Abstract
Overexpression of MAPK/MAK/MRK overlapping kinase (MOK) has been found in various tumors. However, the mechanism underlying MOK upregulation remains unclear. A CpG island was identified in MOK promoter. In this study, we evaluated the expression and methylation status of MOK gene in acute myeloid leukemia (AML). Hypomethylation of MOK promoter was detected in 31.0% (45/145) of AML patients. The degree of MOK hypomethylation was significantly correlated with MOK expression in AML patients. MOK-hypomethylated patients had a trend towards lower WBCs. Receiver operating characteristic curve (ROC) analysis showed a good performance in distinguishing AML patients from controls with an area under the ROC curve (AUC) of 0.820 (P < 0.001). In summary, our results suggest MOK promoter hypomethylation is a common event and contributes to MOK overexpression in AML.
Keywords: MOK, hypomethylation, acute myeloid leukemia
Introduction
Acute myeloid leukemia (AML) is an aggressive malignant disorder caused by uncontrolled proliferation of myeloid precursor cells and marked by genetic and epigenetic abnormalities leading to a block in differentiation and accumulation of leukemic blasts in blood and bone marrow. Genetic alterations are recognized as responsible for pathogenesis and progression of AML. In addition, those genetic abnormalities present in leukemic cells in the majority of AML patients have been already clinically applied as important prognostic factors in AML [1].
In addition to the genetic abnormalities, epigenetic lesions also play essential roles in the pathogenesis of AML. Compared with genetic alterations, epigenetic lesions appear to be more frequent and recurrent [2]. Aberrant DNA methylation including hypermethylation and hypomethylation is one of the most common and the most studied epigenetic feature in human cancers. It is now well known that molecular events such as global DNA hypomethylation, gene-specific hypermethylation occur during the transformation of malignant cells [3]. Aberrant promoter hypermethylation of a great many genes such as p15, p73, ID4, E-cadherin, RARβ2 has been observed in AML [4,5]. However, most researches have focused on the roles and mechanisms of hypermethylation of tumor suppressor genes (TSGs) in cancer so far, much less attention has been paid to the significance of the cancer-linked DNA hypomethylation.
MAPK/MAK/MRK overlapping kinase (MOK), also known as RAGE-1 (renal tumor antigen-1) was firstly identified in a renal carcinoma cell line [6]. To date, MOK protein has been considered as a tumor-associated antigen (TAA) for its wide expression in various tumors including renal carcinoma, melanoma, head and neck cancer, mesothelioma, hepatocellular carcinoma and AML but absent in normal tissues other than retina, which was considered as an immunoprivileged site due to the lack of HLA expression [6-13]. MOK was likewise regarded as a potential target for cancer-specific immunotherapy because it could be recognized by cytotoxic T lymphocytes (CTLs) to induce immune response [6,14]. However, the underlying mechanism of MOK overexpression in these tumors has not been understood. In reviewing the structure of MOK promoter, we found the existence of a large CpG island. Whether there is the aberrant methylation of MOK promoter in leukemia has not been reported until now. The primary aim of the present study is to explore the methylation status of MOK promoter and determine its clinical implications in AML patients.
Materials and methods
Patients and samples
This study was authorized by the Ethics Committee Board of Affiliated People’s Hospital of Jiangsu University. 145 primary AML patients in the present study were gathered based on the availability of obtained leukemic cells. The diagnosis and classification of AML patients was based on morphology and cytochemistry by French-America-British (FAB) and World Health Organization (WHO) criteria [15,16]. Karyotype risk was classified according to reported previously [17]. The bone marrow (BM) samples from patients were obtained at the time of diagnosis after the written informed consent given. BM specimens collected from 21 patients with iron deficiency anemia (IDA) without the evidence of cancers were used as controls. The main clinical and laboratory features of AML patient cohort were summarized in Table 1.
Table 1.
Correlation of MOK promoter methylation with clinical features in AML patients
| Patient’s parameters | The status of MOK methylation | ||
|---|---|---|---|
|
| |||
| Hypomethylated (n = 45) | Methylated (n = 100) | P value | |
| Sex (male/female) | 26/19 | 58/42 | 0.980 |
| Age (years)a | 53 (3-93) | 54 (10-87) | 0.801 |
| WBC (× 109/L)a | 9.55 (1.50-249.30) | 20.60 (0.50-528.00) | 0.056 |
| Hemoglobin (g/L)a | 76.50 (32-147) | 74 (31-142) | 0.963 |
| Platelet counts (× 109/L)a | 41 (3-447) | 42 (4-264) | 0.843 |
| FAB subtypes | 0.978 | ||
| M0 | 0 | 1 | |
| M1 | 4 | 7 | |
| M2 | 17 | 45 | |
| M3 | 6 | 12 | |
| M4 | 12 | 21 | |
| M5 | 4 | 10 | |
| M6 | 2 | 4 | |
| WHO | 0.964 | ||
| AML with t (8; 21) | 3 | 11 | |
| APL with t (15; 17) | 6 | 12 | |
| Minimally differentiated AML | 0 | 1 | |
| AML without maturation | 3 | 7 | |
| AML with maturation | 15 | 34 | |
| Acute myelomonocytic leukemia | 12 | 21 | |
| Acute monoblastic and monocytic leukemia | 4 | 10 | |
| Acute erythroleukemia | 2 | 4 | |
| Cytogenetics classification | 0.427 | ||
| Favorable | 7 | 22 | |
| Intermediate | 26 | 60 | |
| Poor | 5 | 11 | |
| No data | 7 | 7 | |
| Karyotype | 0.657 | ||
| normal | 20 | 46 | |
| T (8; 21) | 3 | 12 | |
| T (15; 17) | 4 | 9 | |
| 11q23 | 1 | 0 | |
| complex | 3 | 9 | |
| others | 7 | 17 | |
| Gene Mutation | |||
| C-KIT (+/-) | 1/42 | 9/82 | 0.167 |
| C/EBPA (+/-) | 5/38 | 12/79 | 0.800 |
| NPM1 (+/-) | 2/41 | 10/81 | 0.337 |
| FLT3 ITD (+/-) | 2/41 | 1/90 | 0.241 |
| IDH1 (+/-) | 1/42 | 3/88 | 1.000 |
| IDH2 (+/-) | 3/40 | 4/87 | 0.680 |
| IDH1/IDH2 (+/-) | 4/39 | 7/84 | 0.745 |
| DNMT3A (+/-) | 2/41 | 8/83 | 0.500 |
| MOK transcript (%)a | 1.87 (0.00-16.34) | 0.30 (0.00-4.39) | 0.001 |
Median (range);
WBC, white blood cells; FAB, French-American-British classification; AML, acute myeloid leukaemia.
RNA isolation, cDNA synthetics and real-time quantitative PCR
BMNCs were separated using Ficoll solution. Total RNA was extracted from BMNCs and reverse transcribed into cDNA as described previously [18]. The primers of MOK expression (forward: 5’-GCTTTCGGGAGTGGTCAG-3’; reverse: 5’-TTCTTGCTCGCAGGGATG-3’) were designed with the software Primer Express 2.0 (Applied Biosystems, Foster City, CA, USA). Real-time quantitative PCR (RQ-PCR) was performed on a 7300 Thermo cycler (Applied Biosystems, CA, USA), using 50 ng of cDNA in a 25 μl reaction mixture with 0.2 mmol/L of dNTP, 0.4 μmol/L of each primer, 4 mmol/L of MgCl2, 1.2 μl of EvaGreen, and 1.0 U of Taq DNA Polymerase (MBI Fermentas, Hanover, USA). RQ-PCR conditions were 5 min at 95°C for denaturation, followed by 45 cycles at 94°C for 30 s for denaturation, 60°C for 30 s for annealing, 72°C for 30 s for elongation, and 85°C for 30 s for collecting fluorescence data. The mRNA abundance of MOK gene was calculated relative to that of the housekeeping gene ABL1.
Real-time quantitative methylation-specific PCR
DNA was isolated using Genomic DNA Purification Kit (Gentra, Minneapolis, MN, USA) following the manufacturer’s standard method. 1 μg of genomic DNA was modified using the CpGenomeTM DNA Modification Kit (Chemicon, Ternecula, CA, USA) according to the manufacturer’s instruction.
Methylation status of MOK promoter was detected using real-time quantitative methylation-specific PCR (RQ-MSP). Primers for the methylated (M) MOK reaction were 5’-AAGATGTTTCGTTTATGTACGC-3’ (forward) and 5’-ACGAAC CGAACGAAAATCG-3’ (reverse), and primer sequences for the unmethylated (U) MOK reaction were 5’-TGTAAGATGTTTTGTTTATGTATG-3’ (forward) and 5’-AACAAACCAAACAAAAATCA-3’ (reverse). 25 μL of reaction volume contained 0.2 mmol/L of dNTP, 0.2 μmol/L of each primer, 2.0 mmol/L of MgCl2, 1.2 μL of EvaGreen, and 1.0 U of Taq DNA Polymerase and 2 μL of modified DNA. RQ-MSP conditions consisted of an initial denaturation step of 95°C for 5 min, followed by an amplification program of 45 cycles for 30 s at 94°C, 30 s at 59°C (M) or 61°C (U) 30 s at 72°C and 85°C for 30 s to collect data before a melting program of one cycle at 95°C for 15 s, 60°C for 60 s, 95°C for 15 s and finally 60°C for 15 s. Negative (distilled water without DNA) and positive controls (recombined methylated and unmethylated MOK plasmids) were integrated in all PCR reactions. PCR products were run on 2% agarose gels and visualized after staining with ethidium bromide. The normalized ratio (Nunmethylation-MOK) calculated relative to the reference ALU was used to assess the methylation level of MOK promoter in samples. Positive products of M and U reaction from one AML patient were cloned and sequenced (Sangon, Shanghai, China).
Bisulfite sequencing analysis
Bisulfite-modified DNA sequencing PCR (BSP) was conducted to verify the result of RQ-MSPPrimer sequences for BSP were 5’-TAGGAAGGTTGTTTTTTTGTTT-3’ (forward) and 5’-CAAACCCAATTAAAACTCAAA-3’ (reverse) with 382 bp products containing 27 CpG sites. PCR conditions were 94°C for 2 min, 40 cycles for 10 s at 98°C, 30 s at 58°C, 1 min at 68°C and a final extension for 7 min at 72°C. PCR products were then cloned into pMD®.19-T Vector (TaKaRa, Dalian, China) and E.coli DH5a (Life Technologies, Gaithersburg, MD) were transformed following the manufacturer’s recommendations. 5 independent colonies of each sample were sequenced.
Mutations analysis
C/EBPA mutations were detected by direct DNA sequencing. FLT3 internal tandem duplication (ITD), IDH1/IDH2, DNMT3A, NPM1 and C-KIT mutations were detected as reported previously [19-23]. All positive samples were confirmed by direct DNA sequencing.
Statistical analysis
Statistical analysis was performed using the SPSS 17.0 software package (SPSS, Chicago, IL, USA). Mann-Whitney’s U-test was carried out to compare the difference of continuous variables between patient groups. Pearson chi-square analysis and Fisher exact test were carried out to compare the difference of categorical variables between patients group. The correlations between the frequency of MOK promoter hypomethylation and the clinical and hematologic parameters were analyzed with Spearman’s rank correlation. Receiver operating characteristic (ROC) curve and the area under the ROC curve (AUC) were used to assess the feasibility of using MOK unmethylation as a diagnostic tool in discriminating AML patients from controls. The P values were two-tailed, and a P-value of less than 0.05 was considered statistically significant for all analyses.
Results
CpG islands in MOK promoter
The upstream 1000 bp region of MOK gene on chromosome 14 [strand (+), nucleotides (nt) 102,771,306-102,772,305] was surveyed for the presence of CpG islands using cpgplot software (http://emboss.bioinformatics.nl /cgi-bin/emboss/cpgplot). The criteria used were as follows: island size > 100 bp, GC percent > 50.0%, and ratio of observed (Obs) CpG sites to expected (Exp) CpG sites > 0.6). One CpG island was predicted spanning bp -421 to -58 (Figure 1). The University of California and Santa Cruz Genome Browser (UCSC Genome Browser) (http://genome.ucsc.edu/) and CpG Island Searcher (http://www.cpgislands.com/) also confirmed the presence of CpG island.
Figure 1.
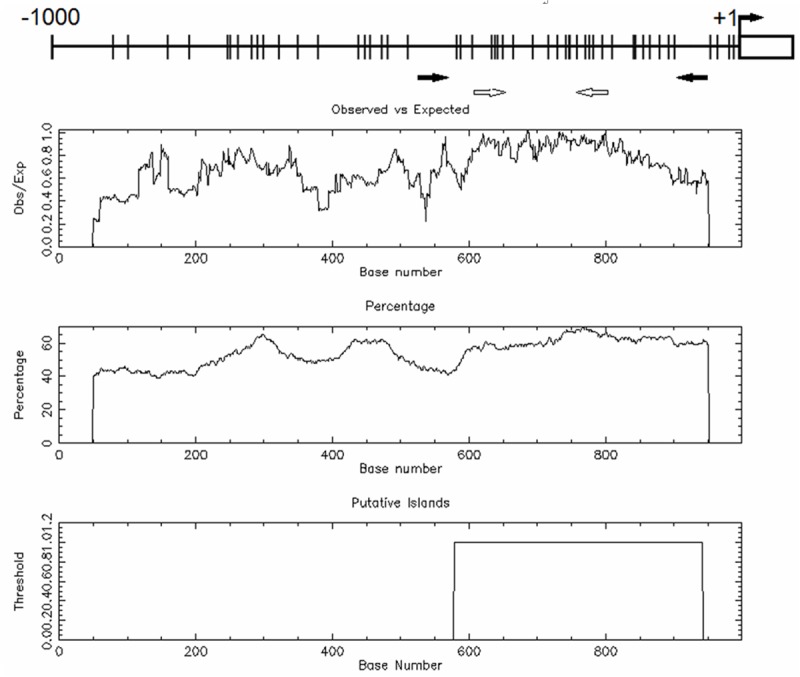
Bioinformatics analysis of MOK promoter on chromosome 14. The vertical lines on the top horizontal line indicate the cytosine resides of CpGs. Numbers in the top panel represent nucleotide positions from MOK translating initial codon. Turning black arrow indicates the translating initial codon of MOK gene; the straight black arrows indicate the locations of primers used for bisulfite sequencing analysis, and the blank arrows indicate the locations of primers used for RQ-MSP analysis. Second panel represents the distribution of observed/expected ratios of CpG dinucleotides; third panel plots the GC content as a percentage of the total; bottom panel represents the putative CpG island within the -1.0 kb of analyzed sequence.
Hypomethylation of MOK promoter in AML
MOK promoter was significantly hypomethylated in AML patients (median 0.64, range 0.00-25.85) compared to controls (median 0.07, range 0.00-1.00) (Figure 2, P < 0.001). The representative electrophoresis results of RQ-MSP products were shown in Figure 3. Nunmethylation-MOK ratio of all controls was 0-100% (15.87 ± 23.11%). AML patients were classified into two groups according to the value of mean plus 4SD obtained in normal controls: hypomethylated (> 108.31%) and methylated (≤ 108.31%).
Figure 2.
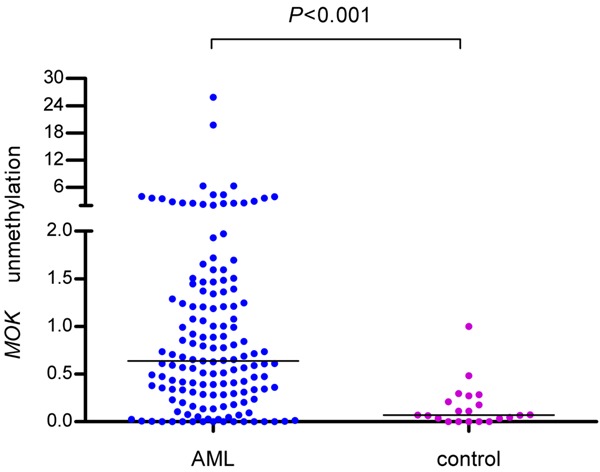
Levels of MOK hypomethylation in AML and control.
Figure 3.
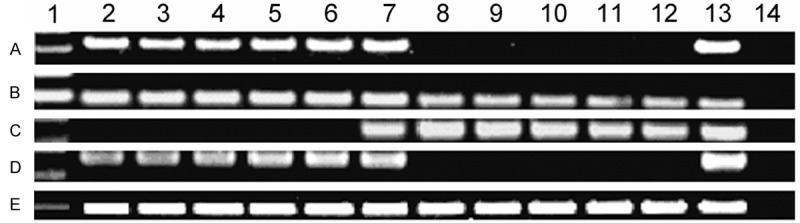
Electropheresis results of RQ-PCR and RQ-MSP products of MOK gene in AML patients. A: MOK expression; B: ABL expression; C: MOK methylation; D: MOK unmethylation; E: ALU. 1: 100 bp DNA Ladder; 2-11: AML; 12: normal control; 13: positive control; 14: negative control.
In order to confirm the results of RQ-MSP, we assessed the methylation density of MOK promoter in two MOK-hypomethylated and two MOK-methylated AML samples according to the results of RQ-MSP (Figure 4). The results of bisulfite-sequencing and RQ-MSP were highly correlated (R = -1.000, P < 0.01).
Figure 4.
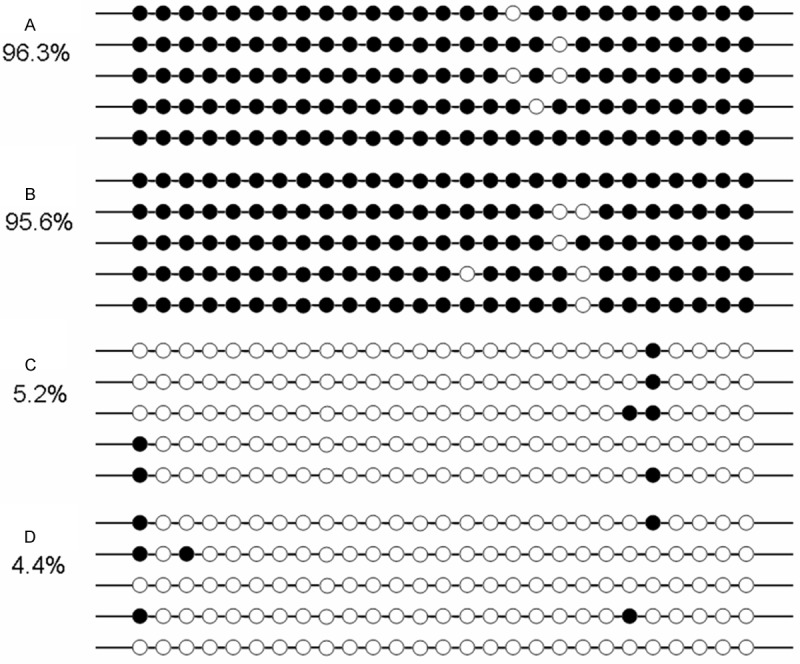
The results of bisulfite-sequencing in two MOK-hypomethylated and two MOK-methylated AML samples according to RQ-MSP. White cycle: unmethylated CpG dinucleotide; Black cycle: methylated CpG dinucleotide. A, B: Two cases with high methylated MOK promoter; C, D: Two cases with low methylated MOK promoter. Percentage was calculated by number of methylated CpG binucleotides divided by that of total CpG binucleotides of all sequenced clones in each sample.
Association of MOK expression and hypomethylation
MOK expression was examined in 68 AML patients with available mRNA. MOK expression was significantly up-regulated in AML patients (median 0.64, range 0.00-16.34) compared to controls (median 0.19, range 0.00-0.27) (P < 0.001). A significantly positive correlation was observed between the degree of MOK hypomethylation and the level of MOK expression (R = 0.401, P < 0.001, Figure 5). Patients with MOK hypomethylation (n = 23) had significantly higher level of MOK transcript (median 1.87, range 0.00-16.34) than those with MOK methylation (n = 45, median 0.30, range 0.00-4.39, P = 0.001) and controls (n = 21, median 0.19, range 0.00-0.27, P < 0.001) (Figure 6).
Figure 5.
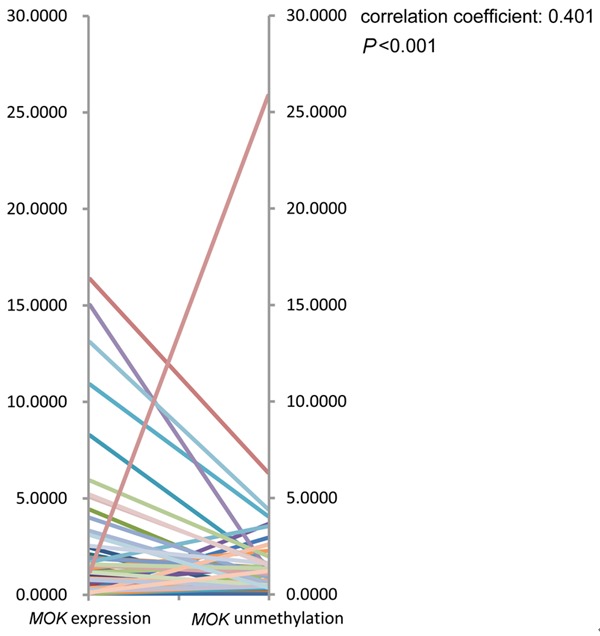
MOK expression correlates with promoter unmethylation levels. Spearman’s rank correlation of MOK expression levels to MOK unmethylation levels shows a significant positive correlation (Spearman correlation coefficient = 0.401, P < 0.001).
Figure 6.
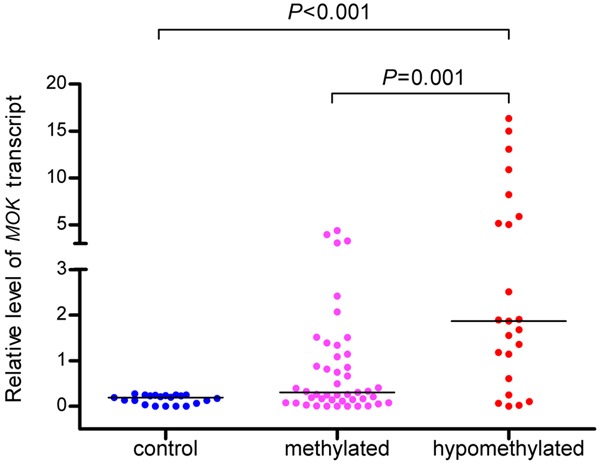
Levels of MOK expression in controls and in AML patients with hypomethylatin and methylation.
Association between MOK hypomethylation and clinical characteristics in AML
MOK hypomethylation was determined in 45 of 145 (31.0%) de novo AML patients. Our results indicated that the frequency of MOK hypomethylation was not associated with the sex, age, peripheral parameters, FAB subtypes, WHO classifications, cytogenetics, and gene mutations (Table 1).
Evaluation of MOK unmethylation as a potential diagnostic marker
ROC curve analysis was conducted to evaluate whether the MOK unmethylation can be used to be a potential marker for diagnosing AML. It was showed that the AUC of AML patients was 0.820 (95% confidence interval: 0.745-0.895) (P < 0.001) (Figure 7). At the cutoff value of 0.3032, the sensitivity and the specificity were 73.8% and 90.5%, respectively.
Figure 7.
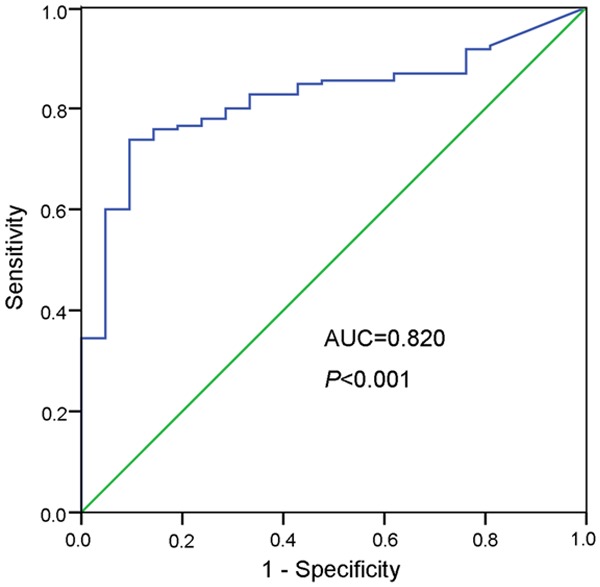
ROC curve of MOK unmethylation for distinguishing AML patients from controls.
Discussion
MOK has been demonstrated to be expressed in different cancers, however, the regulation mechanism of MOK expression is not well established. In the present study, we showed for the first time that the MOK overexpression was associated with hypomethylation of its promoter in AML. Firstly, MOK gene was hypomethylated with a high frequency of 31.0%. Secondly, MOK hypomethylation was correlated with MOK overexpression in AML patients. Similarly, several groups have also demonstrated that the MOK hypomethylation could induce or up-regulate MOK expression by DNA demethylation in renal cell carcinoma cells and malignant mesothelioma cells [9,24,25].
Epigenetic changes are increasingly regarded as key events in the development of cancer. Aberrant DNA methylation profiles, histone modification landscapes and miRNA signatures are early characteristics of carcinogenesis occurring in precancerous lesions and in adjacent tissues [26]. Global DNA hypomethylation and gene-specific hypomethylation were common molecular alterations and play an important role in human cancers [27]. DNA hypomethylation may conduce to the initiation of a tumor cell through generation of chromosomal instability, reactivation of transposable elements, and loss of imprinting [28,29]. In the present study, we provided the first evidence that MOK was hypomethylated in AML patients but absent in controls, suggesting MOK hypomethylation may be associated with the pathogenesis of AML. To date, the function of MOK gene remains largely unknown. Cha et al found that MOK overexpression was associated with the invasion and poor prognosis of hepatocellular carcinoma, suggesting the role of MOK as a tumor-promoting gene [10]. Whether MOK upregulation induced by promoter hypomethylaton contributes to leukemogenesis needs further explored.
MOK hypomethylation was found in each subtype of AML and we did not observe significant difference in MOK hypomethylation frequency either in FAB or WHO subtypes. In addition, there was no significant difference in MOK hypomethylation frequency among cytogenetics risk classification, or between in cytogenetically normal patients and abnormal patients. ROC curve was created to analyze the diagnostic value of MOK hypomethylation, the result clearly indicated that this molecular abnormality may be a helpful marker for distinguishing AML patients from control subjects.
Immunotherapy is an attractive approach to AML patients especially who are over 60 years but resistance to chemotherapy or not ineligible for hematopoietic stem cell transplants. However, the discrepancy of expression of the targeted antigens has been a potential difficulty in developing tumor vaccines [30]. In the present study, we demonstrated that MOK expression was regulated by its promoter hypomethylation in primary leukemic cells. Therefore, MOK may become an ideal target antigen for cancer-specific immunotherapy since it has been identified that MHC class II-binding peptides of MOK could activate T-helper cell response [31] in addition to induce the CTL-initiated immune response. Sequential MOK-targeted immunotherapy may be promising as a novel strategy to enhance the effect of DNA methylation inhibitors, especially in old patients who can not tolerate the toxic reaction of chemotherapy.
Taken together, overexpressed MOK is regulated by promoter hypomethylation in AML.
Acknowledgements
This study was supported by the National Natural Science foundation of China (81270630, 81172592), 333 Project of Jiangsu Province (BRA2013136), Science and Technology Special Project in Clinical Medicine of Jiangsu Province (BL2012056), Clinical Medical Science, Development Foundation of Jiangsu University (JLY20120011, JLY20120012, JLY20120013), Social Development Foundation of Zhenjiang (SH2013042, SH2013082, FZ2011054), Key Medical Talent Program of Zhenjiang City and Jiangsu Government Scholarship for Overseas Studies.
Disclosure of conflict of interest
None.
References
- 1.Grimwade D, Walker H, Oliver F, Wheatley K, Harrison C, Harrison G, Rees J, Hann I, Stevens R, Burnett A, Goldstone A. The importance of diagnostic cytogenetics on outcome in AML: analysis of 1,612 patients entered into the MRC AML 10 trial. The Medical Research Council Adult and Children’s Leukaemia Working Parties. Blood. 1998;92:2322–33. [PubMed] [Google Scholar]
- 2.Melnick AM. Epigenetics in AML. Best Pract Res Clin Haematol. 2010;23:463–8. doi: 10.1016/j.beha.2010.09.017. [DOI] [PubMed] [Google Scholar]
- 3.Simpson AJ, Caballero OL, Jungbluth A, Chen YT, Old LJ. Cancer/testis antigens, gametogenesis and cancer. Nat Rev Cancer. 2005;5:615–25. doi: 10.1038/nrc1669. [DOI] [PubMed] [Google Scholar]
- 4.Oki Y, Issa JP. Epigenetic mechanisms in AML-a target for therapy. Cancer Treat Res. 2010;145:19–40. doi: 10.1007/978-0-387-69259-3_2. [DOI] [PubMed] [Google Scholar]
- 5.Galm O, Wilop S, Lüders C, Jost E, Gehbauer G, Herman JG, Osieka R. Clinical implications of aberrant DNA methylation patterns in acute myelogenous leukemia. Ann Hematol. 2005;84:39–46. doi: 10.1007/s00277-005-0005-0. [DOI] [PubMed] [Google Scholar]
- 6.Gaugler B, Brouwenstijn N, Vantomme V, Szikora JP, Van der Spek CW, Patard JJ, Boon T, Schrier P, Van den Eynde BJ. A new gene coding for an antigen recognized by autologous cytolytic T lymphocytes on a human renal carcinoma. Immunogenetics. 1996;44:323–30. doi: 10.1007/BF02602776. [DOI] [PubMed] [Google Scholar]
- 7.Eichmüller S, Usener D, Jochim A, Schadendorf D. mRNA expression of tumor-associated antigens in melanoma tissues and cell lines. Exp Dermatol. 2002;11:292–301. doi: 10.1034/j.1600-0625.2002.110402.x. [DOI] [PubMed] [Google Scholar]
- 8.Götte K, Usener D, Riedel F, Hörmann K, Schadendorf D, Eichmüller S. Tumor-associated antigens as possible targets for immune therapy in head and neck cancer: comparative mRNA expression analysis of RAGE and GAGE genes. Acta Otolaryngol. 2002;122:546–52. doi: 10.1080/00016480260092381. [DOI] [PubMed] [Google Scholar]
- 9.Sigalotti L, Coral S, Altomonte M, Natali L, Gaudino G, Cacciotti P, Libener R, Colizzi F, Vianale G, Martini F, Tognon M, Jungbluth A, Cebon J, Maraskovsky E, Mutti L, Maio M. Cancer testis antigens expression in mesothelioma: role of DNA methylation and bioimmunotherapeutic implications. Br J Cancer. 2002;86:979–82. doi: 10.1038/sj.bjc.6600174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cha HJ, Kim J, Hong SM, Hong SJ, Park JH, Kim ES, Wang HJ, Choi YJ, Do IG, Joh JW, Kim DS, Choi KY. Overexpression of renal tumor antigen is associated with tumor invasion and poor prognosis of hepatocellular carcinoma. Ann Surg Oncol. 2012;19:S404–11. doi: 10.1245/s10434-011-1856-3. [DOI] [PubMed] [Google Scholar]
- 11.Guinn BA, Gilkes AF, Woodward E, Westwood NB, Mufti GJ, Linch D, Burnett AK, Mills KI. Microarray analysis of tumour antigen expression in presentation acute myeloid leukaemia. Biochem Biophys Res Commun. 2005;333:703–13. doi: 10.1016/j.bbrc.2005.05.161. [DOI] [PubMed] [Google Scholar]
- 12.Guinn BA, Gilkes AF, Mufti GJ, Burnett AK, Mills KI. The tumour antigens RAGE-1 and MGEA6 are expressed more frequently in the less lineage restricted subgroups of presentation acute myeloid leukaemia. Br J Haematol. 2006;134:238–9. doi: 10.1111/j.1365-2141.2006.06135.x. [DOI] [PubMed] [Google Scholar]
- 13.Guinn BA, Tobal K, Mills KI. Comparison of the survival implications of tumour-associated versus cancer-testis antigen expression in acute myeloid leukaemia. Br J Haematol. 2007;136:510–2. doi: 10.1111/j.1365-2141.2006.06454.x. [DOI] [PubMed] [Google Scholar]
- 14.Oehlrich N, Devitt G, Linnebacher M, Schwitalle Y, Grosskinski S, Stevanovic S, Zöller M. Generation of RAGE-1 and MAGE-9 peptide-specific cytotoxic T-lymphocyte lines for transfer in patients with renal cell carcinoma. Int J Cancer. 2005;117:256–64. doi: 10.1002/ijc.21200. [DOI] [PubMed] [Google Scholar]
- 15.Bennett JM, Catovsky D, Daniel MT, Flandrin G, Galton DA, Gralnick HR, Sultan C. Proposed revised criteria for the classification of acute myeloid leukaemia. A report of the French-American-British Cooperative Group. Ann Intern Med. 1985;103:620–5. doi: 10.7326/0003-4819-103-4-620. [DOI] [PubMed] [Google Scholar]
- 16.Vardiman JW, Porwit A, Brunning RD, Tefferi A, Arber DA, Bloomfield CD, Le Beau MM, Thiele J. Introduction and owverview of classification of the myeloid neoplasms. In: Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Vardiman JW, editors. WHO classification of tumours of haematopoietic and lymphoid tissues. Lyon: IARC Press; 2008. pp. 109–48. [Google Scholar]
- 17.Slovak ML, Kopecky KJ, Cassileth PA, Harrington DH, Theil KS, Mohamed A, Paietta E, Willman CL, Head DR, Rowe JM, Forman SJ, Appelbaum FR. Karyotypic analysis predicts outcome of preremission and postremission therapy in adult acute myeloid leukemia: a Southwest Oncology Group/Eastern Cooperative Oncology Group Study. Blood. 2000;96:4075–83. [PubMed] [Google Scholar]
- 18.Chen Q, Lin J, Yao DM, Qian J, Qian W, Yang J, Chai HY, Ma JC, Deng ZQ, Wang CZ, Li Y. Aberrant hypomethylation of DDX43 promoter in myelodysplastic syndrome. Br J Haematol. 2012;158:293–6. doi: 10.1111/j.1365-2141.2012.09138.x. [DOI] [PubMed] [Google Scholar]
- 19.Kottaridis PD, Gale RE, Frew ME, Harrison G, Langabeer SE, Belton AA, Walker H, Wheatley K, Bowen DT, Burnett AK, Goldstone AH, Linch DC. The presence of a FLT3 internal tandem duplication in patients with acute myeloid leukemia (AML) adds important prognostic information to cytogenetic risk group and response to the first cycle of chemotherapy: analysis of 854 patients from the United Kingdom Medical Research Council AML 10 and 12 trials. Blood. 2001;98:1752–9. doi: 10.1182/blood.v98.6.1752. [DOI] [PubMed] [Google Scholar]
- 20.Szankasi P, Jama M, Bahler DW. A new DNA-based test for detection of nucleophosmin exon 12 mutations by capillary electropheresis. J Mol Diagn. 2008;10:236–41. doi: 10.2353/jmoldx.2008.070167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin J, Yao DM, Qian J, Chen Q, Qian W, Li Y, Yang J, Wang CZ, Chai HY, Qian Z, Xiao GF, Xu WR. Recurrent DNMT3A R882 mutations in Chinese patients with acute myeloid leukemia and myelodysplastic syndrome. PLoS One. 2011;6:e26906. doi: 10.1371/journal.pone.0026906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin J, Yao DM, Qian J, Chen Q, Qian W, Li Y, Yang J, Wang CZ, Chai HY, Qian Z, Xiao GF, Xu WR. IDH1 and IDH2 mutation analysis in Chinese patients with acute myeloid leukemia and myelodysplastic syndrome. Ann Hematol. 2012;91:519–25. doi: 10.1007/s00277-011-1352-7. [DOI] [PubMed] [Google Scholar]
- 23.Yang X, Qian J, Sun A, Lin J, Xiao G, Yin J, Chen S, Wu D. RAS mutation analysis in a large cohort of Chinese patients with acute myeloid leukemia. Clin Biochem. 2013;46:579–83. doi: 10.1016/j.clinbiochem.2012.12.022. [DOI] [PubMed] [Google Scholar]
- 24.Coral S, Sigalotti L, Altomonte M, Engelsberg A, Colizzi F, Cattarossi I, Maraskovsky E, Jager E, Seliger B, Maio M. 5-aza-2’-Deoxycytidine-induced expression of functional cancer testis antigens in human renal cell carcinoma: immunotherapeutic implications. Clin Cancer Res. 2002;8:2690–5. [PubMed] [Google Scholar]
- 25.Reu FJ, Bae SI, Cherkassky L, Leaman DW, Lindner D, Beaulieu N, MacLeod AR, Borden EC. Overcoming resistance to interferon-induced apoptosis of renal carcinoma and melanoma cells by DNA demethylation. J. Clin. Oncol. 2006;24:3771–9. doi: 10.1200/JCO.2005.03.4074. [DOI] [PubMed] [Google Scholar]
- 26.Vincent A, Van Seuningen I. On the epigenetic origin of cancer stem cells. Biochim Biophys Acta. 2012;1826:83–8. doi: 10.1016/j.bbcan.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 27.Pogribny IP, Beland FA. DNA hypomethylation in the origin and pathogenesis of human diseases. Cell Mol Life Sci. 2009;66:2249–61. doi: 10.1007/s00018-009-0015-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson AS, Power BE, Molloy PL. DNA hypomethylation and human diseases. Biochim Biophys Acta. 2007;1775:138–62. doi: 10.1016/j.bbcan.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 29.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–59. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 30.Meklat F, Li Z, Wang Z, Zhang Y, Zhang J, Jewell A, Lim SH. Cancer-testis antigens in haematological malignancies. Br J Haematol. 2007;136:769–76. doi: 10.1111/j.1365-2141.2006.06484.x. [DOI] [PubMed] [Google Scholar]
- 31.Stassar MJ, Raddrizzani L, Hammer J, Zöller M. T-helper cell-response to MHC class II-binding peptides of the renal cell carcinoma-associated antigen RAGE-1. Immunobiology. 2001;203:743–55. doi: 10.1016/S0171-2985(01)80003-6. [DOI] [PubMed] [Google Scholar]


