Abstract
Objectives: To investigate the crucial role of miR-26a in breast cancer and to validate whether miR-26a could regulate proliferation of breast cancer cells by targeting high mobility group AT-hook 1 (HMGA1). Methods: Reverse transcription-polymerase chain reaction (RT-PCR) was used to quantify the expression levels of miR-26a in breast cancer and adjacent non-cancerous breast tissues. MTT, cell migration and invasion assay were carried out to characterize the miR-26a function. Finally, to validate the target gene of miR-26a, luciferase reporter assay was employed, followed by RT-PCR and Western blot confirmation. Results: Compared with normal tissues, a significant down-regulation of miR-26a expression was observed in breast cancer tissues (P=0.002). miR-26a suppresses MDA-MB-231 and Mcf-7 breast cancer cell lines proliferation and motility. The luciferase activity was significantly decreased after co-transfection with psiCHECK-2/HMGA1 3’-UTR and miR-26a mimics in comparison with control cells, and qRT-PCR and Western blotting analysis found that HMGA1 expression at the mRNA and protein levels decreased in the miR-26a mimic-treatment group relative to NC. MTT assay showed that down regulation of HMGA1 by siRNA could significantly enhance the tumor-suppressive effect of miR-26a (P < 0.05). Conclusions: The results of the present study indicate that miR-26a may be associated with human breast carcinogenesis, which inhibits tumor cell proliferation by targeting HMGA1.
Keywords: Breast cancer, HMGA1, miR-26a, proliferation
Introduction
Breast cancer is the most common non-skin malignancy in women affecting about 1.2 million women in the world each year [1]. Optimization of treatment with better surgery, cytotoxic agents and endocrine therapy has not altered the prognosis much. We are now in an era where personalized medicine and targeted therapies may give new hope for this patient group. Identification of novel molecular markers which can improve diagnosis and prognostic stratification and serve as possible therapeutic targets will be of great importance in the near future.
MicroRNAs (miRNAs) are small, endogenous, noncoding RNAs which play important gene-regulatory roles in animals via sequence-specific interactions with the 3’UTR of cognate mRNA targets, causing suppression of translation and mRNA decay [2]. It has been firmly established that miRNAs regulate many key cellular processes such as cell growth, differentiation and apoptosis [3-5]. MicroRNA-26a (miR-26a), located in chromosome 3p22, a region with high frequency of loss of heterozigozity (LOH) in cancer, has been proved to be a tumor suppressor [6-8]. However, the role of miR-26a in cancer cells was still controversial as it served as an oncogene in glioma, lung cancer and cholangiocarcinoma [9-11]. However, the mechanism by which miR-26a is implicated in breast cancer tumorigenesis is incompletely understood.
High mobility group AT-hook 1 (HMGA1), a nuclear matrix protein with three AT-hook domains that bind the minor groove of AT-rich DNA sequences, is highly expressed in virtually all aggressive human cancers and could serve as a useful biomarker and therapeutic target in advanced malignancies [12,13]. By inducing a conformational change in DNA or modulating the molecular via protein-protein interaction [13], HMGA1 would participate in various physiological processes, such as proliferation, anchorage-independent cell growth, tumorigenesis, and metastatic progression [14]. HMGA1 was identified as a key transcription factor enriched in poorly differentiated cancers, including breast cancer [15].
We found that miR-26a may modulate HMGA1 using online prediction software Target Scan. In our study, we sought to investigate the crucial role of miR-26a in breast cancer. We identified that miR-26a could regulate proliferation of breast cancer by targeting HMGA1.
Materials and methods
Patients and specimens
This study was approved by the Research Ethics Committee of Shengli Oilfield Central Hospitial. Written informed consent was obtained from all of the patients. All specimens were handled and made anonymous according to the ethical and legal standards. None of the patients recruited in this study had undergone preoperative chemotherapy or radiotherapy. 10 pairs of fresh breast cancer and matched adjacent normal tissue specimens were collected from patients who underwent surgery between June 2012 and July 2013. The fresh tissue specimens were collected and immediately placed in liquid nitrogen and then stored at -80°C until the isolation of RNA.
Cell culture
The MDA-MB-231 and Mcf-7 breast cancer cell lines were obtained from the Chinese Science Institute and cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, USA) supplemented with 10% fetal bovine serum (FBS, Gibco), as well as 100 units of penicillin/ml and 100 μg of streptomycin/ml (Enpromise, China). Cells were incubated at 37°C in a humidified chamber containing 5% CO2.
RNA isolation and real-time PCR
MicroRNA was extracted from cancer cells using RNAiso for Small RNA (Takara, Japan). Total RNA from the cells was isolated with RNAiso Plus (Takara, Japan). The RNA was reverse transcribed with a PrimeScript RT reagent Kit (Takara, Japan). All the procedures were conducted in accordance with the manufacturer’s instructions. The resulting cDNA was quantified by RT-PCR using SYBR Premix Ex Taq (Takara, Japan). The relative expression level of miR-26a and HMGA1 was calculated and quantified with the 2-ΔΔCt method after normalization with reference to expression of U6 small nuclear RNA and GAPDH, respectively.
MTT assay
MDA-MB-231 and Mcf-7 cells (5000/well) were plated in 96-well plates (BD Biosciences, USA) and incubated at 37°C overnight. When the confluence of MDA-MB-231 and Mcf-7 cells reached 50-60%, 100 nmol/L miR-26a mimics or 50 nmol/L HMGA1 siRNA (Genepharma, China) were transfected into cells according to the protocol of lipofectamine 2000 (Invitrogen, USA). The experiment used one negative control transfected with NC or siRNA control. Cell proliferation was assessed at 24, 48, 72, 96 and 120 h, following the instructions of the MTT proliferation assay kit (Sigma, USA).
Cell migration and invasion assay
The 24-well Boyden chamber with 8 um pore size polycarbonate membrane (Corning, NY) was used to analyze the cell motility. For invasion assay, the membrane was pre-coated with matrigel to form a matrix barrier. Equal number of cells, transfected with 150 nmol/L miR-26a mimics or NC for 48 h, was seeded on the upper chamber with serum-free medium. Medium with 20% serum was added to the lower chamber as a chemoattractant. To minimize the effect of proliferation inhibition, 105 cells were added to upper chamber and the membranes were harvested and fixed within 12 h. The membranes were stained with 0.1% crystal violet and five visual fields of x200 magnification of each membrane were randomly selected and counted for the cell numbers under a light microscope.
Protein extraction and Western blotting
Following the RNA duplex and/or plasmid treatment, the cells were harvested and lysed. The protein concentration in the each lysate was determined using the bicinchoninic acid protein assay kit (Pierce Biotechnology, Rockford, IL, USA). Equivalent quantities of protein were separated by 12% SDS-polyacrylamide gels and transferred to nitrocellulose membranes. Membranes were blocked with 5% non-fat milk and then incubated overnight with the appropriate primary antibody. They were next washed and incubated with the corresponding horseradish peroxidase (HRP)-conjugated secondary antibody for 1 h. Bound secondary antibody was visualized using an enhanced chemiluminescence (ECL) system (Pierce Biotechnology, Rockford, IL). The primary antibodies used were: anti-GAPDH, anti-HMGA1 (Epitomics, Burlingame, CA).
Luciferase assays
To construct the luciferase reporter vectors, a wild-type 3’-UTR (untranslated region) fragment of Human HMGA1 containing putative binding sites for miR-26a was amplified from genomic DNA. The amplified fragment was inserted into pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega, USA) within the SacI/SalI sites. The mutant 3’-UTR, carrying a mutated sequence in the seeding region of miR-26a, was mutated with the MutanBest Kit (Takara, Japan). Then, the mutated sequence was subcloned into pmirGLO Dual-Luciferase Vector. Both insertions were verified by sequencing. Breast cancer cells plated in a 24-well plate were co-transfected with 50 nM of either miR-26a mimic or miR-Con or 200 ng reporter comprising wild-type or mutant 3’-UTR. The relative luciferase activity was measured by Dual-Luciferase Reporter Assay System (Promega, USA) 48 h after transfection.
Statistical analysis
SPSS 18.0 software was used for statistical analysis. Data were presented as mean of 3 independent experiments. Two-tailed Student’s t test was used for comparisons of 2 independent groups. P values of < 0.05 were considered statistically significant.
Results
Expression of miR-26a is downregulated in human breast cancer tissues
miR-26a expression levels in 10 pairs of human breast cancer tissues and adjacent normal tissues were quantified by RT-PCR. Compared with normal tissues, a significant down-regulation of miR-26a expression was observed in breast cancer tissues (P=0.002, Figure 1).
Figure 1.
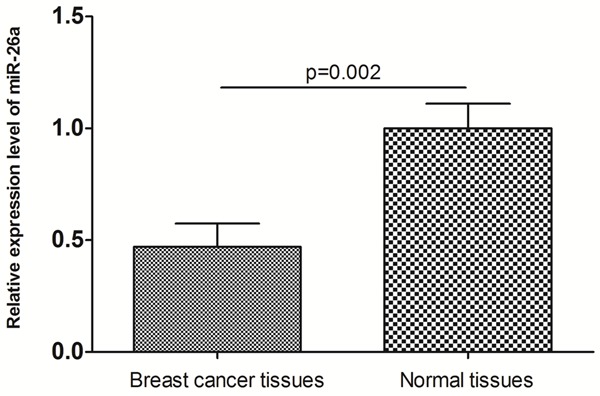
miR-26a expression levels in 10 pairs of human breast cancer tissues and adjacent normal tissues.
miR-26a suppresses MDA-MB-231 and Mcf-7 breast cancer cell lines proliferation
To determine the impact of miR-26a on the proliferation of breast cancer cells, miR-26a mimics were transfected into MDA-MB-231 and Mcf-7 cells. As shown in Figure 2, treatment of cells with miR-26a led to a decrease in MDA-MB-231 and Mcf-7 cell growth at 48 h (17.2% and 16.5%), 72 h (20.3% and 21.5%), 96 h (24.5% and 25.1%) and 120 h (29.9% and 31.8%) (all P < 0.05) compared with the negative control.
Figure 2.
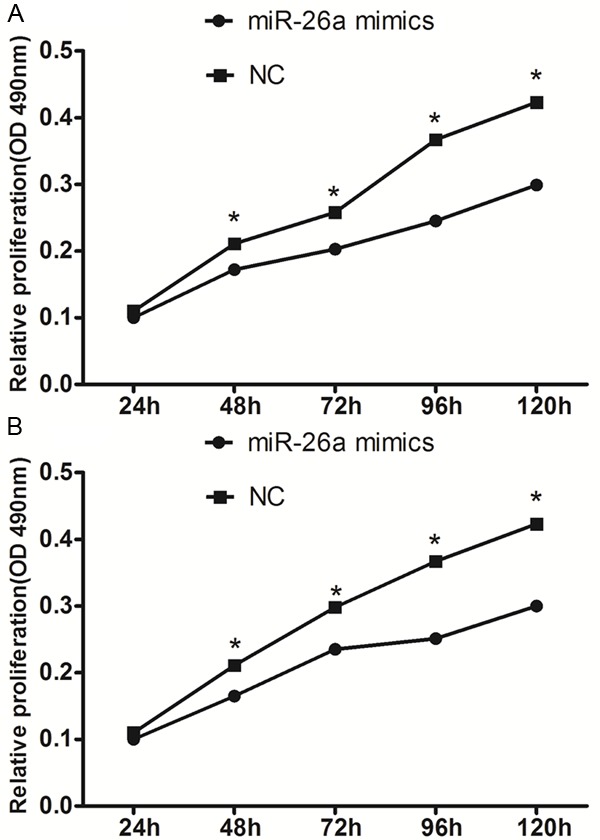
miR-26a suppresses the proliferation of MDA-MB-231 cells (A) and Mcf-7 cells (B). *P < 0.05.
miR-26a impairs MDA-MB-231 and Mcf-7 breast cancer cell lines motility
To further understand the function of miR-26a, we investigated the potential impact of miR-26a on MDA-MB-231 and Mcf-7 breast cancer cell lines motility. As indicated by the transwell assay, forced expression of miR-26a decreased the migration and invasion of MDA-MB-231 and Mcf-7 breast cancer cell lines compared with the control (Figure 3). miR-26a is therefore a negative regulator of migration and invasion of breast cancer cells.
Figure 3.
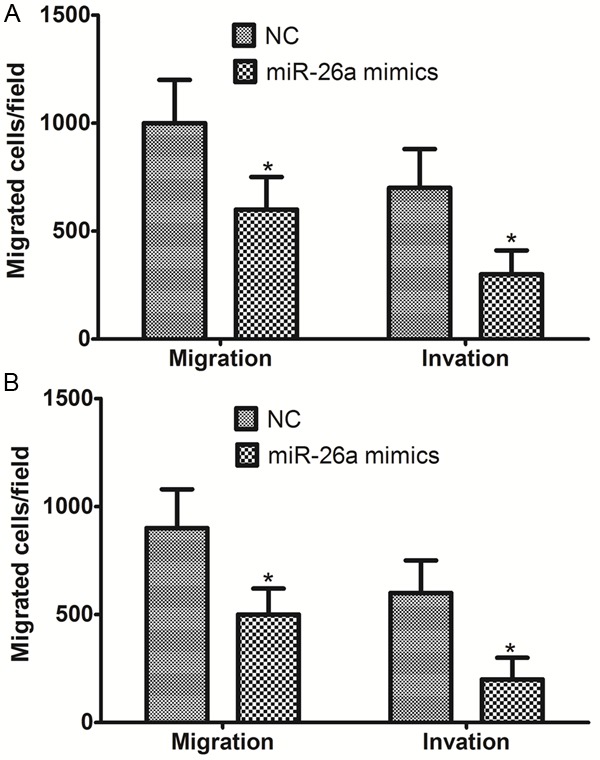
miR-26a impaired the motility of MDA-MB-231 cells (A) and Mcf-7 cells (B). *P < 0.05.
miR-26a directly targets HMGA1 in breast cancer cells
We then investigated the mechanism of miR-26a inhibition of proliferation of MDA-MB-231 cells. Predicted miR-26a targets were identified using the algorithms of TargetScan and microRNA. Among hundreds of potential target genes, we specifically focused on HMGA1, as it has been consistently reported to be oncogene in various cancers. We identified a binding site for miR-26a in the 3’-UTR of HMGA1 mRNA. To confirm that miR-26a can bind to the predicted site, we performed a luciferase reporter assay in the 293T cell line. Figure 4A shows that the luciferase activity significantly decreased after co-transfection with psiCHECK-2/HMGA1 3’-UTR and miR-26a mimics in comparison with control cells. The results demonstrated that miR-26a specifically binds to the 3’-UTR of HMGA1 mRNA. The effect of miR-26a transfection on HMGA1 mRNA and protein expression was respectively assessed using RT-PCR and Western blot in MDA-MB-231 cell line. As shown in Figure 4B and 4C, the qRT-PCR and western blotting analysis found that HMGA1 expression at the mRNA and protein levels decreased in the miR-26a mimic-treatment group relative to NC.
Figure 4.
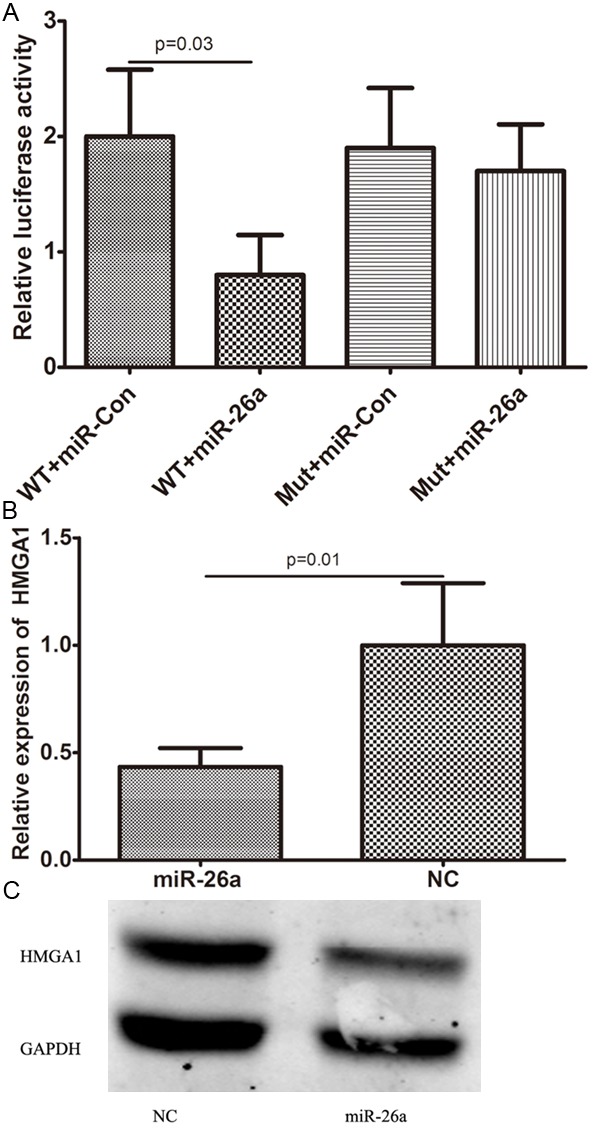
HMGA1 was a direct target of miR-26a. A: the luciferase activity significantly decreased after co-transfection with psiCHECK-2/HMGA1 3’-UTR and miR-26a mimics in comparison with control cells; B: HMGA1 mRNA level was detected by qRT-PCR in MDA-MB-231 cells transfected with miR-26a mimic or the control; C: HMGA1 protein level was detected by Western blotting in MDA-MB-231 cells transfected with miR-26a mimic or the control.
To further investigate whether miR-26a exhibits tumor-suppressive functions by targeting HMGA1, the effect of HMGA1 on miR-26a-mediated cell proliferation was investigated. MTT assay showed that down regulation of HMGA1 by siRNA could significantly enhance the tumor-suppressive effect of miR-26a (P < 0.05, shown in Figure 5), suggesting that miR-26a inhibited the proliferation of breast cancer cells partially by targeting HMGA1.
Figure 5.

MTT assay showed that down regulation of HMGA1 by siRNA could significantly enhance the tumor-suppressive effect of miR-26a (P < 0.05).
Discussion
Breast cancer mortality has substantially decreased since the early 1980s mainly because of the use of adjuvant therapy [16]. However, the last fifteen years demonstrated an urgent need for treatment individualization and finding of adequate therapy for each patient [17,18]. It is generally accepted that the development of breast cancer, like other cancers, involves multiple steps, including the accumulation of genetic and epigenetic changes. However, the precise mechanism underlying breast carcinogenesis remains unclear. Therefore, it has been a global research hotspot to looking for new therapeutic targets for breast cancer treatment.
The discovery of the first miRNA, lin-4, in Caenorhabditis elegans initiated a new era of miRNA biology. Since then, thousands of miRNAs have been identified and annotated. Furthermore, an increasing number of evidences indicate that miRNAs are differentially expressed between normal and malignant tumor tissues, and miRNA deregulation is a critical cause of cancer formation. As one of the most prominent miRNAs implicated in the tumorigenesis, miR-26a has presented with a controversial role during tumor progression. miR-26a was found to be reduced in numerous human cancers, including bladder cancer, hepatic cell carcinoma and nasopharyngeal carcinoma [6-8,19]. EZH2, MTDH and IL-6 were reported to be the targets which mediating tumor suppression effect by miR-26a. However, in glioma, lung cancer and cholangiocarcinoma, forced expression of miR-26a could promote cancer progression by targeting PTEN or GSK-3β [9-11]. The diverse expression pattern and biological function might be owing to unique genetic background in different type of cancer cells. The exact role of miR-26a in breast cancer was still quite unclear. Thus, our current study intended to clarify the biological function of miR-26a in breast cancer.
In the present study, miR-26a expression levels in 10 pairs of human breast cancer tissues and adjacent normal tissues were quantified by RT-PCR. Compared with normal tissues, a significant down-regulation of miR-26a expression was observed in breast cancer tissues. To determine the impact of miR-26a on the proliferation of breast cancer cells, miR-26a mimics were transfected into MDA-MB-231 and Mcf-7 cells, and then we found that miR-26a suppresses MDA-MB-231 and Mcf-7 breast cancer cell lines proliferation and motility. Thus, our data suggest that miR-26a might play an important role in breast cancer development. As we know, miRNA functions through interacting with target gene thus the key to explore the mechanism of miRNA is to study the interaction between miRNA and its target gene. In this study, HMGA1 was predicted to be the target gene of miR-26a by online biological software. HMGA1, a nuclear matrix protein with three AT-hook domains that bind the minor groove of AT-rich DNA sequences, is highly expressed in virtually all aggressive human cancers and could serve as a useful biomarker and therapeutic target in advanced malignancies [12,13]. By inducing a conformational change in DNA or modulating the molecular via protein-protein interaction [6], HMGA1 would participate in various physiological processes, such as proliferation, anchorage-independent cell growth, tumorigenesis, and metastatic progression [14]. Luciferase reporter vectors containing HMGA1 gene 3’-UTR region with miR-26a binding site was constructed and specific binding between miR-26a and HMGA1 was verified. The qRT-PCR and western blotting analysis found that HMGA1 expression at the mRNA and protein levels decreased in the miR-26a mimic-treatment group relative to NC. To further investigate whether miR-26a exhibits tumor-suppressive functions by targeting HMGA1, the effect of HMGA1 on miR-26a-mediated cell proliferation was investigated. MTT assay showed that down regulation of HMGA1 by siRNA could significantly enhance the tumor-suppressive effect of miR-26a, suggesting that miR-26a inhibited the proliferation of breast cancer cells partially by targeting HMGA1.
In conclusion, the results of the present study indicate that miR-26a is associated with human breast carcinogenesis by targeting HMGA1, and may be a potential therapeutic target for human breast cancer.
Disclosure of conflict of interest
None.
References
- 1.Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64:104–17. doi: 10.3322/caac.21220. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Ventura A, Jacks T. MicroRNAs and cancer: short RNAs go a long way. Cell. 2009;136:586–91. doi: 10.1016/j.cell.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schickel R, Boyerinas B, Park SM, Peter ME. MicroRNAs: key players in the immune system, differentiation, tumorigenesis and cell death. Oncogene. 2008;27:5959–74. doi: 10.1038/onc.2008.274. [DOI] [PubMed] [Google Scholar]
- 5.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 6.Yu L, Lu J, Zhang B, Liu X, Wang L, Li SY, Peng XH, Xu X, Tian WD, Li XP. miR-26a inhibits invasion and metastasis of nasopharyngeal cancer by targeting EZH2. Oncol Lett. 2013;5:1223–28. doi: 10.3892/ol.2013.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen L, Zheng J, Zhang Y, Yang L, Wang J, Ni J, Cui D, Yu C, Cai Z. Tumor-specific expression of microRNA-26a suppresses human hepatocellular carcinoma growth via cyclin-dependent and -independent pathways. Mol Ther. 2011;19:1521–8. doi: 10.1038/mt.2011.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu J, He ML, Wang L, Chen Y, Liu X, Dong Q, Chen YC, Peng Y, Yao KT, Kung HF, Li XP. MiR-26a inhibits cell growth and tumorigenesis of nasopharyngeal carcinoma through repression of EZH2. Cancer Res. 2011;71:225–33. doi: 10.1158/0008-5472.CAN-10-1850. [DOI] [PubMed] [Google Scholar]
- 9.Liu B, Wu X, Liu B, Wang C, Liu Y, Zhou Q, Xu K. MiR-26a enhances metastasis potential of lung cancer cells via AKT pathway by targeting PTEN. Biochim Biophys Acta. 2012;1822:1692–704. doi: 10.1016/j.bbadis.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 10.Zhang J, Han C, Wu T. MicroRNA-26a promotes cholangiocarcinoma growth by activating beta-catenin. Gastroenterology. 2012;143:246–56. e8. doi: 10.1053/j.gastro.2012.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huse JT, Brennan C, Hambardzumyan D, Wee B, Pena J, Rouhanifard SH, Sohn-Lee C, le Sage C, Agami R, Tuschl T, Holland EC. The PTEN-regulating microRNA miR-26a is amplified in high-grade glioma and facilitates gliomagenesis in vivo. Genes Dev. 2009;23:1327–37. doi: 10.1101/gad.1777409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shah SN, Resar LM. High mobility group A1 and cancer: potential biomarker and therapeutic target. Histol Histopathol. 2012;27:567–79. doi: 10.14670/HH-27.567. [DOI] [PubMed] [Google Scholar]
- 13.Fusco A, Fedele M. Roles of HMGA proteins in cancer. Nat Rev Cancer. 2007;7:899–910. doi: 10.1038/nrc2271. [DOI] [PubMed] [Google Scholar]
- 14.Fedele M, Fusco A. HMGA and cancer. Biochim Biophys Acta. 2010;1799:48–54. doi: 10.1016/j.bbagrm.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 15.Di Cello F, Shin J, Harbom K, Brayton C. Knockdown of HMGA1 inhibits human breast cancer cell growth and metastasis in immunodeficient mice. Biochem Biophys Res Commun. 2013;434:70–4. doi: 10.1016/j.bbrc.2013.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Early Breast Cancer Trialists’ Collaborative Group (EBCTCG) Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005;365:1687–717. doi: 10.1016/S0140-6736(05)66544-0. [DOI] [PubMed] [Google Scholar]
- 17.Paplomata E, O’Regan R. The PI3K/AKT/mTOR pathway in breast cancer: targets, trials and biomarkers. Ther Adv Med Oncol. 2014;6:154–66. doi: 10.1177/1758834014530023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang W, Liu J, Wang G. The role of microRNAs in human breast cancer progression. Tumour Biol. 2014;35:6235–44. doi: 10.1007/s13277-014-2202-8. [DOI] [PubMed] [Google Scholar]
- 19.Lin Y, Chen H, Hu Z, Mao Y, Xu X, Zhu Y, Xu X, Wu J, Li S, Mao Q, Zheng X, Xie L. miR-26a inhibits proliferation and motility in bladder cancer by targeting HMGA1. FEBS Lett. 2013;587:2467–73. doi: 10.1016/j.febslet.2013.06.021. [DOI] [PubMed] [Google Scholar]


