Abstract
CD8+ cytotoxic T lymphocytes (CTLs) have potent antitumor activity and therefore are leading candidates for use in tumor immunotherapy. The application of CTLs for clinical use has been limited by the susceptibility of ex vivo–expanded CTLs to become dysfunctional in response to immunosuppressive microenvironments. Here, we developed a microRNA-targeting (miRNA-targeting) approach that augments CTL cytotoxicity and preserves immunocompetence. Specifically, we screened for miRNAs that modulate cytotoxicity and identified miR-23a as a strong functional repressor of the transcription factor BLIMP-1, which promotes CTL cytotoxicity and effector cell differentiation. In a cohort of advanced lung cancer patients, miR-23a was upregulated in tumor-infiltrating CTLs, and expression correlated with impaired antitumor potential of patient CTLs. We determined that tumor-derived TGF-β directly suppresses CTL immune function by elevating miR-23a and downregulating BLIMP-1. Functional blocking of miR-23a in human CTLs enhanced granzyme B expression, and in mice with established tumors, immunotherapy with just a small number of tumor-specific CTLs in which miR-23a was inhibited robustly hindered tumor progression. Together, our findings provide a miRNA-based strategy that subverts the immunosuppression of CTLs that is often observed during adoptive cell transfer tumor immunotherapy and identify a TGF-β–mediated tumor immune-evasion pathway.
Introduction
Owing to their unique abilities for specific tumor antigen recognition and efficient cytolysis, CD8+ cytotoxic T lymphocytes (CTLs) represent the primary leukocyte population used for adoptive cell transfer (ACT) cancer treatment. ACT relies on isolation, followed by extensive ex vivo expansion of tumor-infiltrating CTLs (TILs) in the presence of copious amounts of growth factors (e.g., IL-2) in vitro, followed by autologous reinfusion into the patient (1, 2). Recent advances in CTL engineering have allowed the enforced expression of high-affinity and tumor-specific T cell receptors (TCRs) or chimeric antigen receptors, thereby mitigating part of the difficulties in CTL isolation and expansion, and the efficacy of tumor antigen targeting (2–4). Paradoxically, although these strategies are capable of generating highly cytotoxic tumor-specific CTLs in vitro, the clinical success of ACT using ex vivo IL-2–conditioned and TCR-redirected CTLs has been partial at best — the majority of patients fail to respond with complete tumor regressions (5–7).
This apparent discrepancy between the in vitro and in vivo functionality of CTLs in ACT is largely attributed to the presence of immunosuppressive barriers within the tumor microenvironment, which are co-opted by tumors to evade the host immune system (8–11). Of these, TGF-β is a key cytokine during tumor pathogenesis secreted and upregulated by a wide variety of tumors, including melanoma and lung cancer (12–15). In melanoma and lung cancer patients, high plasma TGF-β levels are a negative prognostic indicator of tumor progression, because of their association with increased metastasis and relapse rates, as well as decreased overall patient survival (16–18). Moreover, local expression of TGF-β is further elevated within metastatic melanoma lesions, compared with their primary tumors (19). TGF-β promotes tumor outgrowth and metastasis in various avenues, a critical one of which is to hamper productive antitumor immune responses. Specifically, TGF-β–induced SMAD signaling in both naive and full-fledged effector CTLs represses their expression of key cytotoxic mediators, including granzyme B and IFN-γ, resulting in CTL dysfunction and impaired tumor rejection (20, 21). Engineering tumor-specific CTLs to overcome TGF-β–mediated immune suppression and preserve their cytotoxicity within the tumor microenvironment therefore remains one of the holy grails in the field of cancer intervention.
CTL function and cytotoxicity are governed by several key transcription regulators, including T-bet, EOMES, and BLIMP-1. In effector CTLs, T-bet and EOMES are compensatory and essential transcriptional factors enforcing a type 1 program that instructs their differentiation into highly potent killer CTLs — T-bet and EOMES drive the expression of type 1 cytotoxic mediators (e.g., granzyme B, perforin, and IFN-γ) for the eradication of malignant cells, while simultaneously repressing the acquisition of an unproductive type 17 program that targets extracellular pathogens (22–25). Unsurprisingly, T-bet and EOMES double-deficient CTLs have severely impaired cytotoxicity and antitumor responses (24, 26). Likewise, by repressing the quiescent transcriptional program characteristic of memory CTLs, the transcriptional regulator BLIMP-1 is also essential for promoting CTL cytotoxicity and effector differentiation (27–29). Notably, BLIMP-1–deficient effector CTLs have impaired cytotoxicity, and show reduced expression of multiple type 1 cytotoxic mediators (27, 28).
To enhance the efficacy of current tumor immunotherapy, we became interested in a novel microRNA-based (miRNA-based) approach to augment the cytotoxic capacity of tumor-specific CTLs ex vivo. miRNAs are a group of small noncoding RNAs that have emerged as key regulators of gene expression in plants and animals (30). Importantly, mounting evidence indicates that miRNAs are integral and effective regulatory elements of the adaptive immune system (31–35), making the manipulation of miRNA levels in CTLs an attractive means of enhancing antitumor adaptive responses. In addition, miRNA-based therapy offers two advantages over conventional protein-target-based immune modulation — it is far more straightforward to engineer antisense miRNA inhibitors, and miRNA-based gene therapy can be readily incorporated into conventional ACT (36–39). Unfortunately, little is known of the therapeutic miRNA targets, which are capable of sustaining effector CTL function particularly in the face of tumor-induced immunosuppression.
To address this knowledge gap, we compared the miRNA expression profiles of poorly and highly cytotoxic CTLs generated under different priming conditions, and identified miR-23a as a key inhibitor of antitumor responses in mouse and human CTLs. We demonstrate that miR-23a downregulates its mRNA target BLIMP-1, and simultaneously inhibits the expression of multiple key CTL effector molecules and transcriptional regulators. Additionally, we establish cMYC and tumor-associated TGF-β as key determinants of miR-23a abundance in effector CTLs.
Results
Identification of miR-23a expression as a negative correlate of cytotoxicity of effector CTLs.
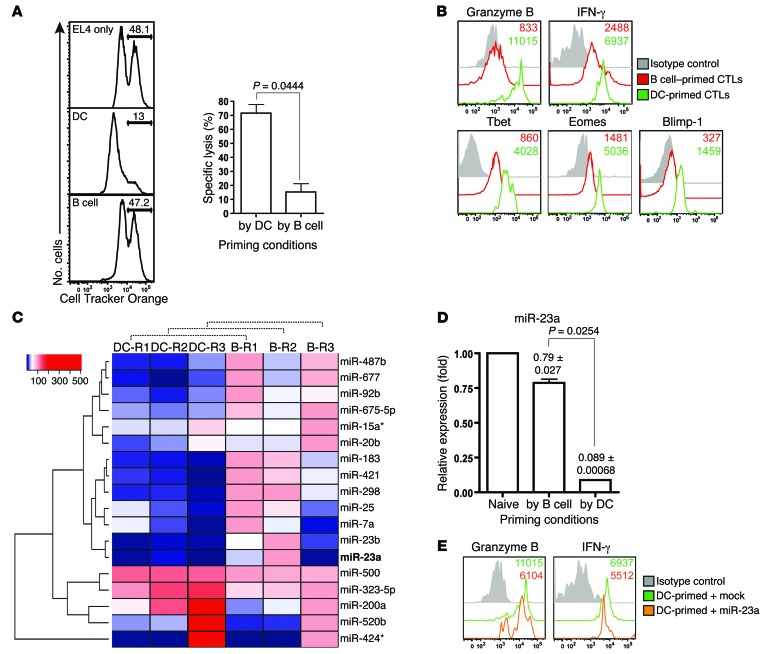
To screen for key miRNA regulators of CTL effector responses, we used different in vitro systems that are well known for generating CTLs with different killing capacities. Naive murine pMel-1 CTLs were primed in vitro with either mature bone marrow–derived DCs (Supplemental Figure 1, A and B; supplemental material available online with this article; doi:10.1172/JCI76561DS1), or splenic B cells pulsed with the melanoma tumor–associated antigen hgp100 (40). pMel-1 CTLs expanded with peptide-loaded syngeneic B cells displayed very poor cytotoxic capacity at an effector/target (E/T) ratio of 5:1; in contrast, CTLs expanded with DCs exhibited 5-fold higher cytotoxic potency (Figure 1A). Accordingly, we found that DC-primed CTLs expressed higher levels of key cytotoxic mediators (granzyme B and IFN-γ), and upstream master regulators (T-bet, EOMES, and BLIMP-1) (Figure 1B). Killing deficiencies observed in B cell priming were previously reported to result from impaired granzyme B expression and increased activation-induced T cell death (AICD) (41, 42), which could be overcome by IL-15 and IL-21 (43, 44). Consistent with earlier reports (41–44), the addition of exogenous IL-15 and IL-21 during B cell priming partially rescued granzyme B expression (Supplemental Figure 1C) and AICD (Supplemental Figure 1D) in CTLs. However, when challenged with a high ratio of antigen-pulsed target cells (E/T = 5:1), these cytokines enhanced their in vitro cytotoxicity insignificantly (Supplemental Figure 1E). Qualitative differences in CTL cytotoxicity induced by DCs and B cells indicate that these 2 priming conditions elicit distinct cytotoxic transcriptomes in CTLs; therefore, as unphysiological as it may be, our in vitro priming system provides us with a comparative platform for discovering master regulator(s) of cytotoxicity.
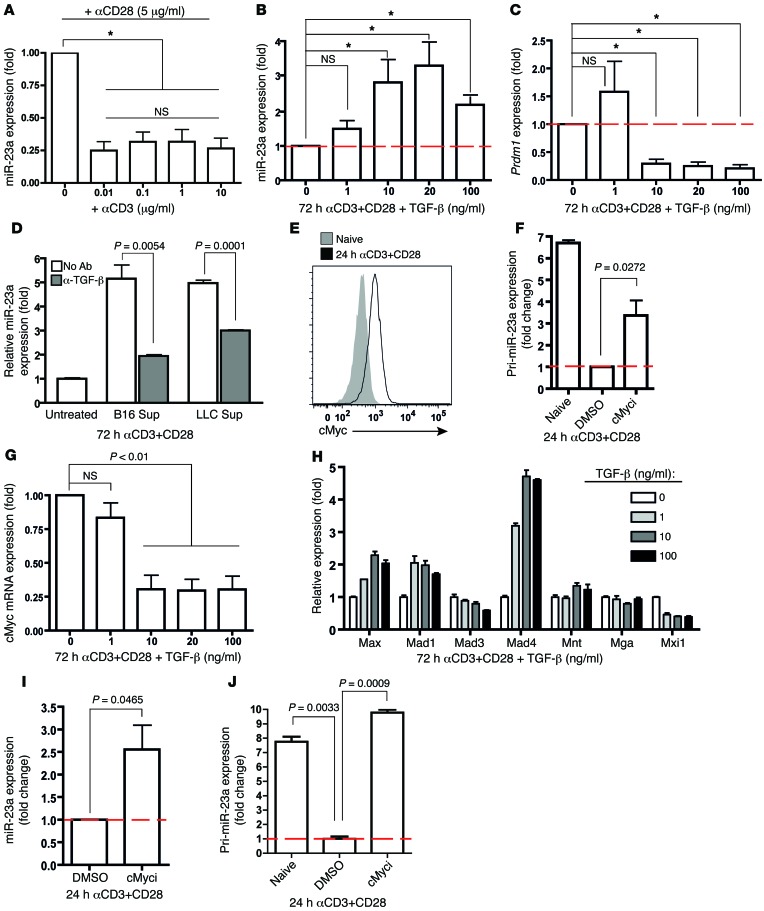
Figure 1. Identification of miR-23a as a negative correlate of CTL effector function.
(A and B) pMel-1 CTLs primed with splenic B cells or LPS-matured bone marrow–derived DCs for 4–6 days were assessed for (A) in vitro cytotoxicity at an E/T ratio of 5:1 and (B) expression of CTL effector molecules. Histograms are representative of n = 3 independent experiments, and the bar graph represents the mean ± SEM of n = 3 independent experiments. (C) After 3 days of in vitro priming by DCs or B cells, pMel-1 CTLs were isolated for miRNA expression profiling. Heat map of miRNAs differentially expressed by DC- and B cell–primed CTLs from n = 3 independent miRNA profiling experiments. Asterisks with miRNAs refer to the passenger strands of the respective miRNA species. (D) Validation of differential miR-23a expression in CTLs under the respective priming conditions with 3 additional batches of samples. Numbers and bar graph represent the mean ± SEM miR-23a expression relative to that of naive CD8+ T cells. (E) DC-primed pMel-1 CTLs were retrovirally transduced with either an empty mock vector or a miR-23a overexpression vector. Three days after transduction, CD8+GFP+ CTL effector molecule expression was assessed by flow cytometry.
As miRNAs can simultaneously regulate the expression of multiple genes post-transcriptionally (31, 45), miRNA-based immuno-therapy holds the potential to bypass the need for complex transcriptional reprogramming of effector CTLs. We therefore sought to identify miRNAs that modulate cytotoxicity using our in vitro priming system. After 3 days of priming with either DCs or B cells, we isolated the differentially primed CTLs for miRNA expression profiling (46). Among the 350 miRNAs screened, 18 were significantly differentially expressed: 13 miRNAs were downregulated and 5 were upregulated in DC-primed CTLs (Figure 1C, Supplemental Figure 2A, and Supplemental Table 1). To determine whether these miRNA candidates directly impacted CTL cytotoxicity, we assessed granzyme B expression in pMel-1 CTLs overexpressing either the respective miRNAs, or a mock-GFP control vector. Only miR-23a was able to inhibit both granzyme B and T-bet expression in CTLs (Supplemental Figure 2C and data not shown). Interestingly, although miR-23b — a paralog of miR-23a — was similarly suppressed in DC-primed CTLs (Figure 1C and Supplemental Table 1), miR-23b overexpression did not affect granzyme B levels (Supplemental Figure 2E). Further validation experiments corroborated that miR-23a expression in CTLs was dramatically suppressed during DC priming (Figure 1D). While miR-23a did not affect CTL proliferation (Supplemental Figure 2F) and AICD (Supplemental Figure 2G), overexpression of miR-23a (~3.4 ± 1.0–fold increase from 3.5 × 104 ± 0.6 × 104 copies per cell; Supplemental Figure 2, B–D) blunted the expression of multiple key CTL effector molecules and transcription factors in vitro (Figure 1E). These data suggest that miR-23a may negatively regulate CTL cytotoxicity.
Forced miR-23a expression compromises antitumor CTL effector responses in vivo.
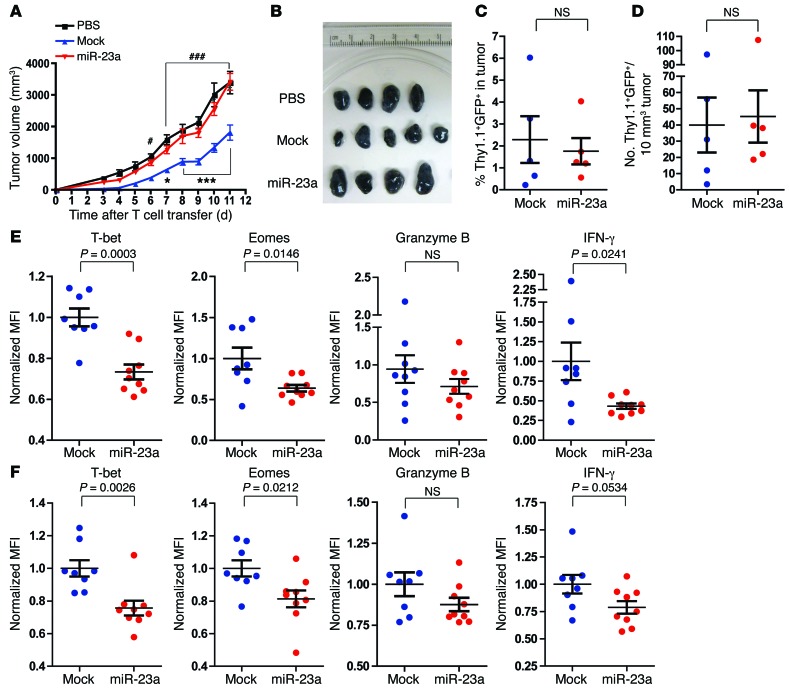
To investigate the impact of miR-23a on CTL antitumor efficacy in vivo, we made use of the poorly immunogenic B16/F10 melanoma tumor model (40). Equal numbers (i.e., 0.6 × 106) of mock pMel-1 CTLs, miR-23a–overexpressing pMel-1 CTLs, or PBS vehicle control were infused into B16/F10 tumor-bearing mice. As previously reported (47, 48), pMel-1 CTLs expressing the mock vector retarded tumor growth substantially. However, this protection was completely abrogated by the forced expression of miR-23a — mice receiving miR-23a–overexpressing CTLs exhibited accelerated tumor progression and higher tumor burdens, comparable to those of untreated (PBS) tumor-bearing mice (Figure 2, A and B). Although miR-23a did not affect CTL accumulation within the tumor (Figure 2, C and D), miR-23a significantly undermined the expression of several key effector molecules in pMel-1 TILs (Figure 2E), and in peripheral pMel-1 CTLs (Figure 2F). Taken together, these results functionally validate that forced miR-23a expression antagonizes antitumor CTL effector responses in vivo.
Figure 2. Forced expression of miR-23a inhibits CTL antitumor responses in vivo.
Three days after s.c. inoculation of 0.2 × 106 B16/F10 melanoma cells, C57BL/6 tumor-bearing mice received adoptive transfers of 0.6 × 106 mock or miR-23a–overexpressing Thy1.1+ pMel-1 CTLs. Tumor progression was monitored and effector responses of Thy1.1+ pMel-1 CTLs were analyzed. (A) Tumor volumes of untreated mice (PBS), and mice adoptively transferred with mock or miR-23a–overexpressing Thy1.1+ pMel-1 CTLs. Graph represents mean ± SEM with n = 9 mice per group, pooled from 2 independent experiments. *P < 0.05 and ***P < 0.001 for mock vs. miR-23a; #P < 0.05 and ###P < 0.001 for mock vs. PBS by 2-way ANOVA and Bonferroni post-test. (B–F) Eleven days after T cell transfer, tumors were excised and Thy1.1+ pMel-1 CTL numbers and effector function were assessed by flow cytometry. Data shown are from 1 representative experiment. (B) Tumor sizes from mice in A. (C) Percentages and (D) densities of tumor-infiltrating mock and miR-23a–overexpressing pMel-1 CTLs. (E and F) Effector molecule expression in (E) tumor-infiltrating and (F) splenic and miR-23a–overexpressing pMel-1 CTLs. Normalized MFI was calculated by division of the MFI of each sample by the average MFIs of the mock group for each experiment.
Functional blockade of miR-23a in CTLs augments their antitumor function in vitro.
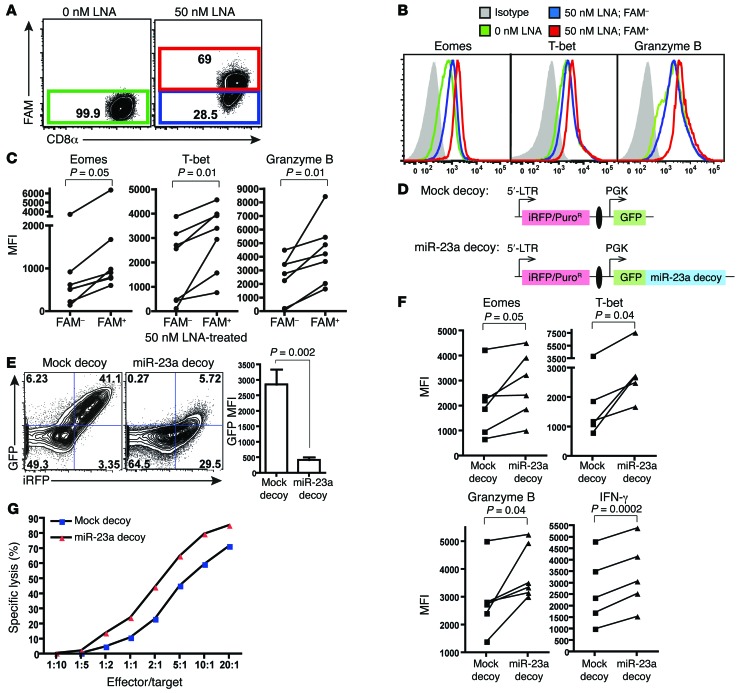
Having identified miR-23a as a repressor of CTL cytotoxicity, we developed 2 strategies for blocking miR-23a function: treatment with an anti–miR-23a locked nucleic acid (LNA) (39) and retroviral transduction of a miR-23a decoy vector (49).
In the first approach, a 6-fluorescein (FAM) fluorescent label conjugated to the 5′-end of the LNA facilitates monitoring the transfection efficiency, enables CTLs that have taken up the LNA to be distinguished as an FAM+ population (Figure 3A), and provides LNA-treated FAM– CTLs as an internal control for the specificity of LNA-mediated miR-23a inhibition. In comparison with the FAM– pMel-1 CTLs, the expression of EOMES, T-bet, and granzyme B was augmented in the miR-23a–inhibited FAM+ population (Figure 3, B and C). To ensure that these observed changes are specific to miR-23a, CTLs were also treated with saturating amounts (Supplemental Figure 3A) of a scrambled antagomir (Supplemental Figure 3B), or an antagomir against the unrelated miR-122 (Supplemental Figure 3C). However, neither antagomir was able to augment CTL effector molecule expression. In spite of this functional enhancement, chemically modified oligonucleotides, such as LNAs, cannot be permanently retained in activated T cells in vivo (data not shown and ref. 39). The short-lived effects of LNA treatment in T cells therefore make it difficult to investigate the long-term antitumor efficacy of miR-23a–inhibited CTLs in vivo.
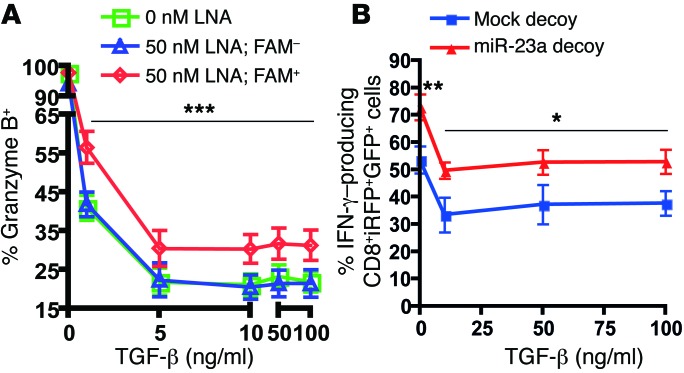
Figure 3. Functional blockade of miR-23a enhances CTL effector responses ex vivo.
(A–C) Purified naive pMel-1 CTLs were activated with anti-CD3/anti-CD28 for 48 hours in vitro, with or without 50 nM FAM-tagged anti–miR-23a LNA. (A) Gating strategy to identify CTLs that have taken up the anti–miR-23a LNA. (B) CTL effector molecule expression in untreated (0 nM LNA), FAM– LNA-treated (50 nM LNA; FAM–), and FAM+ LNA-treated (50 nM LNA; FAM+) CTLs. Representative histograms of n = 6 independent experiments. (C) MFI of CTL effector molecules in FAM– and FAM+ CTLs treated with 50 nM anti–miR-23a LNA; n = 7. P values were determined by 2-tailed paired t test. (D) Schematic of the mock decoy and miR-23a decoy retroviral expression vectors. iRFP was the internal marker for monitoring transfection efficiency; puromycin resistance (PuroR) was the selection marker for enriching engineered cells; GFP was a reporter for miR-23a sequestration and decoy function. The iRFP and GFP decoy expression cassettes were separated by an insulator (51) (black oval). (E) Left: Representative dot plots of iRFP and GFP expression in pMel-1 CTLs transduced with the miR-23a decoy, where iRFP was used as the internal marker. Right: Quenching of GFP intensity by the miR-23a decoy in CD8+iRFP+ CTLs. Bar graph represents mean ± SEM; n = 6. (F) pMel-1 CTLs retrovirally transduced with a mock decoy vector or the miR-23a decoy vector were assessed for CTL effector molecule expression in vitro. P values were determined by 2-tailed paired t test. (G) In vitro cytotoxicity of sorted iRFP+ mock and miR-23a decoy–expressing OT-I CTLs. Representative data of n = 3 independent experiments.
Aimed at achieving long-lasting miR-23a inhibition, we developed a second approach. We retrovirally transduced CTLs with a miR-23a decoy vector capable of sequestering endogenous miR-23a (49). To simultaneously allow the selection of engineered cells, we constructed a bicistronic viral backbone, in which the expression of a selectable marker (iRFP, ref. 50, or puromycin resistance) and the expression of a GFP decoy/reporter are driven independently by the viral 5′-LTR and PGK promoters, respectively (Figure 3D). To maximize their independent expression, we inserted an insulator sequence (51) between the selectable marker and decoy cassettes. A vector omitting miR-23a target sites serves as a mock control. In CTLs transduced with the miR-23a decoy, GFP expression was substantially quenched by 85%, indicating that endogenous miR-23a had been sequestered by our synthetic 3-UTR (Figure 3E). Consistent with LNA-mediated miR-23a suppression, the miR-23a decoy augmented the expression of cytotoxic modulators and effectors in CTLs (Figure 3F), and significantly enhanced their in vitro cytotoxicity over a wide range of E/T ratios (Figure 3G), reiterating our findings that miR-23a inhibition effectively augments CTL functional capacity on a per-cell basis.
miR-23a blunts CTL effector responses by targeting BLIMP-1.
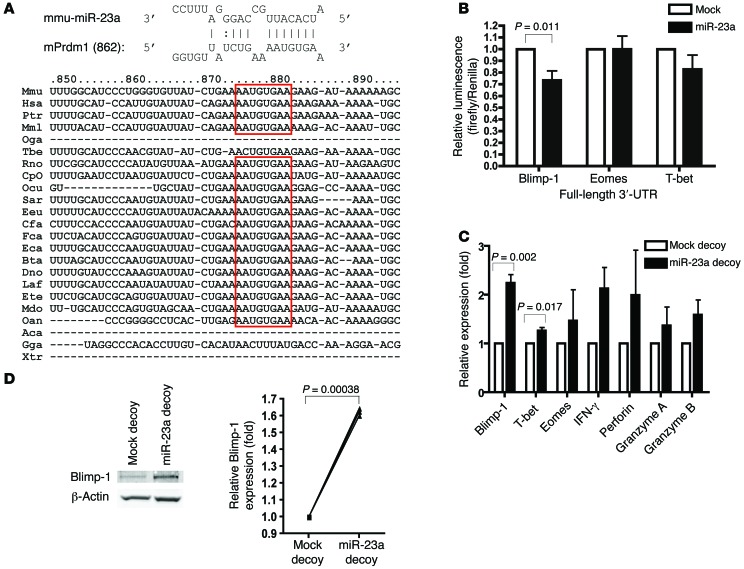
We next sought to investigate the molecular mechanism through which miR-23a modulates CTL effector function. Since glutamine metabolism is central for appropriate T cell activation (52), and miR-23a has previously been shown to directly target glutaminase (GLS) in cancer cell lines (53), we examined the impact of miR-23a on GLS expression in pMel-1 CTLs. However, inhibiting miR-23a by means of the miR-23a decoy failed to upregulate Gls mRNA levels (Supplemental Figure 4A), indicating that miR-23a was unlikely to target GLS in primary effector CTLs. Therefore, we went on to search for miR-23a targets using miRecords (54). miR-23a was computationally predicted to target BLIMP-1 (encoded by the Prdm1 gene) at a highly evolutionarily conserved site (Figure 4A), and to target T-bet and EOMES at weakly conserved sites (Supplemental Figure 4B).
Figure 4. BLIMP-1 is a direct target of miR-23a in CTLs.
(A) Schematic representation of the putative miR-23a binding site within the Prdm1 3′-UTR that is conserved across species. (B) Luciferase assays, in which Jurkat T cells were cotransfected with reporter constructs containing full-length 3′-UTRs of the indicated genes, together with the mock or miR-23a overexpression vector. Graph represents mean ± SEM; n = 5. (C and D) iRFP+ mock and miR-23a decoy–expressing pMel-1 CTLs were sorted for miR-23a target studies. (C) Relative mRNA expression of the predicted miR-23a targets Prdm1, Eomes, and Tbet, as well as other CTL effector molecules. Graph represents mean ± SEM; n = 3. (D) BLIMP-1 protein expression, with relative band intensities normalized to β-actin. Left: Representative Western blot. Right: Pooled data from n = 3 independent experiments. P values in B–D were determined by 2-tailed paired t test.
To assess direct binding of miR-23a to the 3′-UTR of these predicted targets, we constructed luciferase reporters containing the full-length 3′-UTR of the Prdm1, Tbet, or Eomes gene. Each of these luciferase reporters was then cotransfected into Jurkat T cells, together with either a mock vector or the miR-23a overexpression vector. Luciferase activity controlled by the Prdm1 3′-UTR, but not the Tbet or Eomes 3′-UTR, was significantly suppressed by miR-23a (Figure 4B). When the Prdm1 3′-UTR site predicted to interact with the miR-23a seed region was mutated (Supplemental Figure 4C), luciferase reporter activity was no longer controlled by miR-23a (Supplemental Figure 4D). Consistent with this, the miR-23a decoy rescued Prdm1 mRNA (Figure 4C) and protein (Figure 4D) expression in pMel-1 effector CTLs. Interestingly, although T-bet is not a direct target of miR-23a, Tbet mRNA levels were modestly, albeit significantly, increased in miR-23a–inhibited CTLs. Mirroring earlier findings that Tbet transcription is downregulated in Prdm1–/– effector CTLs (28), the observed augmentation in T-bet expression is likely a secondary effect of increased BLIMP-1 abundance in miR-23a–inhibited CTLs. Taken together, these results show that by directly regulating BLIMP-1 expression, silencing miR-23a in CTLs increases their antitumor responses and cytotoxic potency.
In effector CTLs, TCR activation and TGF-β signaling differentially regulate miR-23a expression.
Next, we sought to elucidate how signals received during priming, and within the tumor microenvironment, may reprogram effector CTLs through the alteration of miR-23a levels. We initially identified miR-23a to be differentially regulated in effector CTLs induced by different APCs (Figure 1C), indicating that miR-23a expression in CTLs can be modulated by cell-extrinsic signals. Therefore, we explored multiple pathways known to be differentially influenced by DCs versus B cells. We first explored differences arising from the T/APC interface: TCR signaling strength, coreceptor signals, and NOTCH signaling. Although TCR activation effectively suppressed miR-23a expression in CTLs, increasing the avidity of TCR signaling by varying plate-bound anti-CD3 antibody concentrations from 10 ng/ml to 10 μg/ml did not further alter miR-23a levels (Figure 5A). This 10-ng/ml anti-CD3 threshold indicates that while miR-23a expression is highly sensitive to TCR activation, alteration of TCR signaling strength is not involved in fine-tuning miR-23a abundance in CTLs. Costimulatory signals from CD28 and CD40L, too, had no effect on miR-23a expression (Supplemental Figure 5, A and B). We also investigated the involvement of PD-1, an inhibitory checkpoint molecule exploited by immune-subversive tumors (8, 55–57). Blocking the PD-1 ligands expressed on DCs (Supplemental Figure 5C) had no effect on miR-23a abundance (Supplemental Figure 5D). Conversely, inhibiting miR-23a in CTLs did not alter their surface expression of PD-1 (Supplemental Figure 5E).
Figure 5. TCR and TGF-β signaling converge on cMYC to differentially regulate miR-23a expression in CTLs.
(A) TCR activation, but not stimulation strength, suppresses miR-23a expression in CTLs. miR-23a expression in purified naive CTLs activated in vitro with the indicated concentrations of anti-CD3 (μg/ml) and 5 μg/ml anti-CD28 for 3 days. Data represent mean ± SEM; n = 4. *P < 0.05 by 1-way ANOVA and Bonferroni post-test. (B–D) TGF-β promotes miR-23a expression in CTLs. (B) Mature miR-23a and (C) Prdm1 expression in purified pMel-1 CTLs activated in vitro with varying concentrations of TGF-β for 72 hours. *P < 0.05 by 1-way ANOVA and Bonferroni post-test. (D) miR-23a expression in CTLs activated in vitro with tumor cell–conditioned media (25% in total medium) and neutralizing anti–TGF-β antibody. Data shown in B–D represent mean ± SEM; n = 3. Sup, supernatant from tumor cell–conditioned medium. (E–J) cMYC transcriptionally represses miR-23a in activated CTLs. (E) cMYC protein induction in purified CTLs upon 24 hours of TCR activation. (F) Pri-miR-23a and (I) mature miR-23a expression in purified pMel-1 CTLs treated with or without the cMyc inhibitor 10058-F4. Data in F and I represent mean ± SEM; n = 3 and n = 5, respectively. (G and H) mRNA expression of (G) cMYC and (H) regulators of cMYC activity in activated CTLs upon TGF-β treatment. Data shown in G and H represent mean ± SEM; n = 3. (J) Pri-miR-23a expression in Dicer-deficient CTLs upon cMyc inhibition, expressed as mean ± SEM; n = 3, and this represents 2 independent experiments. Red dashed lines represent expression levels in activated CTLs (control groups) that were set as 1.0, from which relative expression in experimental groups was calculated.
Since NOTCH signaling is known to promote CTL antitumor responses (58, 59), and Notch ligands are differentially expressed on the surface of DCs and B cells (60, 61), we speculated that NOTCH activation may repress miR-23a expression. However, constitutively activating NOTCH in CTLs by forced expression of the NOTCH1 intracellular domain (Supplemental Figure 5F) failed to impact miR-23a expression in CTLs (Supplemental Figure 5G). In reciprocal loss-of-function studies, inhibiting NOTCH signaling with a γ-secretase inhibitor (Supplemental Figure 5H) similarly had no effect on miR-23a expression in CTLs (Supplemental Figure 5I).
In addition to cell/cell interactions, soluble cytokines generated during CTL priming may also influence miR-23a abundance. Therefore, we activated purified naive CTLs in the presence of various DC-derived cytokines for 3 days, before assessing miR-23a expression. Among the panel of cytokines tested, which included type 1 cytokines (IL-2, IL-12, IFN-γ, and TNF-α), inflammasome-derived cytokines (IL-1β and IL-18), and type 1 interferon (IFN-β), none were able to consistently or appreciably regulate miR-23a expression in CTLs (Supplemental Figure 6A).
Finally, we explored the hypothesis that cytokines usually found within the tumor microenvironment — IL-6, IL-10, and TGF-β (57) — may promote miR-23a expression in CTLs. Within this group, IL-6 and IL-10 failed to appreciably impact miR-23a expression (Supplemental Figure 6B). In contrast, TGF-β upregulated miR-23a levels (Figure 5B) and inhibited Prdm1 expression (Figure 5C) in a dose-dependent manner. Importantly, the regulation of Prdm1 by TGF-β closely mirrored that of miR-23a: 1 ng/ml TGF-β altered neither miR-23a nor Prdm1 levels; however, a saturating dose of 10 ng/ml TGF-β significantly stimulated miR-23a expression, while concurrently inhibiting the miR-23a target, Prdm1. This indicates that the suppression of CTL cytotoxicity by TGF-β is, in part, post-transcriptionally mediated by miR-23a and its consequent suppression of the master regulator BLIMP-1.
To investigate whether tumors are capable of driving up miR-23a expression in CTLs, we activated CTLs in the presence of tumor cell–conditioned media. We additionally blocked TGF-β function with an antibody to directly assess the contribution from TGF-β. miR-23a was significantly upregulated in CTLs treated with tumor cell–conditioned media; however, this increase was dampened upon neutralization of TGF-β (Figure 5D). These results demonstrate that in the context of the tumor microenvironment, TGF-β is a primary modulator of miR-23a expression in antitumor CTLs.
TCR and TGF-β signaling converge on cMYC to differentially modulate miR-23a expression in effector CTLs.
We next investigated the signal integration from TCR activation and TGF-β stimulation that controls miR-23a expression. cMYC is one such convergent node impacted by both TCR (52, 62) and TGF-β receptor signaling (63, 64). We examined whether cMYC plays a critical role in regulating pri-miR-23a expression in effector CTLs. Indeed, within 24 hours of activation, naive pMel-1 CTLs rapidly upregulate cMYC expression (Figure 5E), which coincides with a 6.7-fold decrease in pri-miR-23a transcription (Figure 5F). By contrast, during CTL priming, TGF-β repressed Myc mRNA expression (Figure 5G), while augmenting transcription of the cMyc antagonists Mad1 and Mad4 (Figure 5H). This finding in primary T cells parallels an earlier report that cMYC transcriptionally represses the precursor of miR-23a (pri-miR-23a) in cancer cell lines (53). To interrogate the causal relationship between cMYC activity and miR-23a expression, we activated naive pMel-1 CTLs in vitro in the presence of 10058-F4, a specific inhibitor that blocks MYC-MAX dimerization (65). In TCR-activated CTLs, cMYC inhibition increased mature miR-23a expression (Figure 5I), but only resulted in a partial rescue (~50%) of pri-miR-23a transcripts (Figure 5F). At least 2 explanations may account for this incomplete rescue: one possibility is that other inhibitory mechanisms, in parallel to cMYC, may be involved in suppressing pri-miR-23a transcription; alternatively, transcribed pri-miR-23a may have undergone active miRNA processing, which prevents the accumulation and detection of pri-miR-23a transcripts. With Dicer-deleted CTLs, in which miRNA biogenesis was largely blocked, pri-miR-23a transcripts were fully rescued to levels of their naive counterparts (Figure 5J), supporting the latter possibility. Taken together, these results suggested that cMYC is the major repressor of pri-miR-23a transcription in primed CTLs. Our findings thus identified cMYC as a key signaling node that integrates signals transduced through the TCR and TGF-β receptor to consequently govern miR-23a expression levels in effector CTLs. Interestingly, we noted that even with strong TCR signals, exposure of CTLs to TGF-β could effectively override TCR-induced cMYC activation (Figure 5G) and upregulate miR-23a (Figure 5B). Therefore, despite converging on the same signaling node, a TGF-β–enriched tumor microenvironment, but not tumor antigen–elicited TCR signaling, is the dominant regulator of miR-23a expression in CTLs.
Neutralizing miR-23a in CTLs mitigates TGF-β–induced immunosuppression.
The secretion of TGF-β by malignant tumor cells poses a key hurdle to effective CTL antitumor responses (66, 67). Having unveiled a novel regulatory pathway between TGF-β, miR-23a, and BLIMP-1 expression in CTLs, we hypothesized that this may be another mechanism by which TGF-β facilitates immune evasion; and that, by functionally neutralizing this TGF-β–induced accumulation of miR-23a, we can preserve CTL immunocompetence. To directly validate this hypothesis, we interrogated the resilience of CTLs to TGF-β challenge upon LNA- and decoy-mediated miR-23a functional blockade. In CTLs, IFN-γ and granzyme B are known to be suppressed by TGF-β (21), and are also cytotoxic effectors regulated by the miR-23a target BLIMP-1 (27, 28). Therefore, we used these 2 parameters as functional readouts of BLIMP-1 rescue and cytotoxicity. Consistent with an earlier report (21), TGF-β dramatically repressed the acquisition of granzyme B (Figure 6A) and IFN-γ (Supplemental Figure 7) by naive CTLs; however, treatment with the anti–miR-23a LNA partially rescued the expression of these CTL effector molecules, even at high concentrations of TGF-β (Figure 6A and Supplemental Figure 7). As ACT uses activated rather than naive T cells for tumor immunotherapy, we also sought to evaluate the functional competence of effector CTLs expressing the miR-23a decoy upon TGF-β exposure. In agreement with earlier reports (20, 21), TGF-β treatment blocked IFN-γ production in both mock- and miR-23a decoy–transduced CTLs (Figure 6B). Again, even after a 48-hour TGF-β conditioning, miR-23a decoy–transduced CTLs still maintained IFN-γ production at a level similar to TGF-β–untreated mock CTLs. This indicates that miR-23a elevation is a major post-transcriptional mechanism through which TGF-β blunts CTL cytotoxicity; importantly, inhibition of miR-23a in effector CTLs relieves TGF-β–mediated functional suppression.
Figure 6. Neutralizing miR-23a in CTLs mitigates TGF-β–induced immunosuppression.
(A) Purified naive pMel-1 CTLs were activated with anti-CD3/anti-CD28 and the indicated concentrations of TGF-β for 48 hours in vitro, in the presence or absence of 50 nM FAM-tagged anti–miR-23a LNA. The percentage of granzyme B–expressing TCRβ+CD8+FAM– and TCRβ+CD8+FAM+ cells was assessed by flow cytometry. Data shown represent mean ± SEM; n = 8. ***P < 0.001 by 2-way ANOVA and Bonferroni post-test. (B) Mock- and miR-23a decoy–transduced pMel-1 CTLs were cultured with IL-2 for the first 48 hours, then washed and treated with the indicated concentrations of TGF-β for the next 48 hours. CTLs were restimulated with anti-CD3/anti-CD28 (1 μg/ml each) for the final 24 hours, and the percentage of IFN-γ–producing CD8+iRFP+GFP+ cells was assessed by flow cytometry. Data shown represent mean ± SEM; n = 4. *P < 0.05 and **P < 0.01 by 2-way ANOVA and Bonferroni post-test.
miR-23a expression correlates inversely with antitumor potential of mouse and human TILs.
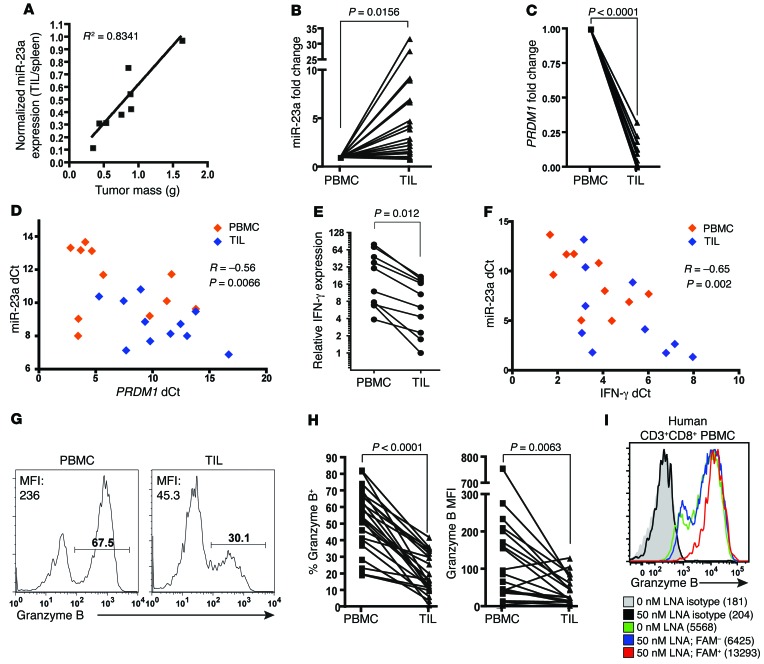
To understand the preclinical relevance of miR-23a expression in CTLs within the tumor microenvironment, we isolated TILs from pMel-1 mice bearing various burdens of B16/F10 melanoma. We found a strong positive correlation between tumor burden and miR-23a expression in TILs (Figure 7A).
Figure 7. miR-23a expression correlates inversely with antitumor potential of mouse and human TILs.
(A) miR-23a expression in TILs isolated from C57BL/6 mice bearing B16/F10 tumors. miR-23a expression in TILs was normalized to basal expression of splenic CTLs in each individual mouse. Linear regression was conducted and the R2 coefficient is shown. (B–I) CD3+CD8+ CTLs from the pleural aspirates (TIL) and peripheral blood (PBMC) of advanced lung cancer patients were assessed for miR-23a and CTL effector molecules. Relative (B) miR-23a (n = 23), (C) PRDM1 (n = 10), and (E) IFNG mRNA (n = 10) expression in patient TILs, expressed as a fold change compared with patient-matched CD3+CD8+ PBMCs. miR-23a was normalized to the average ΔCt of U6 and RNY3 endogenous controls, while mRNA was normalized to the average ΔCt of 18S RNA and RPLP0 endogenous controls (details in Methods). Scatterplots of miR-23a versus (D) PRDM1 (n = 11) or (F) IFNG mRNA (n = 10) expression in CD3+CD8+ T cells from lung cancer patients. The miR-23a ΔCt from each PBMC or TIL sample is plotted against its corresponding mRNA ΔCt. The Pearson correlation coefficients (R) and 2-tailed P values are shown. (G) Representative granzyme B histograms of CD3+CD8+ PBMCs and TILs. (H) Percentage granzyme B+ and granzyme B MFI from CD3+CD8+ PBMCs and TILs (n = 23). (I) Granzyme B expression in untreated (0 nM) or LNA-treated (50 nM) activated human CD3+CD8+ T cells from healthy donor PBMCs. Numbers indicate granzyme B MFI. Histogram is representative of n = 3 independent experiments. P values in B–D were determined by 2-tailed paired t test.
To assess whether miR-23a–mediated CTL suppression is associated with the clinical pathology of human cancers, we explored the relationship between miR-23a expression and TIL cytotoxic potential in a cohort of advanced lung cancer patients. In these patients, pleural effusion CTLs are a good reflection of TILs, as they have received similar conditioning in the tumor microenvironment (e.g., by IL-10 and TGF-β; ref. 68) and are known to be functionally reminiscent of TILs (69, 70). As a control for basal gene expression outside the tumor and to normalize for interindividual variations, we also isolated CTLs obtained from the peripheral blood (PBMCs) of each patient for paired-sample analysis. As compared with CTLs in the periphery, miR-23a in TILs was elevated by a mean of 5.45 ± 1.70–fold (Figure 7B). This upregulation of miR-23a corresponded with a downregulation of PRDM1 mRNA levels in TILs (Figure 7C); additionally, we found an inverse correlation between miR-23a and PRDM1 mRNA expression (Figure 7D). These observations reiterate our findings that miR-23a directly targets BLIMP-1 in CTLs (Figure 4). We also analyzed IFNG mRNA and granzyme B protein expression as more direct readouts of antitumor potential. In these patient samples, IFNG expression was significantly downregulated in TILs (Figure 7E), and correlated inversely with miR-23a levels (Figure 7F); moreover, granzyme B expression on a population level and on a per-cell basis (Figure 7, G and H) was sharply diminished in TILs. Additionally, human PBMCs treated with the anti–miR-23a LNA showed enhanced granzyme B expression (Figure 7I), indicating that functional blockade of miR-23a can boost the cytotoxicity of human CTLs. Taken together, these results demonstrated that miR-23a is a clinically relevant and translatable target for the immunotherapy of human cancers.
Adoptive transfer of miR-23a–inhibited CTLs robustly retards tumor progression.
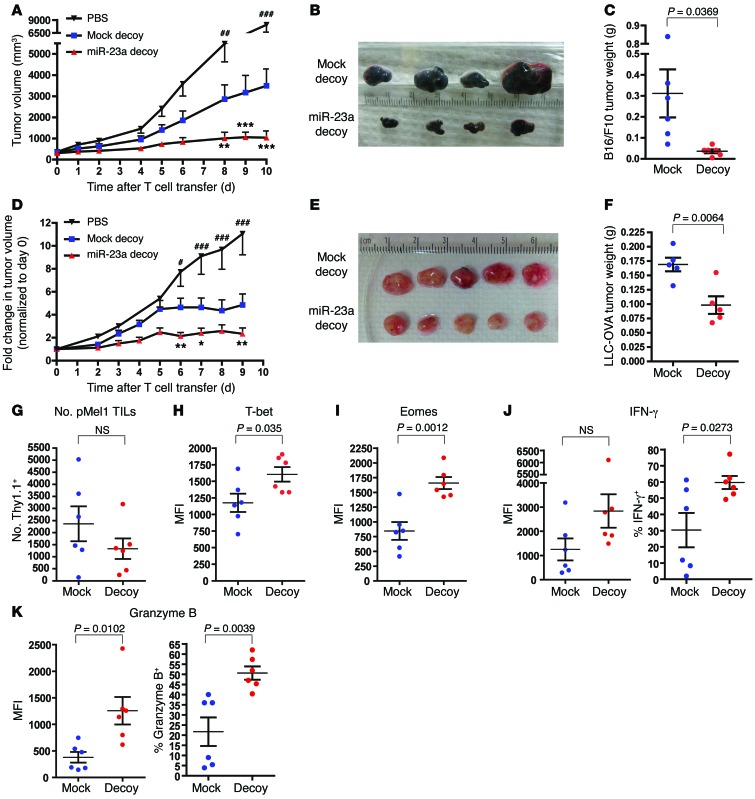
In view of the clinical relevance of miR-23a, we went on to examine the efficacy of our miR-23a targeted therapeutic strategy for cancer intervention. As a novel gene therapy tool, our bicistronic, dual-reporter retroviral construct (Figure 3D) poses several advantages for CTL programming: (a) it can be readily incorporated into conventional ACT, as ex vivo–expanded tumor-specific CTLs can be simultaneously transduced with the retrovirus; (b) the selectable marker enables successfully engineered and functionally robust CTLs to be selected/enriched for reinfusion; (c) GFP reporter activity allows the effectiveness of miR-23a inhibition to be conveniently monitored. With this tool, we mimic human cancer therapy by using 2 mouse models of established tumors: B16/F10 melanoma (Figure 8, A–C) and Lewis lung cancer overexpressing ovalbumin (LLC-OVA) (Figure 8, D–F). When tumor masses reached palpable growth, we sublethally irradiated the tumor-bearing mice, and injected either 0.2 × 106 miR-23a decoy–expressing pMel-1 or OT-I CTLs intratumorally. Compared with equal numbers of mock cells, treatment with miR-23a–inhibited CTLs dramatically retarded tumor progression (Figure 8, A and D), and significantly reduced tumor burdens (Figure 8, B, C, E, and F). Upon examining tumor pathology 10 days after CTL transfer, we found that although CTL persistence within the tumor mass was unaffected (Figure 8F), miR-23a–inhibited CTLs showed augmented expression of the transcription factors T-bet and EOMES (Figure 8, H and I), and the cytolytic molecules IFN-γ and granzyme B (Figure 8, J and K). Additionally, inhibiting granzyme B accelerated tumor progression in vivo, and completely abrogated the antitumor advantage conferred by the miR-23a decoy (Supplemental Figure 8). Therefore, augmented granzyme B expression afforded by miR-23a–inhibited CTLs was functionally essential for enhanced melanoma clearance. Taken together, these results indicated that suppressing miR-23a in CTLs enhances their cytotoxic potency within the tumor microenvironment, thereby attaining optimal tumor eradication.
Figure 8. Adoptive transfer therapy with miR-23a–inhibited CTLs in mouse models of established tumors.
Seven days after s.c. inoculation of 0.2 × 106 B16/F10 melanoma or LLC-OVA cells, C57BL/6 tumor-bearing mice were sublethally irradiated and left untreated (PBS), or treated with intratumoral injections of 0.2 × 106 sorted iRFP+GFP+ mock or miR-23a decoy–expressing pMel-1 or OT-I CTLs. Tumors were excised 10 days after T cell transfer, and mock and miR-23a decoy–expressing Thy1.1+ pMel-1 CTLs within the tumor masses were analyzed by flow cytometry. (A) B16/F10 and (D) LLC-OVA tumor progression after the initiation of CTL therapy. Data represent mean ± SEM, from n = 6 mice per group in 1 representative of 3 independent experiments in A, and from n = 10 mice per group in D. *P < 0.05, **P < 0.01, and ***P < 0.001 indicate mock vs. miR-23a decoy; #P < 0.05, ##P < 0.01, and ###P < 0.001 indicate mock vs. PBS by 2-way ANOVA and Bonferroni post-test. (B and C) B16/F10 and (E and F) LLC-OVA tumor sizes and weight 10 days after CTL therapy. (G) Absolute numbers of Thy1.1+ pMel-1 CTLs present in tumors. (H–K) Expression of the CTL master regulators and effector molecules (H) T-bet, (I) EOMES, (J) IFN-γ, and (K) granzyme B in Thy1.1+ pMel-1 CTLs isolated from B16/F10 tumors. Data represent mean ± SEM.
Discussion
CTL-based immunotherapy is a promising means of achieving durable control over tumor progression. However, its widespread use has been limited by the cost and effort in generating large numbers of antitumor CTLs ex vivo and by the incompetence induced by the tumor microenvironment. In this study, we attempted to redirect the focus of CTL engineering from amplifying the quantity to improving the quality of individual CTLs, which could help to overcome both limitations. We initially identified miR-23a as a hurdle to effector CTL responses by differential priming with B cells or mature DCs. Clearly, this in vitro priming system was not designed to recapitulate the complexities of CTL responses within the tumor microenvironment; however, further investigation into factors that control miR-23a expression in CTLs led to our discovery that miR-23a was in fact a target of the immune-subversive tumor microenvironment. That miR-23a inhibition imparts functional resilience to CTLs, particularly when challenged with immunosuppressive conditions, is supported by 2 pieces of evidence. Firstly, we observed that the enhancement in cytotoxicity provided by CTL-specific miR-23a blockade was consistently more profound in vivo (Figure 8, A and D) than in vitro (Figure 3G). We speculated that in vivo, the susceptibility of WT CTLs to TGF-β–induced suppression might have magnified the functional advantage of miR-23a–inhibited CTLs. Secondly, our in vitro TGF-β challenge experiments directly illustrated that inhibiting miR-23a in CTLs could, at least partially, preserve their immunocompetence in spite of high TGF-β concentrations (Figure 6, A and B).
Our mechanistic studies on miR-23a regulation uncovered a novel mechanism of TGF-β–induced immunosuppression on CTLs: the TGF-β/miR-23a/BLIMP-1 axis. The immunosuppressive effects of TGF-β on CTLs are well established (20, 21, 67). While earlier mechanistic studies focused on SMAD2/3 residing on the cis-elements within regulatory regions of the Ifng and Gzmb genes (21), our data revealed that even after SMAD activation, there still exists a pathway of rescue to preserve cytotoxicity (Figure 9). By targeting miR-23a, our in vitro and in vivo data demonstrated the robustness of this preservation, and we attribute the robustness of miR-23a–mediated suppression to the strength of its target, BLIMP-1. BLIMP-1 is an essential master regulator that turns on the cytotoxic transcriptional program in activated CTLs. Notably, BLIMP-1–deficient CTLs fail to differentiate into cytotoxic effectors, owing to their impaired expression of multiple cardinal cytotoxic molecules, including granzyme B and IFN-γ, as well as the transcription factor T-bet (27, 28). We showed, for the first time to our knowledge, that TGF-β can control BLIMP-1 expression through a miRNA-mediated post-transcriptional mechanism; moreover, abrogating miR-23a ameliorates TGF-β–induced CTL suppression, by rescuing the BLIMP-1 downstream targets granzyme B and IFN-γ (Figure 6, A and B). Our findings identify miR-23a as a TGF-β–responsive rheostat that fine-tunes BLIMP-1 levels in activated CTLs, and highlight the TGF-β/miR-23a/BLIMP-1 axis as a key post-transcriptional determinant controlling CTL cytotoxicity in an immunosuppressive environment. Therefore, TGF-β–mediated immunosuppression is supported by at least 2 pillars: direct SMAD-mediated transcriptional repression on effector molecules, and indirect miR-23a–mediated post-transcriptional controls on the BLIMP-1. Most importantly from the therapeutic point of view, taking down just 1 pillar by blocking miR-23a function is sufficient to maintain CTLs’ cytotoxicity machinery at an adequate level for tumor intervention.
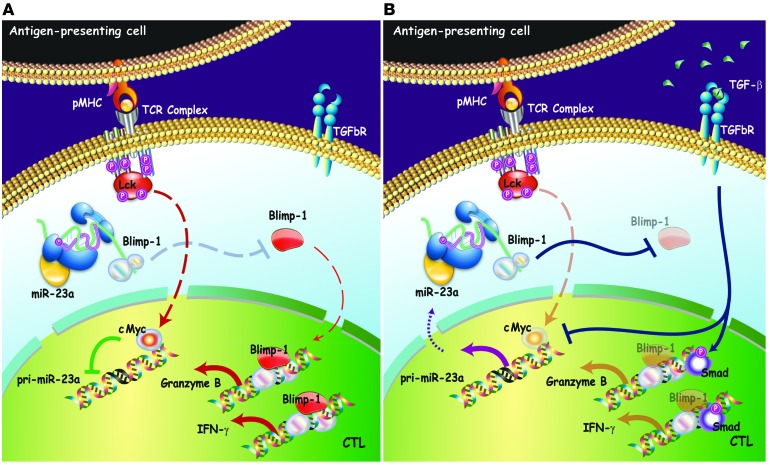
Figure 9. Model of CTL immune modulation by targeting miR-23a.
(A) Under immune-activating conditions, TCR signaling upregulates cMYC in CTLs. Transcriptional repression of pri-miR-23a by cMYC permits accumulation of BLIMP-1, resulting in increased expression of its cytotoxic target genes, granzyme B, and IFN-γ. (B) In the tumor microenvironment, TGF-β suppresses CTL activity via 2 mechanisms — SMAD-mediated transcriptional reprogramming and miRNA-mediated post-transcriptional control. In the former, TGF-β–induced SMADs are recruited to the Gzmb and Ifng gene regulatory regions to repress the transcription of these cytotoxic mediators directly. In the latter, TGF-β antagonizes cMYC activity, thereby derepressing pri-miR-23a transcription. Elevated miR-23a levels in CTLs downregulate BLIMP-1, and consequently its downstream cytotoxic effectors.
Within the tumor microenvironment, TGF-β is a key mediator of tumor immune evasion. Blocking TGF-β signaling in CTLs — by TGF-β neutralization or enforced expression of the dominant-negative TGF-βRII — can reverse their immune-tolerant state to promote tumor regression in vivo (21, 67, 71, 72), making TGF-β and molecules in the TGFBR-mediated signaling pathway druggable targets for tumor therapy (73). However, current preclinical and clinical data indicate that, because of its profound impact on immunosuppression and a wide range of physiological functions, systemic administration of anti–TGF-β reagents can cause severe inflammatory damage and other adverse off-target pathologies (74). By contrast, during the process of ex vivo expansion, ACT provides a window of opportunity to program tumor-specific CTLs with immunocompetence against TGF-β suppression. Notably, this reprogramming is restricted specifically to CTLs before reinfusion. Our findings highlight miR-23a as a clinically relevant target for this purpose, whose functional blockade presents two significant advantages for ACT: it not only augments the cytotoxic potency of tumor-specific CTLs, but also mitigates TGF-β–induced immunosuppression.
Methods
Mice.
pMel-1 mice carrying a transgenic TCR specific for the B16 melanoma antigen gp100 (C57BL/6-Tg(TcraTcrb)8Rest/J) and OT-I mice (C57BL/6-Tg(TcraTcrb)1100Mjb/J) were purchased from The Jackson Laboratory. All mice were housed under pathogen-free conditions, and used between 6 and 10 weeks of age for experimental procedures.
Cell culture.
T cells and EL4 thymoma cells were cultured in RPMI 1640 media supplemented with 10% FBS, 100 U/ml penicillin, 100 U/ml streptomycin, 2 mM l-glutamine, 1 mM sodium pyruvate, 0.1 mM nonessential amino acids, and 50 μM 2-mercaptoethanol (henceforth referred to as complete RPMI) in a humidified 37°C incubator with 7% CO2.
Bone marrow–derived DCs were generated as previously described (75). Briefly, total bone marrow cells were harvested from the femurs of C57BL/6 mice, and cultured in complete RPMI supplemented with 1:30 J558L conditioned media. The J558L cell line, a gift from David Baltimore (California Institute of Technology, Pasadena, CA), had been stably transfected with the murine GM-CSF cDNA, and its cell culture supernatant was used as a source of GM-CSF (76). From day 3 to day 9, cells were fed every other day. Nonadherent cells were collected on day 9 and treated with 1 μg/ml LPS (Sigma-Aldrich) overnight. On day 10, nonadherent mature DCs were collected, and subjected to density gradient centrifugation over Histopaque (Sigma-Aldrich). The viable mature DCs isolated were washed 3 times in complete RPMI before coculture with naive T cells.
Naive T cells were isolated from lymph nodes and/or spleens using the Dynal mouse CD8 negative isolation kit (Invitrogen) according to the manufacturer’s instructions. For T cell priming by APCs, naive pMel-1 CD8+ T cells were cocultured with either mature DCs or sorted immature B220+ splenic B cells at a 1:1 ratio, in the presence of 5 μM hgp10025–33 peptide. For T cell activation by antibodies, naive pMel-1 CD8+ T cells were seeded onto plate-bound anti-CD3 and anti-CD28 antibodies (5 μg/ml each, unless otherwise indicated; Biolegend).
miRNA expression profiling and miRNA quantitative PCR.
Naive pMel-1 CTLs were primed by mature DCs or splenic B cells in vitro, as described above. After 3 days of priming, TCRβ+ pMel-1 CTLs were sorted and lysed using the RNAqueous Micro-kit (for samples containing 105 to 0.5 × 106 cells) or the mirVana miRNA Isolation kit (for samples containing ≥ 0.5 × 106 cells) (both from Ambion) according to the manufacturer’s instructions. To quantify the expression of mature miRNA expression, E. coli poly A polymerase (Epicentre) was first used to generate polyadenylated tails at the 3′-end of all RNA molecules. After annealing oligo-dT primers, cDNA was synthesized using the qScript Flex cDNA synthesis kit (Quanta Biosciences) according to the manufacturer’s instructions for gene-specific priming, with 1 modification: a universal tag that would extend from the 3′-end of cDNA molecules was added during reverse transcription. With the addition of this universal tag, individual miRNAs were detected with miRNA-specific forward primers and a reverse universal primer mix. A SYBR Green–based real-time PCR method was used to quantify the relative expression of mature miRNAs. In the miRNA expression profiling array, a total of 355 mature miRNAs were evaluated in DC- and B cell–primed CTLs (n = 3 independent experiments). miRNA expression was normalized by geometric mean–based global normalization using the Realtime StatMiner (Integromics) analysis software. Differential miRNA expression was determined by paired t test, with significance level set at 0.05. The complete set of miRNA expression profiling data is available on the NCBI Gene Expression Omnibus database under the accession number GSE60884.
mRNA and pri-miRNA quantitative PCR.
Total RNA from cells was isolated using the RNAqueous Micro-kit (for samples containing 105 to 0.5 × 106 cells) or the mirVana miRNA Isolation kit (for samples containing ≥ 0.5 × 106 cells) (both from Ambion) according to the manufacturer’s instructions. cDNA was reverse-transcribed from total RNA using a mixed priming strategy (oligo-dT and random primers) with the qScript Flex cDNA synthesis kit (Quanta Biosciences) according to the manufacturer’s instructions. A SYBR Green–based real-time PCR method was used to quantify the relative expression of mRNAs and pri-miRNAs.
miR-23a decoy construct design and retroviral transduction.
The miR-23a decoy vector is a bicistronic retroviral backbone that encodes 2 independent expression cassettes: the 5′-LTR drives the expression of the selectable marker (either iRFP, ref. 50, or puromycin resistance), whereas the PGK promoter drives the expression of a GFP decoy/reporter. The GFP reporter contains an insertion of 8 tandem miR-23a binding sites (GGAAATCCCTGcgAATGTGATcgttGGAAATCCCTccCAATGTGATactcGGAAATCCCacGCAATGTGATgtacGGAAATCCCaccCAATGTGATccga GGAAATCCCTccgAATGTGATacgcGGAAATCCCTccCAATGTGATcctaGGAAATCCCaccCAATGTGATagctGGAAATCCgacGCAATGTGAT) in its 3′-UTR to monitor the sponge effect of decoy targeting sites. Between these 2 cassettes, we additionally inserted an insulator sequence comprising 2 tandem repeats of the chicken β-globin FII/FIIIΔspacer insulator fragment (FII/FIIIΔspacer: aggcgcgcccccagggatgtaattacgtccctcccccgctagggggccggccagcaccggtccggcgctccccccgcatccccgagccggggcgcgcct) (51) to maximize their independent expression. As a control, a mock decoy vector lacking miR-23a binding sites in its GFP 3′-UTR was also generated.
On day 0, cells from the lymph nodes of pMel-1 and OT-I mice were harvested and seeded into 24-well plates. Non-T cells in the lymph nodes served as APCs, and CTLs were primed in vitro by the addition of 5 μM hgp10025–33 or OVA257–264, respectively. On day 1, 50 U/ml murine IL-2 was added. Six hours later, cells were spin-infected with retroviral supernatants at 1,250 g for 90 minutes at 37°C. CTLs from days 4–6 were used in experiments.
Lymphocyte isolation and miR-23a quantification from lung cancer patients.
Pleural effusion samples were collected from newly diagnosed lung cancer patients with malignant pleural effusion (MPE). Patients included in this study neither underwent any invasive procedures directed into the pleural cavity, nor suffered chest trauma within the 3 months prior to hospitalization. At the time of sample collection, none of the patients had received any anticancer therapy, corticosteroids, or other NSAIDs. Pleural fluid samples were collected in heparin-treated tubes from each subject, using a standard thoracocentesis technique. Twenty milliliters of peripheral blood was drawn simultaneously. MPE and peripheral blood lymphocytes were isolated by density centrifugation using human lymphocyte separation medium (TBD) according to the manufacturer’s instructions. Total RNA extraction, as well as miRNA and mRNA quantitative PCR, was performed as described above. For candidate endogenous controls, hsa-RNY3, hsa-U6, and hsa-U1 were included for miRNA quantitative PCR, while 18S RNA, RPLP0, and RPL13A were included for mRNA quantitative PCR. Using the Realtime StatMiner (Integromics) analysis software, geNorm analysis was performed, and the mean Ct values of RNY3 and U6 were chosen as internal controls for miRNA Ct normalization, while the mean Ct values of 18S RNA and RPLP0 were chosen as internal controls for mRNA Ct normalization. For each patient, the ΔΔCt of miR-23a and PRDM1 was then calculated from the difference between ΔCt values in TIL and PBMC samples, and transformed into a fold change. The relative expression of miR-23a and PRDM1 in PBMCs of each patient was arbitrarily set to 1.0. The ΔΔCt of IFNG mRNA was calculated from the difference between the ΔCt values of each sample and the TIL sample with the lowest ΔCt value, and transformed into a fold change.
Protein quantification by flow cytometry and Western blot.
For cytokine staining, mouse pMel-1 CD8+ T cells and human lymphocytes were restimulated for 4 hours with 0.9 nM PDBu (Sigma-Aldrich) and 0.5 μg/ml ionomycin (Sigma-Aldrich), in the presence of 5 μg/ml brefeldin A (eBioscience) and 2 μM monensin A (eBioscience). Viable cells were stained using the Live/Dead Violet viability kit (Invitrogen) according to the manufacturer’s instructions before intracellular staining. Intracellular staining for cytokines, granzyme B, and transcription factors was performed by fixing of cells in 2% paraformaldehyde, followed by membrane permeabilization in 0.1% saponin. Fc receptors were blocked before incubation with the following staining antibodies: anti–T-bet–PE, anti–granzyme B–Alexa Fluor 647, and anti–IFN-γ–PE (all from Biolegend); and anti-EOMES-Alexa Fluor 647 and anti–granzyme B–PE (both from eBioscience). Samples were acquired on the FACSCanto II flow cytometer (BD), and data were analyzed using FlowJo software.
For Western blot analysis of protein expression, CTLs cultured for the indicated times were lysed in radioimmunoprecipitation assay buffer containing protease inhibitor (Roche) and phosphatase inhibitor cocktails 1 and 2 (Sigma-Aldrich). Samples were run on 10% polyacrylamide gels (BioRad) and transferred onto PVDF membranes. Proteins of interest were probed with the following primary antibodies: mouse anti–BLIMP-1 (Biolegend), rabbit anti-pSMAD2, rabbit anti-SMAD2/3 (both from Cell Signaling), and goat anti–β-actin (Sigma-Aldrich). Anti-mouse Alexa Fluor 680, anti-rabbit Alexa Fluor 680, and anti-goat Alexa Fluor 680 (all from Invitrogen) were used as secondary antibodies. Fluorescence intensity was measured on an Odyssey imaging system (LI-COR Biosciences).
In vitro cytotoxicity assays.
EL4 cells were pulsed with 10 μM hgp10025–33 or 10 μM OVA257–264 overnight. Peptide-pulsed and unpulsed EL4 cells were labeled with 5 μM and 0.5 μM Cell Tracker Orange (Invitrogen), respectively, and mixed in a 1:1 ratio before coculture with CTLs. After 6 days of in vitro culture, viable pMel-1 CTLs were isolated by density gradient centrifugation over Histopaque (Sigma-Aldrich), and cocultured with labeled EL4 target cells at an E/T ratio of 5:1 for 6 hours in a humidified 37°C incubator. iRFP+ and/or GFP+ OT-I CTLs expressing the MSCV-iRFP-2Xins-mG-mock or the MSCV-iRFP-2Xins-mG-miR-23a decoy vector were sorted 48 hours after retroviral transduction, and cocultured with EL4 target cells at the indicated E/T ratios for 6 hours in a humidified 37°C incubator. After 6 hours of coculture, samples were harvested and stained with the Live/Dead Violet viability kit (Invitrogen) and anti-CD8α-FITC (Biolegend). CountBright Absolute counting beads (Invitrogen) were added to samples before acquisition on the FACSCanto II flow cytometer (BD), and data were analyzed using FlowJo software.
In vivo tumor models.
The B16/F10 melanoma and LLC-OVA lung cancer cell lines were gifts from Thomas Tedder (Duke University) and Eckhard Podack (University of Miami, Miami, Florida, USA), respectively. Tumor cells were harvested by trypsinization, and cell viabilities greater than 95% were confirmed by trypan blue exclusion. To study the in vivo antitumor effects of miR-23a–overexpressing pMel-1 CTLs, 0.2 × 106 B16/F10 cells in 200 μl PBS were inoculated s.c. into the shaved right lateral flanks of C57BL/6 recipient mice on day –3. Three days after tumor inoculation, each recipient mouse received an i.v. adoptive transfer of either 0.6 × 106 sorted GFP+7AAD– mock or miR-23a–overexpressing pMel-1 CTLs in 200 μl PBS on day 0. Control mice not treated with CTLs received i.v. injections of 200 μl PBS alone. To study the in vivo therapeutic potential of miR-23a–inhibited pMel-1 and OT-I CTLs, 1 × 106 LLC-OVA cells in 200 μl PBS were inoculated s.c. into the shaved right lateral flanks of C57BL/6 recipient mice on day –7. On days 0 and 5, each recipient mouse received 2 intratumoral injections of either 0.2 × 106 sorted mock or miR-23a decoy–expressing CTLs in 50 μl PBS. Tumor progression was monitored closely, and tumor volumes were calculated using the equation V = 4π (L1 × L2)/3, where V = volume (mm3), L1 = longest radius (mm), and L2 = shortest radius (mm). Mice were sacrificed at the experimental end points and their spleens, draining lymph nodes, and tumors harvested. Tumors were digested using the Papain Dissociation System (Worthington Biochemical) to liberate tumor-infiltrating cells. Effector functions of the transferred pMel-1 or OT-I CTLs were then analyzed by flow cytometry. For granzyme B inhibition studies, transduced pMel-1 CTLs were pretreated with 12.5 μM of the granzyme B inhibitor zAAD-CMK (Enzo Life Sciences) or DMSO vehicle control for 48 hours in vitro, before intratumoral injection. Three days after the transfer of granzyme B–inhibited CTLs, an additional 10 μg zAAD-CMK or DMSO (both solubilized in PBS) was intratumorally administered to sustain granzyme B inhibition in vivo.
Target prediction and luciferase reporter assays.
Candidate targets of miR-23a were derived from the integrated miRNA target prediction resource miRecords (http://mirecords.biolead.org/). The full-length 3′-UTRs of mouse Prdm1, Eomes, and Tbet were amplified from a 3′-RACE-ready cDNA library generated from total mouse T cell RNA, and cloned into the pmirGLO dual-luciferase vector (Promega) downstream of firefly luciferase. Each dual-luciferase reporter vector, together with a mock or miR-23a overexpression vector, was cotransfected into Jurkat T cells using the Amaxa Cell Line Nucleofactor kit (Lonza). Forty-eight hours after transfection, cells were lysed and luciferase reporter activities were determined in a dual-luciferase reporter assay (Promega).
Statistics.
Two-tailed unpaired or paired Student’s t tests were applied for the comparison of 2 means. For multiple comparisons, 1-way or 2-way ANOVA with Bonferroni post-test was performed as indicated. To assess the correlation between miR-23a and mRNA expression in CD8+ T cell samples from lung cancer patients, the Pearson’s correlation coefficient was calculated. P values less than 0.05 were considered statistically significant.
Study approval.
The human study protocol was approved by the Institutional Review Board for human studies of Xinqiao Hospital (Third Military Medical University, Chongqing, China), and written informed consent was obtained from all subjects. All animal studies were performed in accordance with guidelines and protocols approved by the Institutional Animal Care and Use Committee of Duke University.
Supplementary Material
Acknowledgments
We thank David Baltimore (California Institute of Technology), Thomas Tedder (Duke University), and Eckhard Podack (University of Miami) for their kind provision of cell lines used in the study. We are grateful to Feng Feng (Boston University) for his assistance in statistical analyses for miRNA profiling. We thank Peter J.R. Ebert for critical suggestions and reading of the manuscript. J.H. Sampson is supported by the NIH (R01-NS085412, R01-CA177476, P01-CA154291, and P50-CA108786); B. Zhu is supported by the National Natural Science Foundation of China (81222031) and by the National Key Basic Research Program of China (2012CB526603); Q.-J. Li is a Whitehead Family Foundation Scholar and is supported by grants from the American Cancer Society (RSG-10-157-01-LIB) and the National Institute of Allergy and Infectious Diseases (R01-AI091878).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2014;124(12):5352–5367. doi:10.1172/JCI76561.
References
- 1.Rosenberg SA. Progress in human tumour immunology and immunotherapy. Nature. 2001;411(6835):380–384. doi: 10.1038/35077246. [DOI] [PubMed] [Google Scholar]
- 2.Rosenberg SA, Restifo NP, Yang JC, Morgan RA, Dudley ME. Adoptive cell transfer: a clinical path to effective cancer immunotherapy. Nat Rev Cancer. 2008;8(4):299–308. doi: 10.1038/nrc2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park TS, Rosenberg SA, Morgan RA. Treating cancer with genetically engineered T cells. Trends Biotechnol. 2011;29(11):550–557. doi: 10.1016/j.tibtech.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chhabra A. TCR-engineered, customized, antitumor T cells for cancer immunotherapy: advantages and limitations. ScientificWorldJournal. 2011;11:121–129. doi: 10.1100/tsw.2011.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosenberg SA, et al. Durable complete responses in heavily pretreated patients with metastatic melanoma using T-cell transfer immunotherapy. Clin Cancer Res. 2011;17(13):4550–4557. doi: 10.1158/1078-0432.CCR-11-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robbins PF, et al. Tumor regression in patients with metastatic synovial cell sarcoma and melanoma using genetically engineered lymphocytes reactive with NY-ESO-1. J Clin Oncol. 2011;29(7):917–924. doi: 10.1200/JCO.2010.32.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pilon-Thomas S, et al. Efficacy of adoptive cell transfer of tumor-infiltrating lymphocytes after lymphopenia induction for metastatic melanoma. J Immunother. 2012;35(8):615–620. doi: 10.1097/CJI.0b013e31826e8f5f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zou W. Immunosuppressive networks in the tumour environment and their therapeutic relevance. Nat Rev Cancer. 2005;5(4):263–274. doi: 10.1038/nrc1586. [DOI] [PubMed] [Google Scholar]
- 9.Fourcade J, et al. Upregulation of Tim-3 and PD-1 expression is associated with tumor antigen-specific CD8+ T cell dysfunction in melanoma patients. J Exp Med. 2010;207(10):2175–2186. doi: 10.1084/jem.20100637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prado-Garcia H, Romero-Garcia S, Aguilar-Cazares D, Meneses-Flores M, Lopez-Gonzalez JS. Tumor-induced CD8+ T-cell dysfunction in lung cancer patients. Clin Dev Immunol. 2012;2012:741741. doi: 10.1155/2012/741741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woo SR, et al. Immune inhibitory molecules LAG-3 and PD-1 synergistically regulate T-cell function to promote tumoral immune escape. Cancer Res. 2012;72(4):917–927. doi: 10.1158/0008-5472.CAN-11-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Derynck R, et al. Human transforming growth factor-β complementary DNA sequence and expression in normal and transformed cells. Nature. 1985;316(6030):701–705. doi: 10.1038/316701a0. [DOI] [PubMed] [Google Scholar]
- 13.Bennicelli JL, Guerry Dt Production of multiple cytokines by cultured human melanomas. Exp Dermatol. 1993;2(4):186–190. doi: 10.1111/j.1600-0625.1993.tb00031.x. [DOI] [PubMed] [Google Scholar]
- 14.De Jaeger K, Seppenwoolde Y, Kampinga HH, Boersma LJ, Belderbos JS, Lebesque JV. Significance of plasma transforming growth factor-beta levels in radiotherapy for non-small-cell lung cancer. Int J Radiat Oncol Biol Phys. 2004;58(5):1378–1387. doi: 10.1016/j.ijrobp.2003.09.078. [DOI] [PubMed] [Google Scholar]
- 15.Polak ME, et al. Mechanisms of local immunosuppression in cutaneous melanoma. Br J Cancer. 2007;96(12):1879–1887. doi: 10.1038/sj.bjc.6603763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krasagakis K, Tholke D, Farthmann B, Eberle J, Mansmann U, Orfanos CE. Elevated plasma levels of transforming growth factor (TGF)-β1 and TGF-β2 in patients with disseminated malignant melanoma. Br J Cancer. 1998;77(9):1492–1494. doi: 10.1038/bjc.1998.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kong F, Jirtle RL, Huang DH, Clough RW, Anscher MS. Plasma transforming growth factor-β1 level before radiotherapy correlates with long term outcome of patients with lung carcinoma. Cancer. 1999;86(9):1712–1719. [PubMed] [Google Scholar]
- 18.Zhao L, et al. Changes of circulating transforming growth factor-beta1 level during radiation therapy are correlated with the prognosis of locally advanced non-small cell lung cancer. J Thorac Oncol. 2010;5(4):521–525. doi: 10.1097/JTO.0b013e3181cbf761. [DOI] [PubMed] [Google Scholar]
- 19.Van Belle P, Rodeck U, Nuamah I, Halpern AC, Elder DE. Melanoma-associated expression of transforming growth factor-β isoforms. Am J Pathol. 1996;148(6):1887–1894. [PMC free article] [PubMed] [Google Scholar]
- 20.Ahmadzadeh M. TGF-β1 attenuates the acquisition and expression of effector function by tumor antigen-specific human memory CD8 T cells. J Immunol. 2005;174(9):5215–5223. doi: 10.4049/jimmunol.174.9.5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomas DA, Massague J. TGF-β directly targets cytotoxic T cell functions during tumor evasion of immune surveillance. Cancer Cell. 2005;8(5):369–380. doi: 10.1016/j.ccr.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 22.Sullivan BM, Juedes A, Szabo SJ, von Herrath M, Glimcher LH. Antigen-driven effector CD8 T cell function regulated by T-bet. Proc Natl Acad Sci U S A. 2003;100(26):15818–15823. doi: 10.1073/pnas.2636938100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pearce EL, et al. Control of effector CD8+ T cell function by the transcription factor Eomesodermin. Science. 2003;302(5647):1041–1043. doi: 10.1126/science.1090148. [DOI] [PubMed] [Google Scholar]
- 24.Intlekofer AM, et al. Anomalous type 17 response to viral infection by CD8+ T cells lacking T-bet and eomesodermin. Science. 2008;321(5887):408–411. doi: 10.1126/science.1159806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cruz-Guilloty F, et al. Runx3 and T-box proteins cooperate to establish the transcriptional program of effector CTLs. J Exp Med. 2009;206(1):51–59. doi: 10.1084/jem.20081242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu Y, et al. T-bet and eomesodermin are required for T cell-mediated antitumor immune responses. J Immunol. 2010;185(6):3174–3183. doi: 10.4049/jimmunol.1000749. [DOI] [PubMed] [Google Scholar]
- 27.Kallies A, Xin A, Belz GT, Nutt SL. Blimp-1 transcription factor is required for the differentiation of effector CD8(+) T cells and memory responses. Immunity. 2009;31(2):283–295. doi: 10.1016/j.immuni.2009.06.021. [DOI] [PubMed] [Google Scholar]
- 28.Rutishauser RL, et al. Transcriptional repressor Blimp-1 promotes CD8(+) T cell terminal differentiation and represses the acquisition of central memory T cell properties. Immunity. 2009;31(2):296–308. doi: 10.1016/j.immuni.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shin H, et al. A role for the transcriptional repressor Blimp-1 in CD8(+) T cell exhaustion during chronic viral infection. Immunity. 2009;31(2):309–320. doi: 10.1016/j.immuni.2009.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carrington JC, Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301(5631):336–338. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- 31.Li QJ, et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129(1):147–161. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 32.Rodriguez A, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316(5824):608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiao C, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9(4):405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ebert PJ, Jiang S, Xie J, Li QJ, Davis MM. An endogenous positively selecting peptide enhances mature T cell responses and becomes an autoantigen in the absence of microRNA miR-181a. Nat Immunol. 2009;10(11):1162–1169. doi: 10.1038/ni.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang S, et al. Molecular dissection of the miR-17-92 cluster’s critical dual roles in promoting Th1 responses and preventing inducible Treg differentiation. Blood. 2011;118(20):5487–5497. doi: 10.1182/blood-2011-05-355644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krutzfeldt J, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438(7068):685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 37.Thum T, et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456(7224):980–984. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- 38.Kota J, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137(6):1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Obad S, et al. Silencing of microRNA families by seed-targeting tiny LNAs. Nat Genet. 2011;43(4):371–378. doi: 10.1038/ng.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Overwijk WW, et al. Tumor regression and autoimmunity after reversal of a functionally tolerant state of self-reactive CD8+ T cells. J Exp Med. 2003;198(4):569–580. doi: 10.1084/jem.20030590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fuchs EJ, Matzinger P. B cells turn off virgin but not memory T cells. Science. 1992;258(5085):1156–1159. doi: 10.1126/science.1439825. [DOI] [PubMed] [Google Scholar]
- 42.Castiglioni P, Gerloni M, Cortez-Gonzalez X, Zanetti M. CD8 T cell priming by B lymphocytes is CD4 help dependent. Eur J Immunol. 2005;35(5):1360–1370. doi: 10.1002/eji.200425530. [DOI] [PubMed] [Google Scholar]
- 43.Zeng R, et al. Synergy of IL-21 and IL-15 in regulating CD8+ T cell expansion and function. J Exp Med. 2005;201(1):139–148. doi: 10.1084/jem.20041057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Novy P, Huang X, Leonard WJ, Yang Y. Intrinsic IL-21 signaling is critical for CD8 T cell survival and memory formation in response to vaccinia viral infection. J Immunol. 2011;186(5):2729–2738. doi: 10.4049/jimmunol.1003009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang ZN, et al. Transcriptomic analysis of peripheral blood mononuclear cells in rapid progressors in early HIV infection identifies a signature closely correlated with disease progression. Clin Chem. 2013;59(8):1175–1186. doi: 10.1373/clinchem.2012.197335. [DOI] [PubMed] [Google Scholar]
- 47.Klebanoff CA, et al. Determinants of successful CD8+ T-cell adoptive immunotherapy for large established tumors in mice. Clin Cancer Res. 2011;17(16):5343–5352. doi: 10.1158/1078-0432.CCR-11-0503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kerkar SP, et al. Genetic engineering of murine CD8+ and CD4+ T cells for preclinical adoptive immunotherapy studies. J Immunother. 2011;34(4):343–352. doi: 10.1097/CJI.0b013e3182187600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4(9):721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Filonov GS, Piatkevich KD, Ting LM, Zhang J, Kim K, Verkhusha VV. Bright and stable near-infrared fluorescent protein for in vivo imaging. Nat Biotechnol. 2011;29(8):757–761. doi: 10.1038/nbt.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98(3):387–396. doi: 10.1016/S0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- 52.Wang R, et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 2011;35(6):871–882. doi: 10.1016/j.immuni.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gao P, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458(7239):762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xiao F, Zuo Z, Cai G, Kang S, Gao X, Li T. miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res. 2009;37(Database issue):D105–D110. doi: 10.1093/nar/gkn851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Curiel TJ, et al. Blockade of B7-H1 improves myeloid dendritic cell-mediated antitumor immunity. Nat Med. 2003;9(5):562–567. doi: 10.1038/nm863. [DOI] [PubMed] [Google Scholar]
- 56.Blankenstein T, Coulie PG, Gilboa E, Jaffee EM. The determinants of tumour immunogenicity. Nat Rev Cancer. 2012;12(4):307–313. doi: 10.1038/nrc3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gilboa E. The promise of cancer vaccines. Nat Rev Cancer. 2004;4(5):401–411. doi: 10.1038/nrc1359. [DOI] [PubMed] [Google Scholar]
- 58.Cho OH, et al. Notch regulates cytolytic effector function in CD8+ T cells. J Immunol. 2009;182(6):3380–3389. doi: 10.4049/jimmunol.0802598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sugimoto K, et al. Notch2 signaling is required for potent antitumor immunity in vivo. J Immunol. 2010;184(9):4673–4678. doi: 10.4049/jimmunol.0903661. [DOI] [PubMed] [Google Scholar]
- 60.Yoon SO, Zhang X, Berner P, Blom B, Choi YS. Notch ligands expressed by follicular dendritic cells protect germinal center B cells from apoptosis. J Immunol. 2009;183(1):352–358. doi: 10.4049/jimmunol.0803183. [DOI] [PubMed] [Google Scholar]
- 61.Ohishi K, Katayama N, Shiku H, Varnum-Finney B, Bernstein ID. Notch signalling in hematopoiesis. Semin Cell Dev Biol. 2003;14(2):143–150. doi: 10.1016/S1084-9521(02)00183-0. [DOI] [PubMed] [Google Scholar]
- 62.Bandukwala HS, et al. Selective inhibition of CD4+ T-cell cytokine production and autoimmunity by BET protein and c-Myc inhibitors. Proc Natl Acad Sci U S A. 2012;109(36):14532–14537. doi: 10.1073/pnas.1212264109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pietenpol JA, et al. TGF-β1 inhibition of c-myc transcription and growth in keratinocytes is abrogated by viral transforming proteins with pRB binding domains. Cell. 1990;61(5):777–785. doi: 10.1016/0092-8674(90)90188-K. [DOI] [PubMed] [Google Scholar]
- 64.Genestier L, Kasibhatla S, Brunner T, Green DR. Transforming growth factor β1 inhibits Fas ligand expression and subsequent activation-induced cell death in T cells via downregulation of c-Myc. J Exp Med. 1999;189(2):231–239. doi: 10.1084/jem.189.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yin X, Giap C, Lazo JS, Prochownik EV. Low molecular weight inhibitors of Myc-Max interaction and function. Oncogene. 2003;22(40):6151–6159. doi: 10.1038/sj.onc.1206641. [DOI] [PubMed] [Google Scholar]
- 66.Penafuerte C, Galipeau J. TGF β secreted by B16 melanoma antagonizes cancer gene immunotherapy bystander effect. Cancer Immunol Immunother. 2008;57(8):1197–1206. doi: 10.1007/s00262-008-0453-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang L, et al. Inhibition of TGF-β signaling in genetically engineered tumor antigen-reactive T cells significantly enhances tumor treatment efficacy. Gene Ther. 2013;20(5):575–580. doi: 10.1038/gt.2012.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sikora JJ, Dworacki GT, Kaczmarek MT, Jenek RE, Zeromski JO. Immunosuppressive mechanisms in the microenvironment of malignant pleural effusions. Cancer Detect Prev. 2004;28(5):325–330. doi: 10.1016/j.cdp.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 69.Atanackovic D, Block A, de Weerth A, Faltz C, Hossfeld DK, Hegewisch-Becker S. Characterization of effusion-infiltrating T cells: benign versus malignant effusions. Clin Cancer Res. 2004;10(8):2600–2608. doi: 10.1158/1078-0432.CCR-03-0239. [DOI] [PubMed] [Google Scholar]
- 70.Prado-Garcia H, Aguilar-Cazares D, Flores-Vergara H, Mandoki JJ, Lopez-Gonzalez JS. Effector, memory and naive CD8+ T cells in peripheral blood and pleural effusion from lung adenocarcinoma patients. Lung Cancer. 2005;47(3):361–371. doi: 10.1016/j.lungcan.2004.07.046. [DOI] [PubMed] [Google Scholar]
- 71.Terabe M, et al. Synergistic enhancement of CD8+ T cell-mediated tumor vaccine efficacy by an anti-transforming growth factor-beta monoclonal antibody. Clin Cancer Res. 2009;15(21):6560–6569. doi: 10.1158/1078-0432.CCR-09-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ueda R, et al. Systemic inhibition of transforming growth factor-β in glioma-bearing mice improves the therapeutic efficacy of glioma-associated antigen peptide vaccines. Clin Cancer Res. 2009;15(21):6551–6559. doi: 10.1158/1078-0432.CCR-09-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Connolly EC, Freimuth J, Akhurst RJ. Complexities of TGF-β targeted cancer therapy. Int J Biol Sci. 2012;8(7):964–978. doi: 10.7150/ijbs.4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lonning S, Mannick J, McPherson JM. Antibody targeting of TGF-β in cancer patients. Curr Pharm Biotechnol. 2011;12(12):2176–2189. doi: 10.2174/138920111798808392. [DOI] [PubMed] [Google Scholar]
- 75.Yang L, Baltimore D. Long-term in vivo provision of antigen-specific T cell immunity by programming hematopoietic stem cells. Proc Natl Acad Sci U S A. 2005;102(12):4518–4523. doi: 10.1073/pnas.0500600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zal T, Volkmann A, Stockinger B. Mechanisms of tolerance induction in major histocompatibility complex class II-restricted T cells specific for a blood-borne self-antigen. J Exp Med. 1994;180(6):2089–2099. doi: 10.1084/jem.180.6.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.