Abstract
Despite the advances in the management of gastric cancer, the prognosis of advanced gastric cancer remains relatively poor. Thus, it is of urgent need to identify novel prognostic markers and therapeutic targets of gastric cancer. A growing volume of literature has indicated that lncRNAs are differentially expressed in a diverse array of cancer and play an important role in the development of cancer. Linc-UBC1, a recently identified long noncoding RNA, was initially found to be upregulated in bladder cancer. However, the role of linc-UBC1 in gastric cancer remains to be elusive. In this study, we found that linc-UBC1 was significantly upregulated in gastric cancer tissues compared to adjacent normal tissues. Furthermore, high linc-UBC1 expression was associated with lymph-node metastasis, tumor size, TNM stage and poorer prognosis. Inhibition of linc-UBC1 suppressed the proliferation, motility and invasion of gastric cancer cells. Our study suggests that linc-UBC1 may represent a novel diagnostic, prognostic biomarker and a potential therapeutic target of gastric cancer.
Keywords: Gastric cancer, long noncoding RNA, linc-UBC1, prognosis
Introduction
Gastric cancer is fourth most common cancer globally and represents the second most frequent cause of cancer-related mortality [1]. Advances in early detection techniques, the application of more aggressive surgical strategy and postoperative adjuvant therapy have contributed to the survival improvement for early stage gastric cancer, especially in Asian countries [2-4]. Yet, advanced gastric cancer still poses a formidable challenge to the survival of the patients. Therefore, a better understanding of the molecular mechanism underlying the progression of gastric cancer is essential for the development of novel therapeutic strategies.
Recent advances in high-throughput gene sequencing analysis have revealed that the human genome is pervasively transcribed, and the majority (~98%) has limited or no apparent protein-coding capacity, which is defined as noncoding RNAs (ncRNAs) [5,6]. MiRNAs (19-25 nucleotides) have dominated the field of ncRNAs research in the past decade in gastric cancer [7-9]. Another class of ncRNAs, long noncoding RNAs, which are commonly defined as transcripts > 200 nucleotides in length, have emerged as another class of vital regulatory RNAs [10]. A growing volume of literature has indicated that lncRNAs are differentially expressed in a diverse array of cancer and play an important role in the development of cancer [11-14].
Linc-UBC1 was firstly characterized in bladder cancer [15]. It is located on in human chromosome 1q32.1 and has a transcript of about 3 kb with just one exon. Linc-UBC1 was upregulated in bladder cancer tissues compared to adjacent normal tissues and it promoted the oncogenic activity of bladder cancer cells. He et al [15] demonstrated that linc-UBC1 possibly exerted its function via physically associating with PRC2 complex to regulate histone modification status of target genes. However, the role of linc-UBC1 in gastric cancer is elusive.
In this study, we would like to explore the expression pattern of linc-UBC1 and its correlation with clinicopathological factors in gastric cancer. Furthermore, the prognostic significance of linc-UBC1 was assessed. The oncogenic activity of linc-UBC1 was investigated in gastric cancer cell lines.
Materials and methods
Human tissue specimens
The study were undertaken with the understanding and written consent of each subject. This study was approved by the Ethics Committee of Wenzhou People’s Hospital at Wenzhou Medical University (Wenzhou, China). The study methodologies conformed to the standards set by the declaration of Helsinke. Eighty-five gastric cancer and matched adjacent normal non-tumor tissues gastric tissues (> 3 cm away from tumor) were obtained from patients who underwent surgical resection of primary gastric cancer between 2005 and 2009 at Wenzhou People’s Hospital. The diagnosis was based on histopathological examination by two experienced pathologists of Wenzhou People’s Hospital. The tissues were obtained before chemotherapy and radiation therapy. No selection bias was introduced in sample collection. Upon removal of the surgical specimen, each sample was immediately frozen in liquid nitrogen and stored at -80°C prior to RNA isolation and qRT-PCR analysis. All cases were classified according to the World Health Organization’s pathological classification of tumors.
Cell lines
Human gastric cancer cell lines (BGC-823, SGC-7901, AGS, MKN-45, HGC-27) and immortalized normal gastric epithelial cell line, GES-1 were purchased from the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, China). Cells were cultured in DMEM or RPMI 1640 medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS) as well as 100 U/ml penicillin and 100 μg/ml streptomycin (Invitrogen, Carlsbad, CA, USA). All cells were maintained at 37°C under an atmosphere of 5% CO2. All cell lines have been passaged for fewer than 6 months.
RNA extraction and quantitative real-time PCR
RT-qPCR was performed to determine the expression of linc-UBC1 in gastric cancer tissues and cells. Total RNA of cancer tissues or cultured cells was extracted with Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. RNA was reversed transcribed into cDNAs using the Primer-ScriptTM one step RT-PCR kit (TaKaRa, Dalian, China). The resulting cDNA was amplified by PCR using linc-UBC1 and GADPH (endogenous control) specific primers with real-time RT-PCR using the SYBR® Premix Dimmer Eraser kit (TaKaRa, Dalian, China). The quantitative real-time polymerase chain reaction (qRT-PCR) was performed on ABI 7500 system (Applied Biosystems, CA, USA) according to the manufacturer’s instructions. Specific primers for GADPH: 5’-GTCAACGGATTTGGTCTGTATT-3’ (forward), 5’-AGTCTTCTGGGTGGCAGTGAT-3’ (reverse); linc-UBC1: 5’-CCTGCTTGGAAACTAATGACC-3’ (forward), 5’-AGGCTCAACTTCCCAGACTCA-3’ (reverse). Relative expression values were calculated by the 2-ΔΔCt method using GAPDH as a normalizer.
RNA interference
The nucleotide sequences targeting linc-UBC1: (#1 CCUGUCUACAGACUGAAUATT, #2 CCGGAACAAAUGGCUUCAUTT), and nontargeting siRNAs (UUCUCCGAACGUGUCACGUTT) were purchased from GenePharma (Shanghai). Cells were grown on six-well plate to 60% confluency and transfected with 75 nM siRNA as well as Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions.
Cell proliferation assay
Cell proliferation assays were conducted using the CCK-8 assay kits as described by the manufacturer. EdU immunofluorescence staining was performed using an EdU kit (Roche, Mannheim, Germany).
Wound-healing assay
48 h after transfection, gastric cancer cells were scratched in the monolayer and cultured in normal conditions. The mobilized distances were measured at 0, 16 h after scratching for HGC-27 and at 0, 24 h after scratching for SGC-7901, respectively.
Cell invasion assay
48 h after transfection, 1 × 105 cells suspended in 200 μL serum-free media were added into the upper chamber of an insert pre-coated with 50 μL Matrigel (8.0 μm, Millipore, MA). The chambers were then incubated in culture medium supplemented with 10% FBS in the bottom chambers for 24 h before examination. The cells on the upper surface were scraped and washed away, whereas the cells on the lower surface were fixed with 20% methanol and stained with 0.2% crystal violet. Finally, cells were counted under a microscope and the relative number was calculated. Experiments were independently repeated in triplicate.
Statistical analysis
All quantitative data are presented as the mean ± standard deviation (S.D.) from at least three independent experiments. The statistical analysis was performed with SPSS 17.0 software. Unless otherwise specified, the difference between two groups was analyzed with Student’s t test. Metastasis-free survival and overall survival were evaluated using the Kaplan-Meier method, and the long-rank comparison was carried out to assess differences between stratified survival groups using the median value as the cutoff. The correlation between linc-UBC1 expression and clinicopathological characteristics were analyzed using Pearson Chi-square test. A two-sided P value of less than 0.05 was considered to be statistically significant.
Results
Expression of linc-UBC1 in gastric cancer cell lines
To explore the role of linc-UBC1 in gastric cancer, we first examined the expression levels of linc-UBC1 in gastric cancer cell lines. As shown in Figure 1A, gastric cancer cell lines expressed higher levels of linc-UBC1 than normal gastric epithelium cell line (GES-1). The expression of linc-UBC1 was most significantly higher in HGC-27 (P < 0.01) and SGC-7901 (P < 0.01) cells. HGC-27 cell line was established from the metastatic lymph node of gastric cancer patients [16], and SGC-7901 cell was associated with gastric cancer metastasis and invasion, which suggested that the enhanced expression of linc-UBC1 strongly correlated with the migration and invasion of gastric cancer cells.
Figure 1.
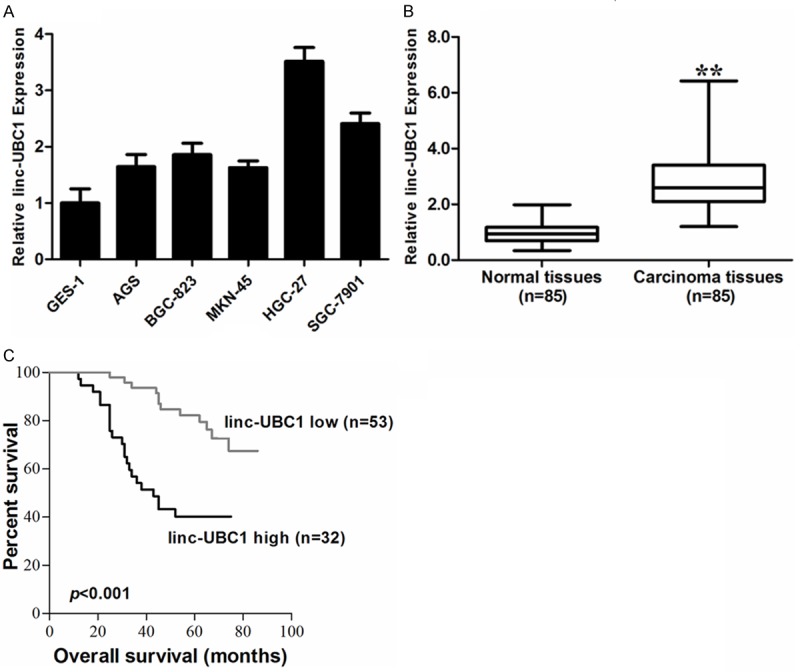
Linc-UBC1 is generally upregulated in gastric cancer, acting as a negative prognostic factor. A. qRT-PCR analysis of linc-UBC1 expression levels in gastric cancer cell lines (SGC-7901, HGC-27, MKN-45, BGC-823 and AGS) compared with normal gastric epithelium cell line (GES-1). B. Difference in expression levels of linc-UBC1 expression levels between gastric cancer tissues and matched non-tumor gastric tissues. The expression of linc-UBC1 was normalized to GADPH. The statistical differences between samples were analyzed with paired samples t-test (n = 85, P < 0.01). C. Patients with high levels of linc-UBC1 expression showed reduced overall survival times compared with patients with low levels of linc-UBC1 expression (P < 0.001, log-rank test). *, P < 0.05; **, P < 0.01.
Linc-UBC1 is upregulated in gastric cancer tissues
The expression of linc-UBC1 in 85 pairs of matched gastric tumor tissues was analyzed utilizing RT-qPCR. We found that linc-UBC1 was significantly upregulated in gastric cancer tissues compared to non-tumor gastric tissues (Figure 1B). To gain further insights into the observation mentioned above, we analyzed the correlation between linc-UBC1 expression and patient clinicopathological characteristics. As shown in Table 1, linc-UBC1 expression was irrelevant with age, sex and tumor differentiation. However, high linc-UBC1 expression was associated with lymph-node metastasis (P < 0.01), tumor size (P = 0.036, P < 0.05) and TNM stage (P = 0.012, P < 0.05).
Table 1.
Correlation between linc-UBC1 expression and clinicopathological characteristics of gastric cancer
| Clinical parameters | Number of patients (%) | Average Fold change | P-value |
|---|---|---|---|
| Age (years) | |||
| < 60 | 46 (54.12%) | 2.91 | 0.440 |
| ≥ 60 | 39 (45.88%) | 2.74 | |
| Gender | |||
| Male | 53 (62.35%) | 3.01 | 0.243 |
| Female | 25 (37.65%) | 2.68 | |
| Size | |||
| ≥ 5 cm | 44 (51.76%) | 3.53 | 0.036 |
| < 5 cm | 41 (48.24%) | 2.37 | |
| Histologic differentiation | |||
| well/moderately | 52 (61.18%) | 2.51 | 0.084 |
| poor | 33 (38.82%) | 3.11 | |
| TNM stage | |||
| I/II | 56 (65.88%) | 2.23 | 0.012 |
| III/IV | 29 (34.12%) | 3.42 | |
| Lymph node metastasis | |||
| Yes | 18 (21.18%) | 3.65 | 0.003 |
| No | 67 (78.82%) | 2.30 |
Enhanced expression of linc-UBC1 was associated with the poor prognosis in gastric cancer
We would like to explore whether linc-UBC1 expression level correlated with outcome of gastric cancer patients after gastrectomy. The median ratio of relative linc-UBC1 expression in gastric cancer (2.81) was used as the cutoff value to stratify high-linc-UBC1 group and low-linc-UBC1-group. Kaplan-Meier survival analysis and log-rank tests were conducted. Remarkably, patients with higher linc-UBC1 expression level had poorer overall survival (Figure 1C, P < 0.01).
Linc-UBC1 promoted the proliferation and invasion of gastric cancer cells in vitro
As demonstrated in Table 1, higher linc-UBC1 expression was associated larger tumor size and lymph node metastasis, suggesting that linc-UBC1 might promote the proliferation and invasion of gastric cancer. To further assess the biological role in gastric cancer, we knocked down linc-UBC1 expression in HGC-27 and SGC-7901 cells using small interfering RNA (Figure 2A). Cell-counting kit-8 assays indicated that linc-UBC1 depletion resulted in decreased tumor cell proliferation in gastric cancer cell line HGC-27 and SGC-7901 (Figure 2B, 2C). Furthermore, the percentage of Edu positive decreased significantly after linc-UBC1 knockdown (Figure 2D, 2E). All these data linc-UBC1 promoted the proliferation of gastric cancer cells.
Figure 2.
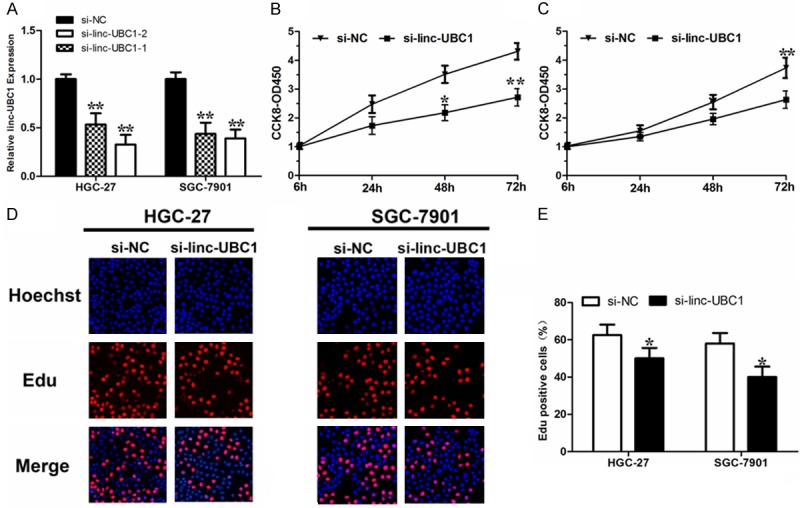
Linc-UBC1 knockdown attenuates gastric cancer cell proliferation. A. Linc-UBC1 expression level was confirmed by qRT-PCR in multiple gastric cancer cell lines. Mean ± S.D. are shown (n = 3). NC denotes siRNA having no homology to any known mammalian genes as a negative control. B, C. Linc-UBC1 knockdown attenuated gastric cancer cell line HGC-27 and SGC-7901 proliferation as determined by CCK8 assay. Mean ± S.D. are shown (n = 3). D. Linc-UBC1 knockdown decreased gastric cancer cell line HGC-27 and SGC-7901 in S phase. Blue color represents the nucleus and red color indicates S phase cells (EdU positive). E. Histological analysis of the percent of EdU positive cells in negative control and linc-UBC1 knockdown in gastric cancer cell lines. Mean ± S.D. are shown (n = 3). *, P < 0.05; **, P < 0.01.
Cancer motility and invasion ability were vital for cancer progression and metastasis. We next examined whether linc-UBC1 had a functional role in facilitating gastric cancer motility and invasion. We performed wound-healing assay and transwell assay with linc-UBC1 knockdown. As illustrated in Figure 3A and 3B, decreased cell motility was observed in both gastric cancer cell lines with linc-UBC1 depletion. Similarly, linc-UBC1 knockdown decreased gastric cancer invasion (Figure 3C, 3D). Taken together, linc-UBC1 may promote the proliferation, motility and invasion of gastric cancer.
Figure 3.
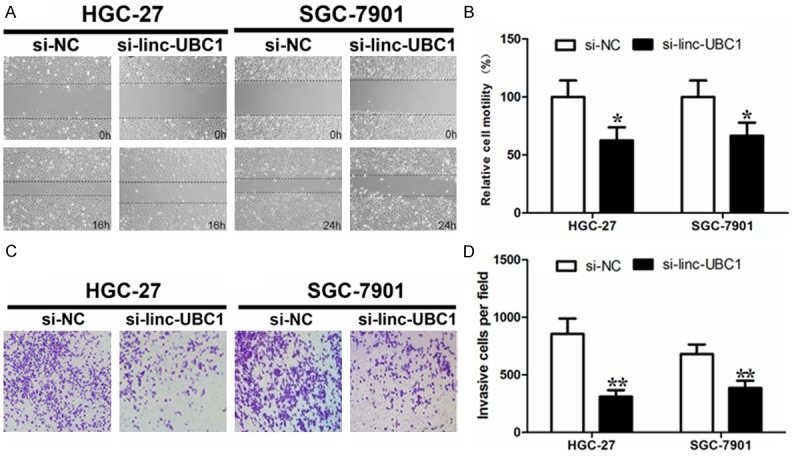
Linc-UBC1 knockdown attenuates gastric cancer cell motility and invasion. A. Representative images of cell motility changes by transfection of linc-UBC1 siRNA. B. Linc-UBC1 siRNA decreased cell motility. C. Representative images of Matrigel invasion assay after linc-UBC1 knockdown in gastric cancer cell line HGC-27 and SGC-7901. D. Histological analysis of OD (570 nm) absorbance of crystal violet-stained cells in transwell assay. Mean ± S.D. are shown (n = 3). *, P < 0.05; **, P < 0.01.
Discussion
In recent years, the research on long noncoding RNA has drawn more and more attention. Although thousands of lncRNAs were identified, only a small fraction of them have been characterized [5]. However, the current studies have demonstrated that lncRNAs are involved in the pathogenesis of cancer [11-15].
Although previous studies have identified a few cancer-associated lncRNAs, only a small number of gastric cancer-associated lncRNAs have been characterized. Wang et al [17] demonstrated that Long noncoding RNA MRUL contributed to the multi-drug resistance of gastric cancer. Zhang et al [18] revealed that lncRNA ANRIL indicates a poor prognosis of gastric cancer. CCAT1, a c-Myc activated lncRNA, promotes the progression of gastric cancer [19].
In this study, we found that linc-UBC1 was generally upregulated in gastric cancer and its overexpression correlates with lymph node metastasis, tumor size, TNM stage and a poorer prognosis of patients with gastric cancer, which was in agreement with the oncogenic role of linc-UBC1 in bladder cancer [15]. The functional studies showed linc-UBC1 depletion decreased the gastric cancer cell proliferation, motility and invasion. The data highlighted the role of linc-UBC1 in gastric cancer.
In summary, we showed that linc-UBC1 may be gastric cancer-specific lncRNA and may play an important role in the development of gastric cancer. Our study may facilitate the development of lncRNA-directed diagnostics and therapeutics against cancers.
Disclosure of conflict of interest
None.
References
- 1.Shah MA, Kelsen DP. Gastric cancer: a primer on the epidemiology and biology of the disease and an overview of the medical management of advanced disease. J Natl Compr Canc Netw. 2010;8:437–47. doi: 10.6004/jnccn.2010.0033. [DOI] [PubMed] [Google Scholar]
- 2.Allum WH, Blazeby JM, Griffin SM, Cunningham D, Jankowski JA, Wong R. Guidelines for the management of oesophageal and gastric cancer. Gut. 2011;60:1449–72. doi: 10.1136/gut.2010.228254. [DOI] [PubMed] [Google Scholar]
- 3.Saka M, Morita S, Fukagawa T, Katai H. Present and future status of gastric cancer surgery. Jpn J Clin Oncol. 2011;41:307–13. doi: 10.1093/jjco/hyq240. [DOI] [PubMed] [Google Scholar]
- 4.Tan HT, Low J, Lim SG, Chung MC. Serum autoantibodies as biomarkers for early cancer detection. FEBS J. 2009;276:6880–904. doi: 10.1111/j.1742-4658.2009.07396.x. [DOI] [PubMed] [Google Scholar]
- 5.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, Lander ES. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–7. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermüller J, Hofacker IL, Bell I, Cheung E, Drenkow J, Dumais E, Patel S, Helt G, Ganesh M, Ghosh S, Piccolboni A, Sementchenko V, Tammana H, Gingeras TR. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–8. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 7.Wu Q, Yang Z, An Y, Hu H, Yin J, Zhang P, Nie Y, Wu K, Shi Y, Fan D. MiR-19a/b modulate the metastasis of gastric cancer cells by targeting the tumour suppressor MXD1. Cell Death Dis. 2014;5:e1144. doi: 10.1038/cddis.2014.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma J, Liu J, Wang Z, Gu X, Fan Y, Zhang W, Xu L, Zhang J, Cai D. NF-kappaB-dependent microRNA-425 upregulation promotes gastric cancer cell growth by targeting PTEN upon IL-1β induction. Mol Cancer. 2014;13:40. doi: 10.1186/1476-4598-13-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He XP, Shao Y, Li XL, Xu W, Chen GS, Sun HH, Xu HC, Xu X, Tang D, Zheng XF, Xue YP, Huang GC, Sun WH. Downregulation of miR-101 in gastric cancer correlates with cyclooxygenase- 2 overexpression and tumor growth. FEBS J. 2012;279:4201–12. doi: 10.1111/febs.12013. [DOI] [PubMed] [Google Scholar]
- 10.Kugel JF, Goodrich JA. Non-coding RNAs. key regulators of mammalian transcription. Trends Biochem Sci. 2012;37:144–151. doi: 10.1016/j.tibs.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang F, Huo XS, Yuan SX, Zhang L, Zhou WP, Wang F, Sun SH. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol Cell. 2013;49:1083–96. doi: 10.1016/j.molcel.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 12.Huang JF, Guo YJ, Zhao CX, Yuan SX, Wang Y, Tang GN, Zhou WP, Sun SH. Hepatitis B virus X protein (HBx)-related long noncoding RNA (lncRNA) down-regulated expression by HBx (Dreh) inhibits hepatocellular carcinoma metastasis by targeting the intermediate filament protein vimentin. Hepatology. 2013;57:1882–92. doi: 10.1002/hep.26195. [DOI] [PubMed] [Google Scholar]
- 13.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–6. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prensner JR, Chen W, Iyer MK, Cao Q, Ma T, Han S, Sahu A, Malik R, Wilder-Romans K, Navone N, Logothetis CJ, Araujo JC, Pisters LL, Tewari AK, Canman CE, Knudsen KE, Kitabayashi N, Rubin MA, Demichelis F, Lawrence TS, Chinnaiyan AM, Feng FY. PCAT-1, a Long Noncoding RNA, Regulates BRCA2 and Controls Homologous Recombination in Cancer. Cancer Res. 2014;74:1651–60. doi: 10.1158/0008-5472.CAN-13-3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He W, Cai Q, Sun F, Zhong G, Wang P, Liu H, Luo J, Yu H, Huang J, Lin T. linc-UBC1 physically associates with polycomb repressive complex 2 (PRC2) and acts as a negative prognostic factor for lymph node metastasis and survival in bladder cancer. Biochim Biophys Acta. 2013;1832:1528–37. doi: 10.1016/j.bbadis.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 16.Akagi T, Kimoto T. Human cell line (HGC-27) derived from the metastatic lymph node of gastric cancer. Acta medica Okayama. 1976;30:215–219. [PubMed] [Google Scholar]
- 17.Wang Y, Zhang D, Wu K, Zhao Q, Nie Y, Fan D. Long noncoding RNA MRUL promotes ABCB1 expression in multidrug-resistant gastric cancer cell sublines. Mol Cell Biol. 2014;34:3182–93. doi: 10.1128/MCB.01580-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang EB, Kong R, Yin DD, You LH, Sun M, Han L, Xu TP, Xia R, Yang JS, De W, Chen JF. Long noncoding RNA ANRIL indicates a poor prognosis of gastric cancer and promotes tumor growth by epigenetically silencing of miR-99a/miR-449a. Oncotarget. 2014;5:2276–92. doi: 10.18632/oncotarget.1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang F, Xue X, Bi J, Zheng L, Zhi K, Gu Y, Fang G. Long noncoding RNA CCAT1, which could be activated by c-Myc, promotes the progression of gastric carcinoma. J Cancer Res Clin Oncol. 2013;139:437–45. doi: 10.1007/s00432-012-1324-x. [DOI] [PMC free article] [PubMed] [Google Scholar]


