Abstract
Autophagy allows cells to adapt to changes in their environment by coordinating the degradation and recycling of cellular components and organelles to maintain homeostasis. Lysosomes are organelles critical for terminating autophagy via their fusion with mature autophagosomes to generate autolysosomes that degrade autophagic materials; therefore, maintenance of the lysosomal population is essential for autophagy-dependent cellular clearance. Here, we have demonstrated that the two most common autosomal recessive hereditary spastic paraplegia gene products, the SPG15 protein spastizin and the SPG11 protein spatacsin, are pivotal for autophagic lysosome reformation (ALR), a pathway that generates new lysosomes. Lysosomal targeting of spastizin required an intact FYVE domain, which binds phosphatidylinositol 3-phosphate. Loss of spastizin or spatacsin resulted in depletion of free lysosomes, which are competent to fuse with autophagosomes, and an accumulation of autolysosomes, reflecting a failure in ALR. Moreover, spastizin and spatacsin were essential components for the initiation of lysosomal tubulation. Together, these results link dysfunction of the autophagy/lysosomal biogenesis machinery to neurodegeneration.
Introduction
Hereditary spastic paraplegias (HSPs) are a group of neurological disorders characterized by a length-dependent axonopathy of corticospinal motor neurons, resulting in progressive spasticity and weakness of the legs. HSPs have traditionally been divided into categories referred to as pure or complex, based upon the presence (complex) or absence (pure) of additional clinical features. More recently, a genetic classification scheme has predominated, with HSPs identified by their spastic gait (SPG) genetic loci in order of identification, SPG1–SPG72 (1–5). Autosomal recessive HSP with thin corpus callosum (AR-HSP-TCC) is a common subtype of complex HSP; SPG11 and SPG15 are the two most prevalent autosomal recessive HSPs (AR-HSPs), comprising about 70% of AR-HSP-TCC (3, 6). Clinically, SPG15 and SPG11 are virtually identical, with distinctive features of early-onset parkinsonism, cognitive impairment, white matter changes, mild cerebellar ataxia, retinal abnormalities, and lens opacities (3, 6).
SPG15 and SPG11 have mutations predicted to lead to elimination or loss of function of the spastizin or spatacsin proteins, respectively. Both spastizin and spatacsin are large proteins, >250 kDa each, and they are present in a common protein complex that also includes the heterotetrameric adaptor protein complex AP-5, which is mutated in SPG48 (7, 8). Their predicted secondary structures include α-solenoids reminiscent of those in the clathrin heavy chain and COP-I subunits (8). Spatacsin also has an N-terminal, β-propeller–like domain, and spastizin harbors a zinc-binding FYVE (present in Fab1, YOTB, Vac1, and EEA1) domain. This spastizin FYVE domain binds to phosphatidylinositol 3-phosphate [PI(3)P], the catalytic product of the class III phosphatidylinositol-3-OH kinase [PI(3)K-III] complex (9). Spastizin in particular has been localized to different and often disparate intracellular locations in published studies (7–11).
Different cellular pathogenic mechanisms have also been suggested for SPG15 and SPG11, including defective DNA repair, cell division abnormalities, endolysosomal dysfunction, and impaired autophagy (7–10, 12, 13). Among these, recent studies in cells from patients SPG15 and SPG11 have favored pathogenic alterations in lysosomes or autophagy (12, 13). Furthermore, AP-5 coprecipitates with spastizin and spatacsin while also colocalizing with the lysosomal protein LAMP1 in cells (7, 8, 14). Studies of a Zfyve26–/– mouse model for SPG15 further support endolysosomal dysfunction in AR-HSP-TCC pathogenesis (10).
Autophagy is an intracellular process for degradation and recycling of cytoplasmic components for survival and homeostasis (15), and the quantity and quality of lysosomes are crucial. Autophagy is initiated by the formation and expansion of the phagophore to form the autophagosome, a double-membrane vesicle that sequesters materials, such as subcellular organelles and long-lived proteins. Autophagosomes then fuse with lysosomes to form autolysosomes, which degrade materials (16). After degradation, new lysosomes can be generated from existing autolysosomes by autophagic lysosome reformation (ALR), a recently described lysosome biogenesis mechanism (17). Autophagic impairment and lysosomal defects are tightly linked to common neurodegenerative disorders, including Parkinson disease, Huntington disease, and Alzheimer disease (18–21).
Here, we have identified critical roles for the SPG15 protein spastizin and the SPG11 protein spatacsin in autophagic lysosome reformation. We have demonstrated that the PI(3)P-binding FYVE domain is responsible for lysosomal targeting of spastizin, which is in a complex with spatacsin. We have also identified lysosomal and autophagic defects in spastizin- and spatacsin-depleted HeLa cells as well as fibroblasts from patients with AR-HSP-TCC. Importantly, the machinery which generates new lysosomes from autolysosomes is impaired in these cells, indicating that spastizin and spatacsin are crucial for lysosomal biogenesis.
Results
SPG15 protein spastizin localizes to lysosomes via its FYVE domain.
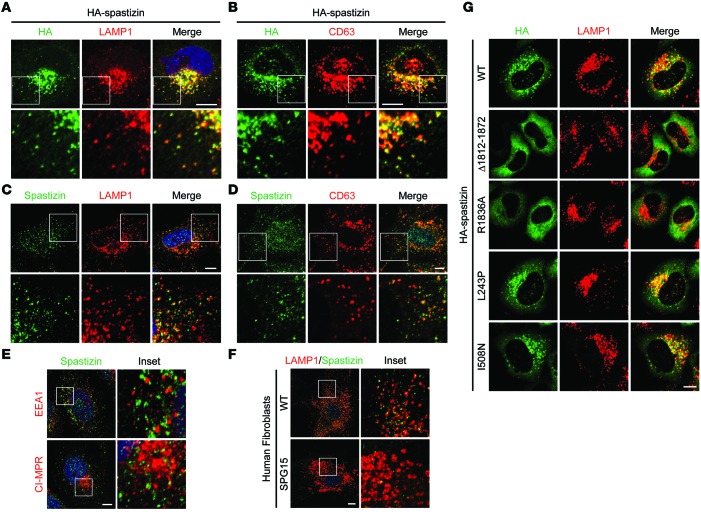
Spastizin is also known as SPG15, ZFYVE26 (for its FYVE domain), and FYVE-CENT (for its localization to centrosomes) (9). The subcellular localization of spastizin has been controversial, with different studies reporting its localization to ER, mitochondria, early/late endosomes, and lysosomes and along microtubules (8, 10–12). We also examined the distributions of endogenous and ectopically expressed spastizin. HA-tagged and endogenous spastizin mainly localized to puncta labeled by markers for lysosomes in HeLa cells (Figure 1, A–D). In contrast, endogenous spastizin immunolabeling overlapped minimally with the early endosomal marker early endosomal antigen 1 (EEA1) and only partially with the late endosomal marker cation-independent mannose-6-phosphate receptor (CI-MPR) (Figure 1E). To confirm the specificity of endogenous spastizin labeling, we compared spastizin immunostaining in control fibroblasts with that in cells from a patient with SPG15 lacking spastizin protein (13). Lysosomal labeling in wild-type fibroblasts was similar to that in HeLa cells, but the immunoreactive signal was nearly undetectable in SPG15 patient cells, indicating that the robust staining represents bona fide spastizin (Figure 1F).
Figure 1. Spastizin localizes to lysosomes through its FYVE domain.
(A and B) HeLa cells transiently expressing HA-spastizin were coimmunostained for HA-epitope (green) and either endogenous (A) LAMP1 or (B) CD63 (red). (C and D) HeLa cells were coimmunostained for endogenous spastizin (green) and either (C) LAMP1 or (D) CD63 (red). (E) HeLa cells were coimmunostained for either EEA1 or CI-MPR (red) with endogenous spastizin (green). (F) Skin fibroblasts derived from a control subject and a patient with SPG15 were coimmunostained for LAMP1 (red) and endogenous spastizin (green). (G) HeLa cells were transfected with wild-type or mutant HA-spastizin constructs as shown and coimmunostained for HA-epitope (green) and LAMP1 (red). Merged images are to the right in A–D and G. Boxed areas in the images in the top rows are enlarged in the bottom rows in A–D (original magnification, ×2.5), and boxed areas in the images in the left columns are enlarged in the right columns in E and F (original magnification, ×4). Scale bar: 10 μm.
FYVE domains typically target proteins to cellular structures enriched in PI(3)P, such as early endosomal compartments (22) and autophagosomes (18), and indeed the FYVE domain of spastizin binds PI(3)P in vitro (9). Furthermore, the PI3 kinase inhibitor wortmannin disperses recombinant spastizin within the cytoplasm, suggesting that PI(3)P is required for the localization of spastizin (8). Although little is known about the lysosomal content of PI(3)P, loss of PI(3)P impairs lysosomal shape and function (23). The production of PI(3)P by class III PI3 kinase is critical for autophagy, especially for autophagosome formation (24). To test whether the spastizin FYVE domain mediates its lysosomal localization, we generated a spastizin construct lacking the FYVE domain (residues 1,812–1,872) and a FYVE domain point mutant (R1836A) that renders it unable to bind PI(3)P (9). Both mutants were dispersed throughout the cytoplasm, whereas wild-type spastizin properly targeted to lysosomes, as assessed by LAMP1 staining (Figure 1G). We also examined two recently reported SPG15 missense mutations outside the FYVE domain (12). Although both mutations impair autophagy and disrupt spastizin binding to beclin-1 (12), they properly localized to lysosomes (Figure 1G). Thus, spastizin localizes prominently to lysosomes through its FYVE domain.
Depletion of spastizin or spatacsin results in enlargement of LAMP1-positive organelles.
The interaction of spastizin with spatacsin was initially identified by immunoprecipitation assays, followed by mass spectrometry analysis (7), and stabilities of these proteins in cells are interdependent (13). We confirmed the interactions between endogenous and recombinant, tagged spastizin and spatacsin (Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/JCI77598DS1), which were consistent with their known colocalization (8).
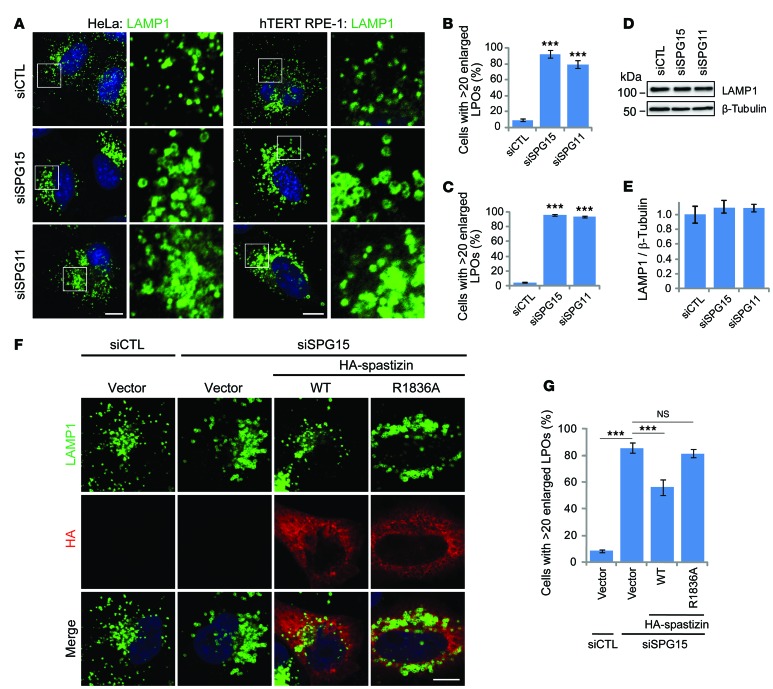
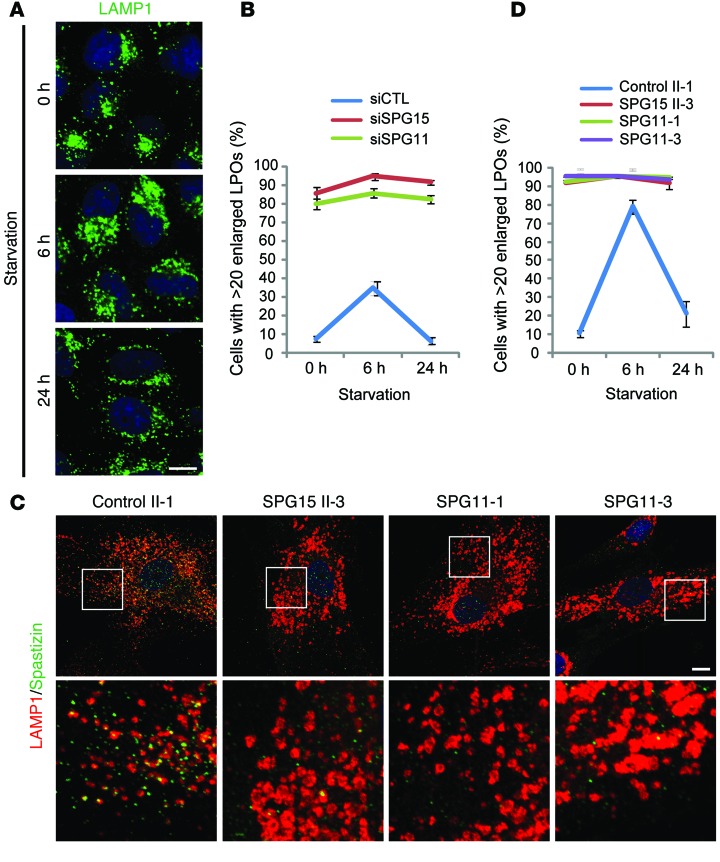
We recently reported that fibroblasts from patients with SPG15 or SPG11 contain enlarged LAMP1-positive organelles (LPOs) (13). The lysosomal localization of spastizin and enlarged LPOs in patient cells prompted us to examine this phenotype. First, we established that, in both HeLa and hTERT-RPE1 cells depleted of spastizin or spatacsin by siRNA, LPOs were significantly enlarged as compared with controls (Figure 2, A–C), without changes in total LAMP1 levels (Figure 2, D and E). The enlarged LPO phenotype was partially restored by expressing wild-type spastizin. However, the spastizin FYVE domain mutant showed no significant recovery at similar levels of expression (Figure 2, F and G), indicating that PI(3)P-dependent localization is critical for this functional role of spastizin at lysosomes. Enlarged LPOs are induced temporarily by short-term starvation, a state that resolves upon prolonged starvation (17). This cycle of enlargement and subsequent restoration of baseline LPO size was similarly induced by starvation in HeLa cells (Figure 3A). Interestingly, cells lacking spastizin or spatacsin failed to restore the normal size of lysosomes under several different long-term starvation conditions (Figure 3B and Supplemental Figure 2). This failure in restoration of LPO enlargement under the same starvation conditions was also observed in fibroblasts from patients with SPG15 and SPG11 (Figure 3, C and D). Lysosomes, in which LAMP1 is concentrated, are a final destination of intracellular trafficking. We did not find notable differences in markers for other organelles by immunofluorescence staining in spastizin- or spatacsin-depleted cells (Supplemental Figure 3). In sum, these results indicate that depletion of spastizin or spatacsin selectively enhances the enlargement of LPOs and prevents the restoration of LPO size induced by long-term nutrient starvation.
Figure 2. Depletion of spastizin or spatacsin causes prominent LPO enlargement.
(A) HeLa (left) or hTERT RPE-1 (right) cells transfected with control (siCTL), spastizin (siSPG15), or spatacsin (siSPG11) siRNAs were immunostained for LAMP1. Nuclei were stained with Hoechst 33342 (blue). Boxes within images in the left columns are enlarged (original magnification, ×4) in the right columns. (B and C) Numbers of (B) HeLa or (C) hTERT RPE-1 cells from A with >20 enlarged LPOs (LPOs with > 0.5 μm diameter) were quantified (n = 3; >200 cells per experiment). (D) HeLa cells were transfected with siCTL, siSPG15, or siSPG11 siRNAs, and cell lysates were immunoblotted. Molecular weight standards (in kDa) are at the left. (E) LAMP1 protein levels from D were normalized to β-tubulin (n = 3). (F) HeLa cells were transfected with siCTL or siSPG15 siRNAs and subsequently with the indicated HA-spastizin constructs or an empty vector and then immunostained for LAMP1 (green) and HA-epitope (red). Merged images are shown in the bottom row. (G) Cells with >20 enlarged LPOs from F were quantified (n = 3; >100 cells per experiment). Scale bar: 10 μm. Mean ± SD are shown. One-way ANOVA followed by Tukey’s multiple comparison test, ***P < 0.001.
Figure 3. Prolonged starvation does not restore normal LPO size in cells lacking spastizin or spatacsin.
(A) Representative images of control HeLa cells immunostained for LAMP1 for the indicated times of starvation. (B) Cells were transfected with siCTL, siSPG15, or siSPG11 siRNAs; starved by serum deprivation; and immunostained for LAMP1. Cells with >20 enlarged LPOs were quantified (n = 3; >200 cells per experiment). (C and D) Fibroblasts derived from a control subject (II-1), a patient with SPG15 (II-3), and 2 patients with SPG11 (SPG11-1 and SPG11-3) were starved by serum deprivation and then coimmunostained for LAMP1 (red) and spastizin (green). (C) Representative images of 0 hour–starved cells. (D) Cells with >20 enlarged LPOs were quantified (n = 3; >200 cells per experiment). Boxes within images are enlarged (original magnification, ×4) in the bottom row. Scale bar: 10 μm. Mean ± SD are shown.
Subunits of heterotetrameric AP-5 complexes, mutated in SPG48 (7), interact with spastizin and spatacsin (7, 8), and depletion of spastizin or spatacsin prominently destabilizes AP-5 (8). We similarly observed a substantial decrease in levels of the AP-5 subunit AP5B1 in cells depleted of spastizin or spatacsin by siRNA as well as in fibroblasts from patients with SPG15 and SPG11 (Supplemental Figure 4A). However, spastizin protein levels were less affected by depletion of AP5B1 (Supplemental Figure 4A), and lysosomal localization of spastizin was not impaired in the AP-5–depleted cells (Supplemental Figure 4B). These results indicate that depletion of spastizin or spatacsin destabilizes AP-5, but the opposite effect appears less prominent. Also, siRNA-mediated depletion of AP5B1 did not cause a prominent enlargement of LPOs, though one siRNA did show a clear trend toward an increase (Supplemental Figure 4, C and D). Together, these data suggest that some functions of spastizin and spatacsin in lysosomes do not depend on binding AP-5, though adaptor protein interactions are likely generally important, particularly if there is some functional redundancy with other orthologous adaptor protein complex subunits in our AP5B1 siRNA experiments.
Depletion of spastizin or spatacsin results in autophagosome accumulation.
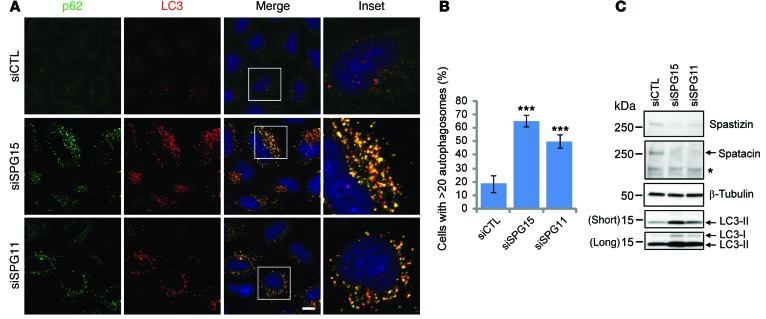
Lysosomes play a pivotal role in degradation and recycling of intracellular substrates by fusing with autophagosomes (16). The accumulation of enlarged LPOs in spastizin- or spatacsin-depleted cells led us to investigate any impairment in lysosome-dependent autophagy. Indeed, accumulation of autophagosomes has been observed in fibroblasts derived from patients with SPG15 and SPG11 (12, 13). Furthermore, mature autophagosomes, as visualized by immunostaining for p62 or LC3, were increased in spastizin- or spatacsin-depleted cells (Figure 4, A and B, and Supplemental Figure 2). During autophagy, cytosolic LC3 (LC3-I) is conjugated to phosphatidylethanolamine to form a LC3-phosphatidylethanolamine conjugate (LC3-II), which is recruited to autophagosomal membranes. Enhanced levels of LC3-II also supported an increase in autophagosome numbers in spastizin- or spatacsin-depleted cells (Figure 4C).
Figure 4. Depletion of spastizin or spatacsin induces accumulation of autophagosomes under nutrient conditions.
(A) HeLa cells cultured in serum-containing media were transfected with siCTL, siSPG15, or siSPG11 siRNAs and then coimmunostained for p62 (green) and LC3 (red). Insets in the merged images are enlarged (original magnification, ×3) to the right. Scale bar: 10 μm. (B) Cells with >20 autophagosomes from A were quantified (n = 3; >200 cells per experiment). One-way ANOVA followed by Tukey’s multiple comparison test, ***P < 0.001. (C) HeLa cells maintained in serum-containing media were transfected with siCTL, siSPG15, or siSPG11 siRNAs, and cell lysates were immunoblotted. Molecular weight standards (in kDa) are shown to the left. Arrows identify specific proteins, while the asterisk denotes a cross-reacting band.
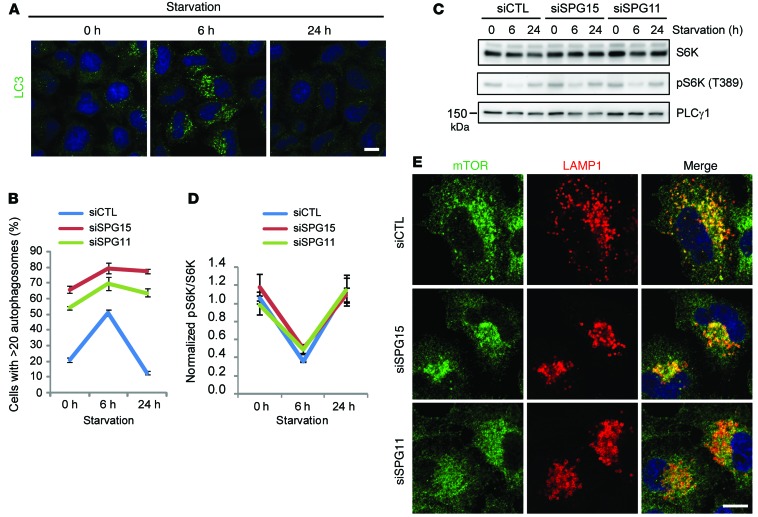
The cycle of assembly and disassembly of autophagosomes is accompanied by enlargement of LPOs in nutrient-starved cells (ref. 17 and Figure 5A). We monitored autophagosomes in spastizin- or spatacsin-depleted cells using LC3 immunostaining. Although assembly of autophagosomes was induced slightly by short-term starvation, the depleted cells failed to disassemble the autophagosomes upon prolonged starvation, in contrast with controls (Figure 5B). Autophagy induced by serum deprivation is mediated by inhibition of the activity of the mammalian target of rapamycin (mTOR) kinase, which is a key negative regulator of autophagy (25). Reactivation of mTOR activity upon prolonged starvation is required for reducing the size of LPOs and clearing autophagosomes (17), but recovery of mTOR activity to normal levels following prolonged starvation was not impaired by spastizin or spatacsin depletion (Figure 5, C and D), and lysosomal localization was not of mTOR affected (Figure 5E). These results indicate that failure of restoration of LPO size and clearance of autophagosomes in spastizin- or spatacsin-depleted cells occur downstream of mTOR activation.
Figure 5. A failure of clearance of autophagosomes in spastizin- or spatacsin-depleted cells occurs downstream of mTOR activation.
(A and B) HeLa cells were transfected with siCTL, siSPG15, or siSPG11 siRNAs; starved by serum deprivation; and immunostained for LC3. (A) Representative images of starved control cells for the indicated time periods. (B) Starved cells with >20 autophagosomes were quantified (n = 3; >200 cells per experiment). (C) Lysates from A were immunoblotted with the indicated antibodies. Molecular weight standards (in kDa) are shown to the left. Molecular weight of S6K is approximately 59 kDa. (D) Phospho-S6K levels from C were normalized to total S6K (n = 3). (E) HeLa cells were transfected with siCTL, siSPG15, or siSPG11 siRNAs and then coimmunostained for mTOR (green) and LAMP1 (red). Scale bar: 10 μm. Mean ± SD are shown.
Enlarged LPOs are functional autolysosomes.
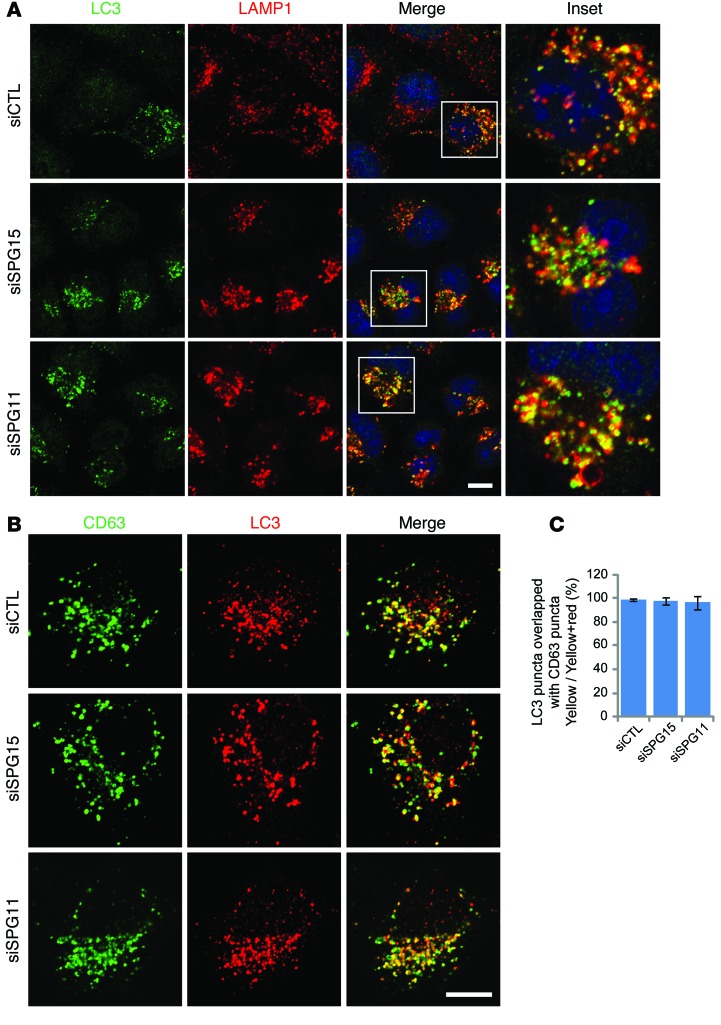
Depletion of spastizin or spatacsin induces an accumulation of both enlarged LPOs and autophagosomes (Figures 2–5 and refs. 12, 13). To examine the relationship of these structures, spastizin- or spatacsin-depleted cells were coimmunostained for LC3 and lysosomal markers. Intriguingly, most autophagosomes labeled by LC3 were also positive for LAMP1 or CD63, indicating that fusion with lysosomes had occurred. Thus, the accumulated vesicles are mostly autolysosomes (Figure 6).
Figure 6. Enlarged LPOs induced by depletion of spastizin or spatacsin in nutrient conditions are autolysosomes.
(A and B) HeLa cells maintained in serum-containing media were transfected with siCTL, siSPG15, or siSPG11 siRNAs and then coimmunostained either for (A) LC3 (green) and LAMP1 (red) or (B) CD63 (green) and LC3 (red). Insets in the images are enlarged (original magnification, ×2.8) to the right in A. (C) LC3 puncta overlapping with CD63 from B were quantified (10 cells; >30 particles per cell). Scale bar: 10 μm. Mean ± SD are shown.
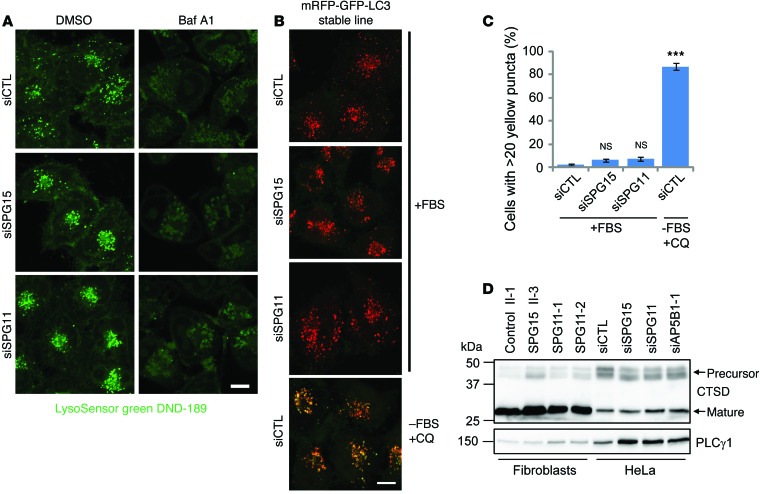
While fusion of autophagosomes with lysosomes is essential for clearance of cellular autophagosomes, the acidic luminal environment of autolysosomes is also crucial for autophagic degradation. Since autophagosomes in spastizin- or spatacsin-depleted cells have successfully fused with lysosomes, we examined the luminal environment of these autolysosomes. LysoSensor Green specifically labels acidic organelles, and this labeling could be abrogated by lysosome inhibitors, such as bafilomycin A1. Compared with that of control cells, spastizin- or spatacsin-depleted cells showed indistinguishable LysoSensor Green signal brightness, which indicated that pH was not significantly impaired by depletion of spastizin or spatacsin (Figure 7A). Tandem fluorescence-tagged LC3 is a useful tool that capitalizes on the pH difference between acidic autolysosomes and neutral autophagosomes and the pH sensitivity differences exhibited by GFP and RFP to monitor progression from the autophagosome to the autolysosome (26). As expected, elevation of lysosomal pH following chloroquine treatment and serum withdrawal resulted in clear visualization of the yellow (combined red and green) LC3 signal (Figure 7, B and C). As in control cells, however, most spastizin- or spatacsin-depleted cells contained <20 yellow puncta, which represent nonacidic autophagosomes (Figure 7, B and C). Thus, depletion of spastizin or spatacsin did not alter the acidic environment of autolysosomes.
Figure 7. Autolysosomes in spastizin- or spatacsin-depleted cells are functional.
(A) HeLa cells were transfected with siCTL, siSPG15, or siSPG11 siRNAs. LysoSensor Green DND-189 (1 mM) was added for 1 hour to stain acidic lysosomes. To neutralize acidic lysosomes, bafilomycin A1 (Baf A1, 40 μM) was added in serum-free media for 16 hours before imaging. (B) HeLa cells stably expressing mRFP-GFP-LC3 were transfected with siCTL, siSPG15, or siSPG11 siRNAs. To elevate the lysosomal pH, chloroquine (CQ, 50 μM) was added to siCTL-transfected, serum-deprived cells for 4 hours. Cells were visualized by confocal microscopy, and images from red and green channels were merged. (C) Cells with >20 yellow puncta from B were quantified (n = 3; >200 cells per experiment). (D) Lysates were prepared from control (II-1), SPG15 (II-3), or SPG11 (SPG11-1 and SPG11-2) fibroblasts. HeLa cell lysates were obtained from cells transfected with siCTL, siSPG15, siSPG11, or AP5B1 (siAP5B1-1) siRNAs. Lysates were immunoblotted as shown. Molecular weight standards (in kDa) are shown to the left. Scale bar: 10 μm. Mean ± SD are shown. One-way ANOVA followed by Tukey’s multiple comparison test, ***P < 0.001.
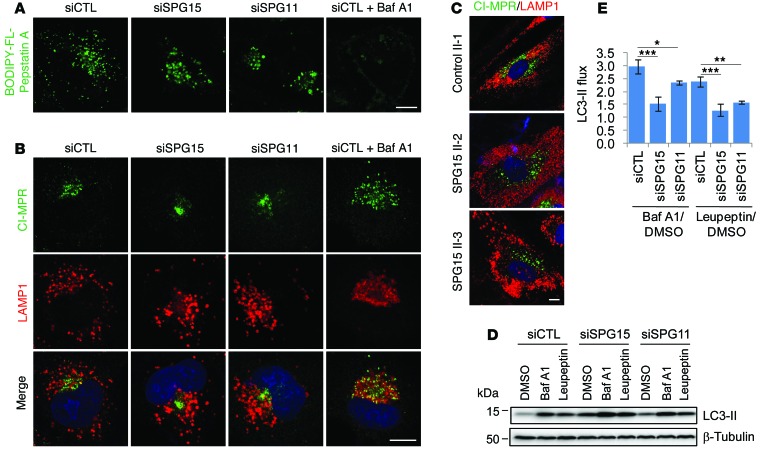
Cathepsins are lysosomal proteases activated at low pH, and a precursor form of cathepsin D (46 kDa; CTSD), one of the major cathepsins, is processed into a mature form (32 kDa) in acidic lysosomes. The ratios of CTSD’s precursor/mature form in fibroblasts from patients with SPG15 or SPG11 were not significantly changed (Figure 7D). Also, depletion of spastizin, spatacsin, or AP-5 did not alter the precursor/mature ratio, indicating that CTSD maturation was not impaired (Figure 7D). To directly visualize CTSD activity, cells were incubated with BODIPY-FL-pepstatin A, which specifically binds active CTSD (Figure 8A). As expected, BODIPY-FL-pepstatin A labeling was nearly abolished by bafilomycin A1, which neutralizes lysosomes by inhibiting proton pumps. By contrast, most lysosomes in control and spastizin- or spatacsin-depleted cells were labeled by BODIPY-FL-pepstatin A, indicating normal CTSD activation (Figure 8A).
Figure 8. Spastizin- and spatacsin-depleted cells exhibit impaired autophagosome clearance.
(A) HeLa cells were transfected with siCTL, siSPG15, or siSPG11 siRNAs and then BODIPY-FL-pepstatin A (1 mM) was added for 30 minutes to stain lysosomes containing active cathepsins. To neutralize the acidic lysosomes, bafilomycin A1 (40 μM) was added in serum-free media to siCTL-transfected cells for 16 hours before imaging. (B) HeLa cells were transfected with siCTL, siSPG15, or siSPG11 siRNAs and coimmunostained for CI-MPR (green) and LAMP1 (red). Bafilomycin A1–treated, siCTL-transfected cells were included for comparison. (C) Fibroblasts derived from a control subject (II-1) or patients with SPG15 (II-2 and II-3) were coimmunostained for CI-MPR (green) and LAMP1 (red). (D) HeLa cells transfected with siCTL, siSPG15, or siSPG11 siRNAs were treated with DMSO, bafilomycin A1 (0.4 μM), or leupeptin (50 μg/mL) for 6 hours before harvesting. Lysates were immunoblotted as shown. Molecular weight standards (in kDa) are shown to the left. (E) LC3-II flux from D was measured (n = 3). Scale bar: 10 μm. Mean ± SD are shown. One-way ANOVA followed by Tukey’s multiple comparison test, *P < 0.05, **P < 0.01, ***P < 0.001.
According to a “hybrid” model for lysosome biogenesis, hybrid organelles are formed by the fusion of late endosomes and lysosomes (27). The low pH of lysosomes induces retrieval of CI-MPR, which is delivered from late endosomes after fusion of late endosomes and lysosomes. Therefore, one characteristic of lysosomes that distinguishes them from hybrid organelles is an absence of CI-MPR (28). To show that the enlarged LPOs are not hybrid organelles that failed to regenerate lysosomes, we examined the colocalization of CI-MPR with LAMP1. Bafilomycin A1 treatment increased lysosomal pH, which abolished retrieval of CI-MPR in control cells, resulting in colocalization of CI-MPR with LAMP1 (Figure 8B). In spastizin- or spatacsin-depleted cells, however, CI-MPR barely colocalized with LAMP1, although they exist in a similar location due to clustering of LAMP1 in the perinuclear region (Figure 8B). SPG15 patient fibroblasts also showed distinct localizations of CI-MPR and LAMP1, indicating that enlarged LPOs are CI-MPR negative (Figure 8C). We conclude that enlarged LPOs in spastizin- or spatacsin-depleted cells are mostly not perturbed late endosome/lysosome hybrid organelles but instead functional autolysosomes.
Spastizin and spatacsin are essential for ALR initiation.
The above results suggest that the accumulated, enlarged LPOs in cells deficient for spastizin or spatacsin are functioning normally. Indeed, treatment of cells with lysosomal inhibitors, such as bafilomycin A1 or leupeptin, elevated levels of membrane-bound LC3 (LC3-II), even in spastizin- or spatacsin-depleted cells (Figure 8, D and E). However, depletion of spastizin or spatacsin alone induced a modest increase in LC3-II levels, indicating that lysosome-dependent clearance of autophagosomes was attenuated but not blocked (Figure 8, D and E). Since fusion of autophagosomes with lysosomes and lysosomal activity were not impaired, we hypothesized that cells lacking spastizin and spatacsin might have impaired lysosome biogenesis.
A canonical model for lysosome generation suggests that newly synthesized lysosomal proteins are targeted to lysosomes via the trans-Golgi network through the endosomal pathway (29). Meanwhile, another model for lysosome reformation, termed ALR, was recently proposed (17). ALR can be induced by nutrient starvation conditions; inhibition of mTOR activity induces autophagy, which results in accumulation of autophagosomes, which become autolysosomes after fusion with lysosomes. After clearance of autophagic materials, protolysosomal tubules emanate from autolysosomes, separate, and mature into functional lysosomes. Subsequent studies have identified potential regulators of ALR, such as Spin, clathrin, phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2], PIP5K1A, PIP5K1B, AP-2, and AP-4 (23, 30–32).
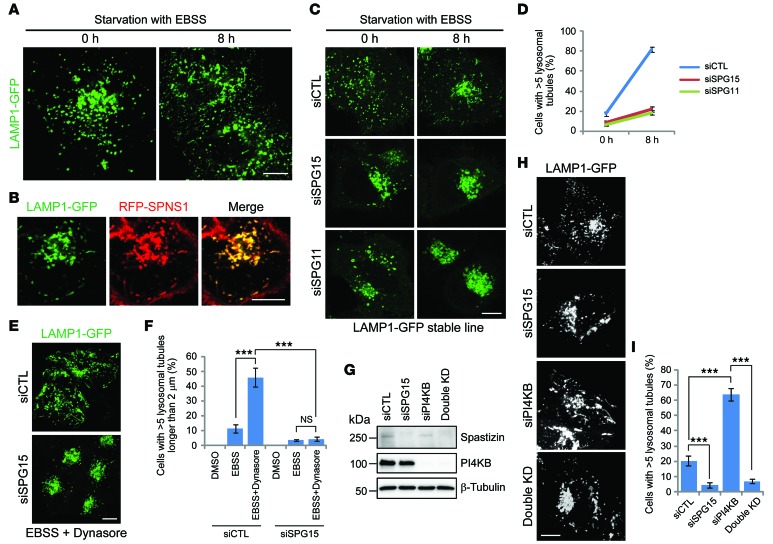
Lysosome tubules induced by starvation have LAMP1 on their surface but little luminal content (17), so we stably expressed LAMP1-GFP in HeLa cells to assess ALR. These cells had a typical lysosome-like distribution of LAMP1 puncta (Figure 9A and Supplemental Video 1), and after 8 hours of starvation with Earle’s Balanced Salts Solution (EBSS), tubules emanated from the puncta (Figure 9A and Supplemental Video 2). These lysosome tubules were also positive for SPNS1, which is known to colocalize with LAMP1 and is required for ALR (Figure 9B and ref. 30). Next, we examined whether spastizin and spatacsin are involved in ALR. Prior to starvation, depletion of spastizin or spatacsin resulted in enlarged LPOs, as expected (Figure 9C). After 8-hour starvation, spastizin- or spatacsin-depleted cells contained numerous clustered and still enlarged LPOs without tubules, whereas about 80% of control cells had >5 lysosomal tubules emanating from LPOs (Figure 9, C and D, and Supplemental Videos 3–5). These results indicate that spastizin and spatacsin are required for ALR induced by starvation.
Figure 9. Spastizin and spatacsin are essential for ALR initiation.
(A) HeLa cells stably expressing LAMP1-GFP were starved in EBSS (0 or 8 hours) and imaged. (B) HeLa cells stably expressing LAMP1-GFP were transfected with RFP-SPNS1, starved in EBSS for 8 hours, and imaged. (C) HeLa cells stably expressing LAMP1-GFP were transfected with siCTL, siSPG15, or siSPG11 siRNAs; starved with EBSS (0 or 8 hours); and imaged. (D) Cells with >5 lysosomal tubules from C were quantified (n = 3; >100 cells per experiment). (E and F) HeLa cells stably expressing LAMP1-GFP were transfected with siCTL or siSPG15 siRNAs and then treated with DMSO, EBSS, or EBSS with Dynasore (40 μM) for 2 hours before imaging. (E) Representative images of Dynasore-treated control or spastizin-depleted (SPG15-depleted) cells. (F) Cells with >5 lysosomal tubules longer than 2 μm were quantified (n = 3; >100 cells per experiment). (G) HeLa cells were transfected with siCTL, siSPG15, PI4KB (siPI4KB), or both spastizin and PI4KB (Double KD) siRNAs, and cell lysates were immunoblotted. Molecular weight standards (in kDa) are shown to the left. (H) HeLa cells stably expressing LAMP1-GFP were transfected with siCTL, siSPG15, siPI4KB, or both spastizin and PI4KB siRNAs and then imaged. (I) Cells with >5 lysosomal tubules from H were quantified (n = 3; >100 cells per experiment). Scale bar: 10 μm. Mean ± SD are shown. One-way ANOVA followed by Tukey’s multiple comparison test, ***P < 0.001.
There are two main possibilities to explain the absence of lysosome tubules in spastizin- or spatacsin-depleted cells: these proteins could be essential for initiation of ALR or they could be suppressors of tubule stabilization and scission/vesiculation. ALR shares key components, such as clathrin, PI(4,5)P2, and AP-2, with endocytosis at the plasma membrane (31, 32). Intriguingly, the large GTPase dynamin 2 (Dyn2), which is crucial for the scission of vesicles from the plasma membrane, has also been implicated in the scission of lysosomal tubules (33). We examined lysosomal tubulation upon single or double inhibition of spastizin and Dyn2. Treatment of cells with Dynasore, an inhibitor of dynamins, resulted in lysosomal tubulation, even upon short-term starvation in control cells (Figure 9, E and F). These tubules were generally longer than those induced by starvation alone (compare Figure 9C and Figure 9E). However, Dynasore-dependent lysosomal tubulation did not occur in spastizin-depleted cells (Figure 9, E and F). In fact, had alterations of lysosomal tubulation upon depletion of spastizin been due to premature scission or vesiculation, inhibition of Dyn2 should have prevented vesiculation and induced tubulation.
Fission of vesicles or tubules from autolysosomes is regulated by PI4KB and phosphatidylinositol 4-phosphate [PI(4)P]. Absence of PI4KB or loss of kinase activity of PI4KB reduces the level of lysosomal PI(4)P, which may have a role in inhibiting tubulation (34). As a result, these cells show lysosomal hypertubulation. Indeed, siRNA-mediated depletion of PI4KB resulted in hypertubulation in HeLa cells (Figure 9, G–I). However, double knockdown of spastizin and PI4KB phenocopied the single knockdown of spastizin, indicating that the reduced levels of PI(4)P did not induce tubulation in the spastizin-depleted background. The above results indicate that spastizin and spatacsin are essential for ALR initiation.
PI(4)P-to-PI(4,5)P2 conversion, catalyzed by PIP5K1B at lysosomes, has been suggested to be required for initiation of ALR; this is followed by conversion via PIP5K1A at the leading edge of lysosomal tubules (31). To determine whether the phenotypes mediated by spastizin and spatacsin depletion result from impairment of this pathway, we examined PIP5K, PI(4)P, and PI(4,5)P2. Protein levels of both PIP5K1A and PIP5K1B were not changed by depletion of spastizin or spatacsin (Supplemental Figure 5A). Also, the localizations of both PI(4)P and PI(4,5)P2 at lysosomes were not altered, indicating that PI(4)P-to-PI(4,5)P2 conversion has occurred at lysosomes in spastizin- and spatacsin -depleted cells (Supplemental Figure 5B). It has been suggested that ALR failure in PIP5K1B-depleted cells is caused not by a decrease of PI(4,5)P2 but instead by accumulation of PI(4)P, which suppresses lysosomal tubulation (34). Although we were unable to establish whether PI(4,5)P2 is a critical component for ALR, our results indicate that the presence of PI(4,5)P2 at autolysosomes is not sufficient for ALR. Spastizin and spatacsin may thus contribute to ALR in a manner independent or downstream of PIP5K1B and PI(4,5)P2.
Spastizin and spatacsin interact with PI4KB.
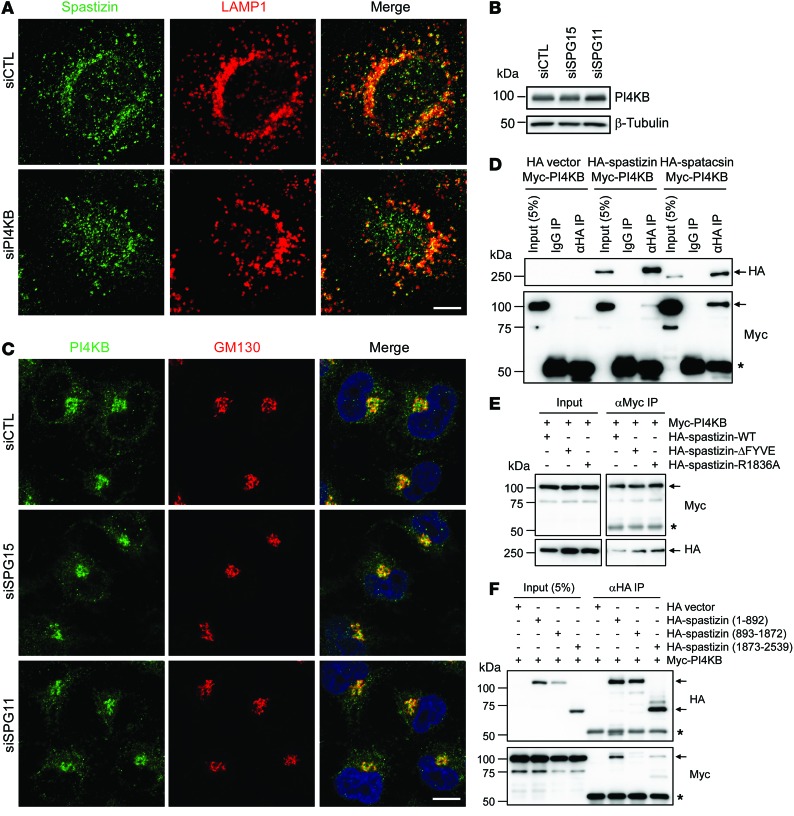
In feeding conditions, autolysosomes usually do not extrude lysosomal tubules. Instead, lysosome reformation mainly occurs via vesiculation mediated by PI4KB (34). Spastizin- or spatacsin-depleted cells showed not only impaired lysosomal tubulation in starvation conditions but also increased numbers of autolysosomes in feeding conditions (Figures 4 and 9). In other words, those cells lack free lysosomes, which are competent to fuse with autophagosomes even under normal conditions, and the defect during starvation could conceivably result from impaired vesiculation. Therefore, we examined the relationship of spastizin and spatacsin with PI4KB. Cellular levels and lysosomal localization of spastizin were not altered upon depletion of PI4KB (Figure 9G and Figure 10A). Also, depletion of spastizin and spatacsin did not change the levels and Golgi localization of PI4KB (Figure 10, B and C). Thus, the opposing phenotypes — enlarged LPOs upon spastizin depletion and hypertubulation upon PI4KB depletion — were not the result of reciprocal changes in their protein levels or intracellular targeting.
Figure 10. Spastizin and spatacsin interact with PI4KB.
(A) HeLa cells were transfected with siCTL or siPI4KB siRNAs and then coimmunostained for spastizin (green) and LAMP1 (red). Merged images are to the right. (B) HeLa cells were transfected with siCTL, siSPG15, or siSPG11 siRNAs and then lysates were immunoblotted as shown. (C) Cells from B were coimmunostained for PI4KB (green) and GM130 (red). (D) Myc-PI4KB was coexpressed with HA vector, HA-spastizin, or HA-spatacsin in HEK293T cells, and lysates were immunoprecipitated and immunoblotted with the indicated antibodies (right). (E) Myc-PI4KB was coexpressed with indicated HA-spastizin wild-type or mutant constructs in HEK293T cells, and lysates were immunoprecipitated and immunoblotted with the indicated antibodies. (F) Myc-PI4KB was coexpressed with HA vector or indicated HA-spastizin truncation mutants in HEK293T cells, and lysates were immunoprecipitated (anti-HA) and immunoblotted for HA- and Myc-epitopes. Scale bar: 10 μm. Arrows identify specific proteins, while asterisks denote the IgG heavy chain. Molecular weight standards (in kDa) are shown to the left in B and D–F.
Some PI4KB localizes to lysosomes, although it is mainly targeted to the Golgi apparatus (34). Interestingly, we detected an interaction of PI4KB with spastizin and spatacsin (Figure 10D). The FYVE domain mutation of spastizin did not interfere with the interaction with PI4KB, indicating that binding with PI4KB is not dependent on lysosomal localization of spastizin (Figure 10E). The middle portion of spastizin containing the FYVE domain did not bind PI4KB (Figure 10F). Rather, the N-terminal region preferentially interacted with PI4KB.
Discussion
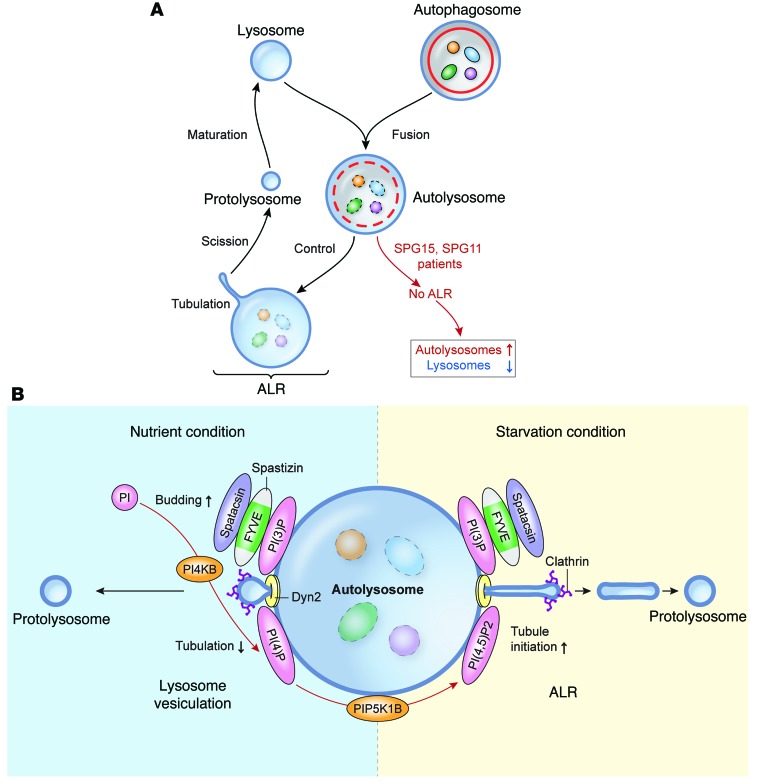
Fusion of autophagosomes and lysosomes enables lysosomal hydrolases to degrade autophagic materials in autolysosomes. While lysosomes are consumed in this scenario, the number of cellular lysosomes is maintained by ALR (32). Here, we demonstrate that proteins mutated in the most common AR-HSPs, spastizin and spatacsin, are required for ALR (Figure 11). Depletion of spastizin or spatacsin does not appear to impair fusion of autophagosomes and lysosomes (as proposed in ref. 12) or alter the acidic environment of the autolysosome. However, ALR via lysosome tubulation is significantly impaired, causing accumulation of autolysosomes (13) and subsequent exhaustion of lysosomes available for fusion with autophagosomes, which are also increased in fibroblasts from patients with SPG15 and SPG11 (12, 13). Thus, autophagic materials accumulate in the cell, which could trigger degeneration in susceptible postmitotic neurons.
Figure 11. Model of effects of spastizin and spatacsin loss in ALR.
(A) In control cells, autophagosomes containing autophagic materials fuse with the lysosome to form the autolysosome. After degrading materials, the lysosomal tubule emanates from the autolysosome and becomes the protolysosome, which is destined to become a lysosome after maturation. This machinery for maintaining the population of cellular lysosomes is called ALR. In cells from patients with SPG15 or SPG11, fusion of the lysosome and the autophagosome occurs normally, but ALR is blocked by impaired initiation of lysosomal tubulation from the autolysosome, which eventually results in accumulation of autolysosomes and exhaustion of free lysosomes. (B) Schematic model of lysosome reformation in starvation and feeding conditions. In feeding conditions, basal autophagy occurs to maintain cell homeostasis. At autolysosomes, PI4KB plays a critical role by converting phosphatidylinositol (PI) to PI(4)P, which suppresses uncontrolled lysosomal tubulation from autolysosomes and facilitates lysosome budding/vesiculation managed by clathrin (or other coat proteins) and Dyn2, reminiscent of endocytosis. The spastizin-spatacsin complex localizes to the lysosome/autolysosome by interaction of the spastizin FYVE domain and PI(3)P, and this complex is also an essential component for lysosome vesiculation. In prolonged starvation conditions, to rapidly generate energy sources cells choose an enhanced pathway, ALR, which reforms lysosomes more efficiently. In this case, PI(4,5)P2 produced from PI(4)P by PIP5K1B is an essential component for tubule initiation. The spastizin-spatacsin complex may function downstream of PI(4,5)P2 or work independently during ALR. The AP-5 protein complex that coprecipitates with spastizin and spatacsin is not shown.
Mechanistically, spastizin and spatacsin are required for initiation of lysosomal tubule formation. Both are large proteins (>250 kDa) in a common complex, with secondary structures predicted to comprise α-solenoids, reminiscent of those in clathrin heavy chain and COP-I (8). This similarity may prefigure a related scaffolding role in the initiation of vesicle and tubule formation. Spastizin appears important for targeting the complex to PI(3)P-rich domains via its FYVE domain (9), and spatacsin has been proposed to have an N-terminal β-propeller–like domain (8). The interaction of spastizin with PI4KB may also prefigure additional lipid and protein interactions to position and locally regulate lysosome vesicle budding. Indeed, PI4KB maintains the amount of lysosomal PI(4)P, which facilitates lysosome vesiculation by preventing abnormal lysosomal tubulation under feeding conditions (34).
The conversion of PI(4)P to PI(4,5)P2 by PIP5K1B at lysosomes has been suggested to be critical for the initiation of ALR (31), and we observed clustered PI(4,5)P2 at enlarged LPOs in spastizin- or spatacsin-depleted cells, which are unable to extrude lysosome tubules. This indicates that PI(4,5)P2 is not sufficient to generate lysosome tubules and instead suggests that spastizin and spatacsin either function downstream of PI(4,5)P2 or else work independently. Since our double-inhibition experiments strongly show that spastizin and spatacsin are initiation factors for ALR, rather than elongation or termination factors, they would likely function just downstream of PI(4,5)P2 if they were on the same pathway. Although lysosomal targeting of spastizin is mediated by binding PI(3)P through its FYVE domain, it will be interesting to establish whether PI(4,5)P2 also harbors targeting or regulatory activity for the spastizin and spatacsin complex at tubule initiation sites.
Last, spastizin and spatacsin interact robustly with the heterotetrameric AP-5 adaptor protein complex, subunits of which are mutated in another form of HSP, SPG48 (7, 8, 14). Though SPG48 clinical data are limited, most patients with SPG48 do not seem to exhibit some of the prominent clinical features of SPG15 and SPG11 or these features are exhibited to a lesser degree. For instance, there appears to be little or no thinning of the corpus callosum, later disease onset, less cognitive impairment, and less extensive white matter changes, as shown by MRI, in patients with SPG48 (35). It will be important to examine comprehensively the role of AP-5 in lysosomal biogenesis, particularly since AP-5 localizes to endolysosomal compartments but does not appear required for spastizin and spatacsin localization to autolysosomes. About half of the patients with AR-HSP-TCC do not harbor mutations in known genes, and as new genes are identified, they may provide further mechanistic insights into these pathways. Since neurons are particularly vulnerable to autophagic defects, and numerous neurological disorders have been linked to autophagy (36, 37), insights into the pathogenesis of AR-HSP-TCC will have broad implications for understanding pathways of neurodegeneration.
Methods
DNA constructs.
Human spastizin (GenBank NM_015346) and spatacsin (GenBank NM_025137) cDNAs were obtained from Kazusa DNA Research Institute. Wild-type or mutant spastizin and spatacsin cDNAs were cloned into the AscI site of the eukaryotic expression vectors pGW1-HA and pGW1-Myc, with epitope tags at the N terminus (38). To generate point mutants for spastizin, PCR and DpnI-based mutagenesis was performed. For siRNA rescue experiments, 3 silent, mismatched nucleotides against the siSPG15 target sequences were introduced into HA-tagged wild-type or R1836A mutant spastizin constructs by mutagenesis, generating siRNA-resistant forms. Human SPNS1 (GenBank NM_001142448) and PI4KB (GenBank NM_001198773) cDNAs were obtained from Open Biosystems. SPNS1 was cloned into the AscI site of pGW1-RFP, and PI4KB was cloned into the AscI site of pGW1-Myc. The mGFP-tagged human LAMP1 (GenBank NM_005561) and mRFP-GFP–tagged rat LC3 (GenBank NM_022867) cDNAs were from Addgene, and they were cloned into the NotI and EcoRI/BamHI sites of pCDH-CMV-MCS-EF1-Puro lentiviral vector (Systems Biosciences), respectively. All constructs were confirmed by DNA sequencing.
Antibodies.
The following antibodies were obtained commercially: anti–β-tubulin (D66, Sigma-Aldrich), anti-CD63 (H5C6, BD Biosciences), anti–CI-MPR (2G11, Abcam), anti-CTSD (BC011, Calbiochem), anti–cyt c (6H2.B4, BD Biosciences), anti-EEA1 (14, BD Biosciences), anti-FAM21 (polyclonal, EMD Millipore), anti-GGA3 (8, BD Biosciences), anti-GM130 (35, BD Biosciences), anti-HA (F-7, Santa Cruz Biotechnology), anti-LAMP1 (H4A3, Abcam), anti-LAMP1 (polyclonal, Abcam), anti-LC3 (polyclonal, Sigma-Aldrich), anti-mTOR (7C10, Cell Signaling Technology), anti–c-Myc (9E10, Santa Cruz Biotechnology), anti-p62 (3, BD Biosciences), anti-PI(4)P (Echelon), anti-PI(4,5)P2 (Echelon), anti-PI4KB (7, BD Biosciences), anti-PIP5K1A (polyclonal, Proteintech), anti-PIP5K1B (polyclonal, Proteintech), anti-PLCγ1 (polyclonal, Cell Signaling Technology), anti-S6K (polyclonal, Cell Signaling Technology), anti–phospho-S6K-T389 (polyclonal, Cell Signaling Technology), anti-SNX1 (51, BD Biosciences), and anti-spatacsin (polyclonal, Proteintech) as well as anti-spastizin (polyclonal, ProSci) for immunoblotting and anti-spastizin (polyclonal, Cell Signaling Technology) for immunofluorescence staining. An anti-AP5B1 antibody was generated commercially (New England Peptide) against a synthetic peptide, NH2-MGPLSRDAWAQRLGC-amide (AP5B1 aa residues 1–14), and affinity purified.
Cell culture, transfection, and RNA interference.
Human fibroblast cell lines were described and named previously (13). HeLa (ATCC), HEK293T (ATCC), and fibroblast cells were cultured in DMEM supplemented with 10% FBS (Gibco), and hTERT RPE-1 cells were cultured in DMEM/F-12 (Gibco) supplemented with 10% FBS at 37°C in the presence of 5% CO2. EBSS was from Sigma-Aldrich. Transfections were performed using GenJet Plus Reagent (SignaGen Laboratories) or Avalanche-Omni Transfection Reagent (EZ Biosystems) for plasmid DNAs and Lipofectamine RNAiMAX (Invitrogen) for siRNAs. ON-TARGETplus siRNA pools against spastizin (SPG15) and spatacsin (SPG11) were obtained from GE Dharmacon. Stealth siRNA against spastizin (SPG15) that was used for the rescue experiment as well as stealth siRNAs against AP5B1 and PI4KB were purchased from Invitrogen. Sequences of siRNAs are as follows: siSPG15 pool: 5′-UGAUGUUCAUGGAGCGCUA-3′, 5′-CCAAGAUGAUGUUCGUCAA-3′, 5′-CCACGUGCCUGACGGAAAU-3′, 5′-CGAAAGGCAUUGCGGGCUU-3′; siSPG11 pool: 5′-CAACUGAAAUUGAUUGAGU-3′, 5′-GCUCAGAGUUGAUAUGAAA-3′, 5′-CCACAUACCUUCAAUAGGA-3′, 5′-CAACAAAUGUGUCAUCCUA-3′; siSPG15: 5′-CGGACAUCAUGAAGCAGCUGCUGUU-3′, siAP5B1-1: 5′-CCCAGAGUGCUACCCUCAACUUUCU-3′; siAP5B1-2: 5′-GCGGCCUUUGGUGAGGCCUUGUUCA-3′; and siPI4KB: 5′-GCUCCUGAGAGAGAAUUCAUCAAGU-3′. Control siRNAs were from Ambion. All primary human cell lines have been described previously (13, 41); SPG11-1, SPG11-2, and SPG11-3 cell lines were provided by William Gahl (NIH, Bethesda, Maryland, USA), Camilo Toro (NIH, Bethesda, Maryland, USA), and Adeline Vanderver (Children’s National Medical Center, Washington, DC, USA).
Generation of stable cell lines.
Stable cell lines were generated as described previously (39). For lentiviral experiments, pCMV-dR8.2 dvpr (Addgene) and pCMV-VSV-G (Addgene) were cotransfected into HEK293T cells with the respective pCDH-CMV-MCS-EF1-Puro–based (System Biosciences) constructs. Eighteen hours later, lentiviral particles were harvested from the culture media and frozen at –80°C. HeLa cells were infected with the virus, and 2 days later, the infected cells were incubated with 2 μg/ml puromycin for at least 2 days for selection.
Immunofluorescence staining.
Human fibroblasts and HeLa and hTERT RPE1 cells cultured on coverslips were washed with PBS and then usually fixed with 4% paraformaldehyde for 15 minutes. Fixed cells were washed 3 times with PBS and blocked and permeabilized with 3% normal donkey serum in PBS with 0.05% saponin for 15 minutes. For autophagosome immunostaining, cells were fixed with cold methanol for 5 minutes and PBS with 0.1% Triton X-100 was used for blocking solution instead of PBS with 0.05% saponin. For phosphatidylinositol phosphate staining, cells were fixed with 2% paraformaldehyde for 15 minutes, washed with PBS containing 50 mM NH4Cl, and permeabilized with 20 μM digitonin in buffer A (20 mM Pipes [pH 6.8], 137 mM NaCl, 2.7 mM KCl) for 5 minutes. Cells were subsequently washed with buffer A and then incubated with blocking solution (buffer A containing 5% normal donkey serum) with 50 mM NH4Cl for 45 minutes. Next, cells were incubated with primary antibodies diluted in blocking solution for 2 hours, washed with buffer A 3 times, incubated with Alexa Fluor–conjugated secondary antibodies (Molecular Probes) diluted in blocking solution for 1 hour, and washed with buffer A 3 times. Cells were counterstained with Hoechst 33342 (Molecular Probes) where indicated, and coverslips were mounted on a glass slide using Fluoromount-G (SouthernBiotech).
Live-cell imaging and confocal microscopy.
Cells were visualized using a Zeiss LSM710 confocal microscope with a ×63 1.4 NA Plan-Apochromat lens. Images were acquired and processed using ZEN 2009 software (Carl Zeiss Microimaging) and ImageJ (NIH). To analyze cells incubated with LysoSensor or BODIPY-FL-pepstatin A, cells were cultured on a glass bottom chamber coverglass system (Lab-Tek) and directly observed by confocal microscopy. For live-cell imaging of lysosomal tubules, LAMP1-GFP–expressing stable lines cultured on glass bottom chambers were placed on a microscope stage equipped with an environmental control system (PeCon GmbH; TempModule S, CO2 Module S1, and Heating Unit XL S) at 37°C in 5% CO2, and images were acquired at 1-second intervals. Microsoft Excel 2007 was used to analyze statistical data and to draw graphs.
Immunoprecipitation and immunoblotting.
Immunoprecipitation and immunoblotting were conducted as described previously (40). Cells were washed with PBS and then lysed in lysis buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 5 mM EGTA [pH 8.0], 1.5 mM EDTA [pH 8.0], 1% Triton X-100, Complete Protease Inhibitor Cocktail [Roche], and PhosSTOP phosphatase inhibitor cocktail [Roche]) on ice for 30 minutes. Lysates were clarified by centrifugation (15,700 g, 4 °C) for 15 minutes, and supernatants were incubated with 0.5 μg of antibodies at 4°C for 16 hours, followed by incubation with Protein A/G PLUS-agarose (Santa Cruz Biotechnology) for 2 hours. Beads were washed 3 times with lysis buffer, and bound proteins were resolved by SDS-PAGE and then electrophoretically transferred onto a nitrocellulose membrane (Hybond ECL; GE Healthcare) for immunoblotting. Membranes were blocked with 5% nonfat milk in Tris-buffered saline with 0.05% Tween-20 (TBST) for 30 minutes and incubated with primary antibodies in 3% BSA/TBST at 4°C for 16 hours. Blots were then washed 3 times with TBST, incubated with horseradish peroxidase–conjugated secondary antibodies (Santa Cruz Biotechnology) for 1 hour, and then washed 3 times with TBST. The immunoreactive proteins were revealed by an Immun-Star WesternC Kit (Bio-Rad) or Amersham ECL Prime Western Blotting Detection Reagent (Amersham). Images were obtained and processed using a ChemiDoc XRS+ system with Image Lab software (Bio-Rad).
Chemicals and dyes.
The following chemicals were obtained commercially: LysoSensor Green DND-189 (L-7535, Molecular Probes); pepstatin A, BODIPY FL conjugate (P12271, Molecular Probes); chloroquine (C6628, Sigma-Aldrich); leupeptin (L2884, Sigma-Aldrich); bafilomycin A1 (196000, Calbiochem); DMSO (D8418, Sigma-Aldrich); and Dynasore (324410, Calbiochem).
Statistics.
Results are expressed as mean ± SD. Statistical analyses were performed using 1-way ANOVA, followed by Tukey’s and Bonferroni’s multiple comparison tests (GraphPad Prism 5), with the same results in all cases. P < 0.05 was considered significant.
Study approval.
Research on human subjects was performed under protocols approved by the NIH Combined Neuroscience Institutional Review Board. Written informed consent was obtained from all study participants or their guardians.
Supplementary Material
Acknowledgments
This work was supported by the Intramural Research Program of the National Institute of Neurological Disorders and Stroke, NIH.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2014;124(12):5249–5262. doi:10.1172/JCI77598.
References
- 1.Blackstone C, O’Kane CJ, Reid E. Hereditary spastic paraplegias: membrane traffic and the motor pathway. Nat Rev Neurosci. 2011;12(1):31–42. doi: 10.1038/nrn2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blackstone C. Cellular pathways of hereditary spastic paraplegia. Annu Rev Neurosci. 2012;35:25–47. doi: 10.1146/annurev-neuro-062111-150400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Finsterer J, Löscher W, Quasthoff S, Wanschitz J, Auer-Grumbach M, Stevanin G. Hereditary spastic paraplegias with autosomal dominant, recessive, X-linked, or maternal trait of inheritance. J Neurol Sci. 2012;318(1):1–18. doi: 10.1016/j.jns.2012.03.025. [DOI] [PubMed] [Google Scholar]
- 4.Novarino G, et al. Exome sequencing links corticospinal motor neuron disease to common neurodegenerative disorders. Science. 2014;343(6170):506–511. doi: 10.1126/science.1247363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fink JK. Hereditary spastic paraplegia: clinico-pathologic features and emerging molecular mechanisms. Acta Neuropathol. 2013;126(3):307–328. doi: 10.1007/s00401-013-1115-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goizet C, et al. SPG15 is the second most common cause of hereditary spastic paraplegia with thin corpus callosum. Neurology. 2009;14(14):1111–1119. doi: 10.1212/WNL.0b013e3181bacf59. [DOI] [PubMed] [Google Scholar]
- 7.SBabicki M, et al. A genome-scale DNA repair RNAi screen identifies SPG48 as a novel gene associated with hereditary spastic paraplegia. PLoS Biol. 2010;8(6):e1000408. doi: 10.1371/journal.pbio.1000408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hirst J, et al. Interaction between AP-5 and the hereditary spastic paraplegia proteins SPG11 and SPG15. Mol Biol Cell. 2013;24(16):2558–2569. doi: 10.1091/mbc.E13-03-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sagona AP, et al. PtdIns(3)P controls cytokinesis through KIF13A-mediated recruitment of FYVE-CENT to the midbody. Nat Cell Biol. 2010;12(4):362–371. doi: 10.1038/ncb2036. [DOI] [PubMed] [Google Scholar]
- 10.Khundadze M, et al. A hereditary spastic paraplegia mouse model supports a role of ZFYVE26/SPASTIZIN for the endolysosomal system. PLoS Genet. 2013;9(12):e1003988. doi: 10.1371/journal.pgen.1003988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murmu RP, et al. Cellular distribution and subcellular localization of spatacsin and spastizin, two proteins involved in hereditary spastic paraplegia. Mol Cell Neurosci. 2011;47(3):191–202. doi: 10.1016/j.mcn.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 12.Vantaggiato C, et al. Defective autophagy in spastizin mutated patients with hereditary spastic paraparesis type 15. Brain. 2013;136(pt 10):3119–3139. doi: 10.1093/brain/awt227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Renvoisé B, et al. Lysosomal abnormalities in hereditary spastic paraplegia types SPG15 and SPG11. Ann Clin Transl Neurol. 2014;1(6):379–389. doi: 10.1002/acn3.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirst J, et al. The fifth adaptor protein complex. PLoS Biol. 2011;9(10):e1001170. doi: 10.1371/journal.pbio.1001170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boya P, Reggiori F, Codogno P. Emerging regulation and functions of autophagy. Nat Cell Biol. 2013;15(7):713–720. doi: 10.1038/ncb2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Settembre C, Fraldi A, Medina DL, Ballabio A. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat Rev Mol Cell Biol. 2013;14(5):283–296. doi: 10.1038/nrm3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu L, et al. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature. 2010;465(7300):942–946. doi: 10.1038/nature09076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nixon RA. The role of autophagy in neurodegenerative disease. Nat Med. 2013;19(8):983–997. doi: 10.1038/nm.3232. [DOI] [PubMed] [Google Scholar]
- 19.Cortes CJ, La Spada AR. The many faces of autophagy dysfunction in Huntington’s disease: from mechanism to therapy. Drug Discov Today. 2014;19(7):963–971. doi: 10.1016/j.drudis.2014.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dehay B, et al. Lysosomal impairment in Parkinson’s disease. Mov Disord. 2013;28(6):725–732. doi: 10.1002/mds.25462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harris H, Rubinsztein DC. Control of autophagy as a therapy for neurodegenerative disease. Nat Rev Neurol. 2012;8(2):108–117. doi: 10.1038/nrneurol.2011.200. [DOI] [PubMed] [Google Scholar]
- 22.Kutateladze T, Overduin M. Structural mechanism of endosome docking by the FYVE domain. Science. 2001;291(5509):1793–1796. doi: 10.1126/science.291.5509.1793. [DOI] [PubMed] [Google Scholar]
- 23.Ktistakis NT, Tooze SA. PIPing on lysosome tubes. EMBO J. 2013;32(3):315–317. doi: 10.1038/emboj.2012.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noda T, Matsunaga K, Taguchi-Atarashi N, Yoshimori T. Regulation of membrane biogenesis in autophagy via PI3P dynamics. Semin Cell Dev Biol. 2010;21(7):671–676. doi: 10.1016/j.semcdb.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 25.Jung CH, Ro S-H, Cao J, Otto NM, Kim D-H. mTOR regulation of autophagy. FEBS Lett. 2010;584(7):1287–1295. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3(5):452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- 27.Luzio JP, Poupon V, Lindsay MR, Mullock BM, Piper RC, Pryor PR. Membrane dynamics and the biogenesis of lysosomes. Mol Membr Biol. 2003;20(2):141–154. doi: 10.1080/0968768031000089546. [DOI] [PubMed] [Google Scholar]
- 28.Peng J, et al. Atg5 regulates late endosome and lysosome biogenesis. Sci China Life Sci. 2014;57(1):59–68. doi: 10.1007/s11427-013-4588-8. [DOI] [PubMed] [Google Scholar]
- 29.Saftig P, Klumperman J. Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat Rev Mol Cell Biol. 2009;10(9):623–635. doi: 10.1038/nrm2745. [DOI] [PubMed] [Google Scholar]
- 30.Rong Y, et al. Spinster is required for autophagic lysosome reformation and mTOR reactivation following starvation. Proc Natl Acad Sci U S A. 2011;108(19):7826–7831. doi: 10.1073/pnas.1013800108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rong Y, et al. Clathrin and phosphatidylinositol-4,5-bisphosphate regulate autophagic lysosome reformation. Nat Cell Biol. 2012;14(9):924–934. doi: 10.1038/ncb2557. [DOI] [PubMed] [Google Scholar]
- 32.Chen Y, Yu L. Autophagic lysosome reformation. Exp Cell Res. 2013;319(2):142–146. doi: 10.1016/j.yexcr.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 33.Schulze RJ, et al. Lipid droplet breakdown requires Dynamin 2 for vesiculation of autolysosomal tubules in hepatocytes. J Cell Biol. 2013;203(2):315–326. doi: 10.1083/jcb.201306140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sridhar S, et al. The lipid kinase PI4KIIIβ preserves lysosomal identity. EMBO J. 2013;32(3):324–339. doi: 10.1038/emboj.2012.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pensato V, et al. Overlapping phenotypes in complex spastic paraplegias SPG11, SPG15, SPG35 and SPG48. Brain. 2014;137(pt 7):1907–1920. doi: 10.1093/brain/awu121. [DOI] [PubMed] [Google Scholar]
- 36.Hara T, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441(7095):885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 37.Komatsu M, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441(7095):880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 38.Zhu P-P, et al. Cellular localization, oligomerization, and membrane association of the hereditary spastic paraplegia 3A (SPG3A) protein atlastin. J Biol Chem. 2003;278(49):49063–49071. doi: 10.1074/jbc.M306702200. [DOI] [PubMed] [Google Scholar]
- 39.Lee S, Chang J, Renvoisé B, Tipirneni A, Yang S, Blackstone C. MITD1 is recruited to midbodies by ESCRT-III and participates in cytokinesis. Mol Biol Cell. 2012;23(22):4347–4361. doi: 10.1091/mbc.E12-04-0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chang J, Lee S, Blackstone C. Protrudin binds atlastins and endoplasmic reticulum-shaping proteins and regulates network formation. Proc Natl Acad Sci U S A. 2013;110(37):14954–14959. doi: 10.1073/pnas.1307391110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vanderver A, et al. Neurotransmitter abnormalities and response to supplementation in SPG11. Mol Genet Metab. 2012;107(1):229–233. doi: 10.1016/j.ymgme.2012.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.