Abstract
Ends-out gene targeting allows seamless replacement of endogenous genes with engineered DNA fragments by homologous recombination, thus creating designer “genes” in the endogenous locus. Conventional gene targeting in Drosophila involves targeting with the preintegrated donor DNA in the larval primordial germ cells. Here we report gene targeting during oogenesis with lethality inhibitor and CRISPR/Cas (Golic+), which improves on all major steps in such transgene-based gene targeting systems. First, donor DNA is integrated into precharacterized attP sites for efficient flip-out. Second, FLP, I-SceI, and Cas9 are specifically expressed in cystoblasts, which arise continuously from female germline stem cells, thereby providing a continual source of independent targeting events in each offspring. Third, a repressor-based lethality selection is implemented to facilitate screening for correct targeting events. Altogether, Golic+ realizes high-efficiency ends-out gene targeting in ovarian cystoblasts, which can be readily scaled up to achieve high-throughput genome editing.
Keywords: gene targeting, homologous recombination, CRISPR, Drosophila, repressor/miRNA-based lethality selection
ALTHOUGH the advent of genome sequencing has given us access to the code of all proteins, RNAs, and transcriptional regulatory elements for many organisms, it gives no insight into how these sequences function. Unraveling the encrypted information requires a method to directly manipulate the genome in a controlled way. Ends-out gene targeting (GT) supplies just such a method by allowing seamless replacement of endogenous sequences with engineered DNA fragments (Thomas and Capecchi 1987). One can therefore place designer “genes” into their native loci or otherwise edit the nucleotide sequences in any genomic region of interest.
GT depends on crossing over between the template DNA (donor) and the genomic target double-strand DNAs, which share extensive sequence homology. For ends-out GT, homologous recombination takes place between flanking homology arms of a linear donor DNA and a target genomic locus, allowing insertion of an arbitrary DNA fragment into the endogenous target. However, homologous recombination occurs at low frequency and the linear donor DNA can be integrated into other genomic regions by nonspecific insertion. Successful GT has therefore relied on effective strategies of recovering the rare GT events. To accomplish such screening requires selection markers both within and outside the homology arms of the donor DNA (Capecchi 2005). However, false positives remain among those that have selectively retained the internal marker. In mice where embryonic stem cells allow cell culture-based GT, researchers can efficiently screen for candidates before proceeding through microinjection and the time-consuming labor-intensive mouse genetics (Capecchi 2005).
The availability of programmable sequence-specific nucleases makes it possible to create DNA breaks at specific genomic loci of interest (e.g., use of zinc-finger nucleases in Bibikova et al. 2002, 2003; Beumer et al. 2008; and Kim and Kim 2014), which can boost homologous recombination around the sites of damage by two to three orders of magnitude (Jasin 1996). The CRISPR/Cas system is particularly useful for targeted genome modifications, as the Cas9 nuclease can be reliably directed by custom-made guide RNAs to make specific DNA cuts based on DNA–RNA base pairing (Jinek et al. 2012; Bassett et al. 2013; Cong et al. 2013; Gratz et al. 2013; Hwang et al. 2013; Kondo and Ueda 2013; Mali et al. 2013; Ren et al. 2013; Sebo et al. 2013; Yu et al. 2013; Port et al. 2014). Within the last 2 years, CRISPR/Cas9 has been fruitfully applied in diverse species to create or correct mutations, insert transgenes at precise locations, and even generate knock-ins via targeted replacement of multikilobase genes (Harrison et al. 2014; Hsu et al. 2014; Sander and Joung 2014). The efficiency of CRISPR/Cas9-mediated genome editing, including knock-ins, is high and often exceeds 10% without selection (e.g., Byrne et al. 2014).
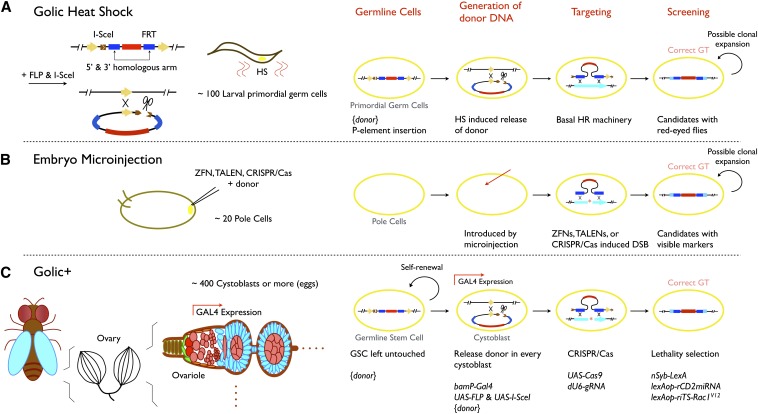
Ends-out GT in Drosophila has conventionally depended on a transgene-based system pioneered by Rong and Golic (2000; Gong and Golic 2003) (Figure 1A). The Golic system utilizes a P-element construct where donor DNA is flanked by FRT sites and contains a rare-cutting I-SceI site, which can therefore be transformed into a linear targeting molecule via the action of FLP and I-SceI. The linear donor DNA can be generated in the female germline by transient induction of hs-FLP and hs-I-SceI during larval development. This transient ubiquitous supply of the donor DNA at the early stage of germline development elicits GT in ∼1% of female founders as demonstrated in a limited number of known cases (Huang et al. 2008). It necessitates single-female crosses to ensure the independence of candidates as one adult female can yield multiple offspring with identical GT by the generation of multiple eggs from the same modified germline stem cell. Selection is further needed to enrich for the rare offspring with correct GT from the ≤1% single-female crosses.
Figure 1.
Comparison between different gene targeting strategies. Ends-out GT steps are generation of linear donor DNA, homologous recombination between donor DNA and targeted sequences, and recovery of correct GT. (A) For the Golic heat-shock strategy, the donor is first inserted in the genome as a P-element transgene and then released (flip-out and linearization) in larval primordial germ cells by heat-shock-induced expression of FLP and I-SceI. Targeting occurs rarely through endogenous homologous recombination machinery. Candidates, possibly carrying the same GT event due to later clonal expansion, are recovered based on the mini-white eye marker in between the 5′ and 3′ homology arms. (B) For embryo microinjection, donor DNA is injected together with the corresponding sequence-specific nuclease to boost GT in embryonic pole cells. Clonal expansion can again lead to multiple offspring carrying identical GT. (C) In Golic+, donor DNA is not released from the transgene until the birth of each cystoblast (CB) from the ovarian germline stem cells, guaranteeing independent GT among candidates. Ends-out GT in CBs requires DNA double-strand breaks made by CRISPR/Cas, and recovery of correct GT is facilitated by a repressor-based lethality selection. The CB-specific induction of FLP, I-SceI, and Cas9 depends on bamP-GAL4; guide RNA for CRISPR/Cas is broadly expressed with the dU6 promoter; strong candidates are recovered based on inheritance of a repressor, miRNA against rCD2, to rescue the pupal lethality caused by nSyb-driven riTS-Rac1V12.
With CRISPR/Cas9, direct embryo injection for ends-out GT in Drosophila has been demonstrated (Baena-Lopez et al. 2013; Gratz et al. 2013; Port et al. 2014; Xue et al. 2014; Yu et al. 2014) (Figure 1B). This saves the ∼2 months needed for the initial transgenesis of the donor DNA and may eliminate the need for complex selections. However, the concern over the independence of candidates from a single founder remains; and the efficiency of CRISPR/Cas9-assisted ends-out GTs in the Drosophila embryonic germline stem cells remains unclear. As GT efficiency could fluctuate significantly for different target loci, we were interested in establishing a GT technology that is scalable and leverages the power of existing Drosophila tools to virtually guarantee targeting success, regardless of genomic loci or insert size.
Here we establish gene targeting during oogenesis with lethality inhibitor and CRISPR/Cas (Golic+) as an enhanced transgene-based GT toolkit (Figure 1C). Golic+ delivers the linear donor DNA in each serially derived cystoblast throughout female germline development. The assembly line of de novo GT allows us to pool unsynchronized organisms and breed them in groups, without concern about resampling the same GT events. Moreover, the CRISPR/Cas system is incorporated to achieve ends-out GT in ∼50% of founder females. We have further engineered a repressor-based pupal lethality selection such that only strong candidates eclose for further breeding and PCR validation of GT. In sum, Golic+ is built upon the existing genetic/transgenic platforms, but made to be scalable such that one fly laboratory can perform multiple GTs simultaneously and easily until recovery of all correct GTs.
Materials and Methods
DNA constructs
The constitutively active Drosophila Rac1V12 was created by amplifying the wild-type Rac1 coding sequence (CDS) (gift of Julian Ng, MRC Laboratory of Molecular Biology, Cambridge, UK) with a forward PCR primer containing the GGA to GTG mutation. Following rules in previous studies (Chen et al. 2007; Yu et al. 2009), six different rCD2 microRNA (miRNA) were created and compared for their effectiveness in suppression (data not shown). rCD2 miRNA 6, with two rCD2 target sequences, 5′-GTAACGGTATACAGCACAAATG-3′ and 5′-GATAAAAGCTTCCAGAATGAGC-3′, was selected as it showed the best performance. 8XLexAop2-ri6TS-Rac1V12-hsp70 was created by cloning ri6TS-Rac1V12 into pJFRC18 (NotI/XbaI) (Pfeiffer et al. 2010) and replacing the original SV40 terminator (SV40T) with hsp70 terminator (hsp70T, XbaI/FseI). 5XLexAop2-ri6TS-Rac1V12-hsp70T was generated subsequently by removing the 3XLexAop2 between the AvrII and NheI sites of 8XLexAop2-ri6TS-Rac1V12-hsp70T (self-ligation with the AvrII/NheI compatible ends). We further created a 3xP3-RFP (instead of mini-white) version of the 5XLexAop2-ri6TS-Rac1V12-hsp70T transgene by replacing the original mini-white marker (AscI, filled in) with a 3xP3-RFP-SV40 fragment (Horn et al. 2000). Additionally, to reduce similarity and chances of recombination between sequences, we ordered a Drosophila codon-optimized coding sequence of Rac1V12 (GeneArt; Invitrogen, Carlsbad, CA) and created the nonrepressible 5XLexAop2-opRac1V12-hsp70T (NotI/XbaI). rCD2 miRNA 6 was cloned into pMLH (NotI/XhoI) (Awasaki et al. 2014) to make lexAop-rCD2i.
pTL1 (targeting with lethality selection), pTL2, Pfife (p10-facilitated indicators of flip excision), and BPfife were constructed following traditional molecular cloning and assembled from smaller DNA fragments. Their sequences and detailed annotations can be found on Addgene (https://www.addgene.org/) once deposited. To assemble pTL1 (the precursor of pTL2), smaller DNA modules were added sequentially by attaching them either on forward or on reverse PCR primers. First, a starting cassette (5XLexaop2-FRT-ISceI-FRT-ri6TS-Rac1V12-hsp70T) was assembled from a triple ligation of two PCR fragments (HindIII-5XLexaop2-FRT-I-SceI, I-SceI-FRT-ri6TS-Rac1V12-hsp70T-EcoRI) and a HindIII/EcoRI-digested pBluescript II KS(+) plasmid. Second, after digesting this starting cassette with I-SceI, two more PCR fragments (I-SceI-5′MCS-I-CreI-attPX-loxP-SpeI and SpeI-hsp70T-opRac1V12-5XLexAop2-I-SceI) were added in to create a bigger intermediate cassette (5XLexAop2-FRT-I-SceI-5′MCS-I-CreI-attPX-loxP-SpeI-hsp70T-opRac1V12-5XLexAop2-I-SceI-FRT-ri6TS-Rac1V12-hsp70T). This intermediate cassette was then transferred (EcoRI and partial HindIII digestion) to pJFRC-MUH (HindIII/EcoRI) (Pfeiffer et al. 2010) after removing the mini-White marker in pJFRC-MUH with AscI cutting and self-ligation. Finally, pTL1 was completed by ligating the final PCR fragment SpeI-lexAop-rCD2i-loxP-I-CeuI-3′MCS-XbaI (SpeI and partial XbaI digestion) into the SpeI site, which was assembled from two smaller PCR fragments, SpeI-lexAop-rCD2i-PacI and PacI-loxP-I-CeuI-3′MCS-XbaI.
To create pTL2, we first removed the I-SceI site downstream of the 3′ multiple-cloning site (MCS) of pTL1. pTL1 was partially digested with I-SceI and recircularized using Gibson assembly [New England BioLabs (Beverly, MA), Gibson Assembly Master Mix, E2611L] with a small PCR fragment containing one AvrII site at each end. This fragment was later removed by AvrII digestion and self-ligation so that the original I-SceI site (TAGGGATAACAGGGTAAT) became TAGGGATAA-CCTAGG-ATAACAGGGTAAT. To add guide RNA (gRNA) backbone, the plasmid was digested with EcoRI, filled in, and then ligated with a HindIII/EcoRI-digested, filled-in dU6-3-gRNA fragment (see below and Supporting Information, Figure S2A). The orientation with the dU6-3 promoter lying adjacent to the suppressible ri6TS-Rac1V12 was selected in pTL2.
To assemble Pfife, a new FRT cassette (NotI-FRT-BglII-HindIII-FseI-BamHI-FRT-XhoI) was first created by annealing two partially overlapping primers, filling in both strands with PCR, and cloning it into pBluescript II KS(+) (NotI/XhoI). Afterward, DNA fragments (HindIII-10XUAS-BglII, HindIII-BPStop-FseI, and BamHI-tdTomato-XbaI-p10-NgoMIV) were sequentially added in to generate a FRT-10XUAS-BPStop-p10-tdTomato-FRT cassette. BPStop is a DNA module that by design should effectively repress/eliminate readthrough of transcription and translation from nearby promoters (Sauer 1993; Zinyk et al. 1998). Finally, this larger FRT cassette was excised (NotI/XhoI) and put into pJFRC28 (Pfeiffer et al. 2012) to create Pfife. To further generate Bfife, we first inserted an ∼5-kb DNA fragment (from plasmid KB700, HindIII/NotI digested, filled in) into the pBluescript II KS(+)-FRT-10XUAS-BPStop-p10-tdTomato-FRT plasmid (SpeI digested, filled in). Then, this ∼9-kb FRT cassette was put into pJFRC28 to generate BPfife (NotI/XhoI).
To create UASt-FLP in this study, we first assembled pJFRC-MUH-IVS-WPRE-hsp70T by replacing the original SV40T of pJFRC-MUH with hsp70T (XbaI/NgoMIV) and further inserted BamHI-IVS-BglII-NotI-XhoI-KpnI-XbaI-WPRE-SpeI into the BglII/XbaI-digested pJFRC-MUH-hsp70T. Then, a BglII-FLP-XhoI PCR fragment was created using pBPopFLP(1GtoD)Uw as the template. Afterward, XhoI-3XGST-securin N50 a.a.-XbaI was added to the fragment with primers and PCR amplification. Finally, the whole BglII-FLP-XhoI-3XGST-securin N50 a.a.-XbaI fragment was inserted into pJFRC-MUH-IVS-WPRE-hsp70T (BglII/XbaI) to create UASt-FLP (UAS-IVS-opFLP(1GtoD)::3XGST::securing N50 a.a.-WPRE-hsp70T). The UASp-FLP used in this study actually contains UASp-FLP and UASp-I-SceI separated by an insulated spacer cassette (Pfeiffer et al. 2010). To assemble it, a blunt-end/EcoRI-digested PCR product (AvrII-UASp promoter-MCS-K10 terminator-FseI-PmeI-NheI-EcoRI) (Rørth 1998) was first inserted into pJFRC-MUH (HindIII, filled-in/EcoRI) to make pUASp-attB. Second, the KpnI/HindIII filled-in fragment from pBPopFLP(1GtoD)Uw was inserted into pUASp-attB (KpnI/XbaI filled in) to make pUASp-attB-FLP. Third, a synthetic Drosophila codon-optimized I-SceI (GeneArt; Invitrogen) was inserted into pUASp-attB (KpnI/XbaI) to make pUASp-attB-I-SceI. Fourth, the insulated spacer cassette flanking with NgoMIV sites was inserted into pUAS-pattB-I-SceI (NgoMIV) to make pUASp-attB-I-SceI-insulator. Finally, the AvrII/PmeI fragment from pUASp-attB-FLP was inserted into NheI/PmeI site of pUASp-attB-I-SceI-insulator to make UASp-FLP (pUASp-attB-I-SceI-insulator-UASp-FLP).
To recover pTL2 transgenesis we generated an eye-specific LexA driver, GMR3-LexA::GADd, which contains three copies of a truncated glass-binding site (Ellis et al. 1993; Hay et al. 1994) to drive LexA::GADd (Pfeiffer et al. 2010). nSyb-LexA::p65 is a neuronal synaptobrevin promoter-fusion LexA::p65 (Pfeiffer et al. 2010; Awasaki et al. 2014) construct created as the essential (neuronal) driver to implement the larval/pupal lethal selection of Golic+. To create bamP-Cas9-2A-FLP-2A-I-SceI, bam 3′-UTR was cloned from BACR06L08 into pJFRC28 (KpnI/EcoRI) with primers (GGGGTACCTCTAGACTAATGCTGTGCACATCGATAAAAG and GGAATTCAGTCCAAACACAAATCGTAAATATTTATTTG). bam 5′-UTR was then cloned (BACR10P10; primers CCAAATCAGTGTGTATAATTGTAGTTAAAATG and GCTCTAGAGGTACCTAAGTTAAATCACACAAATCACTCGATTTTTG) and added into the bam 3′-UTR-carrying vector (SphI digestion, blunt/XbaI). Drosophila codon-optimized CDSs of FLP [opFLP(1GtoD), XhoI/XbaI], I-SceI (GeneArt; Invitrogen, KpnI/XbaI), and Cas9 (gift of Justin Crocker, Janelia Research Campus, HHMI, VA, Kpn/XbaI) were used in this study. For Cas9, we further added in its N terminus a nuclear localization signal (CCAAAGAAAAAGAGAAAGGTT) in the hope of increasing its effectiveness. Additionally, Syn21 was added before the start codon of optimized Cas9 to enhance its expression (Pfeiffer et al. 2012). Finally, Cas9-P2A-FLP-E2A-I-SceI was assembled and added into the previous vector (KpnI/XbaI), using Gibson assembly. P2A and E2A were chosen among 2As for their higher cleavage efficiency (Kim et al. 2011). Also, Gly-Ser-Gly linkers were put in front of these 2As as suggested for improved cleavage efficiency (addendum to Szymczak et al. 2004).
dU6-2 and dU6-3 promoters were cloned from BAC clone BACR47D16. Two SapI sites were put between U6 promoters and the gRNA scaffold for easy gRNA target site insertion. Target sites were identified using the web-based ZiFiT Targeter program (Hwang et al. 2013) or DRSC CRISPR (Ren et al. 2013). The following target sites were used to target yellow, msh, and runt: y1 GCGATATAGTTGGAGCCAGC, y2 GTGCACTGTTCCAGGACAAA, msh1 GGGATAAGTGGCGGCCCAGT, and runt1 GGGGATCAGATGCCCTAGTA.
UAS-GFP::Dbox was created by a three-fragment ligation of pUASt-attB (EcoRI/XbaI), GFP (EcoRI/XhoI, with pUASt-mCD8::GFP as its template) (Lee and Luo 1999), and Drosophila securin N50 a.a., covering the KEN-box and the D-box (XhoI/XbaI; self-amplification from two primers) (Leismann and Lehner 2003).
Fly strains
Fly strains used in this study were as follows: (1) bamP-GAL4::VP16 (gift of Dennis M. Mckearin, K-RITH, Durban, South Africa); (2) 10XUAS-IVS-GFP-p10 in attP2 (pJFRC28) (Pfeiffer et al. 2012); (3) nos.UTR-GAL4::VP16 [Bloomington Drosophila Stock Center (BDSC), no. 4937]; (4) UASt-FLP in su(HW)attP8; (5) UASp-FLP in su(HW)attP8; (6) Pfife in attP40; (7) BPfife in attP40, su(Hw)attP6, VK00002, su(Hw)attP5, attP2, VK00005, su(Hw)attP1, VK00027, VK00020, and VK00040; (8) act5C-Cas9 in attP2A (Port et al. 2014); (9) dU6-gRNAs against yellow in attP2; (10) GMR3-LexA::GADd in attP40 and attP2; (11) nSyb-LexA::p65 in attP16 and VK00027; (12) 5X-ri6TS-Rac1V12 and 5X-ri6TS-Rac1V12(3xP3-RFP) in attP40 and VK00027; (13) bamP-Cas9-2A-FLP-2A-I-SceI in su(Hw)attP8 and attP2; (14) {donor, gRNA} for all targetings in attP40; (15) y1w67c23P{Crey}1b; snaSco/CyO (BDSC, no. 766); and (16) UAS-GFP::Dbox in attP2.
Fly genetics
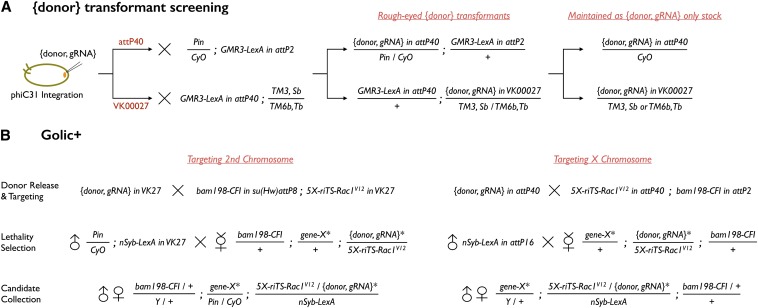
To recover {donor, gRNA} transgenic flies, we raised {donor, gRNA}-injected larvae (Rainbow Transgenic Flies) at room temperature, crossed the eclosed adults with GMR-LexA::GADd, and searched for rough-eyed progeny as successful transformants (Figure 3A).
Figure 3.
Crossing schemes for Golic+. (A) {donor, gRNA} injected embryos were raised and then mated with GMR3-LexA. Transformants with obvious rough bar eye phenotype were identified and maintained as balanced stocks. (B) Schemes for targeting a second or an X chromosome gene are depicted. Targeting a third chromosome gene can be deduced from referencing Table 2. Golic+ consists of two major crosses and three steps (donor release and targeting, lethality selection, and candidate collection). The first cross is to generate linear {donor} with FLP and I-SceI (and additionally express Cas9 and gRNA) and hence create the founder females. The second cross is to perform the lethality selection by mating founder females to male nSyb-LexA::p65. Eclosed candidates are collected for genetic mapping and PCR confirmation.
For Golic+ crosses (Figure 3B), founder females were generated by crossing 5X-riTS-Rac1V12 in attP40; bam198-CFI in attP2 to {donor, gRNA}. Their virgin progeny were then crossed to nSyb-LexA::p65 for lethality selection. The survivors were further subjected to chromosome mapping and genomic PCR confirmation.
Targeting designs and molecular characterization of target loci
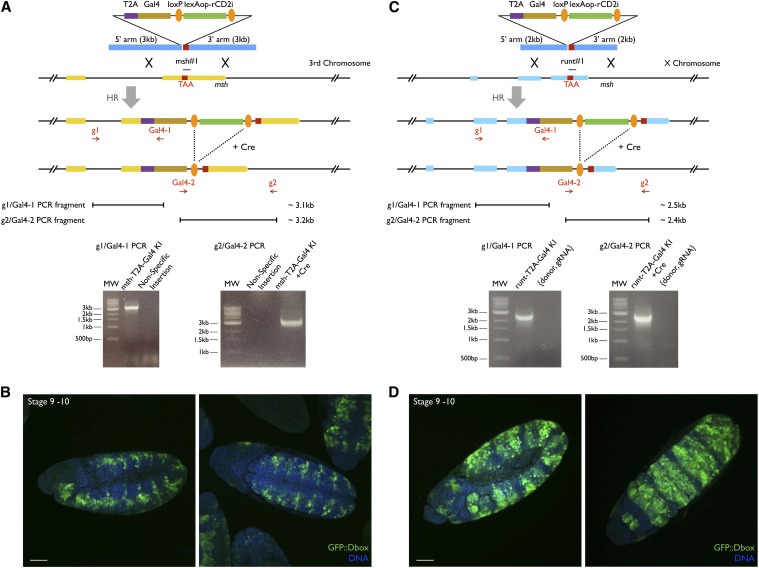
Homology arms of ∼2–3 kb each were designed to knock-in msh-T2A-Gal4 and runt-T2A-Gal4 (Figure 4). The following BAC clones and primers were used for amplifying these arms: BACR10L12 for msh; and BACR50G05 for runt msh_55NgoMIV, TAATTGCCAGCAATTTGCACCG; msh_53AgeI, TACGACCGGTTCCCAGGTGCATCAGGC; msh_35BamHI, CGGGATCCTAAGTGGCGGCCCAGTTG; msh_33PmeI, AACAAATGCCCGCAATCAGCG; runt_55AgeI, ACGTACCGGTAAGTGACCCCCGATAAAGTGAAGTGCATACCGAG; runt_53StuI, ACGTAGGCCTGTAGGGCCGCCACACGGTCTTCTGC; runt_35BamHI, ACGTGGATCCTAGGGCATCTGATCCCCAAAAATCTGGAGGAATGAAG; and runt_33MluI, ACGTACGCGTTCTCAACCGCTTGTAGTCACCATTTAAGTTTTGGAC. T2A-Gal4 was generated by cloning Gal4 from pBPGAL4.2Uw-2 (Pfeiffer et al. 2010) into pTL2 (StuI/SacI) with a forward primer containing T2A coding sequence. The following primers were used in genomic PCR for msh-T2A-Gal4 and runt-T2A-Gal4 knock-in candidate confirmation: msh_g1, CATCCACTGCATCCAATCCTAGTG; Gal4-1, CACACGCTTGTTCAATACTACTCAG; Gal4-2, GATACTCCACCGAACCCAAAGAAG; msh_g2, GGCGTTAATATCAAGCTGTGATTTCG; runt_g1, AATGGTGGTTGCTCGATATACCGATATATAC; and runt_g2, CGGATTCGGATTGGACGAGTTAAATTC.
Figure 4.
Targeting msh and runt with Golic+. (A and C) Donor designs and PCR confirmation for generating msh-T2A-Gal4 and runt-T2A-Gal4 knock-ins. Genomic sequences (2–3 kb) just upstream and downstream of the msh or runt stop codons were used as homology arms (TAA was included in the 3′ arm). Coding sequence of T2A-Gal4 was placed in between so that GAL4 and Msh (or GAL4 and Runt) can be translated from the same mRNA transcript. gRNA target sites around the stop codons were selected. For each gene, two sets of primers (msh_g1 and Gal4-1, msh_g2 and Gal4-2 and runt_g1 and Gal4-1, runt_g2 and Gal4-2) were chosen to confirm T2A-Gal4 knock-ins at both ends. g1 and g2 primers were chosen farther upstream or downstream of the homology arms so that PCR amplicons (3.1 and 3.2 kb for msh and 2.5 and 2.4 kb for runt) were possible only when T2A-Gal4 is correctly situated at the msh or runt locus. The g2/Gal4-2 PCRs were performed after Cre removed the lexAop-rCD2i loxP cassette. (B and D) Bilateral patterned GFP expression seen in embryos of msh-T2A-Gal4 and runt-T2A-Gal4 knock-ins matches msh and runt BDGP in situ data (http://insitu.fruitfly.org/cgi-bin/ex/report.pl?ftype=1&ftext=CG1897 and http://insitu.fruitfly.org/cgi-bin/ex/report.pl?ftype=1&ftext=CG1849). Nuclear DNA was revealed by Hoechst 33342 staining. Bars, 50 μm. In all panels illustrations are not to scale.
Immunostaining and fluorescence microscopy
Ovaries were dissected in 1× Grace’s insect medium, supplemented [GIBCO (Grand Island, NY), Life Technology], and fixed for 10 min in 4% paraformaldehyde fixation solution. Fixation solution was prepared by mixing 20% w/v paraformaldehyde solution (Electron Microscopy Sciences) 1:4 to 1× PBS solution. After fixation, ovaries were first rinsed three times, then washed three times (5, 15, and 30 min) in PBST (0.2% Triton X-100 in 1× PBS), and finally incubated with primary antibodies (diluted in PBST + 5% normal goat serum) overnight at 4°. Afterward, samples were further incubated at room temperature for 1 hr before being rinsed three times and washed three times (20 min each) in PBST. Ovaries were then incubated with secondary antibodies (diluted in PBST + 5% normal goat serum) for 3 hr at room temperature, rinsed three times, and washed three times over 1 hr. For mounting, ovaries were transferred on glass slides, separated into ovariole strings, rinsed one time with 1× PBS, rinsed again in SlowFade Gold antifade reagent (Invitrogen), and soaked in SlowFade for fluorescence imaging.
Washed, collected embryos were dechorionated in 50% bleach for 3 min. Then, they were thoroughly rinsed with water, dried, and transferred into 4% paraformaldehyde fixative (1 ml) plus an equal volume of heptane for 30-min fixation on a shaker. Afterward, the bottom aqueous phase was replaced with 1 ml of methanol, and embryos were devitellinized on a shaker for 1 min. The top heptane phase was aspirated away followed by three rinses of the bottom phase embryos in gradually diluted methanol/PBS solution (3:1, 1:1, and 1:3; v/v). These embryos went through an additional round of 4% paraformaldehyde fixation. Next, they were rinsed three times, further washed in PBST for 30 min, and finally incubated with anti-GFP primary antibody overnight at 4°. The next day, they were rinsed and washed through the same procedures and incubated with secondary bodies overnight at 4° before being rinsed, washed, stained with Hoechst 33342 for 10 min, and mounted in SlowFade. Fluorescent signals of ovarioles and embryos were collected by confocal serial scanning, using a Carl Zeiss (Thornwood, NY) LSM710 microscope. Images were processed with Fiji and then rotated and cropped with Keynote.
The following primary antibodies were used in this study: rat monoclonal anti-GFP, 1:100 (MBL International, D153-3) in Figure S2A; rat monoclonal anti-GFP, 1:1000 (Nacalai Tesque, 04404-84) in Figure S2, C and D; rabbit polyclonal anti-GFP, 1:1500 (Molecular Probes, Eugene, OR; A11122) in Figure 4, B and D; mouse monoclonal anti-Fasciclin III, 1:50 [Developmental Studies Hybridoma Bank (DSHB), antibody 7G10]; rabbit polyclonal anti-DsRed, 1:1000 (Clontech, Living Colors; 632496); and mouse anti-α-spectrin, 1:25 (DSHB, antibody 3A9). Secondary antibodies from Molecular Probes were used in a 1:200 dilution: Alexa Fluor 488 goat anti-rat IgG (H+L), Alexa Fluor 568 goat anti-rabbit IgG (H+L), and Alexa Fluor 647 goat anti-mouse IgG (H+L). For embryos, Cy3-conjugated goat anti-rabbit (1:200; Jackson ImmunoResearch, 111-165-144) and Hoechst 33342 (1:1000; Invitrogen) were used.
Results
To improve GT independence, efficiency, and selection
All previous Golic-based ends-out GT approaches have used donor DNAs that were randomly inserted into the fly genome by P-element-mediated germline transformation. To excise the donor DNA for GT requires tedious heat shocks on synchronized larvae, and the efficiency of flipping out the donor DNA can vary drastically depending on the insertion sites. Furthermore, GT in the female germline targets stem cells that exist at the midlarval stage that later go through clonal expansion leading to multiple identical targeting events.
To eliminate these limitations, we explored the possibility of restricting GT to the serially derived cystoblasts (CBs) of adult ovaries, which each develop into a single female germ cell and thus guarantee independent GT trials among individual offspring. At the tip of each germarium in ovaries, two to three germline stem cells (GSCs) exist. These cells divide alternately, self-renewing themselves while generating a series of CBs (Spradling 1993). A CB-specific promoter has been isolated from bam, a gene essential for female germ cell differentiation (Chen and Mckearin 2003). We tested the available bamP-GAL4 with UAS-GFP carrying the p10 terminator (Brand and Perrimon 1993; Chen and Mckearin 2003; Pfeiffer et al. 2012) and confirmed its selective expression in newborn CBs but not the GSCs (Figure S1A). We next made Pfife, a dual-reporter flip-out construct that carries an FRT cassette, excision of which results in fusion of UAS with GFP on the residual transgene and concomitantly reconstitutes UAS-tdTomato in the circularized FRT cassette (Figure S1B). Induction of UAS-FLP with bamP-GAL4 vs. nosP-GAL4 revealed that bamP-GAL4 elicited coexpression of GFP and tdTomato in newborn CBs of most germaria, but that nosP-GAL4 only led to GFP expression in both GSCs and CBs (Figure S1C). The earlier onset of nosP-GAL4 in GSCs apparently led to complete loss of FRT cassettes prior to adult oogenesis. By contrast, bamP-GAL4 maintained the FRT cassette in GSCs while specifically releasing it in newborn CBs, meeting our need for de novo production of “donor DNA” through adult oogenesis. We further made BPfife, which carries a 9-kb FRT cassette mimicking the donor DNA in size. We inserted BPfife into various attP sites, using the phiC31 integration system, and identified attP40 on the second chromosome and VK00027 on the third chromosome as ideal sites for the initial transgenesis and subsequent release of donor DNAs via flip-out (Figure S1D).
To improve GT efficiency, we explored the possibility of creating double-strand DNA breaks at specific loci in CBs, using the CRISPR/Cas system. We generated various gRNAs targeting the yellow body color gene, using two endogenous Drosophila U6 promoters, dU6-2 and dU6-3 (Wakiyama et al. 2005; Hernandez et al. 2007) (Figure S2A). We identified dU6-3 as a more potent gRNA promoter based on somatic mutation of yellow with act5C-Cas9 (Figure S2B). Next we used bamP-GAL4 to drive expression of UAS-Cas9 in the CBs of female flies carrying the same gRNAs. We recovered yellow mutant offspring and confirmed the efficacy of CRISPR/Cas in manipulating CB genomes (data not shown).
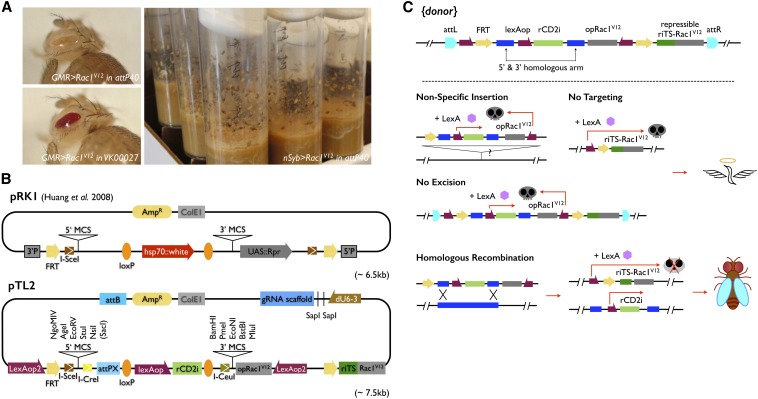
To follow and enrich for correct GT events, we envisioned a repressor-based lethality selection that allows elimination of offspring that no longer carry the donor DNA or retain the entire donor DNA either at the original site or at a new site via nonspecific insertion. Such a selection scheme requires a repressible toxic gene as well as a nonrepressible toxic gene, which can be induced to give visible phenotypes or cause pupal lethality, depending on drivers (see below). Briefly, Rac1V12, encoding a constitutive active small GTPase, was chosen as the toxic gene (Luo et al. 1994). We made Rac1V12 repressible by inserting at its 5′-untranslated region the target sequences of a proved potent transgenic miRNA against rat rCD2, establishing the rCD2 miRNA (rCD2i) as the repressor (Chen et al. 2007; Yu et al. 2009). We placed both the repressible riTS-Rac1V12 and nonrepressible Rac1V12 transgenes as well as rCD2i under the control of lexAop promoters of various strengths (Lai and Lee 2006; Pfeiffer et al. 2010). We explored the use of eye-specific GMR-LexA::GAD and pan-neuronal nSyb-LexA::p65 for inducing visible phenotypes and pupal lethality, respectively. We settled on the weakened 5XLexAop2 for the control of either repressible or nonrepressible Rac1V12 while reserving the original lexAop (Lai and Lee 2006) to drive rCD2i strongly. In that combination, we could consistently elicit a rough bar eye phenotype with GMR > Rac1V12 and generate pupal lethality without escapers in the presence of nSyb > Rac1V12 (Figure 2A). Both phenotypes were completely suppressed when rCD2i was co-induced with repressible Rac1V12.
Figure 2.
Repressor-based selection and GT plasmids. (A) Rough to bar eye and dead pupa phenotypes were elicited by GMR > Rac1V12 and nSyb > Rac1V12, respectively. (B) pTL2 has a similar organization to pRK1. Note the presence of lexAop-rCD2i, instead of hsp70::white, as the marker residing between the 5′ and 3′ multiple-cloning sites (MCS) and use of LexAop2-opRac1V12, rather than UAS::Rpr, for eliminating nonspecific insertions. Two more modules, LexAop2 and riTS-Rac1V12, are separated by the FRT cassette for reconstitution of a suppressible LexAop2-riTS-Rac1V12 following flip-out. A dU6-3 promoter-driven gRNA was further added with two SapI sites for easy gRNA target site cloning. (C) {donor} is first integrated at attP sites on either the second or the third chromosome. To enrich for correct GT following induction of donor DNA flip-out, three scenarios of “unwanted” events are eliminated by lethality selection. Nonsuppressible Rac1V12 is expressed under syb-LexA::p65 to kill organisms experiencing “no excision” or “nonspecific insertion.” In the case of “no targeting,” the reconstituted LexAop2-riTS-Rac1V12 drives lethality. By design, only after ends-out GT leading to loss of the nonsuppressible LexAop2-opRac1V12 can the organism overcome the suppressible LexAop2-riTS-Rac1V12 with lexAop-rCD2i and survive the selection.
Golic+: A genetic toolkit for D. melanogaster HR gene targeting
We assembled a GT backbone (pTL2) from scratch by adding the above selection transgenes in specific orientations plus various features, including FRT/loxP sequences, I-SceI/I-CreI/I-CeuI cutting sites, an attPX integration site, and multiple cloning sites for 5′ and 3′ homology arms, into a basic plasmid (Figure 2B). Briefly, lexAop-rCD2i, flanked by direct repeats of loxP, resides between the 5′ and 3′ multiple-cloning sites, and the nonrepressible 5XLexAop2-Rac1V12 sits in an opposite orientation following the 3′ cloning sites. In addition, another 5XLexAop2 promoter and the repressible Rac1V12 sequence lie before and after the first and second FRTs, respectively, such that upon flip-out, a 5XLexAop2-FRT-riTS-Rac1V12 module is reconstituted to serve as the inducible and suppressible toxicity background. Besides, an I-SceI cutting site is positioned next to one of the FRTs for linearizing circularized donor DNA, and two rare-cutting sites (I-CreI and I-CeuI sites) plus an attPX integration site are included for potential retargeting (Huang et al. 2011). Finally, the gRNA backbone was incorporated into pTL2 that allows integration of donor DNA and gRNA (abbreviated as {donor, gRNA}) into the same attP site via one transgenesis (Figure 2B).
The placement of a nonrepressible 5XLexAop2-Rac1V12 in the FRT cassette outside the recombination region permits tracking of the original transgene based on the bar eye phenotype induced by GMR-LexA::GADd (Figure 3A). During GT screening, it further allows us to kill those offspring, which fail to lose the nonrepressible RacV12 due to lack of excision or nonspecific insertion, at the pupal stage with nSyb-LexA::p65. Among those that have lost the nonrepressible Rac1V12 following excision of the FRT cassette, a repressible 5XLexAop2-riTS-Rac1V12 is automatically reconstituted and only the offspring with an insertion of the repressor, lexAop-rCD2i, contained within the 5′ and 3′ homology arms could survive. Viable candidates likely represent correct GT given loss of the nonrepressor Rac1V12 and presence of the repressor transgene (Figure 2C). Hence, with pTL2, we achieved lethality selection for easy GT candidate recovery.
For co-induction of Cas9, FLP, and I-SceI in CBs, we employed 2A peptides to express all three enzymes in one transcript under the control of a minimal, but appropriately expressed, bam promoter to introduce CRISPR/Cas and donor DNA in most, if not all, CBs (Chen and Mckearin 2003; Szymczak et al. 2004; Diao and White 2012). Additional transgenes involved in Golic+ include GMR-LexA::GAD, nSyb-LexA::p65, and 5XLexAop2-riTS-Rac1V12. To simplify the genetic crosses, all common transgenes required (Table 1) were purposefully preassembled in two of the three major chromosomes so that the target chromosome is left untouched (Table 2).
Table 1. List of transgenic lines required for implementing Golic+.
| Full name | Abbreviation | Integration site | Note |
|---|---|---|---|
| Donor DNA plus gRNA in pTL | {donor, gRNA} | attP40, VK00027 | |
| GMR3-LexA::GADd | GMR3-LexA | attP40, attP2 | Cross with {donor, gRNA} injected adults to create rough eyes for {donor} transformant screening. |
| bamP(198)-Cas9-P2A-FLP-E2A-I-SceI | bam198-CFI | su(Hw)attP8, attP2 | Expressing Cas9, FLP, and I-SceI under the bamP control to release donor DNA and introduce DSB at the target locus in every cystoblast. |
| 5XLexAop2-rCD2miRNATS#6-Rac1V12 (3xP3-RFP) | 5X-riTS-Rac1V12 | attP40, VK00027 | Together with {donor, gRNA}*, providing a homozygous suppressible “toxic” background. |
| Residual {donor, gRNA} | {donor, gRNA}* | After donor release, it will reconstitute as a suppressible toxic module, 5X-FRT-riTS-Rac1V12. | |
| nSyb-LexA::p65 | nSyb-LexA | attP16, VK00027 | Larval/pupal lethality selection. |
Table 2. Quick reference for using Golic+.
| Targeting | |||
|---|---|---|---|
| Transgenes set | X chromosome | Second chromosome | Third chromosome |
| {donor, gRNA} in | attP40 | VK00027 | attP40 |
| 5X-riTS-Rac1V12 in | attP40 | VK00027 | attP40 |
| nSyb-LexA in | attP16 | VK00027 | attP16 |
| bam198-CFI in | attP2 | su(Hw)attP8 | su(Hw)attP8 |
GT with Golic+ starts with construction of {donor, gRNA} and integration of {donor, gRNA} at specific attP sites. The {donor, gRNA} transformants are identified based on the GMR-LexA::GAD-induced rough-eyed phenotypes. To carry out GT in CBs then requires generation of female flies carrying {donor, gRNA} and 5XLexAop2-riTS-Rac1V12 at the same attP sites of the homologous chromosomes plus bamP-Cas9-2A-FLP-2A-I-SceI on a heterologous chromosome. The offspring that survive in the presence of nSyb-LexA::P65 are recovered as GT candidates (Figure 3B). Two categories of false-positive candidates potentially exist and can be identified through locating the repressor-marked donor DNA genetically (Figure S3A). “Escapers” refer to those whose survival no longer depends on lexAop-rCD2i due to defects in the reconstitution of the repressible 5XLexAop2-FRT-Rac1V12 during excision of the donor DNA FRT cassette. “Local integrations” include those that have lost the nonrepressible 5XLexAop2-Rac1V12 part of the donor DNA but maintained lexAop-rCD2i on the original chromosome (Figure S3B). Only the candidates with lexAop-rCD2i mapped onto the targeted chromosome are subjected to genomic PCR confirmation. To recover pure GT lines, one can remove unwanted transgenes by selecting against their mini-white markers in crosses with w− flies.
Generating T2A-GAL4 knock-ins using Golic+
To validate our design of Golic+ and demonstrate its efficiency in ends-out GT, we explored the possibility of knocking T2A-Gal4 into two spatial patterning genes: msh, the dorsal columnar gene in the ventral neuroectoderm (Isshiki et al. 1997), and runt, a pair-rule class segmentation gene (Kania et al. 1990) (Table 3). We engineered the necessary gRNAs as well as msh-T2A-GAL4 and runt-T2A-Gal4 to target the 3′ end of their open reading frames (Diao and White 2012) (Figure 4, A and C). The msh-T2A-GAL4 donor DNA contains 3-kb homologous arms, while the runt-T2A-GAL4 donor DNA carries 2-kb homologous arms. They were both integrated at attP40, on the second chromosome, for targeting genes residing on the third and X chromosomes, respectively.
Table 3. Summary of targeting msh and runt with Golic+.
| Transgenic {donor, gRNA} | No. FFa | Correct targeting | Nonspecific insertion | Escapers |
|---|---|---|---|---|
| {msh-T2A-Gal4 KI, gRNA} in attP40 | 95 | 47 (36)b | 8 (8) | 15 (14) |
| {runt-T2A-Gal4 KI, gRNA} in attP40 | 50 | 12 | 3 | 20 |
Golic+ was used to knock in T2A-Gal4 in both msh and runt. The first available Golic+ set with transgenes on the second and third chromosomes was used although not ideal for targeting msh on the third chromosome. Since one FF could sometimes yield multiple candidates, the number of candidates may exceed the number of candidate-producing FF.
FF, founder females.
Number of candidates (number of candidate-producing FF) in the specific category.
For GT of msh, we carried out 95 single-founder female crosses, 50 of which yielded 70 viable adults in total. By locating the rCD2i-marked donor DNA genetically, we identified 15 escapers and 8 false candidates with local integration. The remaining 47 candidates carry lexAop-rCD2i on the third chromosome where endogenous msh resides and were subsequently confirmed as correct msh knock-ins by genomic PCR. For GT of runt, we carried out a group culture for 50 founder females and recovered 35 viable adults in total. Subsequent genetic mapping revealed 12 candidates carrying lexAop-rCD2i on the targeted X chromosome. Genomic PCR confirmed all these 12 candidates as correct runt knock-ins. Notably, all the correct runt knock-ins were initially recovered as heterozygous females and later found to be homozygous as well as hemizygous lethal. This phenomenon indicates that knocking in T2A-GAL4 somehow impairs endogenous runt function or drives GAL4 expression in a toxic pattern or level. Moreover, it implies that a comparable number of hemizygous male candidates that carry correct runt knock-ins should have existed but died precociously during the initial lethality screen. Taking this into consideration, Golic+ has achieved a consistent efficiency in the ends-out GT at the success rate of recovering ∼50, likely independent, correct GTs from 100 founder females.
We further examined the knocked-in GAL4s’ activity patterns using UAS-GFP::Dbox, a cell cycle labile reporter that drastically reduces the GFP perdurance derived from parental cells’ expressions. Both msh-T2A-Gal4 and runt-T2A-Gal4 show patterned GAL4 activities in the embryonic neuromeres, roughly reflecting msh’s expression in the lateral-most column of neural progenitors and runt’s expression in specific rows of neuroblasts plus additional cells (Figure 4, B and D).
In conclusion, Golic+ has improved the conventional Golic system of ends-out GT in three aspects, including automatic independent induction of GT in CBs, promotion of GT with CRISPR/Cas, and recovery of correct GT based on suppression of pupal lethality (Figure S4). The “+” in Golic+ indicates our improvements to the pioneering Golic GT system.
Discussion
In the original and all previous modified Golic systems, the donor DNA is transiently released by heat shock and subsequently lost in most, if not all, cells at the midlarval stage when the developing ovaries carry female germline stem cells plus a limited number of CBs. By contrast, Golic+ ensures a continuous supply of the linear donor DNA to the serially derived CBs throughout female reproduction. This creates an assembly line with each newborn CB experiencing an independent trial of GT. Given the independent nature of GT occurring in CBs, we can further pool unsynchronized Golic+ founder females and breed them together in one bottle per GT. Golic+ thus converts the once highly involved and often unpredictable GT process into standard straightforward genetic crosses, such that one can perform multiple GT experiments simultaneously. If needed, one can scale crosses up indefinitely to recover targeting events even at loci with significantly lower targeting efficiency.
Fortunately, the bam promoter permits appropriately timed induction of transgenes selectively in newborn CBs. We can therefore maintain the resident donor DNA in the female germline stem cells and then excise it only in the serially derived CBs. Using a reporter construct mimicking the donor DNA in both length and the arrangement of FRTs and the I-SceI site, we have identified the attP sites where donor DNA can be efficiently flipped out. We have further optimized the bam promoter-dependent induction to maximize the percentage of CBs that have received the linear donor DNA during active oogenesis.
Golic+ further employs a repressor-based pupal lethality selection to facilitate the recovery of potential GT events. Our optimization of the supply of linear donor DNAs to CBs has led to the recovery of many more false-positive offspring, compared to the conventional midlarval pulse induction (data not shown). False positives appeared at a rather constant level with ratios over founder females being ∼30%, but the frequencies of correct GT could be drastically increased with potent Cas9 and gRNA transgenes. The promotion of correct GT by CRISPR/Cas did not reduce false positives, suggesting independent sources of escapers, nonspecific insertion, and correct GT. We suspect that local integrations, which all reside on the original chromosome, arose by a common mechanism involving local rearrangement or hopping (Figure S3B). This would imply that the correct GT events enabled by CRISPR/Cas are derived through recruitment of an otherwise lost pool of liberated donor DNAs.
The recent introduction of CRISPR/Cas has made GT via direct injection of donor DNAs plus supporting reagents (e.g., guide RNA) into early embryos possible in Drosophila (Baena-Lopez et al. 2013; Gratz et al. 2013; Bassett and Liu 2014; Port et al. 2014; Xue et al. 2014; Yu et al. 2014). In addition to rapid turnover, direct embryo injection allows easy adoption of diverse GT strategies. Can Golic+ secure the female germline as the preferred site for GT in Drosophila melanogaster? First, it is more efficient and scalable to generate independent trials of genome modification in the continually generated CBs than within the fixed/small pool of embryonic primordial germ cells. Second, the reliability of direct injection remains unclear. Can one consistently recover correct GT from an affordable scale of embryo injection? As large inserts or difficult loci may reduce GT efficiency by orders of magnitude, injection may need to be scaled up to prohibitive levels. To repeat microinjections with freshly prepared DNAs and RNAs is further costly, labor intensive, and time consuming. In addition, the great scalability of Golic+ may allow further reduction in the length of homology arms required for efficient GT (Beumer et al. 2013).
Despite the current success in Golic+, several issues remain to be addressed. First, to eliminate escapers, we have deliberately labeled 5X-riTS-Rac1V12 with 3xP3-RFP, hoping that we can preselect for red-fluorescent-eyed candidates that must carry the repressor, lexAop-rCD2i. Yet, candidates with 3xP3-RFP were a minority among correctly targeted candidates, possibly because the residual donor (5X-FRT-riTS-Rac1V12) is less toxic than 5X-riTS-Rac1V12. To reduce this bias, we lessen the selection toxicity from 5XLexAop2- to 3XLexAop2-riTS-Rac1V12. Second, we note that nonspecific insertion was not reported with direct embryo injection. We suspect that using circular vs. linear donor DNA might underlie this stunning difference. Additionally, it has been argued that circular donor DNA outperforms the linearized form in targeting efficiency (Beumer et al. 2008; Gratz et al. 2014). Hence, we will generate and explore the effectiveness of bamP-Cas9-2A-FLP in eliciting GT in CBs. Finally, while several features (I-CreI, I-CeuI, and attPX) have been built in for retargeting, recombinase-mediated cassette exchange (RMCE) (Schlake and Bode 1994) appears to be a superior approach. We will therefore add the ability for RMCE into our next version of Golic+.
In sum, using the widely available phiC31 integration system one can reliably insert donor DNA and the corresponding guide RNA, in one construct, into pretested attP sites. The remaining transgenes are then supplied via common fly stocks as follows: (i) detecting and balancing the initial transformants (GMR-LexA::GADd), (ii) conducting cystoblast-specific GT (bamP-Cas9-2A-FLP-2A-I-SceI and 5XLexAop2-riTS-Rac1V12), and (iii) implementing the pupal-lethal selection (nSyb-LexA::p65). Only strong candidates can eclose and be PCR validated immediately after breeding. The entire procedure will take just two rounds of en masse crosses after the establishment of the starter line carrying donor DNA and guide RNA (Figure 3). Using only well-established genetic/transgenic techniques, the relatively effortless Golic+ should empower all fly laboratories to perform sophisticated ends-out GT.
Supplementary Material
Acknowledgments
H.-M.C. designed and conducted experiments, collected and analyzed data, and prepared the manuscript. Y.H., B.D.P., and X.Y. provided molecular biology support for a subset of DNA constructs. T.L. conceived the project and wrote the paper. The authors thank Jon-Michael Knapp for the suggestion of bamP, Dennis M. Mckearin for sharing the critical bamP-GAL4 driver, Julian Ng for Rac1 cDNA, Justin Crocker and David Stern for sharing codon-optimized Cas9, Fillip Port and Simon L. Bullock for discussions about constructing transgenic CRISPR/Cas9, Hung-Hsiang Yu and Chun-Hong Chen for miRNA/target sequence design, and Mark Schroeder for comments on the manuscript. This work is supported by the Howard Hughes Medical Institute.
Footnotes
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.114.173716/-/DC1.
Communicating editor: J. Sekelsky
Literature Cited
- Awasaki T., Kao C. F., Lee Y. J., Yang C. P., Huang Y., et al. , 2014. Making Drosophila lineage-restricted drivers via patterned recombination in neuroblasts. Nat. Neurosci. 17: 631–637. [DOI] [PubMed] [Google Scholar]
- Baena-Lopez L. A., Alexandre C., Mitchell A., Pasakarnis L., Vincent J. P., 2013. Accelerated homologous recombination and subsequent genome modification in Drosophila. Development 140: 4818–4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett A. R., Liu J. L., 2014. CRISPR/Cas9 and genome editing in Drosophila. J. Genet. Genomics 41: 7–19. [DOI] [PubMed] [Google Scholar]
- Bassett A. R., Tibbit C., Ponting C. P., Liu J. L., 2013. Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Rep. 4: 220–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beumer K. J., Trautman J. K., Bozas A., Liu J. L., Rutter J., et al. , 2008. Efficient gene targeting in Drosophila by direct embryo injection with zinc-finger nucleases. Proc. Natl. Acad. Sci. USA 105: 19821–19826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beumer, K. J., J. K. Trautman, K. Mukherjee, and D. Carroll, 2013 Donor DNA utilization during gene targeting with zinc-finger nucleases. G3 3: 657–664. [DOI] [PMC free article] [PubMed]
- Bibikova M., Golic M., Golic K., Carroll D., 2002. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics 161: 1169–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M., Beumer K., Trautman J. K., and D. Carroll, 2003. Enhancing gene targeting with designed zinc finger nucleases. Science 300: 764. [DOI] [PubMed] [Google Scholar]
- Brand A. H., Perrimon N., 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415. [DOI] [PubMed] [Google Scholar]
- Byrne S. M., Ortiz L., Mali P., Aach J., Church G. M., 2014. Multi-kilobase homozygous targeted gene replacement in human induced pluripotent stem cells. Nucleic Acids Res. pii: gku1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capecchi M. R., 2005. Gene targeting in mice: functional analysis of the mammalian genome for the twenty-first century. Nat. Rev. Genet. 6: 507–512. [DOI] [PubMed] [Google Scholar]
- Chen C. H., Huang H., Ward C. M., Su J. T., Schaeffer L. V., et al. , 2007. A synthetic maternal-effect selfish genetic element drives population replacement in Drosophila. Science 316: 597–600. [DOI] [PubMed] [Google Scholar]
- Chen D., Mckearin D. M., 2003. A discrete transcriptional silencer in the bam gene determines asymmetric division of the Drosophila germline stem cell. Development 130: 1159–1170. [DOI] [PubMed] [Google Scholar]
- Cong L., Ran F. A., Cox D., Lin S., Barretto R., et al. , 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao F., White B. H., 2012. A novel approach for directing transgene expression in Drosophila: T2A-Gal4 in-frame fusion. Genetics 190: 1139–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis M. C., O’Neill E. M., Rubin G. M., 1993. Expression of Drosophila glass protein and evidence for negative regulation of its activity in non-neuronal cells by another DNA-binding protein. Development 119: 855–865. [DOI] [PubMed] [Google Scholar]
- Gong W. J., Golic K. G., 2003. Ends-out, or replacement, gene targeting in Drosophila. Proc. Natl. Acad. Sci. USA 100: 2556–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz S. J., Cummings A. M., Nguyen J. N., Hamm D. C., Donohue L. K., et al. , 2013. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics 194: 1029–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz S. J., Ukken F. P., Rubinstein C. D., Thiede G., Donohue L. K., et al. , 2014. Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics 196: 961–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison M. M., Jenkins B. V., O’Connor-Giles K. M., Wildonger J., 2014. A CRISPR view of development. Genes Dev. 28: 1859–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay B. A., Wolff T., Rubin G. M., 1994. Expression of baculovirus P35 prevents cell death in Drosophila. Development 120: 2121–2129. [DOI] [PubMed] [Google Scholar]
- Hernandez G., Valafar F., Stumph W. E., 2007. Insect small nuclear RNA gene promoters evolve rapidly yet retain conserved features involved in determining promoter activity and RNA polymerase specificity. Nucleic Acids Res. 35: 21–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn C., Jaunich B., Wimmer E. A., 2000. Highly sensitive, fluorescent transformation marker for Drosophila transgenesis. Dev. Genes Evol. 210: 623–629. [DOI] [PubMed] [Google Scholar]
- Hsu P. D., Lander E. S., Zhang F., 2014. Development and applications of CRISPR-Cas9 for genome engineering. Cell 157: 1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Zhou W., Watson A. M., Jan Y.-N., Hong Y., 2008. Efficient ends-out gene targeting in Drosophila. Genetics 180: 703–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Ghosh P., Hatfull G. F., Hong Y., 2011. Successive and targeted DNA integrations in the Drosophila genome by Bxb1 and φC31 integrases. Genetics 189: 391–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang W. Y., Fu Y., Reyon D., Maeder M. L., Tsai S. Q., et al. , 2013. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 31: 227–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isshiki T., Takeichi M., Nose A., 1997. The role of the msh homeobox gene during Drosophila neurogenesis: implication for the dorsoventral specification of the neuroectoderm. Development 124: 3099–3109. [DOI] [PubMed] [Google Scholar]
- Jasin M., 1996. Genetic manipulation of genomes with rare-cutting endonucleases. Trends Genet. 12: 224–228. [DOI] [PubMed] [Google Scholar]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J. A., et al. , 2012. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kania M. A., Bonner A. S., Duffy J. B., Gergen J. P., 1990. The Drosophila segmentation gene runt encodes a novel nuclear regulatory protein that is also expressed in the developing nervous system. Genes Dev. 4: 1701–1713. [DOI] [PubMed] [Google Scholar]
- Kim H., Kim J.-S., 2014. A guide to genome engineering with programmable nucleases. Nat. Rev. Genet. 15: 321–334. [DOI] [PubMed] [Google Scholar]
- Kim J. H., Lee S.-R., Li L.-H., Park H.-J., Park J.-H., et al. , 2011. High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS ONE 6: e18556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S., Ueda R., 2013. Highly improved gene targeting by germline-specific cas9 expression in Drosophila. Genetics 195: 715–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai S.-L., Lee T., 2006. Genetic mosaic with dual binary transcriptional systems in Drosophila. Nat. Neurosci. 9: 703–709. [DOI] [PubMed] [Google Scholar]
- Lee T., Luo L., 1999. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22: 451–461. [DOI] [PubMed] [Google Scholar]
- Leismann O., Lehner C. F., 2003. Drosophila securin destruction involves a D-box and a KEN-box and promotes anaphase in parallel with Cyclin A degradation. J. Cell Sci. 116: 2453–2460. [DOI] [PubMed] [Google Scholar]
- Luo L., Liao Y. J., Jan L. Y., Jan Y. N., 1994. Distinct morphogenetic functions of similar small GTPases: Drosophila Drac1 is involved in axonal outgrowth and myoblast fusion. Genes Dev. 8: 1787–1802. [DOI] [PubMed] [Google Scholar]
- Mali P., Yang L., Esvelt K. M., Aach J., Guell M., et al. , 2013. RNA-guided human genome engineering via Cas9. Science 339: 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer B. D., Ngo T.-T. B., Hibbard K. L., Murphy C., Jenett A., et al. , 2010. Refinement of tools for targeted gene expression in Drosophila. Genetics 186: 735–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer B. D., Truman J. W., Rubin G. M., 2012. Using translational enhancers to increase transgene expression in Drosophila. Proc. Natl. Acad. Sci. USA 109: 6626–6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Port F., Chen H. M., Lee T., Bullock S. L., 2014. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc. Natl. Acad. Sci. USA 111: E2967–E2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X., Sun J., Housden B. E., Hu Y., Roesel C., et al. , 2013. Optimized gene editing technology for Drosophila melanogaster using germ line-specific Cas9. Proc. Natl. Acad. Sci. USA 110: 19012–19017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong Y. S., Golic K. G., 2000. Gene targeting by homologous recombination in Drosophila. Science 288: 2013–2018. [DOI] [PubMed] [Google Scholar]
- Rørth P., 1998. Gal4 in the Drosophila female germline. Mech. Dev. 78: 113–118. [DOI] [PubMed] [Google Scholar]
- Sander J. D., Joung J. K., 2014. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 32: 347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer B., 1993. Manipulation of transgenes by site-specific recombination: use of cre recombinase. Methods Enzymol. 225: 890–900. [DOI] [PubMed] [Google Scholar]
- Schlake T., Bode J., 1994. Use of mutated FLP recognition target (FRT) sites for the exchange of expression cassettes at defined chromosomal loci. Biochemistry 33: 12746–12751. [DOI] [PubMed] [Google Scholar]
- Sebo Z. L., Lee H. B., Peng Y., Guo Y., 2013. A simplified and efficient germline-specific CRISPR/Cas9 system for Drosophila genomic engineering. Fly 8: 52–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling, A. C., 1993 Developmental genetics of oogenesis, pp. 1–70 in The Development of Drosophila melanogaster, Vol. 1 edited by Michael Bate and Alfonso Martinez Arias, Cold Spring Harbor Laboratory Press, Long Island, New York.
- Szymczak A. L., Workman C. J., Wang Y., Vignali K. M., Dilioglou S., et al. , 2004. Correction of multi-gene deficiency in vivo using a single ‘self-cleaving’ 2A peptide–based retroviral vector. Nat. Biotechnol. 22: 589–594. [DOI] [PubMed] [Google Scholar]
- Thomas K. R., Capecchi M. R., 1987. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell 51: 503–512. [DOI] [PubMed] [Google Scholar]
- Wakiyama M., Matsumoto T., Yokoyama S., 2005. Drosophila U6 promoter-driven short hairpin RNAs effectively induce RNA interference in Schneider 2 cells. Biochem. Biophys. Res. Commun. 331: 1163–1170. [DOI] [PubMed] [Google Scholar]
- Xue, Z., M. Ren, M. Wu, J. Dai, Y. S. Rong et al., 2014 Efficient gene knock-out and knock-in with transgenic Cas9 in Drosophila. G3 4: 925–929. [DOI] [PMC free article] [PubMed]
- Yu H.-H., Chen C. H., Shi L., Huang Y., Lee T., 2009. Twin-spot MARCM to reveal the developmental origin and identity of neurons. Nat. Neurosci. 12: 947–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z., Ren M., Wang Z., Zhang B., Rong Y. S., et al. , 2013. Highly efficient genome modifications mediated by CRISPR/Cas9 in Drosophila. Genetics 195: 289–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z., Chen H., Liu J., Zhang H., Yan Y., et al. , 2014. Various applications of TALEN-and CRISPR/Cas9-mediated homologous recombination to modify the Drosophila genome. Biol. Open 3: 271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinyk D. L., Mercer E. H., Harris E., Anderson D. J., Joyner A. L., 1998. Fate mapping of the mouse midbrain–hindbrain constriction using a site-specific recombination system. Curr. Biol. 8: 665–672. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.