Abstract
Multiple myeloma (MM) is a plasma cell neoplasm that results from clonal expansion of an Ig-secreting terminally differentiated B cell. Advanced MM is characterized by tissue damage that involves bone, kidney, and other organs and is typically associated with recurrent genetic abnormalities. IL-6 signaling via the IL-6 signal transducer GP130 has been implicated as an important driver of MM pathogenesis. Here, we demonstrated that ectopic expression of constitutively active GP130 (L-GP130) in a murine retroviral transduction-transplantation model induces rapid MM development of high penetrance. L-GP130–expressing mice recapitulated all of the characteristics of human disease, including monoclonal gammopathy, BM infiltration with lytic bone lesions, and protein deposition in the kidney. Moreover, the disease was easily transplantable and allowed different therapeutic options to be evaluated in vitro and in vivo. Using this model, we determined that GP130 signaling collaborated with MYC to induce MM and was responsible and sufficient for directing the plasma cell phenotype. Accordingly, we identified Myc aberrations in the L-GP130 MM model. Evaluation of human MM samples revealed recurrent activation of STAT3, a downstream target of GP130 signaling. Together, our results indicate that deregulated GP130 activity contributes to MM pathogenesis and that pathways downstream of GP130 activity have potential as therapeutic targets in MM.
Introduction
Multiple myeloma (MM; also referred to as plasma cell myeloma) is a B-lineage cancer characterized by the expansion of malignant plasma cells predominantly within the BM. Clinical characteristics of the disease consist of osteolytic bone destruction, cytopenia caused by BM infiltration, and nephropathy. Despite the introduction of high-dose chemotherapy followed by hematopoietic stem cell transplantation (SCT), and, more recently, immunomodulatory drugs and proteasome inhibitors, the disease remains incurable (1–3).
Extensive research has revealed numerous genetic events contributing to the pathogenesis of MM. IgH translocations position an oncogene under control of an IgH enhancer, most frequently involving CCND1, MAF, or FGFR3. Nearly half of all MM tumors are hyperdiploid, with various chromosomes involved. Hyperdiploid MM has a primary IgH translocation in about 10% of cases, whereas nonhyperdiploid MM has an IgH translocation in approximately 70%. Additional oncogenic events in MM pathogenesis involve chromosome 13 deletions, activating mutations of RAS or B-RAF, mutations activating NF-κB, chromosome 17p loss and abnormalities of TP53, chromosome 1 aberrations, and MYC deregulation. MYC amplifications and translocations seem to be responsible for progression and are strongly associated with poor prognosis (4–7).
Among the best-described signal transduction pathway implicated in MM cell growth and survival is the IL-6/JAK/STAT pathway. IL-6 binds to the specific IL-6 receptor (IL-6R), and this complex associates with 2 molecules of the ubiquitously expressed GP130. This complex formation induces activation of the JAK and STAT molecules. Subsequently, STATs are released from the receptor, homo- or heterodimerize, translocate to the nucleus, and induce transcription of the respective target genes (8–10). High amounts of IL-6 are secreted by BM stromal cells in a paracrine manner, and — to a lesser extent — by MM cells themselves in an autocrine fashion. Blocking IL-6 or IL-6R or inhibiting JAK/STAT3 results in growth arrest and apoptosis of MM cells. However, there is also evidence that IL-6 signaling might be dispensable for MM cell survival in the context of BM stromal cells (6, 11–15).
Efforts to generate mouse models of MM have led to various genetically defined models. However, these are characterized by rather low penetrance and/or late disease onset. Il6 transgenic Balb/C mice develop MM with a penetrance of 40% at 12 months. Analysis of the malignant plasma cells revealed a t(12;15) translocation involving c-Myc (16). While Eμ-Myc transgenic mice (in which MYC expression is under control of the Eμ enhancer) develop aggressive pro-/pre-B cell lymphomas early in life (17), activation-induced deaminase–dependent expression of MYC in germinal center B cells leads to a high incidence of MM with a median survival of approximately 2 years (4). Transgenic mice expressing abnormally high levels of XBP-1, a protein involved in the terminal differentiation of B cells, develop monoclonal gammopathy of undetermined significance (MGUS), and some develop MM later in life (18).
In the present study, we showed that constitutive activation of GP130/JAK/STAT3 signal transduction in a retroviral murine BM transduction-transplantation model was sufficient to induce or facilitate MM development in mice. This model was characterized by very high penetrance and relatively short latency. Importantly, constitutive GP130 activation efficiently cooperated with MYC overexpression by driving cell growth and differentiation of malignant plasma cells. Our data indicate that constitutive GP130 signal transduction is a critical early step in myelomagenesis.
Results
STAT3 phosphorylation and target gene activation is a hallmark of human MM.
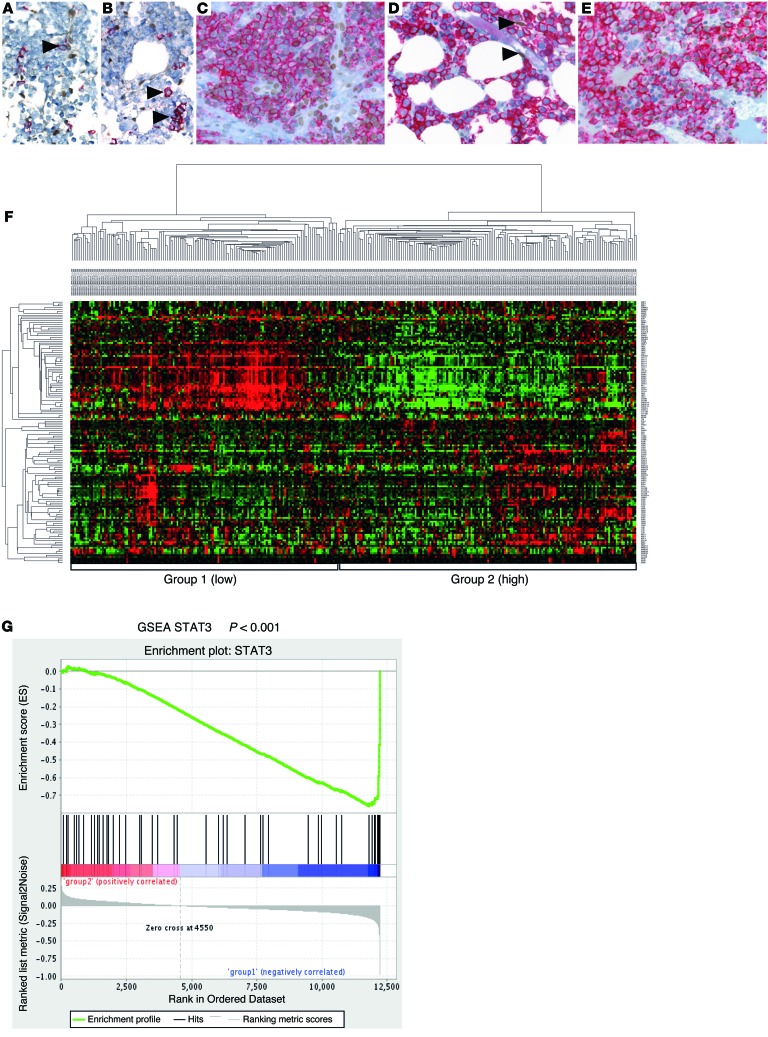
Activation of the IL-6/IL-6R/GP130 complex is crucial for survival and proliferation of human myeloma (6, 11, 13), and the JAK/STAT3 pathway is a major target downstream of IL-6R/GP130 signaling (8, 14). We therefore evaluated a series of BM biopsies from patients with MM, as well as biopsies from normal BM and MGUS, for expression of phosphorylated STAT3 (P-STAT3) in the plasma cell compartment of the BM. Whereas plasma cells in normal BM and in MGUS usually lacked P-STAT3 staining, 99 of 283 (35%) evaluable MM cases showed substantial nuclear P-STAT3 reactivity (Figure 1, A–E). We next mined publicly available gene expression datasets to analyze the expression of STAT3 target genes in MM. Hierarchical clustering analysis for established STAT3 target genes (19) identified a considerable subgroup of patient samples (20) showing an activated STAT3 pathway signature (Figure 1F). Similar results were obtained when analyzing a second dataset (ref. 21 and Supplemental Figure 1A; supplemental material available online with this article; doi:10.1172/JCI69094DS1). Gene set enrichment analysis (GSEA) showed a significantly different distribution of the STAT3 activation–associated genes among the hierarchical clustering–defined gene expression groups (normalized enrichment score [NES], –1.97; nominal P < 0.001; Figure 1G). In addition, mining the public repository Oncomine (www.oncomine.org) showed that STAT3 target genes were elevated in MM versus control tissue (Supplemental Figure 1B). Furthermore, we found the STAT3 signaling gene expression pattern to be associated with low bone disease, the MMSET groups, and the presence of a gain of the 1q21 locus, whereas we identified an inverse correlation with hyperdiploid disease (Supplemental Figure 1, C–E). Thus, STAT3 phosphorylation and target gene activation seems to be a major hallmark of a large subgroup of human MM.
Figure 1. Expression of activated STAT3 and STAT3 target genes is a hallmark of human MM.
(A–E) Immunohistochemical double staining of human BM biopsies for P-STAT3 (brown nuclear stain) and CD138 (red membrane stain). Original magnification, ×400. Plasma cells (arrowheads) in normal BM (A) and MGUS (B) were negative for P-STAT3, whereas positive endothelial cells were clearly visible. (C and E) MM biopsy with nuclear P-STAT3 positivity in the majority of tumor cells. (D) MM biopsy negative for P-STAT3. 2 positive endothelial cells (arrowheads) are illustrated as internal positive control. (F) Hierarchical cluster analysis of n = 304 human MM samples (GEO accession no. GSE26760) based on a STAT3 activation–associated gene expression signature (n = 67 genes; ref. 19). Group 2 correlated with high STAT3 expression (i.e., STAT3 activation). (G) GSEA showed a significantly different distribution of the STAT3 activation–associated genes, with group 1 being negatively correlated (NES, –1.97; P < 0.001).
Constitutive GP130 signaling induces myeloma formation in a murine BM transduction-transplantation model.
To test whether constitutive activation of GP130 signaling enables B cells to proliferate independently of cytokine stimulation, the IL-3–dependent pre-B cell line BA/F3, which lacks GP130 expression (22), was infected with a retrovirus encoding either WT GP130 or the constitutively active form (L-GP130; ref. 23 and Supplemental Figure 2A). In contrast to WT GP130–expressing BA/F3 cells, cultures of L-GP130–expressing cells grew independently of IL-3 with moderately reduced growth kinetics (Supplemental Figure 2, B and C). Furthermore, L-GP130–expressing BA/F3 cells formed colonies when grown in methylcellulose, regardless of growth factor support (Supplemental Figure 2D). Importantly, in BA/F3 cells, the major IL-6R/GP130 downstream signaling pathway, JAK/STAT3 (8, 14), was activated either by ectopic L-GP130 expression or by IL-6 plus soluble IL-6R (sIL-6R) in a dose-dependent manner in the presence of WT GP130 (Supplemental Figure 2E). Thus, constitutive GP130 activation results in cytokine-independent proliferation, colony formation, and STAT3 phosphorylation in a B cell line in vitro.
Stimulation of WT GP130 as well as L-GP130–mediated STAT3 activation have been shown to suppress differentiation of murine embryonic stem cells (23). We therefore reasoned that ectopic L-GP130 expression could lead to a loss of differentiation and immature hematopoietic disease, or possibly to a plasma cell disorder, when overexpressed in hematopoietic stem/progenitor cells. To test this hypothesis, BM of 5-FU–treated donor mice was infected with L-GP130, WT GP130, or GFP control virus and transplanted into lethally irradiated syngeneic recipients (Figure 2A). After stable hematopoietic reconstitution (Supplemental Figure 3A), we analyzed GP130 and L-GP130 expression and activation of downstream signaling pathways in unselected white blood cells and found increased P-STAT3 as well as P-ERK in the primary L-GP130 graft recipients (Supplemental Figure 3B). Mice followed for changes in white blood counts and leucocyte lineages showed an increase of GFP+ B cells upon ectopic L-GP130 expression, but no significant alterations in B cell subsets (Supplemental Figure 3, C and D). In vitro experiments revealed that constitutive JAK/STAT3 activation by means of L-GP130 did not provide a growth advantage or protection from IL-7 withdrawal (Supplemental Figure 4). Thus, L-GP130 overexpression in BM results in activation of GP130 downstream signaling pathways and expansion of B cells in vivo, indicating a role of GP130 activation in B cell development, survival, or differentiation, most likely in collaboration with additional stimuli.
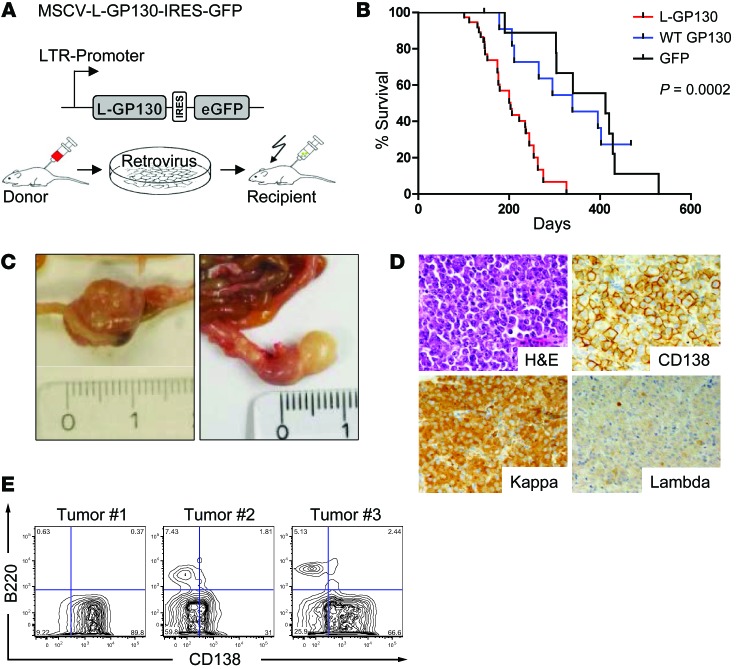
Figure 2. Plasma cell disorder with features of human myeloma.
(A) Top: Scheme of the retroviral expression plasmid used. Bottom: Transduction-transplantation model. 5-FU–mobilized donor BM (Balb/C) was infected with retrovirus and transplanted i.v. into lethally irradiated syngeneic recipients. (B) Survival analysis of lethally irradiated mice that received GFP (n = 15), WT GP130 (n = 17), or L-GP130 (n = 37) virus-infected BM grafts. Note that GFP and GP130 graft recipients did not succumb to GFP+ disease, but rather to other radiation-induced tumors or organ failure. (C) Representative images of mesenterial tumors that arose in primary L-GP130 BM graft recipients. 1 cm is shown for scale. (D) Histological and immunohistochemical analysis of a representative mesenterial tumor from a primary L-GP130 graft recipient. Kappa, Igκ light chain; Lambda, Igλ light chain. Original magnification, ×400. (E) Flow cytometric analysis of tumors from 3 L-GP130 mice. Contour blots are shown from gated viable lymphocytes. Numbers in the quadrants show percent cells rated negative or positive for the indicated antibody staining.
To evaluate the functional relevance of constitutive GP130 signaling in vivo, primary L-GP130, GP130, and GFP graft recipient mice were followed for a prolonged time. Starting around 4 months after transplantation, primary L-GP130 graft recipients began to deteriorate in their general condition and were sacrificed in accordance with the endpoints of the study (weight loss >10% body weight, reduced activity, etc.), as defined in the animal ethics vote; their median survival was 203 days (Figure 2B). In contrast, primary GP130 and GFP graft recipient controls lived significantly longer without evidence of disease (median survival, 339 and 412 days, respectively; Figure 2B). Whole-animal necropsy revealed the formation of extramedullar tumors in the Peyer’s patches within the gut, reflecting a plasma cell disorder (Figure 2, C and D). These tumors were not detectable in control GP130 or GFP graft recipients, which eventually succumbed to radiation-induced organ failure, as we observed previously (data not shown and ref. 24). Details on organ involvement and phenotype of the L-GP130–induced disease are shown in Supplemental Figure 5. In summary, 9 of 11 primary L-GP130 graft recipients that underwent full necropsy developed CD138+ disease with classical morphologic features of myeloma, with disruption of the tissue architecture and invasive growth (25) that involved intestine, spleen, BM, or >1 organ (Figure 2, C and D, and data not shown). Diseased primary L-GP130 graft recipients were also analyzed for the immunophenotype of the CD138+ disease by flow cytometry. The extent of CD138 positivity ranged from highly positive to moderate (Figure 2E and Supplemental Figure 6), as previously described for human MM (26, 27). In concordance with human MM, all CD138+ cells of primary L-GP130 graft recipients were negative for the B cell marker B220 (Supplemental Figure 6). To assess clonality, we subcloned and sequenced IgH rearrangements from individual tumors. Diseased L-GP130 graft recipients contained monoclonal and oligoclonal B cell expansions. Of the 4 mice analyzed, 3 harbored clones with significant somatic mutations (up to 10%), demonstrating a germinal center passage (Supplemental Figure 7). We conclude that constitutive GP130 activation by means of L-GP130 results in clonal CD138+ plasmacytoma with genetic characteristics of a post–germinal center phenotype.
L-GP130–induced plasma cell disorder shows characteristic features of human MM.
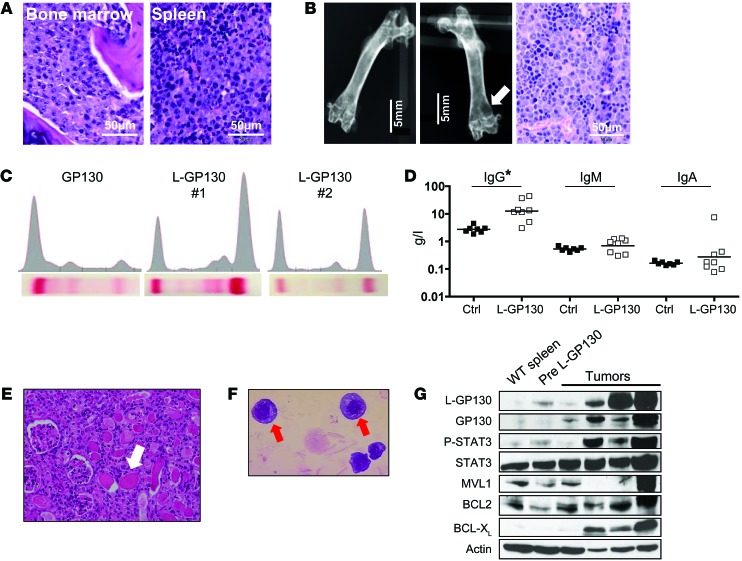
Primary L-GP130 graft recipient mice were next analyzed for the distribution and phenotype of the disease, with focus on features regularly observed in human MM. Moderate to intermediate myeloma involvement of BM and spleen was found in basically all mice analyzed (Figure 3A and Supplemental Figure 5). Corresponding to the focal bone destruction found in human MM, X-ray analysis revealed lytic bone lesions in L-GP130 graft recipients (Figure 3B). In further accord with human MM, serum electrophoresis revealed monoclonal spikes and elevated class-switched IgG levels in L-GP130 graft recipients that were not detected in age-matched control GFP or GP130 graft recipients (Figure 3, C and D). Notably, these serum changes were also associated with renal disease resembling human cast nephropathy (Figure 3E). The morphology of tumor cells resembled human myeloma, with eccentrically located, enlarged nuclei (Figure 3F). Immunoblotting revealed overexpression of antiapoptotic proteins (Figure 3G). In summary, the L-GP130–induced plasma cell disorder bears all key features of human myeloma with organ damage, including lytic bone lesions, monoclonal gammopathy, and kidney injury.
Figure 3. L-GP130–induced myeloma is characterized by gammopathy with kidney damage, BM infiltration with lytic bone lesions, and high expression of antiapoptotic proteins.
(A) BM and spleen histology (H&E staining). Scale bars: 50 μm. (B) X-ray analysis of a control femur (left) and a L-GP130 graft recipient femur (middle) with a lytic bone lesion (white arrow). Right: Histology from the lytic bone lesion (H&E). Scale bars: 5 mm (left and middle); 50 μm (right). (C) Serum electrophoresis from representative GP130 and L-GP130 graft recipients, indicative of monoclonal gammopathy. (D) Ig levels, assessed by ELISA, from diseased L-GP130 graft recipient mice and age-matched control GP130 graft recipients. Shown is the geometric mean. *P < 0.05. (E) Representative H&E staining of cast nephropathy in the tubuli (white arrows) of an L-GP130 graft recipient mouse. Original magnification, ×400. (F) Ex vivo–cultured plasma cells from the BM of primary L-GP130 graft recipients (Pappenheim staining). Original magnification, ×63. (G) Immunoblot analysis for expression of the indicated proteins was performed on mesenterial tumors and spleen control cells. Pre, precancerous spleen.
The syngeneic transplantation-transduction model also allows for testing the ability of established disease from primary BM graft recipients to reestablish myeloma in a secondary recipient. Irradiated secondary syngeneic recipient mice that were transplanted with the BM of primary L-GP130 graft recipients of moderate tumor load and without overt disease deteriorated significantly faster in their general condition compared with the primary recipients. The latency of disease onset was dramatically reduced to a median of 81 days (Supplemental Figure 8A). Full necropsy followed by histological, flow cytometric, and immunohistochemical analyses revealed a high grade of BM involvement in all serially transplanted mice, with some involvement of the spleen and other organs (Supplemental Figure 8B and data not shown). Again, most tested secondary transplanted mice presented with lytic bone lesions and monoclonal gammopathy (Supplemental Figure 8, C and D). In vitro cultured BM GFP+CD138+ cells from secondary L-GP130 graft recipients rapidly engrafted in syngeneic nonirradiated tertiary recipients and caused disease with mean latency of 34 days (Supplemental Figure 8, E and F). Thus, the L-GP130–induced plasma cell disorder is serially transplantable and reflects common features of human myeloma in secondary and tertiary recipients.
L-GP130–induced myeloma is sensitive to established myeloma treatments in vitro and in vivo, thereby providing a novel preclinical MM model.
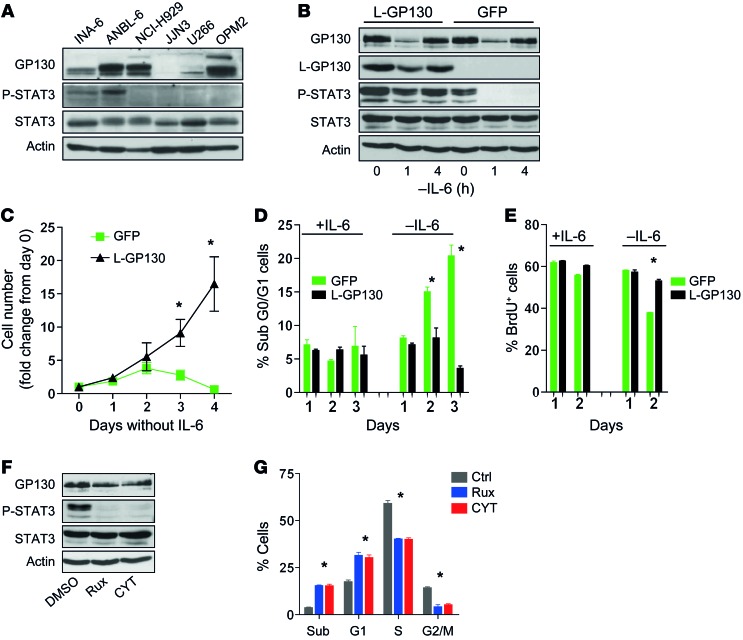
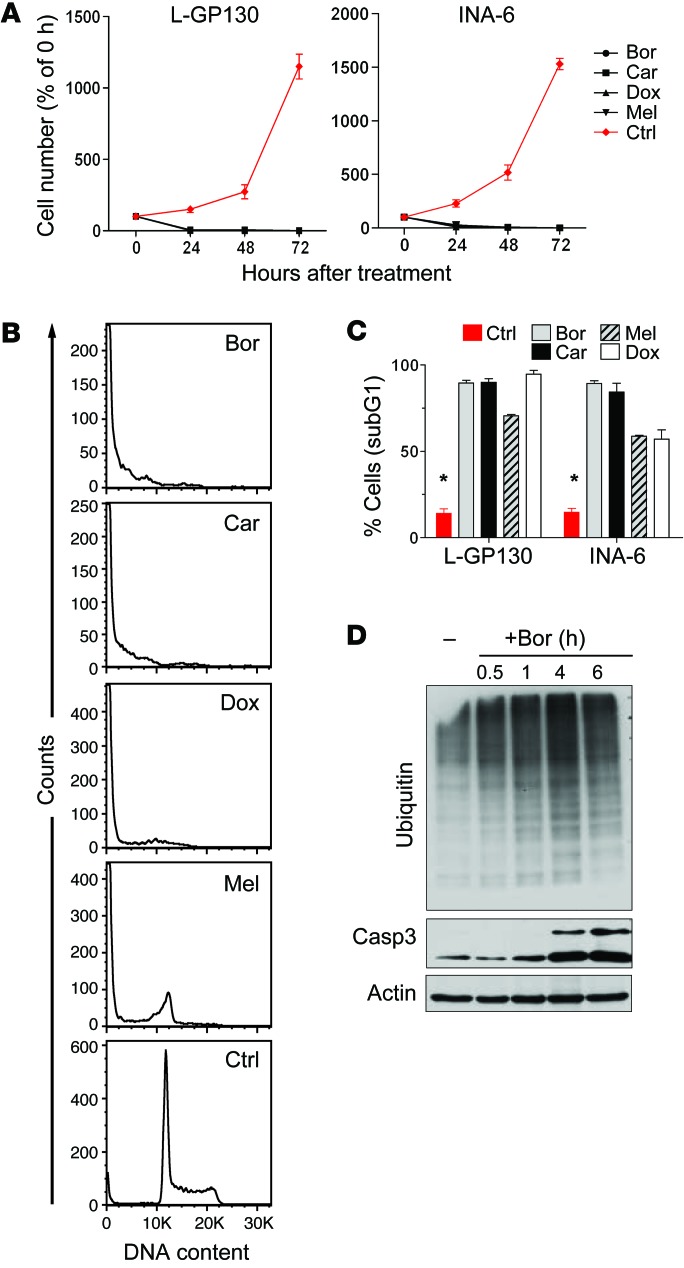
To further establish that constitutive GP130 activation represents a model of IL-6–activated MM (11, 14), we infected the human IL-6–dependent MM line INA-6, which shows IL-6–engaged STAT3 phosphorylation, with L-GP130 and GFP control virus (Figure 4, A and B). L-GP130 rendered INA-6 cells cytokine independent and protected them from apoptosis, a finding that correlated with STAT3 phosphorylation (Figure 4, B–E). Furthermore, treatment with JAK inhibitors (13, 28) blocked STAT3 phosphorylation and caused cell cycle arrest and increased cell death (Figure 4, F and G). We next investigated our novel L-GP130 MM mouse model in terms of its applicability as a preclinical model for myeloma treatment. Established CD138+ MM cell lines derived from diseased L-GP130 secondary graft recipients using a stroma coculture system (EL08-1D2 stromal cells; ref. 29) were generated and then treated with classical chemotherapeutics and clinically used proteasome inhibitors in vitro. These L-GP130–infected myeloma cells exhibited high sensitivity to melphalan and doxorubicin, resulting in G1 arrest and apoptosis (Figure 5, A–C). The effects observed in the L-GP130 MM model were comparable to those observed upon treatment of the human MM cell line INA-6, which was supported with IL-6 (Figure 5, A–C). The potential usefulness of the L-GP130 MM model was further supported by experiments testing its sensitivity to the approved proteasome inhibitors bortezomid and carfilzomib (Figure 5, A–C). As expected, bortezomib treatment resulted in accumulation of ubiquitylated proteins (Figure 5D). Again, the treatment effects were comparable to those found in INA-6 cells supported with IL-6.
Figure 4. Constitutive GP130/JAK/STAT3 activation by means of L-GP130 overexpression protects IL-6–dependent MM cell lines from growth factor withdrawal.
(A) Immunoblot analysis of IL-6–dependent (INA-6 and ANBL-6) and IL-6–independent (NCI-H929, JJN3, U266, and OPM2) MM cell lines for STAT3 phosphorylation. (B) INA-6 cells were infected with retrovirus encoding L-GP130-GFP or GFP only and FACS-sorted for GFP positivity. Immunoblot analysis for the indicated proteins was performed upon IL-6 withdrawal. Actin served as a loading control. (C) Growth of INA-6 cells expressing the indicated proteins upon IL-6 withdrawal. n = 3, mean ± SEM. *P < 0.05 between groups. (D) PI and (E) BrdU flow cytometry analysis for infected INA-6 cells upon IL-6 withdrawal. n = 3 experiments, mean ± SEM. *P < 0.05. (F) IL-6–supported INA-6 cells were treated with the JAK inhibitors ruxolitinib (Rux; 10 nM) or CYT387 (CYT; 50 nM) for 1 hour and assessed for inhibition of STAT3 phosphorylation by immunoblotting. (G) INA-6 cells were treated with 10 nM ruxolitinib or 50 nM CYT387 for 72 hours, then pulsed for 10 minutes with BrdU. n = 3 experiments, mean ± SEM. *P < 0.05. sub, subG1.
Figure 5. L-GP130–induced myeloma is responsive to classical myeloma treatments in vitro.
(A) Growth kinetics of primary in vitro–cultured L-GP130–expressing myeloma cells and the human MM cell line INA-6 treated with 200 nM bortezomib (Bor), 20 nM carfilzomib (Car), 100 μg/ml doxorubicin (Dox), or 100 μg/ml melphalan (Mel). n = 3 independent experiments; mean ± SD. (B) Primary L-GP130–expressing myeloma cells were treated for 24 hours with drugs as in A and assessed for DNA content using PI to estimate apoptosis and cell cycle state. Histograms show results of 1 representative experiment. (C) Quantitative PI analysis of primary L-GP130 myeloma cells and human INA-6 cells after treatment with drugs as in A. Mean ± SD is shown. *P < 0.05, control vs. treated groups. (D) Accumulation of mono- and polyubiquitinylated proteins and cleaved caspase 3 (Casp 3) in primary L-GP130–expressing cells treated with 400 nM bortezomib for the indicated times. 1 representative immunoblot is shown.
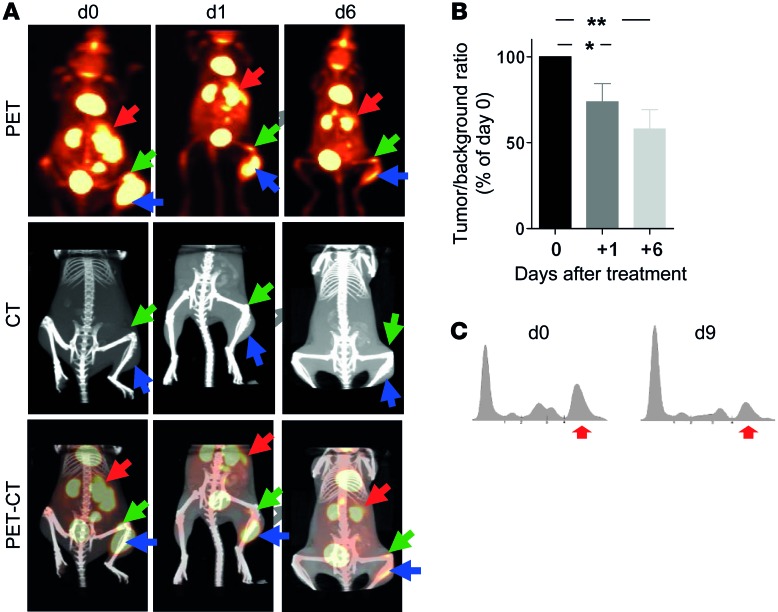
Ex vivo–cultured L-GP130 cells caused rapid disease, including bone destruction and gammopathy, when injected i.v. into syngeneic recipients (Figure 6A and Supplemental Figure 8E). Mice bearing tumors caused by i.v. injected L-GP130 myeloma cells were highly sensitive to a single treatment of doxorubicin (10 mg/kg body weight), as assessed by FDG-PET-CT and serum electrophoresis (Figure 6, A–C). Thus, the L-GP130 MM model represents a valuable tool for evaluating novel treatments against myeloma in vitro and in vivo in the preclinical setting. Based on the serial transplantable nature of the disease in our model, applications may also include evaluations of the myeloma-microenvironment interaction, myeloma cell homing, and the myeloma-initiating cell compartment.
Figure 6. L-GP130–induced myeloma is responsive to classical myeloma treatment in vivo.
(A and B) FDG-PET-CT analysis of tumor response in syngeneic recipient mice injected i.v. 4 weeks earlier with 5 × 104 in vitro–cultured L-GP130 myeloma cells. Animals were examined before (d0) and on days 1 and 6 after a single doxorubicin treatment (10 mg/kg body weight i.p.). (A) Representative PET, CT, and PET-CT scans of 1 mouse. Arrows denote spleen and tumors. (B) Quantification of tumor size, relative to pretreatment. Bars represent mean ± SEM from 9 different tumors of 5 mice. **P < 0.01, *P < 0.05. (C) Serum electrophoresis from a secondary L-GP130 BM graft recipient treated with a single injection of doxorubicin (10 mg/kg body weight i.p.) before (d0) and 9 days after (d9) treatment. 1 representative experiment is shown. Red arrows denote gammopathy.
Constitutive GP130 signaling collaborates with MYC and is a critical determinant of the plasma cell phenotype.
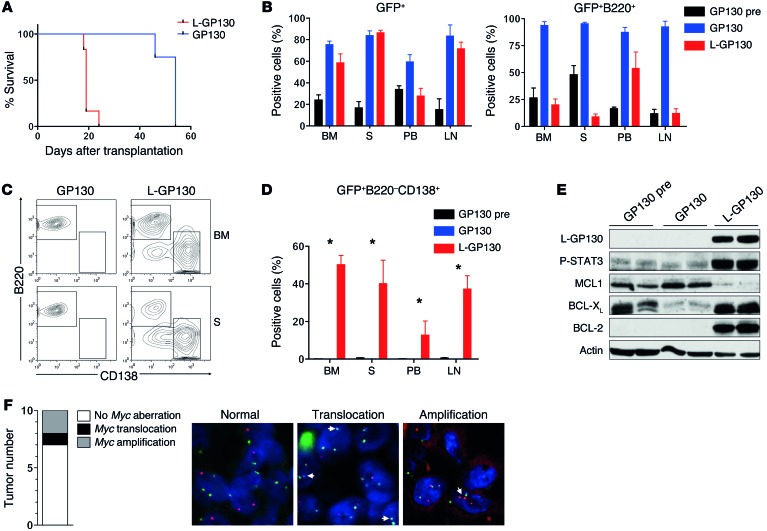
Mouse models allow faithful recapitulation of genetic aberrations that are relevant and of predictive or prognostic importance in human disease (30). Human MM is characterized by recurrent genetic aberrations, including amplification and translocation of MYC, a negative prognostic marker (5–7). Since L-GP130 induced differentiation into plasma cells in virtually all transplanted mice, we sought to further examine the role of constitutive GP130 signaling with regard to the tumor phenotype. Eμ-Myc transgenic mice develop pro- and pre-B cell lymphomas (17) that stain positive for the B cell marker B220 (31–33). When Eμ-Myc fetal liver cells (FLCs) were infected with retrovirus encoding L-GP130 and transplanted into lethally irradiated syngeneic C57BL/6 recipient mice, we found a dramatic acceleration of disease onset (median, 19 days) compared with recipients of GP130-infected Eμ-Myc FLCs (median, 54 days) and the historic Eμ-Myc control (34) (Figure 7A), which indicates that MYC and L-GP130 signaling collaborate to induce lymphoid malignancies. Despite comparable engraftment (assessed as GFP+ cells), we observed a reduction of GFP+B220+ cells in basically all organs of diseased L-GP130 graft recipient mice compared with control GP130 graft recipients (Figure 7B). Further analysis revealed that constitutive GP130 signaling mediated by L-GP130 caused a shift from B cell lymphoma toward a plasma cell disorder, despite the extremely short latency of disease onset (Figure 7, A, C, and D). In addition, the expression of antiapoptotic BCL2 family members also shifted from MCL1 toward BCL-XL (Figure 7E). Thus, constitutive GP130 signaling appears to induce a plasma cell phenotype in this lymphoma model and also dramatically accelerates disease development, potentially by modulating the expression pattern of antiapoptotic proteins.
Figure 7. Constitutive GP130 signaling collaborates with MYC to induce plasmacytoma and is responsible for the plasma cell phenotype.
(A) Eμ-Myc FLCs were infected with the indicated viruses and transplanted into lethally irradiated syngeneic mice. Recipients were then followed for disease onset. Median tumor latency was 19 days for L-GP130 and 54 days for GP130 (n = 5 mice per group). P < 0.05. (B) Flow cytometry to assess the percentage of GFP+ cells and GFP+B220+ B cells in BM, spleen (S), peripheral blood (PB), and lymph node (LN) of the indicated recipient mice. Pretumorous GP130 graft recipients (GP130pre) were included as a control. Bars represent mean ± SEM from n = 5 (GP130 and L-GP130) or n = 3 (GP130pre) mice. (C) Representative and (D) quantitative analysis of organ infiltration by GFP+B220–CD138+ cells in the indicated recipient mice. Bars represent mean ± SEM from n = 5 (GP130 and L-GP130) or n = 3 (GP130pre) mice. *P < 0.05. (E) Immunoblot analysis of BM B cells from the indicated recipient mice using the listed antibodies. (F) FISH for Myc aberrations in plasmacytomas of primary L-GP130 graft recipients. Shown are number and type of Myc aberrations and representative images (arrows indicate Myc aberrations). Original magnification, ×1,000.
Having observed this putative collaboration between transgenic MYC overexpression and enforced GP130/JAK/STAT3 activity by means of L-GP130, we next performed FISH analysis to retrieve Myc aberrations in the primary single L-GP130 myelomas. Of 10 independent tumors analyzed, 2 showed Myc amplifications and 1 an Ig/Myc translocation (Figure 7F), genetic features frequently observed in advanced human MM (5, 6). This finding further underscored that constitutive GP130 signaling induces a disease in mice that faithfully recapitulates many aspects of human MM.
Discussion
In this study, we provided genetic and correlative evidence that signal transduction via the common receptor subunit for the IL-6 family, GP130, constitutes a critical step during MM pathogenesis. Despite recent progress in understanding the molecular pathogenesis of MM, many important questions remain unanswered. New preclinical models, like the L-GP130 MM mouse presented herein, could provide an adequate means of determining the significance of signaling pathways and evaluating treatment approaches.
Biology and clinical behavior of MM are determined not only by the genetic background, but also by the BM microenvironment (6, 14). Bidirectional MM-BM interactions involve IL-6, which is predominantly secreted by BM stromal cells (35) and constitutes a potent growth and survival factor in MM (36). IL-6 binding to IL-6R subsequently induces phosphorylation and heterodimerization of GP130 and activates the JAK/STAT signaling cascade (8). Importantly, serum IL-6 and soluble IL-6R are prognostic factors in MM and reflect the proliferative fraction of MM within patients (37, 38). Previously, the critical role of the GP130/JAK/STAT axis in MM pathogenesis has been appreciated in different studies. IL-6R superantagonists, including SANT7, revealed strong antimyeloma activity, and ectopic expression of STAT3β, a dominant-negative STAT3 construct, potently induced apoptosis in MM cells (39). Likewise, BCL-XL, cyclin D, and c-MYC have been identified as critical STAT3-regulated target genes (40). In the era of small-molecule inhibitors, numerous groups showed the efficacy of JAK inhibitors at inducing growth arrest and cell death (12, 13, 15). In contrast, other investigators demonstrated that MM cells are resistant to SANT7 treatment in the presence of stromal cells, presumably as a result of stroma-dependent activation of the MAPK/ERK1/2 pathway (41, 42). Against this background, our novel murine retroviral transduction-transplantation model of MM showed that constitutive GP130 activation, in collaboration with other events, was sufficient to reproducibly induce MM. The L-GP130 MM model thus represents a valuable tool with which to further investigate this clinically relevant signal transduction pathway in vivo.
Several genetically defined mouse models of MM have recently been generated. These models are characterized by low penetrance and/or late disease onset (4, 16, 18). The L-GP130 MM model circumvents several of these limitations. In particular, our model is characterized by high penetrance and short latency, as well as easy transplantability of tumor cells into syngeneic recipients. These features make it a suitable tool for preclinical in vivo drug studies, for investigating the tumor-niche interaction and MM homing, and for studying new genes with regard to their effect on disease initiation and progression. At the same time, the L-GP130 MM model closely resembles the human disease. Key characteristics of human MM that are present in our L-GP130 MM model include lytic bone destruction, monoclonal gammopathy, and Ig deposition in the kidney. In addition, the finding that STAT3 activation, the likely driving pathway of tumor development upon constitutive GP130 signaling, is a hallmark of human MM (present study and refs. 43, 44) makes our L-GP130 MM model an interesting tool with which to study this subentity of human MM.
Antiapoptotic BCL2 family members are targets of STAT3 and frequently overexpressed in MM (40, 45). We found a shift of expression from antiapoptotic MCL1 (in reconstituted premalignant or diseased mice when WT GP130 virus was used to infect Eμ-Myc FLCs) toward the expression of BCL2 and BCL-XL (in mice reconstituted with L-GP130–infected hematopoietic stem cells). In precancerous Myc-transgenic B cells, both BCL2 and BCL-XL are suppressed (46), whereas transgenic coexpression of BCL-XL with MYC results in murine plasma cell tumors (47). It is thus conceivable that constitutive activation of STAT3 and subsequent BCL-XL expression overcomes MYC-induced BCL-XL suppression and results in dramatic acceleration of MM onset in the L-GP130–infected Eμ-Myc mouse. Likewise, the IL-6 downstream effector molecule STAT3 is a key regulator of c-MYC expression in numerous cell types, including MM cells, leading to upregulation of c-MYC even in the absence of MYC translocations (48).
The tumor cell phenotype CD138+B220– found in our present studies closely resembled what is seen in human MM (CD138+CD20–). Human CD138+ MM cells, unlike CD138– cells, can proliferate and induce lytic bone lesions when transplanted into SCID-hu or SCID-rab immunodeficient mice (49). We showed that constitutive GP130 activation was indeed sufficient to overcome the common B220+ pro- and pre-B cell phenotype that usually occurs in Eμ-Myc transgenic mice (17). Genetic Myc alterations seem to be a secondary event in murine MM pathogenesis (50). This finding is supported by our current data showing dramatically accelerated onset of a CD138+B220– plasma cell disorder when constitutive GP130 signaling collaborates with B cell–targeted MYC overexpression. Accordingly, our analysis of primary L-GP130 myelomas revealed acquired genomic Myc aberrations. Thus, the L-GP130 MM model seems to accurately recapitulate human MM, in which where MYC aberrations are clearly considered secondary genetic events that lower the probability of long survival (4–7).
In summary, our present findings established activation of IL-6/IL-6R downstream signaling as an early event in MM pathogenesis. Constitutive activation of GP130 downstream signaling in mice was sufficient to reproducibly induce a disease strongly resembling human MM. Importantly, GP130-dependent signal transduction critically modulated MYC-driven oncogenesis by regulating plasma cell differentiation and survival. Serial transplantability of myeloma cells not only simplified its application, but also provided evidence for a myeloma-initiating cell compartment. Finally, our L-GP130 MM mouse model displayed virtually all clinically relevant features of the human disease, thereby representing a valuable preclinical tool.
Methods
Cell culture and colony formation assay.
BA/F3 cells were obtained from DSMZ and cultured in RPMI (Gibco) with 10% FCS (PAA Laboratories) and 1% penicillin/streptomycin (Gibco), with or without 2 ng/ml IL-3 (R&D Systems). The human MM cell lines NCI-H929, JJN3, U266, and OPM2 were obtained from DSMZ, and INA-6 and ANBL-6 cells were gifts from M. Gramatzki (Christian-Albrechts-University, Kiel, Germany) and D.F. Jelinek (Mayo Clinic, Rochester, Minnesota, USA). INA-6, ANBL-6, and NCI-H929 cells were cultured in RPMI 1640 supplemented with 20% FCS, 1% penicillin/streptomycin, 2 mM l-glutamine, and 50 μM 2-β mercaptoethanol. U266, OPM2, and JJN3 cells were grown in RPMI 1640 with 10% FCS and 1% penicillin/streptomycin. IL-6–dependent cell lines (INA-6 and ANBL-6) were maintained in medium supplemented with 2 ng/ml IL-6 (R&D Systems). L-GP130 myeloma cells were established from single-cell suspensions of tumors arising in L-GP130 graft recipient mice. Myeloma cells were cocultured on stromal cells (cell line EL08-1D2; ref. 29) in RPMI (Gibco) supplemented with 10% FCS (PAA Laboratories), 1% penicillin/streptomycin (Gibco), 1% l-glutamine (Gibco), 0.1% mercaptoethanol (Gibco), and 10 ng/ml IL-7 (mIL-7; R&D Systems). When steady growth conditions were achieved, myeloma cells were deprived of EL08-1D2 cells and IL-7. Primary BM cells were cultured on S17 stromal cells as described previously (51). Phoenix ecotropic helper-free retroviral producer cells (provided by G. Nolan, Stanford University Medical Center, Stanford, California, USA) and NIH/3T3 cells (DSMZ) were maintained in DMEM (Gibco) supplemented with 10% FCS. Colony formation analysis was performed in MethoCult M3234 (STEMCELL Technologies) at 37°C with 5% CO2 and scored after 7 days.
GP130- and L-GP130–expressing BA/F3 cells were treated with IL-6 (1 ng/ml up to 10 ng/ml) and sIL-6R (100 ng/ml up to 1,000 ng/ml). In vitro treatment of L-GP130–derived myeloma cell lines was performed with 100 μg/ml melphalan (GlaxoSmithKline), 100 μg/ml doxorubicin (Adrimedac), 200 nM bortezomib (Janssen), or 20 nM carfilzomib (SelleckChem). INA-6 cells were treated with 10 nM ruxolitinib (SelleckChem) or 50 nM CYT387 (SelleckChem).
Plasmids.
L-GP130, the constitutively active form of GP130, was described previously (23). MSCV-L-GP130-IRES-GFP (or MSCV-GP130-IRES-GFP) was cloned by digesting the MSCV-IRES-GFP plasmid with EcoRI and BglII (Fermentas) following a PCR for L-GP130 (or GP130) with primers containing overhangs for EcoRI and NcoI restriction (Omniscript RT Kit; Qiagen). Ligation was performed using the Quick T4 DNA Ligase (New England Biolabs).
Mice, tumor surveillance, and treatment.
Balb/C and C57BL/6 mice were purchased from Charles River Laboratories or Harlan Laboratories. Eμ-Myc transgenic mice (C57BL/6; ref. 17) were bred in the animal facility of Technische Universität München. FLCs were obtained on day 14.5 after mating. Recipient Balb/C or C57BL/6 mice for transplantation experiments were purchased from Charles River Laboratories or Harlan Laboratories and were monitored for signs of morbidity and tumor development by daily observation and physical examination. Treatment with doxorubicin (10 mg/kg body weight) was performed by i.p. injection of mice that had received 5 × 104 ex vivo–cultured L-GP130 myeloma cells. Tumors were harvested after mouse sacrifice, and tumors or cells were snap-frozen or frozen viable.
Generation of retrovirus and BM transplantation.
Retrovirus was obtained through transient infection of Phoenix E or Phoenix A cells with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. BA/F3 and INA-6 cells were infected 3 times at 12-hour intervals with retrovirus supplemented with 8 μg/ml polybrene (Sigma-Aldrich). BM and FLC infection and transplantation were performed as described previously (52, 53). Female BM or FLC recipient mice received myeloablative irradiation (9 Gy). Balb/C mice were transplanted with approximately 2.5–3.5 × 105 syngeneic BM cells in total. Infection efficacy (measured as GFP+ cells) was 7%–10%. C57BL/6 mice were transplanted with 1–4 × 106 GFP+ Eμ-Myc FLCs, with 15%–50% infection efficacy (measured as GFP+ cells).
Flow cytometry and cell sorting.
Cells were stained in PBS (PAA) containing 0.5% BSA (Roth), and fluorescently labeled antibodies were used at 1:200–1:1,000 dilution. Viability distinction was realized by staining with the Fixable Viability Dye eF506 (eBioscience). For extracellular staining, fluorescently labeled antibodies (all from BD Pharmingen or eBioscience) were used against the following murine surface proteins: B220 (clone RA3_6B2), CD11b (clone M1/70), CD19 (clone 1D3), CD25 (clone PC61.5), c-kit (clone 2B8), CD56 (clone 809220), CD138 (clone 281-2), IgM (clone II/41), IgD (clone 11-26), TCRβ (clone H57-597). For detection of cytoplasmic Igκ and Igλ light chains, intracellular staining was performed using the Intracellular Fixation and Permeabilization Buffer Set (eBioscience) according to the manufacturer’s instructions, with the following antibodies: Igκ light chain (clone 187.1), Igλ light chain (clone RML-42). All antibodies were purchased from eBioscience, BD Pharmingen, or R&D Systems. For propidium iodide (PI) cell cycle analysis, cells were fixed in 70% ice-cold ethanol and stained in PI staining solution (50 μg/ml PI, 100 μg/ml RNase, and PBS). To analyze apoptosis in GFP+ B cells, the AnnexinV Apoptosis Detection Kit APC (eBioscience) was used according to the manufacturer’s instructions. BrdU cell cycle analysis was performed using the BrdU labeling kit according to the manufacturer’s instructions (BD Biosciences). Data were acquired on a FACSCanto II flow cytometer (BD) or a Cyan ADP Lx P8 (Coulter-Cytomation). FlowJo Software (Tree Star Inc.) was used for data analysis.
Immunoblotting, serum electrophoresis, and ELISA.
Cell lysis for immunoblotting was performed with lysis buffer containing 50 mM HEPES (pH 7.5), 150 mM NaCl, 1 mM EDTA, 2.5 mM EGTA, 0.1% Tween-20 (all from Sigma-Aldrich), and protease inhibitors (Roche) followed by sonication (33). After SDS-PAGE, proteins were transferred to PVDF membranes (Millipore Corp.) and probed with antibody. The following antibodies were used: anti-GP130 (Santa Cruz Biotechnology Inc.), anti–c-MYC (Santa Cruz Biotechnology Inc.), anti-STAT3 (Cell Signaling), anti–P-STAT3 (Cell Signaling), anti-ERK1/2 (Cell Signaling), anti–P-ERK1/2 (Cell Signaling), anti-p53 (Cell Signaling), anti–BCL-XL (Cell Signaling), anti–cleaved caspase 3 (Cell Signaling), anti-monoubiquitinylated and anti-polyubiquitinylated conjugates (Enzo Lifescience), anti–β-actin (Sigma-Aldrich), anti-MCL1 (Santa Cruz Biotechnology Inc), anti-BCL2 (BD Pharmingen).
Serum protein electrophoresis was performed on an Elphoscan ES2000 Plus device (Sarstedt). Briefly, 25 μl mouse serum was applied to a cellulose acetate strip. After separation, the resulting protein fractions were stained with Ponceau S and quantified by densitometric scanning. Quantitative determination of serum IgG, IgM, and IgA levels was performed by ELISA using a SBA Clonotyping System/AP (SouthernBiotech).
Histological analyses, cytology, immunohistochemistry, and FISH.
Mouse organ tissue samples were fixed in formalin for 24 hours and embedded in paraffin. 2-μm sections were deparaffinized, dehydrated, and stained with H&E (DAKO). Immunohistochemistry was performed as described previously (33). The following antibodies were used: anti-B220 (Pharmingen, BD Bioscience), anti-CD138 (Pharmingen), anti-CD3 (DCS), anti-Ki67 (Neomarkers, Thermo Fisher Scientific), anti–Igκ light chain (DAKO), anti–Igλ light chain (DAKO). Ex vivo–cultured myeloma cells were subjected to cytospin, air dried, incubated with Pappenheim staining solution, and washed with aqua dest.
EDTA-decalcified, paraffin-embedded BM trephine biopsies of MM patients were obtained from the archive of the Institute of Pathology and Neuropathology, University Hospital Tübingen. Cases with >20% plasma cell infiltrate were incorporated into a tissue array by transferring tumor-containing areas of the biopsy into a recipient block with predrilled holes. Immunohistochemical double staining for P-STAT3 and the plasma cell marker CD138 was performed on an automated immunostainer (Ventana Medical Systems) following the manufacturer’s protocols, with modifications. In brief, nuclear P-STAT3 was detected with the P-STAT3 Tyr705 antibody (Cell Signaling Technology), using DAB as chromogen, followed by incubation with anti-CD138 (DAKO) and detection with Fast Red chromogen (DAKO). Staining of endothelial cells served as internal positive control. Clear-cut nuclear positivity for P-STAT3 in >10% of CD138+ tumor cells was considered positive for STAT3 activation. Cases with absence of endothelial cell nuclear staining or lack of sufficient tumor cells for evaluation were excluded from analysis.
For FISH analysis, 2-μm-thick unstained histological sections were deparaffinized. Antigen retrieval was performed by heating the slides in a pressure cooker for 5 minutes in citrate buffer (pH 6). After washing, slides were incubated with Pronase E (0.05%; Sigma-Aldrich) at 37°C for 8 minutes. Next, the slides were fixed in 4% paraformaldehyde for 5 minutes. Denaturation was performed by incubating the slides in 70% formamide/SSC (Sigma-Aldrich) for 15 minutes. After dehydration, FISH probes (c-MYC-dig and IgH-biotin; Cegat) were added to the tissue microarray slides and incubated overnight at 37°C. Probes were detected by incubation with Cy3-conjugated rabbit anti-dig (Molecular Probes) and FITC-conjugated rabbit streptavidin (Roche) secondary antibodies. Stringency washes were then performed by heating the slides in 2× SSC with 0.3% NP40, and slides were mounted with Vectastain DAPI Mounting Medium (Vector Laboratories). FISH analysis was performed using a fluorescence microscope equipped with a triple bandpass filter set. Signals were scored manually (×100 oil immersion objective).
Gene expression analysis.
Expression of STAT3 target genes was analyzed in 2 previously published MM datasets (GEO accession nos. GSE2658 and GSE26760; refs. 20, 21). Following standard data normalization and filtering (54), a previously reported STAT3 activation–associated gene expression signature was retrieved from the respective datasets (19). Based on the overlapping STAT3 activation signature, data were then clustered using average linkage hierarchical clustering, and results were visualized using Treeview as previously reported (54). Oncomine-derived expression data were analyzed by Student’s t test directly with Oncomine 4.4.3 software (www.oncomine.org).
GSEA was performed across the list of genes ranked by t statistic as previously reported (55), using the GSEA online version accessible through the GenePattern tool (http://www.broadinstitute.org/cancer/software/genepattern/).
PET-CT and X-ray analysis.
FDG (2-deoxy-2-[18F]fluoro-d-glucose) was obtained from the radiopharmacy unit of Technische Universität München (56) and administered via tail vein injection (100 μl) at an activity dose of 5–10 MBq per mouse. Radiotracer accumulation in the tumor was allowed for 60 minutes. Imaging was performed using a μPET-CT system (Inveon; SIEMENS Preclinical Solutions) with a 15-minute static acquisition. X-ray analysis was performed using a Mammomat 3000 system (Siemens AG).
Analysis of IgH rearrangements.
See Supplemental Methods.
Statistics.
Statistical analyses were performed using the statistical functions of Excel (Microsoft) or GraphPad Prism (GraphPad Software). Graphical data are presented as mean ± SD or mean ± SEM. For continuous variables, statistical analyses were performed using unpaired t test or Mann-Whitney test; for categorical variables, Fisher exact test or χ2 test was performed. Outcome data were visualized by Kaplan-Meier curves, and survival analyses were performed using log-rank test. 2-sided P values less than 0.05 were considered statistically significant.
Study approval.
BM trephine biopsies of patients with MM were obtained from the archive of the Institute of Pathology and Neuropathology, University Hospital Tübingen. The analyses were approved by the responsible ethics committee (University Hospital Tübingen). The animal experiments performed were approved by the responsible regional authorities (Regierung von Oberbayern).
Supplementary Material
Acknowledgments
We thank Sabine Pirsig and the imaging facility of the Nuclear Medicine Department of Technische Universität München for expert technical assistance, Ruth Eichner and Katharina Engel (Technische Universität München) for support analyzing ubiquitylation, and the Zentrum Präklinische Forschung (Technische Universität München) for animal care. We thank Susanne Maas, Ulrich Vogel, and Michaela Rockenstiehl for assembly of the myeloma tissue array and Anne Adam for performing the double staining (all from Tübingen University, Tübingen, Germany). This work was supported by the Deutsche Krebshilfe (grant 111305), the Deutsche Forschungsgemeinschaft (grants SFB 824, SFB841, and TRR 54; Emmy Noether program grant BA 2851/3-1; and Heisenberg-Stipendium BU 1339/3-1), the Sander Stiftung (grants 2005.158.1 and 2012.096.1), the Cluster of Excellence “Inflammation at Interfaces,” and a research grant from Celgene Germany.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2014;124(12):5263–5274. doi:10.1172/JCI69094.
References
- 1.Mahindra A, Laubach J, Raje N, Munshi N, Richardson PG, Anderson K. Latest advances and current challenges in the treatment of multiple myeloma. Nat Rev Clin Oncol. 2012;9(3):135–143. doi: 10.1038/nrclinonc.2012.15. [DOI] [PubMed] [Google Scholar]
- 2.Kapoor P, Rajkumar SV. Update on risk stratification and treatment of newly diagnosed multiple myeloma. Int J Hematol. 2011;94(4):310–320. doi: 10.1007/s12185-011-0947-z. [DOI] [PubMed] [Google Scholar]
- 3.Palumbo A, Anderson K. Medical Progress Multiple Myeloma. New Engl J Med. 2011;364(11):1046–1060. doi: 10.1056/NEJMra1011442. [DOI] [PubMed] [Google Scholar]
- 4.Chesi M, Bergsagel PL. Many multiple myelomas: making more of the molecular mayhem. Hematology Am Soc Hematol Educ Program. 2011;2011:344–353. doi: 10.1182/asheducation-2011.1.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fonseca R, et al. International Myeloma Working Group molecular classification of multiple myeloma: spotlight review. Leukemia. 2009;23(12):2210–2221. doi: 10.1038/leu.2009.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuehl WM, Bergsagel PL. Molecular pathogenesis of multiple myeloma and its premalignant precursor. J Clin Invest. 2012;122(10):3456–3463. doi: 10.1172/JCI61188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Affer M, et al. Promiscuous MYC locus rearrangements hijack enhancers but mostly super-enhancers to dysregulate MYC expression in multiple myeloma. Leukemia. 2014;28(8):1725–1735. doi: 10.1038/leu.2014.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taga T, Kishimoto T. Signaling mechanisms through cytokine receptors that share signal transducing receptor components. Curr Opin Immunol. 1995;7(1):17–23. doi: 10.1016/0952-7915(95)80024-7. [DOI] [PubMed] [Google Scholar]
- 9.Heinrich PC, et al. Membrane-bound and soluble interleukin-6 receptor: studies on structure, regulation of expression, and signal transduction. Ann N Y Acad Sci. 1995;762:222–236. doi: 10.1111/j.1749-6632.1995.tb32328.x. [DOI] [PubMed] [Google Scholar]
- 10.Imada K, Leonard WJ. The Jak-STAT pathway. Mol Immunol. 2000;37(1–2):1–11. doi: 10.1016/s0161-5890(00)00018-3. [DOI] [PubMed] [Google Scholar]
- 11.Hideshima T, Mitsiades C, Tonon G, Richardson PG, Anderson KC. Understanding multiple myeloma pathogenesis in the bone marrow to identify new therapeutic targets. Nat Rev Cancer. 2007;7(8):585–598. doi: 10.1038/nrc2189. [DOI] [PubMed] [Google Scholar]
- 12.Scuto A, et al. The novel JAK inhibitor AZD1480 blocks STAT3 and FGFR3 signaling, resulting in suppression of human myeloma cell growth and survival. Leukemia. 2011;25(3):538–550. doi: 10.1038/leu.2010.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Monaghan KA, Khong T, Burns CJ, Spencer A. The novel JAK inhibitor CYT387 suppresses multiple signalling pathways, prevents proliferation and induces apoptosis in phenotypically diverse myeloma cells. Leukemia. 2011;25(12):1891–1899. doi: 10.1038/leu.2011.175. [DOI] [PubMed] [Google Scholar]
- 14.Yasui H, Hideshima T, Richardson PG, Anderson KC. Novel therapeutic strategies targeting growth factor signalling cascades in multiple myeloma. Br J Haematol. 2006;132(4):385–397. doi: 10.1111/j.1365-2141.2005.05860.x. [DOI] [PubMed] [Google Scholar]
- 15.Pedranzini L, et al. Pyridone 6, a pan-Janus-activated kinase inhibitor, induces growth inhibition of multiple myeloma cells. Cancer Res. 2006;66(19):9714–9721. doi: 10.1158/0008-5472.CAN-05-4280. [DOI] [PubMed] [Google Scholar]
- 16.Kovalchuk AL, et al. IL-6 transgenic mouse model for extraosseous plasmacytoma. Proc Natl Acad Sci U S A. 2002;99(3):1509–1514. doi: 10.1073/pnas.022643999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adams JM, et al. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318(6046):533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- 18.Carrasco DR, et al. The differentiation and stress response factor XBP-1 drives multiple myeloma pathogenesis. Cancer Cell. 2007;11(4):349–360. doi: 10.1016/j.ccr.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dauer DJ, et al. Stat3 regulates genes common to both wound healing and cancer. Oncogene. 2005;24(21):3397–3408. doi: 10.1038/sj.onc.1208469. [DOI] [PubMed] [Google Scholar]
- 20.Chapman MA, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471(7339):467–472. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhan F, et al. The molecular classification of multiple myeloma. Blood. 2006;108(6):2020–2028. doi: 10.1182/blood-2005-11-013458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gearing DP, et al. Proliferative responses and binding properties of hematopoietic cells transfected with low-affinity receptors for leukemia inhibitory factor, oncostatin M, and ciliary neurotrophic factor. Proc Natl Acad Sci U S A. 1994;91(3):1119–1123. doi: 10.1073/pnas.91.3.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stuhlmann-Laeisz C, et al. Forced dimerization of gp130 leads to constitutive STAT3 activation, cytokine-independent growth, and blockade of differentiation of embryonic stem cells. Mol Biol Cell. 2006;17(7):2986–2995. doi: 10.1091/mbc.E05-12-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miething C, et al. The oncogenic fusion protein nucleophosmin-anaplastic lymphoma kinase (NPM-ALK) induces two distinct malignant phenotypes in a murine retroviral transplantation model. Oncogene. 2003;22(30):4642–4647. doi: 10.1038/sj.onc.1206575. [DOI] [PubMed] [Google Scholar]
- 25. Swerdlow SHC, et al., eds. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Lyon, France: International Agency for Research on Cancer; 2008. [Google Scholar]
- 26.Rawstron AC, et al. Report of the European Myeloma Network on multiparametric flow cytometry in multiple myeloma and related disorders. Haematologica. 2008;93(3):431–438. doi: 10.3324/haematol.11080. [DOI] [PubMed] [Google Scholar]
- 27.Guikema JEJ, Hovenga S, Vellenga E, Bos NA. Heterogeneity in the multiple myeloma tumor clone. Leuk Lymphoma. 2004;45(5):857–871. doi: 10.1080/10428190310001593201. [DOI] [PubMed] [Google Scholar]
- 28.Li J, et al. INCB16562, a JAK1/2 selective inhibitor, is efficacious against multiple myeloma cells and reverses the protective effects of cytokine and stromal cell support. Neoplasia. 2010;12(1):28–38. doi: 10.1593/neo.91192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oostendorp RA, et al. Stromal cell lines from mouse aorta-gonads-mesonephros subregions are potent supporters of hematopoietic stem cell activity. Blood. 2002;99(4):1183–1189. doi: 10.1182/blood.V99.4.1183. [DOI] [PubMed] [Google Scholar]
- 30.Rangarajan A, Weinberg RA. Opinion - Comparative biology of mouse versus human cells: modelling human cancer in mice. Nat Rev Cancer. 2003;3(12):952–959. doi: 10.1038/nrc1235. [DOI] [PubMed] [Google Scholar]
- 31.Keller U, et al. Myc suppression of Nfkb2 accelerates lymphomagenesis. BMC Cancer. 2010;10:348. doi: 10.1186/1471-2407-10-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Old JB, et al. Skp2 directs Myc-mediated suppression of p27Kip1 yet has modest effects on Myc-driven lymphomagenesis. Mol Cancer Res. 2010;8(3):353–362. doi: 10.1158/1541-7786.MCR-09-0232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.den Hollander J, et al. Aurora kinases A and B are up-regulated by Myc and are essential for maintenance of the malignant state. Blood. 2010;116(9):1498–1505. doi: 10.1182/blood-2009-11-251074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bouchard C, Lee S, Paulus-Hock V, Loddenkemper C, Eilers M, Schmitt CA. FoxO transcription factors suppress Myc-driven lymphomagenesis via direct activation of Arf. Genes Dev. 2007;21(21):2775–2787. doi: 10.1101/gad.453107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chauhan D, et al. Multiple myeloma cell adhesion-induced interleukin-6 expression in bone marrow stromal cells involves activation of NF-κB. Blood. 1996;87(3):1104–1112. [PubMed] [Google Scholar]
- 36.Kawano M, et al. Autocrine generation and requirement of BSF-2/IL-6 for human multiple myelomas. Nature. 1988;332(6159):83–85. doi: 10.1038/332083a0. [DOI] [PubMed] [Google Scholar]
- 37.Pulkki K, Pelliniemi TT, Rajamaki A, Tienhaara A, Laakso M, Lahtinen R. Soluble interleukin-6 receptor as a prognostic factor in multiple myeloma. Finnish Leukaemia Group. Br J Haematol. 1996;92(2):370–374. doi: 10.1046/j.1365-2141.1996.d01-1470.x. [DOI] [PubMed] [Google Scholar]
- 38.Bataille R, Jourdan M, Zhang XG, Klein B. Serum levels of interleukin 6, a potent myeloma cell growth factor, as a reflect of disease severity in plasma cell dyscrasias. J Clin Invest. 1989;84(6):2008–2011. doi: 10.1172/JCI114392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sporeno E, et al. Human interleukin-6 receptor super-antagonists with high potency and wide spectrum on multiple myeloma cells. Blood. 1996;87(11):4510–4519. [PubMed] [Google Scholar]
- 40.Catlett-Falcone R, et al. Constitutive activation of Stat3 signaling confers resistance to apoptosis in human U266 myeloma cells. Immunity. 1999;10(1):105–115. doi: 10.1016/S1074-7613(00)80011-4. [DOI] [PubMed] [Google Scholar]
- 41.Chatterjee M, Stuhmer T, Herrmann P, Bommert K, Dorken B, Bargou RC. Combined disruption of both the MEK/ERK and the IL-6R/STAT3 pathways is required to induce apoptosis of multiple myeloma cells in the presence of bone marrow stromal cells. Blood. 2004;104(12):3712–3721. doi: 10.1182/blood-2004-04-1670. [DOI] [PubMed] [Google Scholar]
- 42.Chatterjee M, et al. In the presence of bone marrow stromal cells human multiple myeloma cells become independent of the IL-6/gp130/STAT3 pathway. Blood. 2002;100(9):3311–3318. doi: 10.1182/blood-2002-01-0102. [DOI] [PubMed] [Google Scholar]
- 43.Brocke-Heidrich K, et al. Interleukin-6-dependent gene expression profiles in multiple myeloma INA-6 cells reveal a Bcl-2 family-independent survival pathway closely associated with Stat3 activation. Blood. 2004;103(1):242–251. doi: 10.1182/blood-2003-04-1048. [DOI] [PubMed] [Google Scholar]
- 44.Quintanilla-Martinez L, et al. Analysis of signal transducer and activator of transcription 3 (Stat 3) pathway in multiple myeloma: Stat 3 activation and cyclin D1 dysregulation are mutually exclusive events. Am J Pathol. 2003;162(5):1449–1461. doi: 10.1016/S0002-9440(10)64278-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009;9(11):798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maclean KH, Keller UB, Rodriguez-Galindo C, Nilsson JA, Cleveland JL. c-Myc augments gamma irradiation-induced apoptosis by suppressing Bcl-XL. Mol Cell Biol. 2003;23(20):7256–7270. doi: 10.1128/MCB.23.20.7256-7270.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheung WC, et al. Novel targeted deregulation of c-Myc cooperates with Bcl-X(L) to cause plasma cell neoplasms in mice. J Clin Invest. 2004;113(12):1763–1773. doi: 10.1172/JCI20369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheung WC, Van Ness B. Distinct IL-6 signal transduction leads to growth arrest and death in B cells or growth promotion and cell survival in myeloma cells. Leukemia. 2002;16(6):1182–1188. doi: 10.1038/sj.leu.2402481. [DOI] [PubMed] [Google Scholar]
- 49.Yata K, Yaccoby S. The SCID-rab model: a novel in vivo system for primary human myeloma demonstrating growth of CD138-expressing malignant cells. Leukemia. 2004;18(11):1891–1897. doi: 10.1038/sj.leu.2403513. [DOI] [PubMed] [Google Scholar]
- 50.Chesi M, et al. AID-dependent activation of a MYC transgene induces multiple myeloma in a conditional mouse model of post-germinal center malignancies. Cancer Cell. 2008;13(2):167–180. doi: 10.1016/j.ccr.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Keller UB, et al. Myc targets Cks1 to provoke the suppression of p27Kip1, proliferation and lymphomagenesis. EMBO J. 2007;26(10):2562–2574. doi: 10.1038/sj.emboj.7601691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miething C, et al. The Bcr-Abl mutations T315I and Y253H do not confer a growth advantage in the absence of imatinib. Leukemia. 2006;20(4):650–657. doi: 10.1038/sj.leu.2404151. [DOI] [PubMed] [Google Scholar]
- 53.Kratzat S, et al. Cks1 is required for tumor cell proliferation but not sufficient to induce hematopoietic malignancies. PLoS One. 2012;7(5):e37433. doi: 10.1371/journal.pone.0037433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Luck SC, et al. Deregulated apoptosis signaling in core-binding factor leukemia differentiates clinically relevant, molecular marker-independent subgroups. Leukemia. 2011;25(11):1728–1738. doi: 10.1038/leu.2011.154. [DOI] [PubMed] [Google Scholar]
- 55.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Machulla HJ, Blocher A, Kuntzsch M, Piert M, Wei R, Grierson J. Simplified labeling approach for synthesizing 3-deoxy-3-[18F] fluorothymidine ([18F] FLT). J Radioanal Nucl Chem. 2000;243(3):843–846. doi: 10.1023/A:1010684101509. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.