Abstract
MiR-130a has been demonstrated to play important roles in many types of cancers. Nevertheless, its biological function in breast cancer remains largely unknown. In this study, we found that the expression level of miR-130a was down-regulated in breast cancer tissues and cells. Overexpression of miR-130a was able to inhibit cell proliferation, invasion and migration in MCF7 and MDA-MB-435 cells. With the bioinformatics analysis, we further identified that RAB5A was a directly target of miR-130a, and its mRNA and protein level was negatively regulated by miR-130a. Immunohistochemistry verified RAB5A was upregulated in breast cancer tissues. Therefore, the data reported here demonstrate that miR-130a is an important tumor suppressor in breast cancer, and imply that miR-130a/RAB5A axis have potential as therapeutic targets for breast cancer.
Keywords: miR-130a, RAB5A, cell proliferation, cell invasion, cell migration, breast cancer
Introduction
Breast cancer is the most common cancer in women around the world, which can be influenced by a number of environmental factors and is characterized by molecular heterogeneity [1]. Although the implementation of screening programs and the development of new therapeutics in the last 20 years have significantly reduced mortality rates, the molecular mechanisms underlying breast cancer pathogenesis are only partially understood [2].
MicroRNAs (miRNAs) are a class of small non-coding RNAs, which play an important role in regulating gene function through targeting mRNAs for translational repression or degradation [11]. It has been shown that miRNAs play a crucial role in carcinogenesis and tumor progression serving as oncogenes or tumor suppressor genes [3]. miRNAs are implied to influence cell proliferation, apoptosis, invasion, migration and EMT [4,5]. With deregulated expression in several cancers including breast cancer, miRNAs are evolving as potential diagnostic and therapeutic markers [6]. Overexpression of oncogenic miRNAs or underexpression of tumor suppressor miRNAs plays a critical role in tumorigenesis [15,16]. MiR-130a has been described participating in different pathogenesis, involving hepatocellular carcinoma [7], cervical cancer [8], ovarian cancer [9], glioblastoma [10], prostate carcinoma [11], leukemia [12], etc. To date, a cohort of genes related to different cancer pathways have been identified and validated as targeted genes of miR-130a, such as TNF-α [8], Cx43 [13], Runx3 [14], XIAP [9], ATG2B [12], DICER1 [12], MAFB [15], Smad4 [15], GAX [16,17], MET [18], FOG-2 [19], HOXA5 [17]. However, the role of miR-130a in human oral carcinogenesis remains largely unknown.
In the present study, we have investigated the role of miR-130a in the regulation of RAB5A expression in breast cancer cells. We determined that RAB5A was up-regulated and miR-130a was down-regulated in breast cancer tissues and cells. Our findings also demonstrated that miR-130a over-expression leads to inhibited growth, invasion and migration in breast cancer cells. Our findings demonstrated that the 3’UTR of RAB5A contains a putative binding site for miR-130a. Furthermore, we experimentally showed that miR-130a directly targets the 3’UTR of RAB5A to suppress its mRNA and protein expression. Therefore, miR-130a may mediate its tumor suppressor function in breast cancer, at least in part, by suppressing the expression of RAB5A. Altogether, our study characterized a novel microRNA-mediated mechanism of RAB5Aregulation and suggests tumor inhibiting actions of miR-130a in breast cancer cells.
Materials and methods
Patient samples
Breast cancer tissues and adjacent normal tissues were collected in the First Affiliated Hospital of Lanzhou University from January 2009 to December 2013. All the patients recruited into the present study did not receive radiotherapy or chemotherapy or any other treatment before and after operation. Surgical specimens of the tumor resection were collected, and lumps of tumors as well as adjacent normal tissues, which were at least 2 cm distal to tumor margins, were snap-frozen in liquid nitrogen for miR-130a and RAB5A assay. Written informed consent was obtained from all study participants. The use of tissue samples were approved by the ethical committees of the First Affiliated Hospital of Lanzhou University.
Cell culture and transfection
The breast cancer cell lines (MCF-7 and MDA-MB-435) were obtained from the ATCC and maintained in RPMI 1640 with 10% fetal bovine serum (FBS) and 1% antibiotics (Invitrogen, USA). Transfection of the cells with miR-130a mimics or miR-Control (Genepharma, China) was performed using Lipofectamine 2000 (Invitrogen, USA) according to the manufacturer’s instructions.
Cell viability assay
MCF-7 and MDA-MB-435 cells were seeded in 24-well plates overnight and then transfected with miR-130a or miR-NC) (final concentration 200 nM). Afterwards, cells were trypsinized and counted, seeded in 96-well plates (in triplicate) for an MTT assay at a density of 8,000 cells/well (MCF-7) or 10,000 cells/well (MDA-MB-435), and incubated at 37°C for 24 h. Then, at 24, 48 and 72 h after cell seeding, 10 μl MTT (0.5 mg/ml; Sigma-Aldrich, USA) was added to each well (20 μl/well), and the cells were maintained at 37°C for another 4 h. The medium was removed, and the precipitate was dissolved in 100 μl DMSO (Sigma, USA). After shaking for 15 min, the absorbance at 570 nm (A570) was measured using an ELISA reader at a wavelength of 570 nm.
Colony formation assay
Following transfection in 24-well plates as described above, MCF-7 and MDA-MB-435 cells were counted and seeded in 12-well plates (in triplicate) at a density of 200 cells/well. The plates were incubated at 37°C in a 5% CO2 humidified incubator. The culture medium was replaced every 3 days. After 14 days in culture, cells were stained with crystal violet and counted. Colonies with at least 50 cells were considered for quantification.
Transwell invasion and migration assay
Invasion assay was performed with the Transwell chamber with 8 μm pores (Corning, USA). Fifty microliters diluted matrigel (2 mg/ml, BD Biosciences, Bedford, MA) was placed on the inner surface. MCF-7 and MDA-MB-435 cells were transfected for 24 h and isolated to make a final concentration at 2 × 105/ml, which then placed on the top chamber. RMPI1640 with 20% FBS was added to the bottom chamber. After 24 h, non-invading cells were removed from the top of the Matrigel with a cotton-tipped swab. Invading cells at the bottom of the Matrigel were fixed in methanol and stained with Crystal violet. The invasiveness was determined by counting the penetrated cells under a microscope at × 200 magnification of 5 random fields in each well. Each experiment was performed in triplicate. For the transwell migration assay, the process was similar with the transwell invasion assay except the inner surface of the chamber without matrigel.
Wound healing migration assay
MCF-7 and MDA-MB-435 cells were transfected for 24 h, and isolated to make a final concentration at 2 × 105/ml, which then plated in twelve-well plates (2 × 105/well) for 24 h. When the cells reached 90% confluence, sterile pipette tips was used to scratch the wound uniformly. Cell motility was assessed by measuring the movement of cells into a scraped wound. The speed of wound closure was monitored after 72 h by measuring the distance of the wound from 0 h. Each experiment was conducted in triplicate.
Western blotting
The cells were washed twice with cold PBS and total cellular protein was extracted using a modified RIPA buffer with 0.5% sodium dodecyl sulfate (SDS) in the presence of proteinase inhibitor cocktail (Complete mini, Roche). The protein concentration in the supernatants was determined using Bradford protein dye reagent (Bio-Rad, Hercules, CA) and equal amounts of protein lysates were separated on SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a polyvinylidene difluoride membranes (Bio-Rad, Hercules, CA, USA). The membranes were blocked with 5% non-fat milk, followed by incubation with antibodies against RAB5A (1:1000, Proteintech, China) or GAPDH (1:1000, Proteintech, China). GAPDH was used as an internal control. As a secondary antibody, horseradish peroxidase (HRP) conjugated secondary antibody was used, then visualized with enhanced chemiluminescence (ECL) reagents (Amersham Pharmacia; Buckinghamshire, UK) according to the manufacturer’s protocol.
Quantitative real-time polymerase chain reaction (qRT-PCR)
RNA extraction of the cells or tissue samples was performed using the mirVana miRNA Isolation Kit (Ambion, USA) according to the manufacturer’s instructions. For quantitative real-time PCR, RNA was first reversely transcribed into cDNA using SuperScript III reverse transcriptase (Invitrogen). Large RNAs (larger than 200 nt) and small RNAs (smaller than 200 nt) were separated and purified in this procedure. For miRNA detection, 2 μg of small RNA was reverse transcribed to cDNA using M-MLV (Promega, USA). RT-qPCR analysis for miR-130a was performed in triplicate with an SYBR Green I real-time PCR kit (GenePharma, Shanghai, China) according to the manufacturer’s instructions with the ABI 7300 Real-Time PCR System. The expression of miR-130a was normalized to the levels of U6 RNA. To detect the RAB5A gene, 5 μg of large RNA was reverse transcribed to cDNA using oligo (dT) primers and M-MLV. The expression level of RAB5A mRNA was normalized to that of GAPDH mRNA. The change in expression level was calculated using the 2-ΔΔCt method. A two-tailed t-test (P < 0.05 was identified to indicate a statistically significant difference) was performed to identify the differentially expressed miR-130a or RAB5A.
Construction of 3’UTR reporter plasmid and luciferase assay
The human RAB5A 3’UTR harboring miR-130a target sequence as well as the seed-sequence mutated version (RAB5A-3’UTR-mut) were synthesized by GenPharm (Shanghai, China), The RAB5A 3’UTR reporter was generated by inserting the entire 3’UTR or 3’UTR-mut of human RAB5A mRNA into XhoI/NotI sites of psiCHECK-2 vector (Promega) downstream of the Renilla luciferase gene. For the luciferase assay, 1 × 105 cells were transfected along with the RAB5A-3’UTR (or RAB5A-3’UTR-mut) reporter and the miR-130a mimics (or miR-NC) in a 24-well plate using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. After 24 h, firefly and Renilla luciferase activities were measured consecutively using Dual Luciferase Assay (Promega).
Immunohistochemistry
The immunohistochemical analysis for RAB5A in clinical samples was performed as follows. The breast cancer tissues or paraffin-embedded and cut with a thickness of 4 μm. Slides were deparaffinized in xylene twice for 10 min and rehydrated through descending concentration of ethanol. Antigen retrieval was performed in 0.01 mol/L citrate buffer (pH 6.0) by microwave oven for 10 min at 98°C to 100°C. Endogenous peroxidase activity was blocked using 3% hydrogen peroxide (in fresh methanol) for 10 min at room temperature. After washing with phosphate-buffered saline (PBS), the sections were incubated with blocking serum for one hour. Then, tissues were incubated with rabbit RAB5A antibody at 4°C overnight. As a secondary antibody, horseradish peroxidase (HRP) labeled rabbit anti-mouse IgG (Dako Envision plus System) was used. Positive staining was visualized with DAB. Slides were evaluated by two blinded observers.
Statistical analysis
A Student’s test was performed to analyze the significance of differences between the sample means obtained from three independent experiments. Differences were considered statistically significant at P < 0.05.
Results
Relative expression of miR-130a in breast cancer tissue and cell lines
Here, miRNAMap 2.0 (http://miRNAMap.mbc.nctu.edu.tw/) was used to search the miR-130a expression profiles in human cancers, and we found that miR-130a was generally downregulated in various cancers, such as prostate cancer, liver cancer, lung cancer and melanoma, etc (Figure 1A). Furthermore, the result showed miR-130a was also downregulated in breast cancer tissues. To further confirm the result, 10 pairs of breast cancer tissues and adjacent normal tissues were collected and qRT-PCR was performed, which verified that miR-130a was generally downregualted in these cancer tissues (Figure 1C). In addition, we detected the expression of miR-130a in breast cancer cells. As shown is Figure 1B, miR-130a expression was markedly downregulated in MCF-7 and MDA-MB-435 cells compared with the non-malignant breast epithelial cell MCF-10A, and the expression of which was significantly higher in highly metastatic cells, MDA-MB-435, compared with low metastatic MCF-7 cells.
Figure 1.
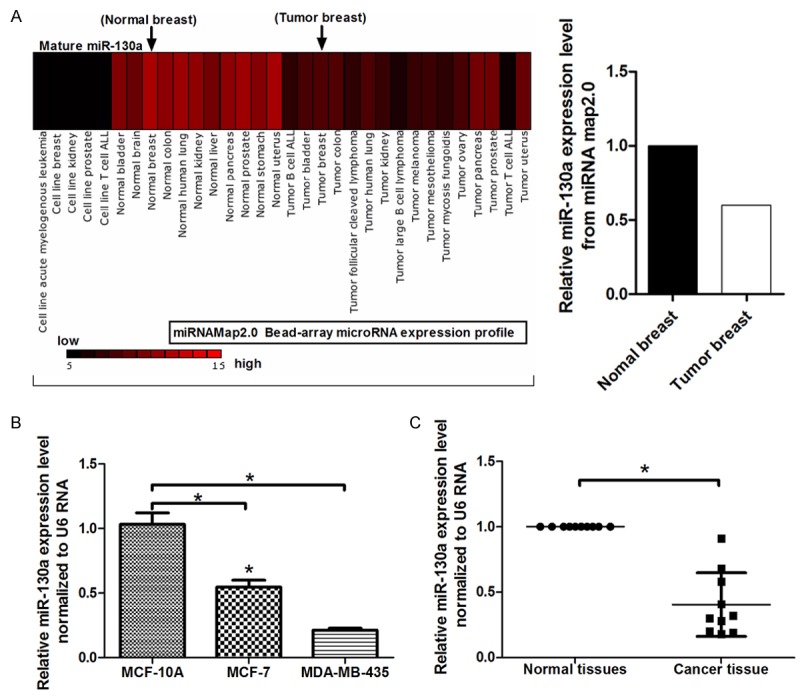
Expression miR-130a in breast cancer tissues and cells. A. miRNAMap 2.0 was used to search the miR-130a expression profiles in human cancers; B. Relative expression of miR-130a (normalized to U6) was detected using qRT-PCR in breast cancer cells, MCF-7 and MDA-MB-435, compared with the non-malignant breast epithelial cell MCF-10A; C. Relative expression of miR-130a in breast cancer tissues and adjacent normal tissues; *P < 0.05.
MiR-130a can suppress breast cancer cells proliferation
To investigate the function of miR-130a in breast cancer cells lines, MCF-7 and MDA-MB-435, were transfected with miR-130a mimics. As shown in Figure 2A, we confirmed that miR-130a mimics can increase the expression of miR-130a by 3-fold and 4-fold in MCF-7 and MDA-MB-435 cells, respectively. MTT assays were then performed to examine the effects of miR-130a on cell growth in vitro. Our data demonstrated that relative cell growth was inhibited in miR-130a mimics transfected cells (Figure 2B, 2C). To assess the effect of miR-130a on the long-term proliferation capacity, colony formation assay was performed and showed that miR-130a can markedly inhibit the cells proliferation capacity (Figure 2D, 2E).
Figure 2.
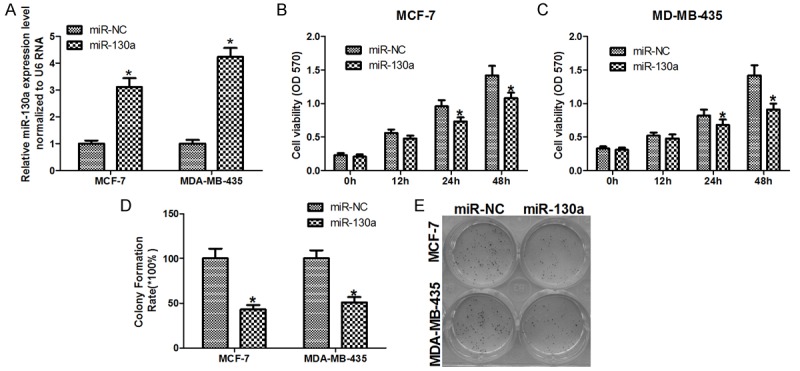
miR-130a inhibits the proliferation of breast cancer cells. A. QRT-PCR was used to detect the transfection efficiency of miR-130a mimics in MCF-7 and MDA-MB-435 cells; B and C. Cell survival was determined by MTT assay; D and E. The long term cell growth as determined by colony formation assay *P < 0.05.
MiR-130a can suppress breast cancer cells invasion and migration
To further investigate whether miR-130a affects cell metastasis, transwell assay and wound healing assay were performed. Transwell invasion assay with matragel demonstrated that miR-130a mimics can inhibit the invasion of MCF-7 and MDA-MB-435 cells by 80% (Figure 3A, 3B), and transwell migration assay without matragel further showed that miR-130a can suppress MCF-7 and MDA-MB-435 cells migration by 78% (Figure 3C, 3D). Furthermore, we used wound healing assay to detect the function of miR-130a on cell migration. As shown in Figure 3E, 3F, the miR-130a mimics inhibited the potential of MCF-7 and MDA-MB-435 cells migration. These results suggested that miR-130a can suppress breast cancer cells invasion and migration.
Figure 3.
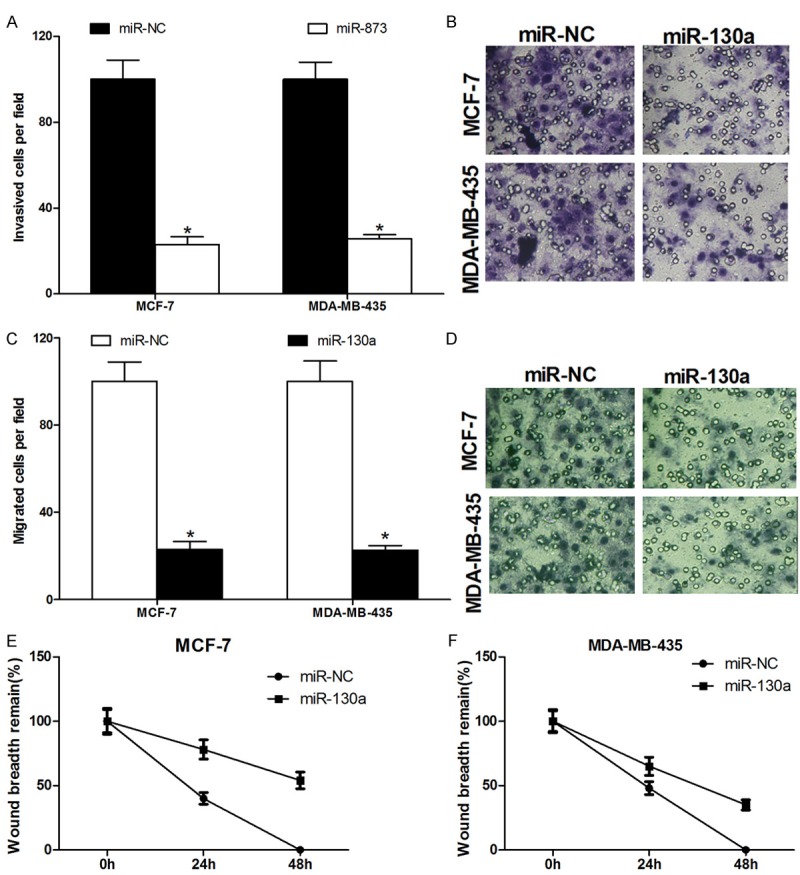
miR-130a represses the invasion and migration of breast cancer cells. A and B. Transwell invasion assay with matragel were performed in miR-130a or miR-NC transfected MCF-7 and MDA-MB-435 cells; C and D. Transwell migration assay without matragel were performed in miR-130a or miR-NC transfected MCF-7 and MDA-MB-435 cells; E and F. Wound healing assay was performed in MCF-7 and MDA-MB-435 cells transfected with miR-130a or miR-NC *P < 0.05.
miR-130a directly targets the 3’TUR of RAB5A
In order to explore the potential target for miR-130a, five computational algorithms, DIANA, miRDB, TargetScan, miRwalk and miRanda, were used and a large number of different target genes were predicted. Among these candidate miRNAs, RAB5A attracted our attention immediately which was predicted by all five algorithms (Figure 4A). To understand whether the effect of miR-130a on RAB5A is specific, we employed the 3’UTR luciferase reporter assay, which showed miR-130a had an obvious effect on inhibiting the luciferase intensity of wild-type 3’UTR luciferase reporter in MCF-7 and MDA-MB-435 cells. However, the inhibitory effect of miR-130a was abrogated in the presence of mutant 3’UTR luciferase reporter (Figure 4B). These results provide evidence that miR-130a can directly target the 3’UTR of RAB5A mRNA.
Figure 4.
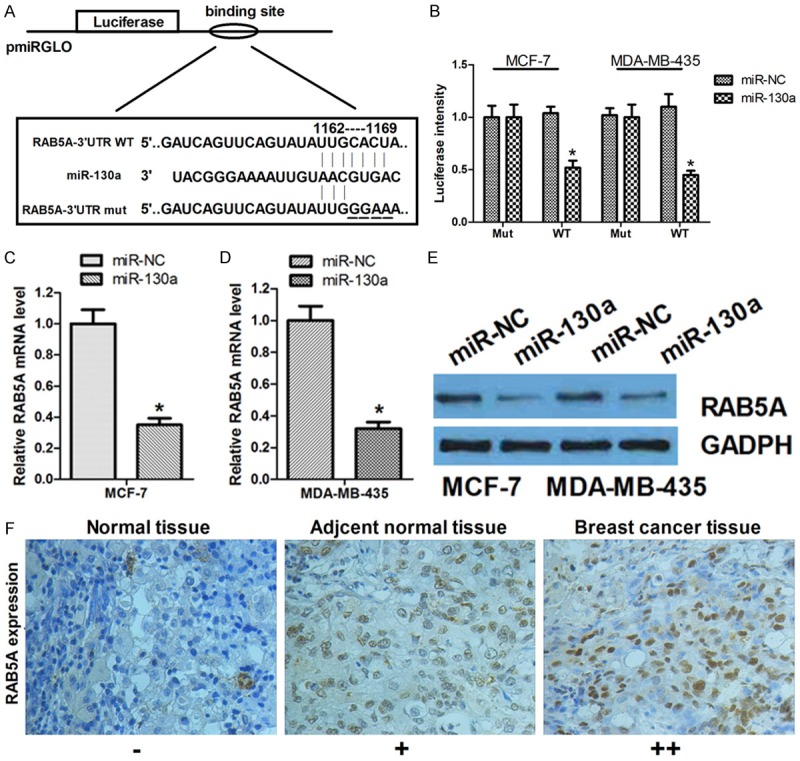
miR-130a can directly target the 3’UTR of RAB5A and regulate RAB5A expression. A. An illustration of pmiRGLO/3’UTR-luciferase plasmid in which the RAB5A-3’UTR (wild-type or mutated version) were ligated downstream of the luciferase gene; B. MCF-7 cells was transfected with miR-130a or miR-NC to analyze the effect on the luciferase intensity of pGL3/Luciferase-RAB5A-3’UTR-WT or RAB5A-3’UTR-mut reporter *P < 0.05; C and D. qRT-PCR was used to detect the mRNA expression of RAB5A in MCF-7 and MDA-MB-435 cells transfected miR-130a or miR-NC; E. Western blot was used to detect the protein expression of RAB5A in MCF-7 and MDA-MB-435 cells transfected miR-130a or miR-NC; F. The protein expression of RAB5A in Normal tissue, adjacent normal tissue and breast cancer tissue was detected by immunohistochemistry *P < 0.05.
Endogenous level of RAB5A can be inhibited by the overexpression of miR-130a
To further test whether miR-130a could influence the endogenous RAB5A expression, qRT-PCR was performed and enforced expression of miR-130a induced 70% reduction of endogenous RAB5A mRNA expression in MCF-7 and MDA-MB-435 cells (Figure 4C, 4D). Furthermore, Western blot assay confirmed that overexpression of miR-130a can significantly suppress the protein level of RAB5A (Figure 4E). In addition, we determined the protein expression RAB5A was significantly higher in breast cancer tissues compared with adjacent normal tissues (Figure 4F), which was generally inversely correlated with the expression of miR-130a in breast cancer tissues (Figure 1). Thus, these results provide evidence that miR-130a directly targets the 3’UTR of RAB5A mRNA to suppress its mRNA and protein expression.
Discussion
MiRNAs have been found to be involved in physiological and pathological processes. Deregulated expression of miRNAs is a common feature of many tumor entities, including breast cancer. The biological role of these miRNAs in tumor development and progression is however still unclear. The aim of this study was therefore a functional analysis of miR-130a in breast cancer, which were shown to be underrepresented in miR-130a in other types of cancer, such as lung cancer [18], prostate carcinoma [11] and hepatocellular carcinoma [7].
In this study, we found that miR-130a was downregulated in breast cancer tissues and cells. These findings pushed us into considering whether miR-130a have the potential to regulate cellular phenotypes. To date, a series of miRNAs have been experimentally verified to be associated with cellular proliferation, invasion and migration, including miR-7 [20], miR-15a [21], miR-34a [22], miR-146a [23], miR-191 [24], miR-204 [25], miR-210 [26], miR-214 [27], miR-335 [28], etc. We experimentally verified that miR-130a is a candidate tumor suppressor of breast cancer cells, as we found that overexpression of miR-130a induced the inhibition of proliferation, invasion and migration in MCF-7 and MDA-MB-435 cells. Our finding expanded the list of miRNA members involved in breast cancer pathogenesis.
miRNA possesses diverse roles that up- or down-regulate target gene expression [29,30]. To identify the miR-130a target genes responsible for its effects on breast cancer cells, we used bioinformatics and functional knowledge associated with miR-130a and chose RAB5A as a candidate gene for further study. In the 3’UTR luciferase reporter assay, the expression of the luciferase reporter plasmid containing the RAB3A 3’UTR was repressed by miR-130a, and the mutated RAB5A 3’UTR abolished this effect. Furthermore, qRT-PCR and Western blot analysis showed that miR-130a decreased RAB5A mRNA and protein expression levels in breast cancer cells compared with the control. Together, these data suggest that miR-130a downregulates RAB5A expression by binding to its 3’UTR. It has been described that Rab5a is significantly overexpressed in ovarian cancer, lung cancer, hepatocellular carcinoma and cervical carcinomas [31]. Furthermore, RAB5A was found to be upregulated in breast cancer tissues by immunohistochemistry analysis in our study. This fact further provides us demonstrated that the downregulation of miR-130a may be the reason for the upregulation of RAB5A in breast cancer tissues.
The finding that miR-130a targets the protein RAB5A for repression highlights an important facet of miRNA-mediated regulation of critical cellular events in breast cancer. RAB5A (RAB GTPase 5A), a member of the Rab subfamily of small GTPases, has been demonstrated to act as an oncogene and has been associated with various key cellular functions, including proliferation and differentiation, gene expression, signal transduction, vesicle trafficking, nuclear assembly, and reorganization of cytoskeleton. Recent findings have revealed that the overexpression of RAB5A gene was correlated with the metastatic potential and malignant degree of lung and stomach cancer [32]. It was also reported that RAB5A was involved in EGF signaling pathway and migration in hepatocellular carcinomas [33]. Moreover, Rab5a was proved to be required for the activation of Rac, a member of the Rho GTPases subfamily, and involve in the regulation of actin cytoskeleton remodeling [34-36]. Therefore, our findings reveal another layer of posttranscriptional RAB5A regulation involving miR-130a, and it needs our further investigation of the miR-130a/RAB5A axis in the regulation of EGF and Rac pathway in breast cancer pathogenesis.
Taken together, our results show that miR-130a plays a inhibitory role in breast cancer cell proliferation, invasion and migration. We identified RAB5A, as a direct target gene of miR-130a. The understanding of the miR-130a/RAB5A regulatory network offers a great potential for the intelligent multitargeted design of new breast cancer cancer therapies.
Disclosure of conflict of interest
None.
References
- 1.Polyak K. Heterogeneity in breast cancer. J Clin Invest. 2011;121:3786–3788. doi: 10.1172/JCI60534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Youlden DR, Cramb SM, Dunn NA, Muller JM, Pyke CM, Baade PD. The descriptive epidemiology of female breast cancer: an international comparison of screening, incidence, survival and mortality. Cancer Epidemiol. 2012;36:237–248. doi: 10.1016/j.canep.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 3.Wang D, Qiu C, Zhang H, Wang J, Cui Q, Yin Y. Human microRNA oncogenes and tumor suppressors show significantly different biological patterns: from functions to targets. PLoS One. 2010:5. doi: 10.1371/journal.pone.0013067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tang J, Ahmad A, Sarkar FH. The Role of MicroRNAs in Breast Cancer Migration, Invasion and Metastasis. Int J Mol Sci. 2012;13:13414–13437. doi: 10.3390/ijms131013414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Y, Zhang J, Wang H, Zhao J, Xu C, Du Y, Luo X, Zheng F, Liu R, Zhang H, Ma D. miRNA-135a promotes breast cancer cell migration and invasion by targeting HOXA10. BMC Cancer. 2012;12:111. doi: 10.1186/1471-2407-12-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He ML, Luo MX, Lin MC, Kung HF. MicroRNAs: potential diagnostic markers and therapeutic targets for EBV-associated nasopharyngeal carcinoma. Biochim Biophys Acta. 2012;1825:1–10. doi: 10.1016/j.bbcan.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 7.Li B, Huang P, Qiu J, Liao Y, Hong J, Yuan Y. MicroRNA-130a is down-regulated in hepatocellular carcinoma and associates with poor prognosis. Med Oncol. 2014;31:230. doi: 10.1007/s12032-014-0230-2. [DOI] [PubMed] [Google Scholar]
- 8.Zhang J, Wu H, Li P, Zhao Y, Liu M, Tang H. NF-kappaB-modulated miR-130a targets TNF-alpha in cervical cancer cells. J Transl Med. 2014;12:155. doi: 10.1186/1479-5876-12-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang X, Huang L, Zhao Y, Tan W. Downregulation of miR-130a contributes to cisplatin resistance in ovarian cancer cells by targeting X-linked inhibitor of apoptosis (XIAP) directly. Acta Biochim Biophys Sin (Shanghai) 2013;45:995–1001. doi: 10.1093/abbs/gmt113. [DOI] [PubMed] [Google Scholar]
- 10.Qiu S, Lin S, Hu D, Feng Y, Tan Y, Peng Y. Interactions of miR-323/miR-326/miR-329 and miR-130a/miR-155/miR-210 as prognostic indicators for clinical outcome of glioblastoma patients. J Transl Med. 2013;11:10. doi: 10.1186/1479-5876-11-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boll K, Reiche K, Kasack K, Morbt N, Kretzschmar AK, Tomm JM, Verhaegh G, Schalken J, von Bergen M, Horn F, Hackermuller J. MiR-130a, miR-203 and miR-205 jointly repress key oncogenic pathways and are downregulated in prostate carcinoma. Oncogene. 2013;32:277–285. doi: 10.1038/onc.2012.55. [DOI] [PubMed] [Google Scholar]
- 12.Kovaleva V, Mora R, Park YJ, Plass C, Chiramel AI, Bartenschlager R, Dohner H, Stilgenbauer S, Pscherer A, Lichter P, Seiffert M. miRNA-130a targets ATG2B and DICER1 to inhibit autophagy and trigger killing of chronic lymphocytic leukemia cells. Cancer Res. 2012;72:1763–1772. doi: 10.1158/0008-5472.CAN-11-3671. [DOI] [PubMed] [Google Scholar]
- 13.Osbourne A, Calway T, Broman M, McSharry S, Earley J, Kim GH. Downregulation of connexin43 by microRNA-130a in cardiomyocytes results in cardiac arrhythmias. J Mol Cell Cardiol. 2014;74:53–63. doi: 10.1016/j.yjmcc.2014.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu Q, Meng S, Liu B, Li MQ, Li Y, Fang L, Li YG. MicroRNA-130a regulates autophagy of endothelial progenitor cells through Runx3. Clin Exp Pharmacol Physiol. 2014;41:351–357. doi: 10.1111/1440-1681.12227. [DOI] [PubMed] [Google Scholar]
- 15.Gaken J, Mohamedali AM, Jiang J, Malik F, Stangl D, Smith AE, Chronis C, Kulasekararaj AG, Thomas NS, Farzaneh F, Tavassoli M, Mufti GJ. A functional assay for microRNA target identification and validation. Nucleic Acids Res. 2012;40:e75. doi: 10.1093/nar/gks145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu WH, Hu CP, Chen XP, Zhang WF, Li XW, Xiong XM, Li YJ. MicroRNA-130a mediates proliferation of vascular smooth muscle cells in hypertension. Am J Hypertens. 2011;24:1087–1093. doi: 10.1038/ajh.2011.116. [DOI] [PubMed] [Google Scholar]
- 17.Chen Y, Gorski DH. Regulation of angiogenesis through a microRNA (miR-130a) that down-regulates antiangiogenic homeobox genes GAX and HOXA5. Blood. 2008;111:1217–1226. doi: 10.1182/blood-2007-07-104133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Acunzo M, Visone R, Romano G, Veronese A, Lovat F, Palmieri D, Bottoni A, Garofalo M, Gasparini P, Condorelli G, Chiariello M, Croce CM. miR-130a targets MET and induces TRAIL-sensitivity in NSCLC by downregulating miR-221 and 222. Oncogene. 2012;31:634–642. doi: 10.1038/onc.2011.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim GH, Samant SA, Earley JU, Svensson EC. Translational control of FOG-2 expression in cardiomyocytes by microRNA-130a. PLoS One. 2009;4:e6161. doi: 10.1371/journal.pone.0006161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou X, Hu Y, Dai L, Wang Y, Zhou J, Wang W, Di W, Qiu L. MicroRNA-7 inhibits tumor metastasis and reverses epithelial-mesenchymal transition through AKT/ERK1/2 inactivation by targeting EGFR in epithelial ovarian cancer. PLoS One. 2014;9:e96718. doi: 10.1371/journal.pone.0096718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jia LF, Wei SB, Mitchelson K, Gao Y, Zheng YF, Meng Z, Gan YH, Yu GY. miR-34a Inhibits Migration and Invasion of Tongue Squamous Cell Carcinoma via Targeting MMP9 and MMP14. PLoS One. 2014;9:e108435. doi: 10.1371/journal.pone.0108435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin SL, Chiang A, Chang D, Ying SY. Loss of mir-146a function in hormone-refractory prostate cancer. RNA. 2008;14:417–424. doi: 10.1261/rna.874808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng WZ, Ma R, Wang F, Yu J, Liu ZB. Role of miR-191/425 cluster in tumorigenesis and diagnosis of gastric cancer. Int J Mol Sci. 2014;15:4031–4048. doi: 10.3390/ijms15034031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xia Y, Zhu Y, Ma T, Pan C, Wang J, He Z, Li Z, Qi X, Chen Y. miR-204 functions as a tumor suppressor by regulating SIX1 in NSCLC. FEBS Lett. 2014;588:3703–3712. doi: 10.1016/j.febslet.2014.08.016. [DOI] [PubMed] [Google Scholar]
- 26.Wang Z, Yin B, Wang B, Ma Z, Liu W, Lv G. MicroRNA-210 promotes proliferation and invasion of peripheral nerve sheath tumor cells targeting EFNA3. Oncol Res. 2014;21:145–154. doi: 10.3727/096504013X13841340689573. [DOI] [PubMed] [Google Scholar]
- 27.Yang TS, Yang XH, Wang XD, Wang YL, Zhou B, Song ZS. MiR-214 regulate gastric cancer cell proliferation, migration and invasion by targeting PTEN. Cancer Cell Int. 2013;13:68. doi: 10.1186/1475-2867-13-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng Q, Cao H, Chen Z, Ma Z, Wan X, Peng R, Jiang B. PAX6, a novel target of miR-335, inhibits cell proliferation and invasion in glioma cells. Mol Med Rep. 2014;10:399–404. doi: 10.3892/mmr.2014.2150. [DOI] [PubMed] [Google Scholar]
- 29.Dell’aversana C, Altucci L. miRNA-mediated deregulation in leukemia. Front Genet. 2012;3:252. doi: 10.3389/fgene.2012.00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmitt MJ, Philippidou D, Reinsbach SE, Margue C, Wienecke-Baldacchino A, Nashan D, Behrmann I, Kreis S. Interferon-gamma-induced activation of Signal Transducer and Activator of Transcription 1 (STAT1) up-regulates the tumor suppressing microRNA-29 family in melanoma cells. Cell Commun Signal. 2012;10:41. doi: 10.1186/1478-811X-10-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao Z, Liu XF, Wu HC, Zou SB, Wang JY, Ni PH, Chen XH, Fan QS. Rab5a overexpression promoting ovarian cancer cell proliferation may be associated with APPL1-related epidermal growth factor signaling pathway. Cancer Sci. 2010;101:1454–1462. doi: 10.1111/j.1349-7006.2010.01558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu L, Hui-chen F, Chen Y, Zou R, Yan S, Chun-xiang L, Wu-ru W, Li P. Differential expression of RAB5A in human lung adenocarcinoma cells with different metastasis potential. Clin Exp Metastasis. 1999;17:213–219. doi: 10.1023/a:1006617016451. [DOI] [PubMed] [Google Scholar]
- 33.Fukui K, Tamura S, Wada A, Kamada Y, Igura T, Kiso S, Hayashi N. Expression of Rab5a in hepatocellular carcinoma: Possible involvement in epidermal growth factor signaling. Hepatol Res. 2007;37:957–965. doi: 10.1111/j.1872-034X.2007.00143.x. [DOI] [PubMed] [Google Scholar]
- 34.Palamidessi A, Frittoli E, Garre M, Faretta M, Mione M, Testa I, Diaspro A, Lanzetti L, Scita G, Di Fiore PP. Endocytic trafficking of Rac is required for the spatial restriction of signaling in cell migration. Cell. 2008;134:135–147. doi: 10.1016/j.cell.2008.05.034. [DOI] [PubMed] [Google Scholar]
- 35.Lanzetti L, Palamidessi A, Areces L, Scita G, Di Fiore PP. Rab5 is a signalling GTPase involved in actin remodelling by receptor tyrosine kinases. Nature. 2004;429:309–314. doi: 10.1038/nature02542. [DOI] [PubMed] [Google Scholar]
- 36.Liu SS, Chen XM, Zheng HX, Shi SL, Li Y. Knockdown of Rab5a expression decreases cancer cell motility and invasion through integrin-mediated signaling pathway. J Biomed Sci. 2011;18:58. doi: 10.1186/1423-0127-18-58. [DOI] [PMC free article] [PubMed] [Google Scholar]


