Abstract
Cap dependent translation is mainly regulated at the level of the eukaryotic initiation factor 4E (eIF4E), the activity of which is controlled by phosphorylation and sequestration by its well established regulator, 4E binding protein 1 (4E-BP1). Both eIF4E and 4E-BP1 have been shown to be involved in the malignant progression of multiple human cancers, including colorectal cancer. However, the data on determining the expression of eIF4E, 4E-BP1 and their phosphorylated forms simultaneously in a single patient with colorectal cancer is lacking. Therefore the aim of our study was to explore the roles of these factors in colorectal carcinogenesis by immunohistostaining colorectal tissues (normal, low grade adenoma, high grade adenoma, and adenocarcinoma). Our results showed that the expression levels of eIF4E increased steadily as the cancer progressed from the case of benign dysplasia to an adenocarcinoma; all the while maintaining an unphosphorylated form. On the other hand, total expression levels of 4E-BP1 increased only in the premalignant state of the disease and decreased (but highly phosphorylated or inactivated) or abolished upon malignancy. Taken together, our findings suggest that strong correlations exist between the expression of eIF4E (not p-eIF4E) and tumor grade providing evidence that eIF4E expression plays a pivotal role in the malignant progression of colorectal cancer. Moreover, 4E-BP1 showed a bi-phasic level of expression during carcinogenesis, which is expressed only in hyperplasic or dysplastic tissues as an endogenous tumor suppressor molecule.
Keywords: Colorectal cancer, translational control, eIF4E, 4E-BP1, immunuhistochemistry
Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed cancer in the world in terms of both incidence and frequency and it is the third most common cause of cancer related deaths in USA [1]. CRC starts out as an adenomatous polyp in the internal lining of the wall of the large intestine which progresses through intermediate stages of dysplasia (low and high grade) to an invasive adenocarinoma. As long as the neoplastic growth is confined to the wall of the large bowel it is curable and can be removed by surgical excision. The progression to malignancy requires years to manifest, thus proper screening could allow for early detection and removal of the polyp before it becomes incurable. Nevertheless diagnosis most commonly occurs in the later stages of the disease thereby decreasing the survival rate. As such, it is mandatory that we unravel new factors involved in the initiation and progression of CRC which have diagnostic or prognostic potential.
Proteins catalyze most of the reactions on which life depends. The final pathway that leads to the expression of proteins is translation. Translation is a multistep, multifactorial process consisting of 3 major parts, initiation, elongation and termination. It requires the exertion of regulation at many levels, especially at the level of initiation which is, until recently, considered as the rate limiting step [2]. There isn’t one single standard mechanism for the initiation of translation of all cellular mRNAs; however since almost all nucleus-transcribed mRNA have a 5’ terminal m7G [5’] ppp [5’] N cap and a 50 to 300 nucleotide long 3’poly (A) tail, cap-dependent translation is the most common mechanism of initiation. The presence of secondary structures in the 5’ UTR (5’ untranslated region) will decrease the initiation efficiency and require an additional complex for proper initiation (eukaryotic initiation factor 4F). eIF4F is a trimeric complex composed of the eukaryotic initiation factors 4A, an ATP dependant RNA helicase which is required for the unwinding of the secondary structures; 4G, a scaffolding molecule and 4E, the 5’ cap mRNA binding protein. Since eIF4E is the least abundant component of this complex, its recruitment serves as the rate limiting step in the initiation of cap dependent translation. In fact changes in the levels of eIF4E profoundly affect translation rates where eIF4E hyperactivity preferentially enhances the selective synthesis of potent growth promoting and oncogenic proteins in which the translation is presumably repressed without affecting global protein synthesis rates [3]. In this way it seems to affect cancer relevant processes such as apoptosis, transformation, angiogenesis, invasion and metastasis [4]. There has been both in vitro and in vivo evidence supporting this oncogenic potential of eIF4E hyperactivity where the overexpression of eIF4E in mammalian cells (including fibroblasts and epithelial cells) induces their malignant transformation [5]. Not only that but also in vivo ectopic eIF4E overexpression leads to increased cancer risk; where eIF4E transgeneic mice developed lymphomas, angiosarcomas, lung carcinomas and hepatomas [6].
Therefore the regulation of eIF4E is crucial and it is accomplished at two levels [3]. The first involves its phoshporylation on Ser209 by MNK kinase which is a downstream target of the RAS/RAF/MAPK signaling pathway. This phosphorylation is thought to enhance cap binding, to be involved in eIF4E’s ability to promote tumorigenesis, and to be dispensable for mouse viability [7]. The second and more prolific mode of regulation is via sequestration by 4E binding proteins (4E-BPs) 1, 2 and 3; of which 4E-BP1 is the most common isoform. This phosphoprotein is the downstream target of another signaling pathway, the PI3K/AKT/mTOR pathway which leads to the hierarchal phosphorylation of 4E-BP1 and its consequent release from eIF4E allowing the initiation of translation. As such the hyposhosphorylated form of 4E-BP1 is actively suppressing cap dependent translation while the hyperphosphorylated form is activating it. Studies have shown that 4E-BP can be viewed as a tumor suppressor since at the cellular level; ectopic expression of 4E-BPs can partially revert eIF4E-transformed cells to nonmalignant phenotype and induce apoptosis in cells transformed by other oncogenes such as Ras [5].
The ubiquitous expression of eIF4E or 4E-BP1 has been linked to multiple human cancers, including those of the stomach, colon, breast, esophagus, head and neck, lung, cervix, prostate, bladder, and ovary. However, to our knowledge, the expression levels of eIF4E and 4E-BP1 with their phosphorylation status has never been evaluated all at once in colorectal cancer. In this study, we immunohistochemically investigated the variation of the activity and expression levels of eIF4E and its inhibitor 4E-BP1 to draw a conclusion about the correlation between these factors and the genesis of CRC leading us to define a potential diagnostic marker or therapeutic target.
Materials and methods
Cases and classification
For this study, 72 cases of colon paraffin-embedded tissues were obtained from specialized medical laboratory. Histological diagnosis, differentiation, and stage were evaluated according to The World Health Organization Classifications [8]. Tissues consisted of 27 well-differentiated or adenocarcinoma, 12 high grade adenoma or dysplasia, 20 low grade or hyperplasia, and 13 with normal structure of cells. In 10 adenocarcinoma cases, different differentiation stages were observed. This project was approved by the institutional ethic committee.
Immunohistochemistry
Serial sections from each case were used for immunohistochemistry (IHC) and all samples were routinely processed as described previously [9]. After sections rehydration and antigen retrieval using citrate buffer, samples were incubated with primary rabbit polyclonal antibodies: eIF4E (9742), phosphor-eIf4E ser209 (9741), 4E-BP1 (9452), phosphor-4E-BP1 ser65 (9451) obtained from Cell signalling using 1:50 dilution. Reactions were amplified with peroxidase-conjugated goat antibodies to rabbit IgG, followed by peroxidase-conjugated rabbit antibodies to goat IgG and revealed by AEC (3-Amino-9-ethylcarbazole) substrate-chromagen purchased from Dako. Sections were counterstained with Mayer’s hematoxylin solution (Sigma-Aldrich).
The IHC expression was evaluated by two persons separately. The intensity of immunoreactivity was scored using a conventional 4-tier system as follows: 0, no discernible staining; 1+, moderate staining; 2+, important staining; and 3+, strong staining. Samples exhibiting 2+ or 3+ staining were considered as “positive for overexpression”.
Results
Expression level of eIF4E in colorectal tissue
To study the role of eIF4E in colorectal carcinogenesis the presence and pattern of total protein expression level of eIF4E was first determined in a patient colonic tissue presenting the different stage of carcinogenesis from normal to adenocarcinoma. The results from the immunohistochemical staining showed that eIF4E was barely detectable in the normal colonic epithelial cells (Figure 1A). Next, the examination of the graded colorectal adenomatous showed a significant increase in the level of expression of this translation initiation factor (Figure 1B, 1C). This increase was slightly higher in the tissue with a higher degree of dysplasia (Figure 1C); however the maximum level of expression can be associated with the malignant invasive adenocarcinoma that is the final stage of disease development (Figure 1D). The study of all tissues showed a correlation between the expression of eIF4E and carcinogenesis stages with 66.67% of cases considered highly stained in adenocarcinoma tissues with a score of 2 or 3 (Table 1).
Figure 1.
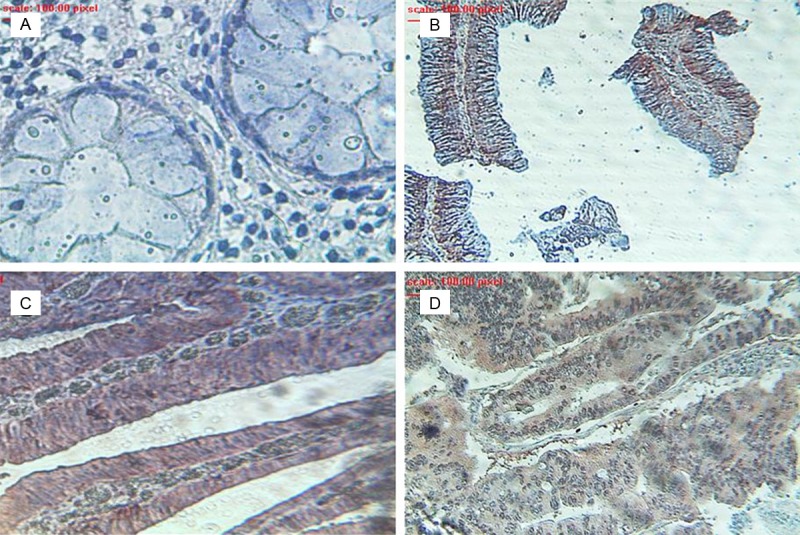
Expression levels of eIF4E in a prototype colorectal tissue with different stage of lesion. A: Normal; B: Low grade adenoma; C: High grade adenoma; D: Adenocarcinoma.
Table 1.
Staining intensity of the total and phosphorylated forms of eIF4E and 4E-BP1 on the different histologically classified colonic tissue sections
| Protein | Score | Normal (N=13) | Hyperplasia (Low grade) (N=20) | Dysplasia (High Grade) (N=12) | Adenocarcinoma (N=27) |
|---|---|---|---|---|---|
| eIF4E | 0 | 13 (100%) | 6 (30%) | 4 (33.33%) | 3 (11.11%) |
| 1 | 0 (0%) | 10 (50%) | 0 (0%) | 6 (22.22%) | |
| 2 | 0 (0%) | 4 (20%) | 8 (66.67%) | 6 (22.22%) | |
| 3 | 0 (0%) | 0 (0%) | 0 (0%) | 12 (44.45%) | |
| 4E‐BP1 | 0 | 13 (100%) | 5 (25%) | 1 (8.33%) | 15 (55.56%) |
| 1 | 0 (0%) | 8 (40%) | 2 (16.67%) | 8 (29.63%) | |
| 2 | 0 (0%) | 6 (30%) | 6 (50%) | 4 (14.81%) | |
| 3 | 0 (0%) | 1 (5%) | 3 (25%) | 0 (0%) | |
| p‐eIF4E | 0 | 13 (100%) | 20 (100%) | 12 (100%) | 27 (100%) |
| 1 | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | |
| 2 | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | |
| 3 | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | |
| p‐4E‐BP1 | 0 | 13 (100%) | 6 (30%) | 3 (25%) | 15 (55.56%) |
| 1 | 0 (0%) | 8 (40%) | 1 (8.33%) | 2 (7.41%) | |
| 2 | 0 (0%) | 6 (30%) | 5 (41.67%) | 4 (14.81%) | |
| 3 | 0 (0%) | 0 (0%) | 3 (25%) | 6 (22.22%) |
Expression level of p-eIF4E in colorectal tissues
The phosphorylation of eIF4E on Ser 209 has been thought to be mandatory for the tumorigenic properties of eIF4E. As such the normal colorectal tissue did not present any expression of this factor; however neither did the other graded tissue specimens (Table 1).
Expression levels of 4E-BP1 in colorectal tissues
The activity of eIF4E is under the tight control of its inhibitor 4E-BP1. Therefore we decided to determine the presence and pattern of the total protein expression level of this protein in a serial section from same patients.
To study the dynamic expression of 4E-BP1 in the process of carcinogenesis, we have analyzed 4E-BP1 expression in a tissue with different stages of lesions. In normal cuboid epithelial cells 4E-BP1 was not detected (Figure 2A) as it was shown for eIF4E. In adenoma lesions, 4E-BP1 protein appeared transiently overexpressed, peaking in high grade stage (Figure 2C) by comparison with low grade (Figure 2B), and returning to basal low levels or becoming less detected as the pathology further progresses to adenocarcinoma (Figure 2D). Analysis of the 72 cases revealed that 4E-BP1 is either not or poorly expressed in more than half (23/27) of the adenocarcinoma samples and only 14.81% of cases had important staining level (Table 1). The highly level of staining was observed for High grade adenoma with 9/12 tissues scored as important to strong, whereas 40% low grade tissues presented moderate staining (Table 1).
Figure 2.
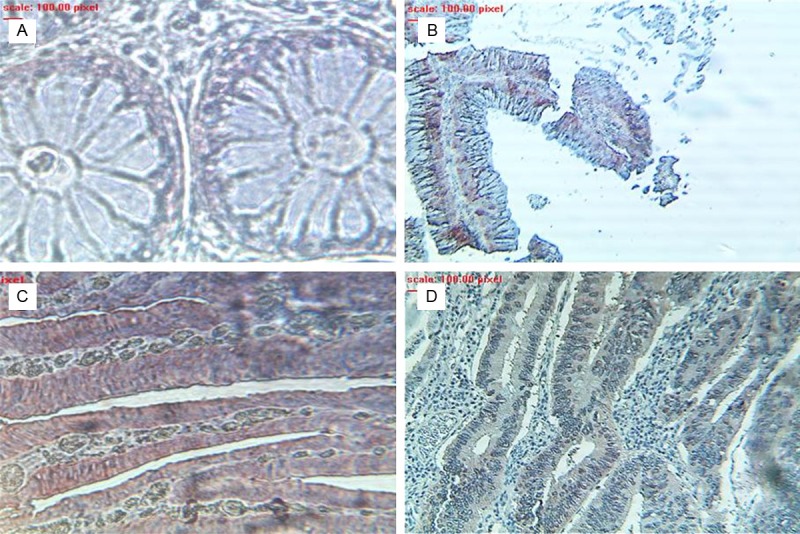
Expression levels of 4E-BP1 in a prototype colorectal tissue with different stage of lesion. A: Normal; B: Low grade adenoma; C: High grade adenoma; D: Adenocarcinoma.
Expression levels of p4E-BP1 in colorectal tissues
Since the ser65 phosphorylation of 4E-BP1 limits its interaction with eIF4E to control translation, the tissues were stained with a specific antibody. Immunohistochemical images of a colonic specimen showed that in accordance with eIF4E the maximum levels were detected in the case of malignancy (adenocarcinoma) (Figure 3D); while the lowest levels seemed to be correlated to the low grade colorectal adenomatous tissue (Figure 3B). On the other hand, the high grade colorectal adenomatous tissue showed expression levels comparable to those of the malignant tissue (Figure 3C). Moreover, p4E-BP1 was absent or barely expressed in the normal colorectal cells (Figure 3A). The data collected from all specimens revealed a staining profile similar to eIF4E during the different stages of carcinogenesis (Table 1). Note that p4E-BP1 staining was positive only in tissues where 4E-BP1 was detected. By comparing 4E-BP1 staining to its phosphorylated form we noticed that when 4E-BP1 is present in adenocarcinoma tissues it’s highly phosphorylated or inactivated (Figure 4).
Figure 3.
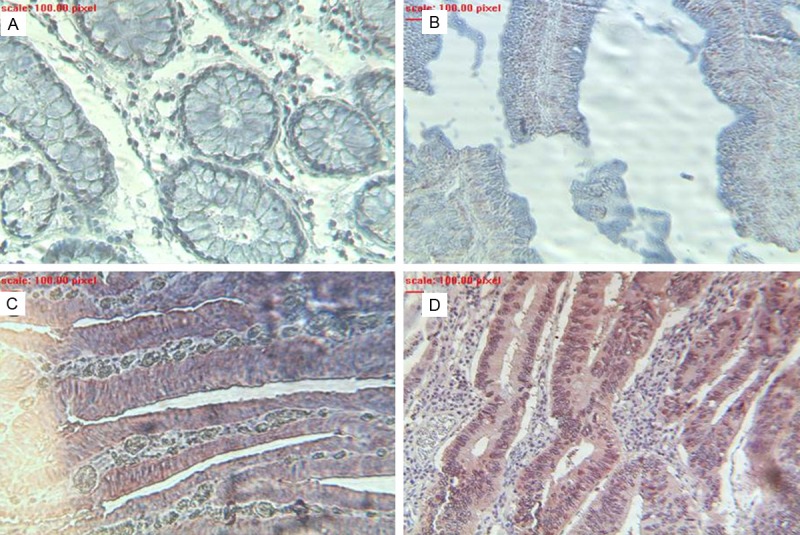
Expression levels of p4E-BP1 in a prototype colorectal tissue with different stage of lesion. A: Normal; B: Low grade adenoma; C: High grade adenoma; D: Adenocarcinoma.
Figure 4.
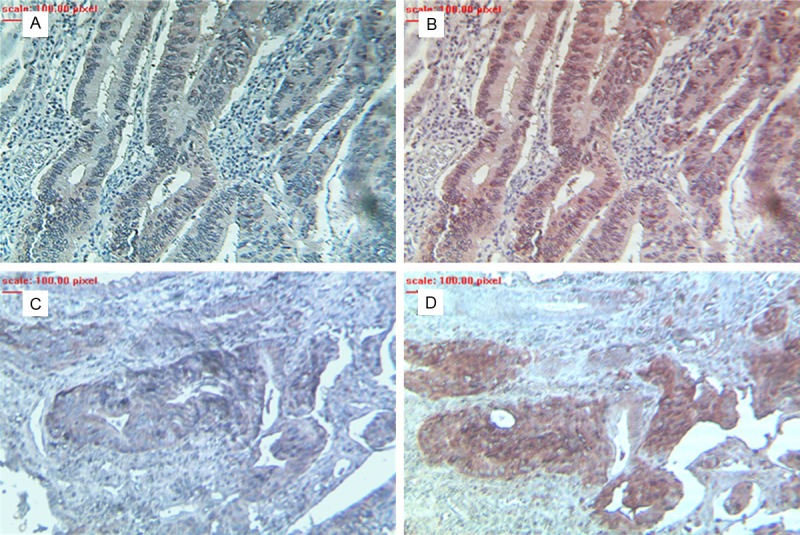
Expression levels of 4E-BP1 and p4E-BP1 in adenocarcinoma tissues. The expression levels of 4E-BP1 (A, C) and the phosphorylated ser65 form (B, D) were showed from two patient samples.
Discussion
Colorectal cancer is a multistep cancer that in 95% of the cases evolves from a preexisting epithelial neoplasia, the adenoma which progresses via intermediate stages, showing increased degrees of dysplasia, to an adenocarcinoma. This pathway to carcinogenesis is driven by a series of mutations giving the cancer cells a survival advantage to grow, accumulate and invade the surrounding tissue [10,11]. The most common of these mutations leads to the inactivation of the tumor suppressor gene APC; which is an initiating event in carcinogenesis in 60% of adecnocarcinomas. On the other hand, LOH of 17p (where the tumor suppressor gene p53 resides) and the inactivation of the TGFβ signaling pathway are thought to occur as late events in the progression to malignancy. The repercussions of genetic mutations on cancer development have been under investigation for decades, however for some time now the involvement of cap-dependent translation initiation in the genesis and maintenance of multiple types of cancer is becoming evident. Cap-dependent translation is controlled mainly by regulating the abundance and activity of the eukaryotic initiation factor 4E (eIF4E) which is the rate limiting component of the cap binding complex (eIF4F) required for the initiation of this process. eIF4E in itself is regulated by phosphorylation at Ser209 and sequestration by 4E-BP1. Therefore in our study we decided to view this genetic disease from another angle and focused on determining the involvement of regulatory factors of cap dependent translation, in specific eIF4E and its inhibitor 4E-BP1, in the progression of CRC.
Our results showed that the expression levels of eIF4E increased steadily as the cancer progressed from the case of benign dysplasia to an adenocarcinoma. This is in agreement with the results of the studies done by Rosenwald et al. and Berkel et al.; who proved that eIF4E overexpression is an early event during colorectal carcinogenesis [12] and that eIF4E expression is strongly associated with the histological type of the lesion [13]. The positive association we drew between eIF4E and the histological type in colorectal cancer has also been observed in head and neck squamous cancer (HNSC) [14], prostate cancer [15] and peripheral lung adenocarcinoma [16]; where the expression levels of eIF4E were lower in the premalignant lesions compared to the cancerous lesions which had the highest expression levels. The increase in the expression levels of eIF4E can be due to multiple mechanisms. It can be the result of gene amplification, like it was found in breast and HNSC carcinoma [17], alterations in transcription or mRNA stability. In fact, elevated eIF4E levels may result from any combination of these [18]. Hu antigen R (HuR) proteins have been shown to regulate the stability of eIF4E transcripts by binding to the 3’UTR, hence increasing its protein expression levels [19]. Therefore the increase of the expression and cytoplasmic abundance of the RNA-binding protein HuR with increased malignancy of colon cancer [20] might account for the observed increased in eIF4E levels. With respect to transcriptional regulation, it was shown that in colorectal cancer the deregulation of the APC/β-catenin/Tcf-4 signaling pathway, leads to the transcriptional activation of c-myc which will in turn result in the transcriptional increase of eIF4E [12]. In addition, immunohistochemical analysis of breast cancer biopsies have found increased eIF4E expression during hypoxic conditions [21] and more recently the implication of hypoxia inducible factor 1α (HIF-1α) was elucidated [22]. In this way, the hypoxia that accompanies tumor growth may stimulate eIF4E expression.
The eIF4E expressed is subject to phosphorylation as a downstream target of the MAPK signaling pathway [3]. The results of the immunohistochemical staining for the detection of p-eIF4E failed to reveal the presence of phosphorylation in any of the cases studied. However these results are not likely to be due to an error in manipulation since a positive control, the pancreatic tissue [9], showed positive staining for this factor. In colorectal tissues, it was found that p-eIF4E positive staining rates increase during carcinogenesis but this was seen in only 37 out of 60 cases studied [23]. It is important to note here that even in the literature the role of the phosphorylation of eIF4E on Ser 209 is highly controversial with respect to the physiological role as well as its involvement in the oncogenic characteristics of eIF4E [24]. Even though there is increasing evidence on the involvement of the phosphorylation of eIF4E in tumorigenesis [25], we cannot ignore the contradicting evidence that p-eIF4E binds with lower affinity to the cap structure [26] and that this phosphorylation is not required for protein synthesis in vitro and in vivo [27]. In fact phosphorylation alone cannot drive the formation of eIF4E/eIF4G complex and therefore is insufficient to activate the translation machinery [24]. Therefore more work should be done in this field to better elucidate, without doubt, the role of p-eIF4E in cancer in general and colorectal cancer in specific.
With respect to 4E-BP1, the total expression levels increased only in the premalignant state of the disease and decreased or stabilized upon malignancy. Also this 4E-BP1, when expressed was in the phosphorylated form. In all cases the expression patterns of each of these factors were significantly higher in the dysplastic and cancerous tissue when compared to the normal colorectal tissue. In literature, 4E-BP1 has received much less attention than eIF4E, especially when it comes to studying its pattern of expression in the initiation and progression of cancer. In fact, in the aforementioned studies on colorectal tissue [12,13], they failed to test for 4E-BP1 expression levels; and in the one study where they did [28] they used western blot analysis on a small cohort, including only malignant colon and gastric tissue and adjacent normal tissue. Their results showed 4E-BP1 to be negatively associated to the progression of gastrointestinal cancer, which is in accordance with our findings. This was also found to be true for breast cancer [29,30], hence further backing up our results. In the case of p-4E-BP1, evidence of its positive association with higher grade in endometrial [31], breast [29,30], ovarian [32] and prostate cancer [15] add credibility to our results.
In the case of 4E-BP1 the increase in its expression in the cytoplasm of the dysplastic cells which are simultaneously expressing increased levels of eIF4E suggests that this 4E-BP1 is just the cells’ physiological response to counter the increased expression of eIF4E where the transcriptional regulation of 4E-BP1 appears to be vital for resistance to stress [33-36]; as well for mediating the antiproliferative effects of TGFβ (at least in pancreatic cancer cell lines) [37]. The consequent deregulation of the TGFβ pathway (by loss of expression of TGFβRII or SMAD2/4), which is commonly a late event in colorectal carcinogenesis, could account for the failure of the protein expression levels of 4E-BP1 to further increase in the adenocarcinoma state. 4E-BP1 is subject to phosphorylation, in a hierarchal manner, at a set of conserved serine and threonine residues by kinases such as mTOR, ERK1/2, and PI3K [3]; therefore the multiple oncogenic alterations that are activated in malignant cells could lead to an increase in the phosphorylation of this protein [29]. In fact PI3K signaling pathway has been found to be mutated in up to 40% of CRC [38] and involved in 4E-BP1 downregulation [39]. Therefore we suggest that the activation of PI3K during CRC carcinogenesis may inactivate the translational control by 4E-BP1, either by downregulating its expression or, when expressed, by phosphorylating it.
The results are the first of their kind in terms of determining the expression level of eIF4E, 4E-BP1 and their phosphorylated forms simultaneously in the same normal, dysplastic and malignant colorectal tissue. We showed that the increase in eIF4E expression levels is being accompanied by an increase in the levels of p-4E-BP1; which means that as the colorectal tumor is progressing, the availability of eIF4E is increasing. This free and available eIF4E will lead to an increase in the efficiency of cap dependant translation of potent growth promoting proteins and oncogenic proteins, including growth factors such as c-myc and cyclin D1, angiogenesis factors such as VEGF and FGF-2, degradative enzymes such as matrix metalloproteinase-9 and heparinase and antiapoptotic proteins such as survivin and Bcl-2 [40]; all of which contribute to every aspect of malignancy. Regardless of the exact mechanism by which these factors are involved in the initiation and progression of CRC, their potential to serve as prognostic markers for this disease is highly likely. That is especially since eIFF4E is a proven biomarker that predicts survival in many cancers; where high eIF4E expression levels have been linked to increased risk of recurrence and poorer overall survival. Similarly the presence of higher levels of p-4E-BP1 have been associated to a worse prognosis in both breast [29] and ovarian cancer [30]; and to be a predictor of post-recurrence and poor overall survival in melanoma [41]. On the other hand, 4E-BP1 has been shown to possess prognostic potential mainly when combined with the expression levels of eIF4E in esophageal [42] and lung [43]; where the higher the ratio of 4E-BP1/eIF4E, the better the prognosis. Therefore our study introduces the possibility of considering the relationship between the presence of eIF4E or p-4E-BP1 and survival in CRC. Also since a number of studies are working on targeting cap dependent translation at multiple levels, including those directly related to eIF4E [3,4] with higher efficacy when the ratio of eIF4E/4E-BP1 increased [44], our results support the idea of determining the efficacy of such targeted therapy in the treatment of colorectal cancer.
Collectively, our data showed that eIF4E expression is elevated significantly with the progression of colorectal cancer and its inhibitor, 4E-BP1, when expressed is in the inactive, phosphorylated form. This thereby implicates enhanced eIF4E activation in colorectal cancer progression and suggests that eIF4E may be an attractive target for colorectal cancer therapy.
Acknowledgements
We acknowledge the work of all technicians in Specialized Medical Laboratory and EDST for assistance. The work was supported by Lebanese University grant for research.
Disclosure of conflict of interest
None.
References
- 1.Siegel R, Desantis C, Jemal A. Colorectal cancer statistics 2014. CA Cancer J Clin. 2014;64:104–117. doi: 10.3322/caac.21220. [DOI] [PubMed] [Google Scholar]
- 2.Merrick WC, Harris ME. Control not at initiation? Bah, humbug! EMBO J. 2014;33:3–4. doi: 10.1002/embj.201387388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martineau Y, Azar R, Bousquet C, Pyronnet S. Anti-oncogenic potential of the eIF4E-binding proteins. Oncogene. 2013;32:671–6777. doi: 10.1038/onc.2012.116. [DOI] [PubMed] [Google Scholar]
- 4.Hsieh AC, Ruggero D. Targeting eukaryotic translation initiation factor 4E (eIF4E) in cancer. Clin Cancer Res. 2010;16:4914–4920. doi: 10.1158/1078-0432.CCR-10-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Avdulov S, Li S, Michalek V, Burrichter D, Peterson M, Perlman DM, Manivel JC, Sonenberg N, Yee D, Bitterman PB, Polunovsky VA. Activation of translation complex eIF4F is essential for the genesis and maintenance of the malignant phenotype in human mammary epithelial cells. Cancer Cell. 2004;5:553–563. doi: 10.1016/j.ccr.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 6.Ruggero D, Montanaro L, Ma L, Xu W, Londei P, Cordon-Cardo C, Pandolfi PP. The translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nat Med. 2004;10:484–486. doi: 10.1038/nm1042. [DOI] [PubMed] [Google Scholar]
- 7.Hou J, Lam F, Proud C, Wang S. Targeting Mnks for cancer therapy. Oncotarget. 2012;3:118–131. doi: 10.18632/oncotarget.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamilton S, Aaltonen L. World Health Organization Classification of Tumours International Agency for Research on Cancer (IARC) Lyon: IARC Press; 2000. Pathology and Genetics of Tumours of the Digestive System. [Google Scholar]
- 9.Martineau Y, Azar R, Müller D, Lasfargues C, El Khawand S, Anesia R, Pelletier J, Bousquet C, Pyronnet S. Pancreatic tumours escape from translational control through 4E-BP1 loss. Oncogene. 2014;33:1367–1374. doi: 10.1038/onc.2013.100. [DOI] [PubMed] [Google Scholar]
- 10.Colussi D, Brandi G, Bazzoli F, Ricciardiello L. Molecular pathways involved in colorectal cancer: implications for disease behavior and prevention. Int J Mol Sci. 2013;14:16365–16385. doi: 10.3390/ijms140816365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Markowitz SD, Bertagnolli MM. Molecular origins of cancer: Molecular basis of colorectal cancer. N Engl J Med. 2009;361:2449–2460. doi: 10.1056/NEJMra0804588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosenwald IB, Chen JJ, Wang S, Savas L, London IM, Pullman J. Upregulation of protein synthesis initiation factor eIF-4E is an early event during colon carcinogenesis. Oncogene. 1999;18:2507–2517. doi: 10.1038/sj.onc.1202563. [DOI] [PubMed] [Google Scholar]
- 13.Berkel HJ, Turbat-Herrera EA, Shi R, de Benedetti A. Expression of the translation initiation factor eIF4E in the polyp-cancer sequence in the colon. Cancer Epidemiol Biomarkers Prev. 2001;10:663–666. [PubMed] [Google Scholar]
- 14.Nathan CO, Franklin S, Abreo FW, Nassar R, De Benedetti A, Glass J. Analysis of surgical margins with the molecular marker eIF4E: a prognostic factor in patients with head and neck cancer. J. Clin. Oncol. 1999;17:2909–2914. doi: 10.1200/JCO.1999.17.9.2909. [DOI] [PubMed] [Google Scholar]
- 15.Graff JR, Konicek BW, Lynch RL, Dumstorf CA, Dowless MS, McNulty AM, Parsons SH, Brail LH, Colligan BM, Koop JW, Hurst BM, Deddens JA, Neubauer BL, Stancato LF, Carter HW, Douglass LE, Carter JH. eIF4E activation is commonly elevated in advanced human prostate cancers and significantly related to reduced patient survival. Cancer Res. 2009;69:3866–3873. doi: 10.1158/0008-5472.CAN-08-3472. [DOI] [PubMed] [Google Scholar]
- 16.Seki N, Takasu T, Mandai K, Nakata M, Saeki H, Heike Y, Takata I, Segawa Y, Hanafusa T, Eguchi K. Expression of eukaryotic initiation factor 4E in atypical adenomatous hyperplasia and adenocarcinoma of the human peripheral lung. Clin Cancer Res. 2002;8:3046–3053. [PubMed] [Google Scholar]
- 17.Sorrells DL, Meschonat C, Black D, Li BD. Pattern of amplification and overexpression of the eukaryotic initiation factor 4E gene in solid tumor. J Surg Res. 1999;85:37–42. doi: 10.1006/jsre.1999.5653. [DOI] [PubMed] [Google Scholar]
- 18.Culjkovic B, Borden KL. Understanding and Targeting the Eukaryotic Translation Initiation Factor eIF4E in Head and Neck Cancer. J Oncol. 2009;2009:981679. doi: 10.1155/2009/981679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Topisirovic I, Siddiqui N, Orolicki S, Skrabanek LA, Tremblay M, Hoang T, Borden KL. Stability of eukaryotic translation initiation factor 4E mRNA is regulated by HuR, and this activity is dysregulated in cancer. Mol Cell Biol. 2009;29:1152–1162. doi: 10.1128/MCB.01532-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.López de Silanes I, Fan J, Yang X, Zonderman AB, Potapova O, Pizer ES, Gorospe M. Role of the RNA-binding protein HuR in colon carcinogenesis. Oncogene. 2003;22:7146–7154. doi: 10.1038/sj.onc.1206862. [DOI] [PubMed] [Google Scholar]
- 21.DeFatta RJ, Turbat-Herrera EA, Li BD, Anderson W, De Benedetti A. Elevated expression of eIF4E in confined early breast cancer lesions: possible role of hypoxia. Int J Cancer. 1999;80:516–522. doi: 10.1002/(sici)1097-0215(19990209)80:4<516::aid-ijc6>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 22.Yi T, Papadopoulos E, Hagner PR, Wagner G. Hypoxia-inducible factor-1α (HIF-1α) promotes cap-dependent translation of selective mRNAs through up-regulating initiation factor eIF4E1 in breast cancer cells under hypoxia conditions. J Biol Chem. 2013;288:18732–18742. doi: 10.1074/jbc.M113.471466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan S, Ramalingam SS, Kauh J, Xu Z, Khuri FR, Sun SY. Phosphorylated eukaryotic translation initiation factor 4 (eIF4E) is elevated in human cancer tissues. Cancer Biol Ther. 2009;8:1463–1469. doi: 10.4161/cbt.8.15.8960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scheper GC, Proud CG. Does phosphorylation of the cap-binding protein eIF4E play a role in translation initiation? Eur J Biochem. 2002;269:5350–5359. doi: 10.1046/j.1432-1033.2002.03291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Diab S, Kumarasiri M, Yu M, Teo T, Proud C, Milne R, Wang S. MAP Kinase-Interacting Kinases-Emerging Targets against Cancer. Chem Biol. 2014;21:441–452. doi: 10.1016/j.chembiol.2014.01.011. [DOI] [PubMed] [Google Scholar]
- 26.Slepenkov SV, Darzynkiewicz E, Rhoads RE. Stopped-flow kinetic analysis of eIF4E and phosphorylated eIF4E binding to cap analogs and capped oligoribonucleotides: evidence for a one-step binding mechanism. J Biol Chem. 2006;281:14927–14938. doi: 10.1074/jbc.M601653200. [DOI] [PubMed] [Google Scholar]
- 27.McKendrick L, Morley SJ, Pain VM, Jagus R, Joshi B. Phosphorylation of eukaryotic initiation factor 4E (eIF4E) at Ser209 is not required for protein synthesis in vitro and in vivo. Eur J Biochem. 2001;268:5375–5385. doi: 10.1046/j.0014-2956.2001.02478.x. [DOI] [PubMed] [Google Scholar]
- 28.Martín ME, Pérez MI, Redondo C, Alvarez MI, Salinas M, Fando JL. 4E binding protein 1 expression is inversely correlated to the progression of gastrointestinal cancers. Int J Biochem Cell Biol. 2000;32:633–642. doi: 10.1016/s1357-2725(00)00007-8. [DOI] [PubMed] [Google Scholar]
- 29.Rojo F, Najera L, Lirola J, Jiménez J, Guzmán M, Sabadell MD, Baselga J, Ramon y Cajal S. 4E-binding protein 1, a cell signaling hallmark in breast cancer that correlates with pathologic grade and prognosis. Clin Cancer Res. 2007;13:81–89. doi: 10.1158/1078-0432.CCR-06-1560. [DOI] [PubMed] [Google Scholar]
- 30.Coleman LJ, Peter MB, Teall TJ, Brannan RA, Hanby AM, Honarpisheh H, Shaaban AM, Smith L, Speirs V, Verghese ET, McElwaine JN, Hughes TA. Combined analysis of eIF4E and 4E-binding protein expression predicts breast cancer survival and estimates eIF4E activity. Br J Cancer. 2009;100:1393–1399. doi: 10.1038/sj.bjc.6605044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Castellvi J, Garcia A, Ruiz-Marcellan C, Hernández-Losa J, Peg V, Salcedo M, Gil-Moreno A, Ramon y Cajal S. Cell signaling in endometrial carcinoma: phosphorylated 4E-binding protein-1 expression in endometrial cancer correlates with aggressive tumors and prognosis. Hum Pathol. 2009;40:1418–1426. doi: 10.1016/j.humpath.2008.12.019. [DOI] [PubMed] [Google Scholar]
- 32.Castellvi J, Garcia A, Rojo F, Ruiz-Marcellan C, Gil A, Baselga J, Ramon y Cajal S. Phosphorylated 4E binding protein 1: a hallmark of cell signaling that correlates with survival in ovarian cancer. Cancer. 2006;107:1801–1811. doi: 10.1002/cncr.22195. [DOI] [PubMed] [Google Scholar]
- 33.Yamaguchi S, Ishihara H, Yamada T, Tamura A, Usui M, Tominaga R, Munakata Y, Satake C, Katagiri H, Tashiro F, Aburatani H, Tsukiyama-Kohara K, Miyazaki J, Sonenberg N, Oka Y. ATF4-mediated induction of 4E-BP1 contributes to pancreatic beta cell survival under endoplasmic reticulum stress. Cell Metab. 2008;7:269–276. doi: 10.1016/j.cmet.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 34.Tettweiler G, Miron M, Jenkins M, Sonenberg N, Lasko PF. Starvation and oxidative stress resistance in Drosophila are mediated through the eIF4E-binding protein, d4E-BP. Genes Dev. 2005;19:1840–1843. doi: 10.1101/gad.1311805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Azar R, Lasfargues C, Bousquet C, Pyronnet S. Contribution of HIF-1α in 4E-BP1 gene expression. Mol Cancer Res. 2013;11:54–61. doi: 10.1158/1541-7786.MCR-12-0095. [DOI] [PubMed] [Google Scholar]
- 36.Azar R, Susini C, Bousquet C, Pyronnet S. Control of contact-inhibition by 4E-BP1 upregulation. Cell Cycle. 2010;9:1241–1245. doi: 10.4161/cc.9.7.11047. [DOI] [PubMed] [Google Scholar]
- 37.Azar R, Alard A, Susini C, Bousquet C, Pyronnet S. 4E-BP1 is a target of Smad4 essential for TGFbeta-mediated inhibition of cell proliferation. EMBO J. 2009;28:3514–3522. doi: 10.1038/emboj.2009.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parsons DW, Wang TL, Samuels Y, Bardelli A, Cummins JM, DeLong L, Silliman N, Ptak J, Szabo S, Willson JK, Markowitz S, Kinzler KW, Vogelstein B, Lengauer C, Velculescu VE. Colorectal cancer: mutations in a signalling pathway. Nature. 2005;436:792. doi: 10.1038/436792a. [DOI] [PubMed] [Google Scholar]
- 39.Azar R, Najib S, Lahlou H, Susini C, Pyronnet S. Phosphatidylinositol 3-kinase-dependent transcriptional silencing of the translational repressor 4E-BP1. Cell Mol Life Sci. 2008;65:3110–3117. doi: 10.1007/s00018-008-8418-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Graff JR, Konicek BW, Carter JH, Marcusson EG. Targeting the eukaryotic translation initiation factor 4E for cancer therapy. Cancer Res. 2008;68:631–644. doi: 10.1158/0008-5472.CAN-07-5635. [DOI] [PubMed] [Google Scholar]
- 41.O’Reilly KE, Warycha M, Davies MA, Rodrik V, Zhou XK, Yee H, Polsky D, Pavlick AC, Rosen N, Bhardwaj N, Mills G, Osman I. Phosphorylated 4E-BP1 is associated with poor survival in melanoma. Clin Cancer Res. 2009;15:2872–2878. doi: 10.1158/1078-0432.CCR-08-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salehi Z, Mashayekhi F. Expression of the eukaryotic translation initiation factor 4E (eIF4E) and 4E-BP1 in esophageal cancer. Clin Biochem. 2006;39:404–409. doi: 10.1016/j.clinbiochem.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 43.Seki N, Takasu T, Sawada S, Nakata M, Nishimura R, Segawa Y, Shibakuki R, Hanafusa T, Eguchi K. Prognostic significance of expression of eukaryotic initiation factor 4E and 4E binding protein 1 in patients with pathological stage I invasive lung adenocarcinoma. Lung Cancer. 2010;70:329–334. doi: 10.1016/j.lungcan.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 44.Alain T, Sonenberg N, Topisirovic I. mTOR inhibitor efficacy is determined by the eIF4E/4E-BP ratio. Oncotarget. 2012;3:1491–1492. doi: 10.18632/oncotarget.799. [DOI] [PMC free article] [PubMed] [Google Scholar]


