Abstract
MiR-1179, a new identified miRNA highly associated with metastasis of colorectal cancer which was never reported in esophageal squamous cell carcinoma (ESCC). Here we measured the expression levels of miR-1179 and the candidate target gene in tissues from 40 patients with ESCC. Transwell, Dual-luciferase reporter assay and immunocytochemistry assay were employed to detect the function role of miR-1179 in vitro. We found that miR-1179 was up-regulated in human ESCC tumor tissues. Bioinformatics analysis indicated that SLIT2 acting as a new potential target of miR-1179 which was confirmed by luciferase reporter assay. Down-regulation of miR-1179 suppressed cell invasion in vitro with an increasing level of SLIT2 and ROBO1, besides, the up-regulation of SLIT2 decreased cell invasion through ROBO1. Taken together, these findings will shed light the role to mechanism of miR-1179 in regulating cell invasion via SLIT2/ROBO1 axis.
Keywords: microRNA, metastasis, 3’UTR, SLIT2-N, ROBO1
Introduction
Esophageal squamous cell carcinoma (ESCC) is the major histologic subtype of esophageal cancer [1]. It has become the sixth leading cause of death and the eighth most frequently diagnosed cancer [2]. Esophageal carcinoma includes two main types: esophageal adenocarcinoma (EADC) and esophageal squamous cell carcinoma [3]. The metastasis of ESCC clinically was one of the most factors inducing the poor prognosis [4]. A large amount of genes have been identified to be associated with tumor metastasis including SLIT2/ROBO1 axis [5]. Along with an evolutionary conserved role in axon guidance, SLIT2/ROBO1 pathway played a key role in tumors by acting as the tumor suppressor, especially in cell invasion [6,7]. The full length of the secreted protein SLIT2 can be cleaved into two smaller fragments, a 140 kDa N-terminal product (N-SLIT2) and a 50-60 kDa C-terminal product (C-SLIT2). N-SLIT2 is the fragment which could bind with ROBO1, a single-pass transmembrane receptor of SLIT2 [8].
MicroRNAs (miRNAs) are a class of small non-coding RNAs that regulate gene expression at the post-transcriptional level [9]. MiRNAs play an important role in biological and pathologic procession including cell differentiation, proliferation, apoptosis and metabolism [10]. They can acted as oncogene or tumor suppressor [11]. Aberrant miRNA expression may correlate with many types of human disease and participating in every aspect of tumorigenesis. A recent study has shown that differential expression of miRNA was correlated with esophageal carcinoma survival [12].
The potential target gene of miRNA could be predicted by online computational algorithm such as TargetScan (http://www.targetscan.org/vert_50/) or PicTar (http://pictar.mdc-berlin.de/). In this study, TargetScan and PicTar both indicated that SLIT2 as a potential target gene of miR-1179. MiR-1179 has been identified to be associated with metastasis of colorectal cancer. Based on these observations, we aimed to detect the expression and the mechanism of miR-1179 in ESCC and analyze its correlation with clinicopathological factors or prognosis in our clinical samples.
Materials and methods
Patient and tissue samples
Between July 2011 and November 2012, 40 patients received resection for ESCC at The First Affiliated Hospital of Nanjing Medical University, Nanjing City, Jiangsu Province, and China. As a result, all the patients were retrospectively reviewed. None of these 40 patients received neoadjuvant therapy before operation. Fresh cancer tissues and paired normal adjacent tissues were obtained from these patients. The differentiation grade, TNM stage, and metastasis were classified according to the UICC/AJCC TNM classification (seventh edition). The Institutional Ethics Committee approved this project and written informed consents were obtained from the patients. The ESCC cell lines (TE-1 and Eca-109) were obtained from the Cell Bank of Shanghai (China) and cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), 100 units/mL penicillin, and 100 g/mL streptomycin (Invitrogen, Carlsbad, USA) at 37°C in a 5% CO2 incubator.
Transfection
MiR-1179 inhibitor and normal control were obtained from Genepharma (Shanghai, China). The transfection assay was conducted by using Lipofectamine 2000 (Invitrogen Corp, CA, USA).
Quantitative RT-PCR
Quantitative real time polymerase chain reaction (qRT-PCR) was performed to determine the expression levels of miR-1179 and mRNAs of all related genes. Total RNA was obtained from tissues using TRIzol reagent as described by the manufacturer (Invitrogen Life Technologies Co, CA, and USA). The detailed was described previously [12].
Protein analysis and immunocytochemistry (ICC)
For immunoblot analyses, 100 μg total proteins were electrophoresed on a 10% SDS-PAGE gel, transferred to PVDF membrane, blocked, and then incubated with primary antibody. The blots were developed using ECL reagent (Millpore, MASS, USA). Equal amount of protein loading in each lane was confirmed using GAPDH antibody. For the cell undergoing immunocytochemistry, 6-well-plate with glass-bottom was applied. Cells were fixed with 95% methyl alcohol and incubated with 10% normal goat serum for 30 min to block non-specific antibody binding. After washing, the samples were incubated with primary anti-rabbit antibody at 4°C overnight, and then washed in PBS for three times and then incubated with secondary antibodies. After that, the cells were stained with DAB according to manufacturer’s protocols and mounted and photographed using a digitalized microscope camera (Nikon Tokyo, Japan).
Cell invasion assays
For those cells treated with miRNA, after transfection for 48 h, cells were seeded at 1×106 cells/ml with serum-free medium, For the cells treated with SLIT2-N, after 48 h treatment, 100 ul cell suspension with serum-free medium was seeded to the upper chamber, cells were stained with crystal violet staining solution (Beyotime, Nantong, China) then counted and photographed under 40× magnification (five views per well). Migrated cells were counted by using Image-pro Plus 6.0 while cell numbers of normal control group were normalized to 1. All experiments were performed in triplicate.
Dual-luciferase reporter assay
The 3’-UTR sequence of SLIT2 predicted to interact with miR-1179 or a mutated sequence with the predicted target sites were inserted into pGL3 promoter vector (Genscript, Nanjing, China). For reporter assay, cells were plated onto 24-well plates and transfected with 100 ng of pGL3-SLIT2 or pGL3-SLIT2-mut, respectively by using Lipofectamine 2000 (Invitrogen Corp, CA, and USA). A Renilla luciferase vector pRL-SV40 (5 ng) was also co-transfected to normalize the differences in transfection efficiency. Transfection was repeated three times in triplicate.
Statistical analysis
The method of 2-ΔCt was used to analyze the results of RT-PCR in all the experiments performed in this study. Statistical analysis was performed using STATA 9.2, and presented with Graph PAD prism software. Experimental data of tissue samples are presented as box of the median and range of log-transformed relative expression level which was analyzed by Wilcoxon rank-sum (Mann-Whitney) test. Pearson analysis was applied for correlation. While the results obtained from experiment in vitro assays are presented as mean ± SEM from three separate experiments in triplicates per experiment, and the data was analyzed by double-sided Student’s t-test. Results were considered statistically significant at P < 0.05.
Results
Expression of miR-1179 and candidate gene SLIT2/ROBO1 in ESCC
We examined the expression of miR-1179 in a set of 40 paired samples using qRT-PCR. The results showed that miR-1179 was significantly up-regulated in ESCC tissues when compared to the adjacent tumor tissues (Figure 1A). Next, the correlation of miR-1179 with the clinicopathological factors was examined. We found that the aberrant expression level of miR-1179 was highly associated with the metastasis of patients, indicating that miR-1179 may play an important role the pathogenesis of ESCC (Table 1). As was predicted by bioinformatical analysis, we detected the expression level of SLIT2 and the receptor ROBO1 in the tissues. The results indicated that both SLIT2 and ROBO1 were decreased in ESCC tumor tissues (Figure 1A, 1D). Furthermore, we analyzed the relationship between the miR-1179 expression level and SLIT2 expression level and the correlation between SLIT2 and ROBO1 in the specimens of the patients. The result demonstrated that miR-1179 negatively correlated with the SLIT2 protein expression while SLIT2 indicated a positive correlation with ROBO1 (Figure 1B, 1C).
Figure 1.
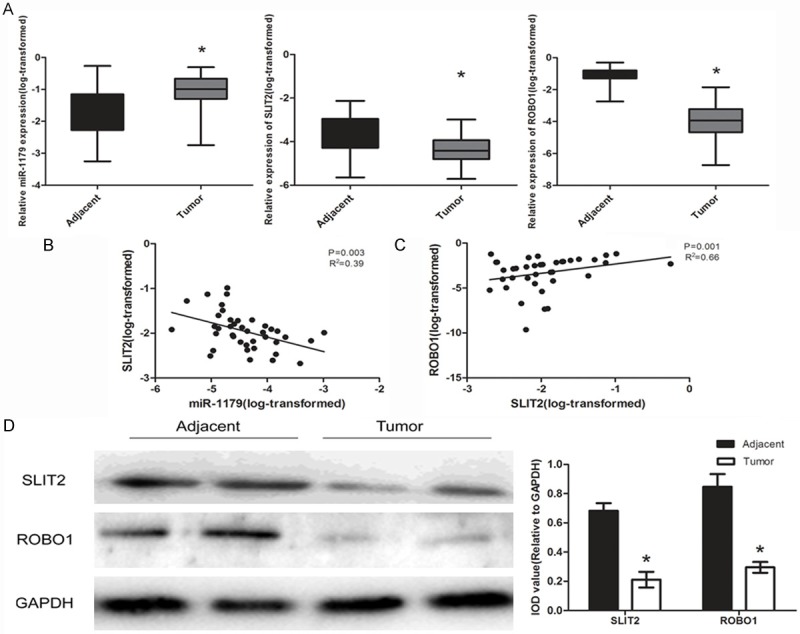
Expression levels of miR-1179, candidate gene SLIT2 and the receptor ROBO1 in ESCC tissues. A: The expression levels of miR-1179, SLIT2 and ROBO1 in human ESCC tissues and adjacent corresponding tissues (n = 40) were evaluated by qRT-PCR. Data were presented as box plot of the median and range of log-transformed relative expression level. *indicates significant difference (P < 0.05). B, C: Pearson analysis of miR-1179 and SLIT2, SLIT2 and ROBO1. D: The protein level of SLIT2 and ROBO1 in ESCC tissues, IOD was measured by normalized with GAPDH.
Table 1.
Clinicopathological features of patients with the expression of miR-1179
| miR-1179 | |||
|---|---|---|---|
|
|
|||
| Feather | Low | High | P value |
| All cases | 20 | 20 | |
| Age | |||
| < 60 | 14 | 11 | 0.327 |
| ≥ 60 | 6 | 9 | |
| Gender | 0.705 | ||
| Male | 15 | 16 | |
| Female | 5 | 4 | |
| Tumor Size (cm) | 0.752 | ||
| ≤ 5 cm | 10 | 9 | |
| > 5 cm | 10 | 11 | |
| Tumor Location | 0.429 | ||
| Middle | 5 | 3 | |
| Lower | 15 | 17 | |
| Tumor Capsular | 0.212 | ||
| Incomplete | 5 | 2 | |
| Complete | 15 | 18 | |
| TNM stage (I-II:III-IV) | 15:5 | 5:15 | 0.002 |
| Metastasis | |||
| Yes | 5 | 15 | 0.002 |
| No | 15 | 5 | |
Cell invasion was suppressed by the decreased level of miR-1179
TE-1 and Eca-106 were chosen for following experiments. After confirming that miR-1179 inhibitor could significantly change the expression level of miR-1179 using qRT-PCR, we then tested whether miR-1179 modulated cell invasion in esophageal cancer cells by transwell assay. As shown in Figure 2A and 2B, cells treated with miR-1179 inhibitor had a significant decreased ability of cell invasion compared with the control transfected cells (P < 0.05).The number of migrated cells was calculated. Cells in control group were normalized to 100%.
Figure 2.
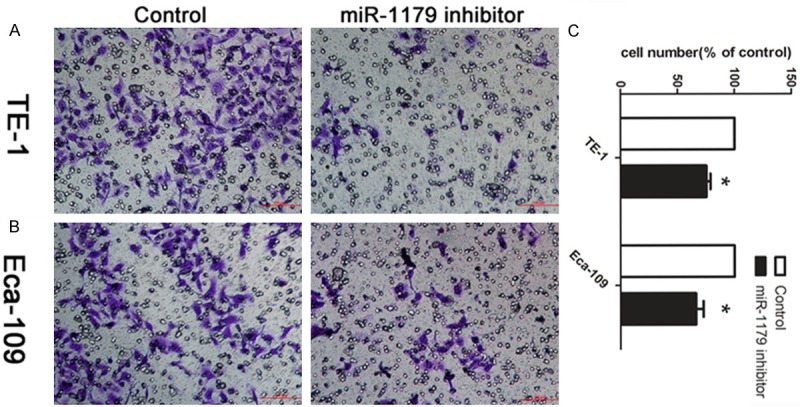
Decreased miR-1179 suppressed cell invasion in vitro. A: Cells were treated with miR-1179 inhibitor and control. The representative images of invasive cells of TE-1 at the bottom of the membrane stained with crystal violet were visualized as shown. B: The invasive affection in Eca-109 cells. C: The quantifications of cell migration were presented as percentage migrated cell numbers. *indicates significant difference compared with control group (P < 0.05).
SLIT2/ROBO1 was activated by the down-regulation of miR-1179
Based on the regulation pattern of miRNAs in human carcinoma, we applied the experiment to investigate whether miR-1179 could regulate the expression of SLIT2 in vitro. The affection of miR-1179 on the expression level of SLIT2 was further examined by ICC. We found that the down-regulation of miR-11179 caused a significant increase level of SLIT2 (Figure 3A). We further detected the expression level of ROBO1, the receptor of SLIT2, in cell treated with miR-1179 inhibitor and control. We found that cells treated with miR-1179 inhibitor presented an up-regulation of ROBO1 in TE-1 and Eca-109 cell lines (Figure 3B).
Figure 3.
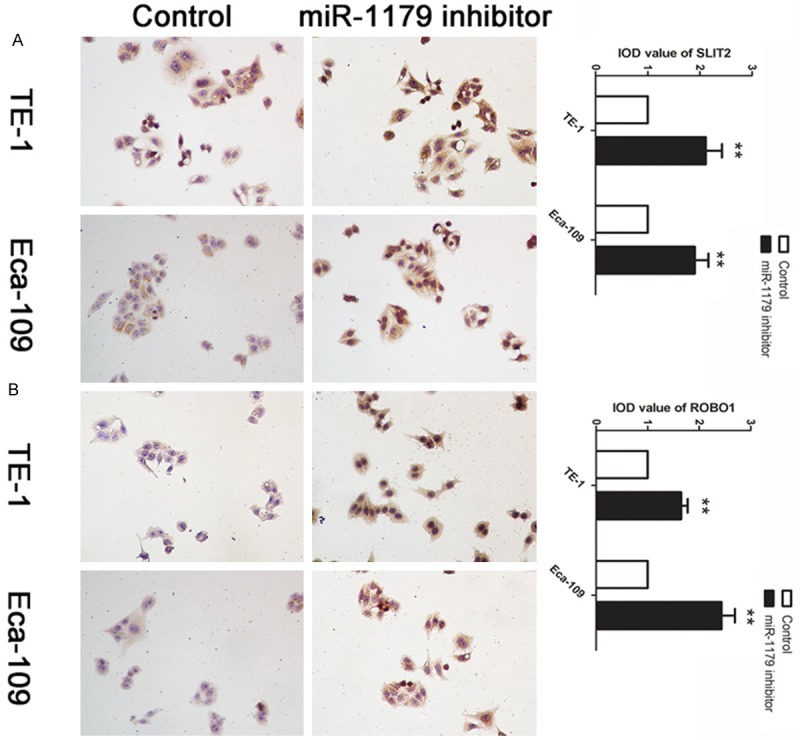
Decreased level of miR-1179 up-regulated SLIT2 and ROBO1 in TE-1 and Eca-109 cells. A: Cells were treated with miR-1179 inhibitor and control. ICC was applied to detect the expression level of SLIT2. IOD was calculated, data was presented with Mean ± SE. **indicates significant difference compared with control group P < 0.01. B, A: Cells were treated with miR-1179 inhibitor and control. ICC was applied to detect the expression level of ROBO1. IOD was calculated, data was presented with Mean ± SE. **indicates significant difference compared with control group P < 0.01.
Next, we performed miRNA luciferase reporter assay by constructing the wild type and mutant type luciferase reporter plasmids containing the binding region of the 3’UTR of SLIT2 (Figure 4A, 4B). We found that co-transfection of miR-1179 mimics and pGL3-SLIT2 3’UTR reporter plasmids significantly decreased the luciferase activity in cell lines, as compared with the control and the mutant type, indicating that miR-1179 directly binding with the 3’UTR of SLIT2 (Figure 4C).
Figure 4.
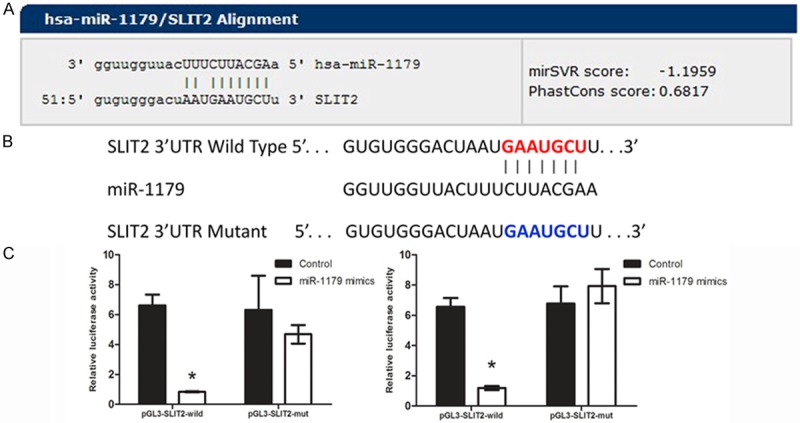
MiR-1179 down-regulated SLIT2 by binding the 3’UTR of SLIT2. A: The binding site of miR-1179 with SLIT2. B: Sequence alignment of human miR-1179 with 3’UTR of SLIT2. Bottom: mutations in the 3’-UTR of SLIT2 in order to create the mutant luciferase reporter construct. C: Cells were co-transfected with miR-1179 mimics or miR-control, renilla luciferase vector pRL-SV40 and SLIT2 3’UTR luciferase reporters for 48 h. Both firefly and Renilla luciferase activities are measured in the same sample. The left panel indicated TE-1 cell line while the right indicated Eca-109 Firefly luciferase signals were normalized with Renilla luciferase signals. *indicates significant difference compared with that of control cells (P < 0.05). All tests were performed in triplicate and presented as mean ± SE.
SLIT2/ROBO1 axis promoted cell invasion in ESCC cell lines
Based on the results above, we aimed to investigate the potential mechanism involved in the invasion suppression of miR-1179. SLIT2 was reported with as an exactions protein. Thus, we conducted the expression to detect whether SLIT2 could cause the abnormal invasion of cells by treating with SLIT2-N. As presented in Figure 5A, cell treated with SLIT2-N showed a decreased ability of invasion comparing with cell treated with placebo. In addition, we further investigated whether the SLIT2 suppressed the cell invasion through the combination of the receptor ROBO1. ICC was applied to detect the expression level of ROBO1 in cells treated with SLIT2-N. We found that ROBO1 was up-regulated in cells treated with SLIT-N (Figure 5B).
Figure 5.
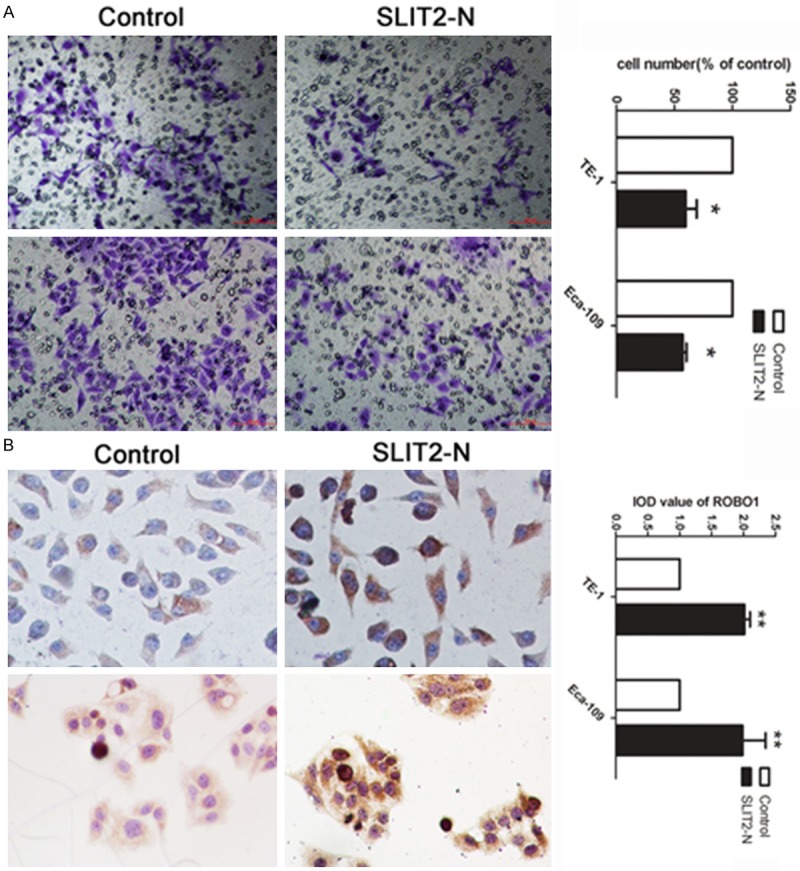
SLIT2 inhibited cell invasion by combining with ROBO1. A: Transwell assay was performed as described in Materials and Methods. The representative images of invasive cells at the bottom of the membrane stained with crystal violet were visualized as shown (left). The quantifications of cell migration were presented as percentage migrated cell numbers (right). B: The expression level of ROBO1 was detected by ICC in cells treated with SLIT2-N and control. IOD was calculated by presenting as mean ± SE. *indicates significant difference compared with that of control cells (P < 0.05). **indicated P < 0.01. All tests were performed in triplicate and presented as mean ± SE.
Discussion
Previous studies have reported that miRNAs play important roles in regulating gene expression. MiRNAs are a group of small non-coding RNAs with 18-25 nucleotides in length that negatively regulate gene expression by imprecisely binding to complementary sequence in the 3’-UTR of their target mRNAs [13,14]. Researchers have found that a large number of aberrant expression level of genes participant in the progression of ESCC [15]. The interaction between these genes mostly exists in the transcription level and post-transcription level [16]. MicroRNAs are defined as one of the most important member in the post-transcription level regulation [17]. MiR-375 have been proved to inhibit tumor growth and metastasis in ESCC by down-regulating the IGF1R, acting as a strong tumor-suppression factor [18]. Besides, miR-138 could suppress the level of NF-κB which promotes lipid raft formation in ESCC [19]. All these evidence indicated that miRNAs play a crucial role in the progression of ESCC.
Based on the important role of miRNAs, we found that miR-1179 was involved in the metastasis of human colorectal cancers. However, the potential mechanism of miR-1179 was only explored by predicting through bioinformatic techniques. In this study, we tried to found the undergoing mechanism of miR-1179 in the pathogenesis of ESCC. Bioinformatics was applied to get the target gene of miR-1179. By combining the binding ability with the gene function, we selected the SLIT2 as the candidate gene of miR-1179. 3’-UTR of SLIT2 was confirmed binding with miR-1179 thus, caused the suppression of cell invasion with the down-regulation of miR-1179.
SLIT2/ROBO1 signaling has been demonstrated to be angiogenic factors under certain circumstances and SLIT2/ROBO1 interaction could inhibit cell migration [5,8]. Researchers have confirmed that SLIT2-N, the functional subunit of SLIT2 suppressed cell migration whereas knockdown of ROBO1 reversed such inhibitory effect [20], suggesting SLIT2/ROBO1 axis was involved in the pathogenesis of cancer by regulating cell invasion.
In conclusion, this was the first study to evaluate the relationship between SLIT2 and miR-1179 in ESCC. Our findings demonstrated that miR-1179 was up-regulated in ESCC, and expression of SLIT2 was correlated with tumor metastasis. We found that miR-1179 post-transcriptional down-regulated SLIT2 expression in vitro. We also concluded that miR-1179 promoted invasive ability of ESCC cells, and an SLIT2/ROBO1 mediated axis may be involved in this function.
Acknowledgements
This work was supported by Grants: Wujin Science and Technique Development Foundation (Social development) [WS201414]; Clinical Science and Technique Development Foundation of Jiangsu University [20140039].
Disclosure of conflict of interest
None.
References
- 1.Zhu S, Wang Z, Zhang Z, Wang J, Li Y, Yao L, Mei Q, Zhang W. PTPLAD2 is a tumor suppressor in esophageal squamous cell carcinogenesis. FEBS Lett. 2014;588:981–989. doi: 10.1016/j.febslet.2014.01.058. [DOI] [PubMed] [Google Scholar]
- 2.Wang ZW, Zhang W, Dong W, Li BS, Mu DB, Huang W, Zhang J, Li HS, Zhang ZC, Lin HQ, Yi Y. Pathological analysis of extracapsular extension of metastatic lymph node and its potential impact on nodal clinical target volume in the radiotherapy of esophageal squamous cell carcinoma. Neoplasma. 2014;61:324–330. doi: 10.4149/neo_2014_042. [DOI] [PubMed] [Google Scholar]
- 3.Zhang X, Zhou L, Fu G, Sun F, Shi J, Wei J, Lu C, Zhou C, Yuan Q, Yang M. The identification of an ESCC susceptibility SNP rs920778 that regulates the expression of lncRNA HOTAIR via a novel intronic enhancer. Carcinogenesis. 2014;35:2062–7. doi: 10.1093/carcin/bgu103. [DOI] [PubMed] [Google Scholar]
- 4.Lee KW, Sung CO, Kim JH, Kang M, Yoo HY, Kim HH, Um SH, Kim SH. CD10 expression is enhanced by Twist1 and associated with poor prognosis in esophageal squamous cell carcinoma with facilitating tumorigenicity in vitro and in vivo. Int J Cancer. 2014;136:310–21. doi: 10.1002/ijc.29006. [DOI] [PubMed] [Google Scholar]
- 5.Gohrig A, Detjen KM, Hilfenhaus G, Korner JL, Welzel M, Arsenic R, Schmuck R, Bahra M, Wu JY, Wiedenmann B, Fischer C. Axon guidance factor SLIT2 inhibits neural invasion and metastasis in pancreatic cancer. Cancer Res. 2014;74:1529–1540. doi: 10.1158/0008-5472.CAN-13-1012. [DOI] [PubMed] [Google Scholar]
- 6.Zhou WJ, Geng ZH, Spence JR, Geng JG. Induction of intestinal stem cells by R-spondin 1 and Slit2 augments chemoradioprotection. Nature. 2013;501:107–111. doi: 10.1038/nature12416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang PH, Hwang-Verslues WW, Chang YC, Chen CC, Hsiao M, Jeng YM, Chang KJ, Lee EY, Shew JY, Lee WH. Activation of Robo1 signaling of breast cancer cells by Slit2 from stromal fibroblast restrains tumorigenesis via blocking PI3K/Akt/beta-catenin pathway. Cancer Res. 2012;72:4652–4661. doi: 10.1158/0008-5472.CAN-12-0877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alajez NM, Lenarduzzi M, Ito E, Hui AB, Shi W, Bruce J, Yue S, Huang SH, Xu W, Waldron J, O'Sullivan B, Liu FF. MiR-218 suppresses nasopharyngeal cancer progression through downregulation of survivin and the SLIT2-ROBO1 pathway. Cancer Res. 2011;71:2381–2391. doi: 10.1158/0008-5472.CAN-10-2754. [DOI] [PubMed] [Google Scholar]
- 9.Tie J, Pan Y, Zhao L, Wu K, Liu J, Sun S, Guo X, Wang B, Gang Y, Zhang Y, Li Q, Qiao T, Zhao Q, Nie Y, Fan D. MiR-218 inhibits invasion and metastasis of gastric cancer by targeting the Robo1 receptor. PLoS Genet. 2010;6:e1000879. doi: 10.1371/journal.pgen.1000879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar P, Johnston BH, Kazakov SA. miR-ID: a novel, circularization-based platform for detection of microRNAs. RNA. 2011;17:365–380. doi: 10.1261/rna.2490111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao L, Sun Y, Hou Y, Peng Q, Wang L, Luo H, Tang X, Zeng Z, Liu M. miRNA expression analysis of cancer-associated fibroblasts and normal fibroblasts in breast cancer. Int J Biochem Cell Biol. 2012;44:2051–2059. doi: 10.1016/j.biocel.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 12.Zhu Y, Xia Y, Niu H, Chen Y. miR-16 induced the suppression of cell apoptosis while promote proliferation in esophageal squamous cell carcinoma. Cell Physiol Biochem. 2014;33:1340–1348. doi: 10.1159/000358701. [DOI] [PubMed] [Google Scholar]
- 13.Guo CJ, Pan Q, Li DG, Sun H, Liu BW. miR-15b and miR-16 are implicated in activation of the rat hepatic stellate cell: An essential role for apoptosis. J Hepatol. 2009;50:766–778. doi: 10.1016/j.jhep.2008.11.025. [DOI] [PubMed] [Google Scholar]
- 14.Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26:2799–2803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 15.Chen Z, Li J, Tian L, Zhou C, Gao Y, Zhou F, Shi S, Feng X, Sun N, Yao R, Shao K, Li N, Qiu B, Tan F, He J. miRNA expression profile reveals a prognostic signature for esophageal squamous cell carcinoma. Cancer Lett. 2014;350:34–42. doi: 10.1016/j.canlet.2014.04.013. [DOI] [PubMed] [Google Scholar]
- 16.Turato C, Vitale A, Fasolato S, Ruvoletto M, Terrin L, Quarta S, Ramirez Morales R, Biasiolo A, Zanus G, Zali N, Tan PS, Hoshida Y, Gatta A, Cillo U, Pontisso P. SERPINB3 is associated with TGF-beta1 and cytoplasmic beta-catenin expression in hepatocellular carcinomas with poor prognosis. Br J Cancer. 2014;110:2708–2715. doi: 10.1038/bjc.2014.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Q, Xie D, Wang S, You C, Monroig O, Tocher DR, Li Y. miR-17 is involved in the regulation of LC-PUFA biosynthesis in vertebrates: effects on liver expression of a fatty acyl desaturase in the marine teleost Siganus canaliculatus. Biochim Biophys Acta. 2014;1841:934–943. doi: 10.1016/j.bbalip.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 18.Ye XM, Zhu HY, Bai WD, Wang T, Wang L, Chen Y, Yang AG, Jia LT. Epigenetic silencing of miR-375 induces trastuzumab resistance in HER2-positive breast cancer by targeting IGF1R. BMC Cancer. 2014;14:134. doi: 10.1186/1471-2407-14-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gong H, Song L, Lin C, Liu A, Lin X, Wu J, Li M, Li J. Downregulation of miR-138 sustains NF-kappaB activation and promotes lipid raft formation in esophageal squamous cell carcinoma. Clin Cancer Res. 2013;19:1083–1093. doi: 10.1158/1078-0432.CCR-12-3169. [DOI] [PubMed] [Google Scholar]
- 20.Ning Y, Sun Q, Dong Y, Xu W, Zhang W, Huang H, Li Q. Slit2-N inhibits PDGF-induced migration in rat airway smooth muscle cells: WASP and Arp2/3 involved. Toxicology. 2011;283:32–40. doi: 10.1016/j.tox.2011.01.026. [DOI] [PubMed] [Google Scholar]


