Abstract
The aim of the present study was to evaluate the influence of polymorphisms in NER and HRR pathways on the response to cisplatin-based treatment and clinical outcome in osteosarcoma patients. 214 osteosarcoma patients treated with cisplatin-based chemotherapy were collected between January 2008 and January 2011. Genotypes of ERCC1 rs11615, ERCC2 rs1799793 and rs13181, NBN rs709816, RAD51 rs1801320, and XRCC3 rs861539 were conducted by Polymerase Chain Reaction Restriction Fragment Length Polymorphism (PCR-RFLP) assay. By conditional logistic regression analysis, patients carrying CC genotype of ERCC1 rs11615 showed a significant more good responder than TT genotype, and the OR (95% CI) was 2.51 (1.02-6.85). In the Cox proportional hazards model, after adjusting for potential confounding factors, we found that individuals carrying CC genotype of ERCC1 rs11615 was associated with decreased risk of death from osteosarcoma, and the HR (95% CI) was 0.43 (0.15-0.93). In conclusion, our results suggest that ERCC1 rs11615 polymorphism in the DNA repair pathways play an important role in the response to chemotherapy and overall survival of osteosarcoma.
Keywords: ERCC1 rs11615, polymorphism, response to chemotherapy, overall survival, osteosarcoma
Introduction
Osteosarcoma is a rare bone cancer, and is a more common in children and adolescents, with an annual incidence of about 3/105 [1,2]. Despite recent advances in the therapies, the survival of osteosarcoma has significantly is still low and unsatisfactory [3]. Although many patients received similar therapies, interindividual differences in treatment outcome are observed between patients. It is estimated that about 50% of patients have poor clinical outcome, and around 30% of patients relapse locally or develop metastases [3,4]. Genetic polymorphisms involved in response to chemotherapeutic agents could influence both survival and treatment toxicity, therefore identification of predictive markers could lead to improve the drug selection and treatment outcomes.
Cisplatin is a platinum analog and is frequently used as a common therapy method for many cancers. It binds to DNA and forms DNA adducts, and inhibits DNA replication [5]. Therefore, DNA repair mechanisms play an important role in determining response to cisplatin. NER is a main DNA repair mechanism, which has a key role in repairing various distorting helix-distorting lesions [6]. Two important enzymes in the NER pathway are associated with resistance to cisplatin, including the excision repair cross-complementation group 2 (ERCC2) gene and excision repair crosscomplementation group 1 (ERCC1).
Homologous recombination repair (HRR) is part of the complex, which is involved in recognizing DNA damage. In the HRR pathway, nibrin (NBN) is involved in recognizing DNA damage, while RAD51 recombinase (RAD51) catalyses homologous search and strand invasion with the aid of other proteins, including Xray complementing defective repair in Chinese hamster cells 3 (XRCC3).
Only a few studies have investigated the influence of SNPs in NER and HRR pathway on cisplatin response in many cancers, but not in osteosarcoma [7-9]. Therefore, we aimed to evaluate the influence of polymorphisms in NER and HRR pathways on the response to cisplatin-based treatment and clinical outcome in osteosarcoma patients.
Patients and methods
Patients
Between January 2008 and January 2010, 214 osteosarcoma patients treated with cisplatin-based chemotherapy were included in our study from the Affiliated Sixth People’s Hospital of Shanghai Jiao Tong University between January 2008 and January 2011. The study was approved by the the Affiliated Sixth People’s Hospital of Shanghai Jiao Tong University.
Assessment of treatment outcome
Demographic and clinical characteristics of included patients were obtained from the medical records. Tumor response was evaluated histologically based on percentage of necrosis and more than 90% necrosis was considered good response, otherwise the response to chemotherapy was regarded as poor responders. Overall survival (OS) was defined as time from the beginning of treatment to an event or death, respectively. Patients without an event or death at the time of the analysis were censored at the date of the last follow-up.
All the patients were followed up until January 2014, with a median follow-up time of 36.5 months (ranged: 4 and 60 months). All patients were followed up by telephone every four weeks until death or the end of study.
Blood samples and genotyping
Each patient was asked to provide 5 ml peripheral blood and kepted in -70°C until use. Genomic DNA was isolated from peripheral blood using the TIANamp Blood DNA Kit (Tiangen, Beijing, China) according to the manufacturer’s instructions. Genotypes of ERCC1 rs11615, ERCC2 rs1799793 and rs13181, NBN rs709816, RAD51 rs1801320, and XRCC3 rs861539 were conducted by Polymerase Chain Reaction Restriction Fragment Length Polymorphism (PCR-RFLP) assay. Probes and primers for ERCC1 rs11615, ERCC2 rs1799793 and rs13181, NBN rs709816, RAD51 rs1801320, and XRCC3 rs861539 were designed using Sequenom Assay Design 3.1 software (Sequenom®) according to the manufacturer instructions. The cycling programme involved preliminary denaturation at 94°C for 5 min, followed by 35 cycles of denaturing at 94°C for 45s, annealing at 62°C for 60s and extending at 72°C for 60s, and performing a final extension at 72°C for 10 minutes. For quality control, 5% of subjects were randomly selected, and the results of repeated samples showed 100% concordance.
Statistical analysis
Continuous variables were expressed as the mean ± SD and analyzed by student t test. Categorical variables were shown as N (%) and analyzed by χ2-test. The association between ERCC1 rs11615, ERCC2 rs1799793 and rs13181, NBN rs709816, RAD51 rs1801320, and XRCC3 rs861539 polymorphisms and response to chemotherapy were described as odds ratio (ORs) and 95% confidence interval (CI) using conditional logistic regression analysis. The prognostic value of eight gene polymorphisms for the OS was estimated by multivariate analysis using the Cox proportional hazards models, describing as the hazard ratio (HR) and 95% CI. Overall survival (OS) was calculated as the time between the first day of treatment and death or last known follow-up. Survival probabilities were estimated by using the Kaplan-Meier method. Two-tailed P values < 0.05 with were considered statistical difference. All statistical analyses were conducted using the STATA version 9.0 statistical software.
Results
Patients and clinical characteristics
The distributions of selected general characteristics of study subjects were shown in Table 1. The mean age of the osteosarcoma subjects was 18.7 ± 11.5 years old (ranging from 11 to 39 years old). Of 214 osteosarcoma patients, 133 (62.15%) were males, 141 (65.89%) had tumor stage of I-II, 158 (73.83%) had tumor location of long tubular bones, 163 (76.17%) received limb salvage, and 54 (25.23%) showed metastasis.
Table 1.
Demographic and clinical characteristics of osteosarcoma patients
| Characteristics | N | % |
|---|---|---|
| Age | ||
| ≤ 20 | 123 | 57.48 |
| > 20 | 91 | 42.52 |
| Gender | ||
| Male | 133 | 62.15 |
| Female | 81 | 37.85 |
| Stage | ||
| I-II | 141 | 65.89 |
| III-IV | 73 | 34.11 |
| Tumor location | ||
| Long tubular bones | 158 | 73.83 |
| Axial skeleton | 56 | 26.17 |
| Therapy | ||
| Amputation | 51 | 23.83 |
| Limb salvage | 163 | 76.17 |
| Metastasis | ||
| Yes | 54 | 25.23 |
| No | 161 | 75.23 |
Association between DNA repaired gene polymorphisms and response to chemotherapy
At the end of the follow-up, 133 osteosarcoma patients showed good response to cisplatin-based chemotherapy, with a response rate of 62.15%. By conditional logistic regression analysis, patients carrying CC genotype of ERCC1 rs11615 showed a significant more good responder than TT genotype, and the OR (95% CI) was 2.51 (1.02-6.85) (Table 2). However, we observed no significant difference between ERCC2 rs1799793 and rs13181, NBN rs709816, RAD51 rs1801320, and XRCC3 rs861539 polymorphisms and response to cisplatin-based chemotherapy.
Table 2.
Analysis of the association between DNA repaired gene polymorphisms and response to cisplatin-based chemotherapy in osteosarcoma patients
| Genotypes | Good response N = 133 | % | Poor response N = 81 | % | Adjusted OR (95% CI)1 | P value |
|---|---|---|---|---|---|---|
| ERCC1 rs11615 | ||||||
| TT | 52 | 38.83 | 41 | 50.82 | 1.0 (Ref.) | - |
| TC | 54 | 40.78 | 31 | 37.70 | 1.37 (0.72-2.62) | 0.31 |
| CC | 27 | 20.39 | 9 | 11.48 | 2.51 (1.02-6.85) | 0.03 |
| ERCC2 rs1799793 | ||||||
| GG | 74 | 55.34 | 54 | 67.21 | 1.0 (Ref.) | - |
| GA | 39 | 29.13 | 20 | 24.59 | 1.42 (0.72-2.87) | 0.28 |
| AA | 21 | 15.53 | 7 | 8.20 | 2.19 (0.82-6.52) | 0.09 |
| ERCC2 rs13181 | ||||||
| AA | 76 | 57.28 | 49 | 60.66 | 1.0 (Ref.) | - |
| AC | 41 | 31.07 | 24 | 29.51 | 1.10 (0.57-2.15) | 0.76 |
| CC | 15 | 11.65 | 8 | 9.84 | 1.21 (0.44-3.55) | 0.69 |
| NBN rs709816 | ||||||
| GG | 47 | 35.34 | 33 | 40.98 | 1.0 (Ref.) | - |
| GC | 56 | 41.75 | 32 | 39.34 | 1.22 (0.63-2.40) | 0.52 |
| CC | 30 | 22.91 | 16 | 19.67 | 1.32 (0.58-3.02) | 0.47 |
| RAD51 rs1801320 | ||||||
| GG | 62 | 46.60 | 41 | 50.82 | 1.0 (Ref.) | - |
| GC | 56 | 41.75 | 31 | 37.70 | 1.19 (0.64-2.25) | 0.55 |
| CC | 15 | 11.65 | 9 | 11.48 | 1.10 (0.41-3.14) | 0.84 |
| XRCC3 rs861539 | ||||||
| CC | 63 | 47.57 | 42 | 52.46 | 1.0 (Ref.) | - |
| CT | 57 | 42.72 | 33 | 40.98 | 1.15 (0.62-2.15) | 0.63 |
| TT | 13 | 9.71 | 5 | 6.56 | 1.73 (0.53-6.65) | 0.32 |
Ajusted for age, tumor size, clinical stage, lymph mode metastasis and ER and PR status.
Association between DNA repaired gene polymorphisms and overall survival
At the end of January 2014, 66 died from all causes, and the five-year survival rate is 69.16%. In the Cox proportional hazards model, after adjusting for potential confounding factors, we found that individuals carrying CC genotype of ERCC1 rs11615 was associated with decreased risk of death from osteosarcoma, and the HR (95% CI) was 0.43 (0.15-0.93) (Table 3). By Kaplan-Meier method, individuals with CC genotype of ERCC1 rs11615 had a longer overall survival of osteosarcoma when compared with TT genotype (Figure 1). However, we observed no association between ERCC2 rs1799793 and rs13181, NBN rs709816, RAD51 rs1801320, and XRCC3 rs861539 polymorphisms and overall survival in osteosarcoma patients.
Table 3.
Association between included gene polymorphisms and overall survival in osteosarcoma patients
| Genotypes | Death N = 66 | % | Alive N = 148 | % | Adjusted HR (95% CI)1 | P value |
|---|---|---|---|---|---|---|
| ERCC1 rs11615 | ||||||
| TT | 34 | 51.52 | 61 | 41.22 | 1.0 (Ref.) | - |
| TC | 24 | 36.36 | 58 | 39.19 | 0.74 (0.37-1.47) | 0.36 |
| CC | 8 | 12.12 | 29 | 19.59 | 0.43 (0.15-0.93) | 0.03 |
| ERCC2 rs1799793 | ||||||
| GG | 43 | 65.15 | 85 | 57.43 | 1.0 (Ref.) | - |
| GA | 19 | 28.79 | 45 | 30.41 | 0.83 (0.41-1.67) | 0.59 |
| AA | 4 | 6.06 | 18 | 12.16 | 0.44 (0.10-1.46) | 0.15 |
| ERCC2 rs13181 | ||||||
| AA | 39 | 59.09 | 82 | 55.41 | 1.0 (Ref.) | - |
| AC | 20 | 30.30 | 48 | 32.43 | 0.88 (0.43-1.75) | 0.69 |
| CC | 7 | 10.61 | 18 | 12.16 | 0.82 (0.27-2.27) | 0.68 |
| NBN rs709816 | ||||||
| GG | 27 | 40.91 | 57 | 38.51 | 1.0 (Ref.) | - |
| GC | 25 | 37.88 | 59 | 39.86 | 0.89 (0.44-1.81) | 0.74 |
| CC | 13 | 19.70 | 32 | 21.62 | 0.86 (0.35-2.01) | 0.7 |
| RAD51 rs1801320 | ||||||
| GG | 34 | 51.52 | 70 | 47.30 | 1.0 (Ref.) | - |
| GC | 25 | 37.88 | 59 | 39.86 | 0.87 (0.45-1.70) | 0.67 |
| CC | 7 | 10.61 | 19 | 12.84 | 0.76 (0.25-2.12) | 0.57 |
| XRCC3 rs861539 | ||||||
| CC | 34 | 51.52 | 69 | 46.62 | 1.0 (Ref.) | - |
| CT | 26 | 39.39 | 61 | 41.22 | 0.86 (0.44-1.67) | 0.64 |
| TT | 6 | 9.09 | 18 | 12.16 | 0.68 (0.20-2.00) | 0.45 |
Ajusted for age, tumor size, clinical stage, lymph mode metastasis and ER and PR status.
Figure 1.
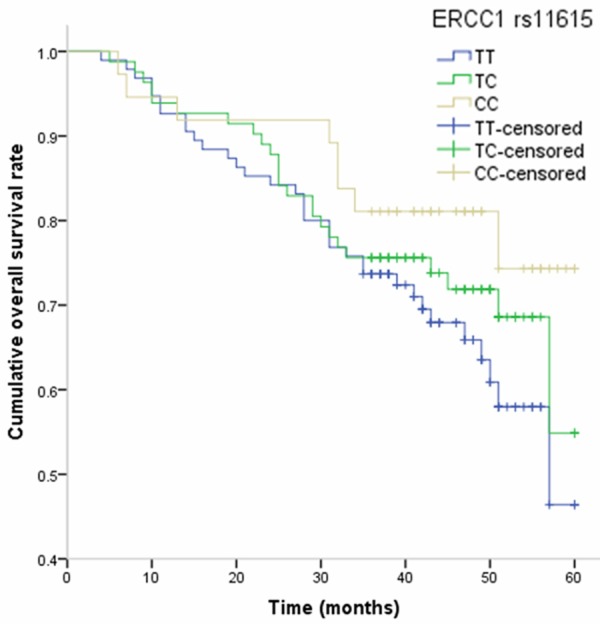
Kaplan-Meier analysis on the influence of ERCC1 rs11615 polymorphism on overall survival of osteosarcoma.
Discussion
In the present study, we investigated the influence of polymorphisms in NER and HRR pathways on treatment response and overall survival in osteosarcoma patients treated with cisplatin-based chemotherapy. Our study found that CC genotype of ERCC1 rs11615 was associated with better response to chemotherapy when compared with TT genotype, and this genotype could influence the OS of osteosarcoma patients.
Cisplatin is the current standard chemotherapy treatment for osteosarcoma, however different cisplatin-based treatment regimens are used in clinical practice. It is well known that cisplatin has a cytotoxic role through formation of different kinds of DNA lesions. Previous study reported that DNA repair mechanisms such as NER enzymes have a key important role in response to cisplatin, and mechanisms, such as HRR, can play a key role in repairing many complex forms of DNA damage, and help to cause interindividuals differences in response to cisplatin between patients [10]. Our study showed that CC genotype of ERCC1 rs11615 was associated with better response to cisplatin-based chemotherapy and overall survival of osteosarcoma patients, which suggests that ERCC1 rs11615 polymorphism can influence the clinical outcome of osteosarcoma patients.
Previous studies showed that ERCC1 rs11615 was often associated with response and cisplatin-based chemotherapy [11-15]. Metzger et al. conducted a study to identify association between ERCC1 rs11615 polymorphism and squamous esophageal cancer receiving a neoadjuvant radiochemotherapy, and found that ERCC1 rs11615 can influence the response to chemotherapy and survival of squamous esophageal cancer [11]. However, some studies did not find ERCC1 rs11615 did not influence the response to chemotherapy. Rumiato et al. conducted a cohort study with 143 esophageal cancer patients, and this study did not find ERCC1 rs11615 can be a predictive marker in the cisplatin/5-FU-based neoadjuvant setting [12]. Mathiaux et al. investigated the association between ERCC1, ERCC2 and ERCC5 polymorphisms and response to chemotherapy in NSCLC, and this study did not find the ERCC1 rs11615 polymorphism can influence the platinum-based chemotherapy treatment of advanced NSCLC [13]. The discrepancy of these results may be caused by differences in ethnicities, study design, tumor types, and sample size.
For the association between ERCC1 rs11615 and treatment outcome of osteosarcoma, several previous studies report their association between them, but the results are inconsistent [16-19]. Hao et al. conducted a study with 267 consecutive osteosarcoma patients, and found that ERCC1 rs11615 can influence the response to chemotherapy and clinical outcome of osteosarcoma patients [16], which is consistent with our findings. However, the other three studies which were conducted in Spanish and Chinese populations found that ERCC1 rs11615 cannot influence the response to chemotherapy and clinical outcome of osteosarcoma [17-19]. Therefore, the further studies with more subjects are needed to confirm our results.
Several limitations should be considered in our study. First, cases were selected from one hospital, which may not be representative of the general osteosarcoma cases. Selection bias may exist in this study. Genetic variability of DNA repair mechanisms could also influence the response to other chemotherapeutics, not only cisplatin, which makes the interpretation of our findings difficult. As osteosarcoma is a rare disease, the number of cases analyzed in the present study was relatively small, which may reduce the statistical power to detect the association between ERCC1 rs11615 polymorphism and clinical outcome of osteosarcoma. Therefore, further studies are greatly needed to verify our results.
In conclusion, our results suggest that ERCC1 rs11615 polymorphism in the DNA repair pathways play an important role in the response to chemotherapy and clinical outcome of osteosarcoma. In the future, these SNPs could contribute to identification of patients, less likely to achieve better response to cisplatin chemotherapy. Further multicenter studies involving various populations are required to confirm our results.
References
- 1.Mirabello L, Troisi RJ, Savage SA. Osteosarcoma incidence and survival rates from 1973 to 2004: Data from the surveillance, epidemiology, and end results program. Cancer. 2009;115:1531–43. doi: 10.1002/cncr.24121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moore DD, Luu HH. Osteosarcoma. Cancer Treat Res. 2014;162:65–92. doi: 10.1007/978-3-319-07323-1_4. [DOI] [PubMed] [Google Scholar]
- 3.Chou AJ, Gorlick R. Chemotherapy resistance in osteosarcoma: current challenges and future directions. Expert Rev Anticancer Ther. 2006;6:1075–85. doi: 10.1586/14737140.6.7.1075. [DOI] [PubMed] [Google Scholar]
- 4.Quartuccio N, Treglia G, Salsano M, Mattoli MV, Muoio B, Piccardo A, Lopci E, Cistaro A. The role of Fluorine-18-Fluorodeoxyglucose positron emission tomography in staging and restaging of patients with osteosarcoma. Radiol Oncol. 2013;47:97–102. doi: 10.2478/raon-2013-0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marsh S, McLeod H, Dolan E, Shukla SJ, Rabik CA, Gong L, Hernandez-Boussard T, Lou XJ, Klein TE, Altman RB. Platinum pathway. Pharmacogenet Genomics. 2009;19:563–4. doi: 10.1097/FPC.0b013e32832e0ed7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Costa RM, Chiganças V, Galhardo Rda S, Carvalho H, Menck CF. The eukaryotic nucleotide excision repair pathway. Biochimie. 2003;85:1083–99. doi: 10.1016/j.biochi.2003.10.017. [DOI] [PubMed] [Google Scholar]
- 7.Erčulj N, Kovač V, Hmeljak J, Franko A, Dodič-Fikfak M, Dolžan V. DNA repair polymorphisms and treatment outcomes of patients with malignant mesothelioma treated with gemcitabine- platinum combination chemotherapy. J Thorac Oncol. 2012;7:1609–17. doi: 10.1097/JTO.0b013e3182653d31. [DOI] [PubMed] [Google Scholar]
- 8.Shen XY, Lu FZ, Wu Y, Zhao LT, Lin ZF. XRCC3 Thr241Met polymorphism and clinical outcomes of NSCLC patients receiving platinum-based chemotherapy: a systematic review and meta-analysis. PLoS One. 2013;8:e69553. doi: 10.1371/journal.pone.0069553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu JL, Hu LM, Huang MD, Zhao W, Yin YM, Hu ZB, Ma HX, Shen HB, Shu YQ. Genetic variants of NBS1 predict clinical outcome of platinum-based chemotherapy in advanced non-small cell lung cancer in Chinese. Asian Pac J Cancer Prev. 2012;13:851–6. doi: 10.7314/apjcp.2012.13.3.851. [DOI] [PubMed] [Google Scholar]
- 10.Wang QE, Milum K, Han C, Huang YW, Wani G, Thomale J, Wani AA. Differential contributory roles of nucleotide excision and homologous recombination repair for enhancing cisplatin sensitivity in human ovarian cancer cells. Mol Cancer. 2011;10:24. doi: 10.1186/1476-4598-10-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Metzger R, Warnecke-Eberz U, Alakus H, Kütting F, Brabender J, Vallböhmer D, Grimminger PP, Mönig SP, Drebber U, Hölscher AH, Bollschweiler E. Neoadjuvant radiochemotherapy in adenocarcinoma of the esophagus: ERCC1 gene polymorphisms for prediction of response and prognosis. J Gastrointest Surg. 2012;16:26–34. doi: 10.1007/s11605-011-1700-x. [DOI] [PubMed] [Google Scholar]
- 12.Rumiato E, Cavallin F, Boldrin E, Cagol M, Alfieri R, Basso D, Castoro C, Ancona E, Amadori A, Ruol A, Saggioro D. ERCC1 C8092A (rs3212986) polymorphism as a predictive marker in esophageal cancer patients treated with cisplatin/5-FU-based neoadjuvant therapy. Pharmacogenet Genomics. 2013;23:597–604. doi: 10.1097/FPC.0b013e3283653afc. [DOI] [PubMed] [Google Scholar]
- 13.Mathiaux J, Le Morvan V, Pulido M, Jougon J, Bégueret H, Robert J. Role of DNA repair gene polymorphisms in the efficiency of platinum-based adjuvant chemotherapy for non-small cell lung cancer. Mol Diagn Ther. 2011;15:159–66. doi: 10.1007/BF03256406. [DOI] [PubMed] [Google Scholar]
- 14.Gao R, Reece K, Sissung T, Reed E, Price DK, Figg WD. The ERCC1 N118N polymorphism does not change cellular ERCC1 protein expression or platinum sensitivity. Mutat Res. 2011;708:21–7. doi: 10.1016/j.mrfmmm.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park SR, Kong SY, Nam BH, Choi IJ, Kim CG, Lee JY, Cho SJ, Kim YW, Ryu KW, Lee JH, Rhee J, Park YI, Kim NK. CYP2A6 and ERCC1 polymorphisms correlate with efficacy of S-1 plus cisplatin in metastatic gastric cancer patients. Br J Cancer. 2011;104:1126–34. doi: 10.1038/bjc.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hao T, Feng W, Zhang J, Sun YJ, Wang G. Association of four ERCC1 and ERCC2 SNPs with survival of bone tumour patients. Asian Pac J Cancer Prev. 2012;13:3821–4. doi: 10.7314/apjcp.2012.13.8.3821. [DOI] [PubMed] [Google Scholar]
- 17.Yang LM, Li XH, Bao CF. Glutathione S-transferase P1 and DNA polymorphisms influence response to chemotherapy and prognosis of bone tumors. Asian Pac J Cancer Prev. 2012;13:5883–6. doi: 10.7314/apjcp.2012.13.11.5883. [DOI] [PubMed] [Google Scholar]
- 18.Li J, Liu S, Wang W, Zhang K, Liu Z, Zhang C, Chen S, Wu S. ERCC polymorphisms and prognosis of patients with osteosarcoma. Tumour Biol. 2014;35:10129–36. doi: 10.1007/s13277-014-2322-1. [DOI] [PubMed] [Google Scholar]
- 19.Caronia D, Patiño-García A, Milne RL, Zalacain-Díez M, Pita G, Alonso MR, Moreno LT, Sierrasesumaga-Ariznabarreta L, Benítez J, González-Neira A. Common variations in ERCC2 are associated with response to cisplatin chemotherapy and clinical outcome in osteosarcoma patients. Pharmacogenomics J. 2009;9:347–53. doi: 10.1038/tpj.2009.19. [DOI] [PubMed] [Google Scholar]


