Abstract
Maize has a long history of genetic and genomic tool development and is considered one of the most accessible higher plant systems. With a fully sequenced genome, a suite of cytogenetic tools, methods for both forward and reverse genetics, and characterized phenotype markers, maize is amenable to studying questions beyond plant biology. Major discoveries in the areas of transposons, imprinting, and chromosome biology came from work in maize. Moving forward in the post-genomic era, this classic model system will continue to be at the forefront of basic biological study. In this review, we outline the basics of working with maize and describe its rich genetic toolbox.
Keywords: genetics, genomics, maize, tools
Working with Maize
THE term maize is often used synonymously with corn, particularly in the United States and in reference to its agricultural use. While both terms are correct, maize is a name that refers uniquely to this plant. Maize is a large grain plant that evolved from its wild-grass ancestors by the direct intervention of human agriculture. Many varieties or “races” differ in physical properties (Goodman and Brown 1988), but generally maize is a single-stalk plant that grows to approximately 8 feet tall with about 20 long, narrow leaves growing individually from nodes along the stalk (Figure 1A) (Kiesselbach 1999). Several characteristics make it an attractive genetic system (Strable and Scanlon 2009). It is easy to culture on any scale, from a few plants in pots to many acres (Figure 1B). It can be grown successfully year round in greenhouses and growth chambers with proper lighting; it is also quite hardy and can be grown outdoors under a range of conditions, from tropical to temperate climates (Shaw 1988). Maize is a naturally outcrossing species, which makes its genetic architecture (diversity, linkage, recombination, etc.) more similar to other outcrossing organisms such as humans rather than self-pollinating plants (Rafalski and Morgante 2004; Wallace et al. 2013). While its genetics are similar to humans, maize retains the major strength of plant genetics: the ability to self-cross and quickly produce homozygotes or F2 populations.
Figure 1.
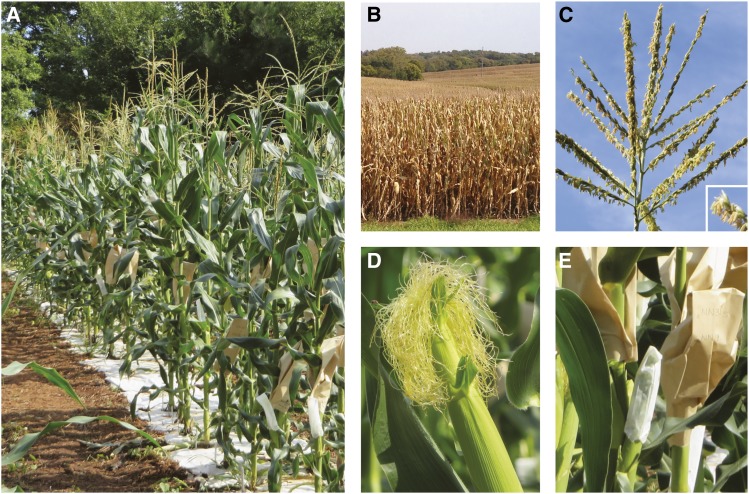
(A) The maize plant (Zea mays ssp. mays). Maize is generally grown in local fields in the summer and either in greenhouses or tropical outdoor locations in the winter. (B) Maize fields ready for harvest. (C) The male reproductive organs are located in the tassel; pollen released from the tassel bears the sperm. The inset shows a closeup of florets splitting open to reveal yellow anthers, the structure that stores and releases pollen. (D) The young ear produces female structures called silks, which receive pollen. The pollen germinates and grows down the silk to the ovule which develops into a kernel. (E) After performing a cross, the fertilized ear is covered by a brown bag to prevent contamination by other pollen; the bag is marked with relevant parent lineages and date of the cross. The small white bags protect developing ears from pollen prior to crossing. Images in A, D, and E are courtesy of Carolina Chavarro, and B is courtesy of Bill and Connie Funk.
The male and female reproductive organs are accessible and separable, making controlled crosses easy to perform. The male germ cells are produced in the tassel found at the top of the plant (Figure 1C). Tassels contain anthers that open upon maturation, releasing up to 107 wind-dispersed pollen grains (Coe et al. 1988). The female germ cells are located in one or more ears, which grow from the base of leaves in the midsection of the plant (Figure 1, A and D). An ear generally contains several hundred egg cells that will develop into kernels after fertilization (Neuffer et al. 1997). Each young kernel contains a silk, an elongated stigma, which emerges out of the husk leaves of the ear (Figure 1D). Pollen grains land on the silk and produce a pollen tube that grows down through the length of the silk, ultimately delivering two sperm to the female gametophyte (Figure 2A). Double fertilization occurs as one sperm fertilizes the egg to create the embryo (2n), and the other sperm fertilizes the central cell with two polar nuclei that gives rise to the aleurone and the starchy endosperm (3n). The aleurone and endosperm nourish the young embryo during germination (Figure 2, D and E, and Figure 4A) (Dresselhaus et al. 2011).
Figure 2.
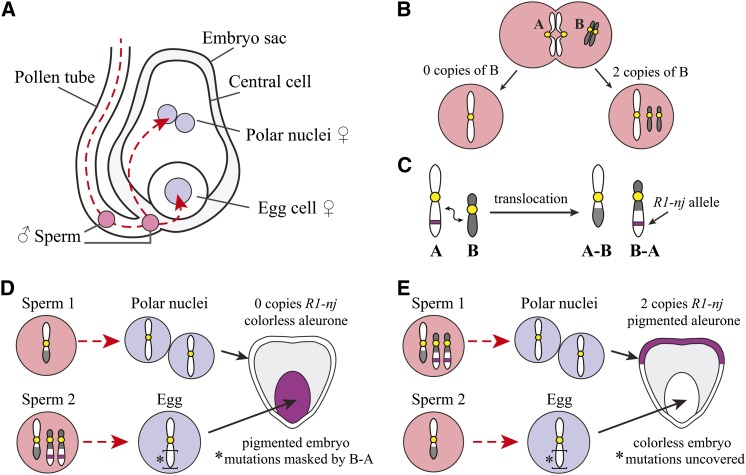
Double fertilization and chromosome tools in maize gametogenesis. (A) The female gametophyte consists of an embryo sac that contains an egg cell (1n) and a central cell with two polar nuclei (1n + 1n). When a pollen grain lands on a silk, it produces a pollen tube that grows down to the embryo sac and fuses with it to deliver two sperm. The two sperm perform double fertilization. One sperm fertilizes the egg cell to create an embryo (2n) and the other fertilizes the central cell to produce the endosperm and aleurone (3n). (B) Two sperm are produced by the second pollen mitosis. The A chromosomes segregate correctly with each sperm receiving one copy, but B chromosomes typically nondisjoin, which results in one sperm receiving two copies and the other receiving no copies. (C) Translocation events that occur between A and B chromosomes yield an A–B chromosome and a B–A chromosome. The B–A chromosome is segregated via the B centromere that causes the nondisjunction behavior. An allele found on the A chromosome segment of a B–A translocation, such as R1-nj, which pigments the embryo and aleurone, can be used as a marker of nondisjunction. (D) Translocations with the B chromosome can be used to generate hypoploids, where only one copy of a chromosome region is present in an embryo. This allows recessive mutations to be uncovered. If the sperm bearing the two copies of the B–A chromosome (sperm 2) and thus the R1-nj alleles fuse with the egg, a maternal recessive mutation will be masked. The colorless aleurone but pigmented embryo reveals that the B–A bearing sperm fertilized the egg. E) If the kernel has a pigmented aleurone and a colorless embryo, the sperm carrying the B–A chromosomes (sperm 1) fused with the polar nuclei. The colorless embryo has only the maternal copy of the A chromosome arm because sperm 2 lacks this region. If the maternal chromosome arm is carrying a recessive mutation, it will be uncovered and the embryo (or resulting plant) will display the associated phenotype.
Figure 4.
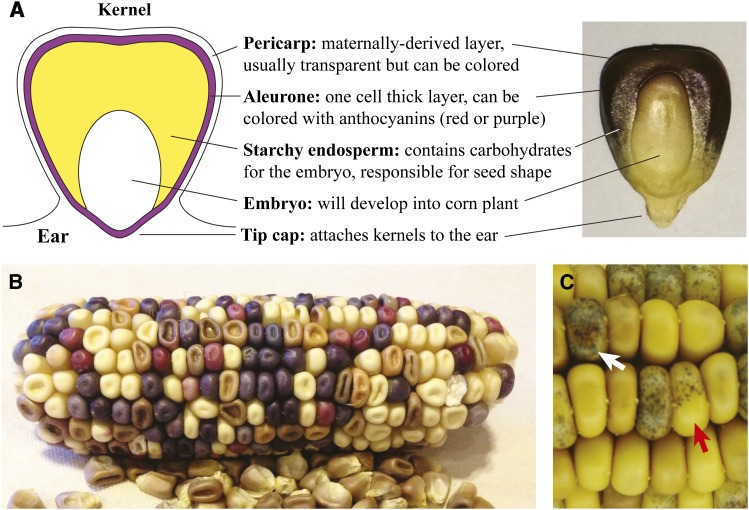
Kernel biology and phenotype markers. (A) The maize kernel is an excellent platform on which to track trait segregation and mutagenesis due to collections of pigmentation and shape markers. Both the pericarp and the aleurone layers can reveal transposon activity, chromosome breaks, or other mutagenesis as colored sectors. The endosperm provides kernel structure; different shape phenotypes like shrunken or wrinkled can be used to identify mutants. (B) A self-crossed ear segregating many different kernel traits. The parent of this plant was y1/y1 Pr1/pr1 R1/r1 C1/c1 Bz1/bz1 Sh1/sh1 and Wx1/wx1. Easily seen are shrunken endosperm kernels (sh1/sh1), red aleurone kernels (pr1/pr1), bronze aleurone kernels (bz1/bz1), and colorless aleurone kernels (either r1/r1 or c1/c1). Plump, purple kernels are wild type. (C) An ear grown by Barbara McClintock showing transposition of Ds from the a1-m3 locus in the aleurone to give purple sectors (sector is A1, marked by white arrow). Ds activity is driven by Ac in the wx1-m7 locus. The red arrow shows a sector in which Ac jumped away from wx1-m7 (sector is Wx1, glassy appearance) rendering a1-m3 somatically stable, and thus there are no purple sectors within this region. Image in B is courtesy of Elizabeth Lee and Jeff Ross-Ibarra.
Controlled crosses are made by placing a bag over the tassel and shaking gently to collect the pollen, which is then sprinkled onto silks (Neuffer 1994a; Neuffer 1994b). The ears are covered before silks emerge to prevent contamination (Figure 1E). Crosses can be made over a large time window, as a normal tassel will produce new pollen for up to 7 days and silks can be receptive to fertilization for 10 days. Ear development can also be hastened by removing tassels, and pollen shedding can be delayed by cool temperatures or accelerated by warm temperatures (Coe et al. 1988). Although pollen is short lived, it can be kept viable by refrigeration for 24 hr and by liquid nitrogen freezing for up to 1 year (Barnabas and Rajki 1976, 1981). The physiology of maize makes it an excellent system for large-scale progeny screens. The vast amounts of pollen produced by a single plant allows many crosses to be performed; one tassel can produce enough pollen to fertilize 50+ ears in a single day. Experienced maize researchers can perform 300–500 crosses in a day, and with each cross yielding several hundred seeds, maize can quickly generate large numbers of offspring for genetic analysis (Neuffer 1994a). Progeny output can also be amplified many fold by growing plants in isolated plots and removing tassels from female parents with alternating rows of the male parent.
Despite the ease of growing and crossing maize, there are some drawbacks to working with this model system. Maize has a relatively long life cycle compared to fast-growing single-cell systems. Maize can be crossed ∼60 days after planting and requires another 30–45 days post-pollination for seed maturation. This 13-week generational time puts maize on par with zebrafish (12–14 weeks) (Ulloa et al. 2011) and mice (11–12 weeks) (Hartwell et al. 2011), but significantly longer than the prominent plant model system, Arabidopsis thaliana (6–8 weeks) (Meyerowitz and Pruitt 1985). While maize can grow in a broad range of climates and in greenhouses, it is not amenable to growth in small chambers due to its size and high clearance required for growth (Neuffer 1994a). To facilitate quicker generation, the laboratory of James Birchler developed a fast flowering “mini-maize” that can go from seed to seed in 60 days that is available from Maize Genetics Stock Center (see below for more information on the Stock Center).
Online Data and Germplasm Resources
The Maize Genetic and Genomics Database (MaizeGDB) (http://www.maizegdb.org) (Lawrence et al. 2008) is the online home for maize researchers. It contains maize community news, genetic maps, information on mutations and alleles, and direct access to requesting seeds for genetic stocks. It also contains a well-curated and annotated genome browser complete with Mu and Ac/Ds insertional mutations (see below) and multiple expression analysis tools. It contains listings of many refereed publications on maize, including those published in the Maize Genetics Cooperation Newsletter (http://mnl.maizegdb.org/), an annual publication of informal communication about ongoing work in maize laboratories around the world. Contact information for researchers who have attended the Annual Maize Genetics Conference (http://www.maizegdb.org/maize_meeting/) as well as abstracts and refereed papers associated with those scientists are also listed. Maize is one of several major cereal crops with advanced sequencing and mapping resources, and comparative mapping can be extremely useful when interpreting physical maps, mutant phenotypes, and quantitative traits. Gramene (http://www.gramene.org) (Monaco et al. 2014) is an online resource dedicated to such genome-wide comparisons.
Obtaining seeds to initiate research projects is simple and free. MaizeGDB provides interlinked access to the Maize Genetics Cooperation Stock Center (http://maizecoop.cropsci.uiuc.edu/), which houses the great wealth of classical and modern genetics resources. The majority of mutants and chromosome variants described in this review can be obtained at the Stock Center. Searching for seed stocks begins at MaizeGDB (http://www.maizegdb.org/stock.php), where an online request form is created and forwarded to the Stock Center, which will mail the seeds to your address. Maize breeding and diversity germplasm resources, such as inbreds and landraces that are not available at the Stock Center, can be obtained from Germplasm Resources Information Network (GRIN; http://www.ars-grin.gov/) and International Maize and Wheat Improvement Center (CIMMYT, http://www.cimmyt.org/). A comprehensive list of the online resources discussed in this review can be found in the Supporting Information section.
Chromosomes and Mutant Collections
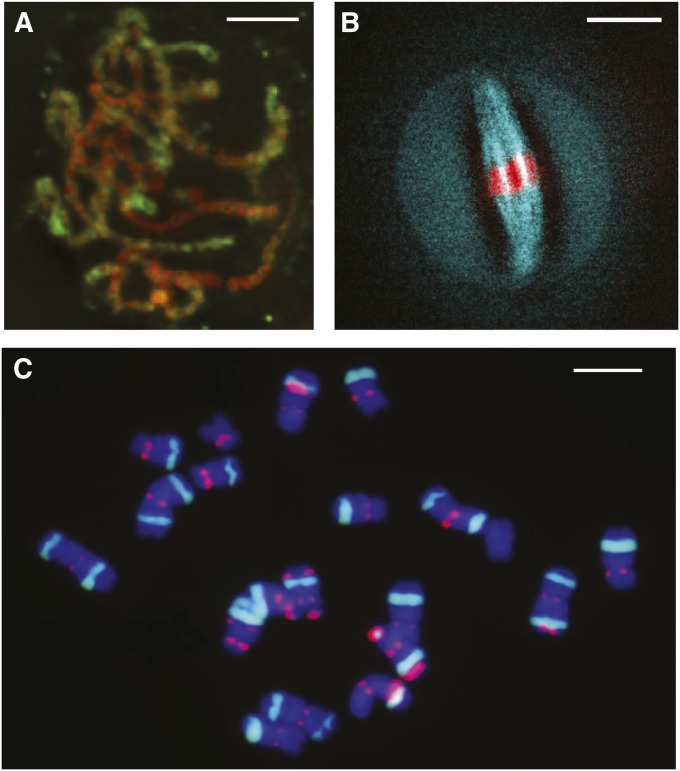
Maize was noted early on for its large and easily interpreted meiotic chromosomes. A famous group at Cornell led by Rollins A. Emerson, including Barbara McClintock, George Beadle, Charles Burnham, and Marcus Rhoades, used these chromosomes as their visual aids for studying genes. They developed cytogenetic tools that include long lists of meiotic mutants, variations in chromosome type and number, and ultimately over a thousand translocation lines (Anderson 1956). Mutants affecting all stages of meiosis, ranging from the initiation to the regulation of chromosome pairing and control of the second division, have been carefully described and many of the causal genes have been cloned (Cande et al. 2009). Modern microscopy has been employed to expand the utility of meiotic chromosomes to include immunolocalization (Figure 3A) (Shi and Dawe 2006) and powerful fluorescent in situ hybridization methods (FISH; Figure 3C) (Kato et al. 2004) have been developed to quickly identify both mitotic and meiotic chromosomes (Kato et al. 2011). These FISH methods are sufficiently powerful to rapidly localize any form of unique sequence, including transgenes, to chromosome arms (Lamb et al. 2007).
Figure 3.
Maize cytogenetics. (A) Meiotic chromosomes can be harvested from immature tassels and visualized via immunostaining. Pachytene chromosomes are labeled with two antibodies, one highlighting heterochromatin (H3K27me2, red) and the other euchromatin (H3K4me2, green). Scale bar, 5 µm. (B) Fluorescent lines are available for a range of tagged proteins; shown here is a meiosis I cell with both tubulin-CFP (courtesy of Anne Sylvester) and histone H2B-mCherry (courtesy of Hank Bass). Scale bar, 15 µm. (C) Nearly all of the 10 maize chromosomes can be differentiated from each other by simple two-color FISH. Chromosome spreads are typically performed on mitotic (diploid) cells from root tips, as shown here. Chromosome size, arm ratio, intensity of CentC staining (centromere repeat, green dots in the middle of chromosomes), and the location and type of knobs (green and red regions located toward chromosome ends) are key identifying features. Scale bar, 10 µm.
As a large plant with distinctive parts and features, maize is ready made for identifying mutants. Although there are mutations affecting nearly every organ and many cell types, corn kernels are particularly well studied. Not only do they express colors in the maternal pericarp tissue surrounding kernels, they express purple or red anthocyanin in the aleurone, the outer layer of the endosperm (Figure 4A). The starchy endosperm within the kernel contains complex carbohydrates and proteins that affect the shape and texture of the kernels. With a brief tutorial or online resources (http://mutants.maizegdb.org/doku.php), new users can discern at least a dozen common phenotypes involving pigmentation and kernel shape (Figure 4B). The hundreds of progeny kernels per cross are conveniently held on the ear, which can be rapidly assayed for chromosome segregation defects and mutant characteristics. Further, a corn kernel demonstrates mitotic mutations and chromosome loss in striking visual fashion. Barbara McClintock famously used color traits to great advantage when describing the breakage–fusion–bridge cycle (McClintock 1938b, 1939, 1941) that led to the discovery of transposable elements (McClintock 1948, 1949). She further used the visual power of maize kernel pigmentation to demonstrate that transposons jump in and out of genes (McClintock 1954) (Figure 4C).
Mutants that affect plant color and organelle function are also important tools in maize genetics. There are hundreds of genes in the nuclear genome that control chloroplast biogenesis or function, and mutants in these genes manifest as albino, pale, or yellow-leaf phenotypes. Nearly all photosynthetic mutants are inherited as simple recessives and the homozygotes die as seedlings. Saturation-level mutant screens for chloroplast phenotypes have been performed (Stern et al. 2004), and >80 of the corresponding genes have been identified (for list, see http://pml.uoregon.edu/photosyntheticml.html). These include genes that control chloroplast transcription, splicing, and protein translation, as well as those involved in electron transport and thylakoid membrane structure. Nuclear mutations that affect mitochrondria are rare; however, there are several maternally inherited mitochondrial mutations that cause failure to produce male gametes (Hanson 1991). These cytoplasmic male sterile mutants (cms) were heavily used in the 1950–’60s to facilitate hybrid corn production but an epidemic in 1969–‘70 revealed that certain cms lines were susceptible to the fungal disease Southern corn leaf blight (Levings 1990). Due to continued concerns about disease susceptibility, use of these lines has greatly diminished. The cms mutations are caused by deletions or rearrangements of the mitochrondrial genome and cause phenotypes only in some backgrounds (Schnable and Wise 1998).
There are hundreds of chromosome variants and unusual introgression lines available in maize. These include a full trisomic series (Birchler 1994), stocks that contain chromosomes from the related wild grass species Tripsacum (Leblanc et al. 2009), and oat varieties that contain single maize chromosomes (called oat-maize addition lines) (Rines et al. 2009). These materials have primarily been used in cytological studies for chromosome identification (Koumbaris and Bass 2003; Jin et al. 2004) but are also potential sources of new genetic variation. The most heavily used chromosomal variant is the B chromosome and its derivatives (Carlson 1978). The B chromosome differs from the normal (A) chromosome set by being supernumerary, lacking known genes, and displaying an accumulation mechanism when crossed as the male. The two sperm cells found in each pollen grain are generated by a mitotic event (second pollen mitosis). The B chromosome generally nondisjoins in this mitotic division (Figure 2B) such that two copies of the chromosome are delivered to either the egg or the central cell (Figure 2, D and E). Segments of A chromosomes that are linked by translocation to the B centromere will also show this property (Figure 2C). Many different B–A translocations are available, and these have been used to rapidly map genes (Beckett 1994) and manipulate gene dosage (Sheridan and Auger 2008; Brunelle and Sheridan 2014). Figure 2 illustrates how this tool can be used with a color marker on chromosome arm 10L called R1-nj, which pigments both the embryo and aleurone. The special properties of B chromosomes are also being developed as artificial chromosome platforms to introduce transgenes (Birchler et al. 2010). A truncated version of the B chromosome has been engineered with lox recombination sites so that transgenes can be inserted and manipulated without affecting the A chromosomes (Yu et al. 2007).
In addition to these traditional genetic resources, there is a large collection of fluorescent protein marker lines (Mohanty et al. 2009; Wu et al. 2013). Among these is a suite of cell biological markers (tubulin, chromosomes, ER, cell wall, etc.) as well as many developmentally regulated genes such as ramosa1 (discussed below). At least 105 such lines have been characterized, labeled with GFP, CFP, and RFP derivatives and can be obtained through the Maize Cell Genomics database (http://maize.jcvi.org/cellgenomics/index.php). These lines are being used in live-cell studies of cell division (Figure 3B) and, for example, to interpret the expression of a gene that suppresses branching (tillering) at the base of the plant thereby promoting the single-stalk growth pattern typical of agricultural corn (Whipple et al. 2011). Two-component trans-activating fluorescent lines have also been generated for tissue-specific control of gene expression (Wu et al. 2013).
Genomics, Genetic Diversity, and Quantitative Genetics
Maize genome sequence and annotation
Maize has a large, transposon-rich genome that is roughly the size of the human genome at 2.5 Gb. A full maize genome sequence of the B73 inbred was assembled by a BAC-by-BAC Sanger-based method (Schnable et al. 2009) (Figure 5) and has since been updated such that the current draft is reference version 3 (http://www.maizegdb.org). The order of genes is largely correct because of the integration of excellent recombination maps (McMullen et al. 2009; Ganal et al. 2011) (http://www.maizegdb.org/map.php). There are ∼40,000 annotated protein-coding genes (Law et al. 2015) and extensive RNA-seq resources have enabled accurate gene model prediction (Li et al. 2010; Campbell et al. 2014). NCBI is actively annotating genes in the version 3 maize genome with comprehensive details of gene structure, neighboring loci, expression, and relatedness to other model species (http://www.ncbi.nlm.nih.gov/genome/?term=maize). Most of the sequences from intergenic spaces are also present in the assembly, including two centromeres (Wolfgruber et al. 2009), but these regions are transposon rich (Baucom et al. 2009) and contain gaps and assembly errors. Retrotransposons are the most common type of transposon between genes, and they are difficult to assemble accurately because they insert into each other in nested arrangements. The cut-and-paste, DNA-type transposons studied by McClintock are generally found at the interface of the retroelement-rich intergenic spaces and genes (Han et al. 2013). The most abundant DNA elements have been reduced in size to only a few hundred bases and are referred to as miniature inverted-repeat elements (MITEs) (Feschotte et al. 2002). Helitrons are another type of DNA transposon that can cause the movement of large pieces of DNA by a rolling circle mechanism (Morgante et al. 2005).
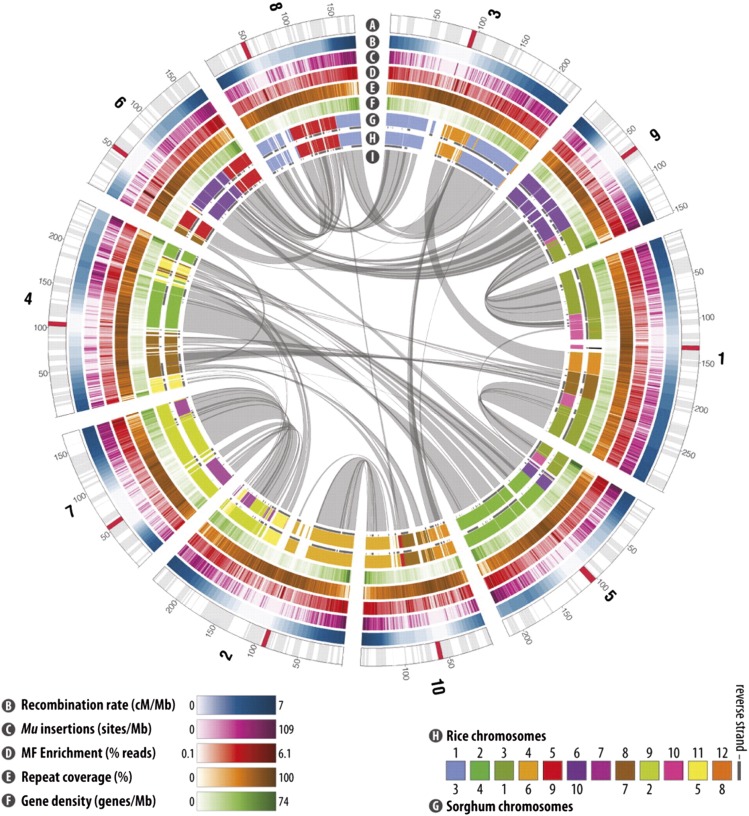
Figure 5.
The B73 maize reference genome, from Schnable et al. (2009), reprinted with permission from AAAS (license number 3525420931165). Circles represent aspects of the genome: (A) chromosome structure with centromeres indicated in red, (B) recombination rates, (C) Locations of 40,000 UniformMu insertions, (D) methyl-filtration enrichment showing genetically active regions, (E) repeat coverage showing transposon enrichment in pericentromeric regions, (F) gene density, (G) synteny with sorghum, a close relative of maize, and (H) synteny with rice a more distant relative. The gray lines tracing through the middle indicate regions of homology within maize that are remnants of the last tetraploidization event.
The evolutionary history of flowering plants includes multiple rounds of tetraploidization followed by genome shrinkage (fractionation) and rediploidization (Doyle et al. 2008). The last tetraploidization event in the maize lineage occurred between ∼4.8 and 11.9 million years ago, and while maize behaves as a simple diploid, large portions of the genome are effectively tetraploid (Swigonova et al. 2004). Sequence comparisons within the genome reveal fascinating patterns of duplication (Schnable et al. 2009) that can predict areas of genetic redundancy (Schnable and Freeling 2011) (Figure 5). Maize is also closely related to other major cereal grains such as rice, wheat, and sorghum, and clear patterns of genome colinearity are evident (Figure 5). Comparative mapping among the cereal species can be highly informative for identifying candidate genes and confirming their general importance in multiple species (Gale and Devos 1998) (http://www.gramene.org).
There is also a growing body of information on maize epigenetics. Extensive DNA methylation and histone profiling maps are available (Wang et al. 2009b; Regulski et al. 2013; Gent et al. 2014). In addition, mutant screens based on an epigenetic phenomenon known as paramutation (where a mutant allele can heritably silence another) have identified a suite of genes that control epigenetic states including genes required for RNA-dependent DNA methylation (Hollick 2012). The centromeres of maize and other species are also epigenetically defined, and several studies have demonstrated that maize centromeres can be inactivated and reactivated and can move positions (Han et al. 2009; Fu et al. 2013).
Genetic diversity
Beyond the array of induced and characterized mutations in cultivated maize, which number well into the thousands, there is great natural diversity among maize races and lines. Maize was domesticated from a wild grass species known as teosinte that is endemic to Mexico. Roughly five major loci account for the very dramatic morphological differences between teosinte and modern maize (Doebley and Stec 1993). Unique forms of maize were developed in hundreds of locations by Native Americans (Goodman and Brown 1988) and the majority of these were carefully collected and cataloged early in the 20th century. These lines, along with other varieties, populations, and inbred lines from around the world, are bred to maintain their diversity and are available free of charge from the National Plant Germplasm System via GRIN and CIMMYT. The advent of high-throughput sequencing has ushered in an era of maize genetics that seeks to identify and catalog this diversity information for use by geneticists and breeders. High-density SNP maps for 103 inbreds (called Hapmap2) (Chia et al. 2012), and additional SNP data for > 2000 more inbreds are available (Romay et al. 2013). Thousands of other accessions and lines are currently being genotyped (http://www.panzea.org).
Inbreds and ploidy
Maize is an outcrossing species. Purely homozygous inbreds such as B73 are specially derived lines that have been selected to maintain reasonable vigor while performing well in commercial hybrids. Most hybrids display a dramatic phenomenon known as heterosis (“hybrid vigor”), which is a level of productivity that exceeds that of either parent. Most of the hundreds of inbreds currently available were developed by continued self-crossing, which can take several years. However, modern homozygous lines are generally created in two generations using stocks called haploid inducers (Prigge et al. 2012). When a haploid inducer is crossed as a male to any other maize line, a high percentage of the progeny are haploids derived from the female. These haploids are subjected to chemical treatments that double the genome and produce homozygous progeny. Paternally derived haploids can be produced via the indeterminate gametophyte1 mutation that affects female gametophyte development (Kermicle 1969). Other techniques (Randolph 1941; Kato and Birchler 2006; Yao et al. 2011) or meiotic mutants (elongate1) can be used to create full ploidy dosage series ranging from 1n to 8n (Rhoades and Dempsey 1966; Guo et al. 1996).
Quantitative genetics
Quantitative trait locus (QTL) mapping is a classical means by which to identify loci underlying complex traits. When multiple genes contribute to a phenotype, QTL mapping can estimate the location of the contributing genes and their relative contribution to the phenotype. Some of the earliest methodologies for QTL mapping were developed using maize as a model (Stuber et al. 1992) and many studies mapping QTL for agronomic traits have been published. For instance, five QTL were identified as contributing to aluminum toxicity on acidic soils, which is a major limitation to growing maize in many parts of the world (Ninamango-Cárdenas et al. 2003). QTL mapping was also used in classic studies defining the major loci responsible for maize domestication (Doebley and Stec 1993), including the teosinte branched1 (tb1) gene (Doebley et al. 2006). The tb1 mutation is required to propagate maize as a crop plant and was subjected to intense selection by humans in the early stages of domestication, resulting in a selective sweep that dramatically reduced local nucleotide variation (Purugganan and Fuller 2009).
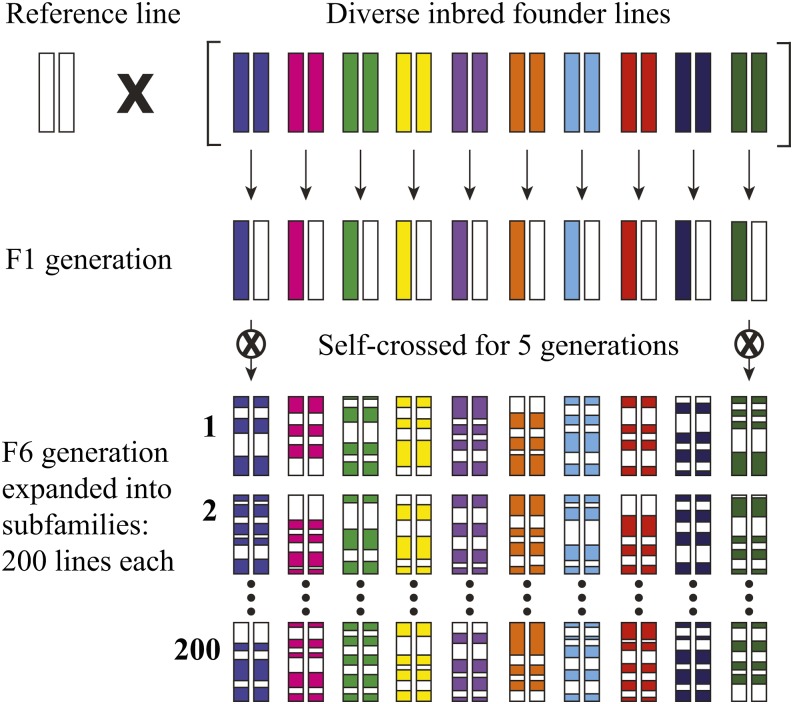
Other more advanced tools to identify genes associated with quantitative traits have also recently gained wide usage. Buckler and colleagues created a set of 25 breeding populations known as the Nested Association Mapping (NAM) panel (McMullen et al. 2009) that samples the diversity of maize. The sequenced inbred reference line (B73) was crossed to a set of diverse inbred founder lines derived from various natural domesticated lines. The 25 hybrids where then repeatedly self-crossed to generate >5000 recombinant inbred lines that represent much of the diversity in maize (Figure 6). The strength of the NAM method is that it combines the power of linkage mapping with the resolution of association mapping. Linkage data are obtained directly from the individual mapping populations, and sequence comparisons among the 25 populations can make it possible to identify gene candidates. Although the outcome is limited by the allelic diversity within the starting materials, the method has extraordinary power to resolve complex traits such as flowering time (Buckler et al. 2009) and resistance to Southern corn leaf blight (Kump et al. 2011). This and other genetic diversity information can be accessed through the Panzea website (http://www.panzea.org).
Figure 6.
Nested association mapping maize lines. The inbred reference maize line (B73) whose genome has been sequenced and assembled (Schnable et al. 2009) was crossed to 25 diverse inbred maize lines from around the world, including popcorn, sweet corn, tropical indigenous corn, and elite agricultural corn (Mcmullen et al. 2009). For simplicity, only 10 founder lines are shown in the figure. The hybrids were first self-crossed to allow recombination to occur between B73 and the other inbred. From these F2 ears, at least 200 progeny were self-crossed by single seed decent for an additional four generations to create recombinant inbred lines for linkage mapping.
Transgenics
Transformation of maize to produce stable, fertile transgenic lines is an essential technology for both basic biological studies and for engineering improved crop lines. Since their commercial release in 1996, the use of genetically engineered maize has grown to dominate US corn production. The primary methods are Agrobacterium-mediated transformation and biolistic transformation. Protocols for both methods (Frame et al. 2000; Frame et al. 2002; Zhao et al. 2002; Sidorov and Duncan 2009; Lee and Zhang 2014) are available, but many researchers take advantage of transformation services offered by public universities such as the University of Missouri (http://www.plantsci.missouri.edu/muptcf/), the University of Nebraska—Lincoln (http://www.biotech.unl.edu/plant-transformation), and Iowa State University (http://agron-www.agron.iastate.edu/ptf/).
Agrobacterium-mediated transformation
Agrobacterium tumefaciens is a soil pathogen that inserts a segment of its DNA (T-DNA) located on a tumor-inducing plasmid (Ti plasmid) into the chromosomes of plants (Lacroix and Citovsky 2013). Because Agrobacterium does not naturally infect maize, parameters such as recipient plant tissue and genotype, transfer vector, Agrobacterium strain, and culture conditions have only a narrow range for successful transformation (Wang et al. 2009a). Only a few genotypes can be transformed, including a subset of elite commercial lines (Cho et al. 2014). Agrobacterium-mediated methods use similar tissue (immature embryos) as biolistic methods, and both have similar transformation rates (5–40%) (Wang et al. 2009a). However, Agrobacterium has a distinct advantage with transgene integration. It yields a high frequency of transformations with one or a few transgene integrations per genome; these transgenes are also rarely rearranged and tend to show more stable expression (Shou et al. 2004). Agrobacterium-mediated transformation requires that transgenes be cloned into specific transfer vectors, which are large low-copy plasmids.
Biolistic transformation
Biolistic transformation uses fine particles coated with DNA to directly deliver transgenes into plant tissue (Gordon-Kamm et al. 1990). It has been successful in a range of genotypes, with DNA in various forms (plasmids, linear molecules, PCR products), and can be used for stable transformation (Wang et al. 2009a) as well as for testing constructs in somatic cells (Du et al. 2010; Kirienko et al. 2012). In addition, multiple transgenes can be introduced at once by biolistic transformation, and the size of the constructs is not a major limiting factor. A synthetic sequence array of 1100 kb was recovered in stable transgenic lines following biolistic tranformation (Zhang et al. 2012). Although it has clear advantages for some applications, biolistic transformation is not as heavily used as T-DNA transformation because it also yields high incidences of multiple transgene insertions per genome (>90% of transformations), transgene structural rearrangements, and can result in unstable, low or silenced transgene expression (Pawlowski and Somers 1996, 1998).
Forward Genetics
Chemical mutagenesis
Maize is highly amenable to forward genetic screens, and many methods exist including chemical, radiation, and transposon-based mutagenesis. Ethyl methanesulfonate (EMS) is the most efficient and widely used chemical mutagen; it alkylates guanine, causing a G-to-A transition point mutation that results in both dominant and recessive mutants. Pollen is mixed with an EMS/paraffin oil emulsion, and the mutagenized pollen is painted onto silks (Neuffer 1994b; Neuffer et al. 1997). First-generation offspring (M1) are then self-crossed to reveal recessive mutations in the second generation (M2). EMS-based mutagenesis has yielded many important maize mutants that have advanced the understanding of plant architecture (Freeling and Hake 1985; Kerstetter et al. 1997; Gallavotti et al. 2010), meiosis (Golubovskaya et al. 2003), kernel and embryo development (Neuffer and Sheridan 1980; Neuffer et al. 1986), photosynthesis (Hopkins et al. 1980; Miles 1982), and disease symptoms (Neuffer and Calvert 1975; Hoisington et al. 1982; Neuffer et al. 1986). EMS-mutagenesis has also been used in a reverse genetics strategy called TILLING, which involves pooling mutagenized lines and screening for mutants in specific genes using enzymes that detect sequence mismatches (Weil and Monde 2007). These methods are currently being improved to incorporate high-throughput sequencing (Weil and Monde 2009).
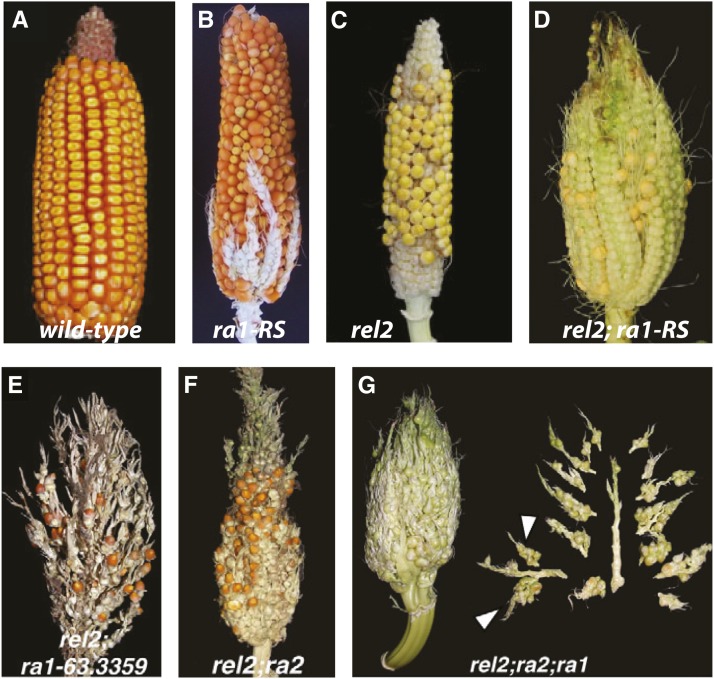
EMS-induced mutagenesis is a particularly useful approach when screening for enhancers or suppressors of a mutant phenotype because it can be performed in any genetic background. For example, in a recent study on the ramosa pathway, EMS mutagenesis was used to find enhancers of the weak ramosa1 phenotype (ra1-RS) (Gallavotti et al. 2010) (Figure 7, A and B). The ramosa pathway regulates the differentiation of specific plant stem cell populations into reproductive organs. By EMS treating ra1-RS homozygous pollen, Gallavotti and colleagues identified ramosa1 enhancer locus2 (rel2); the double mutant gives a more severe phenotype than either single mutant (Figure 7, B–D). The rel2 mutant also enhanced branching in other ra1 alleles (Figure 7E), other members of the pathway (ra2) (Figure 7F), and triple mutants (Figure 7G). Rel2 was identified as a transcriptional corepressor of the indeterminant branching pathway and important in enforcing the differentiated fate of reproductive maize organs (Gallavotti et al. 2010). The work is both interesting and important, as tassel branch number is correlated with maize yield.
Figure 7.
An EMS screen was used by Gallavotti et al. (2010) to find enhancers of the ramosa1 gene. (A) Wild-type (B) ra1-RS: the screen was performed in the ra1-RS mutant background, a weak ramosa1 allele (C) rel2: the screen identified rel2 (ramosa1 enhancer locus2) (D) rel2; ra1-RS: the double mutant phenotype is stronger than either individual mutant (E) rel2; ra1-63.3359 (a second allele of ra1) (F) rel2; ra2 (a mutation in another gene that causes the ramosa phenotype) (G) rel2; ra2; ra1 ear. The data show that rel2 mutations enhance the branching phenotype in various ramosa backgrounds. All images are reproduced with permission from Gallavotti et al. (2010) ©The Company of Biologists, license to reprint ID 3478410008188.
Radiation mutagenesis
The use of radiation as a mutagen has a long history in maize. Loss of dominant kernel phenotypes was one of the first indications that ionizing radiation (X and γ rays) can lead to gross phenotypic alterations and embryo lethality (Stadler 1928, 1930). Cytological studies by Barbara McClintock revealed that these phenotypes were the result of large scale chromosomal aberrations such as deletions, breaks, translocations, and initiation of breakage–fusion cycles that lead to ring chromosomes, dicentrics, and other unstable chromosome features (McClintock 1931, 1932, 1938a, 1939; Rhoades and McClintock 1935; Stadler and Roman 1948). Next-generation sequencing makes it possible to study such events much more efficiently, as illustrated by a recent screen for enhancers of the opaque2 gene where exon capture and RNA-seq were used to identify novel deletion mutants (Yuan et al. 2014).
Ionizing radiation has also been used to generate mutants in somatic tissues to study cell lineage, cell autonomy, and lethal mutations. While chromosome breaks can be highly damaging to an organism generated from irradiated gametes, such lesions can be very useful in somatic tissue. Irradiation of somatic cells at specific developmental times allows researchers to study mutations that would normally be lethal (Fu and Scanlon 2004), mark cells for lineage studies (McDaniel and Poethig 1988), investigate cell autonomous/nonautonomous gene functions (Becraft et al. 2001; Frank et al. 2003), and map genes through chromosomal deletions (Kynast and Riera-Lizarazu 2011). Researchers can take advantage of pigmentation mutants to mark sectors that have lost chromosome arms by radiation-induced breakage. The two following examples made use of photosynthetic mutants, which show as white streaks after irradiation.
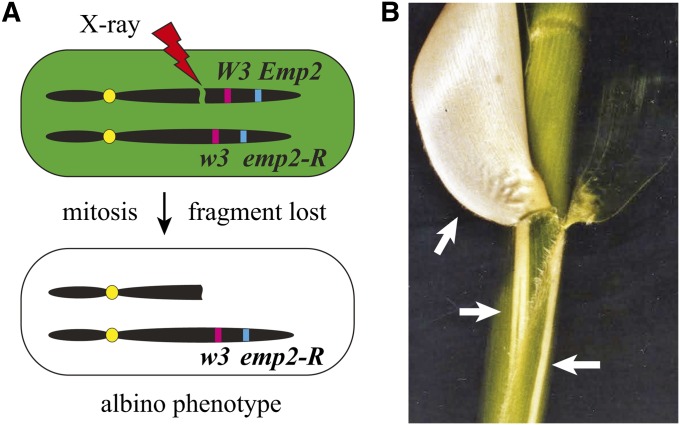
Fu and Scanlon (2004) used γ irradiation to delete the Empty Pericarp2 (Emp2) gene by breaking off the distal arm of chromosome 2 where the gene resides (Figure 8A). Mutants of this gene (emp2-R) are embryonic lethal, making later developmental study impossible. They therefore irradiated heterozygous seedlings (Emp2/emp2-R) to break off the wild-type allele and expose the mutant allele. The emp2-R mutant was linked to an albino mutant (w3) such that the loss of Emp2 could be seen as white somatic sectors (Figure 8B). Using this approach, they were able to dissect different functions for Emp2 throughout development. A similar approach was used to determine whether the Crinkly4 (Cr4) receptor-like kinase gene functions cell autonomously or if it transmits developmental cues to neighboring cells (Becraft et al. 2001). Heterozygous Cr4+/cr4 lines were irradiated, and the linked albino marker oy was used to identify mutant cr4 sectors for further characterization. Their results showed that Cr4 functions primarily through cell autonomous signaling to regulate differentiation.
Figure 8.
Radiation-induced chromosome breakages were used by Fu and Scanlon (2004) to study the developmental role of an essential gene, Emp2. (A) Heterozygous plants are phenotypically wild type until treatment with X rays, which break off the chromosome arm containing dominant W3 and Emp2 alleles. The acentric fragment is lost during mitosis, leaving only one intact chromosome bearing the unmasked recessive alleles w3 and emp2-R. All cells derived from this broken chromosome lineage are marked by the albino phenotype conferred by w3 and can be studied for emp2-R effects. (B) Cell lineages containing the broken chromosome become white sectors within the developing plant, marked by white arrows. Image in B is courtesy of Mike Scanlon.
Transposon mutagenesis
We discuss transposon mutagenesis below as a reverse genetics tool, but it is useful to note that for most of the past 3 decades, transposon mutagenesis was viewed as a forward genetics strategy. Transposons are excellent mutagens, and when a mutation is created by a known transposon, it is possible to clone and sequence the causal gene (Walbot 1992). By 1997, >50 maize genes had been cloned by transposon tagging using a variety of transposons, including Ac/Ds, Robertson’s Mutator, and Spm/En (discussed below) (Neuffer et al. 1997). Methods evolved from using Southern blots to identify transposons linked to genes, to pooled-sample PCR strategies (Brutnell 2002), and finally to methods involving high-hroughput sequencing, such as Mu-Illumina (Williams-Carrier et al. 2010). While a purely forward genetic strategy involving transposons may still be wise in some situations, most such efforts will involve transposon-generated reverse genetics resources as well (Hunter et al. 2013).
Reverse Genetics
Reverse genetics in maize has been greatly facilitated by the release of a complete and annotated genome (Schnable et al. 2009) as well as resulting resources that link transposon insertions to known and predicted genes (McCarty et al. 2013; Williams-Carrier et al. 2010). Additional methods of reverse genetics are also available in maize, including RNAi (McGinnis et al. 2007) and genome-editing technologies such as CRISPR-Cas9 (Liang et al. 2014).
Transposon mutagenesis
Transposable elements were first discovered in maize by Barbara McClintock through her cytological studies on chromosomal breakage and rejoining (McClintock 1984). Many families of transposons have been characterized in maize, and two of these, the Ac/Ds system and Robertson’s Mutator, have been harnessed to create sequence-indexed transposon insertion databases for mutant screening (Settles et al. 2007; Vollbrecht et al. 2010).
Ac/Ds:
The Activator/Dissociation transposon family consists of autonomous elements, called Activator or Ac elements, which are capable of inducing their own movement and the movement of nonautonomous elements called Dissociation or Ds elements. Both Ac and Ds move by a cut-and-paste mechanism to cause new insertions that can potentially disrupt gene function. The Ac/Ds system has a low mutagenesis rate, which can be advantageous because it eliminates confounding effects of multiple insertion events but also disadvantageous because large numbers of plants are needed to recover mutants. Ac/Ds has a strong bias to transpose to locations proximal to their initial location (Greenblatt 1984; Dooner and Belachew 1989), which makes this system particularly well suited to generating allelic series and saturating genes in a defined region (Kolkman et al. 2005). Because this system is limited by the initial location of Ac/Ds elements, a population that distributed 1500 Ds elements across the maize genome was created (Vollbrecht et al. 2010). With all Ds locations mapped, researchers can request seeds that contain Ds elements located near their regions of interest (http://acdstagging.org/). The Ac/Ds system can also be manipulated to create chromosome breaks at Ds insertion sites (Neuffer 1995). The breakage property of the Ac/Ds system has been used to study the embryonic lethal mutant dek1, revealing essential developmental roles in aleurone specification (Becraft and Asuncion-Crabb 2000).
The Suppressor-mutator (McClintock 1957) or Enhancer/Inhibitor (Peterson 1953) family of transposons is similar to Ac/Ds with a low mutation rate and preference to transpose to linked regions (Gierl and Saedler 1989). Like Ac/Ds, it has been used to tag and clone important maize genes, including opaque2 (Schmidt et al. 1987), a transcription factor that regulates seed development and storage, and ramosa1 (Vollbrecht et al. 2005), described above. However, to date, there have been no efforts to develop the Spm/En system for sequence-indexed mutation screening.
Robertson’s Mutator:
Robertson’s Mutator (Mu) has become the most widely used transposon system for both forward and reverse genetics due to the extensive resources and freely available mutant collections. Mutator was originally discovered in 1978, and similar to other transposons, it has autonomous (MuDR) and nonautonomous elements (Robertson 1978; Chomet 1994). Unlike Ac/Ds, Mu elements readily jump to unlinked sites and due to high copy number, transpose at much higher rates than other transposons in maize, making the family highly mutagenic (Chomet 1994; Lisch 2002; McCarty et al. 2013). Any plant from a Mu-active family contains multiple new insertions and those insertion sites can be rapidly identified using high-throughput sequencing strategies. The ability to (1) cheaply identify all the Mu insertion sites in a plant; (2) integrate those data with the genome browser; (3) bulk and store seeds from the same plant; and (4) link the genome browser to a seed distribution facility (the Maize Genetics Cooperation Stock Center) through the maize community database opened the door to creating powerful sequence-tagged Mu-induced mutation resources (McCarty et al. 2013; Williams-Carrier et al. 2010).
Although several Mu resources have been created (McCarty and Meeley 2009), two collections, called UniformMu (McCarty et al. 2013) and Mu-Illumina (Williams-Carrier et al. 2010) are currently indexed to the maize genome browser (http://www.maizegdb.org/). UniformMu was designed to minimize genetic background effects by backcrossing Mu activity into an inbred line (W22). In the current release, 57,000 UniformMu insertion sites that target 15,814 genes have been identified, and this number will increase in the coming years (D. McCarty, personal communication; see Figure 5 for the distribution of Mu elements in the genome). These Mu insertion lines are heritably stable because the active transposable element MuDR has either been outcrossed away or silenced (McCarty et al. 2005). UniformMu insertion lines can be identified and requested through MaizeGDB (see http://www.maizegdb.org/documentation/uniformmu/). A second, complementary resource is Mu-Illumina (Williams-Carrier et al. 2010), which is a catalog of Mu insertions from a smaller set of lines. Mu-Illumina is used in a similar manner by searching for insertions based on the genome browser and requesting seeds from the Maize Genetics Cooperation Stock Center or from the laboratory of Alice Barkan, who developed Mu-Illumina (http://teosinte.uoregon.edu/mu-illumina/). The Mu-Illumina resource includes insertions at or near 6228 loci, many of which are also identified in the UniformMu database.
RNA interference
The utility of RNAi in plants has been demonstrated as a means of knocking down gene function and engineering desired traits (Small 2007). McGinnis et al. (2007) created a large collection of maize RNAi lines targeting genes that regulate chromatin structure and gene silencing. They found a high degree of variability in effectiveness among constructs targeting different genes and among independent replicates of the same construct. Some constructs were able to knock down expression of not only the targeted gene, but related genes as well. Multigene knockdown by a single construct was also demonstrated for the conserved kinetochore protein Mis12. Maize contains two copies of mis12, mis12-1, and mis12-2; a single RNAi construct with an inverted shared sequence produced knockdown of both genes resulting in chromosome segregation defects (Li and Dawe 2009).
Researchers have used RNAi constructs containing viral DNA to create maize lines resistant to maize dwarf mosaic virus (Zhang et al. 2010) and sugarcane mosaic virus (Gan et al. 2010). RNAi has also been used to engineer maize with higher nutritional content. Maize is a major food staple, but compared to other crops, it is nutrient poor due to low lysine content. To combat worldwide malnutrition, breeders have tried to create high-lysine corn. Increasing the lysine content requires eliminating certain storage proteins (zeins) that give maize its hard kernel; high-lysine maize with altered zein content has soft kernels which makes them highly susceptible to pests and disease. A recent study used RNAi to knock down a subset of 22- and 19-α zeins, and the result was nutritionally rich, high-lysine maize with hard kernels (Wu and Messing 2012). These maize lines demonstrate the potential for RNAi-mediated crop engineering.
Genome editing technologies: ZFN, TALENS, CRISPR-Cas9
The ability to make targeted gene modifications is the driving force behind reverse genetics. While Mu mutants that disrupt gene activity can be found and RNAi can be used to reduce mRNA levels, maize and other plant model systems have suffered from the inability to directly target a gene for knockout or nucleotide modification. Plants are not amenable to targeted genome modification by homologous recombination due to low rates of recombination (Puchta 2002). New genome editing technologies have recently emerged that utilize targeted double and single-strand breaks to modify genes, and these systems (ZFN, TALENs, and CRISPR-Cas9) hold great potential for maize and plant genetics. ZFN (zinc finger nucleases) and TALENs (transcription activator-like effector nucleases) make use of engineered chimeric nucleases that are designed to bind and cleave a specific DNA sequence (Gaj et al. 2013). The CRISPR-Cas9 (clustered regularly interspaced short palindromic repeats) system uses a guide RNA to recognize a specific sequence and recruit the nuclease Cas9 (Ran et al. 2013b). CRISPR has several advantages over the protein-based recruitment methods including the ease and low cost of synthesizing new guide RNAs, increased specificity of cleavage sites, and the ability to simultaneously edit multiple loci (Ran et al. 2013b). Recent modification of the CRISPR system to use double nicking of single strands has dramatically decreased off target cleavage events (Ran et al. 2013a).
All three of these systems have been used successfully in maize. ZFN was used to both introduce an herbicide resistance gene to a targeted location and knockout an endogenous gene, Ipk1 (Shukla et al. 2009). Engineered disruption of Ipk1 is significant because this gene is part of the biosynthetic pathway that creates phytic acid, an antinutritional component and environmental pollutant that corn breeders have tried to reduce in seeds. TALENs and CRISPR systems have also been used to knock out Ipk1 with the CRISPR system being more efficient (Liang et al. 2014). The CRISPR technique is remarkable in producing first-generation homozygous mutations at fairly high frequencies in rice and other species (Zhang et al. 2014).
Conclusions
Maize is a classic model system that is undergoing a renaissance in the molecular era. The lure of maize for most scientists is the great beauty of the plant and extraordinary wealth of genetic tools, developed and passed on by generations of earlier geneticists. Those working on maize can look to the past for inspiration and to the future for what promises to be an exciting new era in which quantitative tools and natural diversity are integrated into the common toolbox.
Supplementary Material
Acknowledgments
We are grateful to Philip Becraft, Carolyn Lawrence, Jonathan Gent, and Jim Jensen for comments on the manuscript, and we thank the following people for images: Carolina Chavarro (Figure 1, A, D, E), Bill and Connie Funk (Figure 1B), Elizabeth Lee and Jeff Ross-Ibarra (Figure 4B), Bob Schmidt (Figure 7), and Mike Scanlon (Figure 8B). We also thank Beth Richardson for help with photography and John Fowler for advice on Figure 2. Our work is supported by the National Science Foundation: fellowship IOS-1400616 to Nannas and grants MCB-1412063 and IOS-0922703 to Dawe.
Footnotes
Available freely online through the author-supported open access option.
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.114.165183/-/DC1.
Communicating editor: J. A. Birchler
Literature Cited
- Anderson, E., 1956 The application of chromosomal techniques to maize improvement, pp. 23–36 in Genetics in Plant Breeding, Brookhaven Symposia in Biology. Brookhaven National Laboratory, Upton, NY. [Google Scholar]
- Barnabas B., Rajki E., 1976. Storage of maize (Zea mays L.) pollen at −196°C in liquid nitrogen. Euphytica 25: 747–752. [Google Scholar]
- Barnabas B., Rajki E., 1981. Fertility of deep-frozen maize (Zea mays L.) pollen. Ann. Bot. 48: 861–864. [Google Scholar]
- Baucom R. S., Estill J. C., Chaparro C., Upshaw N., Jogi A., et al. , 2009. Exceptional diversity, non-random distribution, and rapid evolution of retroelements in the B73 maize genome. PLoS Genet. 5: e1000732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckett J., 1994. Locating recessive genes to chromosome arm with BA translocations, pp. 315–327 in The Maize Handbook, edited by Freeling M., Walbot V. Springer, New York. [Google Scholar]
- Becraft P. W., Asuncion-Crabb Y., 2000. Positional cues specify and maintain aleurone cell fate in maize endosperm development. Development 127: 4039–4048. [DOI] [PubMed] [Google Scholar]
- Becraft P. W., Kang S. H., Suh S. G., 2001. The maize CRINKLY4 receptor kinase controls a cell-autonomous differentiation response. Plant Physiol. 127: 486–496. [PMC free article] [PubMed] [Google Scholar]
- Birchler J. A., 1994. Trisomic manipulation, pp. 307–308 in The Maize Handbook, edited by Freeling M., Walbot V. Springer, New York. [Google Scholar]
- Birchler J. A., Krishnaswamy L., Gaeta R. T., Masonbrink R. E., Zhao C., 2010. Engineered minichromosomes in plants. Crit. Rev. Plant Sci. 29: 135–147. [Google Scholar]
- Brunelle D. C., Sheridan W. F., 2014. The effects of varying chromosome arm dosage on maize plant morphogenesis. Genetics 198: 171–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutnell T. P., 2002. Transposon tagging in maize. Funct. Integr. Genomics 2: 4–12. [DOI] [PubMed] [Google Scholar]
- Buckler E. S., Holland J. B., Bradbury P. J., Acharya C. B., Brown P. J., et al. , 2009. The genetic architecture of maize flowering time. Science 325: 714–718. [DOI] [PubMed] [Google Scholar]
- Campbell M. S., Law M., Holt C., Stein J. C., Moghe G. D., et al. , 2014. MAKER-P: A tool kit for the rapid creation, management, and quality control of plant genome annotations. Plant Physiol. 164: 513–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cande W., Golubovskaya I., Wang C., Harper L., 2009. Meiotic genes and meiosis in maize, pp. 353–375 in Handbook of Maize: Genetics and Genomics, edited by Bennetzen J., Hake S. Springer, New York. [Google Scholar]
- Carlson W. R., 1978. The B chromosome of corn. Annu. Rev. Genet. 12: 5–23. [DOI] [PubMed] [Google Scholar]
- Chia J., Song C., Bradbury P. J., Costich D., de Leon N., et al. , 2012. Maize HapMap2 identifies extant variation from a genome in flux. Nat. Genet. 44: 803–807. [DOI] [PubMed] [Google Scholar]
- Cho M., Wu E., Kwan J., Yu M., Banh J., et al. , 2014. Agrobacterium-mediated high-frequency transformation of an elite commercial maize (Zea mays L.) inbred line. Plant Cell Rep. 33: 1767–1777. [DOI] [PubMed] [Google Scholar]
- Chomet P. S., 1994. Transposon tagging with mutator, pp. 243–249 in The Maize Handbook, edited by Freeling M., Walbot V. Springer, New York. [Google Scholar]
- Coe E., Jr, Neuffer M. G., Hoisington D., 1988. The genetics of corn, pp. 81–236 in Corn and Corn Improvement, edited by Sprague G. F., Dudley J. W. American Society of Agronomy, Madison, WI. [Google Scholar]
- Doebley J., Stec A., 1993. Inheritance of the morphological differences between maize and teosinte: comparison of results for two F2 populations. Genetics 134: 559–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J. F., Gaut B. S., Smith B. D., 2006. The molecular genetics of crop domestication. Cell 127: 1309–1321. [DOI] [PubMed] [Google Scholar]
- Dooner H. K., Belachew A., 1989. Transposition pattern of the maize element ac from the bz-M2(ac) allele. Genetics 122: 447–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle J. J., Flagel L. E., Paterson A. H., Rapp R. A., Soltis D. E., et al. , 2008. Evolutionary genetics of genome merger and doubling in plants. Annu. Rev. Genet. 42: 443–461. [DOI] [PubMed] [Google Scholar]
- Dresselhaus T., Lausser A., Marton M. L., 2011. Using maize as a model to study pollen tube growth and guidance, cross-incompatibility and sperm delivery in grasses. Ann. Bot. 108: 727–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y., Topp C. N., Dawe R. K., 2010. DNA binding of centromere protein C (CENPC) is stabilized by single-stranded RNA. PLoS Genet. 6: e1000835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C., Jiang N., Wessler S. R., 2002. Plant transposable elements: where genetics meets genomics. Nat. Rev. Genet. 3: 329–341. [DOI] [PubMed] [Google Scholar]
- Frame B. R., Zhang H., Cocciolone S. M., Sidorenko L. V., Dietrich C. R., et al. , 2000. Production of transgenic maize from bombarded type II callus: effect of gold particle size and callus morphology on transformation efficiency. In Vitro Cell. Dev. Biol. Plant 36: 21–29. [Google Scholar]
- Frame B. R., Shou H., Chikwamba R. K., Zhang Z., Xiang C., et al. , 2002. Agrobacterium tumefaciens-mediated transformation of maize embryos using a standard binary vector system. Plant Physiol. 129: 13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank M. J., Cartwright H. N., Smith L. G., 2003. Three brick genes have distinct functions in a common pathway promoting polarized cell division and cell morphogenesis in the maize leaf epidermis. Development 130: 753–762. [DOI] [PubMed] [Google Scholar]
- Freeling M., Hake S., 1985. Developmental genetics of mutants that specify knotted leaves in maize. Genetics 111: 617–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu S., Scanlon M. J., 2004. Clonal mosaic analysis of EMPTY PERICARP2 reveals nonredundant functions of the duplicated heat shock factor binding proteins during maize shoot development. Genetics 167: 1381–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu S., Lv Z., Gao Z., Wu H., Pang J., et al. , 2013. De novo centromere formation on a chromosome fragment in maize. Proc. Natl. Acad. Sci. USA 110: 6033–6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaj T., Gersbach C. A., Barbas C. F., III, 2013. ZFN, TALEN, and CRISPR/cas-based methods for genome engineering. Trends Biotechnol. 31: 397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale M. D., Devos K. M., 1998. Plant comparative genetics after 10 years. Science 282: 656–659. [DOI] [PubMed] [Google Scholar]
- Gallavotti A., Long J. A., Stanfield S., Yang X., Jackson D., et al. , 2010. The control of axillary meristem fate in the maize ramosa pathway. Development 137: 2849–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan D., Zhang J., Jiang H., Jiang T., Zhu S., et al. , 2010. Bacterially expressed dsRNA protects maize against SCMV infection. Plant Cell Rep. 29: 1261–1268. [DOI] [PubMed] [Google Scholar]
- Ganal M. W., Durstewitz G., Polley A., Bérard A., Buckler E. S., et al. , 2011. A large maize (Zea mays L.) SNP genotyping array: development and germplasm genotyping, and genetic mapping to compare with the B73 reference genome. PLoS ONE 6: e28334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent J. I., Madzima T. F., Bader R., Kent M. R., Zhang X., et al. , 2014. Accessible DNA and relative depletion of H3K9me2 at maize loci undergoing RNA-directed DNA methylation. Plant Cell 26: 4903–4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierl A., Saedler H., 1989. The en/spm transposable element of Zea mays. Plant Mol. Biol. 13: 261–266. [DOI] [PubMed] [Google Scholar]
- Golubovskaya I., Sheridan W., Harper L., Cande W., 2003. Novel meiotic mutants of maize identified from mu transposon and EMS mutant screens. Maize Genet. Coop. News Lett. 77: 10–12. [Google Scholar]
- Goodman M. M., Brown W. L., 1988. Races of corn, pp. 39–74 in Corn and Corn Improvement, edited by Sprague G. F., Dudley J. W. American Society of Agronomy, Madison, WI. [Google Scholar]
- Gordon-Kamm W. J., Spencer T. M., Mangano M. L., Adams T. R., Daines R. J., et al. , 1990. Transformation of maize cells and regeneration of fertile transgenic plants. Plant Cell 2: 603–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblatt I. M., 1984. A chromosome replication pattern deduced from pericarp phenotypes resulting from movements of the transposable element, modulator, in maize. Genetics 108: 471–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M., Davis D., Birchler J. A., 1996. Dosage effects on gene expression in a maize ploidy series. Genetics 142: 1349–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han F., Gao Z., Birchler J. A., 2009. Reactivation of an inactive centromere reveals epigenetic and structural components for centromere specification in maize. Plant Cell 21: 1929–1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, Y., S. Qin and S. R. Wessler, 2013 Comparison of class 2 transposable elements at superfamily resolution reveals conserved and distinct features in cereal grass genomes. BMC Genomics 14: 71. [DOI] [PMC free article] [PubMed]
- Hanson M. R., 1991. Plant mitochondrial mutations and male sterility. Annu. Rev. Genet. 25: 461–486. [DOI] [PubMed] [Google Scholar]
- Hartwell, L. H., L. Hood, M. L. Goldberg, A. E. Reynolds, and L. M. Silver, 2011 Mus musculus: genetic portrait of the house mouse, pp. 109–131 in Genetics: From Genes to Genomes. McGraw–Hill, New York. [Google Scholar]
- Hoisington D., Neuffer M., Walbot V., 1982. Disease lesion mimics in maize. I. effect of genetic background, temperature, developmental age, and wounding on necrotic spot formation with Les1. Dev. Biol. 93: 381–388. [DOI] [PubMed] [Google Scholar]
- Hollick J. B., 2012. Paramutation: a trans-homolog interaction affecting heritable gene regulation. Curr. Opin. Plant Biol. 15: 536–543. [DOI] [PubMed] [Google Scholar]
- Hopkins W., German J., Hayden D., 1980. A light-sensitive mutant in maize (Zea mays L.). II. Photosynthetic properties. Z. Pflanzenphysiol. 100: 15–24. [Google Scholar]
- Hunter C. T., Suzuki M., Saunders J., Wu S., Tasi A., et al. , 2013. Phenotype to genotype using forward-genetic mu-seq for identification and functional classification of maize mutants. Front. Plant Sci. 4: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W., Melo J. R., Nagaki K., Talbert P. B., Henikoff S., et al. , 2004. Maize centromeres: organization and functional adaptation in the genetic background of oat. Plant Cell 16: 571–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato A., Birchler J. A., 2006. Induction of tetraploid derivatives of maize inbred lines by nitrous oxide gas treatment. J. Hered. 97: 39–44. [DOI] [PubMed] [Google Scholar]
- Kato A., Lamb J. C., Birchler J. A., 2004. Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proc. Natl. Acad. Sci. USA 101: 13554–13559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, A., J. C. Lamb, P. S. Albert, T. Danilova, F. Han et al., 2011 Chromosome painting for plant biotechnology, pp. 67–96 in Plant Chromosome Engineering. Springer, New York. [DOI] [PubMed] [Google Scholar]
- Kermicle J. L., 1969. Androgenesis conditioned by a mutation in maize. Science 166: 1422–1424. [DOI] [PubMed] [Google Scholar]
- Kerstetter R. A., Laudencia-Chingcuanco D., Smith L. G., Hake S., 1997. Loss-of-function mutations in the maize homeobox gene, knotted1, are defective in shoot meristem maintenance. Development 124: 3045–3054. [DOI] [PubMed] [Google Scholar]
- Kiesselbach T. A., 1999. The Structure and Reproduction of Corn, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Kirienko D. R., Luo A., Sylvester A. W., 2012. Reliable transient transformation of intact maize leaf cells for functional genomics and experimental study. Plant Physiol. 159: 1309–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolkman J. M., Conrad L. J., Farmer P. R., Hardeman K., Ahern K. R., et al. , 2005. Distribution of activator (ac) throughout the maize genome for use in regional mutagenesis. Genetics 169: 981–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koumbaris G. L., Bass H. W., 2003. A new single-locus cytogenetic mapping system for maize (Zea mays L.): overcoming FISH detection limits with marker-selected sorghum (S. propinquum L.) BAC clones. Plant J. 35: 647–659. [DOI] [PubMed] [Google Scholar]
- Kump K. L., Bradbury P. J., Wisser R. J., Buckler E. S., Belcher A. R., et al. , 2011. Genome-wide association study of quantitative resistance to southern leaf blight in the maize nested association mapping population. Nat. Genet. 43: 163–168. [DOI] [PubMed] [Google Scholar]
- Kynast, R. G., and O. Riera-Lizarazu, 2011 Development and use of oat–maize chromosome additions and radiation hybrids, pp. 259–284 in Plant Chromosome Engineering. Springer, New York. [DOI] [PubMed] [Google Scholar]
- Lacroix B., Citovsky V., 2013. The roles of bacterial and host plant factors in agrobacterium-mediated genetic transformation. Int. J. Dev. Biol. 57: 467–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb J. C., Danilova T., Bauer M. J., Meyer J. M., Holland J. J., et al. , 2007. Single-gene detection and karyotyping using small-target fluorescence in situ hybridization on maize somatic chromosomes. Genetics 175: 1047–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law M., Childs K. L., Campbell M. S., Stein J. C., Olson A. J., et al. , 2015. Automated update, revision, and quality control of the maize genome annotations using MAKER-P improves the B73 RefGen_v3 gene models and identifies new genes. Plant Physiol. 167: 25–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence C. J., Harper L. C., Schaeffer M. L., Sen T. Z., Seigfried T. E., et al. , 2008. MaizeGDB: the maize model organism database for basic, translational, and applied research. Int. J. Plant Genomics 2008: 496957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leblanc O., Grimanelli D., Hernandez-Rodriguez M., Galindo P. A., Soriano-Martinez A. M., et al. , 2009. Seed development and inheritance studies in apomictic maize-tripsacum hybrids reveal barriers for the transfer of apomixis into sexual crops. Int. J. Dev. Biol. 53: 585. [DOI] [PubMed] [Google Scholar]
- Lee, H., and Z. J. Zhang, 2014 Agrobacterium-mediated transformation of maize (Zea mays) immature embryos, pp. 273–280 in Cereal Genomics. Springer, New York. [DOI] [PubMed] [Google Scholar]
- Levings C. S., III, 1990. The texas cytoplasm of maize: cytoplasmic male sterility and disease susceptibility. Science 250: 924–927. [DOI] [PubMed] [Google Scholar]
- Li P., Ponnala L., Gandotra N., Wang L., Si Y., et al. , 2010. The developmental dynamics of the maize leaf transcriptome. Nat. Genet. 42: 1060–1067. [DOI] [PubMed] [Google Scholar]
- Li X., Dawe R. K., 2009. Fused sister kinetochores initiate the reductional division in meiosis I. Nat. Cell Biol. 11: 1103–1108. [DOI] [PubMed] [Google Scholar]
- Liang Z., Zhang K., Chen K., Gao C., 2014. Targeted mutagenesis in Zea mays using TALENs and the CRISPR/cas system. J. Genet. Genomics 41: 63–68. [DOI] [PubMed] [Google Scholar]
- Lisch D., 2002. Mutator transposons. Trends Plant Sci. 7: 498–504. [DOI] [PubMed] [Google Scholar]
- McCarty D. R., Meeley R. B., 2009. Transposon resources for forward and reverse genetics in maize, pp. 561–584 in Handbook of Maize: Genetics and Genomics, edited by Bennetzen J. L., Hake S. Springer, New York. [Google Scholar]
- McCarty D. R., Mark Settles A., Suzuki M., Tan B. C., Latshaw S., et al. , 2005. Steady-state transposon mutagenesis in inbred maize. Plant J. 44: 52–61. [DOI] [PubMed] [Google Scholar]
- McCarty D. R., Latshaw S., Wu S., Suzuki M., Hunter C. T., et al. , 2013. Mu-seq: sequence-based mapping and identification of transposon induced mutations. PLoS ONE 8: e77172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B., 1931. Cytological observations of deficiencies involving known genes, translocations and an inversion in Zea mays. Missouri Agricultural Exp. Station Res. Bull. 163: 1–30. [Google Scholar]
- McClintock B., 1932. A correlation of ring-shaped chromosomes with variegation in Zea mays. Proc. Natl. Acad. Sci. USA 18: 677–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B., 1938a Fusion of broken ends of sister half-chromatids following chromatid breakage at meiotic anaphases. Missouri Agricultural Experimental Station Research Bulletin 290: 1–48. [Google Scholar]
- McClintock B., 1938b The production of homozygous deficient tissues with mutant characteristics by means of the aberrant mitotic behavior of ring-shaped chromosomes. Genetics 23: 315–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B., 1939. The behavior in successive nuclear divisions of a chromosome broken at meiosis. Proc. Natl. Acad. Sci. USA 25: 405–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B., 1941. The stability of broken ends of chromosomes in Zea mays. Genetics 26: 234–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B., 1948. Mutable loci in maize: nature of the Ac action. The mutable c loci. The mutable wx loci. Conclusions. Carnegie Institute of Washington Year Book 47: 155–169. [Google Scholar]
- McClintock B., 1949. Mutable loci in maize: the mechanism of transposition of the Ds locus. The origin of Ac-controlled mutable loci. Transposition of the Ac locus. The action of Ac on the mutable loci it controls. Mutable loci c m-2 and wx m-1. Conclusions. Carnegie Institute of Washington Year Book 48: 142–154. [Google Scholar]
- McClintock B., 1954. Mutations in maize and chromosomal aberrations in neurospora. Carnegie Institute of Washington Year Book 53: 254–260. [Google Scholar]
- McClintock B., 1957. The suppressor-mutator system of control of gene action in maize. Carnegie Institute of Washington Year Book 57: 415–431. [Google Scholar]
- McClintock B., 1984. The significance of responses of the genome to challenge. Science 226: 792–801. [DOI] [PubMed] [Google Scholar]
- McDaniel C. N., Poethig R. S., 1988. Cell-lineage patterns in the shoot apical meristem of the germinating maize embryo. Planta 175: 13–22. [DOI] [PubMed] [Google Scholar]
- McGinnis K., Murphy N., Carlson A. R., Akula A., Akula C., et al. , 2007. Assessing the efficiency of RNA interference for maize functional genomics. Plant Physiol. 143: 1441–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullen M. D., Kresovich S., Villeda H. S., Bradbury P., Li H., et al. , 2009. Genetic properties of the maize nested association mapping population. Science 325: 737–740. [DOI] [PubMed] [Google Scholar]
- Meyerowitz E. M., Pruitt R. E., 1985. Arabidopsis thaliana and plant molecular genetics. Science 229: 1214–1218. [DOI] [PubMed] [Google Scholar]
- Miles, C. D., 1982 The use of mutations to probe photosynthesis in higher plants, pp. 75–107 in Methods in Chloroplast Molecular Biology, edited by M. Edelman. Elsevier, Amersterdam. [Google Scholar]
- Mohanty, A., Y. Yang, A. Luo, A. W. Sylvester, and D. Jackson, 2009 Methods for generation and analysis of fluorescent protein-tagged maize lines, pp. 71–89 in Transgenic Maize. Springer, New York. [DOI] [PubMed] [Google Scholar]
- Monaco M. K., Stein J., Naithani S., Wei S., Dharmawardhana P., et al. , 2014. Gramene 2013: comparative plant genomics resources. Nucleic Acids Res. 42: D1193–D1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgante M., Brunner S., Pea G., Fengler K., Zuccolo A., et al. , 2005. Gene duplication and exon shuffling by helitron-like transposons generate intraspecies diversity in maize. Nat. Genet. 37: 997–1002. [DOI] [PubMed] [Google Scholar]
- Neuffer M. G., 1994a Growing maize for genetic studies, pp. 197–209 in The Maize Handbook, edited by Freeling M., Walbot V. Springer, New York. [Google Scholar]
- Neuffer M. G., 1994b Mutagenesis, pp. 212–219 in The Maize Handbook, edited by Freeling M., Walbot V. Springer, New York. [Google Scholar]
- Neuffer M., 1995. Chromosome breaking sites for genetic analysis in maize. Maydica 40: 99–116. [Google Scholar]
- Neuffer M., Calvert O., 1975. Dominant disease lesion mimics in maize. J. Hered. 66: 265–270. [Google Scholar]
- Neuffer M. G., Sheridan W. F., 1980. Defective kernel mutants of maize. I. genetic and lethality studies. Genetics 95: 929–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuffer M. G., Chang M. T., Clark J., Sheridan W. F., 1986. The genetic control of maize kernel development, pp. 35–50 in Regulation of Carbon and Nitrogen Reduction and Utilization in Maize, edited by Shannon J. American Society of Plant Physiology, Rockville, MD. [Google Scholar]
- Neuffer M. G., Coe E. H., Wessler S. R., 1997. Mutants of Maize. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Ninamango-Cárdenas F. E., Guimarães C. T., Martins P. R., Parentoni S. N., Carneiro N. P., et al. , 2003. Mapping QTLs for aluminum tolerance in maize. Euphytica 130: 223–232. [Google Scholar]
- Pawlowski W. P., Somers D. A., 1996. Transgene inheritance in plants genetically engineered by microprojectile bombardment. Mol. Biotechnol. 6: 17–30. [DOI] [PubMed] [Google Scholar]
- Pawlowski W. P., Somers D. A., 1998. Transgenic DNA integrated into the oat genome is frequently interspersed by host DNA. Proc. Natl. Acad. Sci. USA 95: 12106–12110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson P. A., 1953. A mutable pale green locus in maize. Genetics 38: 682–683. [Google Scholar]
- Prigge V., Xu X., Li L., Babu R., Chen S., et al. , 2012. New insights into the genetics of in vivo induction of maternal haploids, the backbone of doubled haploid technology in maize. Genetics 190: 781–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchta H., 2002. Gene replacement by homologous recombination in plants. Plant Mol. Biol. 48: 173–182. [PubMed] [Google Scholar]
- Purugganan M. D., Fuller D. Q., 2009. The nature of selection during plant domestication. Nature 457: 843–848. [DOI] [PubMed] [Google Scholar]
- Rafalski A., Morgante M., 2004. Corn and humans: recombination and linkage disequilibrium in two genomes of similar size. Trends Genet. 20: 103–111. [DOI] [PubMed] [Google Scholar]
- Ran F., Hsu P. D., Lin C., Gootenberg J. S., Konermann S., et al. , 2013a Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154: 1380–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F. A., Hsu P. D., Wright J., Agarwala V., Scott D. A., et al. , 2013b Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8: 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randolph L., 1941. An evaluation of induced polyploidy as a method of breeding crop plants. Am. Nat. 75: 347–363. [Google Scholar]
- Regulski M., Lu Z., Kendall J., Donoghue M. T., Reinders J., et al. , 2013. The maize methylome influences mRNA splice sites and reveals widespread paramutation-like switches guided by small RNA. Genome Res. 23: 1651–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades M. M., Dempsey E., 1966. Induction of chromosome doubling at meiosis by the elongate gene in maize. Genetics 54: 505–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades M. M., McClintock B., 1935. The cytogenetics of maize. Bot. Rev. 1: 292–325. [Google Scholar]
- Rines H. W., Phillips R. L., Kynast R. G., Okagaki R. J., Galatowitsch M. W., et al. , 2009. Addition of individual chromosomes of maize inbreds B73 and Mo17 to oat cultivars starter and sun II: maize chromosome retention, transmission, and plant phenotype. Theor. Appl. Genet. 119: 1255–1264. [DOI] [PubMed] [Google Scholar]
- Robertson, D. S., 1978 Characterization of a mutator system in maize. Mutat. Res. Fundam. Mol. Mech. Mutagen 51: 21–28.
- Romay M. C., Millard M. J., Glaubitz J. C., Peiffer J. A., Swarts K. L., et al. , 2013. Comprehensive genotyping of the USA national maize inbred seed bank. Genome Biol. 14: R55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt R. J., Burr F. A., Burr B., 1987. Transposon tagging and molecular analysis of the maize regulatory locus opaque-2. Science 238: 960–963. [DOI] [PubMed] [Google Scholar]
- Schnable J. C., Freeling M., 2011. Genes identified by visible mutant phenotypes show increased bias toward one of two subgenomes of maize. PLoS ONE 6: e17855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable P. S., Wise R. P., 1998. The molecular basis of cytoplasmic male sterility and fertility restoration. Trends Plant Sci. 3: 175–180. [Google Scholar]
- Schnable P. S., Ware D., Fulton R. S., Stein J. C., Wei F., et al. , 2009. The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115. [DOI] [PubMed] [Google Scholar]
- Settles A. M., Holding D. R., Tan B. C., Latshaw S. P., Liu J., et al. , 2007. Sequence-indexed mutations in maize using the UniformMu transposon-tagging population. BMC Genomics 8: 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw R. H., 1988. Climate requirement, pp. 609–638 in Corn and Corn Improvement, edited by Sprague G. F., Dudley J. W. American Society of Agronomy, Madison, WI. [Google Scholar]
- Sheridan W. F., Auger D. L., 2008. Chromosome segmental dosage analysis of maize morphogenesis using B-A-A translocations. Genetics 180: 755–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Dawe R. K., 2006. Partitioning of the maize epigenome by the number of methyl groups on histone H3 lysines 9 and 27. Genetics 173: 1571–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shou H., Frame B. R., Whitham S. A., Wang K., 2004. Assessment of transgenic maize events produced by particle bombardment or agrobacterium-mediated transformation. Mol. Breed. 13: 201–208. [Google Scholar]
- Shukla V. K., Doyon Y., Miller J. C., DeKelver R. C., Moehle E. A., et al. , 2009. Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature 459: 437–441. [DOI] [PubMed] [Google Scholar]
- Sidorov, V., and D. Duncan, 2009 Agrobacterium-mediated maize transformation: immature embryos vs. callus, pp. 47–58 in Transgenic Maize. Springer, New York. [DOI] [PubMed] [Google Scholar]
- Small I., 2007. RNAi for revealing and engineering plant gene functions. Curr. Opin. Biotechnol. 18: 148–153. [DOI] [PubMed] [Google Scholar]
- Stadler L. J., 1928. Genetic effects of X-rays in maize. Proc. Natl. Acad. Sci. USA 14: 69–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler L. J., 1930. Some genetic effects of X-rays in plants. J. Hered. 21: 3–20. [Google Scholar]
- Stadler L. J., Roman H., 1948. The effect of X-rays upon mutation of the gene A in maize. Genetics 33: 273–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern D. B., Hanson M. R., Barkan A., 2004. Genetics and genomics of chloroplast biogenesis: maize as a model system. Trends Plant Sci. 9: 293–301. [DOI] [PubMed] [Google Scholar]
- Strable, J., and M. J. Scanlon, 2009 Maize (Zea mays): a model organism for basic and applied research in plant biology. Cold Spring Harb Protoc. 2009: pdb.emo132. [DOI] [PubMed]
- Stuber C. W., Lincoln S. E., Wolff D. W., Helentjaris T., Lander E. S., 1992. Identification of genetic factors contributing to heterosis in a hybrid from two elite maize inbred lines using molecular markers. Genetics 132: 823–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swigonova Z., Lai J., Ma J., Ramakrishna W., Llaca V., et al. , 2004. Close split of sorghum and maize genome progenitors. Genome Res. 14: 1916–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulloa P. E., Iturra P., Neira R., Araneda C., 2011. Zebrafish as a model organism for nutrition and growth: towards comparative studies of nutritional genomics applied to aquacultured fishes. Rev. Fish Biol. Fish. 21: 649–666. [Google Scholar]
- Vollbrecht E., Springer P. S., Goh L., Buckler E. S., IV, Martienssen R., 2005. Architecture of floral branch systems in maize and related grasses. Nature 436: 1119–1126. [DOI] [PubMed] [Google Scholar]
- Vollbrecht E., Duvick J., Schares J. P., Ahern K. R., Deewatthanawong P., et al. , 2010. Genome-wide distribution of transposed dissociation elements in maize. Plant Cell 22: 1667–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walbot V., 1992. Strategies for mutagenesis and gene cloning using transposon tagging and T-DNA insertional mutagenesis. Annu. Rev. Plant Biol. 43: 49–78. [Google Scholar]
- Wallace J., Larsson S., Buckler E., 2013. Entering the second century of maize quantitative genetics. Heredity 112: 30–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Frame B., Ishida Y., Komari T., 2009a Maize transformation, pp. 609–639 in Maize Handbook: Genetics and Genomics, edited by Bennetzen J. L., Hake S. Springer, New York. [Google Scholar]
- Wang X., Elling A. A., Li X., Li N., Peng Z., et al. , 2009b Genome-wide and organ-specific landscapes of epigenetic modifications and their relationships to mRNA and small RNA transcriptomes in maize. Plant Cell 21: 1053–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil C., Monde R., 2009. TILLING and point mutation detection, pp. 585–595 in Handbook of Maize: Genetics and Genomics, edited by Bennetzen J. L., Hake S. Springer, New York. [Google Scholar]
- Weil C. F., Monde R., 2007. Getting the point: mutations in maize. Crop Sci. 47: S-60–S-67. [Google Scholar]
- Whipple C. J., Kebrom T. H., Weber A. L., Yang F., Hall D., et al. , 2011. Grassy Tillers1 promotes apical dominance in maize and responds to shade signals in the grasses. Proc. Natl. Acad. Sci. USA 108: E506–E512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams-Carrier R., Stiffler N., Belcher S., Kroeger T., Stern D. B., et al. , 2010. Use of illumina sequencing to identify transposon insertions underlying mutant phenotypes in high-copy mutator lines of maize. Plant J. 63: 167–177. [DOI] [PubMed] [Google Scholar]
- Wolfgruber T. K., Sharma A., Schneider K. L., Albert P. S., Koo D., et al. , 2009. Maize centromere structure and evolution: sequence analysis of centromeres 2 and 5 reveals dynamic loci shaped primarily by retrotransposons. PLoS Genet. 5: e1000743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q., Luo A., Zadrozny T., Sylvester A., Jackson D., 2013. Fluorescent protein marker lines in maize: generation and applications. Int. J. Dev. Biol. 57: 535–543. [DOI] [PubMed] [Google Scholar]
- Wu Y., Messing J., 2012. RNA interference can rebalance the nitrogen sink of maize seeds without losing hard endosperm. PLoS ONE 7: e32850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H., Kato A., Mooney B., Birchler J. A., 2011. Phenotypic and gene expression analyses of a ploidy series of maize inbred Oh43. Plant Mol. Biol. 75: 237–251. [DOI] [PubMed] [Google Scholar]
- Yu W., Han F., Gao Z., Vega J. M., Birchler J. A., 2007. Construction and behavior of engineered minichromosomes in maize. Proc. Natl. Acad. Sci. USA 104: 8924–8929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L., Dou Y., Kianian S. F., Zhang C., Holding D. R., 2014. Deletion mutagenesis identifies a haploinsufficient role for gamma-zein in opaque2 endosperm modification. Plant Physiol. 164: 119–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Phan B. H., Wang K., Artelt B. J., Jiang J., et al. , 2012. Stable integration of an engineered megabase repeat array into the maize genome. Plant J. 70: 357–365. [DOI] [PubMed] [Google Scholar]
- Zhang H., Zhang J., Wei P., Zhang B., Gou F., et al. , 2014. The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol. J. 12: 797–807. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Fu F., Gou L., Wang H., Li W., 2010. RNA interference-based transgenic maize resistant to maize dwarf mosaic virus. J. Plant Biol. 53: 297–305. [Google Scholar]
- Zhao Z., Gu W., Cai T., Tagliani L., Hondred D., et al. , 2002. High throughput genetic transformation mediated by agrobacterium tumefaciens in maize. Mol. Breed. 8: 323–333. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.