Abstract
Background: Lung cancer is becoming the leading cause of cancer-related deaths with high mortality worldwide and in China as well. Non-small-cell lung cancer (NSCLC) is the most common type of lung cancer accounting for approximately 85% of all cases. Over 70% of cases are at loco-regionally advanced stages or have distant metastasis at the time of presentation with subsequently poor prognosis. MiRNAs are stable molecules in blood and used as biomarkers for the early diagnosis of various malignancy. The purpose of this study was to evaluate whether circulating miR-125a-5p, miR-145 and miR-146a could be used as biomarkers for the diagnosis of NSCLC through measuring their expression and assess their relationship with clinical pathological factors. Methods: Expression levels of serum miR-125a-5p, miR-145 and miR-146a were detected in 70 pairs of NSCLC patients and healthy controls using quantitative real-time PCR analysis. Results: Serum miR-125a-5p, miR-145 and miR-146a were overexpressed in NSCLC patients compared with healthy controls. Their values of the area under the receiver –operating characteristic curve (AUC-ROC) were 0.71, 0.84 and 0.78. Optimal sensitivity and specificity were 73.53% and 55.71%, 92.75% and 61.43%, 84.06% and 58.57%, respectively in differentiating NSCLC patients from healthy controls. Conclusions: These preliminary data suggest that serum miR-125a-5p, miR-145 and miR-146a may be useful noninvasive biomarkers for the clinical diagnosis of NSCLC.
Keywords: MicroRNAs, non-small cell lung cancer, serum, diagnosis
Introduction
Lung cancer is becoming the leading cause of cancer-related deaths with high mortality worldwide [1] and in China [2] as well. Non-small-cell lung cancer (NSCLC) is the most common type of lung cancer accounting for approximately 85% of all cases, which is subdivided into three major types: adenocarcinoma (ADC), squamous-cell carcinoma (SCC) and large-cell carcinoma (LCC) [3,4]. Over 70% of cases are at loco-regionally advanced stages or have distant metastasis at the time of presentation with subsequently poor prognosis [5]. Additionally for the intrinsic resistance and recurrence, the survival rate for NSCLC still remains at low level, including early-stage group. The overall 5-year survival rate is only 17.1% in the United States [6]. It is urgent to explore the pathogenesis and identify more effective biomarkers, especially using noninvasive or minimal invasive methods to detect NSCLC as early as possible and develop effective individualized treatment programs to improve the quality of life and survival.
MicroRNAs (miRNAs) are a type of endogenous non-protein-coding, evolutionally preserved single-stranded small-sized RNA molecules with about 19-24 nucleotides in length that regulate nearly 60% of protein-coded genes [7]. More than half of the miRNA genes are located in cancer-associated genomic regions. MiRNAs are stable molecules that can be found and measured in peripheral blood, and may be associated with cancer diagnosis, treatment, and prognosis [8,9]. Epidermal growth factor receptor (EGFR) signal pathway is an important link in pathogenesis and plays an important role in cell migration and invasion [10]. Some investigations have validated that there is a relationship between miRNAs and the EGFR signaling pathway. For instance, miR-125a-5p was reported to inhibit migration and invasion of lung cancer cells by regulating several downstream genes involved in EGFR signaling pathway [11], restoration of miR-145 can inhibit cancer cell growth in EGFR mutant lung adenocarcinoma [12] and miR-146a can target EGFR and its downstream gene with potential functional significance in lung cancer [13]. These conclusions from above-mentioned studies were drawn in vitro or detecting tissue samples aiming at one single miRNA. Recently circulating miR-145 and miR-146a were found to be overexpressed in breast cancer and might be used for detection of breast cancer [14,15]. In the present study we performed the miRNAs expression profiling in NSCLC serum compared with healthy controls using quantitative RT-PCR and constructed the receiver operating characteristic (ROC) curves in order to identify whether specific miRNAs could discriminate between NSCLC patients and healthy controls.
Material and methods
Patients and samples
Seventy cases were collected from primary NSCLC patients who were diagnosed by histology or cytology from March 2013 to May 2014 in our hospital. Histological diagnoses were independently formulated by two pathologists. Disease stage classifications were determined according to the world health organization (WHO) classification and the international association for the study of lung cancer staging system [3,4]. Clinic pathological characteristics were collected whenever available for all the patients. Peripheral blood samples were collected before they received any anticancer treatment, including surgery, chemotherapy, radiotherapy and hormonal treatment, etc. 70 samples from healthy individuals with same age and gender were also collected. Blood samples (3.5 ml) were drawn from subjects using tubes without anticoagulant. These samples were processed by centrifugation at 3500 g for 5 minutes at room temperature. Serum was transferred into RNA-free EP tube every 500 μl aliquot and stored at -80°C before RNA extraction. This study was approved by the Ethics Committee of 306 Hospital of PLA. All specimens were collected after obtaining the patients’ informed consents. The characteristics with respect to age, gender and smoking status of NSCLC patients and control subjects are summarized in Table 1.
Table 1.
Clinical characteristics of 70 NSCLC patients and 70 healthy individuals
| Patients | Individuals | P-value | |
|---|---|---|---|
| Age (years), median | 64.41 ± 10.77 | 63.74 ± 10.32 | 0.7189 |
| ≥ 60 | 48 | 46 | |
| < 60 | 22 | 24 | |
| Gender | 1.0000 | ||
| Male | 43 | 43 | |
| Female | 27 | 27 | |
| Smoking status | 0.1756 | ||
| Yes | 37 | 29 | |
| No | 33 | 41 |
Total RNA extraction
Total RNA was isolated from 300 μl serum specimens of patients and controls by using miRNA isolation Kit (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s protocol. RNA concentration was measured using Nanodrop 2100. Then the RNA samples were preserved at -80°C for use. Repeated freeze-thawing was avoided to ensure the quality of the samples during storage,
Quantitative reverse transcription-PCR (qRT-PCR)
TaqMan MicroRNA Kit (Applied Biosystems) was used to perform the expression profiling of serum miRNA of all patients and controls. All reagents, primers and probes were also bought from Applied Biosystems. The cDNA reverse transcription was first carried out according to the manufacture’s instruction. The 15 μl reverse transcription reaction system consisted of 0.15 μl of dNTPs mix (100 mM total), 1.0 μl of reverse transcriptase (50 U/μl), 1.5 μl of RT Buffer (10×), 0.19 μl of RNase inhibitor (20 U/μl), 4.16 μl of nuclease free water, 3.0 μl of gene-specific miRNA primer and 5 μl of RNA extract sample. For synthesis of cDNA, reaction conditions of upper mixtures were: incubated at 16°C for 30 min, at 42°C for 30 min, at 85°C for 5 min, and at 4°C for any length of time. Then cDNA was amplified. The 20 μl qRT-PCR system contained: 1.0 μl of TaqMan Small RNA Assay (20×), 1.33 μl of cDNA solution, 10 μl of TaqMan Universal PCR Master Mix (2×) and 7.67 μl of nuclease free water. Quantitative PCR were performed on Applied Biosystems 7500 Real Time PCR system (Applied Biosystems) and the reaction conditions were: 10 min at 95°C, and 40 cycles of 95°C for 15 sec and 60°C for 60 sec. All qRT-PCR assays were undertaken in duplicate, using miR-39 as the internal reference gene to normalize the sample-to-sample variation during the RNA isolation procedure. The fold change of gene relative expression was calculated by using the equation of comparative cycle threshold (2-ΔΔCt) method.
Statistical analysis
Statistical analysis was performed with SPSS 17.0 statistical software (SPSS, Inc., Chicago, IL, USA) and graphs were generated using Graphpad Prism 6.0. Two-tailed unpaired t-test was used to evaluate the differential expression of serum miRNA level between NSCLC patients and normal controls. Receiver operating characteristic (ROC) curves were generated to assess the diagnostic accuracy of each serum miRNA, and the area under the ROC curve (AUC) were used to assess the sensitivity and specificity. Chi-squared test was used to assess the correlation between the expression level of serum miRNAs and clinic pathological factors of NSCLC patients. P-value < 0.05 was defined as statistical significance.
Results
Patient characteristics
There were 43 males and 27 females among the 70 NSCLC patients, including 48 ADCs, 20 SCCs and 2 LCCs. The mean age and gender distribution were similar between the NSCLC patients and the healthy controls. Smoking rate was higher in NSCLC patients than that in controls but without statistical difference (P = 0.1756).
Serum miRNAs expression in NSCLC patients and healthy controls
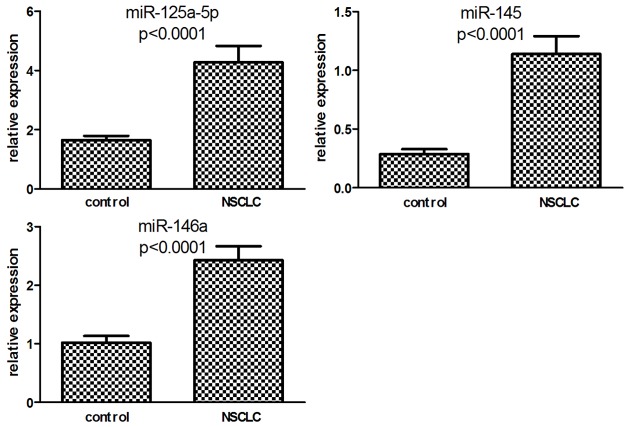
The serum level of miR-125a-5p, miR-145 and miR-146a were compared between the two groups. As shown in Figure 1, the expression of all three miRNAs was significantly higher in NSCLC patients than that in healthy controls (P < 0.0001).
Figure 1.
Serum expression levels of miR-125a-5p, miR-145 and miR-146a in healthy controls and NSCLCs.
Serum miRNAs as diagnosis signatures for NSCLCs
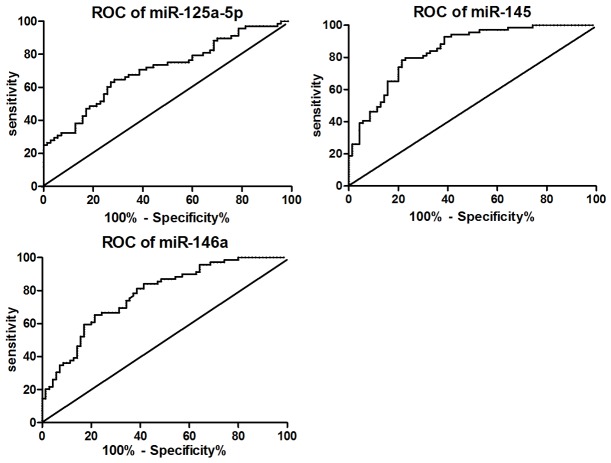
The ROC curve was plotted to identify a cut-off value that could distinguish NSCLCs from healthy controls. Analysis revealed that the area under the ROC curves (AUC) for serum miR-125a-5p was 0.71 (95% confidence interval 0.62-0.79). At an optimal cut-off value of 1.41, the sensitivity and specificity was 73.53% and 55.71%, respectively. Similarly, for miR-145 and miR-146a, results were shown in Table 2 and Figure 2.
Table 2.
AUC ROC values of different miRNAs in NSCLC patients
| miRNA | AUC ROC | 95% CI | Cut-off | Sensitivity | Specificity |
|---|---|---|---|---|---|
| miR-125a-5p | 0.71 | 0.62-0.79 | 1.41 | 73.53 | 55.71 |
| miR-145 | 0.84 | 0.78-0.91 | 0.20 | 92.75 | 61.43 |
| miR-146a | 0.78 | 0.70-0.85 | 0.87 | 84.06 | 58.57 |
Figure 2.
ROC curve of miRNAs in distinguishing healthy controls and NSCLC patients.
Correlation between miRNAs expression levels and clinic pathological factors
The correlation between these three miRNAs expression levels and various clinic pathological factors was examined. These factors included gender, age, smoking status, lymph node metastasis and TNM stage. NSCLC patients were divided into miRNA high and low groups according to the median fold-change values. There were no significant associations between the analyzed miRNAs and the main clinic pathological characteristics of the NSCLC patients. Nevertheless, we found that expression of miR-145 in serum was higher in patients in stage IV than that in other stages, even though without statistical significance.
Discussion
MiRNAs are involved in the whole biological and physiological processed by regulating target mRNA. Severe dysfunctions and aberrant expressions are present by acting in certain cases as oncogenes while in others as tumor suppressor genes through different targets in human malignancy [16]. MiRNAs are released from primary tumor into peripheral circulation, which are stable molecules and resistant to RNase digestion and other harsh conditions in serum. Since circulating miRNAs were first detected in 2008, accumulating studies have shown that they may be potential biomarkers for several human cancers, including NSCLC [14,17-21]. However the technique of detecting miRNAs from tissue is limited owing to its complication and need of invasive method to obtain.
In the current study we compared the profile of three miRNAs in serum from 70 healthy controls with 70 NSCLCs and demonstrated that serum expression of miR-125a-5p, miR-145 and miR-146a strongly differentiated the NSCLC patients from healthy controls. A highly significant increase was found in these three miRNAs in serum of NSCLC compared with that of healthy individuals. Moreover, receiver operating curve analysis indicated that the AUC of these three miRNAs were all greater than 0.7, indicating that they might be potential biomarkers in the diagnosis of NSCLC. Especially miR-145 was proved to be the most accurate tumor marker in distinguishing NSCLC patients from healthy controls with AUC ROC reaching 0.84 and the sensitivity and specificity at optimal cutoff being 92.75% and 61.43%.
We also analyzed the serum expression levels of these miRNAs in relation to the different clinical pathologic characteristics in NSCLC patients. Unfortunately, no significant relationships were found between them.
MiR-125a-5p, miR-145 and miR-146a were known as tumor suppressors and down-regulated in a variety of malignant tumor tissues. But in some malignant tumors they might be up-regulated and the serum expressions of these miRNAs were not identical, even in the same tumor. For instance, miR125a-5p was down-regulated in gastric cancer and suppressed cell proliferation by targeting ERBB2 [22]. In our study we found miR-125a-5p was overexpressed in serum of NSCLC, unlike a prior report which found no difference between NSCLCs and healthy controls [23]. MiR-145 inhibits proliferation of NSCLC cells by targeting c-Myc [24] and plays an inhibitory role in tumor angiogenesis, cell growth and invasion and tumor growth through the post-transcriptional regulation through N-RAS and VEGF-A in breast cancer [25]. Nevertheless conclusions from several studies which focused on the serum expression of miR-145 were not consistent, such as in breast cancer [14,26,27]. For miR-146a, its role can also vary in different types of cancer. It worked as tumor suppressor in prostate cancer [28] and gastric cancer [29] expressing lower regulation whereas in anaplastic thyroid cancer [30] and cervical cancer [31] it was up-regulated as an oncogene. Even converse expression levels of miR-146a in tissue were tested in some malignancy like gastric cancer [29,32]. All of these studies indicate that the targets and expressions are varied in different tumors, especially the expression profiling in peripheral blood. Meanwhile, few literatures were related to their serum expression in NSCLC to date.
In view of these three miRNAs targeting EGFR and its downstream gene to play their roles, we may be able to investigate the relationship between the EGFR mutation status and serum expression level in future studies.
In conclusion, the high stability of serum miRNAs makes them become noninvasive biomarkers for NSCLC detection. However, the sensitivity and specificity are not satisfied in distinguishing NSCLCs from healthy controls and their expression is different in various tumors, and therefore their roles and possible targets in NSCLC require further investigation.
Acknowledgements
This work was supported by the Capital Medical Development Funds for Special Research Project (SF2011-5005-01) and the Medical Research Funds of 306 Hospital (2013ZD06).
Disclosure of conflict of interest
None.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Guo P, Huang ZL, Yu P, Li K. Trends in cancer mortality in China: an update. Ann Oncol. 2012;23:2755–62. doi: 10.1093/annonc/mds069. [DOI] [PubMed] [Google Scholar]
- 3.Travis WD, Brambilla E, Noguchi M. International association for the study of lung cancer/american thoracic society/european respiratory society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol. 2011;6:244–85. doi: 10.1097/JTO.0b013e318206a221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Travis WD, Brambilla E, Noguchi M, Nicholson AG, Geisinger K, Yatabe Y, Ishikawa Y, Wistuba I, Flieder DB, Franklin W, Gazdar A, Hasleton PS, Henderson DW, Kerr KM, Petersen I, Roggli V, Thunnissen E, Tsao M. Diagnosis of lung cancer in small biopsies and cytology: implications of the 2011 International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society classification. Arch Pathol Lab Med. 2013;137:668–84. doi: 10.5858/arpa.2012-0263-RA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reungwetwattana T, Weroha SJ, Molina JR. Oncogenic pathways, molecularly targeted therapies, and highlighted clinical trials in non-small-cell lung cancer (NSCLC) Clin Lung Cancer. 2012;13:252–66. doi: 10.1016/j.cllc.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 6.Siegel R, DeSantis C, Virgo K, Mariotto A, Smith T, Cooper D, Gansler T, Lerro C, Fedewa S, Lin C, Leach C, Cannady RS, Cho H, Scoppa S, Hachey M, Kirch R, Jemal A, Ward E. Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin. 2012;62:220–41. doi: 10.3322/caac.21149. [DOI] [PubMed] [Google Scholar]
- 7.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mitchell PS, Parkin RK, Kroh EM. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–8. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen X, Ba Y, Ma L. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 10.Prenzel N, Fischer OM, Streit S, Hart S, Ullrich A. The epidermal growth factor receptor family as a central element for cellular signal transduction and diversification. Endocr Relat Cancer. 2001;8:11–31. doi: 10.1677/erc.0.0080011. [DOI] [PubMed] [Google Scholar]
- 11.Wang G, Mao W, Zheng S, Ye J. Epidermal growth factor receptor-regulated miR-125a-5p--a metastatic inhibitor of lung cancer. FEBS J. 2009;276:5571–8. doi: 10.1111/j.1742-4658.2009.07238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho WC, Chow AS, Au JS. Restoration of tumour suppressor hsa-miR-145 inhibits cancer cell growth in lung adenocarcinoma patients with epidermal growth factor receptor mutation. Eur J Cancer. 2009;45:2197–206. doi: 10.1016/j.ejca.2009.04.039. [DOI] [PubMed] [Google Scholar]
- 13.Chen G, Umelo IA, Lv S, Teugels E, Fostier K, Kronenberger P, Dewaele A, Sadones J, Geers C, De Grève J. miR-146a inhibits cell growth, cell migration and induces apoptosis in non-small cell lung cancer cells. PLoS One. 2013;8:e60317. doi: 10.1371/journal.pone.0060317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mar-Aguilar F, Mendoza-Ramirez JA, Malagon-Santiago I. Serum circulating microRNA profiling for identification of potential breast cancer biomarkers. Dis Markers. 2013;34:163–9. doi: 10.3233/DMA-120957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar S, Keerthana R, Pazhanimuthu A, Perumal P. Overexpression of circulating miRNA-21 and miRNA-146a in plasma samples of breast cancer patients. Indian J Biochem Biophys. 2013;50:210–4. [PubMed] [Google Scholar]
- 16.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–14. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang C, Wang C, Chen X, Yang C, Li K, Wang J, Dai J, Hu Z, Zhou X, Chen L, Zhang Y, Li Y, Qiu H, Xing J, Liang Z, Ren B, Yang C, Zen K, Zhang CY. Expression profile of microRNAs in serum: a fingerprint for esophageal squamous cell carcinoma. Clin Chem. 2010;56:1871–9. doi: 10.1373/clinchem.2010.147553. [DOI] [PubMed] [Google Scholar]
- 18.Shen J, Todd NW, Zhang Hl. Plasma microRNAs as potential biomarkers for non-small-cell lung cancer. Lab Invest. 2011;91:579–87. doi: 10.1038/labinvest.2010.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen J, Hruby GW, McKiernan JM. Dysregulation of circulating microRNAs and prediction of aggressive prostate cancer. Prostate. 2012;72:1469–77. doi: 10.1002/pros.22499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giraldez MD, Lozano JJ, Ramirez G, Hijona E, Bujanda L, Castells A, Gironella M. Circulating microRNAs as biomarkers of colorectal cancer: results from a genome-wide profiling and validation study. Clin Gastroenterol Hepatol. 2013;11:681–688. e3. doi: 10.1016/j.cgh.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 21.Zhu C, Ren C, Han J, Ding Y, Du J, Dai N, Dai J, Ma H, Hu Z, Shen H, Xu Y, Jin G. A five-microRNA panel in plasma was identified as potential biomarker for early detection of gastric cancer. Br J Cancer. 2014;110:2291–9. doi: 10.1038/bjc.2014.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nishida N, Mimori K, Fabbri M, Yokobori T, Sudo T, Tanaka F, Shibata K, Ishii H, Doki Y, Mori M. MicroRNA-125a-5p is an independent prognostic factor in gastric cancer and inhibits the proliferation of human gastric cancer cells in combination with trastuzumab. Clin Cancer Res. 2011;17:2725–33. doi: 10.1158/1078-0432.CCR-10-2132. [DOI] [PubMed] [Google Scholar]
- 23.Lin Q, Mao W, Shu Y, Lin F, Liu S, Shen H, Gao W, Li S, Shen D. A cluster of specified microRNAs in peripheral blood as biomarkers for metastatic non-small-cell lung cancer by stem-loop RT-PCR. J Cancer Res Clin Oncol. 2012;138:85–93. doi: 10.1007/s00432-011-1068-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Z, Zeng H, Guo Y. miRNA-145 inhibits non-small cell lung cancer cell proliferation by targeting c-Myc. J Exp Clin Cancer Res. 2010;29:151. doi: 10.1186/1756-9966-29-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zou C, Xu Q, Mao F. MiR-145 inhibits tumor angiogenesis and growth by N-RAS and VEGF. Cell Cycle. 2012;11:2137–45. doi: 10.4161/cc.20598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu W, Qin W, Atasoy U, Sauter ER. Circulating microRNAs in breast cancer and healthy subjects. BMC Res Notes. 2009;2:89. doi: 10.1186/1756-0500-2-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ng EK, Li R, Shin VY. Circulating microRNAs as specific biomarkers for breast cancer detection. PLoS One. 2013;8:e53141. doi: 10.1371/journal.pone.0053141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun Q, Zhao X, Liu X, Wang Y, Huang J, Jiang B, Chen Q, Yu J. miR-146a functions as a tumor suppressor in prostate cancer by targeting Rac1. Prostate. 2014;74:1613–21. doi: 10.1002/pros.22878. [DOI] [PubMed] [Google Scholar]
- 29.Kogo R, Mimori K, Tanaka F, Komune S, Mori M. Clinical significance of miR-146a in gastric cancer cases. Clin Cancer Res. 2011;17:4277–84. doi: 10.1158/1078-0432.CCR-10-2866. [DOI] [PubMed] [Google Scholar]
- 30.Pacifico F, Crescenzi E, Mellone S, lannetti A, Porrino N, Liguoro D, Moscato F, Grieco M, Formisano S, Leonardi A. Nuclear factor-{kappa} B contributes to anaplastic thyroid carcinomas through up-regulation of miR-146a. J Clin Endocrinol Metab. 2010;95:1421–30. doi: 10.1210/jc.2009-1128. [DOI] [PubMed] [Google Scholar]
- 31.Wang X, Tang S, Le SY, Lu R, Rader JS, Meyers C, Zheng ZM. Aberrant expression of oncogenic and tumor-suppressive microRNAs in cervical cancer is required for cancer cell growth. PLoS One. 2008;3:e2557. doi: 10.1371/journal.pone.0002557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiao B, Zhu ED, Li N, Lu DS, Li W, Li BS, Zhao YL, Mao XH, Guo G, Yu PW, Zou QM. Increased miR-146a in gastric cancer directly targets SMAD4 and is involved in modulating cell proliferation and apoptosis. Oncol Rep. 2012;27:559–66. doi: 10.3892/or.2011.1514. [DOI] [PubMed] [Google Scholar]