Abstract
Objectives: The aim was to evaluate the clinical significance and prognostic value of tissue miR-150 expression in non-small cell lung cancer (NSCLC) patients. Materials and methods: Quantitative real-time PCR was used to analyze the expression of miR-150. Overall survival (OS) was estimated using the Kaplan-Meier method, and the differences in survival were compared using the log-rank test. A Cox proportional hazards model was used for multivariate analysis. Results: Mean miR-150 levels were significantly higher in NSCLC tissues compared with matched non-cancerous tissues (4.07 ± 2.33 vs. 1.00 ± 0.46, P < 0.0001). The level of miR-150 in NSCLC was strongly correlated with lymph node metastasis (P = 0.04), distant metastasis (P = 0.01) and clinical TNM stage (P = 0.02). Kaplan-Meier analysis showed that the cumulative 5-year OS rate was 40.8% in the high expression group, and 69.2% in the low expression group. The log-rank test showed that the OS rate of patients with high miR-150 expression was significantly poorer than that of the remaining cases (P = 0.007). Conclusion: Our data indicated that overexpression of miR-150 in NSCLC tissues has prognostic value.
Keywords: microRNA, miR-150, non-small-cell lung cancer, RT-PCR, prognosis, biomarker
Introduction
Lung cancer is one of the most common causes of cancer-related deaths worldwide, and majority of lung cancers are the non-small cell lung cancer (NSCLC), which comprises approximately 80% of all lung cancers [1]. Surgical resection, when possible, remains the only curative treatment for early-stage NSCLC. However, nearly 50% of resected patients experience recurrence and have a dismal prognosis [2]. Therefore, clinical indicators that accurately predict NSCLC progression and prognosis are essential for improving patient survival.
microRNAs (miRNAs) are small noncoding RNAs of ~22 nucleotides in the length that regulate the expression of their target mRNAs through translational repression or mRNA cleavage [3]. They are involved in crucial biological processes, including development and differentiation [4,5]. The dysregulation of miRNAs is correlated to play an important role in cancer development and progression by regulating the cell proliferation, differentiation, apoptosis and carcinogesis [6,7]. The miR-150 gene is on chromosome 19q13 and expressed at low levels in most solid tumors [8,9]. Downregulation of miR-150 expression acts as an anti-apoptotic factor in human diffuse gastric cancer and adrenocorticotrophic hormone-secreting pituitary tumors [8,9]. Overexpression of miR-150 inhibits tumor cell growth in vitro and inhibits tumor growth in animal models through direct downregulation of DKC1 and AKT2, reduction of phosphorylated AKTser473/4 and an increase in tumor suppressors such as Bim and p53, leading to telomerase activation and immortalization of cancer cells [10]. However, Cao et al found that the expression levels of miR-150 were significantly higher in lung cancer samples. Furthermore, their findings provided the first clues regarding the role of miR-150 as an oncogene in lung cancer through the inhibition of SRC kinase signaling inhibitor 1(SRCIN1) translation [11]. Gu et al found that down-regulation of miR-150 induced cell proliferation inhibition and apoptosis in NSCLC by targeting BAK1 in vitro [12]. However, until now, the prognostic value of miR-150 in NSCLC has not been investigated.
Materials and methods
Patients and clinical samples
Between 2005 and 2011, 157 patients with NSCLC were diagnosed at Qilu Hospital of Shandong University, and treated with surgery. None of the patients had received radiation therapy or chemotherapy before surgery. All surgical tissues were examined by a pathologist and final surgical pathology reports were obtained and recorded. Patient participation in the study was concluded once the final surgical pathology reports were obtained. Histological type and tumor-node-metastasis (TNM) classifications were made according to the criteria of the American College of Chest Physicians evidence-based clinical practice guidelines. The baseline patient characteristics were shown in Table 1. We obtained written and oral informed consent from all participants. This study was approved by the institutional review board of Qilu Hospital of Shandong University.
Table 1.
The relationship between miR-150 and clinicopathological characteristics in 157 patients with NSCLC
| miR-150 expression | ||||
|---|---|---|---|---|
|
|
||||
| Parameters | Number of cases | High | Low | P value |
| Sex | 157 | 81 | 76 | |
| Male | 89 | 49 | 40 | 0.31 |
| Female | 68 | 32 | 36 | |
| Age | ||||
| < 60 years | 79 | 40 | 39 | 0.25 |
| ≥ 60 years | 78 | 41 | 37 | |
| Smoking | ||||
| Never | 11 | 1 | 10 | 0.38 |
| Current | 75 | 43 | 32 | |
| Former | 71 | 37 | 34 | |
| Tumor Size | ||||
| < 3 cm | 64 | 22 | 42 | 0.08 |
| ≥ 3 cm | 93 | 59 | 34 | |
| Lymphatic invasion | ||||
| Positive | 56 | 37 | 19 | 0.04 |
| Negative | 101 | 44 | 57 | |
| Distant metastasis | ||||
| Positive | 35 | 26 | 9 | 0.01 |
| Negative | 122 | 55 | 67 | |
| TNM stage | ||||
| I/II | 88 | 39 | 49 | 0.02 |
| III/IV | 69 | 42 | 27 | |
| Tumor differentiation | ||||
| High/moderate | 106 | 59 | 47 | 0.09 |
| Poor | 51 | 22 | 29 | |
| Surgical margins | ||||
| Free | 131 | 66 | 65 | 0.21 |
| Not free | 26 | 15 | 11 | |
RNA isolation and qRT-PCR
Total RNA was extracted with Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. The concentration and purity of all RNA samples were detected by NanoDrop ND-2000 spectrophotometer (NanoDrop Technologies, Houston, TX, USA). NCodeTM SYBR® Green miRNA qRT-PCR Kit (Invitrogen, Carlsbad, CA, USA) was used to synthesize specific cDNA of miR-150 and U6B(as an internal control), and perform qRT-PCR, which was analyzed with the DNA Engine Opticon 2 Real-Time Cycler (MJ Research Inc., Waltham, MA, USA) according to the manufacturer’s instructions. Each sample was examined in triplicate and analyzed by the comparative threshold cycle (Ct) method. The expression levels of miR-150 were normalized to U6B.
Statistical analysis
Differences between two groups were estimated using Student’s t test and the Chi-square test. Overall survival (OS) was measured up to the date of death due to any cause or, for living patients, the date of last contact. OS was estimated using the Kaplan-Meier method, and the differences in survival were compared using the log-rank test. A Cox proportional hazards model was used for multivariate analysis. Significant differences were accepted at P < 0.05. Statistical analyses were performed with SPSS for Windows version 17.0 (SPSS Inc., Chicago, USA).
Results
Increased expression of miR-150 in NSCLC
To reveal the role of miR-150 in NSCLC, qRT-PCR was performed to measure miR-150 levels in 157 pairs of NSCLC tissues and adjacent non-cancerous tissues. Mean miR-150 levels were significantly higher in NSCLC tissues compared with matched non-cancerous tissues (4.07 ± 2.33 vs. 1.00 ± 0.46, P < 0.0001, shown in Figure 1). We divided the 157 patients with NSCLC into a high expression group (n = 81) and a low expression group (n = 76), according to the median expression level of miR-150.
Figure 1.
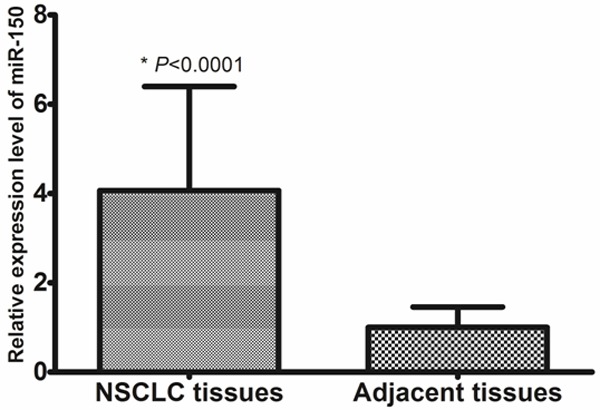
Increased expression of miR-150 in NSCLC.
Correlations between miR-150 expression and NSCLC clinicopathologic characteristics
The relationship between miR-150 expression and clinicopathologic parameters was evaluated. As shown in Table 1, the level of miR-150 in NSCLC was strongly correlated with lymph node metastasis (P = 0.04), distant metastasis (P = 0.01) and clinical TNM stage (P = 0.02). However, there were no significant associations between miR-150 expression and other clinical features including sex, age, smoking status, tumor differentiation, tumor size, and surgical margins. Taken together, these observations indicate that miR-150 expression is upregulated in NSCLC and is associated with the disease progression.
Independent prognostic indicators for patients with NSCLC
Kaplan-Meier analysis showed that the cumulative 5-year OS rate was 40.8% in the high expression group, and 69.2% in the low expression group (Figure 2). The log-rank test showed that the OS rate of patients with high miR-150 expression was significantly poorer than that of the remaining cases (P = 0.007).
Figure 2.
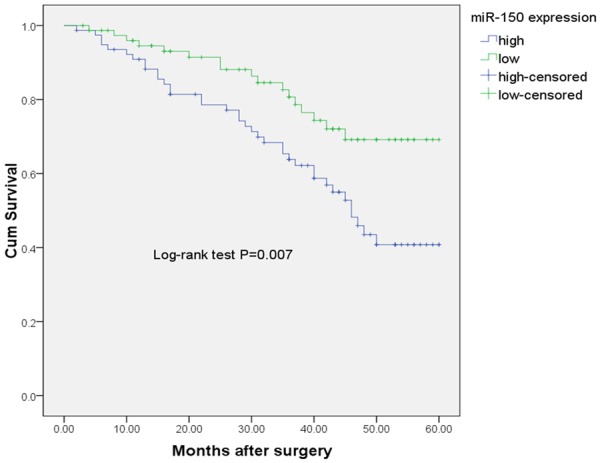
Kaplan-Meier curves for overall-survival in 157 NSCLC patients.
A Cox proportional hazards regression model analysis was performed to determine the independent prognostic indicators for patients with NSCLC. A multivariate analysis of these factors showed that miR-150 level (HR = 3.18, 95% CI: 1.79-10.22, P = 0.006), distant metastasis (HR = 4.57, 95% CI: 3.29-11.55, P = 0.002) and TNM stage (HR = 3.02, 95% CI: 1.38-5.82, P = 0.02) maintained their significance as independent prognostic factors for OS (shown in Table 2).
Table 2.
Multivariate analyses for overall survival in NSCLC patients
| Variable | Hazard ratio | 95% CI | P value |
|---|---|---|---|
| Sex | 1.12 | 0.82-1.97 | 0.56 |
| Age | 1.06 | 0.53-1.73 | 0.75 |
| Smoking | 1.42 | 0.74-2.15 | 0.15 |
| Tumor Size | 1.49 | 0.87-3.24 | 0.09 |
| Lymphatic invasion | 2.78 | 0.89-5.81 | 0.06 |
| Distant metastasis | 4.57 | 3.29-11.55 | 0.002 |
| TNM stage | 3.02 | 1.38-5.82 | 0.02 |
| Tumor differentiation | 2.19 | 0.22-4.87 | 0.12 |
| Surgical margins | 0.68 | 0.39-1.36 | 0.78 |
| miR-150 expression level | 3.18 | 1.79-10.22 | 0.006 |
Discussion
Although recent advances have been made in clinical diagnosis and therapeutic treatment, the overall 5-year mortality rate has remained unfavorable since the 1970s [13]. We are now in an era where personalized medicine and targeted therapies may give new hope for this patient group [14,15]. Identification of novel molecular markers which can improve diagnosis and prognostic stratification and serve as possible therapeutic targets will be of great importance in the near future. Several clinical prognostic markers for lung cancer have been recognized to dichotomize the risk, such as age, sex, tumor stage, cellular differentiation, and vascular invasion. However, since several biomarkers are not sufficiently accurate to predict the prognosis of lung cancer, the search of novel biomarker will be needed for prediction of prognosis in lung cancer.
Recent studies have shown that dysregulation of miRNAs contributes to the initiation, progression, metastasis, and drug resistance of cancer [16,17]. Several upregulated and downregulated miRNAs have been identified in lung cancer, the most frequently diagnosed cancer and the most common cause of cancer-related death worldwide [18,19]. The miR-150 gene is on chromosome 19q13 and expressed at low levels in most solid tumors [8,9]. Downregulation of miR-150 expression acts as an anti-apoptotic factor in human diffuse gastric cancer and adrenocorticotrophic hormone-secreting pituitary tumors [8,9]. Overexpression of miR-150 inhibits tumor cell growth in vitro and inhibits tumor growth in animal models through direct downregulation of DKC1 and AKT2, reduction of phosphorylated AKTser473/4 and an increase in tumor suppressors such as Bim and p53, leading to telomerase activation and immortalization of cancer cells [10]. Chang et al showed that miR-150 was downregulated by c-Myc and that it might function as a tumor suppressor. Injection of mouse lymphoma cell lines into mice expressing miR-150 produced fewer tumor cells in vivo [20]. Yokobori et al found that miR-150 expression was significantly lower in esophageal squamous cell carcinoma (ESCC) tissues compared to adjacent non-cancerous tissues (P < 0.001). Low expression of miR-150 in ESCC contributed to malignant potential, such as tumor depth, lymph node metastasis, lymphatic invasion, venous invasion, clinical staging, and poor prognosis (P < 0.05) [21]. However, the function of miR-150 was found to be different in lung cancer. Cao et al found that the expression levels of miR-150 were significantly higher in lung cancer samples. Furthermore, their findings provided the first clues regarding the role of miR-150 as an oncogene in lung cancer through the inhibition of SRC kinase signaling inhibitor 1 (SRCIN1) translation [11]. Gu et al found that down-regulation of miR-150 induced cell proliferation inhibition and apoptosis in NSCLC by targeting BAK1 in vitro [12]. However, until now, the prognostic value of miR-150 in NSCLC has not been investigated. In the present study, qRT-PCR was performed to measure miR-150 levels in 157 pairs of NSCLC tissues and adjacent non-cancerous tissues. Mean miR-150 levels were significantly higher in NSCLC tissues compared with matched non-cancerous tissues. We found that the level of miR-150 in NSCLC was strongly correlated with lymph node metastasis, distant metastasis and clinical TNM stage. Taken together, these observations indicated that miR-150 expression was upregulated in NSCLC and was associated with the disease progression. Kaplan-Meier analysis showed that the cumulative 5-year OS rate was 40.8% in the high expression group, and 69.2% in the low expression group. The log-rank test showed that the OS rate of patients with high miR-150 expression was significantly poorer than that of the remaining cases. Furthermore, a Cox proportional hazards regression model analysis was performed to determine the independent prognostic indicators for patients with NSCLC. A multivariate analysis of these factors showed that high miR-150 expression predicted a worse prognosis, suggesting that miR-150 should be considered as a potential biomarker associated with the prognosis in NSCLC.
In conclusion, our data indicated that overexpression of miR-150 in NSCLC tissues has prognostic value. Further studies and more samples will be required to confirm the prognostic value of miR-150 in NSCLC.
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Goldstraw P, Ball D, Jett JR, Le Chevalier T, Lim E, Nicholson AG, Shepherd FA. Non-small-cell lung cancer. Lancet. 2011;378:1727–1740. doi: 10.1016/S0140-6736(10)62101-0. [DOI] [PubMed] [Google Scholar]
- 3.Furuta M, Kozaki K, Tanimoto K, Tanaka S, Arii S, Shimamura T, Niida A, Miyano S, Inazawa J. The tumor-suppressive miR-497-195 cluster targets multiple cell-cycle regulators in hepatocellular carcinoma. PLoS One. 2013;8:e60155. doi: 10.1371/journal.pone.0060155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu X, Li Z, Shen J, Wu WK, Liang J, Weng X, Qiu G. MicroRNA-10b promotes nucleus pulposus cell proliferation through RhoC-Akt pathway by targeting HOXD10 in intervetebral disc degeneration. PLoS One. 2013;8:e83080. doi: 10.1371/journal.pone.0083080. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5.Hummel R, Hussey DJ, Haier J. MicroRNAs: predictors and modifiers of chemo- and radiotherapy in different tumour types. Eur J Cancer. 2010;46:298–311. doi: 10.1016/j.ejca.2009.10.027. [DOI] [PubMed] [Google Scholar]
- 6.Li Z, Lei H, Luo M, Wang Y, Dong L, Ma Y, Liu C, Song W, Wang F, Zhang J, Shen J, Yu J. DNA methylation downregulated mir-10b acts as a tumor suppressor in gastric cancer. Gastric Cancer. 2015;18:43–54. doi: 10.1007/s10120-014-0340-8. [DOI] [PubMed] [Google Scholar]
- 7.Wang HJ, Ruan HJ, He XJ, Ma YY, Jiang XT, Xia YJ, Ye ZY, Tao HQ. MicroRNA-101 is down-regulated in gastric cancer and involved in cell migration and invasion. Eur J Cancer. 2010;46:2295–2303. doi: 10.1016/j.ejca.2010.05.012. [DOI] [PubMed] [Google Scholar]
- 8.Katada T, Ishiguro H, Kuwabara Y, Kimura M, Mitui A, Mori Y, Ogawa R, Harata K, Fujii Y. microRNA expression profile in undifferentiated gastric cancer. Int J Oncol. 2009;34:537–542. [PubMed] [Google Scholar]
- 9.Amaral FC, Torres N, Saggioro F, Neder L, Machado HR, Silva WA Jr, Moreira AC, Castro M. MicroRNAs differentially expressed in ACTH-secreting pituitary tumors. J Clin Endocrinol Metabol. 2009;94:320–323. doi: 10.1210/jc.2008-1451. [DOI] [PubMed] [Google Scholar]
- 10.Watanabe A, Tagawa H, Yamashita J, Teshima K, Nara M, Iwamoto K, Kume M, Kameoka Y, Takahashi N, Nakagawa T, Shimizu N, Sawada K. The role of microRNA-150 as a tumor suppressor in malignant lymphoma. Leukemia. 2011;25:1324–1334. doi: 10.1038/leu.2011.81. [DOI] [PubMed] [Google Scholar]
- 11.Cao M, Hou D, Liang H, Gong F, Wang Y, Yan X, Jiang X, Wang C, Zhang J, Zen K, Zhang CY, Chen X. miR-150 promotes the proliferation and migration of lung cancer cells by targeting SRC kinase signalling inhibitor 1. Eur J Cancer. 2014;50:1013–1024. doi: 10.1016/j.ejca.2013.12.024. [DOI] [PubMed] [Google Scholar]
- 12.Gu XY, Wang J, Luo YZ, Du Q, Li RR, Shi H, Yu TP. Down-regulation of miR-150 induces cell proliferation inhibition and apoptosis in non-small-cell lung cancer by targeting BAK1 in vitro. Tumour Biol. 2014;35:5287–5293. doi: 10.1007/s13277-014-1688-4. [DOI] [PubMed] [Google Scholar]
- 13.Marshall E. Cancer research and the $90 billion metaphor. Science. 2011;331:1540–1541. doi: 10.1126/science.331.6024.1540-a. [DOI] [PubMed] [Google Scholar]
- 14.Drilon A, Rekhtman N, Ladanyi M, Paik P. Squamous-cell carcinomas of the lung: emerging biology, controversies, and the promise of targeted therapy. Lancet Oncol. 2012;13:e418–426. doi: 10.1016/S1470-2045(12)70291-7. [DOI] [PubMed] [Google Scholar]
- 15.Ma PC. Personalized targeted therapy in advanced non-small cell lung cancer. Cleveland Clin J Med. 2012;79(Electronic Suppl 1):eS56–60. doi: 10.3949/ccjm.79.s2.12. [DOI] [PubMed] [Google Scholar]
- 16.Chen PS, Su JL, Hung MC. Dysregulation of microRNAs in cancer. J Biomed Sci. 2012;19:90. doi: 10.1186/1423-0127-19-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suzuki H, Maruyama R, Yamamoto E, Kai M. DNA methylation and microRNA dysregulation in cancer. Mol Oncol. 2012;6:567–578. doi: 10.1016/j.molonc.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abd-El-Fattah AA, Sadik NA, Shaker OG, Aboulftouh ML. Differential microRNAs expression in serum of patients with lung cancer, pulmonary tuberculosis, and pneumonia. Cell Biochem Biophys. 2013;67:875–84. doi: 10.1007/s12013-013-9575-y. [DOI] [PubMed] [Google Scholar]
- 19.Vosa U, Vooder T, Kolde R, Vilo J, Metspalu A, Annilo T. Meta-analysis of microRNA expression in lung cancer. Int J Cancer international du cancer. 2013;132:2884–2893. doi: 10.1002/ijc.27981. [DOI] [PubMed] [Google Scholar]
- 20.Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, Dang CV, Thomas-Tikhonenko A, Mendell JT. Widespread microRNA repression by Myc contributes to tumorigenesis. Nature genetics. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yokobori T, Suzuki S, Tanaka N, Inose T, Sohda M, Sano A, Sakai M, Nakajima M, Miyazaki T, Kato H, Kuwano H. MiR-150 is associated with poor prognosis in esophageal squamous cell carcinoma via targeting the EMT inducer ZEB1. Cancer Sci. 2013;104:48–54. doi: 10.1111/cas.12030. [DOI] [PMC free article] [PubMed] [Google Scholar]


