A transcription factor increases plant productivity by delaying leaf senescence and stimulating leaf cell division, chloroplast division, photosynthesis, and tolerance to nitrogen deprivation.
Abstract
Arabidopsis (Arabidopsis thaliana) leaf development relies on subsequent phases of cell proliferation and cell expansion. During the proliferation phase, chloroplasts need to divide extensively, and during the transition from cell proliferation to expansion, they differentiate into photosynthetically active chloroplasts, providing the plant with energy. The transcription factor GROWTH REGULATING FACTOR5 (GRF5) promotes the duration of the cell proliferation period during leaf development. Here, it is shown that GRF5 also stimulates chloroplast division, resulting in a higher chloroplast number per cell with a concomitant increase in chlorophyll levels in 35S:GRF5 leaves, which can sustain higher rates of photosynthesis. Moreover, 35S:GRF5 plants show delayed leaf senescence and are more tolerant for growth on nitrogen-depleted medium. Cytokinins also stimulate leaf growth in part by extending the cell proliferation phase, simultaneously delaying the onset of the cell expansion phase. In addition, cytokinins are known to be involved in chloroplast development, nitrogen signaling, and senescence. Evidence is provided that GRF5 and cytokinins synergistically enhance cell division and chlorophyll retention after dark-induced senescence, which suggests that they also cooperate to stimulate chloroplast division and nitrogen assimilation. Taken together with the increased leaf size, ectopic expression of GRF5 has great potential to improve plant productivity.
Arabidopsis (Arabidopsis thaliana) leaves initiate as primordia at the flank of the shoot apical meristem by extensive cell divisions. Later during leaf development, cell proliferation ceases with the arrest of the mitotic cell cycle, and cell expansion starts, concomitant with the onset of endoreduplication (i.e. genome replication without cell division; Donnelly et al., 1999; Beemster et al., 2005). The mitotic arrest front, where cells exit proliferation and start expansion, initiates at the tip of the leaf and migrates in the basipetal direction. It is maintained around the middle of the leaf for a few days, after which it proceeds rapidly toward the leaf base to disappear (Kazama et al., 2010; Andriankaja et al., 2012). The transcription factor GROWTH REGULATING FACTOR5 (GRF5) promotes leaf growth and functions partially redundantly with eight other members of the GRF family in Arabidopsis (Kim et al., 2003; Horiguchi et al., 2005; Kim and Lee, 2006). Recently, several Arabidopsis and rice (Oryza sativa) GRFs were shown to bind DNA to repress or activate the expression of their targets genes, which are not only involved in leaf formation but also in stress responses and floral organ development (Kim et al., 2012; Kuijt et al., 2014; Liu et al., 2014a). GRF5 acts most likely within a complex with the transcriptional coactivator GRF-INTERACTING FACTOR1/ANGUSTIFOLIA3 (AN3) that regulates transcription by means of recruitment of SWITCH/SUCROSE NONFERMENTING chromatin-remodeling complexes (Vercruyssen et al., 2014). It has been suggested that GRF5 and AN3 delay the exit from the cell proliferation phase, because they are expressed in dividing cells of leaf primordia and overexpression increases final leaf size due to an increase in cell division (Horiguchi et al., 2005; Gonzalez et al., 2010; Vercruyssen et al., 2014).
It has long been known that the application of kinetin, a synthetic cytokinin, enhances the photosynthetic rate measured as CO2 assimilation and stimulates chloroplast differentiation, callus greening, and redifferentiation into shoot tissue (Mok, 1994; Kieber and Schaller, 2014). Furthermore, exogenous cytokinin application and endogenously induced increases in cytokinin levels enhance the abundance of chloroplast proteins and the expression of chloroplast-encoded genes as well as nuclear genes encoding chloroplast constituents, such as the small subunit of Rubisco or chlorophyll (Chl) biosynthesis genes (Brenner et al., 2005; Boonman et al., 2007; Lochmanová et al., 2008; Zubo et al., 2008). In addition, cytokinins are well known to promote leaf blade expansion while negatively affecting senescence (Hwang et al., 2012; Kieber and Schaller, 2014).
Cytokinin signaling is mediated by a multistep phosphorelay that initiates with the autophosphorylation of the ARABIDOPSIS HISTIDINE KINASE receptors (AHKs) after cytokinin perception, followed by phosphotransfer to ARABIDOPSIS HISTIDINE PHOSPHOTRANSFER proteins (AHPs) and finally leading to the phosphorylation of ARABIDOPSIS RESPONSE REGULATORS (ARRs), of which two types can be distinguished: the A-type and B-type ARRs (Hwang and Sheen, 2001; Hutchison et al., 2006; Dortay et al., 2008; Hwang et al., 2012). The B-type ARRs function as transcription factors that induce the expression of the primary cytokinin response genes, including the A-type ARRs (Mason et al., 2005; Kim et al., 2006; Taniguchi et al., 2007; Argyros et al., 2008; Brenner et al., 2012). The latter are negative feedback regulators of cytokinin signaling (Kiba et al., 2003; To et al., 2004; Dortay et al., 2006; Lee et al., 2008). Moreover, several CYTOKININ RESPONSE FACTORS (CRFs) were identified as immediate-early cytokinin response targets, which interact with the AHPs and in turn regulate the transcription of a large portion of cytokinin response genes, many of which are also differentially regulated by B-type ARRs (Rashotte et al., 2006; Cutcliffe et al., 2011; Brenner et al., 2012).
The generation of mutants with compromised cytokinin metabolism or signaling confirmed the positive function of cytokinins in chloroplast development. For example, single ahk2 or ahk3, double ahk2 ahk3, and triple ahk2 ahk3 ahk4 mutants showed progressively reduced Chl contents (Riefler et al., 2006; Argyros et al., 2008). Chl levels were also decreased in the shoot of the B-type arr1 arr10 arr12 triple mutant, affected in the majority of cytokinin-activated responses during vegetative plant development (Argyros et al., 2008; Ishida et al., 2008). In addition, ectopic expression of the bacterial cytokinin biosynthetic ISOPENTENYLTRANSFERASE (ipt) gene increased Chl levels, enhanced photosynthetic activity, affected the ultrastructure of chloroplasts, and delayed leaf senescence (Synková et al., 2006; Procházková et al., 2008; Cortleven and Valcke, 2012).
Chloroplasts are inherited as proplastids, usually from the mother plant, and reside in meristematic tissue to differentiate into photosynthetically active chloroplasts in the leaves (Sakamoto et al., 2008). They obtain a more elongated shape, while the thylakoid membranes increase in number and start forming granal stacks (Kim et al., 2012). Chloroplast differentiation is tightly linked with the onset of cell expansion in developing leaves. Transcript profiling during the transition from cell proliferation to cell expansion has demonstrated a tight link with chloroplast development, given the enrichment for genes involved in photosynthesis and chloroplast retrograde signaling (Andriankaja et al., 2012). Chloroplasts not only need to differentiate when mesophyll cells develop, they also need to divide extensively until they reach maturity.
The current knowledge of the regulatory network that stimulates chloroplast development includes three transcription factor families. The GOLDEN2-LIKE (GLK) transcription factors, encoded in Arabidopsis by two homologous genes named GLK1 and GLK2, are essential for the transition from proplastids to functional chloroplasts (Waters et al., 2008). They were proposed to act as nuclear regulators that optimize photosynthetic capacity in response to environmental conditions (Waters et al., 2009). The division of chloroplasts was shown to be promoted by CRF2, belonging to the CRF family of transcription factors (Okazaki et al., 2009), and two GATA transcription factors: GATA, NITRATE-INDUCIBLE, CARBON-METABOLISM INVOLVED (GNC) and GNC-LIKE/CYTOKININ-RESPONSIVE GATA FACTOR1 (CGA1; Hudson et al., 2011; Chiang et al., 2012). Their expression is stimulated by cytokinins, and similarly, cytokinin treatment increases chloroplast division (Rashotte et al., 2006; Naito et al., 2007; Okazaki et al., 2009). Moreover, GNC and CGA1 further regulate proplastid differentiation into chloroplasts, at least in part by inducing an increase in transcript and protein levels of Chl biosynthesis enzymes, such as PROTOCHLOROPHYLLIDE OXIDOREDUCTASES (PORs), that catalyze the second to last step of Chl production. (Richter et al., 2010; Hudson et al., 2011; Tanaka et al., 2011). Remarkably, GLKs also promote the expression of POR genes (Waters et al., 2009).
Previously, combined ectopic expression of GRF5 and CKX3, one of seven Arabidopsis catabolic CYTOKININ OXIDASES/DEHYDROGENASES (Werner et al., 2003), revealed that the 35S:GRF5-driven increase in leaf size was suppressed by enhanced cytokinin degradation. Therefore, it was hypothesized that GRF5 and cytokinins work together to stimulate cell proliferation during leaf development (Vercruyssen et al., 2011). Interestingly, the leaves of GRF5-overexpressing plants show more intense greening, suggesting an increase in Chl levels, a phenotypic trait that could be caused by altered cytokinin signaling.
Here, we show that GRF5 positively regulates leaf development not only by stimulating cell division but also by promoting chloroplast division, leaf longevity, and nitrogen assimilation. The observed synergistic effects of cytokinin on cell division, on the one hand, and Chl retention during dark-induced senescence, on the other hand, demonstrate cross talk between GRF5 and cytokinin functions. Furthermore, GRF5 affects the expression of PORA, GLK1, and ARRs. These new insights are discussed in light of a role for GRF5 to synchronize chloroplast division with cell division in relation to cytokinin and nitrogen signals.
RESULTS
GRF5 Stimulates Chloroplast Proliferation
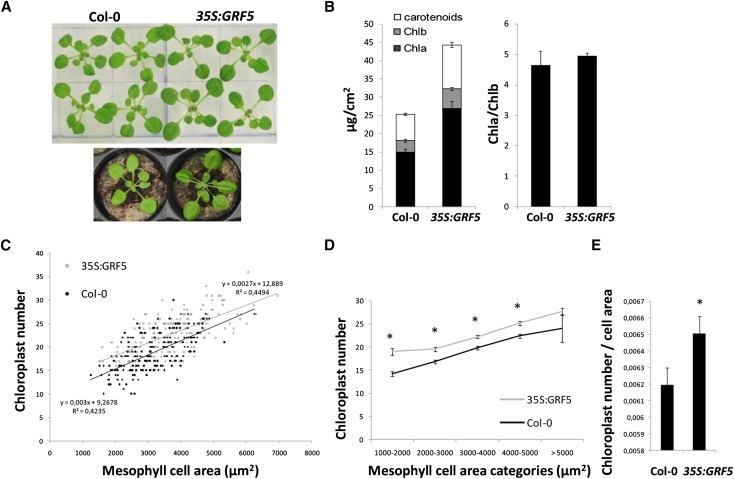
Besides being larger (Horiguchi et al., 2005; Gonzalez et al., 2010), a remarkable feature of the leaves of Arabidopsis plants constitutively overexpressing GRF5 (35S:GRF5) is their darker green color compared with wild-type leaves, which can be observed when the plants are grown both in vitro and in soil (Fig. 1A). Therefore, the photosynthetic pigment concentration was determined in leaves of 21-d-old plants, revealing a strong increase in chlorophyll a (Chla) and chlorophyll b (Chlb) as well as carotenoids per cm2 of leaf surface in 35S:GRF5 plants, while the Chla-Chlb ratio was not altered compared with wild-type plants (Fig. 1B). Significant increases in total Chl content were also measured in independent transgenic lines overexpressing GRF5 in the an3-4 mutant background (Supplemental Fig. S1; Debernardi et al., 2014). Different processes can be the cause of the enhanced chloroplastic pigment levels, such as an increase in leaf thickness due to more or larger mesophyll cells, a larger number of chloroplasts per cell, an increase in chloroplast size, or an elevated pigment biosynthesis per chloroplast.
Figure 1.
Overexpression of GRF5 increases chloroplast number. A, Rosettes of 21-d-old wild-type Columbia-0 (Col-0) and 35S:GRF5 plants grown in vitro and in soil. B, Chl and carotenoid contents and Chla-Chlb ratio, measured in the fifth leaf of 21-d-old wild-type (Col-0) and 35S:GRF5 plants, grown in vitro under long-day conditions (16 h of light/8 h of dark). Error bars indicate sd (n = 4). C, Chloroplast number plotted as a function of mesophyll cell area. Microscopic differential interference contrast images were taken from perpendicular transverse sections of leaves 1 and 2 of wild-type and 35S:GRF5 plants grown for 21 d. The area of 200 mesophyll cells flanking the epidermis was measured with ImageJ, and the corresponding chloroplast number was determined. D and E, Average chloroplast number as a function of mesophyll cell area categories (D) and relative average chloroplast number per mesophyll cell area (E). Chloroplast numbers were determined as described in C. Error bars indicate se. *, Significantly different from the wild type (P < 0.05, Student’s t test).
To assess the cause of the increased pigment levels, transverse sections were made of wild-type and 35S:GRF5 leaves 1 and 2, harvested at 21 d after stratification (DAS), of soil-grown plants. Leaf epidermal and subepidermal palisade cell numbers were shown previously to be strongly increased at this stage, whereas cell size was moderately reduced and unchanged, respectively (Horiguchi et al., 2005; Gonzalez et al., 2010). Measurements of the area of 200 wild-type and 200 35S:GRF5 transverse-sectioned palisade cells adjacent to the adaxial epidermis and determination of the corresponding chloroplast number revealed that, besides the small increase in average cell size, 35S:GRF5 cells equal in size to wild-type cells contained more chloroplasts (Figs. 1C and 2, A and C). Subdivision of mesophyll cell areas in different categories of a multiple of 1,000 µm2, and calculation of the average chloroplast number per category, showed significant increases in 35S:GRF5 chloroplast numbers in mesophyll cell area categories ranging from 1,000 to 5,000 µm2 (Fig. 1D). As a result, the average chloroplast number per transverse two-dimensional cell area was significantly increased by 4% (Fig. 1E). No obvious differences in leaf thickness and organization of cell layers or chloroplast size could be observed between GRF5-overexpressing and wild-type leaves.
Figure 2.
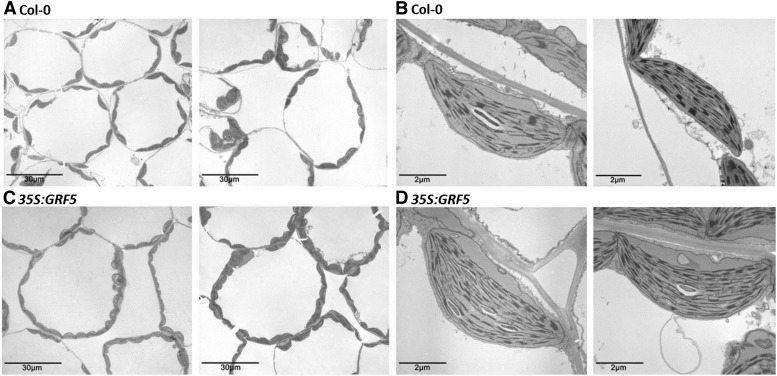
GRF5 promotes chloroplast division. Transmission electron micrographs are shown for sections of 21-d-old leaves 1 and 2 of wild-type (Col-0; A and B) and 35S:GRF5 (C and D) plants grown in soil. Representative sections are shown at magnifications of 800× (A and C) or 10,000× (B and D).
Furthermore, the unaltered Chla-Chlb ratio (Fig. 1B), which reflects a general overview of the photosynthetic apparatus, suggests that the chloroplast ultrastructure is unaffected. Because Chla is a core component of the photosystems while Chlb in addition seems to be required for protein accumulation in the photosystem-associated light-harvesting complexes (Tanaka and Tanaka, 2011), unaltered Chla-Chlb ratios suggest the absence of modifications in the light-harvesting complexes and, hence, in the PSII-PSI ratio. Since both photosystems are physically separated, with PSI mainly located in stromal lamellae and PSII in the closely stacked grana of the thylakoid (Dekker and Boekema, 2005), similar Chla-Chlb ratios also point toward unchanged conformations of the thylakoid membrane system in 35S:GRF5 plastids. Indeed, the ultrastructure of chloroplasts of 35S:GRF5 plants did not seem to differ from that of wild-type plants, as imaged by transmission electron microscopy (Fig. 2, B and D).
Taken together, GRF5 overexpression increases the number of chloroplasts per cell rather than promoting changes in the development of palisade tissue or chloroplast structure.
GRF5 Influences Photosynthetic Capacity
To investigate if the increased amount of chloroplasts in the leaves of GRF5-overexpressing plants can lead to increased photosynthesis rates, PSII fluorescence parameters were determined (Fig. 3, A–E). One-month-old wild-type and 35S:GRF5 leaves displayed a similar maximum photochemical efficiency of PSII in the dark-adapted state (Fv/Fm) and fluorescence quantum yield of PSII photochemistry [Y(II)], although 35S:GRF5 plants showed a slight tendency for an increased electron transport rate (ETR) at higher light intensities (Fig. 3, A–C). Also, a mild increase in photochemical quenching could be observed in 35S:GRF5 leaves, while nonphotochemical quenching was the same as in wild-type leaves (Fig. 3, D and E). On the other hand, analysis of PSII fluorescence parameters of grf5-1 mutant leaves (Horiguchi et al., 2005) did not reveal differences from the wild type at different light intensities (Supplemental Fig. S2). These data indicate a more efficient electron transport beyond PSII in the 35S:GRF5 transgenic plants but a similar loss in energy by heat dissipation processes compared with the wild type.
Figure 3.
Altered photosynthetic capacity in 35S:GRF5 plants. A to E, Photosynthetic parameters were determined by measurements of Chl fluorescence in wild-type and 35S:GRF5 leaves grown for 1 month in long-day conditions in vitro. A, Y(II) as a function of the time. B, ETR through PSII. PAR, Photoactive radiation (µmol photons m−2 s−1). C, Y(II) as a function of photoactive radiation. D, Nonphotochemical quenching (qN). E, Photochemical quenching (qP). Data are means ± sd from three independent experiments (n = 10). F and G, Photosynthetic activity was determined from 21-d-old plants grown in short-day conditions (8 h of light/16 h of dark). F, CO2 assimilation measured for two different leaves from wild-type (Col-0) and 35S:GRF5 (GRF5) plants. G, WUE calculated from maximum CO2 assimilation (µmol CO2 m−2 s−1) divided by transpiration (µmol water m−2 s−1). Error bars in F and G are sd (n = 3). *, Significantly different from the wild type (P ≤ 0.002, Student’s t test).
Furthermore, 35S:GRF5 plants could sustain a higher maximum rate of CO2 assimilation compared with the wild type, from light intensities higher than 200 µmol photons m−2 s−1 upward (10.60 ± 0.14 versus 7.34 ± 0.64 µmol CO2 m−2 s−1), as measured in two different leaves for both wild-type and 35S:GRF5 plants (Fig. 3F). However, the light saturation point (500 ± 100 µmol photons m−2 s−1) and apparent quantum efficiency were similar in both lines (Fig. 3F). Likewise, water use efficiency (WUE) was higher than in the wild type, but only at a photoactive radiation of more than 560 µmol photons m−2 s−1 (Fig. 3G), indicating that 35S:GRF5 leaves are able to assimilate higher amounts of CO2 for a given amount of water at high light intensities. Taken together, these data are in agreement with a similar structure and composition of the 35S:GRF5 and wild-type photosynthetic apparatus.
GRF5 Overexpression Increases Tolerance to Nitrogen Deprivation
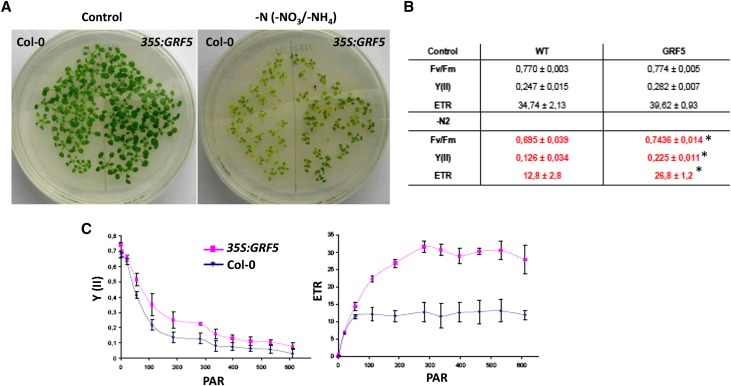
Together with chloroplast development, the leaves gain the ability to assimilate nitrate, which occurs in part in the chloroplasts (Lillo, 2008). Because of the increased chloroplast number in 35S:GRF5 leaves, the response to nitrogen deficiency was studied. One-week-old 35S:GRF5 and wild-type seedlings were transferred to medium without NH4+ and NO3−, lacking any source of nitrogen. As a control, plants were simultaneously transferred to mock medium. After 12 d, enhanced greening of 35S:GRF5 plants compared with control plants was observed, concomitant with an increased photosynthetic capacity (Fig. 4). Fv/Fm, Y(II), and ETR values significantly higher than wild-type values could be measured in 35S:GRF5 plants after growth on medium without nitrogen (Fig. 4, B and C), indicating that 35S:GRF5 plants displayed an increased tolerance against nitrogen deprivation.
Figure 4.
35S:GRF5 plants are more resistant to nitrogen deprivation. A, Wild-type and 35S:GRF5 plants were grown for 7 d on normal one-half-strength Murashige and Skoog medium and subsequently transferred to control medium (left) or medium completely depleted of nitrogen for 12 d (right; −NO3/NH4). B, Photosynthetic parameters at 300 µmol photons m−2 s−1 determined by measurements of Chla fluorescence of wild-type (WT) and 35S:GRF5 plants grown as described in A. *Significantly different from the wild type (P ≤ 0.05, Student’s t test). C, Y(II) and ETR as a function of photoactive radiation (PAR; µmol photons m−2 s−1) in wild-type and 35S:GRF5 leaves after growth on nitrogen-deprived medium. Data are means ± sd of two independent experiments.
Cytokinins Increase the Ability of GRF5 to Promote Cell Division
The positive effects of GRF5 overexpression on chloroplast development, Chl content, photosynthetic rate, and nitrogen status resemble cytokinin-induced responses (Mok, 1994; Riefler et al., 2006; Sakakibara et al., 2006; Argyros et al., 2008), suggesting that GRF5 and cytokinins are interconnected, as postulated previously for leaf growth (Vercruyssen et al., 2011). To further investigate this, the sensitivity of 35S:GRF5 leaf primordia to cytokinin treatment was tested, since both GRF5 and cytokinins are known to stimulate leaf cell proliferation (Werner et al., 2003; Horiguchi et al., 2005; Werner and Schmülling, 2009; Gonzalez et al., 2010; Holst et al., 2011). 35S:GRF5 was combined with the quantitative mitotic marker CYCB1;1:D-Box-GUS-GFP (CYCB1;1:DB-GUS; Eloy et al., 2011), which allows the identification of actively dividing cells (Colón-Carmona et al., 1999). Double homozygous 35S:GRF5/CYCB1;1:DB-GUS plants were generated and showed an evenly strong increase in GRF5 transgene expression levels compared with the 35S:GRF5 parent plants (Supplemental Fig. S3). 35S:GRF5/CYCB1;1:DB-GUS and CYCB1;1:DB-GUS control plants were grown for 9 DAS and subsequently transferred to medium with different concentrations of the synthetic cytokinin 6-benzylaminopurine (BAP) for 24 h, after which the first leaves were analyzed for GUS staining.
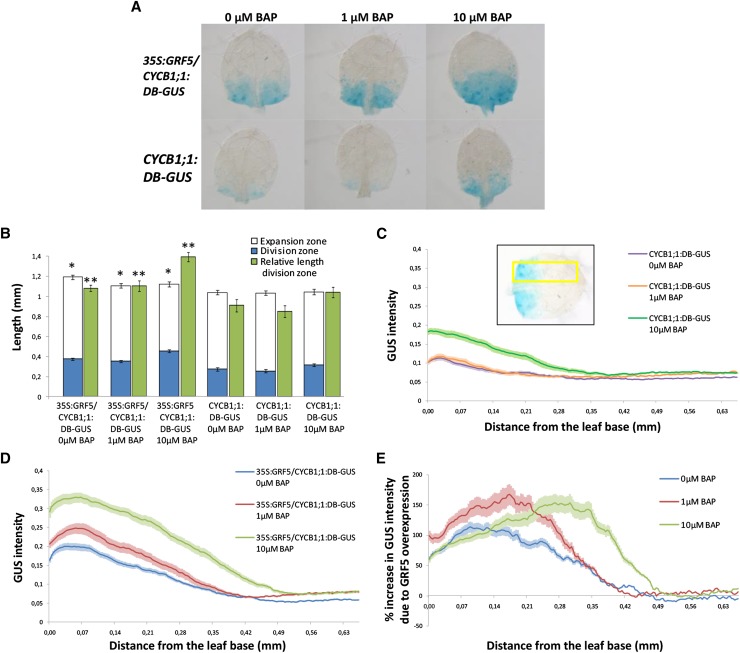
In the absence of BAP, mitotic activity was restricted to the basal part in CYCB1;1:DB-GUS leaves 1 and 2, whereas this GUS-stained region was extended along the length of the leaf in 35S:GRF5/CYCB1;1:DB-GUS plants (Fig. 5, A and B). Although GRF5 overexpression increased leaf length, the relative length of the division zone was significantly larger compared with control leaves (Fig. 5, A and B). In addition to measurement of the length of the GUS-stained region, the intensity of the GUS staining was measured in a defined area along the leaf length, from the base to the tip of the leaf blade (Fig. 5C, inset). This revealed that the GUS intensity was enhanced in 35S:GRF5/CYCB1;1:DB-GUS leaves (Fig. 5, C–E), indicating that GRF5 increases not only the length of the division zone but most likely also the fraction of mitotically active cells for a given distance from the leaf base.
Figure 5.
35S:GRF5 plants show increased CYCB1;1 activity and are more susceptible to cytokinin treatment. 35S:GRF5/CYCB1;1:DB-GUS and CYCB1;1:DB-GUS plants were grown on nylon meshes for 9 d in vitro and subsequently transferred to medium with 0, 1, and 10 µm BAP for 24 h before GUS staining. A, Leaves 1 and 2 were mounted on slides for picture taking. B, GUS-stained and nonstained regions, indicating the division and expansion zones, respectively, measured along the length of the leaf and the relative length of the GUS-stained zone in arbitrary units. *, Significantly different from CYCB1;1:DB-GUS control plants at a similar BAP concentration; **, the relative length of the GUS-stained zone is significantly different from CYCB1;1:DB-GUS control plants at a similar BAP concentration (P < 0.05, Student’s t test). C to E, GUS staining was measured with ImageJ in a defined area along the leaf length, depicted by the yellow rectangle in the inset in C. C, GUS intensity in CYCB1;1:DB-GUS plants after 24 h of growth on 0, 1, and 10 µm BAP. D, GUS intensity in 35S:GRF5/CYCB1;1:DB-GUS plants after 24 h of growth on 0, 1, and 10 µm BAP. E, Percentage increase in GUS intensity in 35S:GRF5/CYCB1;1:DB-GUS compared with CYCB1;1:DB-GUS leaves at the different BAP concentrations. Error bars indicate se (n ≥ 25).
Exogenous application of 1 µm BAP did not affect leaf length or GUS staining in CYCB1;1:DB-GUS plants (Fig. 5, A–C), nor did it further extend the GUS-stained region in 35S:GRF5/CYCB1;1:DB-GUS leaves, but it did increase the intensity of GUS staining in the latter leaves (Fig. 5, A, B, and D). A higher BAP concentration of 10 µm promoted both the length and intensity of the GUS-stained region in CYCB1;1:DB-GUS and 35S:GRF5/CYCB1;1:DB-GUS plants compared with untreated plants (Fig. 5, A–D). Moreover, the percentage of increase in GUS intensity due to 10 µm BAP treatment remained higher in a larger region along the length of the leaves of 35S:GRF5/CYCB1;1:DB-GUS compared with CYCB1;1:DB-GUS plants (Fig. 5E), demonstrating the synergistic effect of GRF5 overexpression and cytokinin treatment.
Taken together, these data show that a BAP concentration as low as 1 µm was able to stimulate cell division when GRF5 was overexpressed, but not in wild-type plants. A higher BAP concentration of 10 µm enhanced the mitotic activity and the length of the cell proliferation zone in both control and 35S:GRF5 plants, but cell division was increased in an extended region along the length of GRF5-overexpressing leaves. Thus, ectopic expression of GRF5 increases the sensitivity to cytokinin-driven stimulation of cell proliferation, demonstrating that GRF5 and cytokinins work together.
GRF5 Stimulates Leaf Longevity Together with Cytokinins
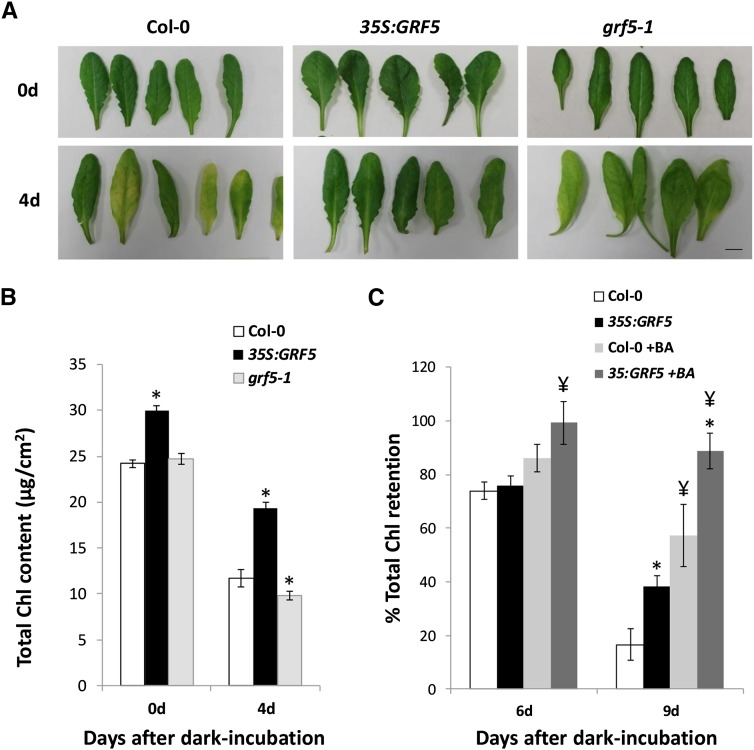
Overexpression of GRF5 not only yields darker green rosettes with larger leaves but also lengthens the vegetative growth period by an average of 10 d under long-day conditions, resulting in increased leaf numbers and rosette fresh weight (Supplemental Fig. S4). Extending the capacity of the plant to photosynthesize and produce assimilates during later developmental stages is proposed to delay senescence (Spano et al., 2003; Zhang et al., 2012). Moreover, leaf senescence is postponed by cytokinins and could serve an additional commonality between GRF5 and cytokinin functions. To investigate this, Chl retention in 35S:GRF5 leaves was measured after dark-induced senescence and cytokinin application, a frequently used assay to determine leaf longevity and cytokinin sensitivity (Riefler et al., 2006).
When detached leaves from 7-week-old plants grown in short-day conditions were incubated for 4 d in the dark, the relative Chl retention in 35:GRF5 leaves was increased compared with wild-type leaves. Whereas 35S:GRF5 leaves retained 65% of the total Chl that was present before incubation in the dark, wild-type leaves retained only around 48%, indicating that GRF5 overexpression delays senescence (Fig. 6, A and B). In addition, leaf Chl contents were determined in grf5-1 mutant plants. Although no significant differences in total Chl were observed compared with control plants just after detachment (0 d), incubation in the dark during 4 d revealed that grf5-1 leaves retained slightly but significantly less Chl than wild-type leaves, suggesting an accelerated senescence (Fig. 6, A and B).
Figure 6.
Overexpression of GRF5 and cytokinin treatment synergistically enhances Chl retention after dark-induced senescence. A, Wild-type (Col-0), 35S:GRF5, and grf5-1 leaves from 7-week-old plants grown in short-day conditions were detached (0d) and subsequently incubated for 4 d in the dark (4d). B, Total Chl content before and after 4 d of dark incubation of the leaves shown in A. Error bars indicate se (n ≥ 20). *, Significantly different from Col-0 (P < 0.05, Student’s t test). C, Chl retention in Col-0 and 35S:GRF5 leaves 6 and 9 d after dark incubation (6d and 9d, respectively) in the absence or presence of 2 µm BA. Values are relative to the Chl content before dark incubation, which was set at 100%. Error bars indicate se (n ≥ 3). *, Significantly different from Col-0; ¥, significant difference between BA-treated and nontreated leaves (P < 0.1, Student’s t test).
Next, the cytokinin sensitivity of 35S:GRF5 leaves was assayed by incubation in water supplemented with 2 µm benzyladenine (BA). After 6 d in the dark, BA treatment did not result in a significant difference in Chl retention in wild-type leaves (Fig. 6C). Chl levels in detached 35S:GRF5 leaves, on the other hand, were significantly higher in the presence of BA, resulting in almost 100% Chl retention after 6 d. Nine days after dark incubation, a significant increase in total Chl content was observed due to BA treatment in both wild-type and 35S:GRF5 leaves compared with nontreated leaves at 9 d. However, 35S:GRF5 leaves incubated in BA retained 51% more Chl compared with mock treatment (89% versus 38%), while wild-type leaves retained only 40% more Chl in the presence of BA (57% versus 17%; Fig. 6C). Mutation of grf5, on the other hand, did not result in a reduced sensitivity toward BA treatment during dark-induced senescence (Supplemental Fig. S5).
These results show that 35S:GRF5 leaves are more sensitive to cytokinin-induced Chl retention during incubation in the dark, indicating that GRF5 overexpression potentiates the senescence-delaying effect of cytokinins.
GRF5 Overexpression Alters the Expression of Marker Genes for Chloroplast Development in Growing Leaves
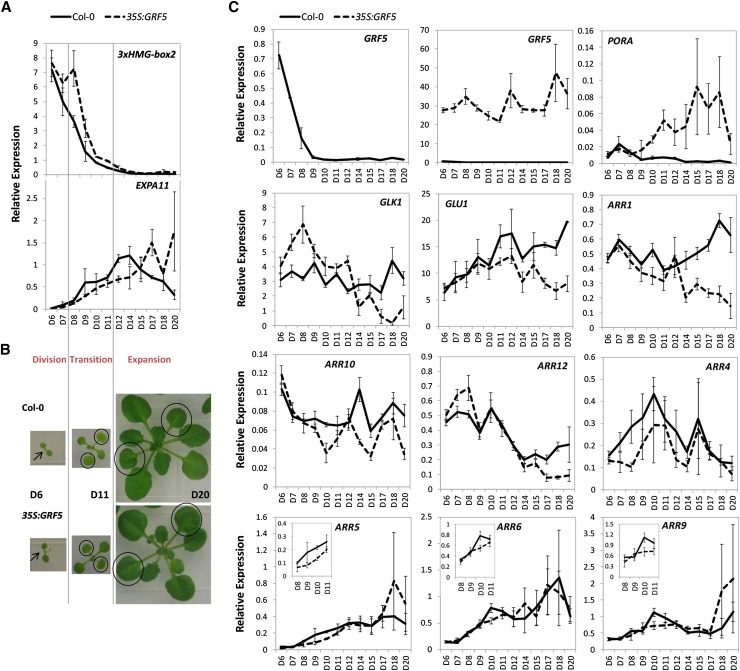
To find an explanation for the phenotype of GRF5-overexpressing plants at the molecular level, the genes identified in a previous study to be differentially expressed in the vegetative part of 35S:GRF5 seedlings at stage 1.03 were investigated (Gonzalez et al., 2010). Although no enrichments of gene categories related to cytokinins or chloroplasts were uncovered, a gene involved in Chl synthesis (i.e. PORA) was found to be strongly induced (Armstrong et al., 1995; Reinbothe et al., 1996; Gonzalez et al., 2010). To investigate the involvement of PORA during leaf development in more detail and to confirm its up-regulation by GRF5 overexpression, PORA expression was quantified by quantitative reverse transcription (qRT)-PCR in wild-type and 35S:GRF5 leaves 1 and 2 harvested at 6 to 20 DAS.
First, to establish the timing of the leaf developmental phases, the expression of marker genes for cell division and cell expansion was verified. Transcript levels of a marker for cell proliferation, 3xHMG-box2 (Pedersen et al., 2011), were highest 6 DAS, started declining on day 7, and disappeared by day 12. Consistent with the function of GRF5 to stimulate cell division, 3xHMG-box2 expression was higher in 35S:GRF5 leaves (Fig. 7A). Similar expression levels were observed for CYCB1;1 (Supplemental Fig. S6), indicating that cell division had ceased completely at 12 DAS. Since expression of the expansion marker EXPA11 started to increase from 8 DAS onward, the cell proliferation phase was defined from 6 to 7 DAS, the transition phase from 8 to 11 DAS, and the cell expansion phase from 12 to 20 DAS (Fig. 7, A and B). Consistently, endogenous GRF5 expression was the highest at 6 DAS, decreased strongly at 7 and 8 DAS, and was virtually absent by 9 DAS and later during leaf development.
Figure 7.
GRF5 affects chloroplast and cytokinin marker gene expression during leaf development. Relative qRT-PCR expression levels are shown for wild-type Col-0 and 35S:GRF5 leaves 1 and 2 dissected at 6 DAS until 20 DAS from plants grown in vitro. A, The expression of a proliferation- and expansion-specific gene marks the subsequent phases of leaf development. B, Rosettes of Col-0 (top) and 35S:GRF5 (bottom) plants at 6, 11, and 20 DAS. Arrows and circles indicate leaves 1 and 2 that were harvested for qRT-PCR. C, Relative expression levels of GRF5 and marker genes for chloroplast development and cytokinin signaling. Error bars indicate se (n = 3). The insets show magnifications of the graphs of the transition phase from 8 to 11 DAS.
In agreement with the microarray data (Gonzalez et al., 2010), PORA was strongly up-regulated in 35S:GRF5 leaves at the end of the transition phase and during cell expansion (Fig. 7C). Interestingly, this PORA up-regulation by GRF5 overexpression was observed only at time points when the endogenous GRF5 expression was close to zero, although the GRF5 transgene expression levels were significantly higher compared with the wild type at each time point (Fig. 7C). The expression levels of the two other Arabidopsis POR genes, PORB and PORC, on the other hand, were more similar to wild-type levels throughout leaf development (Supplemental Fig. S6). Wild-type PORA mRNA levels were very low to almost completely absent, whereas PORB and PORC expression levels were high during leaf development, with the highest levels during the cell proliferation and transition phases, consistent with reports in the literature (Armstrong et al., 1995; Oosawa et al., 2000; Tanaka et al., 2011). Analysis of the transcript levels of genes active more upstream in the tetrapyrrole biosynthesis pathway, GENOMES UNCOUPLED4 (GUN4) and HEMA1, did not reveal obvious differences between wild-type and GRF5-overexpressing plants (Supplemental Fig. S6).
Next, the transcription factors known to be involved in chloroplast development were analyzed. An altered expression pattern in 35S:GRF5 plants was observed for GLK1, which was up-regulated at the end of the cell division phase at 7 and 8 DAS but was down-regulated in expanding leaves from 17 to 20 DAS (Fig. 7C). GLK2 transcription, on the other hand, was unaffected. Although the expression of the transcription factors CRF2, CGA1, and GNC was affected by cytokinin signaling, they were expressed at wild-type levels in 35S:GRF5 leaves during development (Supplemental Fig. S6). Similarly, the mRNA levels of PLASTID DIVISION2, which was described to enhance chloroplast division in 35S:CRF2 plants and after cytokinin treatment (Okazaki et al., 2009), were not affected by overexpression of GRF5. Nevertheless, a down-regulation of GLUTAMATE SYNTHASE1 (GLU1), a target gene of CGA1 and GNC (Hudson et al., 2011), was observed in 35S:GRF5 leaves at later stages of leaf development from 17 to 20 DAS (Fig. 7C).
Steady-State Expression Levels of ARR Genes Are Affected by GRF5
To determine the extent to which the constitutive expression of GRF5 influences cytokinin signaling at different leaf developmental stages, expression levels of the B-type and A-type ARRs were analyzed in dissected leaves 1 and 2. B-type ARR1, ARR10, and ARR12 were selected because they modulate the majority of cytokinin-regulated genes, such as the ones involved in cell division and photosynthesis (Argyros et al., 2008; Ishida et al., 2008; Hwang et al., 2012). Interestingly, all three were expressed at lower levels in 35:GRF5 leaves during the expanding phase of leaf development. ARR1 and ARR10 were down-regulated from 14 DAS and ARR12 from 17 DAS onward (Fig. 7C). Although the expression of the A-type ARRs was more variable, ARR4, ARR5, ARR6, and ARR9 were significantly repressed in 35S:GRF5 plants during the transition from cell proliferation to cell expansion at 8 or 10 DAS (Fig. 7C). ARR3 transcripts were rarely detected in these leaves, and no differences in ARR7 and ARR15 levels were observed in leaves 1 and 2 due to GRF5 overexpression (Supplemental Fig. S6).
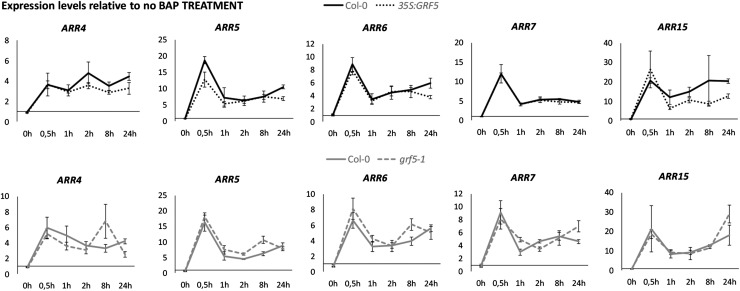
To further characterize the effect of GRF5 on cytokinin signaling, A-type ARR expression was quantified after cytokinin treatment. Since A-type ARRs are rapidly and strongly up-regulated after cytokinin application, they serve as a readout to reveal changes in the primary response to cytokinins (D’Agostino et al., 2000). Therefore, 35S:GRF5, grf5-1 mutant, and Col-0 plants were grown for 9 DAS and subsequently transferred to medium supplemented with 10 µm BAP or mock medium. Shoot tissue was harvested just after transfer (time 0) and after 0.5, 1, 2, 8, and 24 h. Growth on BAP for 0.5 h strongly increased the expression of all A-type ARRs tested, from 3-fold for ARR4 to 26-fold for ARR15 (Fig. 8). After 1 h, A-type ARR expression levels dropped again to be less strongly increased, except for ARR4, for which the same levels were maintained over time. The responses of 35S:GRF5, grf5-1, and Col-0 plants to BAP treatment were largely similar, implying that the primary response to cytokinins is not affected by GRF5 (Fig. 8). However, a significant reduction in A-type ARR expression was observed in 35S:GRF5 as well as grf5-1 plants compared with the wild type, regardless of the BAP treatment (Supplemental Fig. S7). This is in accordance with the observed A-type ARR down-regulation in 35S:GRF5 leaves 1 and 2 during the transition phase (Fig. 7C).
Figure 8.
The primary response to cytokinin treatment is not changed by GRF5. Col-0, 35S:GRF5, and grf5-1 mutant plants were grown for 9 DAS and transferred to medium supplemented with 10 μm BAP or mock medium. A-type ARR expression levels were determined by qRT-PCR in Col-0, 35S:GRF5, and grf5-1 shoots just after transfer (time 0) and after 0.5, 1, 2, 8, and 24 h. Expression levels are relative to mock treatment at each time point. Error bars represent se (n = 3).
Taken together, induction of the primary cytokinin response genes is not changed upon cytokinin application by overexpression or mutation of GRF5. Stable differences in A- and B-type ARR levels rather suggest that an altered steady state has been reached in the cytokinin signaling pathway in 35S:GRF5 and grf5-1 plants.
DISCUSSION
GRF5 and Cytokinins Stimulate Cell Division and Chloroplast Division
GRF5 not only stimulates the division of leaf cells during leaf development but also the division of chloroplasts, similar to cytokinin functions. The region of CYCB1;1:DB-GUS marker gene expression is increased by GRF5 overexpression, resulting in a longer cell division zone, indicating that GRF5 acts to promote the duration of the cell proliferation phase. In addition, the intensity of CYCB1;1:DB-GUS staining is increased, suggesting the presence of a larger fraction of mitotically active cells in the division zone. Cytokinins have also been shown to delay the exit from the proliferation phase and to enhance the expression of mitotic CYCD3 and CYCB1;2 genes (Riou-Khamlichi et al., 1999; Dewitte et al., 2007; Holst et al., 2011; Steiner et al., 2012). In agreement, exogenous cytokinin application enhanced the intensity and region of CYCB1;1:DB-GUS expression, indicating that, similar to GRF5, cytokinins stimulate mitotic activity and the duration of the cell proliferation phase. Moreover, evidence is provided that cytokinins and GRF5 work together to stimulate these processes, since cytokinin treatment and overexpression of GRF5 synergistically increased CYCB1;1:DB-GUS levels.
Overexpression of GRF5 yielded mesophyll cells that contain more chloroplasts. Also, cytokinins stimulated chloroplast division. Given that GRF5 and cytokinins cooperate during leaf cell division, it is very likely that they also work together to promote chloroplast division. Interestingly, the increase in chloroplast number by cytokinin treatment has been associated with a decrease in chloroplast size (Okazaki et al., 2009). In addition, a strong correlation has been observed between the size of mesophyll cells and the number of chloroplasts, indicating that cell area is an important factor to drive chloroplast proliferation (Possingham and Lawrence, 1983; Pyke and Leech, 1992; Kawade et al., 2013). However, such compensation mechanisms were not observed in GRF5-overexpressing leaves. Although a slight increase in average mesophyll cell size was measured, cells equal in size to wild-type cells contained more chloroplasts that were unaltered in size. Compensation has also been observed with respect to final leaf size (e.g. reducing endogenous cytokinin levels results in decreased cell division but enhanced cell expansion; Werner et al., 2003; Holst et al., 2011). Mutation of grf5 also diminishes cell division, but this does not trigger compensated cell enlargement (Horiguchi et al., 2005). Taken together, both GRF5 and cytokinins stimulate cell and chloroplast division, but unlike cytokinins, alterations of GRF5 levels do not lead to compensatory effects on cell or chloroplast size.
Effects of GRF5 and Cytokinins on Senescence and Nitrogen Metabolism
In addition to promoting cell and chloroplast division, GRF5 overexpression also contributes to leaf development by delaying senescence. Consistently, dark incubation of detached grf5-1 leaves revealed an accelerated senescence, which was likewise demonstrated for grf3-1 and an3-1 mutants and plants overexpressing microRNA396 (miR396; Debernardi et al., 2014). The higher Chl levels and increased photosynthetic CO2 uptake in 1-month-old 35S:GRF5 leaves could directly lead to a delay in leaf senescence, since senescence is only initiated when the photosynthetic rate drops below a certain threshold, which is accompanied by chloroplast and Chl breakdown (Lim et al., 2007). Simultaneously, leaf senescence is tightly regulated by genetic programs (Woo et al., 2013) in which GRF5 could actively function beyond the leaf cell division phase. This is supported by the observation that the stimulating effects of a miR396-insensitive version of GRF3 (rGRF3) on cell division and leaf longevity could be uncoupled (Debernardi et al., 2014). Interestingly, GRF5 is one of the two Arabidopsis GRF family members that does not contain an miR396 target site (Jones-Rhoades and Bartel, 2004; Rodriguez et al., 2010); hence, leaf longevity is also promoted by GRFs independently of this posttranscriptional regulation.
Also during senescence, GRF5 and cytokinin functions are interconnected, as demonstrated by the enhanced sensitivity of 35S:GRF5 leaves to cytokinin-driven stimulation of Chl retention after dark-induced senescence. Preventing the decline in cytokinin levels during senescence by expression of the ipt gene has been shown to delay the senescence of tobacco (Nicotiana tabacum) leaves, which was associated with increased antioxidant capacity and ascorbate levels in the chloroplasts (Gan and Amasino, 1995; Procházková et al., 2008). Concomitantly, 35S:GRF5 plants accumulate high levels of ascorbate (Gonzalez et al., 2010). The likely associated increased antioxidant capacity is in favor of the enhanced photochemical quenching, ETR, and CO2 assimilation in 35S:GRF5 plants at high light intensities.
In addition, 35S:GRF5 plants showed tolerance to nitrogen deprivation. This could rely on increased nitrogen storage before transfer to nitrogen-depleted medium, due to more chloroplasts being directly involved in nitrogen assimilation (Lillo, 2008). Alternatively, the increased resistance could result from cross talk between GRF5 and cytokinins, since cytokinins function as secondary messengers that signal nitrogen availability and coordinate its acquisition with the amount required for growth (Sakakibara et al., 2006; Argueso et al., 2009; Kiba et al., 2011). Although further research is necessary to demonstrate this, the tolerance of 35S:GRF5 plants to the lack of nitrogen could be caused by an enhanced cytokinin-dependent nitrogen uptake or assimilation (Brenner et al., 2005; Kiba et al., 2011).
Interestingly, the chloroplast-localized GLU1 was down-regulated during the expansion phase of leaf development in 35S:GRF5 plants, similar to GLK1. GLU1 is key to nitrogen assimilation in leaves (Coschigano et al., 1998; Rachmilevitch et al., 2004; Maurino and Peterhansel, 2010), and also the transcription factor GLK1 has been proposed as an important component in nitrogen signaling (Gutiérrez et al., 2008). This illustrates the close relationship between nitrogen metabolism and chloroplast development, which could be influenced by GRF5 though the modulation of GLU1 and GLK1 expression.
GRF5 and Cytokinins as Coordinators of Chloroplast Division with Cell Division during Leaf Development
The question remains what the putative molecular basis is for the cross talk between cytokinin and GRF5 pathways. Comparison of transcript profiles from 35S:GRF5 seedlings with cytokinin-treated seedlings and the measurement of endogenous cytokinin concentrations in 35S:GRF5 seedlings have suggested that GRF5 transcript levels are not directly influenced by cytokinin signaling, nor that GRF5 affects cytokinin levels (Brenner et al., 2005; Nemhauser et al., 2006; Lee et al., 2007; Argyros et al., 2008; Gonzalez et al., 2010). Rather, the integration is accomplished through the regulation of common target genes.
Overexpression or mutation of GRF5 does not result in rapid changes in the expression of the A-type ARR primary response genes after cytokinin application, suggesting that GRF5 does not impinge on the primary cytokinin response pathway. Nevertheless, stable differences in A- and B-type ARR levels were observed. A-type ARR expression was reduced in 35S:GRF5 leaves during the transition from cell division to expansion as well as in grf5-1 seedlings. Reduced B-type ARR expression also was observed during cell expansion in 35S:GRF5 leaves. This implies that an altered steady state has been reached in the cytokinin signaling pathway in 35S:GRF5 and grf5-1 plants, which could explain the increased sensitivity of 35S:GRF5 plants to cytokinin-driven stimulation of cell division and leaf longevity. Recently, analysis of the root transcript profiles of the grf1 grf2 grf3 triple mutant and plants overexpressing rGRF1 or rGRF3 revealed a significant overlap with a robust set of cytokinin-responsive genes, defined as the golden list, including ARR9 (Bhargava et al., 2013; Liu et al., 2014b). Moreover, GRF1 and GRF3 transcript levels were reduced in 2-week-old ahk2 ahk3 double mutant plants, corroborating that GRFs and cytokinins interact (Liu et al., 2014b).
PORA is one of the three tetrapyrrole pathway enzymes in Arabidopsis that catalyze the light-dependent conversion of the Chl precursor protochlorophyllide (Pchlide) to chlorophyllide, which is subsequently converted to Chl (Armstrong et al., 1995; Tanaka et al., 2011). PORA is mainly active in etiolated seedlings after illumination, which is consistent with the lack of expression in wild-type developing leaves. However, overexpression of PORA rescues Chl levels and the photoautotrophic development of porB porC double mutant plants (Frick et al., 2003; Paddock et al., 2010). The observed higher Chl accumulation in GRF5-overexpressing plants, therefore, likely results from the strong up-regulation of PORA at the end of the transition and expansion phases of leaf development. This is supported by the overexpression of a Brassica napus homolog of AtGRF2 (35S:BnGRF2a) in Arabidopsis leaves, which yields increased Chl contents and photosynthetic rates together with PORA up-regulation (Liu et al., 2012). It is currently unknown, however, if the observed effects are also accompanied by similar changes in active protein levels.
Remarkably, cytokinins stimulate the accumulation of Pchlide in the dark, since Pchlide levels are increased and reduced, respectively, in the ckx quadruple and the ahk2 ahk3 double mutants (Hedtke et al., 2012). In addition, POR mRNA and enzyme levels are increased strongly by cytokinin treatment in lupine (Lupinus luteus; Kusnetsov et al., 1998) and cucumber (Cucumis sativus ‘Aonagajibai’) plants (Kuroda et al., 2000). In Arabidopsis, the regulation of POR transcript and protein levels also seems to depend on cytokinins, in part via induction by cytokinin-responsive CGA1 (Richter et al., 2010; Hudson et al., 2011). As such, PORA could be a putative point of convergence for GRF5 and cytokinin action in promoting Chl synthesis.
Although the expression of CRF2, CGA1, and GNC is positively affected by cytokinin treatment and/or ARR1 and ARR12 function (Okazaki et al., 2009; Chiang et al., 2012), they appeared not to be differentially regulated by the overexpression of GRF5. Likewise, GUN4 and HEMA1 expression were not affected, indicating that PORA might be targeted directly by GRF5.
GLK1 was shown to be up-regulated by GRF5 overexpression during early leaf development. GLKs positively regulate chloroplast development from proplastids (Waters et al., 2008, 2009). Interestingly, GLK1 has been shown to directly modulate the transcription of photosynthetic genes, including PORA, PORB, and PORC (Waters et al., 2009). Reciprocally, GLK expression is affected by chloroplast retrograde signals that likely originate from the tetrapyrrole biosynthesis pathway and communicate the status of the chloroplasts to the nucleus (Waters et al., 2009; Terry and Smith, 2013). Up-regulation of PORA by GRF5 overexpression most likely increases the flux through the tetrapyrrole pathway. Since intermediates of this pathway have been shown to regulate nuclear DNA replication through the activation of cyclin-dependent kinases (Kobayashi et al., 2009), GRF5 provides a means to fine-tune chloroplast division with cell division during leaf development. Therefore, we hypothesize that the observed increased sensitivity of 35S:GRF5 plants to cytokinin-driven stimulation of cell division could be accomplished through the common regulation of PORA and associated changes in retrograde signaling.
CONCLUSION
In order to maintain a balance between the photosynthetic capacity and metabolism, plants must have developed a complex regulatory network to translate inputs such as nitrogen and the developmental stage into responses in the chloroplast. Reciprocally, retrograde signals communicate the chloroplast status to the nucleus. Here, we propose GRF5 as one of the components of this regulatory network, acting as an integrator of cytokinin and developmental signals to synchronize chloroplast division with cell division according to the photosynthetic capacity of the plant cell, intrinsically linked to nitrogen assimilation.
It is tempting to speculate that the enhanced potential for carbon and nitrogen assimilation contributes to the growth increase and delayed senescence in 35S:GRF5 plants. The ability of GRF5 to positively influence cell and chloroplast division without any penalties on chloroplast size could have significant implications with respect to plant yield. Furthermore, enhanced nitrogen use efficiency has become an important biotechnological trait for the genetic improvement of crops, especially due to the detrimental environmental effects and high cost of nitrogen fertilizers (Edgerton, 2009; Kant et al., 2011). Taken together, GRF5 is a highly valuable candidate for genetic engineering and breeding approaches aimed at improving crop productivity by the selection of important traits such as growth and photosynthesis.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) 35S:GRF5 #29, grf5-1, and an3-4 seeds were kindly provided by Dr. Hirokazu Tsukaya (Horiguchi et al., 2005). Independent 35S:GRF5/an3-4 lines were generated by the transformation of homozygous an3-4 inflorescences with a pK7WG2 vector (Karimi et al., 2002), in which the GRF5 coding sequence was introduced by Gateway cloning. Double homozygous 35S:GRF5/CYCB1;1:DB-GUS plants were obtained by crossing 35S:GRF5 plants with CYCB1;1:DB-GUS plants (Eloy et al., 2011), followed by selfing and selection based on hygromycin and kanamycin resistance, respectively. All lines are in the Arabidopsis ecotype Col-0 background.
For in vitro experiments, seeds were sown on sterile plates containing one-half-strength Murashige and Skoog medium (Murashige and Skoog, 1962) supplemented with 1% (w/v) Suc and 0.8% (w/v) agar. The plates were sealed and put in a tissue culture room at 21°C under a 16-h-day/8-h-night regime. For experiments in soil, the plants were grown at 22°C under long-day (16-h-day/8-h-night) or short-day (8-h-day/16-h-night) conditions (50 μmol m−2 s−1).
For cytokinin treatments, the plates containing control medium were overlaid with nylon meshes (Prosep) of 20-µm pore size to prevent roots from growing into the medium, after which seeds were sown. At 9 DAS, seedlings were transferred by gently lifting the nylon mesh with a forceps to plates containing mock medium or medium supplemented with different concentrations of BAP.
Chloroplast Analysis
Soil-grown plants were harvested at 21 DAS, and perpendicular transverse sections were made of leaves 1 and 2 and mounted on slides, according to a previously described protocol (Skirycz et al., 2010). Micropscopic differential interference contrast images were taken, the area of 200 mesophyll cells flanking the epidermis was measured with ImageJ software (http://rsb.info.nih.gov/ij/), and the corresponding chloroplast number was determined. For transmission electron microscopy, ultrathin sections were prepared as described previously (Skirycz et al., 2010).
Intact chloroplasts were isolated from wild-type and transgenic plants using Percoll gradient centrifugation (Wu et al., 1991). Chl in leaves and chloroplasts was determined spectrophotometrically in acetone extracts (Lichtenthaler, 1987).
Photosynthetic Activity Determinations
Chl fluorescence measurements were performed at 25°C on in vitro-grown dark-adapted plants using the Imaging-PAM M-Series Chlorophyll Fluorescence System (Heinz Walz) or on dark-adapted plants grown in soil with the Closed FluorCam FC 800-C (Photon Systems Instruments). Variable PSII fluorescence in the dark-adapted state and maximum PSII fluorescence in the dark-adapted state were determined after 30 min in the dark. Photosynthetic parameters [Fv/Fm, Y(II), ETR, and nonphotochemical and photochemical quenching] were calculated as described (Baker and Rosenqvist, 2004; Baker, 2008).
Light-dependent CO2 assimilation (μmol CO2 m−2 s−1) and transpiration (μmol water m−2 s−1) were determined on fully expanded attached leaves of 3-week-old plants grown in soil under short-day growth conditions (two leaves of three plants of each line) using the GFS-3000 portable photosynthesis system from Heinz Walz. The CO2 concentration of the air entering the leaf chamber and the temperature were adjusted to 360 μL L−1 and 25°C, respectively. Photosynthetic photon flux density ranging from 50 to 1,500 mmol m−2 s−1 was supplied by a controlled halogen light source. WUE (μmol CO2 mol−1 water) was calculated from light-dependent CO2 assimilation divided by transpiration. The data were further analyzed by Photosyn Assistant (http://www.scientific.force9.co.uk/photosyn.htm)
Nitrogen Depletion Assays
For in vitro survival assays under nitrogen-free growth conditions, 7-d-old seedlings grown on control medium were transferred for 12 d to plates without nitrogen, on which ammonium nitrate and potassium nitrate were replaced by 18.79 mm potassium chloride.
GUS Staining and Analysis
Seedlings were harvested at 10 DAS, 24 h after BAP treatment, incubated in heptane for 10 min, and subsequently left to dry for 5 min. Then, they were submersed in 5-bromo-4-chloro-3-indolyl-β-glucuronide buffer {100 mm 2-amino-2-(hydroxymethyl)-1,3-propanediol-HCl, 50 mm NaCl buffer (pH 7), 2 mm K3[Fe(CN)6], and 4 mm 5-bromo-4-chloro-3-indolyl-β-glucuronide}, vacuum infiltrated for 10 min, and incubated at 37°C for 8 h. Seedlings were cleared in 100% and 70% (v/v) ethanol and then kept in 90% (v/v) lactic acid. Leaves 1 and 2 were mounted on slides and photographed with a stereomicroscope.
Leaf length and GUS staining were measured with ImageJ software (http://rsb.info.nih.gov/ij/) according to a method described previously (Vercruyssen et al, 2014). In short, the leaves were imaged in a horizontal position, and the background was subtracted. Next, the color intensity in a defined area along the length of the leaf was measured using the plot profile function, after which the color intensities were normalized to an arbitrary scale of 0 to 1.
Chl Measurements after Dark-Induced Senescence
Leaves 4 and 5 were detached from plants grown for 7 weeks in short-day conditions. The leaves were floated adaxial side up on water or water + 2 µm BA and incubated in the dark. One circular disc 8 mm in diameter was punched per leaf, and total Chl (Chla + Chlb) was extracted in ethanol and measured spectrophotometrically. Total Chl content was normalized to the leaf area.
RNA Extraction and Expression Analysis
Rosettes were harvested in liquid nitrogen. For expression analysis of leaves 1 and 2, harvested rosettes were put in RNAlater solution (AM7021; Ambion) and incubated at 4°C for at least one night, after which leaves 1 and 2 were dissected as such or on a cold plate using a stereomicroscope for the young rosettes. Leaves were frozen in liquid nitrogen and ground, and RNA was extracted according to a combined protocol of TRI reagent RT (Molecular Research Center) and the RNeasy kit with on-column DNase digestion (Qiagen).
The iScript complementary DNA synthesis kit (Bio-Rad) was used to prepare complementary DNA from 1 µg of RNA, and qRT-PCR was done on the LightCycler 480 with SYBR Green I Master (Roche) according to the manufacturer’s instructions. Three technical and three to five biological replicates were done. Relative expression levels were determined by the method of Livak and Schmittgen (2001) and normalized to the housekeeping genes CASEIN KINASE2 and CYCLIN DEPENDENT KINASE A;1. Primer sequences are listed in Supplemental Table S1.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. GRF5 overexpression increases Chl content.
Supplemental Figure S2. Photosynthetic capacity in grf5-1 plants.
Supplemental Figure S3. GRF5 transgene expression levels.
Supplemental Figure S4. GRF5 overexpression delays flowering.
Supplemental Figure S5. Mutation of GRF5 enhances leaf senescence.
Supplemental Figure S6. Chloroplast and cytokinin marker gene expression during leaf development.
Supplemental Figure S7. GRF5 influences A-type ARR expression.
Supplemental Table S1. qRT-PCR primer sequences.
Supplementary Material
Acknowledgments
We thank colleagues in the Systems Biology of Yield group for support and Annick Bleys for help in preparing the article.
Glossary
- Chl
chlorophyll
- Chla
chlorophyll a
- Chlb
chlorophyll b
- DAS
days after stratification
- ETR
electron transport rate
- Y(II)
fluorescence quantum yield of PSII photochemistry
- Fv/Fm
maximum photochemical efficiency of PSII in the dark-adapted state
- WUE
water use efficiency
- BAP
6-benzylaminopurine
- BA
benzyladenine
- qRT
quantitative reverse transcription
- Col-0
Columbia-0
- Pchlide
protochlorophyllide
Footnotes
This work was supported by the European Research Council under the European Union’s Seventh Framework Programme (grant no. FP7/2007–2013, European Research Council grant no. [339341]11); by the Interuniversity Attraction Poles Programme (grant no. IUAP P7/29 “MARS”), initiated by the Belgian State, Science Policy Office; by Ghent University (Bijzonder Onderzoeksfonds Methusalem project grant no. BOF08/01M00408 and Multidisciplinary Research Partnership Biotechnology for a Sustainable Economy grant no. 01MRB510W); by a Marie Curie Intra-European Fellowship for Career Development (grant no. PIEF–GA–2008–221427 to V.B.T.); and by the European Social Fund (CZ.1.07/2.3.00/20.0043 to V.B.T.).
Articles can be viewed without a subscription.
References
- Andriankaja M, Dhondt S, De Bodt S, Vanhaeren H, Coppens F, De Milde L, Mühlenbock P, Skirycz A, Gonzalez N, Beemster GTS, et al. (2012) Exit from proliferation during leaf development in Arabidopsis thaliana: a not-so-gradual process. Dev Cell 22: 64–78 [DOI] [PubMed] [Google Scholar]
- Argueso CT, Ferreira FJ, Kieber JJ (2009) Environmental perception avenues: the interaction of cytokinin and environmental response pathways. Plant Cell Environ 32: 1147–1160 [DOI] [PubMed] [Google Scholar]
- Argyros RD, Mathews DE, Chiang YH, Palmer CM, Thibault DM, Etheridge N, Argyros DA, Mason MG, Kieber JJ, Schaller GE (2008) Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development. Plant Cell 20: 2102–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong GA, Runge S, Frick G, Sperling U, Apel K (1995) Identification of NADPH:protochlorophyllide oxidoreductases A and B: a branched pathway for light-dependent chlorophyll biosynthesis in Arabidopsis thaliana. Plant Physiol 108: 1505–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NR. (2008) Chlorophyll fluorescence: a probe of photosynthesis in vivo. Annu Rev Plant Biol 59: 89–113 [DOI] [PubMed] [Google Scholar]
- Baker NR, Rosenqvist E (2004) Applications of chlorophyll fluorescence can improve crop production strategies: an examination of future possibilities. J Exp Bot 55: 1607–1621 [DOI] [PubMed] [Google Scholar]
- Beemster GTS, De Veylder L, Vercruysse S, West G, Rombaut D, Van Hummelen P, Galichet A, Gruissem W, Inzé D, Vuylsteke M (2005) Genome-wide analysis of gene expression profiles associated with cell cycle transitions in growing organs of Arabidopsis. Plant Physiol 138: 734–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhargava A, Clabaugh I, To JP, Maxwell BB, Chiang YH, Schaller GE, Loraine A, Kieber JJ (2013) Identification of cytokinin-responsive genes using microarray meta-analysis and RNA-Seq in Arabidopsis. Plant Physiol 162: 272–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonman A, Prinsen E, Gilmer F, Schurr U, Peeters AJM, Voesenek LACJ, Pons TL (2007) Cytokinin import rate as a signal for photosynthetic acclimation to canopy light gradients. Plant Physiol 143: 1841–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner WG, Ramireddy E, Heyl A, Schmülling T (2012) Gene regulation by cytokinin in Arabidopsis. Front Plant Sci 3: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner WG, Romanov GA, Köllmer I, Bürkle L, Schmülling T (2005) Immediate-early and delayed cytokinin response genes of Arabidopsis thaliana identified by genome-wide expression profiling reveal novel cytokinin-sensitive processes and suggest cytokinin action through transcriptional cascades. Plant J 44: 314–333 [DOI] [PubMed] [Google Scholar]
- Chiang YH, Zubo YO, Tapken W, Kim HJ, Lavanway AM, Howard L, Pilon M, Kieber JJ, Schaller GE (2012) Functional characterization of the GATA transcription factors GNC and CGA1 reveals their key role in chloroplast development, growth, and division in Arabidopsis. Plant Physiol 160: 332–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colón-Carmona A, You R, Haimovitch-Gal T, Doerner P (1999) Technical advance: spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J 20: 503–508 [DOI] [PubMed] [Google Scholar]
- Cortleven A, Valcke R (2012) Evaluation of the photosynthetic activity in transgenic tobacco plants with altered endogenous cytokinin content: lessons from cytokinin. Physiol Plant 144: 394–408 [DOI] [PubMed] [Google Scholar]
- Coschigano KT, Melo-Oliveira R, Lim J, Coruzzi GM (1998) Arabidopsis gls mutants and distinct Fd-GOGAT genes: implications for photorespiration and primary nitrogen assimilation. Plant Cell 10: 741–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutcliffe JW, Hellmann E, Heyl A, Rashotte AM (2011) CRFs form protein-protein interactions with each other and with members of the cytokinin signalling pathway in Arabidopsis via the CRF domain. J Exp Bot 62: 4995–5002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Agostino IB, Deruère J, Kieber JJ (2000) Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol 124: 1706–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debernardi JM, Mecchia MA, Vercruyssen L, Smaczniak C, Kaufmann K, Inze D, Rodriguez RE, Palatnik JF (2014) Post-transcriptional control of GRF transcription factors by microRNA miR396 and GIF co-activator affects leaf size and longevity. Plant J 79: 413–426 [DOI] [PubMed] [Google Scholar]
- Dekker JP, Boekema EJ (2005) Supramolecular organization of thylakoid membrane proteins in green plants. Biochim Biophys Acta 1706: 12–39 [DOI] [PubMed] [Google Scholar]
- Dewitte W, Scofield S, Alcasabas AA, Maughan SC, Menges M, Braun N, Collins C, Nieuwland J, Prinsen E, Sundaresan V, et al. (2007) Arabidopsis CYCD3 D-type cyclins link cell proliferation and endocycles and are rate-limiting for cytokinin responses. Proc Natl Acad Sci USA 104: 14537–14542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly PM, Bonetta D, Tsukaya H, Dengler RE, Dengler NG (1999) Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev Biol 215: 407–419 [DOI] [PubMed] [Google Scholar]
- Dortay H, Gruhn N, Pfeifer A, Schwerdtner M, Schmülling T, Heyl A (2008) Toward an interaction map of the two-component signaling pathway of Arabidopsis thaliana. J Proteome Res 7: 3649–3660 [DOI] [PubMed] [Google Scholar]
- Dortay H, Mehnert N, Bürkle L, Schmülling T, Heyl A (2006) Analysis of protein interactions within the cytokinin-signaling pathway of Arabidopsis thaliana. FEBS J 273: 4631–4644 [DOI] [PubMed] [Google Scholar]
- Edgerton MD. (2009) Increasing crop productivity to meet global needs for feed, food, and fuel. Plant Physiol 149: 7–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eloy NB, de Freitas Lima M, Van Damme D, Vanhaeren H, Gonzalez N, De Milde L, Hemerly AS, Beemster GTS, Inzé D, Ferreira PCG (2011) The APC/C subunit 10 plays an essential role in cell proliferation during leaf development. Plant J 68: 351–363 [DOI] [PubMed] [Google Scholar]
- Frick G, Su Q, Apel K, Armstrong GA (2003) An Arabidopsis porB porC double mutant lacking light-dependent NADPH:protochlorophyllide oxidoreductases B and C is highly chlorophyll-deficient and developmentally arrested. Plant J 35: 141–153 [DOI] [PubMed] [Google Scholar]
- Gan S, Amasino RM (1995) Inhibition of leaf senescence by autoregulated production of cytokinin. Science 270: 1986–1988 [DOI] [PubMed] [Google Scholar]
- Gonzalez N, De Bodt S, Sulpice R, Jikumaru Y, Chae E, Dhondt S, Van Daele T, De Milde L, Weigel D, Kamiya Y, et al. (2010) Increased leaf size: different means to an end. Plant Physiol 153: 1261–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez RA, Stokes TL, Thum K, Xu X, Obertello M, Katari MS, Tanurdzic M, Dean A, Nero DC, McClung CR, et al. (2008) Systems approach identifies an organic nitrogen-responsive gene network that is regulated by the master clock control gene CCA1. Proc Natl Acad Sci USA 105: 4939–4944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedtke B, Alawady A, Albacete A, Kobayashi K, Melzer M, Roitsch T, Masuda T, Grimm B (2012) Deficiency in riboflavin biosynthesis affects tetrapyrrole biosynthesis in etiolated Arabidopsis tissue. Plant Mol Biol 78: 77–93 [DOI] [PubMed] [Google Scholar]
- Holst K, Schmülling T, Werner T (2011) Enhanced cytokinin degradation in leaf primordia of transgenic Arabidopsis plants reduces leaf size and shoot organ primordia formation. J Plant Physiol 168: 1328–1334 [DOI] [PubMed] [Google Scholar]
- Horiguchi G, Kim GT, Tsukaya H (2005) The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J 43: 68–78 [DOI] [PubMed] [Google Scholar]
- Hudson D, Guevara D, Yaish MW, Hannam C, Long N, Clarke JD, Bi YM, Rothstein SJ (2011) GNC and CGA1 modulate chlorophyll biosynthesis and glutamate synthase (GLU1/Fd-GOGAT) expression in Arabidopsis. PLoS ONE 6: e26765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison CE, Li J, Argueso C, Gonzalez M, Lee E, Lewis MW, Maxwell BB, Perdue TD, Schaller GE, Alonso JM, et al. (2006) The Arabidopsis histidine phosphotransfer proteins are redundant positive regulators of cytokinin signaling. Plant Cell 18: 3073–3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Sheen J (2001) Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 413: 383–389 [DOI] [PubMed] [Google Scholar]
- Hwang I, Sheen J, Müller B (2012) Cytokinin signaling networks. Annu Rev Plant Biol 63: 353–380 [DOI] [PubMed] [Google Scholar]
- Ishida K, Yamashino T, Yokoyama A, Mizuno T (2008) Three type-B response regulators, ARR1, ARR10 and ARR12, play essential but redundant roles in cytokinin signal transduction throughout the life cycle of Arabidopsis thaliana. Plant Cell Physiol 49: 47–57 [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP (2004) Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell 14: 787–799 [DOI] [PubMed] [Google Scholar]
- Kant S, Bi YM, Rothstein SJ (2011) Understanding plant response to nitrogen limitation for the improvement of crop nitrogen use efficiency. J Exp Bot 62: 1499–1509 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kawade K, Horiguchi G, Usami T, Hirai MY, Tsukaya H (2013) ANGUSTIFOLIA3 signaling coordinates proliferation between clonally distinct cells in leaves. Curr Biol 23: 788–792 [DOI] [PubMed] [Google Scholar]
- Kazama T, Ichihashi Y, Murata S, Tsukaya H (2010) The mechanism of cell cycle arrest front progression explained by a KLUH/CYP78A5-dependent mobile growth factor in developing leaves of Arabidopsis thaliana. Plant Cell Physiol 51: 1046–1054 [DOI] [PubMed] [Google Scholar]
- Kiba T, Kudo T, Kojima M, Sakakibara H (2011) Hormonal control of nitrogen acquisition: roles of auxin, abscisic acid, and cytokinin. J Exp Bot 62: 1399–1409 [DOI] [PubMed] [Google Scholar]
- Kiba T, Yamada H, Sato S, Kato T, Tabata S, Yamashino T, Mizuno T (2003) The type-A response regulator, ARR15, acts as a negative regulator in the cytokinin-mediated signal transduction in Arabidopsis thaliana. Plant Cell Physiol 44: 868–874 [DOI] [PubMed] [Google Scholar]
- Kieber JJ, Schaller GE (2014) Cytokinins. The Arabidopsis Book 12: e0168, doi/10.1199/tab.0168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Ryu H, Hong SH, Woo HR, Lim PO, Lee IC, Sheen J, Nam HG, Hwang I (2006) Cytokinin-mediated control of leaf longevity by AHK3 through phosphorylation of ARR2 in Arabidopsis. Proc Natl Acad Sci USA 103: 814–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Choi D, Kende H (2003) The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J 36: 94–104 [DOI] [PubMed] [Google Scholar]
- Kim JH, Lee BH (2006) GROWTH-REGULATING FACTOR4 of Arabidopsis thaliana is required for development of leaves, cotyledons, and shoot apical meristem. J Plant Biol 49: 463–468 [Google Scholar]
- Kim JS, Mizoi J, Kidokoro S, Maruyama K, Nakajima J, Nakashima K, Mitsuda N, Takiguchi Y, Ohme-Takagi M, Kondou Y, et al. (2012) Arabidopsis GROWTH-REGULATING FACTOR7 functions as a transcriptional repressor of abscisic acid- and osmotic stress-responsive genes, including DREB2A. Plant Cell 24: 3393–3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Kanesaki Y, Tanaka A, Kuroiwa H, Kuroiwa T, Tanaka K (2009) Tetrapyrrole signal as a cell-cycle coordinator from organelle to nuclear DNA replication in plant cells. Proc Natl Acad Sci USA 106: 803–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuijt SJ, Greco R, Agalou A, Shao J, ’t Hoen CC, Overnäs E, Osnato M, Curiale S, Meynard D, van Gulik R, et al. (2014) Interaction between the GROWTH-REGULATING FACTOR and KNOTTED1-LIKE HOMEOBOX families of transcription factors. Plant Physiol 164: 1952–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda H, Masuda T, Fusada N, Ohta H, Takamiya K (2000) Expression of NADPH-protochlorophyllide oxidoreductase gene in fully green leaves of cucumber. Plant Cell Physiol 41: 226–229 [DOI] [PubMed] [Google Scholar]
- Kusnetsov V, Herrmann RG, Kulaeva ON, Oelmüller R (1998) Cytokinin stimulates and abscisic acid inhibits greening of etiolated Lupinus luteus cotyledons by affecting the expression of the light-sensitive protochlorophyllide oxidoreductase. Mol Gen Genet 259: 21–28 [DOI] [PubMed] [Google Scholar]
- Lee DJ, Kim S, Ha YM, Kim J (2008) Phosphorylation of Arabidopsis response regulator 7 (ARR7) at the putative phospho-accepting site is required for ARR7 to act as a negative regulator of cytokinin signaling. Planta 227: 577–587 [DOI] [PubMed] [Google Scholar]
- Lee DJ, Park JY, Ku SJ, Ha YM, Kim S, Kim MD, Oh MH, Kim J (2007) Genome-wide expression profiling of ARABIDOPSIS RESPONSE REGULATOR 7 (ARR7) overexpression in cytokinin response. Mol Genet Genomics 277: 115–137 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK. (1987) Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol 148: 350–382 [Google Scholar]
- Lillo C. (2008) Signalling cascades integrating light-enhanced nitrate metabolism. Biochem J 415: 11–19 [DOI] [PubMed] [Google Scholar]
- Lim PO, Kim HJ, Nam HG (2007) Leaf senescence. Annu Rev Plant Biol 58: 115–136 [DOI] [PubMed] [Google Scholar]
- Liu H, Guo S, Xu Y, Li C, Zhang Z, Zhang D, Xu S, Zhang C, Chong K (2014a) OsmiR396d-regulated OsGRFs function in floral organogenesis in rice through binding to their targets OsJMJ706 and OsCR4. Plant Physiol 165: 160–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Hua W, Yang HL, Zhan GM, Li RJ, Deng LB, Wang XF, Liu GH, Wang HZ (2012) The BnGRF2 gene (GRF2-like gene from Brassica napus) enhances seed oil production through regulating cell number and plant photosynthesis. J Exp Bot 63: 3727–3740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Rice JH, Chen N, Baum TJ, Hewezi T (2014b) Synchronization of developmental processes and defense signaling by growth regulating transcription factors. PLoS ONE 9: e98477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-∆∆CT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lochmanová G, Zdráhal Z, Konecná H, Koukalová S, Malbeck J, Soucek P, Válková M, Kiran NS, Brzobohaty B (2008) Cytokinin-induced photomorphogenesis in dark-grown Arabidopsis: a proteomic analysis. J Exp Bot 59: 3705–3719 [DOI] [PubMed] [Google Scholar]
- Mason MG, Mathews DE, Argyros DA, Maxwell BB, Kieber JJ, Alonso JM, Ecker JR, Schaller GE (2005) Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell 17: 3007–3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurino VG, Peterhansel C (2010) Photorespiration: current status and approaches for metabolic engineering. Curr Opin Plant Biol 13: 249–256 [DOI] [PubMed] [Google Scholar]
- Mok M. (1994) Cytokinins and plant development: an overview. InMok D, Mok M, eds, Cytokinins: Chemistry, Activity, and Function. CRC Press, Boca Raton, FL, pp 155–166 [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Naito T, Kiba T, Koizumi N, Yamashino T, Mizuno T (2007) Characterization of a unique GATA family gene that responds to both light and cytokinin in Arabidopsis thaliana. Biosci Biotechnol Biochem 71: 1557–1560 [DOI] [PubMed] [Google Scholar]
- Nemhauser JL, Hong F, Chory J (2006) Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126: 467–475 [DOI] [PubMed] [Google Scholar]
- Okazaki K, Kabeya Y, Suzuki K, Mori T, Ichikawa T, Matsui M, Nakanishi H, Miyagishima SY (2009) The PLASTID DIVISION1 and 2 components of the chloroplast division machinery determine the rate of chloroplast division in land plant cell differentiation. Plant Cell 21: 1769–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oosawa N, Masuda T, Awai K, Fusada N, Shimada H, Ohta H, Takamiya K (2000) Identification and light-induced expression of a novel gene of NADPH-protochlorophyllide oxidoreductase isoform in Arabidopsis thaliana. FEBS Lett 474: 133–136 [DOI] [PubMed] [Google Scholar]
- Paddock TN, Mason ME, Lima DF, Armstrong GA (2010) Arabidopsis protochlorophyllide oxidoreductase A (PORA) restores bulk chlorophyll synthesis and normal development to a porB porC double mutant. Plant Mol Biol 72: 445–457 [DOI] [PubMed] [Google Scholar]
- Pedersen DS, Coppens F, Ma L, Antosch M, Marktl B, Merkle T, Beemster GT, Houben A, Grasser KD (2011) The plant-specific family of DNA-binding proteins containing three HMG-box domains interacts with mitotic and meiotic chromosomes. New Phytol 192: 577–589 [DOI] [PubMed] [Google Scholar]
- Possingham JV, Lawrence ME (1983) Controls to plastid division. Int Rev Cytol 84: 1–56 [Google Scholar]
- Procházková D, Haisel D, Wilhelmová N (2008) Antioxidant protection during ageing and senescence in chloroplasts of tobacco with modulated life span. Cell Biochem Funct 26: 582–590 [DOI] [PubMed] [Google Scholar]
- Pyke KA, Leech RM (1992) Chloroplast division and expansion is radically altered by nuclear mutations in Arabidopsis thaliana. Plant Physiol 99: 1005–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachmilevitch S, Cousins AB, Bloom AJ (2004) Nitrate assimilation in plant shoots depends on photorespiration. Proc Natl Acad Sci USA 101: 11506–11510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashotte AM, Mason MG, Hutchison CE, Ferreira FJ, Schaller GE, Kieber JJ (2006) A subset of Arabidopsis AP2 transcription factors mediates cytokinin responses in concert with a two-component pathway. Proc Natl Acad Sci USA 103: 11081–11085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinbothe S, Reinbothe C, Lebedev N, Apel K (1996) PORA and PORB, two light-dependent protochlorophyllide-reducing enzymes of angiosperm chlorophyll biosynthesis. Plant Cell 8: 763–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter R, Behringer C, Müller IK, Schwechheimer C (2010) The GATA-type transcription factors GNC and GNL/CGA1 repress gibberellin signaling downstream from DELLA proteins and PHYTOCHROME-INTERACTING FACTORS. Genes Dev 24: 2093–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riefler M, Novak O, Strnad M, Schmülling T (2006) Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 18: 40–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riou-Khamlichi C, Huntley R, Jacqmard A, Murray JAH (1999) Cytokinin activation of Arabidopsis cell division through a D-type cyclin. Science 283: 1541–1544 [DOI] [PubMed] [Google Scholar]
- Rodriguez RE, Mecchia MA, Debernardi JM, Schommer C, Weigel D, Palatnik JF (2010) Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development 137: 103–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara H, Takei K, Hirose N (2006) Interactions between nitrogen and cytokinin in the regulation of metabolism and development. Trends Plant Sci 11: 440–448 [DOI] [PubMed] [Google Scholar]
- Sakamoto W, Miyagishima SY, Jarvis P (2008) Chloroplast biogenesis: control of plastid development, protein import, division and inheritance. The Arabidopsis Book 6: e0110, doi/10.1199/tab.0110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skirycz A, De Bodt S, Obata T, De Clercq I, Claeys H, De Rycke R, Andriankaja M, Van Aken O, Van Breusegem F, Fernie AR, et al. (2010) Developmental stage specificity and the role of mitochondrial metabolism in the response of Arabidopsis leaves to prolonged mild osmotic stress. Plant Physiol 152: 226–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spano G, Di Fonzo N, Perrotta C, Platani C, Ronga G, Lawlor DW, Napier JA, Shewry PR (2003) Physiological characterization of ‘stay green’ mutants in durum wheat. J Exp Bot 54: 1415–1420 [DOI] [PubMed] [Google Scholar]
- Steiner E, Efroni I, Gopalraj M, Saathoff K, Tseng TS, Kieffer M, Eshed Y, Olszewski N, Weiss D (2012) The Arabidopsis O-linked N-acetylglucosamine transferase SPINDLY interacts with class I TCPs to facilitate cytokinin responses in leaves and flowers. Plant Cell 24: 96–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Synková H, Schnablová R, Polanská L, Husák M, Siffel P, Vácha F, Malbeck J, Machácková I, Nebesárová J (2006) Three-dimensional reconstruction of anomalous chloroplasts in transgenic ipt tobacco. Planta 223: 659–671 [DOI] [PubMed] [Google Scholar]
- Tanaka R, Kobayashi K, Masuda T (2011) Tetrapyrrole metabolism in Arabidopsis thaliana. The Arabidopsis Book 9: e0145, doi/10.1199/tab.0145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka R, Tanaka A (2011) Chlorophyll cycle regulates the construction and destruction of the light-harvesting complexes. Biochim Biophys Acta 1807: 968–976 [DOI] [PubMed] [Google Scholar]
- Taniguchi M, Sasaki N, Tsuge T, Aoyama T, Oka A (2007) ARR1 directly activates cytokinin response genes that encode proteins with diverse regulatory functions. Plant Cell Physiol 48: 263–277 [DOI] [PubMed] [Google Scholar]
- Terry MJ, Smith AG (2013) A model for tetrapyrrole synthesis as the primary mechanism for plastid-to-nucleus signaling during chloroplast biogenesis. Front Plant Sci 4: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To JPC, Haberer G, Ferreira FJ, Deruère J, Mason MG, Schaller GE, Alonso JM, Ecker JR, Kieber JJ (2004) Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell 16: 658–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercruyssen L, Gonzalez N, Werner T, Schmülling T, Inzé D (2011) Combining enhanced root and shoot growth reveals cross talk between pathways that control plant organ size in Arabidopsis. Plant Physiol 155: 1339–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercruyssen L, Verkest A, Gonzalez N, Heyndrickx KS, Eeckhout D, Han SK, Jégu T, Archacki R, Van Leene J, Andriankaja M, et al. (2014) ANGUSTIFOLIA3 binds to SWI/SNF chromatin remodeling complexes to regulate transcription during Arabidopsis leaf development. Plant Cell 26: 210–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters MT, Moylan EC, Langdale JA (2008) GLK transcription factors regulate chloroplast development in a cell-autonomous manner. Plant J 56: 432–444 [DOI] [PubMed] [Google Scholar]
- Waters MT, Wang P, Korkaric M, Capper RG, Saunders NJ, Langdale JA (2009) GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell 21: 1109–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Motyka V, Laucou V, Smets R, Van Onckelen H, Schmülling T (2003) Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15: 2532–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Schmülling T (2009) Cytokinin action in plant development. Curr Opin Plant Biol 12: 527–538 [DOI] [PubMed] [Google Scholar]
- Woo HR, Kim HJ, Nam HG, Lim PO (2013) Plant leaf senescence and death: regulation by multiple layers of control and implications for aging in general. J Cell Sci 126: 4823–4833 [DOI] [PubMed] [Google Scholar]
- Wu J, Neimanis S, Heber U (1991) Photorespiration is more effective than the Mehler reaction in protecting the photosynthetic apparatus against photoinhibition. Bot Acta 104: 283–291 [Google Scholar]
- Zhang Z, Li G, Gao H, Zhang L, Yang C, Liu P, Meng Q (2012) Characterization of photosynthetic performance during senescence in stay-green and quick-leaf-senescence Zea mays L. inbred lines. PLoS ONE 7: e42936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubo YO, Yamburenko MV, Selivankina SY, Shakirova FM, Avalbaev AM, Kudryakova NV, Zubkova NK, Liere K, Kulaeva ON, Kusnetsov VV, et al. (2008) Cytokinin stimulates chloroplast transcription in detached barley leaves. Plant Physiol 148: 1082–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.