS) production in response to at least three pathogen-associated molecular patterns (PAMPs; Ishikawa, 2009; Liu et al., 2013; Torres et al., 2013). Gα is not involved in PCD or PAMP-triggered ROS production (Liu et al., 2013; Torres et al., 2013). Nonetheless, Arabidopsis Gα plays a positive role in defense against P. syringae, probably by mediating stomatal function and hence physically restricting bacterial entry to the leaf interior (Zhang et al., 2008; Zeng and He, 2010; Lee et al., 2013). Given the small contribution from Gα, the involvement of heterotrimeric G proteins in Arabidopsis resistance could be explained in two ways: either the Gβγ dimer acts independently from Gα, raising a question of how is it activated upon a pathogen attack, or Gα is replaced by another protein for heterotrimer formation.
The Arabidopsis genome contains at least three genes encoding Gα-like proteins that have been classified as extra-large G proteins (XLGs; Lee and Assmann, 1999; Ding et al., 2008). XLGs comprise two structurally distinct regions. The C-terminal region is similar to the canonical Gα, containing the conserved helical and GTPase domains, while the N-terminal region is a stretch of approximately 400 amino acids including a putative nuclear localization signal (Ding et al., 2008). GTP binding and hydrolysis were confirmed for all three XLG proteins, although their enzymatic activities are very slow and require Ca2+ as a cofactor, whereas canonical Gα utilizes Mg2+ (Heo et al., 2012). Several other features differentiate XLGs from Gα subunits. Comparative analysis of XLG1 and Gα at the DNA level showed that the genes are organized in seven and 13 exons, respectively, without common splicing sites (Lee and Assmann, 1999). XLGs have been reported to localize to the nucleus (Ding et al., 2008). Analysis of knockout mutants revealed a nuclear function for XLG2, as it physically interacts with the Related To Vernalization1 (RTV1) protein, enhancing the DNA binding activity of RTV1 to floral integrator gene promoters and resulting in flowering initiation (Heo et al., 2012). Therefore, it appears that XLGs may act independently of G protein signaling. On the other hand, functional similarities between XLGs and the Arabidopsis Gβ subunit (AGB1) were also discovered. For instance, XLG3- and Gβ-deficient mutants were similarly impaired in root gravitropic responses (Pandey et al., 2008). Knockout of all three XLG genes caused increased root length, similarly to the Gβ-deficient mutant (Ding et al., 2008). Furthermore, as observed in Gβ-deficient mutants, xlg2 mutants displayed increased susceptibility to P. syringae, indicating a role in plant defense (Zhu et al., 2009). Nevertheless, a genetic analysis of the possible functional interaction between XLGs and Gβ has not been established.
In this report, we performed in-depth genetic analyses to test the functional interaction between the three XLGs and Gβγ dimers during defense-related responses in Arabidopsis. We also examined physical interaction between XLG2 and the Gβγ dimers using yeast (Saccharomyces cerevisiae) three-hybrid (Y3H) and bimolecular fluorescent complementation (BiFC) assays. Our findings indicate that XLGs function as direct partners of Gβγ dimers in plant defense signaling. To estimate relatedness of XLGs and Gα proteins, we carried out a phylogenetic analysis. Based on our findings, we conclude that plant XLG proteins most probably originated from a canonical Gα subunit and retained prototypical interaction with Gβγ dimers. They function together with Gβγ in a number of processes including plant defense, although they most probably evolved activation/deactivation mechanisms very different from those of a prototypical Gα.
RESULTS
XLGs and Gβγ Provide Similar Defense Responses against Multiple Plant Pathogens
In Arabidopsis, the involvement of the Gβ (AGB1) and two Gγ subunits (AGG1 and AGG2) in plant defense is well established (Llorente et al., 2005; Trusov et al., 2006, 2009, 2010; Delgado-Cerezo et al., 2012). Recently, XLG proteins have been found to play a role in plant defense as well (Zhu et al., 2009). The similar behavior reported for agb1, agg1 agg2, and xlg2 mutants against P. syringae prompted us to directly compare the responses of agb1 and all available xlg mutants, including xlg1, xlg2, and xlg3 single, xlg2 xlg3 double, and xlg1 xlg2 xlg3 triple mutants, to pathogens representing different lifestyles.
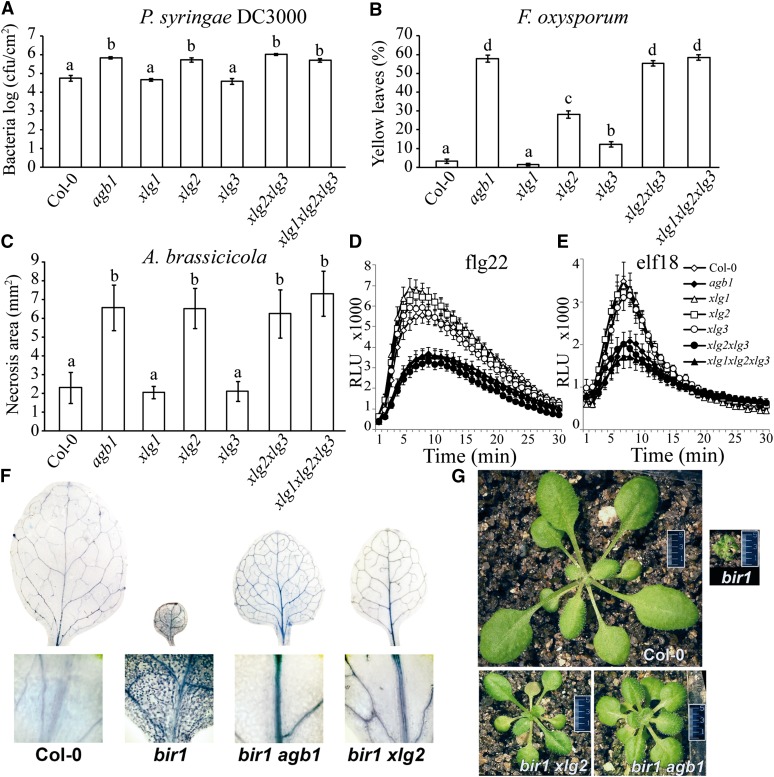
The hemibiotrophic bacterium P. syringae pv tomato (Pst) DC3000 was spray inoculated on wild-type ecotype Columbia (Col-0) and the mutants. As previously reported, bacterial multiplication in xlg1 and xlg3 single mutants was similar to that in the wild type, while agb1 and xlg2 single, xlg2 xlg3 double, and xlg1 xlg2 xlg3 triple mutants supported elevated bacterial populations compared with the wild type (Fig. 1A; Zhu et al., 2009; Torres et al., 2013). The bacterial growth observed in agb1, xlg2, xlg2 xlg3, and xlg1 xlg2 xlg3 mutants was almost identical, indicating very similar levels of susceptibility to this pathogen.
Figure 1.
XLGs and AGB1 have similar contributions in the defense against pathogens, response to PAMPs, and PCD. A, Bacterial growth of Pst DC3000 at 4 d post inoculation (dpi). Five-week-old plants were sprayed with bacterial solution (OD600 = 0.4). Four plants per genotype (seven leaf discs per plant) were harvested for bacterial titer. B, Percentage of yellow leaves 7 d after F. oxysporum infection. Three-week-old plants (n = 20) were inoculated. C, Area of necrosis 7 d after A. brassicicola drop inoculation. Five-week-old plants (n = 10) were assayed. D and E, ROS measurements (relative luminescence units [RLU] plotted against time [min]) after treatment with 1 µm flagellin22 (flg22; D) and 1 µm elongation factor thermo unstable (elf18; E). Leaf discs of 5-week-old plants (n = 12) were assayed. Each dataset shows mean ± sem. Experiments were repeated at least three times with similar results. For A to C, the letters represent statistically significantly different groups based on one-way ANOVA with Tukey’s multiple comparison method. A difference between each group represents P < 0.0001 (A and B) and P < 0.05 (C). F, Trypan blue stain for cell death showing that agb1 and xlg2 mutations suppress PCD caused by the bir1 mutation. Leaves of 3-week-old plants were stained. G, Both agb1 and xlg2 mutations partially suppress the bir1 phenotype. Rosettes of 3-week-old plants are shown.
Subsequently, all mutants and wild-type plants were challenged with the hemibiotrophic vascular pathogen Fusarium oxysporum, and disease progression was evaluated as percentage of leaves with chlorotic veins. Wild-type and xlg1 mutants showed similar disease progression (Fig. 1B). The xlg2 and xlg3 mutants were more susceptible than the wild type, although their susceptibility levels differed significantly from each other (Fig. 1B). The xlg2 xlg3 double mutant and the xlg1 xlg2 xlg3 triple mutant were highly susceptible, showing the same percentage of yellow leaves as the agb1 mutant (Fig. 1B).
Alternaria brassicicola is a necrotrophic airborne fungus requiring high humidity for spore germination and hence infectivity. The area of necrotic lesions caused by the pathogen was low in wild-type plants and xlg1 and xlg3 single mutants, whereas xlg2 single, xlg2 xlg3 double, xlg1 xlg2 xlg3 triple, and agb1 mutants all developed significantly larger lesions (Fig. 1C).
For all three pathogens, the enhanced susceptibility observed in xlg2 xlg3 double and xlg1 xlg2 xlg3 triple mutants was equal to that of the agb1 mutant, indicating similar contributions of XLGs and Gβ to defense.
XLGs Redundantly Control the Oxidative Burst Induced by PAMPs
Arabidopsis Gβ (AGB1) facilitates ROS production induced by three different PAMPs (Ishikawa, 2009; Liu et al., 2013). To compare the contributions of AGB1 and XLGs to ROS induction, agb1 and xlg mutants were treated with the bacterial-derived peptides flg22 and elf18. None of the single xlg mutants showed significant differences to wild-type Col-0 plants in ROS production in response to flg22 or elf18 elicitors (Fig. 1, D and E). However, there was a significant reduction in ROS production to both elicitors in xlg2 xlg3 double and xlg1 xlg2 xlg3 triple mutants, and the reduction was identical to that observed in agb1 mutants, suggesting that XLG2 and XLG3 contribute redundantly to ROS generation in response to these pathogen-associated elicitors (Fig. 1, D and E).
Mutations in AGB1 and XLG2 Suppress bir1 Morphology and Cell Death in a Similar Way
BRASSINOSTEROID INSENSITIVE1-associated receptor kinase1-interacting receptor-like kinase1 (BIR1) is a negative regulator of defense pathways in Arabidopsis, as the knockout mutant bir1 displays constitutive activation of cell death and defense responses, leading to lethality during seedling development (Gao et al., 2009). Recently, it was observed that disruption of the AGB1 gene suppressed seedling lethality and cell death phenotypes of bir1 (Liu et al., 2013). To test whether a knockout mutation in XLG2 can also suppress the bir1 phenotypes, we obtained bir1 xlg2 double mutant. The double mutant bir1 xlg2 was able to grow to maturity and set seeds at 23°C similarly to the bir1 agb1 mutant. Seedlings were stained with trypan blue to determine whether cell death was inhibited in the double mutants. As shown in Figure 1F, cell death (manifested as dark blue spots) in bir1 mutants was completely blocked by knockout of either AGB1 or XLG2. Interestingly, growth inhibition and the small crinkled leaves observed in bir1 were only partially suppressed by either the agb1 or xlg2 mutations (Fig. 1G). The rosette size of the bir1 agb1 and bir1 xlg2 double mutants was similar, larger than bir1 single mutants but smaller than the wild type (Fig. 1G).
XLGs and Gβγ Operate in the Same Defense Signaling Pathway
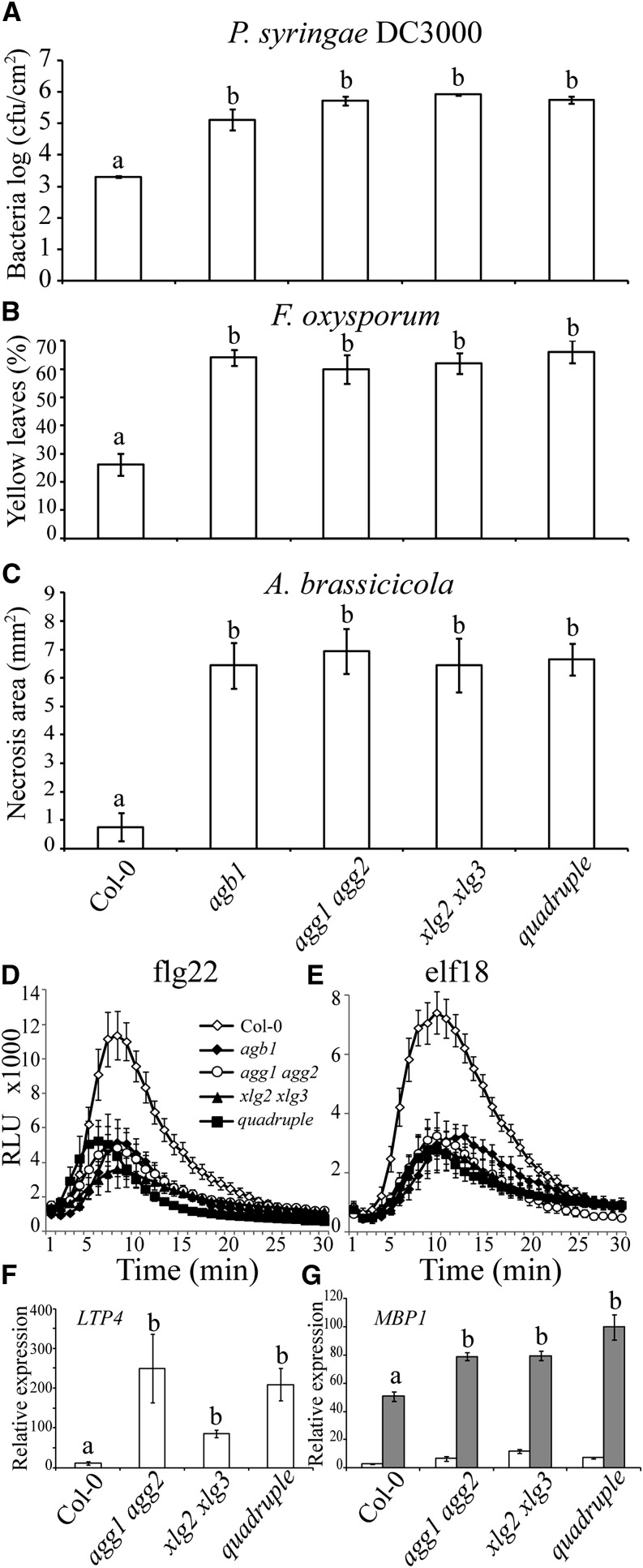
Based on the results described above, we hypothesized that XLG1 did not appear to have a role in defense and that XLG2, XLG3, and Gβγ might control the same signaling defense pathways. The simple way to study functional interaction would have been an analysis of agb1 xlg2 xlg3 triple mutants. Unfortunately, the proximity of the AGB1 and XLG2 genes on the same chromosome precluded production of the triple mutant by conventional crossing. Nevertheless, in all known defense-related functions, AGB1 forms a functional dimer with AGG1 or AGG2 (Mason and Botella, 2000, 2001; Trusov et al., 2007; Delgado-Cerezo et al., 2012). Moreover, the double mutant agg1 agg2 has reduced steady-state levels of AGB1, while in the triple mutant, lacking all three Gγ subunits, AGB1 abundance was even more severely decreased (Wolfenstetter et al., 2014). Therefore, we produced a quadruple mutant agg1 agg2 xlg2 xlg3 to analyze through disease resistance assays.
Quantitative disease assays allow evaluation of additive effects and, hence, determination of dependent versus independent contributions of signaling elements. The agb1 mutant, agg1 agg2 and xlg2 xlg3 double mutants, and the agg1 agg2 xlg2 xlg3 quadruple mutant all showed similarly increased sensitivity above the wild-type controls to Pst DC3000 (Fig. 2A). To confirm that additive effect could be observed in this assay, we evaluated the upper limit of the disease severity. Previously, we demonstrated that G proteins function independently from salicylic acid pathways (Trusov et al., 2009). Therefore, we compared disease progression in agb1, agg1 agg2, NahG (transgenic line overexpressing salicylate hydroxylase), and agb1 NahG. Supplemental Figure S1 shows that disease severity was well below the ceiling in our assay. The absence of an additive effect between agg and xlg mutants indicates that they are involved in the same signaling pathway. Likewise, no additive effect on susceptibility was observed for the quadruple mutant, when tested in assays with F. oxysporum and A. brassicicola (Fig. 2, B and C). The same assays were successfully used for genetic analysis of functional interaction between G proteins and hormonal defense signaling components (Trusov et al., 2009). It was shown that disease severity levels could be considerably higher providing opportunity for detection of the additive effect if such would exist. Additionally, each of the parental double mutants and the quadruple mutant were similarly impaired in ROS production induced by flg22 and elf18, with no additive effect observed (Fig. 2, D and E).
Figure 2.
XLGs and Gβγ act in the same signaling pathway to elicit the defense response. A, Bacterial growth of Pst DC3000 at 3 dpi. Five-week-old plants were spray inoculated with the bacterium (OD600 = 0.4). Three plants per genotype (seven leaf discs per sample) were harvested to measure bacterial titer. B, Percentage of yellow leaves 6 d after F. oxysporum inoculation of 3-week-old plants (n = 20). C, Area of necrotic lesion development in response to A. brassicicola drop inoculation at 5 dpi. Five-week-old plants (n = 10) were assayed. D and E, ROS production in response to PAMP treatment including 1 µm flg22 (D) and 1 µm elf18 (E). Relative luminescence units (RLU) was plotted against time (min). Leaf discs of 5-week-old plants (n = 12) were used. F and G, Defense-related gene expression in quadruple mutants compared with their parental mutants. Three-week-old plants were harvested for total RNA extraction (n = 3). Three biological replicates were used for quantitative real-time PCR. F, Steady-state expression levels of LTP4 in uninfected plants. G, Pathogen-induced expression levels of MBP1. Plants were inoculated with F. oxysporum (gray bars) or mock inoculated (white bars). Samples were collected at 3 dpi. In all sections, wild-type Col-0 and agg1 agg2 double, xlg2 xlg3 double, and agg1 agg2 xlg2 xlg3 quadruple mutants were studied. For A to E, agb1 mutant was included for comparison. The dataset represents mean ± sem. Experiments were repeated twice with similar results. For A to C and F and G, the letters represent groups of statistically significant differences based on one-way ANOVA with Tukey’s multiple comparison method. A difference between each group represents P < 0.001 (A), P < 0.0001 (B and C), and P < 0.01 (F and G). cfu, Colony forming units.
We also analyzed expression of pathogenesis-related genes in agg1 agg2 double, xlg2 xlg3 double, and agg1 agg2 xlg2 xlg3 quadruple mutants. Lipid Transfer Protein4 (LTP4; At5g59310) and Myrosinase-Binding Protein1 (MBP1; At1g52040) that encode defense-related proteins (García-Olmedo et al., 1995; Rask et al., 2000; Brotman et al., 2012) were selected, because their expression levels were found to be altered in the xlg2 mutant in the previous studies (Zhu et al., 2009). Steady-state expression levels of LTP4 in agg1 agg2 double, xlg2 xlg3 double, and agg1 agg2 xlg2 xlg3 quadruple mutants were significantly higher than those in wild-type plants, and no additive effect was observed in the quadruple mutant (Fig. 2F). MBP1 transcript levels significantly increased in wild-type and mutant plants upon F. oxysporum infection compared with uninfected plants (Fig. 2G). The induction levels in agg1 agg2 double and xlg2 xlg3 double mutants were higher than those observed in wild-type plants, and the quadruple mutant did not show any additive effect compared with the parental double mutants (Fig. 2G). Overall, the comparative analysis of the double and quadruple mutants indicates that Gβγ and XLGs share the same signaling pathway leading to PAMP-triggered immune responses to pathogens.
XLG2 Interacts with Gβγ Dimers in Yeast Assays
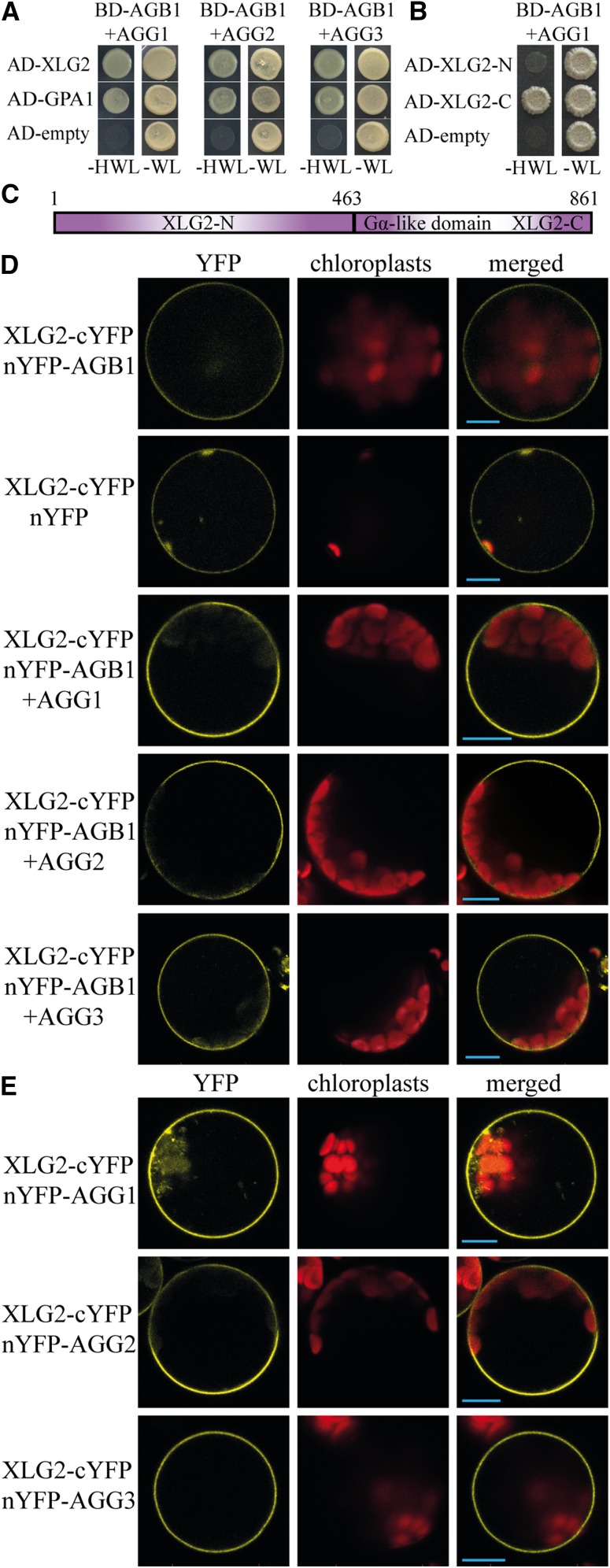
Based on the evidence that XLG2 operates in the same pathway as Gβγ and the sequence similarity existing between the C-terminal region of XLGs and the Gα subunit, we explored the possibility of physical interaction between XLG2 and AGB1. Considering that the Gβ and Gγ subunits form a compulsory dimer, we performed Y3H assays with XLG2 fused to the GAL4 activation domain (AD) and AGB1 fused to the GAL4-binding domain (BD) with each of the three AGG subunits coexpressed without any tags. When yeast was cotransformed using AD-XLG2 and BD-AGB1 with AGG1, growth was observed on a medium lacking His, indicating interaction between XLG2 and the Gβγ1 dimer (Fig. 3A). The interaction between XLG2 and the other two dimers, Gβγ2 and Gβγ3, was also confirmed (Fig. 3A). The canonical Gα subunit (AD-GPA1; positive control) and AD-empty vector (negative control) results corroborated the validity of the Y3H assay (Fig. 3A).
Figure 3.
In vivo interaction between XLG2 and Gβγ. A, Y3H assays showing growth of the yeast cells cotransformed with pACT2 carrying an AD-XLG2 fusion and the double expression pBridge vector carrying BD-AGB1 and one of three AGGs. pACT2 (AD-empty) was used as a negative control. Interaction between GPA1 and the three Gβγ dimers was tested in the similar way. Growth on a synthetic complete (SC) medium lacking His, Trp, and Leu (SC-HWL) indicates positive interaction; growth on an SC medium lacking Trp and Leu (SC-WL) indicates successful cotransformation of the yeast with both vectors. B, Y3H assay showing interaction between the C-terminal region of XLG2 (AD-XLG2-C) and Gβγ1 (BD-AGB1+AGG1), whereas no interaction was observed with the N terminus of XLG2 (AD-XLG2-N). C, A diagram representing XLG2 domain structure. Numbers correspond to amino acids. D, BiFC visualization of the interaction between XLG2 fused to cYFP and AGB1 fused to nYFP in mesophyll protoplasts isolated from an Arabidopsis agg1agg2agg3 mutant. Strong yellow signal indicating interaction was observed only when one of the Gγ subunits was present. E, BiFC assays were conducted for XLG2-cYFP and all three Gγ subunits fused to nYFP. To test if the interaction is possible in the absence of Gβ, mesophyll protoplasts were produced from the agb1 mutant and cotransfected with designated constructs. Fluorescence of the reconstituted YFP was detected by confocal microscopy 16 to 24 h after transfection. The representative protoplasts were photographed with 510- to 550-nm (for YFP) and 640- to 700-nm (for chloroplasts) filters. Bars = 10 μm.
To test whether the Gα-like region of XLG2 is responsible for binding with Gβγ, we individually analyzed the possible interaction between the N-terminal and C-terminal Gα-like region of XLG2 (Fig. 3C) and AGB1 in the presence of AGG1. Yeast growth on the selective medium was detected only for cells expressing the C-terminal Gα-like region (Fig. 3B), indicating that XLG2 interacts with the Gβγ dimer by its Gα-like domain.
Interaction between XLG2 and the Gβγ Dimer Occurs at the Plasma Membrane
To confirm Y3H results and establish the subcellular location of the XLG2-Gβγ interaction in vivo, we used BiFC in Arabidopsis mesophyll protoplasts. To avoid ambiguity in interpretation of the results, we produced protoplasts from the Arabidopsis agg1 agg2 agg3 triple knockout mutant deficient in all Gγ subunits. This approach allowed us to test if XLG2 and AGB1 are able to interact without any of the three Gγ subunits existent in Arabidopsis. When protoplasts were cotransfected using XLG2 fused to the C-terminal region of the yellow fluorescence protein (XLG2-cYFP) and AGB1 fused to the N-terminal region of the yellow fluorescence protein (nYFP-AGB1), weak YFP fluorescence was detected (Fig. 3D). However, the similar level of fluorescence was also detected when XLG2-cYFP was coexpressed with the nYFP empty vector (Fig. 3D), demonstrating possibility of nonspecific false-positive signal. When XLG2-cYFP and nYFP-AGB1 were coexpressed with untagged AGGs, a strong BiFC signal was observed (Fig. 3D). Importantly, the YFP signal was detected at the plasma membrane, but not in the nucleus (Supplemental Fig. S2).
To determine whether XLG2 can interact with AGGs independently of AGB1, we produced mesophyll protoplasts from agb1 mutant leaves and cotransfected those with XLG2-cYFP and each of nYFP-AGG1, nYFP-AGG2, or nYFP-AGG3. Strong fluorescence was observed from the plasma membrane in each case (Fig. 3E), indicating that XLG2 is able to interact with Gγ directly even in the absence of Gβ.
GFP-XLG2 Localizes to the Nucleus and the Plasma Membrane
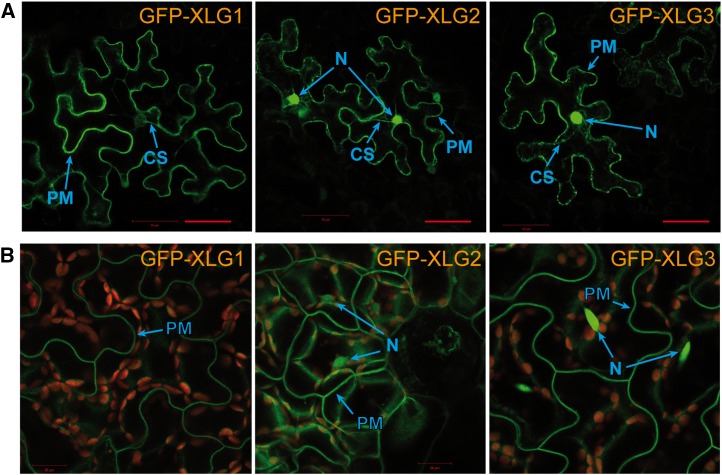
Localization of XLG2-Gβγ to the plasma membrane contrasts with the previously reported nuclear localization of Arabidopsis XLGs and the interaction of XLG2 with the nuclear protein RTV1 (Ding et al., 2008; Heo et al., 2012). This prompted us to reexamine the subcellular localization of all three XLG proteins using transient expression in Nicotiana benthamiana, as well as stably transformed Arabidopsis plants expressing XLGs fused to GFP. Surprisingly, transient expression in N. benthamiana leaves revealed the GFP fluorescence at the cell periphery for all three XLGs, while only GFP-XLG2 and GFP-XLG3 were observed in nuclei (Fig. 4A). GFP signal was also detected in the cytoplasmic strings. To study XLG localization in Arabidopsis, we produced and analyzed a large number of transgenic lines (15–35) expressing GFP fused to each of three XLGs. A silencing-impaired mutant deficient in RNA-dependent RNA polymerase6 (rdr6-11) was used as a background for transformation to improve transgene expression. Interestingly, in these lines, the GFP signal was consistently observed as a sharp line at the cell periphery, suggesting plasma membrane localization of all three XLGs. Consistent with the results in N. benthamiana, nuclear fluorescence was present only in plants transformed with GFP-XLG2 and GFP-XLG3 (Fig. 4B). Notably, no signal was observed in the cytoplasm. To confirm that the signal observed in the cell periphery concurs with the plasma membrane localization of the fusion proteins, we produced protoplasts from transgenic plants expressing GFP-XLG2 and transfected them with AGG2 fused to mCherry. Plasma membrane localization of YFP-AGG2 has been previously established (Adjobo-Hermans et al., 2006; Zeng et al., 2007). GFP and mCherry signals clearly overlapped, indicating the plasma membrane localization of XLG2 (Supplemental Fig. S3A). Upon protoplast rupture, cytosol proteins are released into solution, while membrane-bound proteins retained on the membrane (Serna, 2005). We observed that in ruptured protoplasts, GFP-XLG2 and mCherry-AGG2 remained on the membrane (Supplemental Fig. S3B). As a further test, we performed western-blot analysis of membrane-bound and soluble protein fractions from leaves of rdr6 GFP-XLG2 plants with GFP-specific antibodies. Nuclei were removed before separation of membrane-bound and soluble fractions. Consistent with the fluorescence results, intact GFP-XLG2 was detected only in the membrane fraction (Supplemental Fig. S3C).
Figure 4.
Subcellular localization of XLG proteins. A, Transient expression in N. benthamiana leaves. B, Expression of the designated GFP-XLG fusion proteins in stably transformed Arabidopsis rdr6-11 mutant plants (silencing impaired). PM, Plasma membrane; N, nucleus; CS, cytoplasmic strands. Bar = 20 μm.
XLGs Are Plant-Specific Proteins Originated from Gα
Despite evident sequence similarities between XLGs and Gα proteins, significant differences in functional domains, biochemical characteristics, and exon-intron structure of their genes question the origin of XLG proteins (Lee and Assmann, 1999; Ding et al., 2008; Heo et al., 2012). To establish whether XLG originated from Gα or acquired similarities during convergent evolution, we aimed to reconstruct the evolutionary relationships between XLGs and Gα subunits from animals, plants, and basic eukaryotes.
A recursive search in National Center for Biotechnology Information databases using known XLG sequences as queries identified multiple homologs in all major phyla of land plants, but not in algae. No XLG homologs were found outside the plant kingdom. Analysis of the EST and transcriptome databases showed that the identified XLGs are expressed. In the fully sequenced genome of the moss Physcomitrella patens (Bryophyta), a representative of the most primitive land plants alive today, we found two copies of the XLG gene, one of which contains multiple stop codons, suggesting that it is a pseudogene. We found no canonical Gα in this species, in agreement with a previous report (Urano et al., 2013). Interestingly, in the transcriptome of liverwort (Marchantia polymorpha; Sharma et al., 2014), another Bryophyte representative, we identified both XLG and Gα.
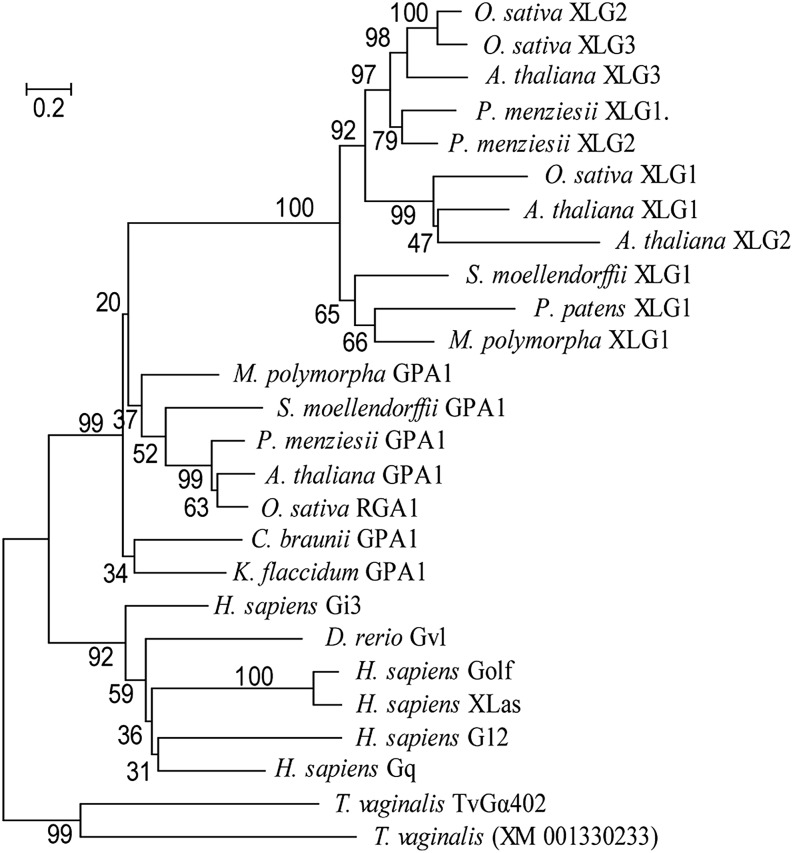
To reconstruct the evolutionary relationships between XLGs and Gα, we performed a phylogenetic analysis. Sequences representing the major plant phyla, five classes of Gα identified in animals (Suga et al., 1999; Oka et al., 2009), and two Gα proteins from the basal eukaryote Trichomonas vaginalis (Hirt et al., 2003) were selected (Supplemental Table S1). The T. vaginalis sequences were used as an outgroup. Several algorithms available from the MEGA6 package (Tamura et al., 2013) were used to reconstruct the phylogeny of the selected sequences. The neighbor-joining method with Poisson correction produced a tree where XLG and Gα subtrees best fit the plant phylogeny (Finet et al., 2010; Fig. 5). The neighbor-joining tree showed that XLGs form a monophyletic subclade within the plant Gα group. Arguably, it branched out just before the land plant emergence, which is consistent with the XLG’s taxonomic distribution. Importantly, the considerably closer relation between XLGs and plant Gαs compared with the relation between animal and plant Gαs was supported by 99% of 1,000 bootstrap trees (Fig. 5). These statistics provide strong evidence supporting the origin of XLG proteins from a plant Gα ancestor, rather than a convergent evolution scenario.
Figure 5.
Phylogenetic analysis of XLG and Gα proteins. Phylogenetic tree (neighbor-joining method) of XLG and Gα proteins of major plant phyla and representatives of five established classes of animal Gαs. Two T. vaginalis Gαs were used as an outgroup. Plant XLG proteins form a distinct subclade within plant Gα group. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) is shown next to the branches. Bar shows the rate of amino acid substitution. Species used in the analysis: Arabidopsis, Chara braunii, Danio rerio, Homo sapiens, Klebsormidium flaccidum, M. polymorpha, rice (Oryza sativa), Physcomitrella patens, Pseudotsuga menziesii, and T. vaginalis. GPA, G protein alpha; RGA, rice G protein alpha; XM_001330233, not characterized G protein alpha. Gi3, Gvl, Golf, XLas, and Gq are representative proteins of animal G protein alpha subunits.
DISCUSSION
Since the discovery of heterotrimeric G proteins in plants, an overwhelming amount of evidence has been accumulated pointing to significant differences between canonical animal G protein subunits and their plant counterparts (Chen et al., 2003, 2004; Jones and Assmann, 2004; Johnston and Siderovski, 2007; Temple and Jones, 2007; Chakravorty et al., 2011; Jones et al., 2011; Urano et al., 2012, 2013; Urano and Jones, 2014). One of the intriguing observations was the autonomous targeting of Gγ subunits and Gβγ dimers to the plasma membrane (Adjobo-Hermans et al., 2006; Zeng et al., 2007; Chakravorty et al., 2011; Li et al., 2012). These observations combined with mutant analyses prompted the hypothesis that the Gβγ dimer could have independent functions, in particular in plant defense (Llorente et al., 2005; Trusov et al., 2006, 2010). Recently, it was suggested that XLG proteins may function downstream of Gβ in defense against P. syringae (Zhu et al., 2009). The XLG proteins were discovered in homology searches for additional Gα subunits in Arabidopsis (Lee and Assmann, 1999; Ding et al., 2008). However, further characterization revealed extensive differences between XLGs and Gα subunits, precluding the consideration of XLGs as a part of the plant heterotrimeric G proteins (Lee and Assmann, 1999; Ding et al., 2008; Pandey et al., 2008; Zhu et al., 2009; Heo et al., 2012).
Earlier studies on the involvement of XLG proteins in plant defense indicated that only XLG2 plays a role in resistance, and it was restricted to the hemibiotrophic bacterium P. syringae, but not against the necrotrophic fungi A. brassicicola and Botrytis cinerea (Zhu et al., 2009). At the same time, the Gβγ dimer was found to play a key role in defense against bacterial and fungal pathogens (Llorente et al., 2005; Trusov et al., 2006, 2010; Lee et al., 2013; Lorek et al., 2013; Torres et al., 2013). In this report, we demonstrate that defense responses of xlg and agb1 mutants are severely compromised in a similar way and to the same degree when infected with pathogens of different life styles, including bacteria (P. syringae), necrotrophic fungi (A. brassicicola), and hemibiotrophic fungi (F. oxysporum). We found that XLG2 is the major contributor to the immunity; XLG3 plays a part in resistance against F. oxysporum infection, while XLG1 does not seem to have any role in defense response. Functional similarities between xlg and agb1 mutants were also found in ROS production, cell death, and pathogenesis-related gene expression. More importantly, the comparative study of the agg1 agg2 double, xlg2 xlg3 double, and agg1 agg2 xlg2 xlg3 quadruple mutants during infection by three pathogens showed similar susceptibility level without an additive effect, hence providing evidence that XLG proteins and Gβγ dimers mediate the same defense pathways, which protect plants from bacterial and fungal pathogens. The ROS production and gene expression results were also consistent with this conclusion.
Another interesting finding was the link between XLG2 and the BIR1 receptor-like kinase, where mutations in XLG2 abolished the cell death and seedling lethal phenotypes observed in bir1 mutants, in a similar way to AGB1 mutations. It is established that BIR1 negatively controls at least two cell death pathways involving Phytoalexin Deficient4 and a receptor-like kinase, Suppressor of BIR1 (SOBIR1; Gao et al., 2009). Recently, it was established that Gβγ1 and Gβγ2 act downstream of SOBIR1, contributing to immune and cell death responses (Liu et al., 2013). Given the similar phenotypes of the bir1 xlg2 and bir1 agb1 double mutants and the altered responses to flg22 and elf18, it is highly plausible that XLG2 acts together with Gβγ dimers in the receptor-like kinase (RLK)-mediated pathways. In maize (Zea mays), the Gα protein was reported to transduce signals downstream from CLAVATA, a Leu-rich repeat RLK, providing another example of the plant G proteins engaged in RLK-initiated signaling (Bommert et al., 2013). Recently, a direct interaction between Gβ subunit and Receptor-like Protein Kinase2, a receptor for the CLAVATA3 (CLV3) peptide, has been reported (Ishida et al., 2014). Intriguingly, this study showed that Gβ and Gγ subunits, but not Gα, are involved in CLV3 signaling (Ishida et al., 2014). Further research is needed to understand a mechanism of action involving RLKs, Gβγ, and XLGs.
In contrast to previous observations (Ding et al., 2008), we found that all three XLGs reside at the plasma membrane, the correct cellular location for the canonical heterotrimeric complexes to form and associate with transmembrane receptors. The nuclear localization was confirmed only for XLG2 and XLG3, but not for XLG1 proteins. This dual localization can account for the plural functions reported for XLGs. It is possible that XLG2, for example, is involved in Gβγ-mediated defense signaling at the plasma membrane but also plays an independent role in flowering initiation interacting with the RTV1 nuclear protein (Heo et al., 2012).
We also demonstrated by Y3H and BiFC assays that XLG2 physically interacts with AGB1 only in the presence of AGG subunits. Noteworthy, in triple AGG mutant protoplasts, lacking all AGG subunits, a weak YFP signal was observed, which can be interpreted as a weak direct interaction between XLG2 and AGB1. However, because similar signals were detected when XLG2 was cotransfected with unfused nYFP, we conclude that XLG2 and AGB1 may not interact directly, but their interaction requires an AGG subunit. This conclusion is in agreement with a recently established fact that AGB1 protein could not be detected in mutant Arabidopsis plants lacking all three AGGs (Wolfenstetter et al., 2014), indicating that it is probably unstable unless engaged with at least one AGG subunit. Importantly, these interactions take place only at the plasma membrane, and not in the nucleus. This observation implies that the complex is formed at the plasma membrane and is in agreement with the autonomous migration of the Gβγ dimer to the membrane (Adjobo-Hermans et al., 2006; Zeng et al., 2007). Interestingly, we observed strong direct binding between XLGs and Gγ subunits even in the absence of Gβ using protoplasts derived from the agb1-2 mutant. By contrast, interaction between Gα and Gγ1 was detected only when Gβ was present (Wang et al., 2008). This fact emphasizes the diversification between XLGs and plant Gαs but is not unprecedented because it has been demonstrated that the human Gα subunit Gαo directly interacts with the Gγ2 subunit (Rahmatullah and Robishaw, 1994).
Gα subunits are conserved proteins with widespread, but sporadic, presence in eukaryotic phyla (Anantharaman et al., 2011), pointing out that functions of heterotrimeric G proteins are not essential for survival of certain species. Canonical Gα family members are reported in land plants and charophyte algae (Urano et al., 2012; Hackenberg and Pandey, 2014), but not in chlorophyte algae (Anantharaman et al., 2011; Hackenberg and Pandey, 2014) and not in the moss P. patens (Urano et al., 2013). We identified XLG sequences in all major land plant phyla starting as early as mosses (Bryophyta), the most primitive group of existing land plants. In agreement, our phylogenetic studies showed that the ancestral XLG gene most likely appeared around the origin of land plants. Despite the relatively late evolutionary origin, the ancestral XLG seems to have diverged substantially from conserved Gα sequences, as indicated by the long branch leading to the XLG subclade (Fig. 5). We speculate that the ancestral XLG acquired unique functions and that is not surprising considering the dramatic environmental change resulting from venturing onto land. In the complex human locus for Gαs proteins, Gnas, one variant is extra-large Gαs (XLαs), which has a bipartite structure (Kehlenbach et al., 1994). The XLαs transcript is generated by an alternative promoter (Plagge et al., 2008). Therefore, a plausible scenario for origin of the plant XLGs is that after duplication, one copy of the Gα gene acquired an alternative promoter, and hence plants had two Gα and one XLG protein, which shared the Gα domain similarly to the contemporary human Gαs and XLαs. Later, the promoter of the shared Gα could have been lost from the complex locus, leaving the XLG ancestor and the second Gα gene. These two genes were passed to the modern-day plants. Curiously, in the moss P. patens, the second Gα gene seems to be lost as well.
Incorporating our data with the previous findings, we propose to consider XLG proteins as a distinct class of heterotrimeric G protein subunits, which form a complex with the Gβγ dimers. This complex most probably is quite different from that formed with the canonical Gα, and many of the characteristics we have learned from studies of the canonical heterotrimers will not be applicable. To highlight how the consideration of XLGs as heterotrimer members profoundly changes understanding of G protein biology, we will consider some examples in detail. In the classic animal model, Gα and the Gβγ dimer act interdependently during activation and later transfer a signal to their specific cellular effectors. Upon ligand recognition, a receptor activates the Gα subunit, causing the subsequent release of Gβγ. Thus, initiation of Gβγ-mediated pathways requires activation of Gα. In turn, Gβγ is essential for functional coupling of Gα to the receptor (McCudden et al., 2005). This dependency implies that in a mutant lacking Gα, Gβγ is present in a free and, hence, active form, increasing Gβγ signal output. By contrast, absence of Gβγ would abolish Gα signal output. Hence, opposite phenotypes observed in Gα- and Gβγ-deficient mutants would imply that Gβγ is a predominant signaling element, while similar phenotypes would suggest a principal signaling role for Gα. Based on this model and assuming a solitary Gα in Arabidopsis, we and others previously postulated that Gβγ is the predominant signaling module in plant innate immunity (Llorente et al., 2005; Trusov et al., 2006). In this report, we demonstrate that mutants lacking Gβγ and XLGs have the same phenotypes and suggest that it is the XLG, and possibly not Gβγ, that play the dominant signaling role.
Another example is auxin-dependent lateral root production in Arabidopsis, where Gα- and Gβ-deficient mutants displayed opposite phenotypes, fewer and more lateral roots, respectively (Ullah et al., 2003). It was suggested that free Gβγ was actively attenuating the auxin pathway, leading to reduced lateral root production (Ullah et al., 2003). However, in a later study, it was observed that xlg mutants produced more lateral roots, somehow resembling Gβγ-deficient mutants (Ding et al., 2008). It is plausible that this response is actually controlled by XLGs rather than Gβγ, although further genetic analyses are essential to test this prediction. For future studies, generation of a quadruple mutant lacking Gα and all XLG subunits is necessary to discriminate between Gα, XLG, and Gβγ functions. The recognition of XLGs as functional Gβγ partners logically increases the number of potential heterotrimer combinations in Arabidopsis from three to 12, uncovering hidden plasticity and selectivity of the G protein signaling in plants. It also will help to explain the existing conundrum of a very limited G protein repertoire regulating a diverse range of processes, coined as a bottleneck issue in plant G signaling (Urano et al., 2013).
On a broader view, XLG subunits add to the overwhelming list of discrepancies between plant and animal heterotrimeric G proteins. These unconventional subunits will join the unconventional extra-large Gγ subunits (Chakravorty et al., 2011; Trusov et al., 2012), the unconventional regulator of G protein signaling1 (RGS1) protein, which combines the seven transmembrane domain and the RGS activity (Chen et al., 2003; Temple and Jones, 2007), the scarcity of G protein-coupled receptors (Urano et al., 2013; Urano and Jones, 2014), the self-activation and slow GTPase activity of the canonical Gα (Johnston et al., 2007; Jones et al., 2011; Urano et al., 2012), and the independent plasma membrane targeting of Gα and Gβγ (Adjobo-Hermans et al., 2006; Zeng et al., 2007).
In conclusion, we want to stress that XLGs originated from plant Gα, and although deviated substantially in structural and biochemical aspects, they still retain the ability to form functional complexes with Gβγ. One of these complexes, XLG2/Gβγ1, is a main contributor to plant defense. XLGs have been reported to bind and hydrolyze GTP in vitro (Heo et al., 2012), although given their highly unusual biochemical characteristics, including affinity for GTP, Km, and choice of cofactors, it is highly debatable whether they can be considered Gα subunits. We believe XLG/Gβγ heterotrimers will almost certainly have different activation/deactivation mechanisms from Gα/Gβγ. They are also likely to mediate signaling from different kinds of receptors. Gα/Gβγ heterotrimers have been proven to mediate signaling from RGS1, an atypical G protein coupled-like receptor (Chen et al., 2003; Temple and Jones, 2007), while we provide evidence here that XLG2, together with Gβγ, is linked to BIR1 signaling, a receptor-like kinase involved in control of cell death and plant defense. Further biochemical studies are required to establish if the XLG-Gβγ interaction is nucleotide dependent (Wall et al., 1995; Digby et al., 2006) or follows the alternative, nondissociable activation mechanism (Bünemann et al., 2003; Galés et al., 2006). Interestingly, while, in rice, the nonhydrolysable nucleotide GTPγS caused full dissociation of Gα from the complex (Kato et al., 2004), only about 30% of Gα was dissociated in Arabidopsis (Wang et al., 2008). Moreover, a mutated GPA1, which is unable to hydrolyze GTP and hence constitutively active, was found in a complex with Gβγ1 (Adjobo-Hermans et al., 2006), suggesting that, in Arabidopsis, this canonical heterotrimer does not dissociate upon activation and therefore the Gα/Gβγ interaction is not entirely nucleotide dependent. Further research is also required to clarify if all possible 12 heterotrimeric combinations exist in plants and to establish their functions. Identification of additional plant G protein signaling elements is also crucial to unravel the complexity of this signal transduction pathway.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) transfer DNA insertion mutants used in this study, xlg1-2 (SALK_119657), xlg2-1 (SALK_062645; Ding et al., 2008), xlg3-2 (SALK_030162; Zhu et al., 2009), xlg2 xlg3 (Ding et al., 2008), xlg1 xlg2 xlg3 (Ding et al., 2008), and agg1 agg2 (Trusov et al., 2007), were obtained from original authors, agb1-2 (SALK_061896) and rdr6-11 (CS24285) were obtained from the Arabidopsis Biological Research Center (Ohio State University). Plants were grown under the short day (8 h of light/16 h of dark) at 23°C.
Pathogen Inoculation Assays
Pst DC3000 was spray inoculated (optical density at 600 nm [OD600] = 0.4) on 4- to 6-week-old plants according to the established protocols (Katagiri et al., 2002). Inoculation experiments with Fusarium oxysporum on 3-week-old plants were conducted as previously described (Trusov et al., 2013). Drop inoculation of Alternaria brassicicola was conducted on 5-week-old plants as described (Trusov et al., 2006).
ROS Measurement
Leaf discs (6 mm in diameter) of 5- to 6-week-old plants were collected and kept in 200 µL of water for overnight. Water was then removed, and 150 µL of reaction buffer (10 mm Tris-HCl, pH 8.5) was added, followed by addition of 30 µL of luminol/peroxidase solution (200 µg mL–1 each) and 20 µL of either 10 µm flg22 or 10 µm elf18 (1 µm final elicitor concentration) into each well of a 96-well plate. Luminescence was measured in a GloMax 96 Microplate Luminometer (Promega).
Quantitative Real-Time PCR
Total RNA was isolated from 3-week-old plants that were either intact or infected with F. oxysporum at 72 h post inoculation. Samples were treated with DNase I (Invitrogen), followed by complementary DNA synthesis using SuperScript First-Strand Synthesis (Invitrogen). Reactions for quantitative real-time PCR were set up with SYBR Green Master (Roche) according to manufacturer’s instructions. Primers used are specified in Supplemental Table S2.
Y3H Assays
XLG2 complementary DNAs were cloned into pACT2 using XmaI/BamHI restriction sites to produce pACT2-AD-XLG2 (full length), pACT2-AD-XLG2-N (N-terminal domain), and pACT2-AD-XLG2-C (C-terminal domain). GPA1 was cloned in pACT2 with NcoI/EcoRI. AGB1 was fused to GAL4-BD in pBridge Multiple Cloning Site I using EcoRI/BamHI sites. AGG1, AGG2, or AGG3 was cloned into pBridge-AGB1 Multiple Cloning Site II with NotI/BglII sites, producing pBridge-BD-AGB1-AGGs constructs, where s is 1, 2, or 3. The yeast (Saccharomyces cerevisiae) strain AH109 was used for transformation following the Matchmaker Yeast Protocols (Clontech). Yeast cotransformed with two plasmid constructs was grown on an SC medium lacking Leu and Trp. For interaction tests, the SC medium lacking His, Leu, and Trp was used. All media were made according to the Clontech protocol.
BiFC Assays
The full-length XLG2 and GPA1 were cloned into pSAT1A-cEYFP-N1 (CD3-1064). Fusion of the cYFP fragment to the C terminus of XLG2 and GPA1 was preferable because the N terminus of GPA1 is posttranscriptionally modified to ensure plasma membrane targeting (Adjobo-Hermans et al., 2006; Zeng et al., 2007). AGB1, AGG1, AGG2, and AGG3 were each cloned into pKannibal-nEYFP produced by cloning N-terminal part of enhanced yellow fluorescent protein (nEYFP) from pSAT1A-nEYFP-N1 (CD3-1066) using XhoI/EcoRI sites with introduction of the NcoI site at the 3′ end of nEYFP. Here, nYFP fragment was fused to the N-termini of the proteins, because C terminus of AGGs is prenylated to ensure plasma membrane targeting (Adjobo-Hermans et al., 2006; Zeng et al., 2007; Chakravorty et al., 2011). Primers are listed in Supplemental Table S2. Arabidopsis plants were grown for 3 to 4 weeks, and mesophyll protoplasts were isolated and transfected with the constructs of interest, according to the established protocol (Yoo et al., 2007). Transfected protoplasts were kept at room temperature with gentle shaking in darkness for 16 to 24 h until observation with the confocal microscope (Zeiss LSM700). The representative protoplasts were photographed with 510 to 550 nm (for YFP), 580 to 630 nm (for mCherry), and 640 to 700 nm (for chloroplasts) filters.
Subcellular Localization
The full-length XLG1, XLG2, and XLG3 reading frame sequences were cloned into pKannibal-GFP using KpnI/BamHI restriction sites. pKannibal-GFP was produced by cloning GFP using XhoI/EcoRI sites with introduction of the NcoI site at the 3′ of the GFP. The cassette of 35S::GFP-XLG2 was transferred to pART27 (Gleave, 1992) using the NotI restriction site. Primers are listed in Supplemental Table S2. Five-week-old Arabidopsis plants were transformed with the Agrobacterium tumefaciens-mediated floral dip method (Bent, 2006). To perform GFP fusions, transient expression in Nicotiana benthamiana and A. tumefaciens (GV3101 strain) harboring the construct was grown in 5 mL of Luria-Bertani media with appropriate antibiotics at 220 rpm at 28°C overnight. The bacteria were harvested and resuspended in 10 mm MgCl2 with 150 μm acetosyringone (3,5-dimethoxy-acetophenone [Fluka]) and 10 mm MES at pH 5.5 to give a final OD600 of 0.2. Leaves of N. benthamiana grown for 2 to 3 weeks were infiltrated using a syringe without a needle. For each analysis, a Zeiss LSM700 confocal microscope was used. Photographs were taken with the 480- to 510-nm (GFP) filter.
Protein Extraction and Immunoblotting
Protein extraction was conducted according to methods described by Zeng et al. (2007). Briefly, 200 mg of 3-week-old plants was ground in liquid nitrogen and homogenized in buffer that contains 20 mm HEPES (pH 7.5), 100 mm NaCl, 5 mm MgCl2, 1 mm dithiothreitol, and the protease inhibitor cocktail (Sigma-Aldrich). The sample was spun at 6,000g for 5 min at 4°C. The postnuclear supernatant was transferred to a new tube, and 0.2 mL was spun at 100,000g for 1 h at 4°C. Supernatant (soluble protein fraction) was transferred to a new tube. The pellet (membrane-bound protein fraction) was resuspended in 100 µL of the buffer. An equal volume of standard 2× SDS-PAGE loading buffer was added into each fractionated solution. Samples were boiled for 10 min and spun for 2 min before loading onto the gel. Immunoblotting was conducted using Hybond-C extra nitrocellulose membrane (Amersham Biosciences), following the manufacturer’s protocol. GFP antibody (Cell Signaling catalog no. 2555) and anti-rabbit IgG alkaline phosphatase-conjugated secondary antibody (Promega) were used. Immunodetection was performed by adding 5 mL of Western Blue Stabilized Substrate for Alkaline Phosphatase (Promega) and incubation at room temperature for color development.
Phylogenetic Analysis
The C-terminal regions of XLG proteins and Gα sequences were aligned with CLUSTAL W using MEGA6 (Tamura et al., 2013). A conserved fragment starting from GTPase domain G-1 (KooooGxxxxGKST, where o is hydrophobic and x is any residue) till the end of the protein was identified within XLGs and Gα sequences. All tested sequences were cut accordingly and realigned. The analysis involved 26 amino acid sequences. The evolutionary history was inferred by using the neighbor-joining method (Saitou and Nei, 1987). The phylogeny was tested by bootstrap test (1,000 replicates; Felsenstein, 1985). The evolutionary distances were computed using the Poisson correction method (Zuckerkandl and Pauling, 1965). The rate variation among sites was modeled with a γ distribution (shape parameter = 1). All ambiguous positions were removed for each sequence pair. There were a total of 492 positions in the final dataset. Evolutionary analyses were conducted in MEGA6 (Tamura et al., 2013). The list of species and corresponding sequence identifications are provided in Supplemental Table S1. The sequences and alignment are available from the authors upon request.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Evaluation of the Pst DC3000 disease severity using independent pathways mediated by Gβγ and salicylic acid.
Supplemental Figure S2. Localization of the XLG2-AGB1 interaction in the presence of AGG2.
Supplemental Figure S3. Plasma membrane localization of GFP-XLG2.
Supplemental Table S1. Gα and XLG proteins used in phylogenetic reconstructions.
Supplemental Table S2. List of primer sequences used for PCR and cloning.
Supplementary Material
Acknowledgments
We thank Dr. Alan Jones, Dr. Yiji Xia, and the Arabidopsis Biological Research Center for providing Arabidopsis mutant seeds; Dr. David Chakravorty for provision of Y3H constructs; Dr. Kirill Alexandrov for experimental advice; Dr. Mark Ragan for help with phylogenetic data analysis; and Dr. Lars Petrasovits, Dr. Kemal Kazan, Dr. Robert Birch, and Dr. Brendan Kidd for critical reading of the article.
Glossary
- XLG
extra-large G protein
- PCD
programmed cell death
- ROS
reactive oxygen species
- PAMP
pathogen-associated molecular pattern
- BiFC
bimolecular fluorescent complementation
- Pst
Pseudomonas syringae pv tomato
- Col-0
ecotype Columbia
- Y3H
yeast three-hybrid
- OD600
optical density at 600 nm
Footnotes
This work was supported by the Australian Research Council (grant no. DP1094152).
Articles can be viewed without a subscription.
References
- Adjobo-Hermans MJ, Goedhart J, Gadella TW Jr (2006) Plant G protein heterotrimers require dual lipidation motifs of Gα and Gγ and do not dissociate upon activation. J Cell Sci 119: 5087–5097 [DOI] [PubMed] [Google Scholar]
- Anantharaman V, Abhiman S, de Souza RF, Aravind L (2011) Comparative genomics uncovers novel structural and functional features of the heterotrimeric GTPase signaling system. Gene 475: 63–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent A. (2006) Arabidopsis thaliana floral dip transformation method. Methods Mol Biol 343: 87–103 [DOI] [PubMed] [Google Scholar]
- Bisht NC, Jez JM, Pandey S (2011) An elaborate heterotrimeric G-protein family from soybean expands the diversity of plant G-protein networks. New Phytol 190: 35–48 [DOI] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH (2004) Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 16: 1667–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bommert P, Je BI, Goldshmidt A, Jackson D (2013) The maize Gα gene COMPACT PLANT2 functions in CLAVATA signalling to control shoot meristem size. Nature 502: 555–558 [DOI] [PubMed] [Google Scholar]
- Brotman Y, Lisec J, Méret M, Chet I, Willmitzer L, Viterbo A (2012) Transcript and metabolite analysis of the Trichoderma-induced systemic resistance response to Pseudomonas syringae in Arabidopsis thaliana. Microbiology 158: 139–146 [DOI] [PubMed] [Google Scholar]
- Bünemann M, Frank M, Lohse MJ (2003) Gi protein activation in intact cells involves subunit rearrangement rather than dissociation. Proc Natl Acad Sci USA 100: 16077–16082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravorty D, Trusov Y, Zhang W, Acharya BR, Sheahan MB, McCurdy DW, Assmann SM, Botella JR (2011) An atypical heterotrimeric G-protein γ-subunit is involved in guard cell K⁺-channel regulation and morphological development in Arabidopsis thaliana. Plant J 67: 840–851 [DOI] [PubMed] [Google Scholar]
- Chen JG, Gao Y, Jones AM (2006) Differential roles of Arabidopsis heterotrimeric G-protein subunits in modulating cell division in roots. Plant Physiol 141: 887–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JG, Pandey S, Huang J, Alonso JM, Ecker JR, Assmann SM, Jones AM (2004) GCR1 can act independently of heterotrimeric G-protein in response to brassinosteroids and gibberellins in Arabidopsis seed germination. Plant Physiol 135: 907–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JG, Willard FS, Huang J, Liang J, Chasse SA, Jones AM, Siderovski DP (2003) A seven-transmembrane RGS protein that modulates plant cell proliferation. Science 301: 1728–1731 [DOI] [PubMed] [Google Scholar]
- Coursol S, Fan LM, Le Stunff H, Spiegel S, Gilroy S, Assmann SM (2003) Sphingolipid signalling in Arabidopsis guard cells involves heterotrimeric G proteins. Nature 423: 651–654 [DOI] [PubMed] [Google Scholar]
- Delgado-Cerezo M, Sánchez-Rodríguez C, Escudero V, Miedes E, Fernández PV, Jordá L, Hernández-Blanco C, Sánchez-Vallet A, Bednarek P, Schulze-Lefert P, et al. (2012) Arabidopsis heterotrimeric G-protein regulates cell wall defense and resistance to necrotrophic fungi. Mol Plant 5: 98–114 [DOI] [PubMed] [Google Scholar]
- Digby GJ, Lober RM, Sethi PR, Lambert NA (2006) Some G protein heterotrimers physically dissociate in living cells. Proc Natl Acad Sci USA 103: 17789–17794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Pandey S, Assmann SM (2008) Arabidopsis extra-large G proteins (XLGs) regulate root morphogenesis. Plant J 53: 248–263 [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (1985) Confidence-limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791 [DOI] [PubMed] [Google Scholar]
- Finet C, Timme RE, Delwiche CF, Marlétaz F (2010) Multigene phylogeny of the green lineage reveals the origin and diversification of land plants. Curr Biol 20: 2217–2222 [DOI] [PubMed] [Google Scholar]
- Galés C, Van Durm JJ, Schaak S, Pontier S, Percherancier Y, Audet M, Paris H, Bouvier M (2006) Probing the activation-promoted structural rearrangements in preassembled receptor-G protein complexes. Nat Struct Mol Biol 13: 778–786 [DOI] [PubMed] [Google Scholar]
- Gao M, Wang X, Wang D, Xu F, Ding X, Zhang Z, Bi D, Cheng YT, Chen S, Li X, et al. (2009) Regulation of cell death and innate immunity by two receptor-like kinases in Arabidopsis. Cell Host Microbe 6: 34–44 [DOI] [PubMed] [Google Scholar]
- García-Olmedo F, Molina A, Segura A, Moreno M (1995) The defensive role of nonspecific lipid-transfer proteins in plants. Trends Microbiol 3: 72–74 [DOI] [PubMed] [Google Scholar]
- Gleave AP. (1992) A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol Biol 20: 1203–1207 [DOI] [PubMed] [Google Scholar]
- Hackenberg D, Pandey S (2014) Heterotrimeric G proteins in green algae: an early innovation in the evolution of the plant lineage. Plant Signal Behav 9: e28457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo JB, Sung S, Assmann SM (2012) Ca2+-dependent GTPase, extra-large G protein 2 (XLG2), promotes activation of DNA-binding protein related to vernalization 1 (RTV1), leading to activation of floral integrator genes and early flowering in Arabidopsis. J Biol Chem 287: 8242–8253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirt RP, Lal K, Pinxteren J, Warwicker J, Healy B, Coombs GH, Field MC, Embley TM (2003) Biochemical and genetic evidence for a family of heterotrimeric G-proteins in Trichomonas vaginalis. Mol Biochem Parasitol 129: 179–189 [DOI] [PubMed] [Google Scholar]
- Ishida T, Tabata R, Yamada M, Aida M, Mitsumasu K, Fujiwara M, Yamaguchi K, Shigenobu S, Higuchi M, Tsuji H, et al. (2014) Heterotrimeric G proteins control stem cell proliferation through CLAVATA signaling in Arabidopsis. EMBO Rep 15: 1202–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa A. (2009) The Arabidopsis G-protein β-subunit is required for defense response against Agrobacterium tumefaciens. Biosci Biotechnol Biochem 73: 47–52 [DOI] [PubMed] [Google Scholar]
- Johnston CA, Siderovski DP (2007) Receptor-mediated activation of heterotrimeric G-proteins: current structural insights. Mol Pharmacol 72: 219–230 [DOI] [PubMed] [Google Scholar]
- Johnston CA, Taylor JP, Gao Y, Kimple AJ, Grigston JC, Chen JG, Siderovski DP, Jones AM, Willard FS (2007) GTPase acceleration as the rate-limiting step in Arabidopsis G protein-coupled sugar signaling. Proc Natl Acad Sci USA 104: 17317–17322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AM, Assmann SM (2004) Plants: the latest model system for G-protein research. EMBO Rep 5: 572–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JC, Duffy JW, Machius M, Temple BR, Dohlman HG, Jones AM (2011) The crystal structure of a self-activating G protein α subunit reveals its distinct mechanism of signal initiation. Sci Signal 4: ra8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri F, Thilmony R, He SY (2002) The Arabidopsis thaliana-Pseudomonas syringae interaction. The Arabidopsis Book 1: e0039, doi/10.1199/tab.0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato C, Mizutani T, Tamaki H, Kumagai H, Kamiya T, Hirobe A, Fujisawa Y, Kato H, Iwasaki Y (2004) Characterization of heterotrimeric G protein complexes in rice plasma membrane. Plant J 38: 320–331 [DOI] [PubMed] [Google Scholar]
- Kehlenbach RH, Matthey J, Huttner WB (1994) XL α s is a new type of G protein. Nature 372: 804–809 [DOI] [PubMed] [Google Scholar]
- Lee S, Rojas CM, Ishiga Y, Pandey S, Mysore KS (2013) Arabidopsis heterotrimeric G-proteins play a critical role in host and nonhost resistance against Pseudomonas syringae pathogens. PLoS ONE 8: e82445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YR, Assmann SM (1999) Arabidopsis thaliana ‘extra-large GTP-binding protein’ (AtXLG1): a new class of G-protein. Plant Mol Biol 40: 55–64 [DOI] [PubMed] [Google Scholar]
- Li L, Wright SJ, Krystofova S, Park G, Borkovich KA (2007) Heterotrimeric G protein signaling in filamentous fungi. Annu Rev Microbiol 61: 423–452 [DOI] [PubMed] [Google Scholar]
- Li S, Liu Y, Zheng L, Chen L, Li N, Corke F, Lu Y, Fu X, Zhu Z, Bevan MW, et al. (2012) The plant-specific G protein γ subunit AGG3 influences organ size and shape in Arabidopsis thaliana. New Phytol 194: 690–703 [DOI] [PubMed] [Google Scholar]
- Liu J, Ding P, Sun T, Nitta Y, Dong O, Huang X, Yang W, Li X, Botella JR, Zhang Y (2013) Heterotrimeric G proteins serve as a converging point in plant defense signaling activated by multiple receptor-like kinases. Plant Physiol 161: 2146–2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorente F, Alonso-Blanco C, Sánchez-Rodriguez C, Jorda L, Molina A (2005) ERECTA receptor-like kinase and heterotrimeric G protein from Arabidopsis are required for resistance to the necrotrophic fungus Plectosphaerella cucumerina. Plant J 43: 165–180 [DOI] [PubMed] [Google Scholar]
- Lorek J, Griebel T, Jones AM, Kuhn H, Panstruga R (2013) The role of Arabidopsis heterotrimeric G-protein subunits in MLO2 function and MAMP-triggered immunity. Mol Plant Microbe Interact 26: 991–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason MG, Botella JR (2000) Completing the heterotrimer: isolation and characterization of an Arabidopsis thaliana G protein γ-subunit cDNA. Proc Natl Acad Sci USA 97: 14784–14788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason MG, Botella JR (2001) Isolation of a novel G-protein γ-subunit from Arabidopsis thaliana and its interaction with Gβ. Biochim Biophys Acta 1520: 147–153 [DOI] [PubMed] [Google Scholar]
- McCudden CR, Hains MD, Kimple RJ, Siderovski DP, Willard FS (2005) G-protein signaling: back to the future. Cell Mol Life Sci 62: 551–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka Y, Saraiva LR, Kwan YY, Korsching SI (2009) The fifth class of Gα proteins. Proc Natl Acad Sci USA 106: 1484–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham WM, Hamm HE (2008) Heterotrimeric G protein activation by G-protein-coupled receptors. Nat Rev Mol Cell Biol 9: 60–71 [DOI] [PubMed] [Google Scholar]
- Pandey S, Chen JG, Jones AM, Assmann SM (2006) G-protein complex mutants are hypersensitive to abscisic acid regulation of germination and postgermination development. Plant Physiol 141: 243–256 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Pandey S, Monshausen GB, Ding L, Assmann SM (2008) Regulation of root-wave response by extra large and conventional G proteins in Arabidopsis thaliana. Plant J 55: 311–322 [DOI] [PubMed] [Google Scholar]
- Plagge A, Kelsey G, Germain-Lee EL (2008) Physiological functions of the imprinted Gnas locus and its protein variants Gα(s) and XLα(s) in human and mouse. J Endocrinol 196: 193–214 [DOI] [PubMed] [Google Scholar]
- Rahmatullah M, Robishaw JD (1994) Direct interaction of the α and γ subunits of the G proteins. Purification and analysis by limited proteolysis. J Biol Chem 269: 3574–3580 [PubMed] [Google Scholar]
- Rask L, Andréasson E, Ekbom B, Eriksson S, Pontoppidan B, Meijer J (2000) Myrosinase: gene family evolution and herbivore defense in Brassicaceae. Plant Mol Biol 42: 93–113 [PubMed] [Google Scholar]
- Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Serna L. (2005) A simple method for discriminating between cell membrane and cytosolic proteins. New Phytol 165: 947–952 [DOI] [PubMed] [Google Scholar]
- Sharma N, Jung CH, Bhalla PL, Singh MB (2014) RNA sequencing analysis of the gametophyte transcriptome from the liverwort, Marchantia polymorpha. PLoS ONE 9: e97497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suga H, Koyanagi M, Hoshiyama D, Ono K, Iwabe N, Kuma K, Miyata T (1999) Extensive gene duplication in the early evolution of animals before the parazoan-eumetazoan split demonstrated by G proteins and protein tyrosine kinases from sponge and hydra. J Mol Evol 48: 646–653 [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temple BR, Jones AM (2007) The plant heterotrimeric G-protein complex. Annu Rev Plant Biol 58: 249–266 [DOI] [PubMed] [Google Scholar]
- Torres MA, Morales J, Sánchez-Rodríguez C, Molina A, Dangl JL (2013) Functional interplay between Arabidopsis NADPH oxidases and heterotrimeric G protein. Mol Plant Microbe Interact 26: 686–694 [DOI] [PubMed] [Google Scholar]
- Trusov Y, Chakravorty D, Botella JR (2012) Diversity of heterotrimeric G-protein γ subunits in plants. BMC Res Notes 5: 608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trusov Y, Chakravorty D, Botella JR (2013) Fusarium oxysporum infection assays in Arabidopsis. Methods Mol Biol 1043: 67–72 [DOI] [PubMed] [Google Scholar]
- Trusov Y, Jorda L, Molina A, Botella JR (2010) G proteins and plant innate immunity. InYalovsky S, Baluska F, Jones AM, eds, Integrated G Protein Signaling in Plants. Springer-Verlag, Berlin Heidelberg, pp 221–250 [Google Scholar]
- Trusov Y, Rookes JE, Chakravorty D, Armour D, Schenk PM, Botella JR (2006) Heterotrimeric G proteins facilitate Arabidopsis resistance to necrotrophic pathogens and are involved in jasmonate signaling. Plant Physiol 140: 210–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trusov Y, Rookes JE, Tilbrook K, Chakravorty D, Mason MG, Anderson D, Chen JG, Jones AM, Botella JR (2007) Heterotrimeric G protein γ subunits provide functional selectivity in Gβγ dimer signaling in Arabidopsis. Plant Cell 19: 1235–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trusov Y, Sewelam N, Rookes JE, Kunkel M, Nowak E, Schenk PM, Botella JR (2009) Heterotrimeric G proteins-mediated resistance to necrotrophic pathogens includes mechanisms independent of salicylic acid-, jasmonic acid/ethylene- and abscisic acid-mediated defense signaling. Plant J 58: 69–81 [DOI] [PubMed] [Google Scholar]
- Ullah H, Chen JG, Temple B, Boyes DC, Alonso JM, Davis KR, Ecker JR, Jones AM (2003) The β-subunit of the Arabidopsis G protein negatively regulates auxin-induced cell division and affects multiple developmental processes. Plant Cell 15: 393–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah H, Chen JG, Young JC, Im KH, Sussman MR, Jones AM (2001) Modulation of cell proliferation by heterotrimeric G protein in Arabidopsis. Science 292: 2066–2069 [DOI] [PubMed] [Google Scholar]
- Urano D, Chen JG, Botella JR, Jones AM (2013) Heterotrimeric G protein signalling in the plant kingdom. Open Biol 3: 120186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano D, Jones AM (2014) Heterotrimeric G protein-coupled signaling in plants. Annu Rev Plant Biol 65: 365–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano D, Jones JC, Wang H, Matthews M, Bradford W, Bennetzen JL, Jones AM (2012) G protein activation without a GEF in the plant kingdom. PLoS Genet 8: e1002756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall MA, Coleman DE, Lee E, Iñiguez-Lluhi JA, Posner BA, Gilman AG, Sprang SR (1995) The structure of the G protein heterotrimer Gi α 1 β 1 γ 2. Cell 83: 1047–1058 [DOI] [PubMed] [Google Scholar]
- Wang S, Assmann SM, Fedoroff NV (2008) Characterization of the Arabidopsis heterotrimeric G protein. J Biol Chem 283: 13913–13922 [DOI] [PubMed] [Google Scholar]
- Wang XQ, Ullah H, Jones AM, Assmann SM (2001) G protein regulation of ion channels and abscisic acid signaling in Arabidopsis guard cells. Science 292: 2070–2072 [DOI] [PubMed] [Google Scholar]
- Warpeha KM, Lateef SS, Lapik Y, Anderson M, Lee BS, Kaufman LS (2006) G-protein-coupled receptor 1, G-protein Gα-subunit 1, and prephenate dehydratase 1 are required for blue light-induced production of phenylalanine in etiolated Arabidopsis. Plant Physiol 140: 844–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warpeha KM, Upadhyay S, Yeh J, Adamiak J, Hawkins SI, Lapik YR, Anderson MB, Kaufman LS (2007) The GCR1, GPA1, PRN1, NF-Y signal chain mediates both blue light and abscisic acid responses in Arabidopsis. Plant Physiol 143: 1590–1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wettschureck N, Offermanns S (2005) Mammalian G proteins and their cell type specific functions. Physiol Rev 85: 1159–1204 [DOI] [PubMed] [Google Scholar]
- Wolfenstetter S, Chakravorty D, Kula R, Urano D, Trusov Y, Sheahan MB, McCurdy DW, Assmann SM, Jones AM, Botella JR (2014) Evidence for an unusual transmembrane configuration of AGG3, a class C Gγ subunit of Arabidopsis. Plant J 81: 388–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zeng Q, Wang X, Running MP (2007) Dual lipid modification of Arabidopsis Gγ-subunits is required for efficient plasma membrane targeting. Plant Physiol 143: 1119–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W, He SY (2010) A prominent role of the flagellin receptor FLAGELLIN-SENSING2 in mediating stomatal response to Pseudomonas syringae pv tomato DC3000 in Arabidopsis. Plant Physiol 153: 1188–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, He SY, Assmann SM (2008) The plant innate immunity response in stomatal guard cells invokes G-protein-dependent ion channel regulation. Plant J 56: 984–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Li GJ, Ding L, Cui X, Berg H, Assmann SM, Xia Y (2009) Arabidopsis extra large G-protein 2 (XLG2) interacts with the Gβ subunit of heterotrimeric G protein and functions in disease resistance. Mol Plant 2: 513–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuckerkandl E, Pauling L (1965) Evolutionary divergence and convergence in proteins. InBryson V, Vogel HJ, eds, Evolving Genes and Proteins. Academic Press, New York, pp 97–166 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.