Abstract
Purpose: CRC (Colorectal cancer) is a lethal cancer for death worldwide and the underlying pathological mechanisms for CRC progression remain unclear. We aimed to explore the regulatory mechanism of CRC and provide novel biomarkers for CRC screening. Methods: Downloading from GEO (Gene Expression Omnibus) database, Microarray data GSE44861 were consisted of 111 colon tissues samples including 55 from adjacent noncancerous tissues and 56 from tumors tissues. After data pre-processing, up- and down regulated DEGs (differentially expressed genes) were identified using Bayes moderated t-test. Then DIVAD (Database for Annotation, Visualization and Integrated Discovery) was recruited to perform functional analysis for DEGs. Thereafter, PPI (protein-protein interaction) network was constructed by mapping DEGs into STRING (Search Tool for the Retrieval of Interacting Genes) database. Further, PPI modules were constructed and the protein domains of DEGs in the modules were analyzed. Moreover, miRNA regulatory network was established through GSEA (gene set enrichment analysis) method. Results: In summary, 96 up- and 212 down-regulated DEGs were identified. Totally, ten DEGs with high degrees in the constructed PPI network were selected, in which COLL1A1, PTGS2 and ASPN were also identified as crucial genes in PPI modules. Furthermore, COLL1A1 was predicted to be targeted by miR-29, while PTGS2 and ASPN were both predicted to be regulated by miR-101 and miR-26. Conclusion: COL11A1 might involve in the progression of CRC via being targeted by miR-29, whereas PTGS2 and ASPN were both regulated by miR-101 and miR-26. Moreover, ASPN may be supposed as a novel biomarker for CRC detection and prevention.
Keywords: Colorectal cancer, COLL1A1, PTGS2, ASPN, miRNA, target genes
Introduction
CRC (Colorectal cancer) is a lethal disease which known as the second leading cause for death worldwide [1] and the third frequent malignancy in US and western countries [2,3]. In 2013, nearly 142, 820 new cases were estimated to be suffered from CRC and approximately 50, 830 people would die from this disease [4]. CRC arises from a benign adenomatous polyp and evolves through multiple pathways [5,6], which makes its classification diversity. According to the molecular features of the pathways CRC involved in, 5 molecular subtypes with morphological correlations were outlined [6].
Early detection of CRC facilitates to prevent its progression [7], therefore contributes to reduce mortality for that most cases in CRC was curable during tumor-node-metastasis stages I to III [5]. The established conventional screening tests for CRC are fecal occult blood testing, flexible sigmoidoscopy and colonoscopy. Among them, though inexpensive, noninvasive and recommended by the American Cancer Society, only small fraction of adults in USA has annual CRC screening by fecal occult blood test, due to the lack of reducing CRC mortality to a large extent [8]. By contrast, except detecting the malignant tumors, colonoscopy could also detect adenomas and other benign precursor lesions, and then remove them, which contribute to reduce the mortality from CRC and decrease the incidence of the disease. Possessing this advantage, colonoscopy is currently used as “gold standard” of CRC screening [9]. However, restrictive factors are inevitably existed such as patient discomfort, invasiveness and expensive cost, which limit the application of this screening method [10]. Besides, the 5-year survival rates were extremely low for these tests are unavailable for patients with advanced CRC due to the distant metastases [11]. Therefore, considerable studies sought for antitumor agents with genomic biomarkers for CRC treatment, in order to provide more effective and accurate intervention for the disease [12-14]. The accumulated genetic alterations such as genetic mutations and epigenetic changes were involved in CRC progression [11,15]. Mutations of oncogene K-ras was considered as a biomarker for CRC and Doolittle et al. provided a potential protocol for CRC screening through detecting these mutations [2]. Recently, a five-gene biomarker was identified as a novel blood-based test for CRC detection using quantitative real-time PCR (polymerase chain reaction) [8]. Nevertheless, the evidence is limited and the underlying pathological mechanisms responsible for CRC progression and metastases remain unclear and need to be further elucidated.
In this study, we performed microarray analysis using bioinformatics methods, which involve computer processes to solve biological problems and have predictive capability [16], to explore the comprehensive regulatory mechanism and thereby, to provide potentially more effective biomarkers for CRC screening and prevention.
Material and methods
Microarray data
The gene expression profile GSE44861 [17] was downloaded from the public database, GEO (Gene Expression Omnibus, http://www.ncbi.nlm.nih.gov/geo/), up to March 12, 2014, which deposited the most comprehensive information of Affimatrix microarray data, EST (expression sequence tag), SAGE (Serial Analysis of Gene Expression) and the next generation sequencing data [18]. According to the expression profile, the samples were consisted of 55 colon tissues from adjacent noncancerous tissues (control group) and 56 colon tissues from tumors tissues (disease group). The platform was GPL3921 ([HT_HG-U133A], Affymetrix HT Human Genome U133A Array) and the annotation file on it was also downloaded.
Data preprocessing and differentially expressed genes (DEGs) screening
After the data was normalized by log2 transformation, Limma (Linear Model for Microarray, http://www.bioconductor.org/packages/release/bioc/html/limma.html) package in R [19] was recruited for linearization and Bayes moderated t-test [20] was used for the DEGs (differentially expressed genes) identification. The screening criterion for DEGs were |log FC (fold change)| > 1 and P-value < 0.01.
Functional and pathway enrichment analysis for DEGs
GO (gene ontology, http://www. geneontology.org/) analysis has been widely used for the functional annotation of genomic data. To explore the functions of DEGs, DAVID (Database for Annotation, Visualization and Integrated Discovery, http://david.abcc.Ncifcrf.gov/) [21] was employed to perform GO enrichment analysis, based on the hypergeometric distribution method. The count number ≥ 2 and P-value < 0.05 were selected as the threshold.
To further identify the metabolic pathways that DEGs involved in, KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway (http://www.genome.jp/kegg/pathway.html) analysis [22] was also performed. The thresholds for the pathway enrichment analysis were with count number > 3 and P-value < 0.05.
Construction of protein-protein interaction (PPI) network
The selected DEGs were put into the STRING (Search Tool for the Retrieval of Interacting Genes) database [23] (http://string-db.org/) to match the interactions of proteins. The interaction with a score of 0.4 was eligible to construct the PPI (protein-protein interaction) network, using the Cytoscape software (http://cytoscape.org/) [24].
Construction of modules of the PPI network
To explore more specific regulatory relationship of proteins, ClusterONE [25] plug-in in the Cytoscape software was applied to dig the cluster modules, under the criterion of P-value < 0.01. Then INTERPRO [26] protein domain database (http://www.ebi.ac.uk/interpro/) was used for functional annotation of DEGs in cluster modules. The screening criterion was also with P-value < 0.05.
MiRNA enrichment analysis
We predicted the miRNAs that have potential to target the selected DEGs using GSEA (gene set enrichment analysis) method, which could statistically determine whether the predefined sets of genes are differentially expressed in different phenotypes [27]. In the present study, we randomized the distribution of the two categories of samples for 1,000 times to estimate the statistical P-value. If a set of genes targeted by one miRNA has significantly differently expressions in control and disease groups, the miRNA was likely associated with the pathological process of the disease. The default parameter with FDR (false discovery rate) less than 25% in GSEA method were the criterion to identify the enriched sets of DEGs, which were significantly targeted by miRNA.
Results
DEGs selection and the functional analysis
Based on the statistical analysis for the microarray data between control and disease groups, a total of 308 DEGs related to CRC were screened, in which 96 were up-regulated and 212 were down-regulated.
As shown in Table 1, GO enrichment analysis indicated that the functions of up-regulated genes were mainly relevant to cellular compartment such as extracellular region part, extracelluar matrix, extracelluar space and proteinaceous extracelluar matrix, while the functions of down-regulated genes were primarily enriched in cell fraction, insoluble fraction and membrane fraction.
Table 1.
Significantly enriched functions for DEGs in the samples of colorectal tumor tissues
| Category | Term | Count | P-value |
|---|---|---|---|
| Up-regulated DEGs | |||
| Cluster 1 | Enrichment Score: 10.769284583391107 | ||
| CC | GO:0044421~extracellular region part | 31 | 2.25E-15 |
| CC | GO:0005578~proteinaceous extracellular matrix | 17 | 2.97E-11 |
| CC | GO:0005576~extracellular region | 37 | 3.17E-11 |
| CC | GO:0031012~extracellular matrix | 17 | 9.12E-11 |
| CC | GO:0005615~extracellular space | 20 | 7.38E-09 |
| Cluster 2 | Enrichment Score: 4.866243173643159 | ||
| CC | GO:0005578~proteinaceous extracellular matrix | 17 | 2.97E-11 |
| CC | GO:0031012~extracellular matrix | 17 | 9.12E-11 |
| MF | GO:0005201~ext matrix structural constituent | 8 | 7.89E-07 |
| CC | GO:0005581~collagen | 6 | 1.61E-06 |
| CC | GO:0044420~extracellular matrix part | 7 | 5.77E-05 |
| BP | GO:0030199~collagen fibril organization | 4 | 6.75E-04 |
| CC | GO:0005583~fibrillar collagen | 3 | 2.10E-03 |
| BP | GO:0030198~extracellular matrix organization | 5 | 3.49E-03 |
| Cluster 3 | Enrichment Score: 2.543911806644643 | ||
| BP | GO:0001501~skeletal system development | 11 | 1.97E-05 |
| Cluster 4 | Enrichment Score: 2.391819902010825 | ||
| BP | GO:0007155~cell adhesion | 13 | 8.88E-04 |
| BP | GO:0022610~biological adhesion | 13 | 8.99E-04 |
| Cluster 5 | Enrichment Score: 2.171321263363379 | ||
| BP | GO:0032963~collagen metabolic process | 4 | 6.08E-04 |
| BP | GO:0044259~multicellular org mac metabolic process | 4 | 8.24E-04 |
| BP | GO:0044236~multicellular org metabolic process | 4 | 1.39E-03 |
| MF | GO:0004175~endopeptidase activity | 9 | 1.81E-03 |
| MF | GO:0008233~peptidase activity | 11 | 2.21E-03 |
| MF | GO:0004222~metalloendopeptidase activity | 5 | 3.52E-03 |
| MF | GO:0008237~metallopeptidase activity | 6 | 4.85E-03 |
| MF | GO:0070011~peptidase activity, act on L- acid pep | 10 | 5.50E-03 |
| BP | GO:0030574~collagen catabolic process | 3 | 6.27E-03 |
| Cluster 6 | Enrichment Score: 2.124875246229077 | ||
| MF | GO:0004857~enzyme inhibitor activity | 8 | 1.17E-03 |
| Cluster 7 | Enrichment Score: 2.121422115415992 | ||
| BP | GO:0009611~response to wounding | 12 | 3.04E-04 |
| MF | GO:0005125~cytokine activity | 7 | 1.11E-03 |
| MF | GO:0008009~chemokine activity | 4 | 2.63E-03 |
| BP | GO:0042330~taxis | 6 | 2.70E-03 |
| BP | GO:0006935~chemotaxis | 6 | 2.70E-03 |
| MF | GO:0042379~chemokine receptor binding | 4 | 3.15E-03 |
| BP | GO:0006954~inflammatory response | 8 | 3.29E-03 |
| BP | GO:0006952~defense response | 11 | 3.59E-03 |
| Down-regulated DEGs | |||
| Cluster 1 | Enrichment Score: 3.3026715504296 | ||
| CC | GO:0000267~cell fraction | 29 | 7.71E-05 |
| CC | GO:0005626~insoluble fraction | 22 | 1.02E-03 |
| CC | GO:0005624~membrane fraction | 21 | 1.57E-03 |
| Cluster 2 | Enrichment Score: 2.958218737815963 | ||
| MF | GO:0004089~carbonate dehydratase activity | 5 | 2.72E-05 |
| MF | GO:0016836~hydro-lyase activity | 5 | 1.58E-03 |
| Cluster 3 | Enrichment Score: 2.3681708741683325 | ||
| BP | GO:0009725~response to hormone stimulus | 14 | 1.39E-04 |
| BP | GO:0031667~response to nutrient levels | 10 | 2.44E-04 |
| BP | GO:0009719~response to endogenous stimulus | 14 | 3.62E-04 |
| BP | GO:0009991~response to extracellular stimulus | 10 | 5.48E-04 |
| BP | GO:0048545~response to steroid hormone stimulus | 9 | 9.89E-04 |
| BP | GO:0007584~response to nutrient | 7 | 3.70E-03 |
| BP | GO:0051384~response to glucocorticoid stimulus | 5 | 9.33E-03 |
| BP | GO:0033189~response to vitamin A | 4 | 9.83E-03 |
| BP | GO:0042594~response to starvation | 4 | 9.83E-03 |
DEGs, differentially expressed genes; Cluster, the classification based on different functions; Count, number of DEGs; BP, biological process; CC, cellular compartment; MF, molecular function.
According to the results of KEGG pathway analysis (Table 2), the up-regulated DEGs were enriched in focal adhesion pathway, and most significantly enriched in pathway of ECM-receptor interaction, whereas the down-regulated DEGs were mainly involved in Steroid hormone biosynthesis, and Androgen and estrogen metabolism pathways.
Table 2.
Significantly enriched pathways for DEGs in the samples of colorectal tumor tissues
| Term | Count | P-value |
|---|---|---|
| Up-regulated DEGs | ||
| Hsa04512: ECM-receptor interaction | 7 | 2.62E-05 |
| Hsa04510: Focal adhesion | 7 | 2.95E-03 |
| Down-regulated DEGs | ||
| Hsa00150: Androgen and estrogen metabolism | 6 | 1.52E-04 |
| Hsa00910: Nitrogen metabolism | 5 | 2.67E-04 |
| Hsa00140: Steroid hormone biosynthesis | 6 | 4.32E-04 |
| Hsa00053: Ascorbate and aldarate metabolism | 4 | 1.61E-03 |
| Hsa00040: Pentose and glucuronate interconversions | 4 | 1.91E-03 |
| Hsa00500: Starch and sucrose metabolism | 5 | 2.76E-03 |
| Hsa00983: Drug metabolism | 5 | 3.01E-03 |
| Hsa00830: Retinol metabolism | 5 | 6.85E-03 |
DEGs, differentially expressed genes; Count, number of DEGs.
Construction of PPI network and functional analysis of DEGs in the network
By separately mapping the identified up- or down-regulated DEGs into STRING database, the PPI networks of up- or down-regulated DEGs were constructed respectively, with the confidence score larger than 0.4. There were 156 edges involving 60 nodes in the PPI network for up-regulated DEGs, in which COL1A2, TGFBI, COL5A2, MMP1, TIMP1, MMP7, PTGS2, COL11A1, ASPN and PLAU were located in the top 10 nodes with high degrees (> 10) (Figure 1A; Table 3). On the other hand, the network for down-regulated DEGs was composed of 151 edges and 110 nodes, among which, GCG and MUC2 with high degrees (> 10) were recognized (Figure 1B; Table 3).
Figure 1.
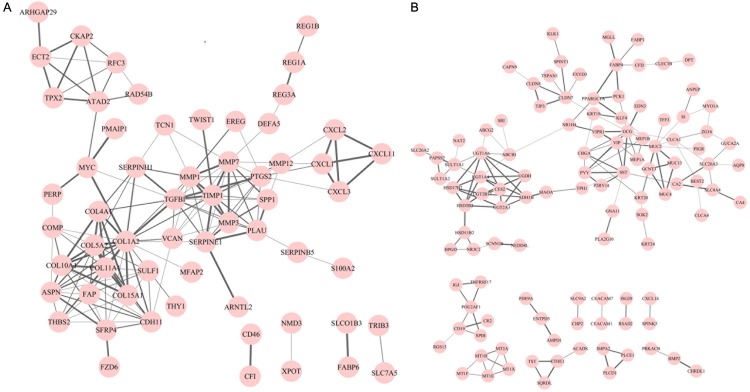
PPI (protein-protein interaction) network constructed for DEGs (differentially expressed genes) in colorectal tumor tissue samples. A. PPI network for up-regulated DEGs; B. PPI network for down-regulated DEGs. The nodes represent DEGs in colorectal tumor tissue samples and the lines represent the degree between DEGs, in which the thicker line refers to the higher degree.
Table 3.
Top ten DEGs with high degrees identified in PPI network
| Up-regulated DEGs | Degree | Down-regulated DEGs | Degree |
|---|---|---|---|
| COL1A2 | 21 | GCG | 10 |
| TGFBI | 13 | MUC2 | 10 |
| COL5A2 | 13 | UGT1A6 | 8 |
| MMP1 | 12 | SST | 8 |
| TIMP1 | 12 | UGT1A4 | 7 |
| MMP7 | 12 | PYY | 6 |
| PTGS2 | 11 | CLDN7 | 6 |
| COL11A1 | 11 | CHGA | 6 |
| ASPN | 10 | HSD3B2 | 6 |
| PLAU | 10 | VIP | 6 |
DEGs, differentially expressed genes; PPI, protein-protein interaction.
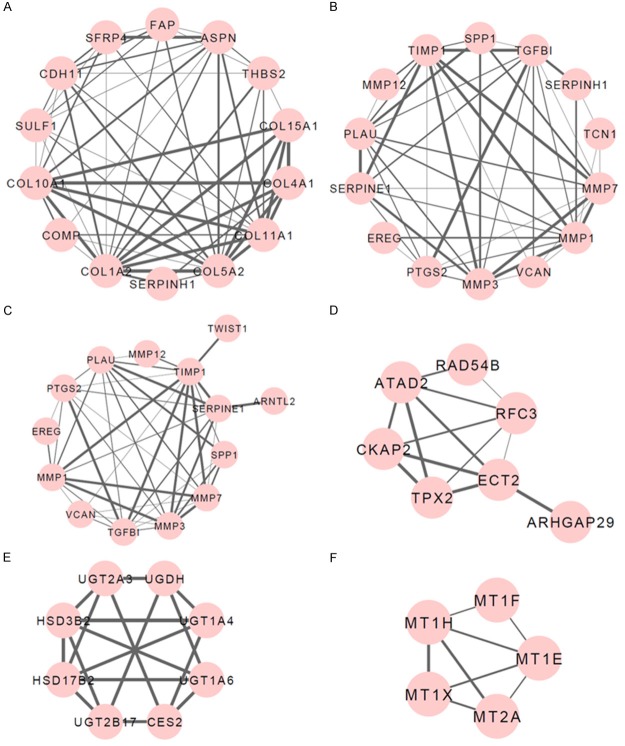
Applying ClusterONE plug-in in the Cytoscape software, we performed the cluster module analysis for PPI networks of up- and down-regulated DEGs respectively, to predict the protein complex. As a result, 4 modules for up-regulated DEGs and 2 modules for down-regulated DEGs were identified with P-values < 0.01 (Figure 2). INTERPRO Protein Domain enrichment analysis for these modules revealed that genes in Module 2 and Module 3 of up-regulated DEGs were predominantly enriched in protein functional domains containing peptidoglycan binding-like (IPR002477), peptidase, metallopeptidases (IPR006026) and Peptidase M10A and M12B, matrixin and adamalysin (IPR001818). Referred to down-regulated DEGs, genes in Module 1 and Module 2 were primarily enriched in UDP-glucuronosyl/UDP-glucosyltransferase (IPR002213), NAD (P)-binding domain (IPR016040), metallothionein, vertebrate, metal binding site (IPR018064), metallothionein, vertebrate (IPR000006) and metallothionein superfamily, eukaryotic (IPR003019) protein domains (Table 4).
Figure 2.
Module clusters identified from the PPI (protein-protein interaction) network in colorectal tumor tissue samples. (A-D) Module clusters for up-regulated DEGs (differentially expressed genes) (A, Module 1; B, Module 2; C, Module 3; D, Module 4); (E, F) Module clusters for down-regulated DEGs (E, Module 1; F, Module 2). The nodes represent DEGs in colorectal tumor tissue samples and the lines represent the degree between DEGs, in which the thicker line refers to the higher degree.
Table 4.
Protein domains that DEGs in the modules of PPI network enriched in
| Term | Count | P-value |
|---|---|---|
| Up-regulated DEGs | ||
| Module 2 | ||
| IPR002477: Peptidoglycan binding-like | 4 | 4.20E-07 |
| IPR006026: Peptidase, metallopeptidases | 4 | 1.34E-06 |
| IPR001818: Peptidase M10A and M12B, matrixin and adamalysin | 4 | 4.50E-06 |
| IPR006025: Peptidase M, neutral zinc metallopeptidases, zinc-binding site | 4 | 5.09E-05 |
| IPR016293: Peptidase M10A, matrix metallopeotidase | 3 | 8.54E-05 |
| IPR018486: Hemopexin/matrixin, conserved site | 3 | 1.41E-04 |
| IPR000585: Hemopexin/matrixin | 3 | 1.41E-04 |
| IPR018487: Hemopexin/matrixin, repeat | 3 | 1.41E-04 |
| IPR000742: EGF-like, type 3 | 4 | 4.08E-04 |
| IPR006210: EGF-like | 4 | 4.53E-04 |
| IPR013032: EGF-like region, conserved site | 4 | 1.35E-03 |
| IPR006209: EGF | 3 | 4.26E-03 |
| Module 3 | ||
| IPR002477: Peptidoglycan binding-like | 4 | 4.20E-07 |
| IPR006026: Peptidase, metallopeptidases | 4 | 1.34E-06 |
| IPR001818: Peptidase M10A and M12B, matrixin and adamalysin | 4 | 4.50E-06 |
| IPR006025: Peptidase M, neutral zinc metallopeptidases, zinc-binding site | 4 | 5.09E-05 |
| IPR016293: Peptidase M10A, matrix metallopeotidase | 3 | 8.54E-05 |
| IPR018486: Hemopexin/matrixin, conserved site | 3 | 1.41E-04 |
| IPR018487: Hemopexin/matrixin, repeat | 3 | 1.41E-04 |
| IPR000585: Hemopexin/matrixin | 3 | 1.41E-04 |
| IPR000742: EGF-like, type 3 | 4 | 4.08E-04 |
| IPR006210: EGF-like | 4 | 4.53E-04 |
| IPR013032: EGF-like region, conserved site | 4 | 1.35E-03 |
| IPR006209: EGF | 3 | 4.26E-03 |
| Down-regulated DEGs | ||
| Module 1 | ||
| IPR002213: UDP-glucuronosyl/UDP-glucosyltransferase | 3 | 8.42E-06 |
| IPR016040: NAD(P)-binding domain | 3 | 1.18E-03 |
| Module 2 | ||
| IPR018064: Metallothionein, vertebrate, metal binding site | 5 | 1.03E-13 |
| IPR000006: Metallothionein, vertebrate | 5 | 1.54E-13 |
| IPR003019: Metallothionein superfamily, eukaryotic | 5 | 1.54E-13 |
DEGs, differentially expressed genes; PPI, protein-protein interaction; Count, number of DEGs.
Construction of regulatory network of miRNAs and targeted DEGs
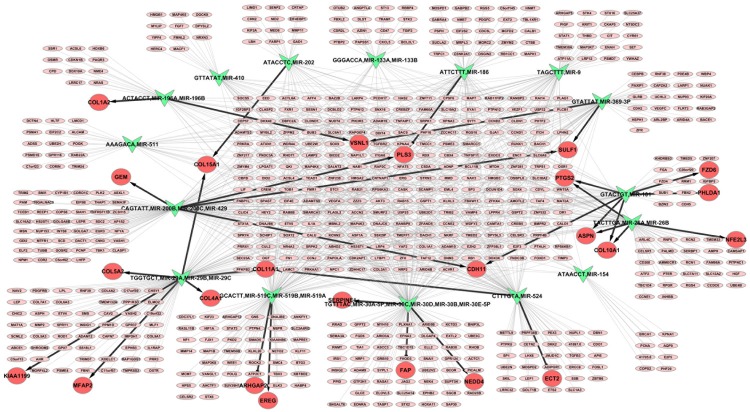
The network which revealed the regulatory relationship of miRNAs and the targeted DEGs were constructed through GSEA method. Following the default setting with FDR < 25%, the miRNAs were differently expressed in the two sample groups. Under the criterion of P < 0.05, a total of 16 significantly enriched miRNAs were found in disease group, by contrast, none were enriched in control group (Table 5), suggesting that these miRNAs have great potential to participate in CRC progression. As presented in Figure 3, 9 DEGs targeted by miRNAs were selected including ASPN, CDH11, COL10A1, COL15A1, PLS3, PTGS2, SULF1, VSNL1 and COL11A1.
Table 5.
MiRNAs that predicted to target DEGs based on GSEA method
| NAME | SIZE | ES | NES | NOM P-value |
|---|---|---|---|---|
| GTATTAT, MIR-369-3P | 143 | -0.42481536 | -1.5778576 | 0 |
| CAGTATT, MIR-200B, MIR-200C, MIR-429 | 341 | -0.34288508 | -1.4310081 | 0 |
| ACTACCT, MIR-196A, MIR-196B | 96 | -0.4422097 | -1.5706176 | 0.002232143 |
| TGCACTT, MIR-519C, MIR-519B, MIR-519A | 310 | -0.3275328 | -1.339217 | 0.006993007 |
| ATTCTTT, MIR-186 | 192 | -0.35181046 | -1.3669945 | 0.013452915 |
| TAGCTTT, MIR-9 | 167 | -0.3572543 | -1.3752931 | 0.014736842 |
| TGTTTAC, MIR-30A-5P, MIR-30C, MIR-30D, MIR-30B, MIR-30E-5P | 412 | -0.30206627 | -1.2713937 | 0.01814059 |
| GTACTGT, MIR-101 | 195 | -0.3322817 | -1.3145992 | 0.025423728 |
| GGGACCA, MIR-133A, MIR-133B | 137 | -0.359421 | -1.3479977 | 0.026373627 |
| CTTTGTA, MIR-524 | 319 | -0.3010239 | -1.23321 | 0.026966292 |
| TGGTGCT, MIR-29A, MIR-29B, MIR-29C | 358 | -0.29058316 | -1.2047044 | 0.03189066 |
| ATACCTC, MIR-202 | 124 | -0.3649094 | -1.3451693 | 0.036480688 |
| ATGTACA, MIR-493 | 234 | -0.3109604 | -1.2353733 | 0.042105265 |
| AAAGACA, MIR-511 | 150 | -0.33384982 | -1.2600027 | 0.045045044 |
| TACTTGA, MIR-26A, MIR-26B | 220 | -0.31613997 | -1.2490146 | 0.04835165 |
| ATAACCT, MIR-154 | 43 | -0.4706566 | -1.4422263 | 0.048523206 |
DEGs, differentially expressed genes; PPI, protein-protein interaction; GSEA, gene set enrichment analysis; Size, the number of the enriched DEGs; ES, enrichment score; NES, normalized enrichment score; NOM P-value, P-values based on hypergeometric distribution method.
Figure 3.
Integrated regulatory network for miRNA and the targeted genes. Symbols in green represent miRNAs and red circles represent significant DEGs (differentially expressed genes) targeted by miRNAs, while circles in pink refer to non-significant DEGs in colorectal tumor tissue samples.
Discussion
CRC development involves multiple genetic alterations and progressive changes in signaling pathways [11]. In the present study, the microarray data that downloaded from GEO database were utilized to identify DGEs between control and disease samples. As a result, 96 up- and 212 down-regulated DEGs were identified. Further analysis of the constructed PPI network revealed that COL11A1, PTGS2, and ASPN were listed in the top-ten nodes with high degrees (> 10), implying they may play important roles in CRC development.
Comprised of trimeric extracellular matrix proteins with structurally similarity, collagens are used for cell structural integrity and involved in cell-matrix interaction and tumor progression as major components of basement membrane [28]. The primary structure of collagens are characterized by three α chains normally consisting of glycine-X-Y repeats [29,30]. There are 28 different types of collagens and the structural characteristics of them have been researched [30]. The gene COL11A1 which encodes the type XI collagen α-1 chain protein, an important factor for connective tissue structure and resistance, is found to be lower expressed in different adult human tissues including lung, parotid gland and colorectal cells [31]. Additionally, overexpressed COL11A1 was suggested as a candidate marker of various cancers such as NSCLC (non-small cell lung), ovarian, oral cavity and CRC [32-34]. Particularly, several studies verified that COL11A1 was up-regulated in CRC [35,36], indicating the important role of this gene in tumorigenic response in epithelial cells. Further confirmed by a recent research, COL11A1 was suggested as a biomarker targeted in stool samples for early detection of CRC at risk patients [31].
The regulatory relationship between COL11A1 and miRNA was also concerned and COL11A1 was predicted to be targeted by miR-29 [37,38], down-regulation of which could induce the expression of collagens [39]. Revealed as our constructed PPI network, COL11A1 was identified as a crucial gene related to CRC with a high degree, giving another evidence for the roles of COL11A1 in the regulation of CRC (Figure 1A; Table 3). Besides, the miRNA regulatory network also predicted that COL11A1 could be targeted by miR-29 (miR-29A, miR-29B, miR-29C) (Figure 3). Taken together, these results provide a clue that COL11A1 might involve in the progression of CRC via being targeted by miR-29.
PTGS2 (also called COX-2) that encodes an inducible isozyme of prostaglandin-endoperoxide synthase (prostaglandin-endoperoxide synthase 2), is triggered by the inflammatory response and responsible for the synthesis of prostaglandins [40]. The expression of PTGS2 is found to be elevated in CRC development [41], whereas the inhibition of PTGS2 could prevent tumor growth and improve overall survival [15]. Additionally, the polymorphisms in PTGS2 is convinced to be associated with increased risk of CRC and it is hypothesized that the genetic polymorphisms in PTGS2 results in alteration of the expression and/or the activity of the protein, which may modulate the inflammatory response, thus modifying the risk of colorectal cancer [40]. All these elucidate that PTGS2 plays a vital role in promoting CRC growth and metastasis [11] and implicate that inflammation might be an important mediator in the carcinogenesis of colorectal tumors [40]. The plausible explanation for the mechanism of PTGS2 in CRC is that PTGS2 enzyme could convert arachidonic acid into an unstable intermediate, PGH2, which could remarkably enhance the AOM (azoxymethane)-induced colon tumor incidence and play predominant role in carcinogenesis [11,42].
The expression of PTGS2 is modulated in both transcriptional and post-transcriptional levels. Several transcription factors such as NFKB1, C/EBP, CREB, NFAT and AP-1, have been verified to regulate the expression of PTGS2 [43,44]. As a class of short, endogenously-initiated non-coding RNAs, miRNAs could mediate gene expression in post-transcriptional level via either translational repression or mRNA degradation [45]. Larger amounts of miRNAs have been established to regulate the expression of PTGS2. It is demonstrated that the PTGS2 mRNA translation was down-regulated by miR-101 in colon cancer cells [46]. Besides, PTGS2 is identified as the target gene of miR-146, which negtively regulates the expression of PTGS2 via mRNA degragation in gastric epithelial cells and macrophages [47,48]. Except that, PTGS2 was also found to be regulated by hsa-miR-143 in amnion mesenchymal cells [49], as well as hsa-miR-137 which acts as a tumor suppressor in human glioma [50]. Our results indicated that PTGS2 was also a vital gene relevant to CRC development and was predicted to be regulated by miR-101 (Figures 1A, 3), which was consisted with previous studies [46]. Notably, according to our result of miRNA network, miR-26 was another candidate miRNA to mediate PTGS2 (Figure 3), however, miR-26 was only reported to influence PTGS2 in the regulation of GSK-3 for cholangiocarcinoma growth [51], implying a novel regulatory pathway that PTGS2 might play pivotal roles in CRC development via being regulated by miR-26. What’s important, based on the protein domain enrichment analysis in modules for up-regulated DEGs (Table 4), PTGS2 was mainly enriched in protein domains relevant to peptidoglycan binding-like, peptidase and metallopeptidases, suggesting that PTGS2 may play a role in protein degradation in CRC progression.
ASPN was another crucial gene selected in the integrated networks of PPI and miRNA in the present study (Figure 1A; Table 3). Belonging to the small secreted leucine-rich proteoglycans (SLRP) family, ASPN encodes a cartilage extracellular protein which may regulate chondrogenesis and induce collagen mineralization [52]. In aggressive endometrial cancer cells, ASPN is predicted to be targeted by miR-101, [53], consisting with our results of the miRNA regulatory network. Moreover, miR-26 was also predicted to modulate ASPN in the current study (Figure 3). To our knowledge, there wasn’t any illustration about the relationship between ASPN and CRC, as well as the regulation of ASPN in CRC progression, therefore ASPN might be supposed as a novel biomarker for CRC screening and prevention.
In conclusion, we infer that COL11A1 might involve in the progression of CRC via being targeted by miR-29, whereas PTGS2 and ASPN may play crucial roles in CRC development both through the regulation of miR-101 and miR-26. Moreover, ASPN may play its roles through protein degradation and be supposed as a novel biomarker for CRC detection and prevention.
Disclosure of conflict of interest
None.
References
- 1.Fodde R. The APC gene in colorectal cancer. Eur J Cancer. 2002;38:867–871. doi: 10.1016/s0959-8049(02)00040-0. [DOI] [PubMed] [Google Scholar]
- 2.Doolittle BR, Emanuel J, Tuttle C, Costa J. Detection of the mutated K-Ras biomarker in colorectal carcinoma. Exp Mol Pathol. 2001;70:289–301. doi: 10.1006/exmp.2001.2364. [DOI] [PubMed] [Google Scholar]
- 3.Grothey A, Sargent D, Goldberg RM, Schmoll HJ. Survival of patients with advanced colorectal cancer improves with the availability of fluorouracil-leucovorin, irinotecan, and oxaliplatin in the course of treatment. J. Clin. Oncol. 2004;22:1209–1214. doi: 10.1200/JCO.2004.11.037. [DOI] [PubMed] [Google Scholar]
- 4.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 5.Markowitz SD, Bertagnolli MM. Molecular basis of colorectal cancer. N Engl J Med. 2009;361:2449–2460. doi: 10.1056/NEJMra0804588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jass J. Classification of colorectal cancer based on correlation of clinical, morphological and molecular features. Histopathology. 2007;50:113–130. doi: 10.1111/j.1365-2559.2006.02549.x. [DOI] [PubMed] [Google Scholar]
- 7.Heitman SJ, Ronksley PE, Hilsden RJ, Manns BJ, Rostom A, Hemmelgarn BR. Prevalence of adenomas and colorectal cancer in average risk individuals: a systematic review and meta-analysis. Clin Gastroenterol Hepatol. 2009;7:1272–1278. doi: 10.1016/j.cgh.2009.05.032. [DOI] [PubMed] [Google Scholar]
- 8.Han M, Liew CT, Zhang HW, Chao S, Zheng R, Yip KT, Song ZY, Li HM, Geng XP, Zhu LX. Novel blood-based, five-gene biomarker set for the detection of colorectal cancer. Clin Cancer Res. 2008;14:455–460. doi: 10.1158/1078-0432.CCR-07-1801. [DOI] [PubMed] [Google Scholar]
- 9.Kanthan R, Senger JL, Kanthan SC. Fecal molecular markers for colorectal cancer screening. Gastroenterol Res Pract. 2011;2012:184343. doi: 10.1155/2012/184343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong Y, Wu W, Wu C, Sung J, Yu J, Ng S. MicroRNA dysregulation in colorectal cancer: a clinical perspective. Br J Cancer. 2011;104:893–898. doi: 10.1038/bjc.2011.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang D, Xia D, DuBois RN. The crosstalk of PTGS2 and EGF signaling pathways in colorectal cancer. Cancers. 2011;3:3894–3908. doi: 10.3390/cancers3043894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu CC, Chen HC, Chen SJ, Liu HP, Hsieh YY, Yu CJ, Tang R, Hsieh LL, Yu JS, Chang YS. Identification of collapsin response mediator protein-2 as a potential marker of colorectal carcinoma by comparative analysis of cancer cell secretomes. Proteomics. 2008;8:316–332. doi: 10.1002/pmic.200700819. [DOI] [PubMed] [Google Scholar]
- 13.Hijova E, Chmelarova A. Short chain fatty acids and colonic health. Bratisl Lek Listy. 2007;108:354. [PubMed] [Google Scholar]
- 14.Kim HJ, Yu MH, Kim H, Byun J, Lee C. Noninvasive molecular biomarkers for the detection of colorectal cancer. BMB Rep. 2008;41:685–692. doi: 10.5483/bmbrep.2008.41.10.685. [DOI] [PubMed] [Google Scholar]
- 15.Wang D, DuBois RN. The role of COX-2 in intestinal inflammation and colorectal cancer. Oncogene. 2009;29:781–788. doi: 10.1038/onc.2009.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kapetanovic IM, Rosenfeld S, Izmirlian G. Overview of commonly used bioinformatics methods and their applications. Ann N Y Acad Sci. 2004;1020:10–21. doi: 10.1196/annals.1310.003. [DOI] [PubMed] [Google Scholar]
- 17.Ryan BM, Zanetti KA, Robles AI, Schetter AJ, Goodman J, Hayes RB, Huang WY, Gunter MJ, Yeager M, Burdette L. Germline variation in NCF4, an innate immunity gene, is associated with an increased risk of colorectal cancer. Int J Cancer. 2014;134:1399–1407. doi: 10.1002/ijc.28457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barrett T, Suzek TO, Troup DB, Wilhite SE, Ngau WC, Ledoux P, Rudnev D, Lash AE, Fujibuchi W, Edgar R. NCBI GEO: mining millions of expression profiles-database and tools. Nucleic Acids Res. 2005;33:D562–D566. doi: 10.1093/nar/gki022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diboun I, Wernisch L, Orengo CA, Koltzenburg M. Microarray analysis after RNA amplification can detect pronounced differences in gene expression using limma. BMC Genomics. 2006;7:252. doi: 10.1186/1471-2164-7-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rouder JN, Speckman PL, Sun D, Morey RD, Iverson G. Bayesian t tests for accepting and rejecting the null hypothesis. Psychon Bull Rev. 2009;16:225–237. doi: 10.3758/PBR.16.2.225. [DOI] [PubMed] [Google Scholar]
- 21.Alvord G, Roayaei J, Stephens R, Baseler MW, Lane HC, Lempicki RA. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome biol. 2007;8:R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aoki-Kinoshita KF, Kanehisa M. Comparative Genomics. Springer; 2007. Gene annotation and pathway mapping in KEGG; pp. 71–91. [DOI] [PubMed] [Google Scholar]
- 23.Von Mering C, Huynen M, Jaeggi D, Schmidt S, Bork P, Snel B. STRING: a database of predicted functional associations between proteins. Nucleic Acids Res. 2003;31:258–261. doi: 10.1093/nar/gkg034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nepusz T, Yu H, Paccanaro A. Detecting overlapping protein complexes in protein-protein interaction networks. Nat Methods. 2012;9:471–472. doi: 10.1038/nmeth.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hunter S, Jones P, Mitchell A, Apweiler R, Attwood TK, Bateman A, Bernard T, Binns D, Bork P, Burge S, de Castro E, Coggill P, Corbett M, Das U, Daugherty L, Duquenne L, Finn RD, Fraser M, Gough J, Haft D, Hulo N, Kahn D, Kelly E, Letunic I, Lonsdale D, Lopez R, Madera M, Maslen J, McAnulla C, McDowall J, McMenamin C, Mi H, Mutowo-Muellenet P, Mulder N, Natale D, Orengo C, Pesseat S, Punta M, Quinn AF, Rivoire C, Sangrador-Vegas A, Selengut JD, Sigrist CJ, Scheremetjew M, Tate J, Thimmajanarthanan M, Thomas PD, Wu CH, Yeats C, Yong SY. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 2011;40:D306–12. doi: 10.1093/nar/gkr948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi J, Walker MG. Gene set enrichment analysis (GSEA) for interpreting gene expression profiles. Curr Bioinform. 2007;2:133–137. [Google Scholar]
- 28.Fenyvesi A. The prognostic significance of type IV collagen expression in colorectal carcinomas. Archive of Oncology. 2003;11:65–70. [Google Scholar]
- 29.Fallahi A, Kroll B, Warner LR, Oxford RJ, Irwin KM, Mercer LM, Shadle SE, Oxford JT. Structural model of the amino propeptide of collagen XI α1 chain with similarity to the LNS domains. Protein Sci. 2005;14:1526–1537. doi: 10.1110/ps.051363105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gordon MK, Hahn RA. Collagens. Cell Tissue Res. 2010;339:247–257. doi: 10.1007/s00441-009-0844-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suceveanu AI, Mazilu L, Suceveanu AP. COL11A1-Genetic Biomarker Targeted in Stool Samples for Early Diagnosis of Colorectal Cancer in Patients at Risk. 2014 [Google Scholar]
- 32.Park HJ, Choe BK, Kim SK, Park HK, Kim JW, Chung JH, Hong IK, Chung DH, Kwon KH. Association between collagen type XI α1 gene polymorphisms and papillary thyroid cancer in a Korean population. Exp Ther Med. 2011;2:1111–1116. doi: 10.3892/etm.2011.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chong IW, Chang MY, Chang HC, Yu YP, Sheu CC, Tsai JR, Hung JY, Chou SH, Tsai MS, Hwang JJ. Great potential of a panel of multiple hMTH1, SPD, ITGA11 and COL11A1 markers for diagnosis of patients with non-small cell lung cancer. Oncol Rep. 2006;16:981–988. [PubMed] [Google Scholar]
- 34.Kim H, Watkinson J, Varadan V, Anastassiou D. Multi-cancer computational analysis reveals invasion-associated variant of desmoplastic reaction involving INHBA, THBS2 and COL11A1. BMC Med Genomics. 2010;3:51. doi: 10.1186/1755-8794-3-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fischer H, Stenling R, Rubio C, Lindblom A. Colorectal carcinogenesis is associated with stromal expression of COL11A1 and COL5A2. Carcinogenesis. 2001;22:875–878. doi: 10.1093/carcin/22.6.875. [DOI] [PubMed] [Google Scholar]
- 36.Lascorz J, Hemminki K, Försti A. Systematic enrichment analysis of gene expression profiling studies identifies consensus pathways implicated in colorectal cancer development. J Carcinog. 2011;10:7. doi: 10.4103/1477-3163.78268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liao M, Zou S, Weng J, Hou L, Yang L, Zhao Z, Bao J, Jing Z. A microRNA profile comparison between thoracic aortic dissection and normal thoracic aorta indicates the potential role of microRNAs in contributing to thoracic aortic dissection pathogenesis. J Vasc Surg. 2011;53:1341–1349. e1343. doi: 10.1016/j.jvs.2010.11.113. [DOI] [PubMed] [Google Scholar]
- 38.Shioya M, Obayashi S, Tabunoki H, Arima K, Saito Y, Ishida T, Satoh J. Aberrant microRNA expression in the brains of neurodegenerative diseases: miR-29a decreased in Alzheimer disease brains targets neurone navigator 3. Neuropathol Appl Neurobiol. 2010;36:320–330. doi: 10.1111/j.1365-2990.2010.01076.x. [DOI] [PubMed] [Google Scholar]
- 39.Van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, Marshall WS, Hill JA, Olson EN. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci. 2008;105:13027–13032. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cox D, Pontes C, Guino E, Navarro M, Osorio A, Canzian F, Moreno V. Polymorphisms in prostaglandin synthase 2/cyclooxygenase 2 (PTGS2/COX2) and risk of colorectal cancer. Br J Cancer. 2004;91:339–343. doi: 10.1038/sj.bjc.6601906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cianchi F, Cortesini C, Bechi P, Fantappiè O, Mazzanti R. Up-regulation of cyclooxygenase 2 gene expression correlates with tumor angiogenesis in human colorectal cancer. Gastroenterology. 2001;121:1339–1347. doi: 10.1053/gast.2001.29691. [DOI] [PubMed] [Google Scholar]
- 42.Wang D, DuBois RN. Pro-inflammatory prostaglandins and progression of colorectal cancer. Cancer Lett. 2008;267:197–203. doi: 10.1016/j.canlet.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taniguchi K, Matsuoka A, Kizuka F, Lee L, Tamura I, Maekawa R, Asada H, Taketani T, Tamura H, Sugino N. Prostaglandin F2α (PGF2α) stimulates PTGS2 expression and PGF2α synthesis through NFKB activation via reactive oxygen species in the corpus luteum of pseudopregnant rats. Reproduction. 2010;140:885–892. doi: 10.1530/REP-10-0240. [DOI] [PubMed] [Google Scholar]
- 44.Tsatsanis C, Androulidaki A, Venihaki M, Margioris AN. Signalling networks regulating cyclooxygenase-2. Int J Biochem Cell Biol. 2006;38:1654–1661. doi: 10.1016/j.biocel.2006.03.021. [DOI] [PubMed] [Google Scholar]
- 45.Cai Y, Yu X, Hu S, Yu J. A brief review on the mechanisms of miRNA regulation. Genomics Proteomics Bioinformatics. 2009;7:147–154. doi: 10.1016/S1672-0229(08)60044-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Strillacci A, Griffoni C, Sansone P, Paterini P, Piazzi G, Lazzarini G, Spisni E, Pantaleo MA, Biasco G, Tomasi V. MiR-101 downregulation is involved in cyclooxygenase-2 overexpression in human colon cancer cells. Exp Cell Res. 2009;315:1439–1447. doi: 10.1016/j.yexcr.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 47.Liu Z, Wang D, Hu Y, Zhou G, Zhu C, Yu Q, Chi Y, Cao Y, Jia C, Zou Q. MicroRNA-146a negatively regulates PTGS2 expression induced by Helicobacter pylori in human gastric epithelial cells. J Gastroenterol. 2013;48:86–92. doi: 10.1007/s00535-012-0609-9. [DOI] [PubMed] [Google Scholar]
- 48.Liu Z, Zhou G, Deng X, Yu Q, Hu Y, Sun H, Wang Z, Chen H, Jia C, Wang D. Analysis of miRNA expression profiling in human macrophages responding to Mycobacterium infection: Induction of the immune regulator miR-146a. J Infect. 2014;68:553–561. doi: 10.1016/j.jinf.2013.12.017. [DOI] [PubMed] [Google Scholar]
- 49.Kim SY, Romero R, Tarca AL, Bhatti G, Lee J, Chaiworapongsa T, Hassan SS, Kim CJ. miR-143 regulation of prostaglandin-endoperoxidase synthase 2 in the amnion: implications for human parturition at term. PLoS One. 2011;6:e24131. doi: 10.1371/journal.pone.0024131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen L, Wang X, Wang H, Li Y, Yan W, Han L, Zhang K, Zhang J, Wang Y, Feng Y. miR-137 is frequently down-regulated in glioblastoma and is a negative regulator of Cox-2. Eur J Cancer. 2012;48:3104–3111. doi: 10.1016/j.ejca.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 51.Zhang J, Han C, Wu T. MicroRNA-26a promotes cholangiocarcinoma growth by activating β-catenin. Gastroenterology. 2012;143:246–256. e248. doi: 10.1053/j.gastro.2012.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tasheva ES, Klocke B, Conrad GW. Analysis of transcriptional regulation of the small leucine rich proteoglycans. Mol Vis. 2004;10:758–72. [PubMed] [Google Scholar]
- 53.Konno Y, Dong P, Xiong Y, Suzuki F, Lu J, Cai M, Watari H, Mitamura T, Hosaka M, Hanley SJ. MicroRNA-101 targets EZH2, MCL-1 and FOS to suppress proliferation, invasion and stem cell-like phenotype of aggressive endometrial cancer cells. Oncotarget. 2014;5:6049–62. doi: 10.18632/oncotarget.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]