Abstract
HMGB3, an X-linked member of the high-mobility group (HMG) superfamily of HMG proteins, has been shown to affect numerous tumorigenic progression. However, the expression and the prognostic role of HMGB3 in esophageal squamous cell carcinoma (ESCC) remained unknown. In this study, we examined the HMGB3 expression in ESCC tissues and adjacent nontumorous tissues by qRT-PCR and immuohistochemistry. Statistical analyses were applied to test for prognostic and diagnostic associations. The mRNA levels of HMGB3 were found to be significantly higher in tumorous tissues than in the adjacent normal tissues. We found that the HMGB3 expression was higher in tumorous tissues than in the adjacent non-tumorous tissues by immunohistochemical analysis of paired tissue specimens (P < 0.001). Moreover, there was a significant correlation between HMGB3 expression and gender (P = 0.037), clinical stage (P = 0.038), T classification (P = 0.013) and N classification (P = 0.017). Patients with higher HMGB3 expression had shorter overall survival than those with lower HMGB3 expression. Multivariate Cox analysis indicated that HMGB3 expression is an independent prognostic factor for overall survival (HR = 0.591, 95% CI = 0.379-0.793, P = 0.039). In summary, these findings demonstrate that HMBG3 may be a potential molecular marker for predicting the prognosis of ESCC patients.
Keywords: HMGB3, esophageal cancer, immunohistochemistry, real-time PCR, prognosis, biomarker
Introduction
Esophageal cancer is the sixth leading cause of cancer mortalities worldwide. ESCC, which is one of the predominant histological subtype of esophageal cancer, occurs at a very high frequency rate in China [1]. Despite the development of rapid advances in surgical techniques, radiotherapy and chemotherapy, the prognosis in patients with ESCC remains poor. The survival rate is being estimated only 15% at 5 years and remains less than 20-30% at 2 years [2]. Esophageal squamous cell carcinogenesis is multifactorial process with influence of genetic predisposition, local environmental conditions and lifestyle [3]. Genetic factors may play a pivotal role in the etiology of esophageal cancer. Studies showed that the regions with high incidence rate of esophageal cancer such as the Shanxi province in north central China have shown a tendency toward familial aggregation [4,5]. The risk was increased by 70% among those whose parents had esophageal cancer while only 20% among those whose spouses had such cancer. Thus, searching for molecular changes that occur during esophageal squamous cell carcinogenesis and progression could discover novel prognostic biomarkers to guide individualized treatments and targeted therapy. However, few genes have been identified to be associated with pathological features of ESCC, implicating that novel genes involved in the carcinogenesis and progression of ESCC need to be identified.
High mobility group-box 3 (HMGB3), a recently discovered member of high mobility group box subfamily, was originally identified in an EST database and has been mapped to X chromosome band q28. HMG-Box subfamily containing HMGB1 and HMGB2 plays an important role in various processes of cancer progression, including cell proliferation, angiogenesis, invasion, and metastasis [6-8]. HMGB3 mRNA is present at relatively high levels in adult mouse bone marrow erythroid and primitive progenitor cells. Recently, studies showed that HMGB3 plays a significant role in regulating the balance between proliferation and differentiation in primitive stages of hematopoiesis [9]. In addition, it was known that HMGB3 and NPU98 fusion protein forms a new oncogenic gene in acute myeloblastic leukemia [10]. Ola ea al. reported that overexpression of HMGB3 mRNA are found in metastatic breast cancers, that knockdown of HMGB3 decreases invasiveness in vitro and patients with increased HMGB3 expression have poor survival [11].
Although HMGB1, HMGB2 and HMGB3 are homologous genes in which their amino acid sequence exhibits 80% homology, and both HMGB1 and HMGB2 are potential oncogenes [12], the role of HMGB3 in cancer is still unclear. Until now, limited studies have been done to evaluate the relationship between HMGB3 expression and cancer progression, such as breast [11], gastric [13] and non-small-cell lung cancer (NSCLC) [14]. Although HMGB3 may play an important role in carcinogenesis, there is no report on its role in tumorigenesis and progression of esophageal carcinoma. To explore the exact role of HMGB3 in ESCC, we investigated whether the expression of HMGB3 protein is different between tumor tissues and normal tissues, whether HMGB3 has any role in the development and progression of ESCC, and whether HMGB3 is a prognostic factor in ESCC after curative surgical treatment.
Materials and methods
Patients and tumor specimens
Twenty-three pairs of fresh ESCC tissue specimens and corresponding nontumorous specimens were obtained from patients with ESCC who underwent surgical esophageal tissue resection at the Cancer Center of Sun Yat-sen University. All excised samples were obtained within 1 h after operation and immediately placed in liquid nitrogen until further analysis. In addition, a total of 194 paraffin-embedded ESCC samples and 95 adjacent normal esophageal tissue samples, which were histologically and clinically diagnosed between 2002 and 2007 at the Cancer Center of Sun Yat-sen University were also included in this study. Prior to the use of these clinical materials for investigation, patients’ consent and approval from the Institute Research Ethics Committee were obtained. Clinical and pathological data of the 194 patients with ESCC were collected, such as age, tumor size, stage, differentiation grade, lymph node metastases, treatment and recurrence. There were 115 male and 79 female, with a median age of 60 years (ranging from 32 to 80 years). The All patients’ disease stages were classified according to the pathological TNM classification. Clinical follow-up information was obtained by telephone or from the outpatients’ records. A total of 137 (70.6%) patients died during follow-up.
Real-time PCR (RT-PCR)
Total RNA from human tissues was extracted using TRIzol solution (Invitrogen, USA) according to the manufacturer’s protocol. Reverse transcription was performed, according to the manufacturer’s recommendations. The cDNA was subjected to real-time quantitative PCR for the evaluation of the relative mRNA levels of HMGB3 and GAPDH (as an internal control) with the corresponding primer pairs (.HMGB3 sense strand: 5’-GACCAGCTAAGGGAGGCAA-3’, HMGB3 antisense strand: 5’-ACAGGAAGAATCCAGACGGT-3’, GAPDH sense strand: 5’-CTCCTCCTGTTCGACAGTCAGC-3’, GAPDH antisense strand: 5’-CCCAATACGACCAAATCCGTT-3’). Real-time PCR and data collection were performed with an ABI PRISM 7900HT sequence detection system. To ensure reproducibility of results, all reactions were run in triplicate in three independent experiments.
Immunohistochemical analysis
Immunohistochemistry was performed as described previously [15]. In brief, paraffin-embedded specimens were cut into 4 mm sections and baked at 65°C for 1 hour. The sections were deparaffined with xylenes and rehydrated, submerged into 0.01M citrate buffer (PH 6.0) antigen retrieval bufferm, and microwaved for antigenic retrieval. They were treated with 0.3% H2O2 for 15 min to block the endogenous peroxidase at RT, and then were treated with normal goat serum for 30 min to reduce the nonspecific binding. Consequently, the sections were incubated with rabbit polyclonal anti-HMGB3 antibody (1:200) overnight at 4°C. After being washed, the sections were treated with MaxVisionTM HRP-Polymer anti-rabbit HIC Kit (Maixin Bio; Fujian, China) at 37°C for 30 min. The tissue sections were immersed in 3-amino-9-ethyl carbazole, counterstained with Mayer’s hematoxylin, dehydrated, and finally mounted in Crystal Mount.
The immunohistochemically stained tissue sections were scored independently by two pathologists blinded to the clinical parameters. The final score for HMGB3 was the average of the scores obtained by the two observers. The intensity and extent of the staining were used as criteria of evaluation. The staining intensity was scored as 0 (no staining), 1 (light yellow), 2 (yellow brown), or 3 (brown). Extent of staining was scored as 0 (0%), 1 (1 to 5%), 2 (6 to 25%), 3 (26 to 50%), 4 (51 to 75%), or 5 (76 to 100%), according to the percentages of the positive staining areas in relation to the whole carcinoma area or entire section for the normal samples. Staining index was calculated as the multiplication of staining intensity score and the proportion of HMGB3-positive tumor cells. We evaluated HMGB3 expression in benign esophageal tissue and malignant lesions on the basis of the staining index values, with scores of 0, 1, 2, 3, 4, 5, 6, 8, 9, 10, 12, and 15. An optimal cutoff value was identified: a staining index score of ≥ 5 was considered as high HMGB3 expression, whereas a staining index score of ≤ 4 was considered as lowHMGB3 expression.
Statistical analysis
All statistical analyses were carried out using the SPSS18.0 statistical software package. In the RT-PCR and immunohistochemical assays, paired-sample t tests were used to analyze the significance of the differences in mRNA and protein expression between ESCCs and the adjacent normal tissues. The Chi-square and Fisher’s exact tests were used to analyze the relationship between HMGB3 expression and clinical-pathological characteristics. Survival curves were plotted by the Kaplan-Meier method and compared by the log-rank test. Univariate and multivariate regression analyses were performed with the Cox proportional hazards regression model to analyze independent factors affecting prognosis. P-value of less than 0.05 was considered to be statistically significant.
Results
Expression of HMGB3 in ESCC tissues
To determine the expression of HMGB3 in esophageal carcinoma biopsies, quantitative RT-PCR was performed in 23 esophageal cancer and nontumorous tissue samples obtained from the same patients. In 23 tumor samples, the mRNA levels of HMGB3 were significantly higher than that in nontumorous samples (P < 0.01, Figure 1). To determine whether the higher level of HMGB3 mRNA expression revealed by Real time PCR analysis was directly linked to increased level of HMGB3 protein expression, we further examined the subcellular expression of HMGB3 protein in 95 paraffin-embedded ESCC samples and 95 paired specimens of adjacent normal tissues by immunohistochemical analysis (Figure 2). In ESCC tissue, HMGB3 protein mainly expressed in the nuclei of cells (Figure 3). The HMGB3 protein expression in the 95 tumor tissue samples was higher than that in the adjacent normal tissue samples (P < 0.01).
Figure 1.
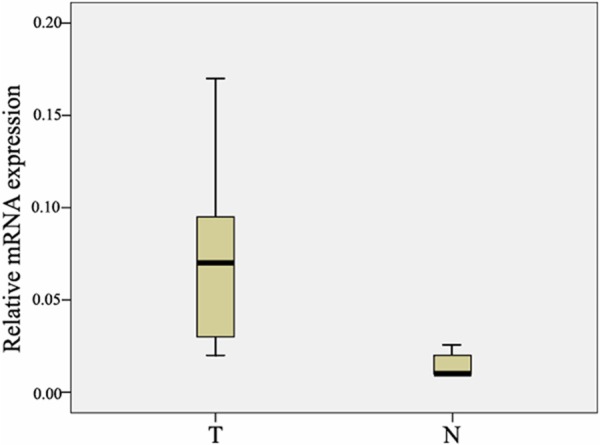
Real-time quantitative RT-PCR analysis of HMGB3 expression. The relative expression of HMGB3 mRNA in ESCC tumor tissues samples was lower than that in the paired adjacent normal (N) tissue samples (n = 23, P < 0.01). The bottom and the top of the box represent whisker represent the 25th and the 75th percentile, respectively, and the band near the middle of the box is the 50th percentile (the median). The ends of the whiskers represent the 2.5th percentile and the 97.5 percentile.
Figure 2.
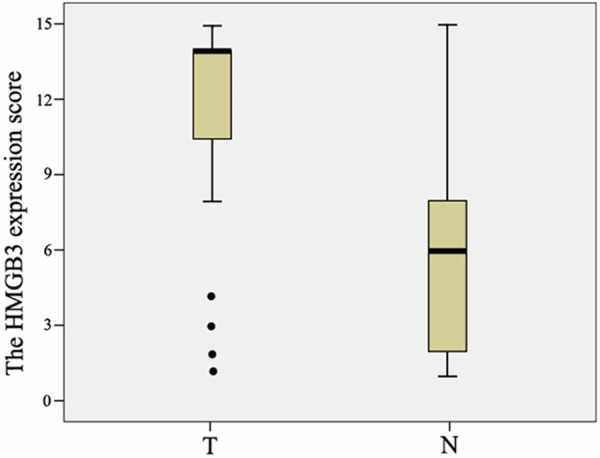
Increased protein expression of HMGB3 in ESCC. The relative protein of HMGB3 in ESCC tumor (T) tissue samples was higher than that in the paired adjacent normal (N) tissue samples (n = 90, P < 0.01). The bottom and the top of the box are the lower and upper quartiles, and the band near the middle of the box is the median. The ends of the whiskers represent the 2.5th and the 97.5th percentile. Four black spots represent the special value outliers.
Figure 3.
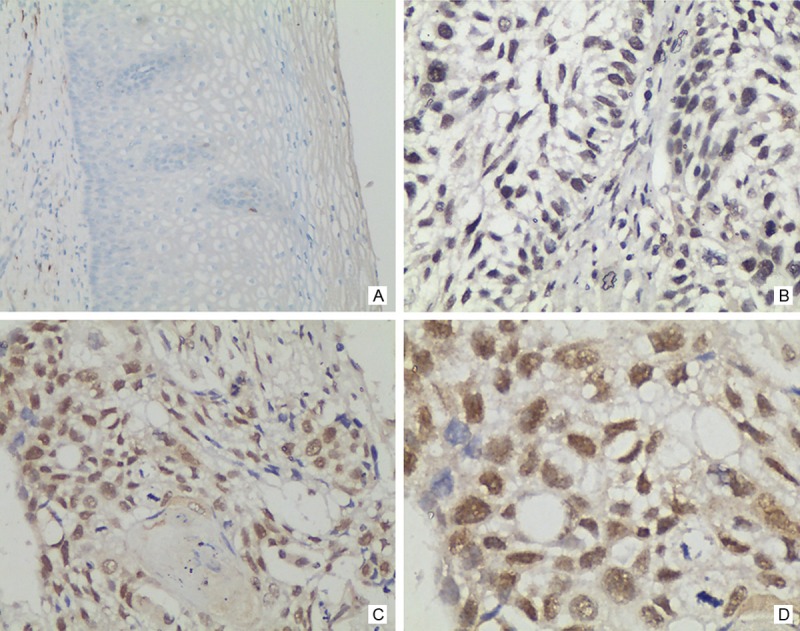
Analysis of HMGB3 protein by immunohistochemistry in ESCC. HMGB3 expression was mainly localized within nuclei of tumor cells, and its expression was observed in tumor cells. A. Negative HMGB3 staining in a non-cancerous tissue sample (400×). B. Weak HMGB3 staining in esophageal carcinoma tissues (400×). C and D. Strong HMGB3 staining in esophageal carcinoma tissues (200× and 400×, respectively).
Relationship between HMGB3 protein expression and clinicopathological parameters
To further investigate the effect and prognostic value of HMGB3, immunohistochemical analysis was performed to assess the expression of HMGB3 in 194 ESCC tissue blocks. Overall, 113 of 194 ESCC cases (58.2%) showed high expression, whereas 81 samples showed low expression. The correlation between the expression of HMGB3 and clinical characteristics were listed in Table 1. The expression of HMGB3 was closely associated with stage of esophageal cancer patients (P = 0.038), N classification (P = 0.017) and T classification (P = 0.013). Higher staging, N classification and T classification correlated with higher HMGB3 expression. However, there was no significant correlation between the expression level of HMGB3 and age and histological differentiation of esophageal cancer patients.
Table 1.
Correlation Between Patient’s Clinicopathologic Features and the Expression of HMGB3 Protein
| No. (n = 194) | HMGB3 | |||
|---|---|---|---|---|
|
|
||||
| High expressions | Low expressions | P | ||
| Age | ||||
| > 60 | 131 | 74 | 57 | |
| < 60 | 63 | 39 | 24 | 0.474 |
| Sex | ||||
| Male | 115 | 71 | 44 | |
| Female | 79 | 42 | 37 | 0.037 |
| Histological differentiation | ||||
| Well/moderate | 135 | 83 | 52 | |
| Poor | 59 | 30 | 29 | 0.234 |
| Clinical stage | ||||
| I | 6 | 3 | 3 | |
| II | 109 | 57 | 52 | 0.038* |
| III | 67 | 42 | 25 | |
| IV | 12 | 11 | 1 | |
| pT classification | ||||
| T1 | 6 | 3 | 3 | |
| T2 | 59 | 25 | 34 | |
| T3 | 104 | 66 | 38 | 0.013# |
| T4 | 15 | 9 | 6 | |
| pN classification | ||||
| No | 93 | 46 | 47 | 0.017 |
| Yes | 101 | 67 | 34 | |
Stage I+II versus III + IV;
pT1-2 versus pT3-4.
Correlation of HMGB3 expression with overall survival
We next evaluated whether the level of HMGB3 expression was associated with patient prognosis. As shown in Figure 4, the expression level of HMGB3 in esophageal carcinoma was significantly correlated with patients’ survival time (P < 0.01), indicating that higher level of HMGB3 expression was correlated with shorter survival time. The low HMGB3 expression group had better survival, whereas the high HMGB3 expression group had shorter survival. The median survival of patients with high HMGB3 expression was much shorter (27 months) than those with low HMGB3 expression (44 months). Furthermore, the relationship of HMGB3 expression with prognosis was determined in 194 patients, which were divided into 2 subgroups depending on the pathologic stage. Patients with tumors exhibiting low HMGB3 expression had significantly longer overall survival than those with high expression of HMGB3 either in the stage I plus II subgroup (n = 115; log-rank, P = 0.023; Figure 5A), the stage III plus IV subgroup (n = 69; log rank, P = 0.036; Figure 5B).
Figure 4.
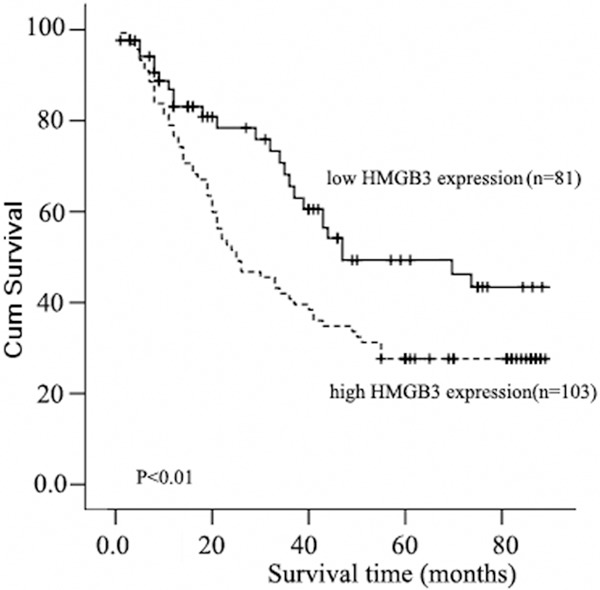
Kaplan-Meier curves with univariate analyses (log-rank) for patients with low HMGB3 expression (bold line) versus high HMGB3 expressing tumors (dotted line). The median survival of patients with high HMGB3 expression was much shorter (23 months) than those with low HMGB3 expression (44 months) (P < 0.01, Log-rank).
Figure 5.
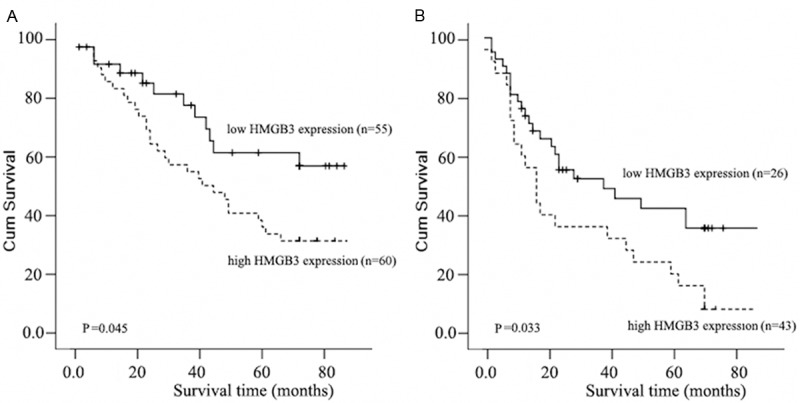
Kaplan-Meier analysis showing the overall survival of esophageal carcinoma patients categorized according to the Pathological stage and status of HMGB3 expression. The statistical significance of the difference between curves of HMGB3 high-expressing and low-expressing patients was compared in I-II (A) and III-IV (B) patient subgroups. Plot shows the survival rate for patients with low HMGB3 expression (bold line) versus high HMGB3 expressing tumors (dotted line). P values were calculated by the log-rank test.
In addition, univariate Cox regression analysis showed that gender, stage and N classification were also significantly correlated with overall survival (for stage, P = 0.013 and for gender, P = 0.017; for N classification, P = 0.009). We thereafter confirmed multivariate survival analysis, which included HMGB3 expression level, stage, N classification and gender, to determine whether HMGB3 expression level is an independent prognostic factor for outcomes. In this analysis, N classification and HMGB3 expression were recognized as independent prognostic factors (Table 2). Thus, our findings indicated that HMGB3 expression exerted an obvious correlation with poor prognosis of ESCC patients.
Table 2.
Univariate and Multivariate Analysis of Different Prognostic Variables in Patients with ESCC by Cox
| Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|
|
|
||||
| P | HR (95% CI) | P | HR (95% CI) | |
| Gender | ||||
| Female vs. Male | 0.017 | 1.425 (1.079-2.018) | 0.054 | 1.436 (1.012-2.157) |
| Clinical stage | ||||
| III-IV vs. I-II | 0.013 | 1.371 (1.006-2.054) | 0.076 | 1.227 (0.953-1.493) |
| pN classification | ||||
| Yes vs. no | 0.009 | 1.547 (1.038-2.158) | 0.004 | 1.675 (1.032-2.317) |
| Expression of HMGB3 | ||||
| low vs. high | 0.010 | 0.743 (0.501-0.964) | 0.039 | 0.591 (0.379-0.793) |
Discussion
HMGB3 is originally identified in an EST database and has been mapped to X chromosome band q28. HMG-Box family plays an important role in DNA replication, transcription, recombination and repair, etc [16,17]. HMGB3 plays a significant role in regulating the balance between proliferation and differentiation in primitive stages of hematopoiesis. A few studies have been done in gastric adenocarcinoma, NSCLC and breast cancer to evaluate the role of HMGB3 in cancer, however, little has been known about the expression of HMGB3 in ESCC tissue. The present study showed the first evidences that the status of HMGB3 expression in tumor tissues is much higher than that in paracarcinoma tissues in ESCC, and HMGB3 is an independent prognostic factor for ESCC patients.
In this study, we showed that HMGB3 mRNA and protein expression were significantly different between the ESCC and the adjacent normal tissue samples. Furthermore, immunohistochemical analysis showed that HMGB3 expression was high in ESCCs, while it was low to moderate in the adjacent normal tissues. Accordingly, we found that HMGB3 expression was increased in a large number of human clinical ESCC samples. The expression of HMGB3 protein in male patients was higher than in female patients. Generally, the male patients had better survival, whereas the female patients had shorter survival. Our analysis showed the same results, in accordance with the relation analysis between the expression of HMGB3 protein and the overall survival (Figure 4). Patients with higher HMGB3 expression had a shorter survival time, and those with lower HMGB3 expression had a longer survival time. There was a significant relationship of HMGB3 expression in patients categorized according to stage (P = 0.038), T classification (P = 0.013) and N classification (P = 0.017), strongly indicating that HMGB3 can be used as a marker to identify subsets of ESCC cancer patients with more aggressive disease. In addition, the relationship of HMGB3 expression with prognosis was determined in the patients, which were divided into 2 subgroups depending on the pathologic stage. We found that HMGB3 could be a valuable prognostic marker for ESCC patients at all disease stages. Furthermore, multivariate Cox regression analyses revealed that HMGB3 expression was an independent predictor for the overall survival of ESCC patients. Thus, our findings indicate that the HMGB3 expression level has a significant correlation with clinicopathological features and is a potential prognostic marker for ESCC patients. Our results are consistent with previous reports in some other cancer types, including breast cancer and lung cancer.
Because HMGB3 is highly homologous to HMGB1, it may have similar effects with regard to neoplastic development. Overexpression of HMGB1 has been observed in several cancers including ESCC [18]. Furthermore, HMGB1 has been shown to stimulate the migration of smooth muscle cells and fibroblasts, whereas anti-HMGB1 antibodies inhibited the migration of neuroblastoma and glioma cells. ESCC tissues have been reported to overexpress receptors for advanced glycation end products that could relay HMGB signals, indicating a potentially important function of HMGBs [19]. HMGB3 has been reported to promote the cell proliferation rate of retinal progenitor cells in Xenopus [20]. They further discovered that this is accompanied by P27 downregulation [21]. Recently, HMGB3 has been shown to functionally interact with Wnt signaling to regulate hematopoietic stem cell self-renewal and differentiation [9]. These possible functional links of HMGB3 to other important cellular regulators in the context of ESCC development and progression warrant further investigation.
In conclusion, this is the first study highlighting the clinical significance of HMGB3 in ESCC, and higher HMGB3 expression is also a significant prognostic marker of poor survival in ESCC patients.
Acknowledgements
This study was supported by grants from National Natural Science Foundation of China (Grant No. 81402187 to Z-Z Zou).
Disclosure of conflict of interest
None.
References
- 1.Li JY. Epidemiology of esophageal cancer in China. Natl Cancer Inst Monogr. 1982;62:113–120. [PubMed] [Google Scholar]
- 2.Bosset JF, Gignoux M, Triboulet JP, Tiret E, Mantion G, Elias D, Lozach P, Ollier JC, Pavy JJ, Mercier M, Sahmoud T. Chemoradiotherapy followed by surgery compared with surgery alone in squamous-cell cancer of the esophagus. N Engl J Med. 1997;337:161–167. doi: 10.1056/NEJM199707173370304. [DOI] [PubMed] [Google Scholar]
- 3.Mir MM, Dar NA, Gochhait S, Zargar SA, Ahangar AG, Bamezai RN. p53 mutation profile of squamous cell carcinomas of the esophagus in Kashmir (India): a high-incidence area. Int J Cancer. 2005;116:62–68. doi: 10.1002/ijc.21002. [DOI] [PubMed] [Google Scholar]
- 4.Chang-Claude J, Becher H, Blettner M, Qiu S, Yang G, Wahrendorf J. Familial aggregation of oesophageal cancer in a high incidence area in China. Int J Epidemiol. 1997;26:1159–1165. doi: 10.1093/ije/26.6.1159. [DOI] [PubMed] [Google Scholar]
- 5.Hu N, Dawsey SM, Wu M, Bonney GE, He LJ, Han XY, Fu M, Taylor PR. Familial aggregation of oesophageal cancer in Yangcheng County, Shanxi Province, China. Int J Epidemiol. 1992;21:877–882. doi: 10.1093/ije/21.5.877. [DOI] [PubMed] [Google Scholar]
- 6.Naghavi MH, Nowak P, Andersson J, Sonnerborg A, Yang H, Tracey KJ, Vahlne A. Intracellular high mobility group B1 protein (HMGB1) represses HIV-1 LTR-directed transcription in a promoter- and cell-specific manner. Virology. 2003;314:179–189. doi: 10.1016/s0042-6822(03)00453-7. [DOI] [PubMed] [Google Scholar]
- 7.Kostova N, Zlateva S, Ugrinova I, Pasheva E. The expression of HMGB1 protein and its receptor RAGE in human malignant tumors. Mol Cell Biochem. 2010;337:251–258. doi: 10.1007/s11010-009-0305-0. [DOI] [PubMed] [Google Scholar]
- 8.Tang D, Kang R, Zeh HJ 3rd, Lotze MT. High-mobility group box 1 and cancer. Biochim Biophys Acta. 2010;1799:131–140. doi: 10.1016/j.bbagrm.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nemeth MJ, Kirby MR, Bodine DM. Hmgb3 regulates the balance between hematopoietic stem cell self-renewal and differentiation. Proc Natl Acad Sci U S A. 2006;103:13783–13788. doi: 10.1073/pnas.0604006103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Somervaille TC, Matheny CJ, Spencer GJ, Iwasaki M, Rinn JL, Witten DM, Chang HY, Shurtleff SA, Downing JR, Cleary ML. Hierarchical maintenance of MLL myeloid leukemia stem cells employs a transcriptional program shared with embryonic rather than adult stem cells. Cell Stem Cell. 2009;4:129–140. doi: 10.1016/j.stem.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elgamal OA, Park JK, Gusev Y, Azevedo-Pouly AC, Jiang J, Roopra A, Schmittgen TD. Tumor suppressive function of mir-205 in breast cancer is linked to HMGB3 regulation. PLoS One. 2013;8:e76402. doi: 10.1371/journal.pone.0076402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moleri S, Cappellano G, Gaudenzi G, Cermenati S, Cotelli F, Horner DS, Beltrame M. The HMGB protein gene family in zebrafish: Evolution and embryonic expression patterns. Gene Expr Patterns. 2011;11:3–11. doi: 10.1016/j.gep.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 13.Gong Y, Cao Y, Song L, Zhou J, Wang C, Wu B. HMGB3 characterization in gastric cancer. Genet Mol Res. 2013;12:6032–6039. doi: 10.4238/2013.December.2.1. [DOI] [PubMed] [Google Scholar]
- 14.Song N, Liu B, Wu JL, Zhang RF, Duan L, He WS, Zhang CM. Prognostic value of HMGB3 expression in patients with non-small cell lung cancer. Tumour Biol. 2013;34:2599–2603. doi: 10.1007/s13277-013-0807-y. [DOI] [PubMed] [Google Scholar]
- 15.Mi YJ, Gao J, Xie JD, Cao JY, Cui SX, Gao HJ, Yao SP, Liu T, Zhang YY, Guo CH, Qiu GQ, Chen YQ. Prognostic relevance and therapeutic implications of centromere protein F expression in patients with esophageal squamous cell carcinoma. Dis Esophagus. 2013;26:636–643. doi: 10.1111/dote.12002. [DOI] [PubMed] [Google Scholar]
- 16.Agresti A, Bianchi ME. HMGB proteins and gene expression. Curr Opin Genet Dev. 2003;13:170–178. doi: 10.1016/s0959-437x(03)00023-6. [DOI] [PubMed] [Google Scholar]
- 17.Stros M. HMGB proteins: interactions with DNA and chromatin. Biochim Biophys Acta. 2010;1799:101–113. doi: 10.1016/j.bbagrm.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 18.Chuangui C, Peng T, Zhentao Y. The expression of high mobility group box 1 is associated with lymph node metastasis and poor prognosis in esophageal squamous cell carcinoma. Pathol Oncol Res. 2012;18:1021–1027. doi: 10.1007/s12253-012-9539-3. [DOI] [PubMed] [Google Scholar]
- 19.Jing RR, Cui M, Sun BL, Yu J, Wang HM. Tissue-specific expression profiling of receptor for advanced glycation end products and its soluble forms in esophageal and lung cancer. Genet Test Mol Biomarkers. 2010;14:355–361. doi: 10.1089/gtmb.2009.0064. [DOI] [PubMed] [Google Scholar]
- 20.Terada K, Kitayama A, Kanamoto T, Ueno N, Furukawa T. Nucleosome regulator Xhmgb3 is required for cell proliferation of the eye and brain as a downstream target of Xenopus rax/Rx1. Dev Biol. 2006;291:398–412. doi: 10.1016/j.ydbio.2005.12.029. [DOI] [PubMed] [Google Scholar]
- 21.Terada K, Furukawa T. Sumoylation controls retinal progenitor proliferation by repressing cell cycle exit in Xenopus laevis. Dev Biol. 2010;347:180–194. doi: 10.1016/j.ydbio.2010.08.023. [DOI] [PubMed] [Google Scholar]


