Abstract
Accounting for gene–environment (G×E) interactions in complex trait association studies can facilitate our understanding of genetic heterogeneity under different environmental exposures, improve the ability to discover susceptible genes that exhibit little marginal effect, provide insight into the biological mechanisms of complex diseases, help to identify high-risk subgroups in the population, and uncover hidden heritability. However, significant G×E interactions can be difficult to find. The sample sizes required for sufficient power to detect association are much larger than those needed for genetic main effects, and interactions are sensitive to misspecification of the main-effects model. These issues are exacerbated when working with binary phenotypes and rare variants, which bear less information on association. In this work, we present a similarity-based regression method for evaluating G×E interactions for rare variants with binary traits. The proposed model aggregates the genetic and G×E information across markers, using genetic similarity, thus increasing the ability to detect G×E signals. The model has a random effects interpretation, which leads to robustness against main-effect misspecifications when evaluating G×E interactions. We construct score tests to examine G×E interactions and a computationally efficient EM algorithm to estimate the nuisance variance components. Using simulations and data applications, we show that the proposed method is a flexible and powerful tool to study the G×E effect in common or rare variant studies with binary traits.
Keywords: binary traits, gene–environment interaction, rare variant association, GLMM, marker-set interaction analysis, variance-component methods
HUMAN complex traits have a multifactor etiology that involves the interplay between genetic susceptibility and environmental exposures. Studies of gene–environment (G×E) interactions can facilitate our understanding of genetic heterogeneity under different environmental exposures (Kraft et al. 2007; Van Os and Rutten 2009), help to identify high-risk subgroups in the population (Murcray et al. 2009), provide insight into the biological mechanisms of complex diseases (Thomas 2010), and improve the ability to discover susceptible genes that interact with other factors but exhibit little marginal effect (Thomas 2010). However, finding significant G×E interactions is not an easy task. Model misspecification, inconsistent definitions of the environmental variable, and insufficient sample sizes are just a few of the issues that often lead to low power and nonreproducible findings in G×E studies (Mechanic et al. 2012; Jiao et al. 2013; Winham and Biernacka 2013). In particular, the sample size needed to detect a G×E effect is usually four times larger than that needed to detect a main effect of similar magnitude (Thomas 2011). Thus, researchers need a robust, powerful G×E test to generate reproducible findings.
Conventionally, researchers search for significant genetic or G×E associations, using single-SNP methods, e.g., the Kraft 2-d.f. test (Kraft et al. 2007) or the simultaneous test of Dai (Dai et al. 2012). More complex methods (e.g., Mukherjee and Chatterjee 2008; Murcray et al. 2009; Sohns et al. 2013) aim to retain the advantages from both the case-only test (high power but sensitive to G–E correlations) and the standard case–control G×E test (low power but robust to G–E correlations). Despite the many efforts to improve single-SNP G×E tests, issues remain; e.g., a large proportion of trait heritability remains unexplained (Manolio et al. 2009) due to false positive and/or false negative findings.
Inflated false positive rates arise when the model used to screen for G×E interactions does not correctly reflect the true underlying genetic (G) and environmental (E) effects (Voorman et al. 2011; Lin et al. 2013; Wang et al. 2013). To address this issue, Voorman et al. (2011) suggested a model-robust estimate of the variance, and Lin et al. (2013) and Wang et al. (2013) suggested a random-effect model to capture the genetic main effect. The false negative (underpower) issues can be addressed by evaluating G×E effects on a set of markers, e.g., on genes, linkage disequilibrium (LD) blocks, or pathways (Tzeng et al. 2011; Lin et al. 2013). Marker–set G×E analysis can improve power by aggregating effects across markers. Such accumulation methods account for LD among markers and reduce the total number of tests to be performed. The improved power is particularly crucial for common variants with subtle individual effects and for rare variants with sparse occurrence (Sham and Cherny 2011). In addition, operating at a gene/pathway level helps increase reproducibility (Sohns et al. 2013).
Several G×E marker-set methods are available to study associations with common variants, where the major task is to avoid a large number of parameters for modeling G, E, and G×E variables. One of the first proposed G×E marker-set methods was Tukey’s 1-d.f. test (Chatterjee et al. 2006), which made significant progress toward fully understanding complex diseases. However, this method makes the often incorrect assumption that a SNP’s interaction effect is proportional to its marginal genetic effect (Winham and Biernacka 2013). Other commonly adopted G×E marker-set methods include minimum P-value (min-P) methods and weighted burden methods, where weights can be obtained from the principal components (PCs) of the SNP genotypes (Winham and Biernacka 2013) or from the G–E correlation (Jiao et al. 2013). In particular, Jiao et al. (2013) showed that the correlation between G and E can serve as an informative indicator for G×E interactions and that incorporating G–E correlations as weights can increase the signal-to-noise ratio in a G×E marker set while avoiding permutations. However, these observations are valid only when the true G–E correlation is in the same direction as the G×E interaction (Jiao et al. 2013). Fan and Lo (2013) proposed a model-free approach based on a summation of partitions to evaluate the interaction effects for rare variants. However, their method evaluates only the combined effect of G and G×E, not the separated effects. Recently, Lin et al. (2013) proposed a generalized linear mixed-effect model (GLMM) for G×E interactions for binary and continuous traits and showed it has superior power and robustness over min-P methods. A similar method, similarity regression (SimReg), proposed by Tzeng et al. (2011) to study marker-set G×E for continuous traits, was shown to be connected to linear mixed-effect models.
In this article, we extend the SimReg G×E framework established in Tzeng et al. (2011) to binary traits with common or rare variants. SimReg, which is inspired by Haseman–Elston regression for linkage analysis (Haseman and Elston 1972; Elston et al. 2000) and haplotype similarity tests for regional association (Tzeng et al. 2003; Beckmann et al. 2005), uses a regression model to correlate trait similarity with genetic similarity across multiple loci and to account for covariates. SimReg has been shown to perform well for common and rare variants (Tzeng et al. 2011). However, unlike similarity-based testing for the genetic main effect (Tzeng et al. 2009) or for G×E with quantitative traits (Tzeng et al. 2011), G×E tests with binary traits have several challenges associated with computation and estimation. In particular, G×E tests require the estimation of nuisance parameters to capture the main effects. Estimating these parameters requires high-dimensional integration and the inversion of a high-dimensional similarity matrix. For quantitative G×E tests, this estimation can be sidestepped using the normality of the phenotype, but no such useful properties exist for binary G×E tests. To overcome these challenges, Lin et al. (2013) proposed using ridge regression to estimate the nuisance main effects, selecting the tuning parameter using generalized cross validation.
In our work, we develop an EM algorithm to approximate the integration and we alleviate the computational burden of maximum-likelihood estimation (MLE) by performing a low-rank approximation of the similarity matrix. We show that the SimReg coefficient can be expressed as a variance component of a working GLMM, which facilitates the derivation of a test statistic and unifies SimReg with other random-effect-based methods (e.g., Lin et al. 2013; Wang et al. 2013). The proposed SimReg method can incorporate covariates and uses a permutation-free procedure to evaluate G×E effects. In addition, the proposed method extends the model from linear effects (e.g., Jiao et al. 2013; Lin et al. 2013) to other complex effects by selecting appropriate similarity metrics, and it avoids the need to select tuning parameters. Unlike current robust marker-set G×E methods that focus on common variant analysis, we investigate the performance of the proposed G×E strategy with rare and common variants. We evaluate the validity and power of the proposed method using simulation studies and illustrate the utility of the proposed method via two data applications: one studies the interactions between PLA2G7 and physical activity on obesity, using Cohorte Lausannoise (CoLaus) sequencing data, and a second assesses the effect modifier role of body mass index (BMI) on the association between TCF7L2 and type 2 diabetes, using the Wellcome Trust Case Control Consortium data.
Materials and Methods
Gene–trait similarity regression for G×E effects
Let be the binary disease indicator for individual i (); i.e., if individual i has the disease of interest and otherwise. Let be the minor allele count for individual i at locus , let be a vector of environmental factors, and let be the vector of confounders. The full covariate vector is with dimension . All covariates are standardized to have a mean of 0 and a variance of 1. For illustration, we consider the case where , but it is straightforward to extend the proposed work to .
We quantify the trait similarity for a pair of individuals i and j, , as the weighted sample covariance between their disease statuses; i.e., , where is the subject-specific trait mean accounting for covariate but assuming no genetic effects and is a weight accounting for the fact that the ’s have difference variances (Tzeng et al. 2009). From this definition, the expected trait similarity is the covariance of and with weights . For binary traits, we assume a logistic model, , where γ is the coefficient vector of the covariate and is the optimal weight for the logistic model (Tzeng et al. 2009).
Genetic similarity is calculated as the weighted sum of single-marker similarities; i.e., where is the genetic similarity at marker m and is the weight. There are several choices for (e.g., Wessel and Schork 2006; Schaid 2010a); a popular one is the identity-by-state (IBS) metric: . Weights are typically based on allele frequencies, the degree of evolutionary conservation, or the functionality of the variants (Wessel and Schork 2006; Price et al. 2010; Schaid 2010a,b). For example, one can use the minor allele frequency (MAF) of marker m, denoted by , to up-weight similarities that are contributed by rare variants: e.g., (Wu et al. 2011) can be used to target rare variants only, or a moderate weight (Pongpanich et al. 2012) can be used to promote similarities attributed to rare alleles while retaining the contributions from common variants.
The proposed G×E gene–trait similarity regression model is
| (1) |
Because incorporates baseline covariate information, model (1) does not contain an intercept or an interaction covariate term (i.e., (Tzeng et al. 2011). Using model (1), one can assess the G×E interaction by testing , or one can perform a joint test for the genetic main effect and G×E interactions simultaneously by testing . The joint test is recommended if either the genetic heterogeneity or the G×E interaction mechanism is unknown (Kraft et al. 2007; Tzeng et al. 2011).
Score test for G×E effects and joint effects
Following a similar procedure to that found in Tzeng et al. (2009), we connect the similarity regression to a working GLMM to derive the score test. Consider the following GLMM,
| (2) |
where is a vector of conditional means and is a link function. Here, we consider a logit link .. Vectors and contain the subject-specific genetic main effect and G×E interaction, respectively. Assume and are random effects; i.e., and with , , and . Then, the marginal covariance of and in this working model is
where (see Appendix A). Recall the expected trait similarity is . Therefore,
where . In other words, we can examine and of model (1) by testing and in model (2), respectively.
To derive the score test statistics, we rewrite model (2) as
| (3) |
where , , L is the rank of matrix , and is a matrix satisfying . Matrix is defined in the same manner as , and because . Following Zhang and Lin (2003), the score statistic to examine the G×E effect (i.e., testing can be calculated as
where is the working vector in model (3) under with , and is the ith entry of and are the MLEs for and γ under , respectively; with , and . As noted in the literature (Zhang and Lin 2003; Tzeng and Zhang 2007), the second term, , is the mean of the first term and its variability is small compared to the first term. Thus, we derive our test statistic using only the first term; i.e.,
We propose an EM algorithm in Appendix B to obtain the MLEs for and γ.
In a similar manner, the score statistic under can be obtained as
and we define the test statistic of the joint effect as
where is the working vector under . Here, , , , , and is the MLE for γ under .
We show in Appendix C that and follow a weighted -distribution asymptotically under and , respectively. P-values can then be calculated numerically using moment-matching approximations (Duchesne and Lafaye de Micheaux 2010).
Low-rank approximation of for computational and statistical efficiency
The calculation of the G×E test statistic involves the inversion of matrices and , both of dimension . When n is large (e.g., >5k), direct inversion of these matrices can be computationally intensive, and the inversion must be performed at every EM iteration to obtain main-effect term b (see Appendix B). To reduce the computational intensity and to facilitate the inversion of these matrices, we consider a low-rank approximation of . The low-rank approximation has been used in the literature to improve power when the number of markers increases and when more noise is incorporated into (Cai et al. 2011). Previous works (Cai et al. 2011; Tzeng and Zhang 2007; Tzeng et al. 2011) indicate that is a positive semidefinite matrix, for which there are a few dominant eigenvalues. Assume that, are the leading eigenvalues that explain the majority of the variance of [i.e., for some ] and have corresponding eigenvectors . Then, we approximate by . For an appropriate choice of p (e.g., , contains most of the information from . Especially with rare variant data, is usually , and the computation is more straightforward.
Miao (2009) indicated that the potential bias caused by a low-rank approximation can be minimized if a high percentage of the variation of can be retained. In our explorations, we found that selecting too small a p did not affect the test size but did lead to power loss because too much genetic information is discarded. We also found that the power loss with a large p (e.g., p = 0.99) was negligible but could stabilize the numerical calculation and boost computational efficiency. The improvement when occurs because has many eigenvalues that are near zero. Using a p slightly <1 removes a large number of near-zero eigenvalues, which stabilizes the numerical computations, shortens the computational time, and yields a type I error rate close to the nominal level (Table 1).
Table 1. Type I error rates of SimReg tests with vs. without low-rank approximation in rare-variant (RV) simulations.
| % variance retained in (denoted by p) | Case–control sampling | Random sampling |
|---|---|---|
| Joint test | ||
| p = 100% | 0.052 (0.0070)a | 0.025 (0.0049) |
| p = 99% | 0.052 (0.0070) | 0.052 (0.0070) |
| G×E test | ||
| p = 100% | 0.036 (0.0059) | 0.024 (0.0048) |
| p = 99% | 0.047 (0.0067) | 0.043 (0.0064) |
| G×E test | ||
| p = 99% with 20 causal G SNPs | 0.064 (0.0077) | 0.046 (0.0066) |
| p = 99% with 40 causal G SNPs | 0.049 (0.0068) | 0.046 (0.0066) |
| p = 99% with 60 causal G SNPs | 0.045 (0.0066) | 0.043 (0.0064) |
| p = 99% with 80 causal G SNPs | 0.050 (0.0069) | 0.042 (0.0063) |
| p = 99% with 100 causal G SNPs | 0.041 (0.0063) | 0.045 (0.0066) |
The corresponding standard errors (SEs) are shown in parentheses. The values in italics are those whose 95% confidence intervals (i.e., rate ) fall below the nominal level. The results are based on 1000 replications. and are the group PARs of the genetic main effect and the G×E effect, respectively.
Standard errors of the type I error rates.
Simulation studies
To investigate the performance of the proposed SimReg G×E method, we conducted simulation studies. The first simulation focuses on rare-variant (RV) analysis using sequence data, and the second simulation focuses on common-variant (CV) analysis using HapMap data. The simulation data and code are available from the Dryad Digital Repository (http://datadryad.org/) at http://doi.org/10.5061/dryad.742gv (i.e., Dryad data identifier:doi:10.5061/dryad.742gv).
RV simulations:
We obtained 10,000 haplotypes for a 1-Mb region simulated by COSI (Schaffner et al. 2005) according to a coalescent model where the LD pattern and population history mimicked those of the European population. We selected the first 100 rare loci [i.e., minor allele frequency (MAF) <5%] for further analyses. We randomly drew 2 haplotypes with replacement from the 10,000 to form each subject’s genotype. We generated the binary phenotype from a Bernoulli distribution, where , , R is the number of causal loci, and is the number of rare alleles at causal locus r, . While we varied the value of R, we controlled the population attributable risk (PAR) at and for the genetic main effect and G×E effect, respectively (Madsen and Browning 2009). Given , , and R, we calculate and using and (Madsen and Browning 2009), where , and is the MAF for the rth locus based on the 10,000 haplotypes. We considered both case–control sampling with 750 cases and 750 controls and random sampling with sample size 1500 and prevalence rate 0.3.
In the type I error analysis, we set for the joint test and considered and for the G×E test. Because the burden-based tests are sensitive to the misspecification of the main-effect model (Voorman et al. 2011), we set a weak main-effect PAR so that the burden-based tests can still serve as a valid benchmark. We performed 10,000 replicates per scenario. In the power analysis, we set for both the G×E test and the joint test and considered , and 100. We performed 500 replicates per scenario. In all analyses, the 100 loci were included in the association tests.
SimReg’s performance was compared to GESAT (Lin et al. 2013) and a burden-based G×E test. GESAT is a GLMM-based G×E test that is closely connected to SimReg: from the GLMM representation in model (2), we see that SimReg assumes , where (calculated through the similarity kernel) determines how the G×E effects are modeled. In contrast, GESAT assumes a linear effect on , i.e., with , which is equivalent to setting (i.e., a linear kernel with ).
For SimReg, we used the weighted IBS kernel with weight . For GESAT, we used R code provided by the authors with the default settings to perform G×E tests (the code does not support joint tests). For the burden-based G×E test, we first summarize the marker-set information of subject i, using the number of rare variants in the set, referred to as mutation burden. Then, we fit a logistic model, , where is the mutation burden for subject i. Under this model, the G×E effect can be detected by testing , and the joint effect can be detected by testing .
CV simulations:
We obtained 234 phased haplotypes of gene TCF7L2 from chromosome 10 of the Utah residents with ancestry from northern and western Europe (CEU) samples in HapMap 3. We focused our analysis on the 29 typed SNPs genotyped in the Wellcome Trust Case Control Consortium (WTCCC) analysis (Wellcome Trust Case Control Consortium 2007). The MAFs of these 29 SNPs ranged from 0.0085 to 0.48. We randomly drew 2 haplotypes with replacement from the 234 phased haplotypes to form an individual genotype. We assumed that 2 of the 29 SNPs were causal and simulated the binary phenotype of individual i from a Bernoulli distribution, where , , and is the number of minor alleles at the causal locus . We generated the ith individual’s environmental covariate, , from a distribution and set , . As in the RV simulations, we considered case–control sampling (with 750 cases and 750 controls) and random sampling (with sample size 1500 and prevalence rate 0.3).
In the type I error analysis, we set for the joint test. For the G×E test, we set and considered and . We considered five pairs of causal SNPs (i.e., ) with different MAFs as shown in Table 3. We performed 1000 replicates per scenario. In the power analysis, we set and for both the G×E test and the joint test. We considered all possible pairs of causal SNPs for a total of = 406 scenarios. We performed 100 replicates per scenario. To mimic the typical CV analysis, we excluded the 2 causal SNPs and analyzed the other 27 SNPs only in the association tests. For SimReg, we set the locus-specific weight . We compared the proposed SimReg method to GESAT and the single-SNP minimum P-value method (referred to as min-P). For the min-P method, we fitted the model for each SNP m to obtain the P-values of the G×E test (i.e., testing ) and the joint test (i.e., testing ). For a given test (e.g., the G×E test), we took the minimum of the 27 G×E P-values and calculated the adjusted P-value as , where is the effective number of independent tests obtained using the method of Moskvina and Schmidt (2008).
Table 3. Type I error rates of the G×E test and the joint test for common-variant (CV) simulations.
| Effect size considered | MAFs of the causal SNPs | SimReg | min-P | GESAT |
|---|---|---|---|---|
| Joint test | NA | 0.044 (0.0065)a | 0.060 (0.0075) | NA |
| G×E test | ||||
| NA | 0.037 (0.0060) | 0.054 (0.0072) | 0.036 (0.0059) | |
| 0.009, 0.094 | 0.042 (0.0068) | 0.040 (0.0062) | 0.053 (0.0071) | |
| 0.009, 0.1966 | 0.040 (0.0062) | 0.043 (0.0064) | 0.055 (0.0072) | |
| 0.094, 0.1966 | 0.040 (0.0062) | 0.045 (0.0066) | 0.070 (0.0081) | |
| 0.1966, 0.2222 | 0.047 (0.0067) | 0.044 (0.0065) | 0.051 (0.0070) | |
| 0.2991, 0.4188 | 0.049 (0.0068) | 0.050 (0.0069) | 0.054 (0.0072) |
The corresponding standard errors (SEs) are shown in parentheses. The values in italics/boldface type are those whose 95% confidence intervals (i.e., rate ± 1.96 × SE) fall below/above the nominal level. The results were obtained based on 1000 replications, and and are the effect sizes of the causal SNPs for the main effect and the G×E effect, respectively.
Standard errors of the type I error rates.
Results and Discussion
Simulation studies
Results of type I error analyses (Table 1, Table 2, and Table 3):
Table 2. Type I error rates of the G×E test and the joint test for rare-variant (RV) simulations.
| Nominal level | SimRega | Burden-based | GESAT |
|---|---|---|---|
| Joint test | |||
| 0.05 | 0.0504 (0.0022)b | 0.0511 (0.0022) | NA |
| 0.01 | 0.0093 (0.0010) | 0.0110 (0.0010) | NA |
| 0.005 | 0.0047 (0.0007) | 0.0056 (0.0007) | NA |
| 0.001 | 0.0010 (0.0003) | 0.0011 (0.0003) | NA |
| G×E test | |||
| 0.05 | 0.0496 (0.0022) | 0.0523 (0.0022) | 0.05090 (0.0024) |
| 0.01 | 0.0085 (0.0009) | 0.0104 (0.0010) | 0.0119 (0.0011) |
| 0.005 | 0.0038 (0.0006) | 0.0044 (0.0007) | 0.0050 (0.0007) |
| 0.001 | 0.0007 (0.0026) | 0.0008 (0.0003) | 0.0007 (0.0003) |
| G×E test c | |||
| 0.05 | 0.0473 (0.0021) | 0.0482 (0.0021) | 0.0602 (0.0024) |
| 0.01 | 0.0099 (0.0010) | 0.0112 (0.0011) | 0.0119 (0.0011) |
| 0.005 | 0.0052 (0.0007) | 0.0055 (0.0007) | 0.0062 (0.0008) |
| 0.001 | 0.0014 (0.0004) | 0.0010 (0.0003) | 0.0009 (0.0003) |
Data were generated using a case–control design. The corresponding standard errors (SEs) are shown in parentheses. The values in italics/boldface type are those whose 95% confidence intervals (i.e., rate ± 1.96 × SE) fall below/above the nominal level. and are the group PARs of the genetic main effect and the G×E effect, respectively. The results were obtained based on 10,000 replications.
Using p (the proportion of variation explained by the leading eigenvalues in ) = 0.99.
Standard errors of the type I error rates.
Assuming 40 SNPs with causal main (G) effect.
The type I errors for the G×E test and the joint test are shown in Table 1 and Table 2 for RV simulations and Table 3 for CV simulations. From Table 1, we see that SimReg can have conservative type I errors when using P = 100%, which can be alleviated by using P = 99%. Table 2 shows that SimReg, burden-based, and GESAT methods all have type I error rates around the nominal level in RV analyses. Table 3 shows that SimReg, min-P, and GESAT all have type I error rates around the nominal level in the CV analyses.
Results of RV power analyses (Figure 1):
Figure 1.
Power of G×E and joint tests for rare-variant simulations. The powers of SimReg, burden-based, and GESAT tests are represented by the solid (─), dashed (- - -), and dotted (⋯) lines, respectively. GESAT is performed only under case–control studies. The results were obtained based on 500 replicates.
The power results for a main-effect group PAR ( of 0.02 and a G×E group PAR () of 0.1 are shown in Figure 1. For the G×E tests and the joint tests, SimReg has higher power than the burden-based test and GESAT (G×E test only) across different numbers of causal SNPs and different study designs. GESAT has the lowest power for the G×E test. Because we assumed a linear G×E effect in the simulation, the power loss may be attributable to the unweighted similarity (i.e., , which resulted in an overall similarity score dominated by less-frequent over rare variants and led to little variations among individual pairs.
We note that for both the SimReg and burden-based tests, the power of the joint test is slightly less than the power of the G×E test. It is likely that this is caused by the weak main-effect signal in the simulation: the majority of the simulated data sets had significant G×E effects but negligible genetic main effects. Consequently, compared to the G×E test statistic, the joint test statistic may have incorporated additional noise from the G test statistic, which can result in power loss. We also observe that the power loss in the joint test appears to be larger for SimReg than for the burden-based tests because the degrees of freedom (d.f.) of a SimReg test spent on the G effect tend to be higher than those of a burden-based test. However, the power of SimReg is still higher than that of the burden-based test, and the additional d.f. consumed by SimReg (compared to the burden-based test) ensure robustness against between-locus etiological heterogeneities (Pongpanich et al. 2012) as well as against model misspecifications.
Results of CV power analyses (Figure 2):
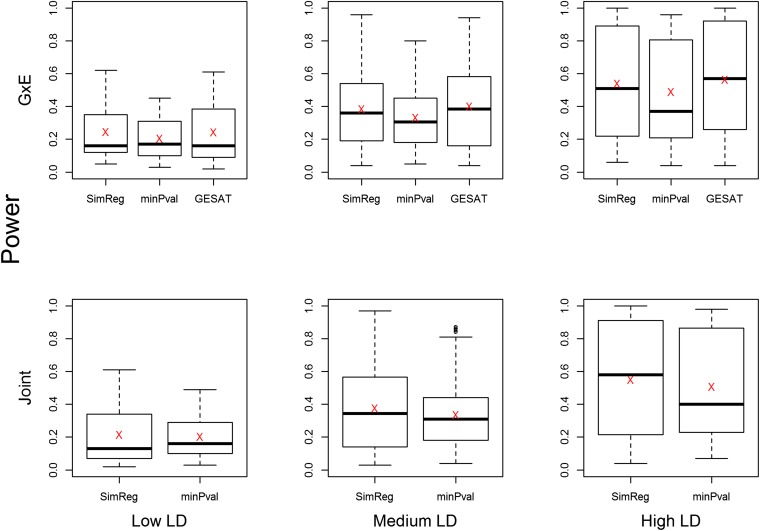
Figure 2.
Power of G×E and joint tests for common-variant simulations. The side-by-side boxplots show the powers of the proposed SimReg method, the minimum P-value method, and GESAT. A total of scenarios were considered (i.e., letting each SNP pair of the 29 SNPs be causal), with 100 replicates per scenario. The 406 scenarios were classified into three groups based on the LD pattern between the 2 causal SNPs and the remaining 27 SNPs. The red “X” in each boxplot represents the average power for each LD group.
To present the power results of the = 406 scenarios, we grouped the scenarios into three categories based on the LD structure between the causal SNPs and the analyzed SNPs. The three LD groups, i.e., the lower one-third (low LD), the middle one-third (medium LD), and the top one-third (high LD), are defined based on the average of 54 values, where each value is the between a causal SNP (2 in total) and an analyzed SNP (27 in total). We present side-by-side boxplots of the power of SimReg, min-P, and GESAT (for G×E tests) as well as the mean power value in Figure 2. We observe that when the LD is lower, the power of all methods is lower. This is expected because under low-LD scenarios the markers contain less information about the 2 causal loci. For the G×E test (Figure 2, top), SimReg and GESAT have very similar power, as expected because both methods set . The powers of SimReg and min-P are similar when LD is low. As the LD increases, SimReg starts to have power improvement over min-P. The difference becomes more obvious when LD is high. For the joint test (Figure 2, bottom), the relative power of SimReg vs. min-P is similar to what was observed for the G×E tests. Furthermore, the relative performance between SimReg and min-P for binary traits is similar to what was observed for quantitative traits (Tzeng et al. 2011).
Data Applications
Analysis of gene-by-physical activity effect on obesity, using CoLaus samples:
We used Sanger sequence data of the PLA2G7 gene for 1961 subjects from the CoLaus (Song et al. 2012) and studied PLA2G7’s association with the levels of lipoprotein-associated phospholipase A2 (Lp-PLA2). The CoLaus study of Firmann et al. (2008) is a population-based study to assess the risk factors of cardiovascular disease (CVD) in Caucasian residents of Lausanne, Switzerland aged 35–75 years. PLA2G7 encodes Lp-PLA2, and the elevated plasma levels of Lp-PLA2 activity have been shown to be associated with increased risk of coronary heart disease (Thompson et al. 2010). We imputed sporadic missing genotypes, using the MaCH software package (Li et al. 2010), and obtained a total of 100 SNPs with MAF < 0.05 (range from 0.000255 to 0.029).
The genetic influence of PLA2G7 on the body mass affected by exercise has been reported in the literature (Wootton et al. 2007; Detopoulou et al. 2009). The potential modulating effect of PLA2G7 on arachidonic acid was hypothesized to be related to the association between the PLA2G7 variants and a reduced risk of coronary artery disease (Ninio et al. 2004; Wootton et al. 2007). Using PLA2G7 as a positive control, we investigated the potential interaction between physical activity and genetic variants on BMI. We defined obesity as BMI > 30 and evaluated the effects of PLA2G7 (G), physical activity (E), and G×E interactions on obesity. We considered three methods: SimReg, GESAT, and the burden-based test. In all analyses, we adjusted for age, sex, ethnic background (five PCs), smoking status, and alcohol consumption. For SimReg, we used weight and a low-rank approximation with the resulting P-values of the joint test and the G×E test were and , respectively, which suggested that PLA2G7 may affect the influence of physical activity on obesity. GESAT, which set , yielded a G×E P-value of 0.637. These results are not unexpected given the simulation results; i.e., the unweighted similarity scores did not have power to detect rare variants because the contribution from rarer variants may be overwhelmed by the less rare variants during collapsing. The P-values of the burden-based tests were 0.013 for the joint test and for the G×E test, which are larger than SimReg P-values but give the same significant conclusions as SimReg. The results agree with the observation from the RV simulations that the proposed method is more powerful in detecting G×E effects.
Analysis of TCF7L2-by-BMI effect on type 2 diabetes, using WTCCC samples:
The data were obtained from the type 2 diabetes (T2D) case–control study conducted by the WTCCC (Wellcome Trust Case Control Consortium 2007). The controls were samples from the 1958 British Birth Cohort. The case samples were collected from various sites across the United Kingdom to be comparable to the controls. The genotyping was conducted on an Affymetrix 500K chip. Previous genome-wide association studies (Timpson et al. 2009) have indicated an interaction between TCF7L2 and BMI on T2D. Treating this TCF7L2×BMI effect on T2D as a true positive, we evaluated the performance of the proposed SimReg test (with weight ) and compared to GESAT and the min-P test.
We fitted a model where the response variable is the T2D status and the explanatory variables include the 29 SNPs in TCF7L2, BMI, TCF7L2×BMI, and sex. After applying sample and SNP quality control filters to remove substantial missing data, the data set contained 1913 cases and 1455 controls. We first performed the joint test and obtained a P-value of for SimReg and for min-P. The gene-level P-value of min-P is obtained as , where is the effective number of independent tests for TCF7L2 estimated by Moskvina and Schmidt (2008). The P-values of the G×E tests are for SimReg, for GESAT, and for min-P (adjusted P-value). The difference between SimReg and GESAT P-values can be attributed to the different choices of kernels (e.g., IBS kernel for SimReg vs. linear kernel for GESAT) and the different algorithm to estimate the nuisance main effects (e.g., EM algorithm vs. ridge penalization). The relatively large P-values of min-P suggest that there may be multiple moderate-effect loci in TCF7L2 contributing to the T2D risk, as opposed to a few strong-effect loci. The magnitude of the P-value difference in the joint tests was relatively small compared to the P-value difference in G×E tests, suggesting a strong main effect of TCF7L2 on T2D as shown in the literature (Helgason et al. 2007; Scott et al. 2007).
Conclusion
In this article we proposed a marker-set method based on similarity regression to examine G×E effects for binary traits and showed it is computationally feasible, powerful, and applicable to both common and rare variants. By demonstrating the equivalence of our gene-similarity regression model to a GLMM framework, we showed that SimReg is robust against model misspecification, like other random-effects-based approaches (e.g., Lin et al. 2013). However, because the structure of is atypical, one cannot apply the general score test of GLMM as implemented in existing statistical software because it often yields invalid estimates of (e.g., negative values). We developed an EM algorithm to address the challenges associated with estimation and computation encountered in GLMM model fitting. The C code that implements the proposed joint and G×E tests is available at http://www4.stat.ncsu.edu/~jytzeng/software_simreg.php. We demonstrated the utility of SimReg in rare variant G×E analysis. We also found that for RVs, the low-rank approximation to the main-effect similarity matrix () is necessary to avoid an overconservative type I error rate.
One possible strategy to apply the proposed SimReg tests is to start with a joint test to detect the overall association induced by the G main effect or the G×E effects. A screening by joint tests may lead to increased flexibility and power to detect a signal because some genes can exhibit negligible marginal effects but strong effects among particular exposure groups (Kraft et al. 2007; Thomas 2010). If the joint test is rejected, a G×E test can then be used to identify whether the effects of the genetic variables are modified by the environmental variables.
One can view the SimReg framework as an implementation of a class of models for modeling , which includes GESAT as a special case. In SimReg, one can determine how the G×E effect is modeled by specifying a certain similarity metric, e.g., linear kernel, IBS kernel, or quadratic kernel, as well as by imposing variant-specific weights when collapsing the information across markers. If a linear kernel is used with , the SimReg G×E test is equivalent to GESAT. However, one subtle difference is that SimReg uses an EM algorithm to estimate the nuisance main effects, whereas GESAT uses a penalized method. Another remark concerns the role of the variant-specific weight based on MAFs. As we observed in the numerical studies, although the unweighted similarity performed satisfactorily in CV analyses, it has little power in RV analyses. This is because the sum of unweighted similarity scores would be dominated by information from nonrare events. Consequently, when rare variants are studied, the multimarker similarity scores would exhibit little variation. The MAF-based weights in essence perform a soft thresholding to downweight or diminish the contribution of less-frequent or common variants in the multimarker similarity score.
The rationale of a collapsing analysis is to detect the amplified effects of rare variants in aggregate. Experience from main-effect testing suggests that variance component-based tests such as SimReg would have better power than burden-based tests if genetic effects vary radically across variants or if many null variants exist in the set (Pongpanich et al. 2012; Lee et al. 2014). However, the presence of many null variants can still unfavorably affect the test performance. For main-effect collapsing tests, efforts have been made to boost power when the signal sparsity is low by adaptively focusing on the subsets enriched with causal variants (e.g., Barnett 2014; Pan et al. 2014). Their extensions to G×E tests will be helpful to further optimize the power to detect G×E effects.
In this work we focused on examining the G×E interaction effect for a single environmental factor. However, a similar model involving multiple G×E interaction effects could be fitted. This method could be easily extended to test for gene–gene interaction in cases where one gene is suspected to interplay with other genes.
Acknowledgments
The authors are grateful to Mark McCarthy, Timothy Frayling, William Rayner, and members of the Warren 2 Consortium for providing the BMI data. This work makes use of data generated by the Wellcome Trust Case Control Consortium (WTCCC). A full list of the investigators who contributed to the generation of the data is available from http://www.wtccc.org.uk. The authors are also grateful to Peter Vollenweider and Gerard Waeber, Principal Investigators of the CoLaus study, and Meg Ehm and Matthew Nelson, collaborators at GlaxoSmithKline, for providing the CoLaus phenotype and sequence data. The authors thank Michael Wu for helping with the COSI simulation design and thank Shannon Holloway for her constructive input to improve the manuscript. The CoLaus study is supported by research grants from GlaxoSmithKline, the Faculty of Biology and Medicine of Lausanne, Switzerland, and by the Swiss National Science Foundation grants 33CSCO-122661 and 33CS30-139468. This work was partially supported by National Institutes of Health grants R01 MH084022 (to G.Z., D.Z., and J.-Y.T.), T32GM081057 (to R.M.), R01 CA85848-12 (to D.Z.), and P01 CA142538 (to J.-Y.T.).
Appendix A: Marginal Trait Covariance
Define . Under GLMM (2),
[by taking the first-order Taylor expansion of with respect to around ]
Appendix B: EM Algorithm to Estimate and σ in the SimReg G×E Test
Under the null hypothesis , model (3) becomes with . Let be the vector of binary traits, and let be the parameter vector. We consider an expectation-maximization algorithm based on observed data Y and missing data b. Let be the complete data log-likelihood. In the expectation step (E-step), we compute as
because . For the first term, we have
| (B1) |
For the second term, note that
where . Therefore,
| (B2) |
By expressing the complete-data log-likelihood in two parts, the fixed effect γ occurs only in the first term and variance component occurs only in the second term, .. Thus, the maximization steps for obtaining and can be discussed separately.
Maximization step for obtaining
To obtain , we can focus on . We take the derivative of (B2) with respect to and get
Setting this equal to zero, we get
| (B3) |
Equation B3 follows because approximately. To derive this approximation, we first reexpress as , i.e., a product of a Gaussian kernel and some function of Y. Finally, because , we have . We provide the details in the next subsection.
Derivation of as well as its mean and variance
where
| (B4) |
Let be the value that maximizes i.e., . By a Taylor expansion of with respect to b around , we have
Therefore, the complete data log-likelihood can be approximated by
| (B5) |
In Equation B5, is a Gaussian kernel with . Thus, the conditional distribution of approximately follows a multivariate normal distribution with mean vector and variance–covariance matrix .
Next we calculate and . In Equation B4, we rewrite as to emphasize that it is a function of b; i.e., with , the ith row of matrix . Note that . Then
where , and
where .
Finally, we obtain , i.e., the maximizer of . First, we rewrite as then we apply the Newton–Raphson method and obtain the iterative estimator of as
which depends on and γ, and we set and . The maximizer, , is obtained at each iteration until it converges, i.e., until the difference falls below a prespecified threshold, e.g., . We denote the maximizer as and also set .
Maximization step for obtaining
To obtain , we focus on the first term of i.e.,
We rewrite as here to emphasize that it is a function of γ; i.e., . We have that .. Then
where μ = (μ1(γ), μ2(γ), … , μn(γ))T = μ(b), and
Recall that . Using the first and second derivatives of , the estimator of , rewritten as , at the th iteration, is given by
which depends on and b. We set and . Then .
Putting it all together, at iteration we have following estimators:
, where and .
and .
Appendix C: Asymptotic Distributions of the Score Test Statistics
Recall that . Because , we have
where .. Therefore, can be rewritten as
| (C1) |
where , and . In addition, the working vector has mean and variance (Zhang and Lin 2003), and thus has mean 0 and variance .
Let , , denote the nonzero eigenvalues of matrix and let denote the corresponding eigenvectors. Then, , where . Therefore, can be approximated by a weighted sum of -distributions . By a similar derivation, the distribution of can be approximated by , where the ’s are the nonzero eigenvalues of matrix , with .
Footnotes
Communicating editor: I. Hoeschele
Literature Cited
- Barnett, I. J., 2014 SNP-set tests for sequencing and genome-wide association studies. Ph.D. Dissertation, Harvard University, Cambridge, MA. Available at: http://nrs.harvard.edu/urn-3:HUL.InstRepos:12274530
- Beckmann L., Fischer C., Obreiter M., Rabes M., Chang-Claude J., 2005. Haplotype-sharing analysis using Mantel statistics for combined genetic effects. BMC Genet. 6(Suppl. 1):S70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai T., Tonini G., Lin X., 2011. Kernel machine approach to testing the significance of multiple genetic markers for risk prediction. Biometrics 67: 975–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee N., Kalaylioglu Z., Moslehi R., Peters U., Wacholder S., 2006. Powerful multilocus tests of genetic association in the presence of gene-gene and gene-environment interactions. Am. J. Hum. Genet. 79(6): 1002–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J. Y., Logsdon B. A., Huang Y., Hsu L., Reiner A. P., et al. , 2012. Simultaneously testing for marginal genetic association and gene-environment interaction. Am. J. Epidemiol. 176(2): 164–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detopoulou P., Nomikos T., Fragopoulou E., Panagiotakos D. B., Pitsavos C., et al. , 2009. Lipoprotein-associated phospholipase A2 (Lp-PLA2) activity, platelet-activating factor acetylhydrolase (PAF-AH) in leukocytes and body composition in healthy adults. Lipids Health Dis. 8: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchesne P., Lafaye De Micheaux P., 2010. Computing the distribution of quadratic forms: further comparisons between the Liu-Tang-Zhang approximation and exact methods. Comput. Stat. Data Anal. 54(4): 858–862. [Google Scholar]
- Elston R., Buxbaum C. S., Jacobs K. B., Olson J. M., 2000. Haseman and Elston revisited. Genet. Epidemiol. 19: 1–17. [DOI] [PubMed] [Google Scholar]
- Fan R., Lo S. H., 2013. A robust model-free approach for rare variants association studies incorporating gene-gene and gene-environmental interactions. PLoS ONE 8(12): e83057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firmann M., Mayor V., Vidal P., Bochud M., Pecoud A., et al. , 2008. The CoLaus study: a population-based study to investigate the epidemiology and genetic determinants of cardiovascular risk factors and metabolic syndrome. BMC Cardiovasc. Disord. 8(1): 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseman J. K., Elston R. C., 1972. The investigation of linkage between a quantitative trait and a marker locus. Behav. Genet. 2(1): 3–19. [DOI] [PubMed] [Google Scholar]
- Helgason A., Pálsson S., Thorleifsson G., Grant S. F. A., Emilsson V., et al. , 2007. Refining the impact of TCF7L2 gene variants on type 2 diabetes and adaptive evolution. Nat. Genet. 39: 218–225. [DOI] [PubMed] [Google Scholar]
- Jiao S., Hsu L., Bézieau S., Brenner H., Chan A. T., et al. , 2013. SBERIA: set based gene environment interaction test for rare and common variants in complex diseases. Genet. Epidemiol. 37: 452–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft P., Yen Y. C., Stram D. O., Morrison J., Gauderman W. J., 2007. Exploiting gene-environment interaction to detect genetic associations. Hum. Hered. 63(2): 111–119. [DOI] [PubMed] [Google Scholar]
- Li Y., Willer C. J., Ding J., Scheet P., Abecasis G. R., 2010. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol. 34(8): 816–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X., Lee S., Christiani D. C., Lin X., 2013. Test for interactions between a genetic marker set and environment in generalized linear models. Biostatistics 14: 667–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Abecasis G. R., Boehnke M., Lin X., 2014. Rare-variant association analysis: study designs and statistical tests. Am. J. Hum. Genet. 95: 5–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen B. E., Browning S. R., 2009. A groupwise association test for rare mutations using a weighted sum statistic. PLoS Genet. 5: e1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao, H., 2009 Model selection and estimation in additive regression models. Ph.D. Dissertation, North Carolina State University, Raleigh, NC. [Google Scholar]
- Manolio T. A., Collins F. S., Cox N. J., Goldstein D. B., Hindorff L. A., et al. , 2009. Finding the missing heritability of complex diseases. Nature 461: 747–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechanic L. E., Chen H.-S., Amos C. I., Chatterjee N., Cox N. J., et al. , 2012. Next generation analytic tools for large scale genetic epidemiology studies of complex diseases. Genet. Epidemiol. 36: 22–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskvina V., Schmidt K. M., 2008. On multiple-testing correction in genome-wide association studies. Genet. Epidemiol. 32: 567–573. [DOI] [PubMed] [Google Scholar]
- Mukherjee B., Chatterjee N., 2008. Exploiting gene-environment independence for analysis of case-control studies: an empirical Bayes-type shrinkage estimator to trade-off between bias and efficiency. Biometrics. 64:685–94. [DOI] [PubMed] [Google Scholar]
- Murcray C. E., Lewinger J. P., Gauderman W. J., 2009. Gene-environment interaction in genome-wide association studies. Am. J. Epidemiol. 169(2): 219–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninio E., Tregouet D., Carrier J. L., Stengel D., Bickel C., et al. , 2004. Platelet-activating factor-acetylhydrolase (PAF-AH) and PAF-receptor gene haplotypes in relation to future cardiovascular events in patients with coronary artery disease. Hum. Mol. Genet. 13(13): 1341–1351. [DOI] [PubMed] [Google Scholar]
- Pan W., Kim J., Zhang Y., Shen X., Wei P., 2014. A powerful and adaptive association test for rare variants. Genetics 197: 1081–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A. L., Kryukov G. V., de Bakker P. I. W., Purcell S. M., Staples J., et al. , 2010. Pooled association tests for rare variants in exon-resequencing studies. Am. J. Hum. Genet. 86(6): 832–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pongpanich M., Neely M. L., Tzeng J.-Y., 2012. On the aggregation of multimarker information for marker-set and sequencing data analysis: genotype collapsing vs. similarity collapsing. Front. Genet. 2: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffner S. F., Foo C., Gabriel S., Reich D., Daly M. J., et al. , 2005. Calibrating a coalescent simulation of human genome sequence variation. Genome Res. 15(11): 1576–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaid D. J., 2010a Genomic similarity and kernel methods I: advancements by building on mathematical and statistical foundations. Hum. Hered. 70(2): 109–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaid D. J., 2010b Genomic similarity and kernel methods II: genomic information. Hum. Hered. 70(2): 132–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott L. J., Mohlke K. L., Bonnycastle L. L., Willer C. J., Li Y., et al. , 2007. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science 316: 1341–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sham, P., and S. Cherny, 2011 Analysis of Complex Disease Association Studies [Electronic Re-Source]: A Practical Guide. Academic Press/Elsevier, London/Burlington, MA. [Google Scholar]
- Sohns M., Viktorova E., Amos C. I., Brennan P., G. Fehringer et al, 2013. Empirical hierarchical Bayes approach to gene–environment interactions: development and application to genome–wide association studies of lung cancer in TRICL. Genet. Epidemiol. 37: 551–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K., Nelson M. R., Aponte J., Manas E. S., Bacanu S. A., et al. , 2012. Sequencing of Lp-PLA2-encoding PLA2G7 gene in 2000 Europeans reveals several rare loss-of-function mutations. Pharmacogenomics J. 12(5): 425–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D., 2010. Methods for investigating gene-environment interactions in candidate pathway and genome-wide association studies. Annu. Rev. Public Health 31: 21–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D., 2011. Response to ‘Gene-by-environment experiments: a new approach to finding the missing heritability’ by Van Ijzendoorn et al. Nat. Rev. Genet. 12(12): 881. [DOI] [PubMed] [Google Scholar]
- Thompson A., Gao P., Orfei L., Watson S., Di A. E., et al. , 2010. Lipoprotein-associated phospholipase A2 and risk of coronary disease, stroke, and mortality: collaborative analysis of 32 prospective studies. Lancet 375: 1536–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timpson N. J., Lindgren C. M., Weedon M. N., Randall J., Ouwehand W. H., et al. , 2009. Adiposity-related heterogeneity in patterns of type 2 diabetes susceptibility observed in genome-wide association data. Diabetes 58: 505–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng J.-Y., Devlin B., Wasserman L., Roeder K., 2003. On the identification of disease mutations by the analysis of haplotype similarity and goodness of fit. Am. J. Hum. Genet. 72(4): 891–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng J.-Y., Zhang D., 2007. Haplotype-based association analysis via variance component score test. Am. J. Hum. Genet. 81: 927–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng J.-Y., Zhang D., Chang S.-M., Thomas D. C., Davidian M., 2009. Gene-trait similarity regression for multimarker-based association analysis. Biometrics 65: 822–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng J.-Y., Zhang D., Pongpanich M., Smith C., McCarthy M. I., et al. , 2011. Studying gene and gene-environment effects of uncommon and common variants on continuous traits: a marker-set approach using gene-trait similarity regression. Am. J. Hum. Genet. 89(2): 277–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Os J., Rutten B., 2009. Gene-environment-wide interaction studies in psychiatry. Am. J. Psychiatry 166(9): 964–966. [DOI] [PubMed] [Google Scholar]
- Voorman A., Lumley T., McKnight B., Rice K., 2011. Behavior of qq-plots and genomic control in studies of gene-environment interaction. PLoS ONE 6: e19416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Elston R. C., 2005. Two-level Haseman-Elston regression for general pedigree data analysis. Genet. Epidemiol. 29: 12–22. [DOI] [PubMed] [Google Scholar]
- Wang X., Morris N. J., Zhu X., Elston R. C., 2013. A variance component based multi-marker association test using family and unrelated data. BMC Genet. 14(1): 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium , 2007. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447: 661–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel J., Schork N. J., 2006. Generalized genomic distance-based regression methodology for multilocus association analysis. Am. J. Hum. Genet. 79: 792–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winham S. J., Biernacka J. M., 2013. Gene–environment interactions in genome–wide association studies: current approaches and new directions. J. Child Psychol. Psychiatry 54(10): 1120–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wootton P. T., Flavell D. M., Montgomery H. E., World M., Humphries S. E., et al. , 2007. Lipoprotein-associated phospholipase A2 A379V variant is associated with body composition changes in response to exercise training. Nutr. Metab. Cardiovasc. Dis. 17(1): 24–31. [DOI] [PubMed] [Google Scholar]
- Wu M. C., Kraft P., Epstein M. P., Taylor D. M., Chanock S. J., et al. , 2010. Powerful SNP-set analysis for case-control genome-wide association studies. Am. J. Hum. Genet. 86: 929–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M. C., Lee S., Cai T., Li Y., Boehnke M., et al. , 2011. Rare variant association testing for sequencing data using the sequence kernel association test (skat). Am. J. Hum. Genet. 89: 82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Lin X., 2003. Hypothesis testing in semiparametric addictive mixed models. Biostatistics 4: 57–74. [DOI] [PubMed] [Google Scholar]