Abstract
Introduction: Long non-coding RNAs (lncRNAs) have been demonstrated to play key roles in tumorgenesis, and the lncRNA LET is down-regulated in several cancers. However, little is known about the function of lncRNA LET in human cervical cancer. The aim of this study was to investigate the clinical significance of lncRNA LET expression in cervical cancer. Methods: We examined the expression of lncRNA LET in 94 cervical cancer tissues and matched adjacent non-tumor tissues using quantitative real-time PCR and analyzed its correlation with the clinicopathological features. Results: The results showed that lncRNA LET expression in cervical cancer tissues was significantly down-regulated compared with the adjacent non-tumor tissues (P < 0.05). Decreased lncRNA LET expression was significantly correlated with FIGO stage, lymph node metastasis, and depth of cervical invasion (P < 0.05), but not other clinical characteristics. Moreover, cervical cancer patients with lncRNA LET lower expression have shown significantly poorer overall survival than those with higher lncRNA LET expression (P < 0.05). Univariate and multivariate analyses suggested that lncRNA LET expression served as an independent predictor for overall survival. Conclusions: Our data provided the first evidence that lncRNA LET may represent a prognostic marker and a potential therapeutic target for cervical cancer.
Keywords: lncRNA LET, cervical cancer, quantitative real-time PCR, prognosis
Introduction
Cervical cancer is the third most common cancer in women worldwide and is one of the main causes of cancer-related death in the developing countries [1]. Cervical cancer accounts for approximately 12% of all cancers in women. Annually, approximately 500,000 women develop cervical cancer worldwide, and approximately 200,000 die of the disease [2]. The clinical stage is very important in relation to the prognosis for patients with cervical cancer. The prognosis depends on the stage of the disease at the time of diagnosis, and the 5-year survival rate for all stages of cervical cancer combined is approximately 70%, however, patients with lymph node metastasis is only 20% to 30% [3]. Therefore, it is still urgently for us to find new and effective biomarkers for early stage diagnosis and potential targets for cervical cancer.
With the development of whole genome sequencing technology, it was determined that less than 2% of the mammalian genome is in protein encoded regions and the remainder is in non-coding RNAs [4]. Among them are long non-coding RNAs (lncRNAs), which are more than 200 nucleotides in length and unable to be translated into proteins [5]. Recently, many lncRNAs are known to play important roles in cellular development, differentiation, and many other biological processes [6-8]. For example, Gupta et al showed lncRNA HOTAIR was increased in expression in primary breast tumors and metastases, and HOTAIR expression level in primary tumors was a powerful predictor of eventual metastasis and death [9]. Shi et al suggested that GAS5 was a tumor suppressor in non small cell lung cancer (NSCLC), and the action of GAS5 was mediated by p53-dependent and p53-independent pathways. GAS5 could serve as a potential diagnostic marker for NSCLC and may be a novel therapeutic target in patients with NSCLC [10]. Lai et al showed MALAT1 was an independent prognostic factor for hepatocellular carcinoma recurrence after liver transplantation. Depletion of MALAT1 in HepG2 cells reduces cell viability, motility, and invasiveness and increases the sensitivity to apoptosis pointing towards a functional role of MALAT1 in liver cancer progression [11]. Zhang et al demonstrated that lncRNA SPRY4-IT1 was up-regulated in renal cancer and associated with advanced clinical stage and poorer prognosis, in vitro assay they showed knockdown SPRY4-IT1 expression could reduce renal cell proliferation, invasion, and migration [12]. However, to our knowledge, the prognostic significance of lncRNA LET in cervical caner is still unknown.
In the present study, qRT-PCR assay was performed to detect the expression of lncRNA LET in cervical cancer and adjacent non-tumor tissues. Moreover, the correlations of lncRNA LET expression with clinicopathologic features of cervical cancer patients were statistically analyzed. Finally, we determined the potential role of lncRNA LET in cervical cancer prognostic prediction. Our data showed that lncRNA LET was significantly down-regulated in cervical cancer and could be served as a potential molecular biomarker for the prediction of poor prognosis.
Materials and methods
Patients and specimens
A total of 94 cervical cancer tumors and matched adjacent non-tumor tissues were collected at the Department of Gynaecology and Obstetrics of The First Affiliated Hospital of Xinxiang Medical University between 2005 and 2008. All patients recruited in this study were not subjected to preoperative radiotherapy and/or chemotherapy and were diagnosed as infiltrating carcinoma by pathology. The tumor stage was classified by two experienced gynecological oncologists according to the International Federation of Gynecology and Obstetrics (FIGO) staging system for cervical cancer [13]. Clinical and pathological variables analyzed are shown in Table 1. All patients were regularly followed up, with a mean observation period of 46 months. After surgical resection, tumor specimens and adjacent non-tumor tissues were collected and stored in liquid nitrogen until use. The study was approved by the Medical ethics committee of The First Affiliated Hospital of Xinxiang Medical University.
Table 1.
Association between lncRNA LET expression and different clinicopathological features of human cervical cancers
| Clinicopathological features | Total | lncRNA LET expression | P value | |
|---|---|---|---|---|
|
| ||||
| Low | High | |||
| Age | ||||
| < 35 | 44 | 23 | 21 | 0.867 |
| ≥ 35 | 50 | 27 | 23 | |
| Tumor size (cm) | ||||
| < 4.0 | 38 | 20 | 18 | 0.929 |
| ≥ 4.0 | 56 | 30 | 26 | |
| Histology | ||||
| Squamous | 72 | 39 | 33 | 0.732 |
| Adenocarcinoma | 22 | 11 | 11 | |
| Differentiation | ||||
| Well + Moderate | 50 | 22 | 28 | 0.057 |
| Poor | 44 | 28 | 16 | |
| FIGO stage | ||||
| Ib~IIa | 48 | 16 | 32 | 0.000 |
| IIb~IIIa | 46 | 34 | 12 | |
| Lymph node metastasis | ||||
| No | 63 | 27 | 36 | 0.004 |
| Yes | 31 | 23 | 8 | |
| Depth of cervical invasion | ||||
| < 2/3 | 56 | 19 | 37 | 0.000 |
| ≥ 2/3 | 38 | 31 | 7 | |
RNA extraction and quantitative real-time PCR analyses
Total RNAs were extracted from tissues using Trizol (Invitrogen) following the manufacturer’s protocol. RT and qPCR kits were used to evaluate expression of lncRNA LET from tissue samples. The 20 μL RT reactions were performed using a PrimeScript® RT reagent Kit (Takara) and incubated for 30 min at 37°C, 5 s at 85°C, and then maintained at 4°C. For real-time PCR, 1 μL diluted RT products were mixed with 10 μL of 2×SYBR® Premix ExTaq™ (Takara), 0.6 μL forward and reverse primers (10 μM), and 8.4 μL Nuclease-free water in a final volume of 20 μL according to manufacturer instructions. The primers used in this study were 5’-CCTTCCTGACAGCCAGTGTG-3’ (forward) and 5’-CAGAATGGAAATACTGGAGCAAG-3’ (reverse). All reactions were run on the Eppendorf Mastercycler EP Gradient S (Eppendorf). Real-time PCR was done in triplicate, including no-template controls. Amplification of the appropriate product was confirmed by melting curve analysis following amplification. Relative expression of lncRNA LET was calculated using 2−ΔΔCT method with GAPDH as the endogenous control to normalize the data.
Statistical analysis
All computations were carried out using the software of SPSS version 18.0 (IBM). Data were expressed as mean ± SD. The analysis of variance (ANOVA) was used to determine the statistical differences among the groups. The Kaplan-Meier method was used to estimate survival rates, and the log-rank and the Wilcoxon rank sum tests were used to assess survival differences between groups. Variables with a value of P < 0.05 in univariate analysis were used in subsequent multivariate analysis on the basis of Cox proportional hazards model. A two-sided P value of less than 0.05 was considered to statistically significant.
Results
lncRNA LET was down-regulated and associated with poor prognosis in patients with cervical cancer
LncRNA LET expression was detected in 94 pairs of human cervical cancer and adjacent non-tumor tissues by qRT-PCR. As shown in Figure 1, the expression level of LncRNA LET in cervical cancer tissues was significantly lower than that in adjacent non-tumor tissues (P < 0.05). To identify the clinical relevance of lncRNA LET expression in cervical cancer, correlation between lncRNA LET expression and clinicopathological features such as age, tumor size, histology, tumor differentiation, FIGO stage, lymph node metastasis and depth of cervical invasion was examined. The mean expression level of LncRNA LET was used as a cutoff point to divide all patients into two groups: cervical cancer patients who express LncRNA LET at levels less than the cutoff value were assigned to the low expression group, and those with expression above the cutoff value were assigned to the high expression group. Our results showed that decreased lncRNA LET expression was significantly correlated with FIGO stage, lymph node metastasis, depth of cervical invasion (P < 0.05), but not associated with other clinicopathologic factors such as age, tumor size, histology, and tumor differentiation (P > 0.05).
Figure 1.
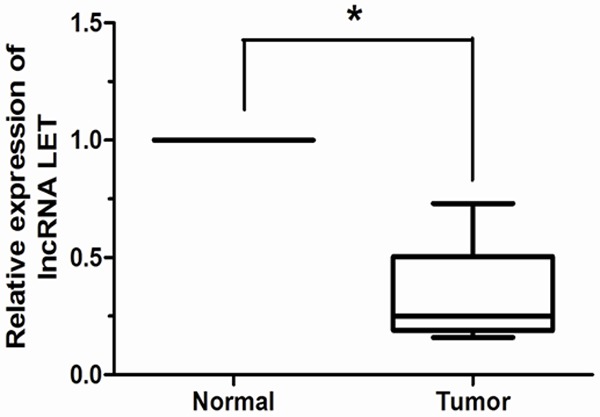
lncRNA LET expression in 94 pairs of cervical cancer and adjacent non-tumor tissues were respectively detected by qRT-PCR. After normalization to GAPDH, the expression level of lncRNA LET in cervical cancer tissues was significantly lower than that in adjacent non-tumor tissues. *P < 0.05.
lncRNA LET down-regulation associated with poor prognosis in patients with human cervical cancer
The association between lncRNA LET expression and survival of cervical cancer patients was investigated by Kaplan-Meier analysis and log-rank test. As shown in Figure 2, our result showed that cervical cancer patients with low lncRNA LET expression tend to have shorter overall survival than those with high lncRNA LET expression (log-rank test, P < 0.05). Univariate analysis demonstrated that FIGO stage, lymph node metastasis, depth of cervical invasion and lncRNA LET expression were significantly associated with overall survival of cervical cancer patients (P < 0.05, Table 2). No significant associations were found for age, tumor size, histology, and tumor differentiation and patient outcome. Multivariate analysis using the Cox proportional hazards model for all variables that were significant in the univariate analysis showed that FIGO stage, lymph node metastasis, depth of cervical invasion, and lncRNA LET expression were independent prognostic factors for patients with cervical cancer (P < 0.05, Table 2).
Figure 2.
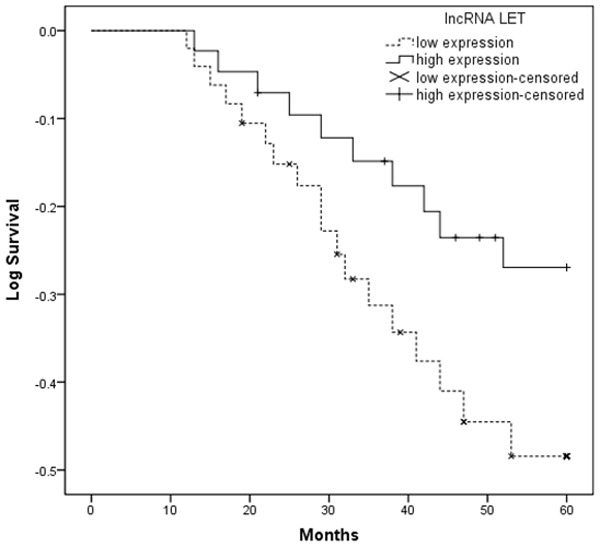
Kaplan-Meier survival curves according to lncRNA LET level. Patients with low lncRNA LET expression had a significantly poorer prognosis than those with high lncRNA LET expression.
Table 2.
Univariate and multivariate analysis of clinicopathological features for overall survival
| Clinicopathological features | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| Risk ratio | 95% CI | P | Risk ratio | 95% CI | P | |
| Age (years) | ||||||
| ≥ 35 vs. < 35 | 1.418 | 0.739-2.016 | 0.374 | |||
| Tumor size | ||||||
| ≥ 4 cm vs. < 4 cm | 1.833 | 0.814-2.916 | 0.179 | |||
| Histology | ||||||
| Squamous vs. Adenocarcinoma | 0.947 | 0.672-1.663 | 0.538 | |||
| Differentiation | ||||||
| Poor vs. Well + Moderate | 2.719 | 0.687-3.918 | 0.097 | |||
| FIGO stage | ||||||
| IIb-IIIa vs. Ib-IIa | 2.614 | 1.373-3.615 | 0.017 | 1.886 | 1.174-2.905 | 0.006 |
| Lymph node metastasis | ||||||
| Yes vs. No | 3.817 | 1.862-7.027 | 0.010 | 3.019 | 1.411-5.739 | 0.004 |
| Depth of cervical invasion | ||||||
| ≥ 2/3 vs. < 2/3 | 2.218 | 1.305-4.647 | 0.021 | 1.975 | 1.174-3.376 | 0.019 |
| LncRNA LET | ||||||
| Low vs. High | 2.894 | 1.615-6.427 | <0.001 | 2.483 | 1.339-4.728 | 0.003 |
Discussion
Cervical cancer remains one of the leading causes of cancer death in women worldwide [14]. In spite of the development of advanced therapeutic strategies, the prognosis in patients with this cancer varies significantly and is hard to predict. Treatment outcome still depends primarily on early detection and diagnosis. Recent studies have demonstrated that some abnormal molecular biology changes may play central roles in the tumorigenesis and the development of cervical cancer [15]. For example, Luo et al found that miR-497 is a potential prognostic marker in human cervical cancer and functions as a tumor suppressor by targeting the insulin-like growth factor 1 receptor [16]. Liao et al demonstrated that low expression of long non-coding XLOC 010588 indicates a poor prognosis and promotes proliferation through up-regulation of c-Myc in cervical cancer [17]. Huang et al reported that lncRNA HOTAIR was frequently up-regulated in cervical cancer tissues, and as an independent prognostic factor in patients with cervical cancer [18]. Therefore, it is critical to identify biomarkers for the early diagnosis and the early identification of patients with a high risk of treatment failures, in order to modify therapeutic methods for improving overall survival of patients with cervical cancer.
lncRNA LET, a newly identified lncRNA, was found to be down-regulated in hepatocellular carcinomas, colorectal cancers, and squamous cell lung carcinomas [20]. Yang et al showed down-regulation of lncRNA-LET was found to be a key step in the stabilization of nuclear factor 90 protein, which leads to hypoxia induced cancer cell invasion [19]. Ma et al found that lncRNA LET was a positive prognostic factor and exhibits tumor-suppressive activity in gallbladder cancer [20]. Therefore, we hypothesized that expression of lncRNA LET might decrease and associate with prognosis in cervical cancer patients.
To test this hypothesis, in the present study we performed a qRT-PCR assay to explore the expression profile of this lncRNA as well as to investigate its association with clinicopathological features of cervical cancer patients. Our results showed that lncRNA LET expression was significantly lower in cervical cancer tissues compared with that in adjacent non-tumor tissues. In addition, lncRNA LET expression was associated with FIGO stage, lymph node metastasis, and depth of cervical invasion which strongly suggested that lncRNA LET was involved in the invasion and metastasis of cervical cancer. More importantly, we found that lncRNA LET expression was significantly associated with overall survival of patients with cervical cancer. In support of this, Kaplan-Meier analysis of overall survival showed that patients with low lncRNA LET expression had a significantly poorer prognosis than those with high lncRNA LET expression. More importantly, both the univariate and multivariate survival analyses demonstrated that low lncRNA LET expression was correlated with shorter overall survival in cervical cancer, which was also consistent with the prognostic significance of lncRNA LET in other human malignancies, such as hepatocellular carcinomas and gallbladder cancer. These results indicated that lncRNA LET might be an important modulator involved in cervical cancer development.
Taken together, the current study indicates that lncRNA LET is down-regulated in cervical cancer tissues and might be an independent molecular biomarker for predicting the prognosis of cervical cancer patients. This new findings indicated that lncRNA LET may be used as a potential target for diagnosis and gene therapy of cervical cancer. However, the precise molecular mechanisms of lncRNA LET that involved in cervical cancer need to be further studied.
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Ellenson LH, Wu TC. Focus on endometrial and cervical cancer. Cancer Cell. 2004;5:533–538. doi: 10.1016/j.ccr.2004.05.029. [DOI] [PubMed] [Google Scholar]
- 3.Shingleton HM, Jones WB, Russell A, Fremgen A, Chmiel JS, Ocwieja K, Winchester DP, Clive R. Hysterectomy in invasive cervical cancer: national patterns of care study of the American College of Surgeons. J Am Coll Surg. 1996;183:393–400. [PubMed] [Google Scholar]
- 4.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 6.Hu W, Alvarez-Dominguez JR, Lodish HF. Regulation of mammalian cell differentiation by long non-coding RNAs. EMBO Rep. 2012;13:971–983. doi: 10.1038/embor.2012.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 9.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi X, Sun M, Liu H, Yao Y, Kong R, Chen F, Song Y. A critical role for the long non-coding RNA GAS5 in proliferation and apoptosis in non-small-cell lung cancer. Mol Carcinog. 2013 doi: 10.1002/mc.22120. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 11.Lai MC, Yang Z, Zhou L, Zhu QQ, Xie HY, Zhang F, Wu LM, Chen LM, Zheng SS. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med Oncol. 2012;29:1810–1816. doi: 10.1007/s12032-011-0004-z. [DOI] [PubMed] [Google Scholar]
- 12.Zhang HM, Yang FQ, Yan Y, Che JP, Zheng JH. High expression of long non-coding RNA SPRY4-IT1 predicts poor prognosis of clear cell renal cell carcinoma. Int J Clin Exp Pathol. 2014;7:5801–5809. [PMC free article] [PubMed] [Google Scholar]
- 13.Pecorelli S. Revised FIGO staging for carcinoma of the vulva, cervix, and endometrium. Int J Gynaecol Obstet. 2009;105:103–104. doi: 10.1016/j.ijgo.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 14.Schiffman M, Castle PE, Jeronimo J, Rodriguez AC, Wacholder S. Human papillomavirus and cervical cancer. Lancet. 2007;370:890–907. doi: 10.1016/S0140-6736(07)61416-0. [DOI] [PubMed] [Google Scholar]
- 15.Lui WO, Pourmand N, Patterson BK, Fire A. Patterns of known and novel small RNAs in human cervical cancer. Cancer Res. 2007;67:6031–6043. doi: 10.1158/0008-5472.CAN-06-0561. [DOI] [PubMed] [Google Scholar]
- 16.Luo M, Shen D, Zhou X, Chen X, Wang W. MicroRNA-497 is a potential prognostic marker in human cervical cancer and functions as a tumor suppressor by targeting the insulin-like growth factor 1 receptor. Surgery. 2013;153:836–847. doi: 10.1016/j.surg.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 17.Liao LM, Sun XY, Liu AW, Wu JB, Cheng XL, Lin JX, Zheng M, Huang L. Low expression of long noncoding XLOC_010588 indicates a poor prognosis and promotes proliferation through upregulation of c-Myc in cervical cancer. Gynecol Oncol. 2014;133:616–623. doi: 10.1016/j.ygyno.2014.03.555. [DOI] [PubMed] [Google Scholar]
- 18.Huang L, Liao LM, Liu AW, Wu JB, Cheng XL, Lin JX, Zheng M. Overexpression of long noncoding RNA HOTAIR predicts a poor prognosis in patients with cervical cancer. Arch Gynecol Obstet. 2014;290:717–23. doi: 10.1007/s00404-014-3236-2. [DOI] [PubMed] [Google Scholar]
- 19.Yang F, Huo XS, Yuan SX, Zhang L, Zhou WP, Wang F, Sun SH. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol Cell. 2013;49:1083–1096. doi: 10.1016/j.molcel.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 20.Ma MZ, Kong X, Weng MZ, Zhang MD, Qin YY, Gong W, Zhang WJ, Quan ZW. Long non-coding RNALET is a positive prognostic factor and exhibits tumor-suppressive activity in gallbladder cancer. Mol Carcinog. 2014 doi: 10.1002/mc.22215. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]


