Abstract
MicroRNAs (miRNAs) are a class of small, non-coding RNAs, which have demonstrated to important gene regulators, and have critical roles in diverse biological processes including cancer cell proliferation. Previous studies suggested microRNA-338-3p (miR-338-3p) was down-regulated and play tumor suppressor roles in gastric cancer, colorectal carcinoma and lung cancer. However, the role of miR-338-3p in hepatocellular carcinoma (HCC) is still unclear. In this study, we analyzed the expression of miR-338-3p in HCC tissues and HCC cell lines. We find that miR-338-3p was downregulated in HCC tissues and cell lines. Then functional studies demonstrate ectopic miR-338-3p expression significantly suppressed the in vitro proliferation and colony formation of HCC cells and cause to cell cycle arrest. Using bio-informatic method and report assay we identified a novel miR-338-3p target, FOXP4 in HCC cells. Furthermore, knockdown of FOXP4 have the similar effects in HCC corrected with miR-338-3p. These findings suggest that miR-338-3p regulates survival of HCC cells partially through the downregulation of FOXP4. Therefore, targeting with the miR-338-3p/FOXP4 axis might serve as a novel therapeutic application to treat HCC patients.
Keywords: miR-338-3p, HCC, FOXP4, cell growth, cell cycle
Introduction
Hepatocellular carcinoma (HCC), which is the sixth most commonly malignant tumor and third leading cause of cancer-related death worldwide [1,2], has a high mortality [3]. The tumorigenesis process of HCC is a complicate and involving many gene alterations including microRNAs (miRNAs) [4]. Although many researchers have demonstrate many signal pathways in HCC proliferation and cell cycle, understanding the molecular mechanism of HCC cell growth is full of challenge. Currently, miRNA offers a novel molecular approach and has been reported to be involved in HCC pathogenesis [5].
MicroRNAs (miRNA) are small, non-coding RNAs, approximate 22nt in length and bind to partially complementary recognition sequences of mRNA, causing either degradation or inhibition of translation, thus effectively silencing their mRNA target [6]. The smaller gene regulator has been reported to be participating in various biological processes such as differentiation, morphogenesis and tumorigenesis [7,8]. Previous studies have indicated miRNAs could play oncogene or tumor suppressor roles in the etiology and pathogenesis of cancer by targeting tumor suppressors or oncogenes [9,10]. Many miRNAs have been identified to participated in HCC cellular transformation and tumorigenesis such as MiR-126-3p [11] and miR-1285-3p [12]. However the characterization of miR-338-3p associated with HCC progression and development is still unclear.
The aim of the present study was to demonstrate the role of miR-338-3p in HCC. And we explored its functions in HCC HepG2 and Hep-3B cells with MTT and Colony formation assays. Then we want to examine the effects of miR-338-3p on the cell cycle of HCC cells. Moreover, the direct target of miR-338-3p was finding in HCC cells. To our knowledge, our study is the first to document the role of miR-338-3p-3p in HCC.
Materials and methods
Tissue samples
A total of 30 patients who were diagnosed as primary HCC in Department of Oncology, Jinan Central hospital were included in this study. None of these patients received chemotherapy and radiotherapy before the surgery. Tumor and corresponding non-tumor lung tissue samples were collected and rapidly frozen in liquid nitrogen and stored at -80°C. Ethical approval was obtained from the hospital and fully informed consent from all patients before sample collection.
Cell culture
Two HCC cell lines (HepG2 and Hep-3B) were cultured in RPMI 1640 (GIBCO-BRL) medium supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 mg/ml streptomycin in humidified air at 37°C with 5% CO2.
Bioinformatics methods
The miRNA targets predicted by computer-aided algorithms were obtained from targetscan (http://www.targetscan.org).
Real-time reverse transcription (qRT) polymerase chain reaction (PCR)
A TaqMan miRNA-assay kit was obtained from Applied Biosystems (Foster City, CA, USA) for the detection of mature miR-338-3p expression. According to the manufacturer’s instructions, the 2-DeltCt method was used in conjunction with the RNU6B gene as a control for normalization. All experiments were performed in triplicate and repeated once. To verify integrity of FOXP4 expression, GAPDH gene was used as an internal control. PCR conditions were 30 cycles consisted of denaturation at 94°C for 30 s, annealing at 56°C (58°C for GAPDH) for 30 s, and extension at 72°C for 30 s. Each PCR product was separated by 1.5% agarose gel electrophoresis and visualized by ethidium bromide staining.
Western blot assay
Cell protein lysates were separated in 10% sodium dodecyl sulfate polyacrylamide gels, electrophoretically transferred to polyvinylidene difluoride membranes (Roche Diagnostics, Mannheim, Germany), then detected with anti-FOXP4. Protein loading was estimated using mouse anti-GAPDH monoclonal antibody. Lab Works Image Acquisition and Analysis Software (UVP, Upland, CA, USA) were used to quantify band intensities.
Assay of luciferase activity
The 3’UTR of FOXP4 was amplified and cloned into the downstream of pGL3/Luciferase vector. Then the mutant 3’UTR of FOXP4 (several nucleotides within the binding sites were mutantant) was amplified using pGL3/Luc-FOXP4 3’UTR as the template, and then was cloned into the downstream of pGL3/Luciferase vector. For the luciferase reporter assay, the cells were co-transfected with miR-338-3p mimics or control and pGL3/Luciferase-FOXP4 3’UTR or the mutant 3’UTR, together with the controls. At 48 h after transfection, the cells were lysed using RIPA buffer, and then Luciferase intensity was measured by an F4500 Fluorescence Spectrophotometer (HITACHI).
Cell proliferation (MTT) assay and colony formation assay
The transfected cells were plated into 96-well plates at a density of 5000 cells/well. At 48 h after transfection, the cells were incubated with MTT (3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) for 4 h at 37°C. Then the cells were agitated with MTT solvent on an orbital shaker for 10 min avoiding light. The absorbance at 490 nm (OD490 nm) was measured with a spectrophometer. The cells were seeded into 12-well plate at a density of 200 cells/well after transfection. The medium was changed every three days. Approximately 10 days later, most of the cell clones contained more than 50 cells. The clones were washed with 1×PBS and stained with crystal violet for about 5 min. Finally the clones were taken pictures and counted. The colony formation rate = (number of clones)/(number of seeded cells) ×100%.
Statistical analysis
All the data were shown as mean + SD and the experiments were repeated three times. The difference was determined by two-tailed students’t-test and P < 0.05 was considered statistically significant.
Results
Decreased miR-338-3p expression in HCC tissues and HCC cell lines
MiR-338-3p could be observed to be significantly downregulated in HCC tissues compared with that in matched adjacent normal lung tissues (Figure 1A). The characteristics of 30 HCC patients were shown (Table 1). Among 30 matched normal and HCC tumor tissues, miR-338-3p was significantly downregulated in 30 cancer tissues compared with the normal tissues. Furthermore, we also examined the expression of miR-338-3p in HCC cells HepG2, QGY7703 and Hep-3B compared to Normal tissue and LO2 cells (Figure 1B). The miR-338-3p downregulation in HCC cell lines was correlated with HCC tumor tissues. Therefore, we concluded that downregulation of miR-338-3p expression might play important roles in HCC progression and development.
Figure 1.
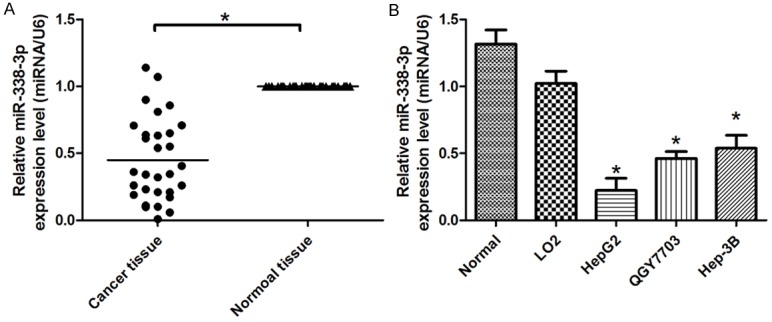
Determining miR-338-3p expression in HCC tissues and cell lines. A. Relative expression of miR-338-3p in HCC tissues in comparison with corresponding non-tumor lung tissues. B. The miR-338-3p expression was significantly lower in HCC cell lines compared with Normal tissue or LO2 cells. MiR-338-3p expression was normalized to U6 expression. And the results were reproducible in three independent experiments. *P < 0.05 was considered as statistic significant.
Table 1.
Characteristics of HCC patients in this study
| Features | Number of cases |
|---|---|
| Tumor size (cm) | |
| < 5 | 12 |
| ≥ 5 | 18 |
| HBV infection | |
| Negative | 5 |
| Positive | 25 |
| Age | |
| ≥ 60 | 13 |
| < 60 | 17 |
| Differentiation | |
| Well and moderate | 15 |
| Poor or no differentiation | 15 |
| TNM stage | |
| I+II | 14 |
| III+IV | 16 |
| Lymphatic metastasis | |
| pN (N0) | 12 |
| pN (N1+N2+N3) | 18 |
Effects of miR-338-3p expression on in vitro proliferation of HCC cells
Based on miR-338-3p was downregulated in HCC tissues and cell lines, we ectopic over-expressed miR-338-3p in HCC cells. As results from TaqMan real-time PCR assay, miR-338-3p could significantly upregulate the level of miR-338-3p expression in HepG2 and Hep-3B cells by 8.5 or 7.2-fold, respectively (Figure 2A). To testify that miR-338-3p may function as a tumor suppressor, the effects of miR-338-3p overexpression on the proliferation of HCC cell were determined in vitro. Thereafter, a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide (MTT) and colony formation assays were performed to detect the cell viability. HepG2 and Hep-3B cells transfected with miR-338-3p was observed to grow more slowly than those cells transfected miR-controls (Figure 2B, 2C). And Figure 2D and 2E shows the colony numbers of HepG2 and Hep-3B cells transfected with miR-338-3p were significantly lower than those transfected with miR-control. To further verify the role of miR-338-3p in HCC cells, cell cycle analysis was performed in HepG2 and Hep-3B cells. The results shows over-express miR-338-3p could caused to the G1 to S spare arrest in HCC cells (Figure 2F) Thus, these results indicated that ectopic miR-338-3p expression could inhibit in vitro proliferation of HCC cells.
Figure 2.
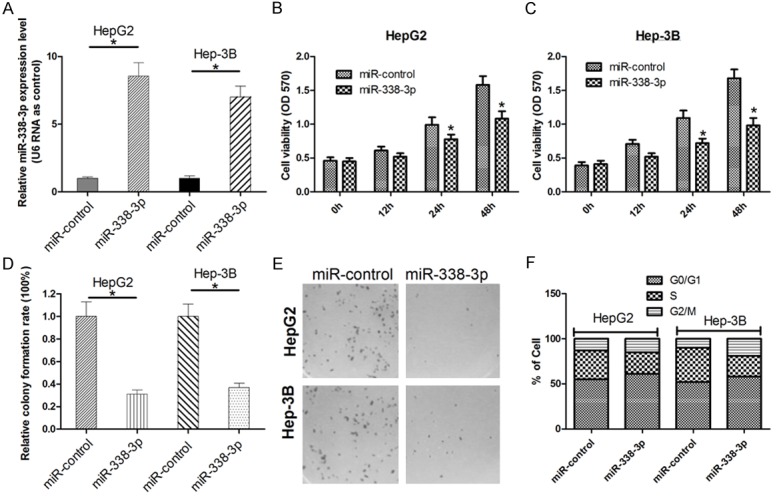
Overexpression of miR-338-3p inhibits in vitro growth of HCC cells. (A) HepG2 and Hep-3B cells were transfected with miR-338-3p or miR-controls, respectively. (B, C) At 12 h, 24 h and 48 h after transfection, MTT assay was performed to determine the proliferation of HepG2 and Hep-3B cells. Data represent the mean ± s.d. from three independent experiments. (D, E) Colony formation rate are shown in each group. Representative results of colony formation of HepG2 and Hep-3B cells transfected with miR-338-3p or miR-control (F) FACS was used to detect the effect of miR-338-3p on cell cycle in HepG2 and Hep-3B cells. The results were reproducible in three independent experiments. *P < 0.05 was considered as statistic significant.
MiR-338-3p directly targets FOXP4 gene by interaction with the binding site in the 3’-UTR
The miRNA target prediction algorithm TargetScan was used to computational screen for genes with complementary sites of miR-338-3p in their 3’-UTR. We found that forkhead box P4 (FOXP4) was a putative target of miR-338-3p. To confirm this possibility, the miR-338-3p binding sequences present at the 3’-UTR of FOXP4 mRNA (WT-3’-UTR) or its mutant site (FOXP4-3’UTR-mut) were subcloned into the downstream of the luciferase reporter gene in pGL3 vector (Figure 3A) and then co-transfected with into HepG2 and Hep-3B cells. The relative luciferase activity of the reporter that contained wildtype 3’-UTR was significantly decreased by 30% when miR-338-3p was co-transfected, but the luciferase activity of FOXP4-3’UTR-mut reporter was unaffected by simultaneous transfection of miR-338-3p (Figure 3B, 3C). These results suggesting that miR-338-3p might suppress FOXP4 expression through the putative binding site in its 3’UTR. Furthermore, real time PCR and Western Blot assays were performed to check whether miR-338-3p expression affects the expression of endogenous FOXP4 at both transcriptional and translational levels. Consistent with the results of Luciferase report assay, the levels of FOXP4 mRNA showed a significant decreasing between miR-338-3p transfected HCC cells and miR-control transfected cells (Figure 3D). And Western blot analysis showed that the level of FOXP4 protein expression in miR-338-3p transfected HCC cells was significantly inhibited by 80% compared with that in miR-control transfected cells (Figure 3E). These data indicated that miR-338-3p could directly target FOXP4 in HCC cells by interaction with the binding site in 3’-UTR of FOXP4 gene.
Figure 3.
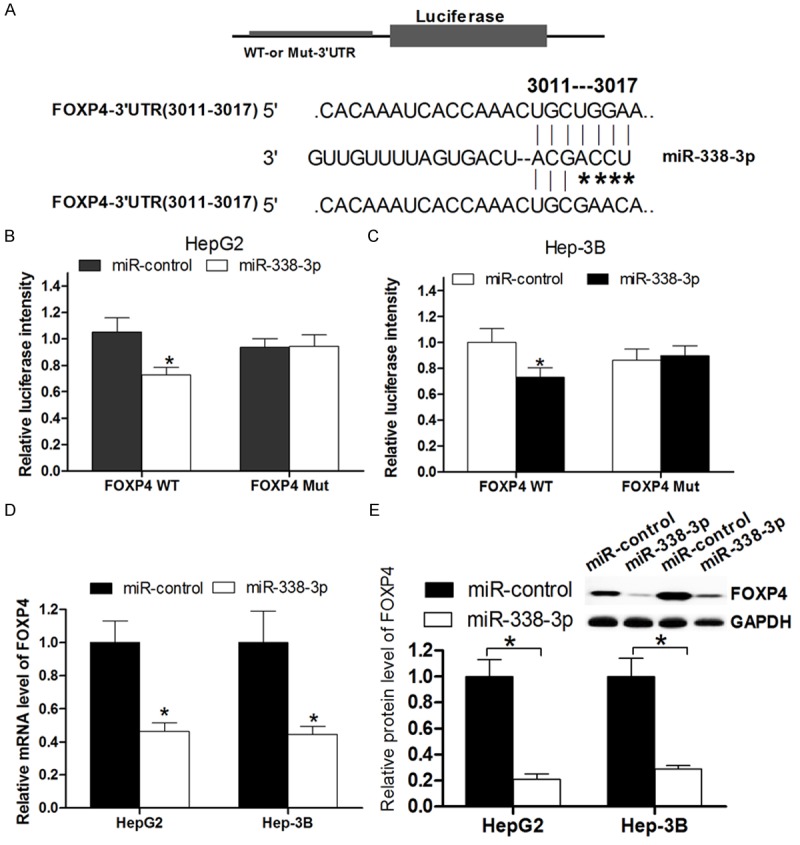
MiR-338-3p directly targets FOXP4. A. The putative miR-338-3p-binding sites in the 3’-UTR of FOXP4 mRNA was shown. Mutation was generated on the FOXP4 3’-UTR sequence in the complementary site for the seed region of miR-338-3p. B, C. The wild type (FOXP4 3’-UTR-WT) or mutant (FOXP4 3’-UTR-mut) reporter plasmids was co-transfected into HepG2 and Hep-3B cells with miR-338-3p or miR-control. The normalized luciferase activity in the control group was set as relative luciferase activity. D. The expression of FOXP4 mRNA was analyzed by real time PCR assay. GAPDH was used as an internal control. E. The expression of FOXP4 protein was analyzed by western blot assay. GAPDH was used as an internal control. All experiments were at least repeated in triplicate with similar results (*P < 0.05 was considered as statistic significant).
Knockdown of FOXP4 inhibits cell proliferation
To investigate the function of FOXP4 in HCC cells, HepG2 and Hep-3B were transfected with FOXP4 siRNA. As shown in Figure 4A and 4B FOXP4 siRNA decreased 75% and 90% of the expression of FOXP4 in HepG2 and Hep-3B cells, respectively. Colony formation and MTT assays were performed to examine the effects of FOXP4 on cell growth in vitro. Our data demonstrated that relative cell growth was significantly inhibited in FOXP4 siRNA transfected cells (Figure 4C-E). These results may partly elucidated the miR-338-3p induced growth inhibition of HCC cells though directly target FOXP4.
Figure 4.
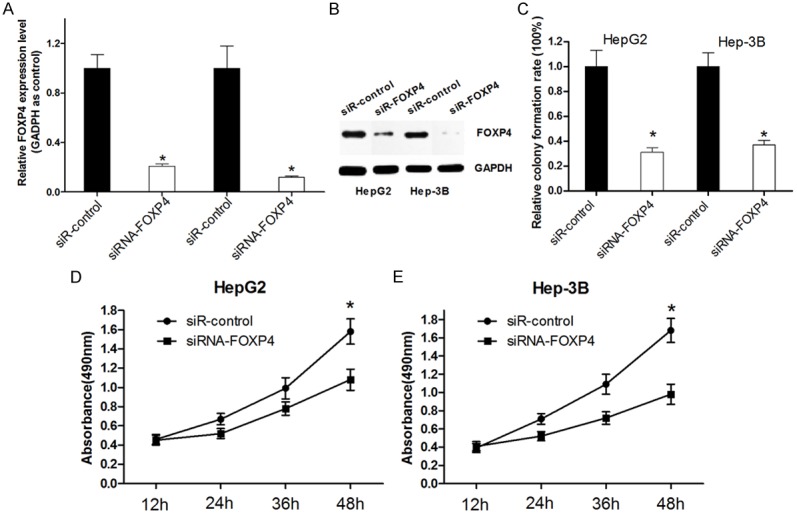
Knockdown of FOXP4 inhibits the cell proliferation of HCC cells. A, B. Western blot was used to detect the transfection efficiency of siRNA FOXP4 in HCC cells. C. The long term cell growth as determined by colony formation assay. D, E. Cell survival was determined by MTT assay. All experiments were at least repeated in triplicate with similar results (*P < 0.05 was considered as statistic significant).
Expression of FOXP4 in HCC tissues and cell lines
Since we have demonstrated the expression levels of miR-338-3p in HCC cell lines and tissues, we then used quantitative real-time PCR (qRT-PCR) to measure FOXP4 mRNA expression levels in the 30 pairs HCC tissues and adjacent normal tissues. Compared to Normal tissues, the expression of FOXP4 was obviously increased in HCC tissues (Figure 5A). Furthermore, the expression of FOXP4 was downregulated in HCC cell lines showed in Figure 5B. These results demonstrate the oncogene role of FOXP4 in HCC and imply the miR-338-3p/FOXP4 may serve as a potential target for HCC therapy.
Figure 5.
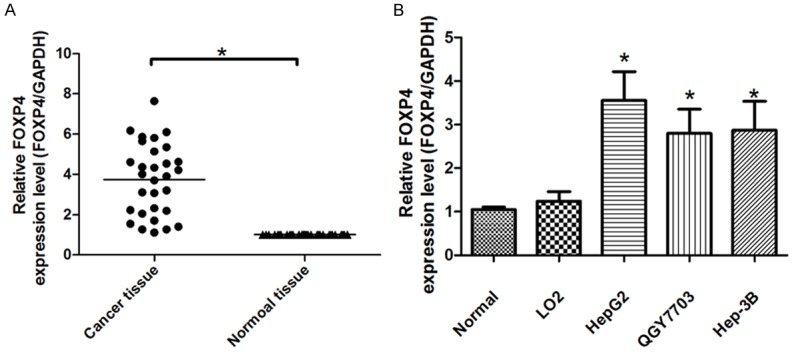
FOXP4 expression levels in HCC tissues. A. qRT-PCR was used to detect the expression of FOXP4 in HCC tissues and adjacent normal tissues (Normal tissue); B. qRT-PCR was used to detect the expression of FOXP4 in HCC cell lines Normal tissue. All experiments were at least repeated in triplicate with similar results (*P < 0.05 was considered as statistic significant).
Discussion
Hepatocelluar carcinoma (HCC) is currently the fifth highest in solid tumor and is the third leading cause of cancer death worldwide, which accounting for approximately one million deaths annually [3,13]. Althogh these years the techniques treated to HCC have been improved, unfortunately, the survival rate of HCC patients remains poor despite recent advances in medical treatment and surgical techniques [14,15]. So it is important to understanding the molecular mechanism of HCC development for effective therapy.
Evidences have been report that numerous of miRNAs expressions were deregulated occurs frequently in HCC, and contributed to tumorigenesis. Therefore, it is crucial to explore the function of deregulated miRNAs in HCC. Previous studies have discovered that miR-338-3p is downregulated in colorectal carcinoma and gastric Cancer [16,17]. However, the role of miR-338-3p in HCC was still poorly explored. In this study, we determined the deregulated expression of miR-338-3p in HCC using real time PCR, and confirmed miR-338-3p was significantly down-regulated in HCC tissues and cell lines compared to the corresponding matched adjacent normal tissues.
Previous study has demonstrated that miR-338-3p act as tumor suppressor role in CRC cells and suppressed cell proliferation and induced apoptosis. This tumor suppressor miRNA also has been investigated to suppress invasion of liver cancer cell [18]. So we speculate miR-338-3p may inhibit HCC cell proliferation. MTT, colony formation assay and cell cycle assay were performed to demonstrate this idea. Indeed, the ectopic expression of miR-338-3p could suppress HCC cells proliferation and induce cell cycle arrest in HepG2 and Hep-3B cells. These results demonstrated miR-338-3p suppress the proliferation of HCC cells, and triggered cell cycle arrest in vitro further support the tumor suppressor role in HCC.
MiRNA could function as tumor suppressor or oncogene through target oncogene or tumor suppressor gene. In this study, we want to explore the targets of miR-338-3p that may explain its cell proliferation inhibitor role in HCC. We used Targetscan bioinformatics for target genes prediction, and the FOXP4 as the potential target for further validation. FOXP4, as a member of the Foxp subfamily of winged-helix transcription factors, encodes a 685-amino-acid protein that is similar to Foxp1 and Foxp2. Foxp4 gene expression is observed primarily in pulmonary, neural, and gut tissues during embryonic development [19]. To the present, Wiehagen KR et al reported Foxp4 is dispensable for T cell development, but required for robust recall responses [20]. Long et al. provide further support for association of the FOXP4 with prostate cancer in Eastern Asian populations [21]. In this study, we performed Luciferase reporter assay, Western blot assay and real time PCR assays to verify that miR-338-3p can directly target FOXP4 though interaction with the biding sites in its 3’UTR. And knockdown of FOXP4 has the similar effects with miR-338-3p overexpression in HCC cells, which may explained the miR-338-3p induced HCC cell growth inhibition.
In conclusion, we have these major findings: 1) miR-338-3p was downregulated in HCC tissues and cell lines compared with the normal tissues. 2) Overexpression of miR-338-3p inhibits HCC cell proliferation and induces cell cycle arrest. 3) miR-338-3p could directly target FOXP4, which as a novel target of miR-338-3p. 4) Knockdown of FOXP4 has the similar effects with miR-338-3p overexpression in HCC.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81472329).
Disclosure of conflict of interest
None.
References
- 1.Waly Raphael S, Yangde Z, Yuxiang C. Hepatocellular carcinoma: focus on different aspects of management. ISRN Oncol. 2012;2012:421673. doi: 10.5402/2012/421673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379:1245–1255. doi: 10.1016/S0140-6736(11)61347-0. [DOI] [PubMed] [Google Scholar]
- 3.Bosch FX, Ribes J, Diaz M, Cleries R. Primary liver cancer: worldwide incidence and trends. Gastroenterology. 2004;127:S5–S16. doi: 10.1053/j.gastro.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 4.Farazi PA, DePinho RA. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat Rev Cancer. 2006;6:674–687. doi: 10.1038/nrc1934. [DOI] [PubMed] [Google Scholar]
- 5.Toffanin S, Hoshida Y, Lachenmayer A, Villanueva A, Cabellos L, Minguez B, Savic R, Ward SC, Thung S, Chiang DY, Alsinet C, Tovar V, Roayaie S, Schwartz M, Bruix J, Waxman S, Friedman SL, Golub T, Mazzaferro V, Llovet JM. MicroRNA-based classification of hepatocellular carcinoma and oncogenic role of miR-517a. Gastroenterology. 2011;140:1618–1628. e1616. doi: 10.1053/j.gastro.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 8.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 9.Shenouda SK, Alahari SK. MicroRNA function in cancer: oncogene or a tumor suppressor? Cancer Metastasis Rev. 2009;28:369–378. doi: 10.1007/s10555-009-9188-5. [DOI] [PubMed] [Google Scholar]
- 10.Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 11.Du C, Lv Z, Cao L, Ding C, Gyabaah OA, Xie H, Zhou L, Wu J, Zheng S. MiR-126-3p Suppresses Tumor Metastasis and Angiogenesis of Hepatocellular Carcinoma by Targeting LRP6 and PIK3R2. J Transl Med. 2014;12:259. doi: 10.1186/s12967-014-0259-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu J, Yan J, Zhou C, Ma Q, Jin Q, Yang Z. miR-1285-3p acts as a potential tumor suppressor miRNA via downregulating JUN expression in hepatocellular carcinoma. Tumour Biol. 2015;36:219–225. doi: 10.1007/s13277-014-2622-5. [DOI] [PubMed] [Google Scholar]
- 13.Jemal A, Ward E, Hao Y, Thun M. Trends in the leading causes of death in the United States, 1970-2002. JAMA. 2005;294:1255–1259. doi: 10.1001/jama.294.10.1255. [DOI] [PubMed] [Google Scholar]
- 14.Hao K, Luk JM, Lee NP, Mao M, Zhang C, Ferguson MD, Lamb J, Dai H, Ng IO, Sham PC, Poon RT. Predicting prognosis in hepatocellular carcinoma after curative surgery with common clinicopathologic parameters. BMC Cancer. 2009;9:389. doi: 10.1186/1471-2407-9-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bosch FX, Ribes J, Borras J. Epidemiology of primary liver cancer. Semin Liver Dis. 1999;19:271–285. doi: 10.1055/s-2007-1007117. [DOI] [PubMed] [Google Scholar]
- 16.Li P, Chen X, Su L, Li C, Zhi Q, Yu B, Sheng H, Wang J, Feng R, Cai Q, Li J, Yu Y, Yan M, Liu B, Zhu Z. Epigenetic silencing of miR-338-3p contributes to tumorigenicity in gastric cancer by targeting SSX2IP. PLoS One. 2013;8:e66782. doi: 10.1371/journal.pone.0066782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun K, Deng HJ, Lei ST, Dong JQ, Li GX. miRNA-338-3p suppresses cell growth of human colorectal carcinoma by targeting smoothened. World J Gastroenterol. 2013;19:2197–2207. doi: 10.3748/wjg.v19.i14.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang XH, Chen JS, Wang Q, Chen XL, Wen L, Chen LZ, Bi J, Zhang LJ, Su Q, Zeng WT. miR-338-3p suppresses invasion of liver cancer cell by targeting smoothened. J Pathol. 2011;225:463–472. doi: 10.1002/path.2877. [DOI] [PubMed] [Google Scholar]
- 19.Lu MM, Li S, Yang H, Morrisey EE. Foxp4: a novel member of the Foxp subfamily of winged-helix genes co-expressed with Foxp1 and Foxp2 in pulmonary and gut tissues. Gene Expr Patterns. 2002;2:223–228. doi: 10.1016/s1567-133x(02)00058-3. [DOI] [PubMed] [Google Scholar]
- 20.Wiehagen KR, Corbo-Rodgers E, Li S, Staub ES, Hunter CA, Morrisey EE, Maltzman JS. Foxp4 is dispensable for T cell development, but required for robust recall responses. PLoS One. 2012;7:e42273. doi: 10.1371/journal.pone.0042273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Long QZ, Du YF, Ding XY, Li X, Song WB, Yang Y, Zhang P, Zhou JP, Liu XG. Replication and fine mapping for association of the C2orf43, FOXP4, GPRC6A and RFX6 genes with prostate cancer in the Chinese population. PLoS One. 2012;7:e37866. doi: 10.1371/journal.pone.0037866. [DOI] [PMC free article] [PubMed] [Google Scholar]


