Abstract
The adaptive response to hypoxia is accompanied by widespread transcriptional changes that allow for prolonged survival in low oxygen. Many of these changes are directly regulated by the conserved hypoxia-inducible factor-1 (HIF-1) complex; however, even in its absence, many oxygen-sensitive transcripts in Caenorhabditis elegans are appropriately regulated in hypoxia. To identify mediators of these non-HIF-dependent responses, we established a hif-1 mutant reporter line that expresses GFP in hypoxia or when worms are treated with the hypoxia mimetic cobalt chloride (CoCl2). The reporter is selective and HIF independent, in that it remains insensitive to a number of cellular stresses, but is unaffected by mutation of the prolyl hydroxylase egl-9, suggesting that the regulators of this response pathway are different from those controlling the HIF pathway. We used the HIF-independent reporter to screen a transcription factor RNA interference (RNAi) library and identified genes that are required for hypoxia-sensitive and CoCl2-induced GFP expression. We identified the zinc finger protein BLMP-1 as a mediator of the HIF-independent response. We show that mutation of blmp-1 renders animals sensitive to hypoxic exposure and that blmp-1 is required for appropriate hypoxic-induced expression of HIF-independent transcripts. Further, we demonstrate that BLMP-1 is necessary for an increase of hypoxia-dependent histone acetylation within the promoter of a non-HIF-dependent hypoxia response gene.
Keywords: hypoxia, HIF independent, BLMP-1, C. elegans
CELLS are routinely challenged with low-oxygen conditions that drive compensatory responses. For instance, in development, low-oxygen conditions induce differentiation of placental cells (Dunwoodie 2009), and in diseased states, hypoxia is often a major contributing factor in cardiovascular disease and in the progression of solid tumor growth (Denko 2008; Semenza 2014). These low-oxygen challenges, whether scripted as in development or as consequences of physiologic dysfunction, are confronted by rapid cellular changes and by slower responses such as those mediated by the hypoxia-inducible factor-1 (HIF-1) transcription pathway.
The conserved HIF-1 complex regulates the expression of a broad set of genes that affect various aspects of metabolism, vascularization, and cell survival across species (Semenza 2011). In normoxia, the HIF-1 pathway is rendered inactive by prolyl hydroxylases (PHDs) that use molecular oxygen to modify conserved proline residues on the HIF-1α subunit of the transcription factor. The proline-hydroxylated HIF-1α is recognized by the von Hippel-Lindau (VHL) tumor suppressor protein, which targets HIF-1α for degradation (Bruick and McKnight 2001; Epstein et al. 2001). In hypoxia, owing to the reduced activity of PHD, unmodified HIF-1α pairs with the oxygen insensitive HIF-1β and the complex is transcriptionally active (Berra et al. 2006).
The nematode Caenorhabditis elegans is adept at enduring periods of low oxygen (Powell-Coffman 2010). Not surprisingly, the core components of the HIF-1 pathway (HIF-1α, HIF-1β, PHD, and VHL) are well conserved in the worm, where HIF-1, AHA-1, EGL-9, and VHL-1 fulfill the necessary roles of their respective mammalian counterparts (Bruick and Epstein et al. 2001; Jiang et al. 2001; McKnight 2001). As in vertebrates, the C. elegans HIF-1 pathway plays a critical role in varied biological processes including aging, neuronal reorganization, and stress responses (Treinin et al. 2003; Bretscher et al. 2008; Pocock and Hobert 2008; Chen et al. 2009; Budde and Roth 2010).
In spite of the pervasive nature of the HIF pathway, evidence from diverse systems demonstrates that hypoxia-induced transcriptional responses arise from multiple pathways that collectively drive adaptation in low-oxygen conditions, some of which do not require HIF-1 (Shen et al. 2005; Lee and Lee 2013; Li et al. 2013). While much progress has been made in defining the mechanisms that support HIF-1-mediated hypoxic actions, the nature and influence of HIF-1-independent hypoxic responses remain ill defined. In C. elegans, there is clear evidence of HIF-independent responses in hypoxia. Shen et al. (2005) showed that of 110 hypoxia-induced genes, 47 were induced in a HIF-independent manner. Additionally, C. elegans can survive for ∼24 hr in anoxic conditions and this response does not require hif-1; instead, this “suspended animation” response is mediated by the spindle checkpoint protein SAN-1 in embryos and the ceramide synthase HYL-2 in adults (Padilla et al. 2002; Nystul et al. 2003; Menuz et al. 2009; Miller and Roth 2009). Additionally, Lee and Lee (2013) described the role of the chromatin-remodeling factor NURF-1 in regulating the expression of a novel HIF-1-independent protein, heat shock protein HSP-16.1 (Lee and Lee 2013).
To better understand the genetic program induced in response to hypoxia, we set out to more concretely understand the HIF-1-independent branch of hypoxic responses. We focused on the regulation of F45D3.4, a previously identified HIF-1-independent hypoxia response gene in C. elegans (Shen et al. 2005). We show here that numerous transcription factors can regulate its hypoxia-sensitive expression, including BLMP-1, which we demonstrate is necessary for expression of select HIF-independent hypoxia transcripts and is essential for hypoxic adaptation.
Materials and Methods
Strains and culture conditions
Worms were maintained as described (Sulston and Hodgkin 1988) with the following modifications: grown on NGMSR plates (Avery 1993) at 20° on Escherichia coli strain HB101 unless indicated differently. NGMSR differs from NGM in containing 200 μg/ml streptomycin sulfate, 10 μg/ml nystatin, and 2% agar instead of 1.7%.
The wild-type strain was C. elegans variant Bristol, N2. The blmp-1 (tm548) mutant was generated by the National Bioresource Project in Japan; it contains an 810-bp deletion that has been described elsewhere (Huang et al. 2014b). We acquired it from H. R. Horvitz then outcrossed it 10 times against N2 (the strain is called YJ55). The hif-1 mutant strain is ZG31 hif-1(ia4), carrying a 1231-bp deletion (Jiang et al. 2001). The egl-9 allele (sa307) contains a 243-bp deletion (Darby et al. 1999; Shao et al. 2009). The 3kb-F45D3.4::GFP reporter plasmid was generated by cloning the promoter of F45D3.4 into pPD95.69 using the following primers: 5′-ataagctcacttgttaggtccaattggc-3′ (forward containing a HindIII site) and 5′-atgagctcgtttatttcgagcggttgtg-3′ (reverse containing a SacI site). The plasmid was injected into either the wild-type or hif-1(ia4) background to generate the respective reporter strains (YJ28 [uyEx3[myo-2::mCherry pF45d3.4::GFP] and YJ36 [hif(ia4);uyEx3[myo-2::mCherry pF45d3.4::GFP]]). mCherry, under the control of a myo-2 promoter, was used as a transgenic marker. The egl-9 reporter strain (YJ204) was generated by crossing YJ36 with CB6088 egl-9(sa307) hif-1(ia4).
For hypoxia treatment, animals were cultured on NGMSR plates in a Coy hypoxia chamber with oxygen analyzer (Coy Laboratory Products, Grass Lake, MI). Oxygen was kept constant using a continuous nitrogen gas source.
For cobalt chloride treatment, a 100-mM stock solution was prepared and filter sterilized using a 0.22-µm filter. A total of 500 μl of the stock solution was added onto a 10-ml plate to make a final concentration of 4.76 mM. The effect of CoCl2 on worms was examined either by a Western blot assay to measure HIF-1 protein, by qRT-PCR to assess transcript level, or by fluorescence microscopy to detect reporter gene expression induced by hypoxia. CoCl2-induced GFP fluorescence lasts for at least 48 hr in our reporter strain.
Hypoxia sensitivity assay
Assays for embryo viability at 0.5 or 2.0% O2 were performed as described previously (Padilla et al. 2012). In short, 15–20 1-day-old adults were allowed to lay eggs on a NGM plate seeded with OP50 for 1 hr and then removed. The eggs were counted and incubated in hypoxia (0.5 or 2.0% O2, 20°) for 96 hr using a hypoxia glovebox workstation (Ruskinn, Inc.). Immediately after treatment, animals were examined to determine the number of animals that survived the hypoxia treatment and the developmental stage of survivors. At least three independent experiments were performed for each condition.
RNA interference screen
A bacteria-mediated feeding RNA interference (RNAi) screen was performed as described (Fraser et al. 2000) with the following modifications: The YJ36 strain was screened with the clones of transcription factor genes from the Ahringer feeding library (Kamath and Ahringer 2003). The plates contained NGM agar with 1 mM IPTG and 100 µg/ml carbenicillin that were inoculated with bacterial cultures grown 16–18 hr for each targeted gene. L1 stage worms were then transferred onto clonal plates and left at 20°. Adults were then subjected to hypoxia or cobalt chloride treatment.
Quantitative RT-PCR
For total RNA preparation, 1-day-old adults were grown on NGMSR plates at 20°, washed with M9 buffer, and resuspended in Trizol (Bioline). RNA extraction was carried out by freeze–thawing in liquid nitrogen and chloroform extraction. The RNA was subjected to DNase I treatment, and after ethanol precipitation, air dried and dissolved in DEPC water. A total of 2 µg of total RNA in a 20-µl reaction was used to synthesize complementary DNA (cDNA) using a kit (Applied Biosystems, catalog no. 438706). Quantitative RT-PCR was carried out in a C-1000 thermal cycler Real-Time PCR system (Biorad) and analyzed using the Ct method (Lee et al. 2009). Messenger RNA (mRNA) levels of ama-1 (RNA polymerase II) were used for normalization. An average of at least three biological replicates was used for each data point. Primers used for qRT-PCR can be viewed in Supporting Information, Table S1.
Western blot analysis and antibodies
Electrophoresis of proteins in sample buffer was performed using standard methods. After trans-blotting, membranes were incubated in blocking buffer (5% nonfat dry milk and 1% BSA in 0.5% TBST) overnight. Membranes were incubated with antibody in 0.5% TBST (1% milk, 0.1% BSA). Antibodies used were: anti-Ce-HIF-1 antibody (a gift from Peter Ratcliffe, Oxford; 1:5000 dilution), AA4.3 antitubulin antibody (developed by Charles Walsh, obtained from the Developmental Studies Hybridoma Bank, created by the National Institute of Child Health and Human Development of the National Institutes of Health and maintained at The University of Iowa, Department of Biology, Iowa City, IA 52242, 1:1000 dilution), antirabbit antibody conjugated with HRP (Santa Cruz SC2030, 1:5000 dilution), and antimouse antibody conjugated with HRP. The bands were detected using ECL Plus kit (GE Healthcare, catalog no. RPN2133).
Microscopy
GFP-expressing worms were observed using a Zeiss Axio A2 Imager. Images were acquired using Zeiss Axiovision software.
Chromatin immunoprecipitation
The chromatin immunoprecipitation (ChIP) assays were performed as described, with minor modification (Zhong et al. 2010; Niu et al. 2011). In brief, L1 stage worms were grown at 20° and then treated to hypoxia at L3 or 1-day adult stages. The worms were cross-linked by 1% formaldehyde at room temperature for 30 min. Formaldehyde was quenched with Tris-HCl, washed, and lysed in FA buffer (50 mM HEPES-KOH, pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.1% sodium deoxycholate, 1% Triton X-100, 0.1% SDS and protease inhibitor cocktail (Roche, 11836170001) by sonication (five times at 10-sec intervals). The lysates were incubated overnight at 4° with either anti-GFP (Abcam ab290) or IgG (Millipore). The precipitates were washed and the cross-links reversed by heating at 65° with proteinase K. DNA was recovered by phenol-chloroform extraction, precipitation, and then eluted. PCR was performed using specific primers listed in Table S1.
Results
Generating a hypoxia-sensitive, HIF-independent reporter line
Previous studies have demonstrated the necessity and importance of non-HIF pathway-mediated transcriptional responses in hypoxia (Shen et al. 2005; Arany et al. 2008; Lee and Lee 2013; Li et al. 2013). We were interested in further elucidating how, and to what extent, oxygen-sensitive regulation of transcripts occurs independent of HIF-1 in C. elegans. To accomplish this, we focused on the transcriptional regulation of F45D3.4, an uncharacterized gene that was previously identified as a HIF-independent hypoxia-induced gene (Shen et al. 2005). We validated that F45D3.4 is normally induced in wild-type larvae (L3) when they are subject to 0.1% O2 treatment for 12 hr (Figure 1A). Further, we confirmed that the response is HIF independent by demonstrating that the induction of F45D3.4 in hypoxia is not diminished in the hif-1 mutant background (Figure 1A).
Figure 1.
HIF-independent transcriptional regulation of F45D3.4 in hypoxia. (A) Total mRNA was collected from wild-type animals and hif-1(ia4) mutants after incubation in normoxia (21% O2) or hypoxia (0.1% O2) for 12 hr and the expression levels of F45D3.4 were analyzed by qRT-PCR. Normalized values are the average of at least three biological replicates. Error bars are SEM. **P-value <0.01. A student’s t-test was used to determine significance. (B) Expression of GFP in wild-type, hif-1(ia4) mutant, and egl-9(sa307); hif(ia4) double-mutant backgrounds carrying the 3kb-F45D3.4::GFP construct after treatment in normoxia and hypoxia for 12 hr. Bars, 10 μm.
We next established transgenic reporter lines (3kb-F45D3.4::GFP) in both the wild-type and hif-1 mutant backgrounds, where GFP was placed under the control of elements controlling F45D3.4 expression. We found that following a 12-hr hypoxic treatment in a chamber containing 0.1% O2, the proximal 3-kb region of the F45D3.4 promoter was sufficient to drive reporter expression in wild-type and hif-1 mutant backgrounds (Figure 1B). These data confirm that F45D3.4 is a HIF-independent hypoxia response gene and suggest that the regulatory elements mediating the response localize to a 3-kb region that flanks the F45D3.4 transcription start site.
To determine if the 3kb-F45D3.4::GFP reporter was regulated similarly to HIF-1 in normoxia by oxygen-dependent hydroxylation via the HIF prolyl hydroxylase EGL-9 (Epstein et al. 2001), we further assayed the reporter in egl-9; hif-1 double mutants. We found that the behavior of the reporter was unchanged in normoxia and indistinguishable from the hypoxic activity observed in the control background (Figure 1B). This suggests that the HIF-independent response is not affected by one of the primary regulators controlling the HIF pathway, the EGL-9 prolyl hydroxylase.
Cobalt chloride induces HIF- and non-HIF-mediated pathways
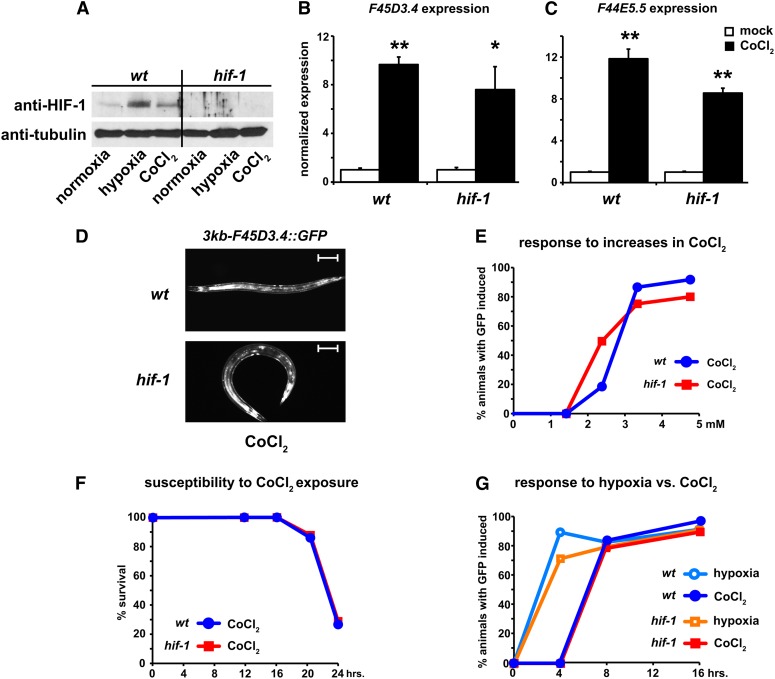
Cobalt chloride is widely used as a hypoxic mimetic, because it can elicit normoxic transcriptional responses that greatly resemble those seen in hypoxia (Ho and Bunn 1996; Vengellur et al. 2003); however, its utility in recapitulating transcriptional hypoxic responses in C. elegans has not been demonstrated. To address this, we grew worms on plates containing 5.0 mM CoCl2 for 12 hr and asked if the treatment affected HIF-1 protein levels. We found that CoCl2 caused a marked increase of HIF-1 protein in control animals, which is similar to the accumulation seen in hypoxia (Figure 2A). The CoCl2 treatment also induced a robust transcriptional upregulation of the F45D3.4 transcript in wild-type animals and hif-1 mutants in a manner that resembled previous hypoxic treatments (Figure 2B). Similarly, a separate HIF-independent transcript, F44E5.5 (Shen et al. 2005), was also upregulated by the treatment (Figure 2C). These results suggest that CoCl2 treatment triggers a transcriptional response that mimics hypoxic treatment, impacting both HIF- and non-HIF-mediated pathways.
Figure 2.
Cobalt chloride mimics hypoxia treatment in C. elegans. (A) Stabilization of HIF-1 protein was monitored by Western analysis following normoxic, hypoxic, and CoCl2 (4.76 mM) treatments for 12 hr in wild type and hif-1(ia4) mutants. Tubulin signal served as a loading control. (B and C) Total mRNA was collected from wild-type animals and hif-1(ia4) mutants after incubation on mock- or CoCl2-treated (4.76 mM) plates for 12 hr to measure the expression of the HIF-independent genes F45D3.4 (B) and F44E5.5 (C) by qRT-PCR. Normalized values are the average of at least three biological replicates. Error bars are SEM. *P-value <0.05 and **P-value <0.01. A Student’s t-test was used to determine significance. (D) Expression of GFP in a wild-type animal and a hif-1(ia4) mutant carrying the 3kb-F45D3.4::GFP construct after treatment in 4.76 mM CoCl2. Bars, 10 μm. (E–G) Transgenic F45D3.4 worms were treated on plates containing an increasing concentration of CoCl2 (0–4.76 mM) for 12 hr (E), a constant level of CoCl2 (4.76 mM) for 0–24 hr (F), or in hypoxia (0.1% O2) or on CoCl2-treated plates (4.76 mM) for 0–16 hr (G) to determine the optimal concentration of CoCl2 that elicits a response in wild type or hif-1(ia4) mutants (E), to measure the effect on lethality of prolonged exposure to CoCl2 (F), and to compare the hypoxic and CoCl2-elicited responses in the indicated backgrounds over time (G). All data points represent the average of at least two different biological pools consisting of at least 20 individual animals.
Consistent with the effects observed on the endogenous F45D3.4 transcript, the 3kb-F45D3.4::GFP reporter animals also responded to CoCl2 in a concentration-dependent manner in wild-type and hif-1 mutant backgrounds (Figure 2, D and E). Although 4 hr of hypoxic exposure was more effective than CoCl2 treatment at inducing GFP expression in either background, by the 8-hr time, there was no distinguishable difference between the treatments and their abilities to elicit a positive GFP response (Figure 2G). Importantly, up to 16 hr of CoCl2 treatment did not diminish survival, but treatments beyond that time tended to negatively impact survival of the wild-type and hif-1 backgrounds equally (Figure 2F), suggesting the loss of HIF action was not an important factor in the susceptibility.
The 3kb-F45D3.4::GFP reporter is not a general readout for stress
As is the case with CoCl2 treatment, stimuli other than low oxygen can transcriptionally upregulate hypoxic response genes. To better understand the upstream mediators of the HIF-independent response and to determine how they may differ from those controlling HIF-dependent responses, we subjected 3kb-F45D3.4::GFP reporter animals to different cellular stresses and compared the responses to results obtained from NHR-57::GFP reporter animals, a previously described HIF-dependent reporter line that also responds to hypoxic treatment (Shen et al. 2006). Unexpectedly, the NHR-57::GFP reporter was not activated by CoCl2 treatment (1.42–4.76 mM) as was the F45D3.4-3kb::GFP reporter (Table 1). In spite of the difference in responsiveness to CoCl2, the two reporters behaved quite similarly when subjected to atmospheric or low levels of oxygen; neither was active in normoxia (∼21% O2) or in anoxic conditions, but both were robustly activated when treated in 0.1–0.4% O2 for 4 hr. Only the NHR-57::GFP reporter was active in 0.5% O2.
Table 1. HIF-1- and non-HIF-1-dependent reporters have differential responses to various stress treatments.
| Stress | Treatment | GFP reporter induction? | |
|---|---|---|---|
| F45D3.4-3kb | NHR-57 | ||
| Anoxia | 0.0% O2 | No | No |
| Hypoxia | 0.1–0.4% O2 | Yes | Yes |
| Hypoxia | 0.5% O2 | No | Yes |
| CoCl2 | 1.42–4.76 mM | Yes | No |
| Osmotic | 300 mM NaCl | No | No |
| Heat | 37° 6 hr | No | No |
| ROS | 4 mM paraquat | No | Yes |
| Starvation | 12 hr | Yes | No |
| Heavy metal | 0.02 mM NiCl2 | No | No |
| Calcium chelation | 0.4.76 mM EGTA | No | No |
We also assayed for GFP induction when reporter animals were subject to a variety of cellular stresses, some of which are known to elicit HIF- and/or non-HIF-pathway responses that resemble aspects of hypoxic treatment through target gene induction. These treatments included hypertonic stress (300 mM NaCl) (Frazier and Roth 2009), heat shock (37° for 6 hr) (Treinin et al. 2003), calcium chelation (0.4.76 mM EGTA) (Lee and Lee 2013), reactive oxygen species (ROS) generation (4 mM paraquat) (Lee et al. 2010), heavy metal exposure (0.02 mM NiCl2), and starvation (12 hr from L4 onset). None of the stress treatments was able to induce the 3kb-F45D3.4::GFP reporter, except for the 12-hr starvation (Table 1). Similarly, the NHR-57::GFP reporter was also unresponsive to the treatments, except when treated with the ROS-inducing agent paraquat. These collective results demonstrate the selective nature of hypoxia-sensitive pathways and suggest that HIF- and non-HIF-dependent pathways have different, but overlapping, threshold responses to various cellular stresses, but have near-identical behaviors in low-oxygen settings at their respective promoters.
A transcription factor RNAi screen identifies candidate mediators of the HIF-independent response
To identify factors that facilitate non-HIF-mediated hypoxia-sensitive transcriptional responses we performed a screen, where each of 387 transcription factors in the C. elegans genome was targeted by RNAi (Kamath and Ahringer 2003) (Table S2). In a hif-1 mutant background, the 3kb-F45D3.4::GFP reporter strain was fed bacteria containing individual RNAi clones and grown to young adulthood in otherwise normal conditions, at which point animals were switched to plates containing 4.76 mM CoCl2 and assayed 12 hr later for GFP expression (Figure S1). All clones were tested in duplicate on individual batches of 40 animals per trial. Initial positive hits were considered those that robustly inhibited GFP signal in at least 75% of animals (average ≥ 30 of 40 animals per trial). We found that 8 transcription factors, or 2.1% of the RNAi clones screened, eliminated GFP expression following CoCl2 treatment (Table 2).
Table 2. Identified RNAi clones that inhibit GFP expression of the HIF-independent pathway driven by cobalt chloride treatment or hypoxic exposure.
| RNAi target | F45D3.4-3kb::GFP expression inhibited in: | ||
|---|---|---|---|
| Function/process | 4.76 mM CoCl2 | 0.1% O2 | |
| tbx-38 | T-box transcription factor, essential for mesordermal induction | Yes | Yes |
| lin-40 | Histone deacetylaction, chromatin remodeling | Yes | Yes |
| blmp-1 | Zinc-finger protein, putative methyl transferase | Yes | Yes |
| nhr-31 | Nuclear hormone receptor, required for excretory cell function | Yes | No |
| npp-12 | Nuclear pore membrane glycoprotein | Yes | No |
| nhr-89 | Nuclear hormone receptor | Yes | No |
| elt-3 | GATA transcription factor, hypodermal cell differentiation | Yes | No |
| taf-11.2 | Transcription initiation factor TFIID subunit | Yes | No |
To each of these initial positive clones a two-pronged secondary check was applied, whereby 200 animals were again tested for GFP suppression in the presence of CoCl2. Additionally, a separate batch of animals was screened for GFP fluorescence following a 6-hr treatment in 0.1% O2. These validation experiments confirmed the suppression of CoCl2-induced GFP in all eight clones. They further demonstrated, however, that only three of the eight RNAi clones—tbx-38, lin-40, and blmp-1—were capable of extinguishing hypoxia-induced GFP expression (Table 2). The differential effectiveness of RNAi on suppressing GFP signal following the two treatments, suggests that cobalt chloride and oxygen deprivation target distinct, but overlapping upstream regulators of non-HIF-mediated pathways.
blmp-1 mediates expression of a distinct subset of hypoxia response genes
Of the three clones that were effective at mitigating GFP signal in both CoCl2 and hypoxia, we specifically focused on the zinc finger gene blmp-1, which we confirmed was necessary for hypoxic-induced expression of the HIF-independent transcript F45D3.4 (Figure 3A). BLMP-1 is a highly conserved factor that acts as an inhibitor of transcription in mammals, where it is known as PRDM1/BLIMP-1, and in C. elegans it has been found to regulate developmental timing via DRE-1-mediated ubiquitylation and destruction (Huang 1994; Gyory et al. 2004; Horn et al. 2014; Huang et al. 2014b). To determine if and to what extent BLMP-1 may participate in the hypoxic induction of other transcripts we treated blmp-1 mutant worms with hypoxia and surveyed the expression of five other HIF-independent hypoxic transcripts described by Shen et al. (2005) (Figure 3, B–F). As expected, control animals responded robustly to hypoxic challenge through upregulation of transcript. Additionally, in each case, mutation of hif-1 failed to extinguish this response (Figure 3, B–F); however, while icl-1 and F44E5.5 were entirely unaffected in this background (Figure 3, B and C), mnk-1, mod-5, and zip-1 exhibited somewhat muted, though still significant, response profiles (Figure 3, D–F), suggesting that the hypoxic regulation of these transcripts may entail a combination of HIF- and non-HIF-mediated events. These results differed from those obtained in the blmp-1 mutant background, where all hypoxic responsiveness was eliminated (Figure 3, B–F).
Figure 3.
Hypoxic regulation of transcripts by BLMP-1 is HIF independent. (A–I) Total mRNA was collected from wild-type animals and blmp-1(tm548) mutants (A) or from wild-type animals, hif-1(ia4) mutants, and blmp-1(tm548) mutants (B–I) after incubation in normoxia or hypoxia (0.1% O2) for 12 hr. The expression levels of HIF-independent genes F45D3.4 (A), icl-1 (B), F44E5.5 (C), mnk-1 (D), mod-5 (E), and zip-1 (F) and HIF-dependent genes nhr-57 (G), egl-9 (H), and fmo-2 (I) were analyzed by qRT-PCR. Normalized values are the average of at least three biological replicates. Error bars are SEM. **P-value <0.01 and *P-value <0.05. A Student’s t-test was used to determine significance.
We also assayed known HIF-dependent hypoxic transcripts and found that the hypoxic expression of nhr-57, egl-9, and fmo-2 was greatly effected (Figure 3, G–I), if not eliminated, in the hif-1 mutant, results that recapitulated published data (Shen et al. 2005). In contrast—and in line with BLMP-1 acting independently from HIF-1—the effect of blmp-1 mutation on the hypoxic expression of these same transcripts was significantly less impactful, and in the case of fmo-2 had no effect on hypoxic expression. These collective results demonstrate that BLMP-1 is essential for the hypoxic regulation of a unique subset of hypoxic response genes that differs from those regulated by HIF-1.
blmp-1 mutants are sensitive to hypoxia
To further investigate the effects of BLMP-1 participation in HIF-independent hypoxic responses, we challenged embryos of control animals and blmp-1 mutants, which were allowed to develop for 4 days in normoxia or hypoxia. We additionally assayed hif-1 mutants and hif-1; blmp-1 double mutants as a comparative benchmark. As expected, the control and hif-1 mutants developed equally well under normoxic conditions (Jiang et al. 2001), while blmp-1 mutants and hif-1; blmp-1 double mutants displayed a mildly penetrant lethal phenotype (Figure 4A). In contrast, only 62.5% of control animals survived in 0.5% O2; however, among surviving animals, the vast majority (∼75%) were able to progress to adulthood, which was 48% of all the embryos surveyed. This was different from hif-1 mutants and hif-1; blmp-1 double mutants, which showed no progression and no survivability in hypoxia. Importantly, blmp-1 mutants were also severely affected by hypoxia; no animals matured to adulthood over the 4-day challenge (Figure 4A). Furthermore, even among the reduced number of surviving blmp-1 mutants, fewer than half progressed out of the L1/L2 stage. We also saw an increased sensitivity phenotype for blmp-1 mutants and hif-1; blmp-1 double mutants under the less severe hypoxic treatment regimen of 2% oxygen (Figure S2). These results demonstrate that BLMP-1 is essential for normal developmental progression in hypoxia.
Figure 4.
blmp-1 mutants are sensitive to hypoxic exposure. A total of 15–20 adult animals [wild type, hif-1(ia4) mutants, blmp-1(tm548) mutants, or hif-1(ia4); blmp-1(tm548) double mutants] per trial were allowed to lay eggs on an OP50-seeded NGM plate for 1–2 hr. The embryos were placed in either normoxia or hypoxia (0.5% O2) at 20°. After 4 days, surviving animals were scored for developmental progression as adults, L3/L4 larvae, or L1/L2 larvae. Note that hypoxia slows development, resulting in young adults, whereas in normoxia adults are 1 day old and gravid. For each genotype and condition, four independent experiments were performed with a total of 200–350 worms. Error bars are SEM.
BLMP-1 mediates hypoxia-dependent histone acetylation in hypoxia
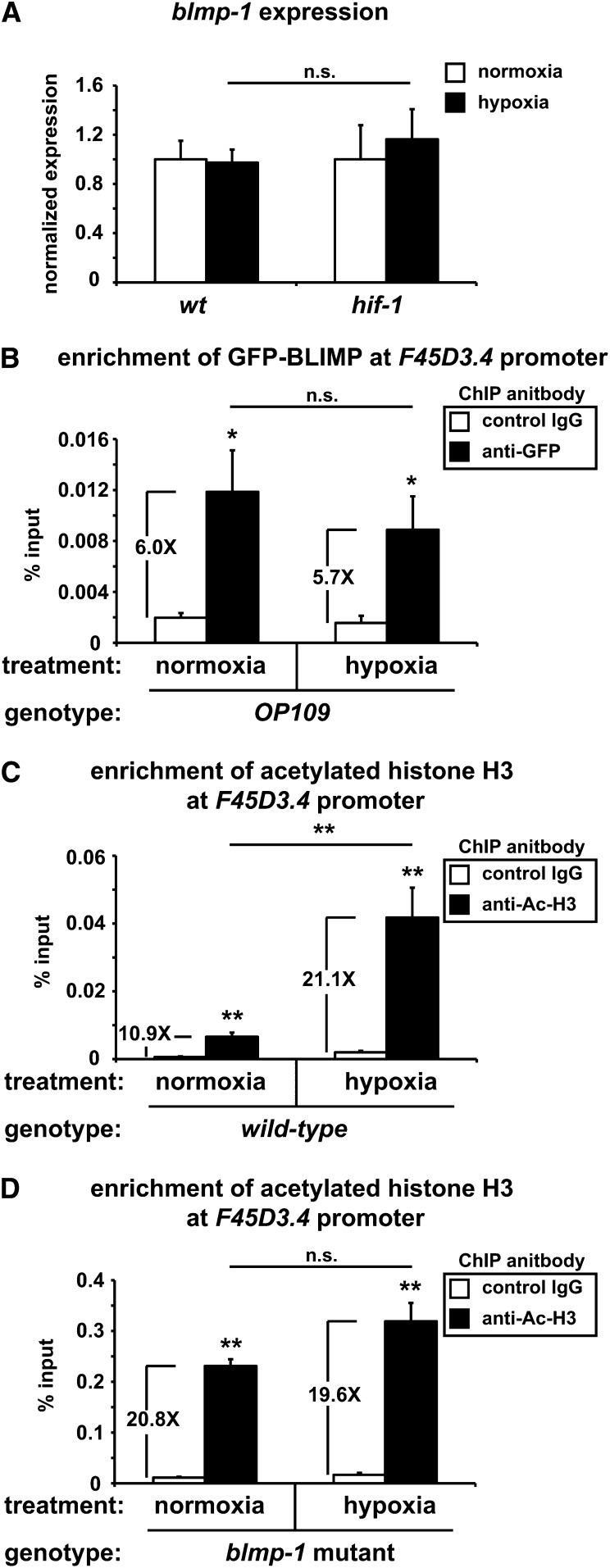
Apart from its nondependency on HIF-1 and lack of EGL-9 regulation, the mechanisms supporting blmp-1-dependent control of hypoxic transcription are unknown. As an initial step toward uncovering a possible mechanism, we looked at several aspects of BLMP-1 biology in hypoxia. First, we determined that blmp-1 expression remains unchanged in hypoxia; this is the case in control animals and hif-1 mutants (Figure 5A). Hence, transcriptional upregulation cannot account for a change in BLMP-1 activity in hypoxia. Further, chromatin immunoprecipitation (ChIP) was used to assess if hypoxic treatment altered recruitment of a GFP-tagged version of BLMP-1 (Niu et al. 2011) to the F45D3.4 promoter. Although we found a significant enrichment of BLMP-1–GFP at the proximal F45D3.4 promoter sequence, our results indicate the localization is unaffected by hypoxia treatment (Figure 5B). These results were not a product of nonspecific antibody binding, since no enrichment was observed in the wild-type background (Figure S3). This suggests that changes in oxygen status are relayed to prelocalized BLMP-1 at respective loci, which subsequently trigger increased transcriptional activity. Consistent with this idea, additional ChIP experiments demonstrated a surge in the acetylation of histone H3 following hypoxia treatment (Figure 5C), indicating an oxygen-sensitive increase in chromatin decondensation in the same region bound by BLMP-1–GFP. Importantly, similar increases were not observed in the blmp-1 mutant (Figure 5D), a result that aligns with a failure to upregulate transcription.
Figure 5.
BLMP-1 mediates hypoxic-induced histone acetylation. (A) The potential hypoxic regulation of blmp-1 transcript was assayed by qRT-PCR from total RNA collected from wild-type or hif-1(ia4) mutant animals treated in normoxia or hypoxia (0.1% O2) for 12 hr. (B–D) Chromatin immunoprecipitation was used to monitor the affects of hypoxia (0.1% O2) for 6 hr on BLMP-1 localization (B) or histone H3 acetylation (C and D) at the F45D3.4 promoter. F45D3.4 promoter sequences were enriched in the OP109 background by IP of BLMP-1–GFP with anti-GFP antibodies compared to control IgG antibodies, but results were unaffected by hypoxic treatment (B). Acetylation of the F45D3.4 promoter increased in a hypoxia-dependent manner in the wild-type background (C), but no such increase was apparent in blmp-1(tm548) mutants (D), which was measured by ChIP assay, using antiacetylated histone H3 antibodies vs. IgG control antibodies. Values are the average of at least three biological replicates and are plotted as a factor of percent input. Fold-change increases between control IgG antibody and anti-AcH3 or anti-GFP signals are shown. Error bars are SEM. **P-value <0.01 and *P-value <0.05. Statistical insignificance between measurements is noted by n.s. One-way and two-way ANOVA were used to compare results within and between groups with a post hoc Tukey honest significant difference test (A–D).
Discussion
Here, we have uncovered regulators of a hypoxia-sensitive pathway that works independently of HIF-1 to facilitate expression of F45D3.4 in C. elegans. Through a targeted RNAi approach in which the elements controlling its expression were tied to GFP in a hif-1 mutant background, we screened for transcription factors that mitigated cobalt chloride responsiveness and subsequently hypoxic induction. We found that this pathway displays important regulatory differences from those that control HIF, despite results that suggest the oxygen-sensitive induction of the two pathways at respective loci are virtually identical. First, and most importantly, hif-1 mutation has no impact on the hypoxic induction of F45D3.4 as evidenced by the induction of GFP fluorescence. Moreover, mutation of the prolyl hydroxylase gene egl-9, which encodes the primary negative regulator of the HIF pathway, also did not affect the hypoxic or normoxic expression of the reporter gene. These results suggest that the triggers for hypoxic-induced transcription are equally sensitive to decreases in oxygen, yet entirely different between the two pathways.
Among the eight factors that were originally identified in our cobalt chloride-based screen, only three were subsequently confirmed to also work in hypoxia when knocked down. This indicates that the targets of increased cobalt chloride and decreased oxygen (probably through the generation of reactive oxygen species; Bell et al. 2007) are separable. These results align with previous findings that have demonstrated differential effects brought about by cobalt chloride and hypoxia treatments (Vengellur et al. 2005; Huang et al. 2014a).
Precisely how either insult affects BLMP-1 to alter transcriptional output in C. elegans will require more investigation; however, it is intriguing to note that hypoxia-initiated and BLMP-1-dependent increases in histone acetylation at the F45D3.4 promoter may point to a shared response pathway that includes the prolongevity factor LIN-40, one of the other two positive hits identified in our screen. LIN-40 is an essential component of the nucleosome remodeling deacetylase (NuRD) complex (Johnsen and Baillie 1991; Solari et al. 1999) that was recently shown to promote stress resistance and longevity in a circuit that involves the germline and the insulin pathway (Zimmerman and Kim 2014). The integration of BLMP-1-mediated action into this stress response is feasible, given that BLMP-1 is already known to direct migration of gonadal precursor cells (Huang et al. 2014b) and our findings that show starvation is equally as effective as hypoxia for induction of the blmp-1-dependent 3kb-F45D3.4::GFP reporter. Indeed, our unpublished results are consistent with LIN-40 and BLMP-1 acting together through direct interaction in stress conditions to regulate common targets for dauer formation (M. Hyun, personal communication). Furthermore, the potential involvement of DRE-1-mediated degradation of BLMP-1 in hypoxia—as was recently reported to occur in normal developmental progression (Horn et al. 2014)—remains to be determined.
Such future investigations will be important, because of the overall significance that the non-HIF-mediated hypoxic program is not limited to F45D3.4 expression. On the contrary, diverse aspects of the BLMP-1-mediated non-HIF-dependent response pathway also include isocitrate lyase-1 (icl-1) and the serotonin transporter gene, mod-5. And, though the breadth of influence that BLMP-1 has on the genomic hypoxic response is not known, our findings indicate that, regardless of the absolute number of transcripts affected, it is essential in hypoxia.
BLMP-1 actions in hypoxia do not require HIF-1, but our data do not exclude that it acts in concert with it. For example, loss of hif-1 can dampen, but not extinguish hypoxic induction of mod-5 and zip-1, while loss of blmp-1 eliminates it altogether. This suggests that BLMP-1 acts as a hypoxic competence factor in certain contexts, which provides a path for further response by other factors, including HIF-1. In this respect, the ability of a BLMP-1-mediated non-HIF-dependent pathway to work apart from and in conjunction with the HIF pathway in C. elegans is similar to results we previously observed in Drosophila for ERR-mediated non-HIF-dependent hypoxic responses (Li et al. 2013).
Given our findings here, and the conserved nature of HIF- and non-HIF-mediated hypoxic signaling pathways in general, it will be interesting to determine if the mammalian equivalent of C. elegans BLMP-1 fulfills similarly important roles in mammalian hypoxic responses. The description of such an intervention has not been made, but it is clear the opportunity exists. For instance, PRDM1/BLIMP-1 is a well-known regulator of plasma cell differentiation and is an important regulator of T cells (Kallies et al. 2006; Nutt et al. 2007), which can be dramatically influenced by the dynamic and sometimes hypoxic microenvironments that are known to affect T cell-mediated cytokine production and inflammatory responses (McNamee et al. 2013). Regardless, further elaboration of oxygen-sensitive signaling networks that work apart from HIF-1 in simple model systems should provide a fruitful platform to elucidate the complexities governing hypoxic responses.
Supplementary Material
Acknowledgments
We thank Leon Avery for helpful comments and Moonjung Hyun, Mi Cheong Cheong, Jeongho Kim, and Valerie Reinke for technical support and helpful discussions. This work was supported in part by funding from the National Institutes of Health (1R56DK090984-01A1 to K.D.B.), the National Science Foundation (NSF CAREER 0747391 to P.A.P.), and the Virginia Commonwealth University School of Medicine (K.D.B. and Y.-J.Y.).
Footnotes
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.114.173989/-/DC1.
Communicating editor: B. Goldstein
Literature Cited
- Arany Z., Foo S. Y., Ma Y., Ruas J. L., Bommi-Reddy A., et al. , 2008. HIF-independent regulation of VEGF and angiogenesis by the transcriptional coactivator PGC-1alpha. Nature 451: 1008–1012. [DOI] [PubMed] [Google Scholar]
- Avery L., 1993. The genetics of feeding in Caenorhabditis elegans. Genetics 133: 897–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell E. L., Klimova T. A., Eisenbart J., Moraes C. T., Murphy M. P., et al. , 2007. The Qo site of the mitochondrial complex III is required for the transduction of hypoxic signaling via reactive oxygen species production. J. Cell Biol. 177: 1029–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berra E., Ginouves A., Pouyssegur J., 2006. The hypoxia-inducible-factor hydroxylases bring fresh air into hypoxia signalling. EMBO Rep. 7: 41–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher A. J., Busch K. E., de Bono M., 2008. A carbon dioxide avoidance behavior is integrated with responses to ambient oxygen and food in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 105: 8044–8049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruick R. K., McKnight S. L., 2001. A conserved family of prolyl-4-hydroxylases that modify HIF. Science 294: 1337–1340. [DOI] [PubMed] [Google Scholar]
- Budde M. W., Roth M. B., 2010. Hydrogen sulfide increases hypoxia-inducible factor-1 activity independently of von Hippel-Lindau tumor suppressor-1 in C. elegans. Mol. Biol. Cell 21: 212–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Thomas E. L., Kapahi P., 2009. HIF-1 modulates dietary restriction-mediated lifespan extension via IRE-1 in Caenorhabditis elegans. PLoS Genet. 5: e1000486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darby C., Cosma C. L., Thomas J. H., Manoil C., 1999. Lethal paralysis of Caenorhabditis elegans by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 96: 15202–15207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denko N. C., 2008. Hypoxia, HIF1 and glucose metabolism in the solid tumour. Nat. Rev. Cancer 8: 705–713. [DOI] [PubMed] [Google Scholar]
- Dunwoodie S. L., 2009. The role of hypoxia in development of the Mammalian embryo. Dev. Cell 17: 755–773. [DOI] [PubMed] [Google Scholar]
- Epstein A. C., Gleadle J. M., McNeill L. A., Hewitson K. S., O’Rourke J., et al. , 2001. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107: 43–54. [DOI] [PubMed] [Google Scholar]
- Fraser A. G., Kamath R. S., Zipperlen P., Martinez-Campos M., Sohrmann M., et al. , 2000. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408: 325–330. [DOI] [PubMed] [Google Scholar]
- Frazier H. N., 3rd, Roth M. B., 2009. Adaptive sugar provisioning controls survival of C. elegans embryos in adverse environments. Curr. Biol. 19: 859–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyory I., Wu J., Fejer G., Seto E., Wright K. L., 2004. PRDI-BF1 recruits the histone H3 methyltransferase G9a in transcriptional silencing. Nat. Immunol. 5: 299–308. [DOI] [PubMed] [Google Scholar]
- Ho V. T., Bunn H. F., 1996. Effects of transition metals on the expression of the erythropoietin gene: further evidence that the oxygen sensor is a heme protein. Biochem. Biophys. Res. Commun. 223: 175–180. [DOI] [PubMed] [Google Scholar]
- Horn M., Geisen C., Cermak L., Becker B., Nakamura S., et al. , 2014. DRE-1/FBXO11-dependent degradation of BLMP-1/BLIMP-1 governs C. elegans developmental timing and maturation. Dev. Cell 28: 697–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B. W., Miyazawa M., Tsuji Y., 2014a Distinct regulatory mechanisms of the human ferritin gene by hypoxia and hypoxia mimetic cobalt chloride at the transcriptional and post-transcriptional levels. Cell. Signal. 26: 2702–2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., 1994. Blimp-1 is the murine homolog of the human transcriptional repressor PRDI-BF1. Cell 78: 9. [DOI] [PubMed] [Google Scholar]
- Huang T. F., Cho C. Y., Cheng Y. T., Huang J. W., Wu Y. Z., et al. , 2014b BLMP-1/Blimp-1 regulates the spatiotemporal cell migration pattern in C. elegans. PLoS Genet. 10: e1004428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Guo R., Powell-Coffman J. A., 2001. The Caenorhabditis elegans hif-1 gene encodes a bHLH-PAS protein that is required for adaptation to hypoxia. Proc. Natl. Acad. Sci. USA 98: 7916–7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsen R. C., Baillie D. L., 1991. Genetic analysis of a major segment [LGV(left)] of the genome of Caenorhabditis elegans. Genetics 129: 735–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallies A., Hawkins E. D., Belz G. T., Metcalf D., Hommel M., et al. , 2006. Transcriptional repressor Blimp-1 is essential for T cell homeostasis and self-tolerance. Nat. Immunol. 7: 466–474. [DOI] [PubMed] [Google Scholar]
- Kamath R. S., Ahringer J., 2003. Genome-wide RNAi screening in Caenorhabditis elegans. Methods 30: 313–321. [DOI] [PubMed] [Google Scholar]
- Lee J., Lee J., 2013. Hypoxia-inducible Factor-1 (HIF-1)-independent hypoxia response of the small heat shock protein hsp-16.1 gene regulated by chromatin-remodeling factors in the nematode Caenorhabditis elegans. J. Biol. Chem. 288: 1582–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. J., Murphy C. T., Kenyon C., 2009. Glucose shortens the life span of C. elegans by downregulating DAF-16/FOXO activity and aquaporin gene expression. Cell Metab. 10: 379–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. J., Hwang A. B., Kenyon C., 2010. Inhibition of respiration extends C. elegans life span via reactive oxygen species that increase HIF-1 activity. Curr. Biol. 20: 2131–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Padmanabha D., Gentile L. B., Dumur C. I., Beckstead R. B., et al. , 2013. HIF- and non-HIF-regulated hypoxic responses require the estrogen-related receptor in Drosophila melanogaster. PLoS Genet. 9: e1003230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamee E. N., Korns Johnson D., Homann D., Clambey E. T., 2013. Hypoxia and hypoxia-inducible factors as regulators of T cell development, differentiation, and function. Immunol. Res. 55: 58–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menuz V., Howell K. S., Gentina S., Epstein S., Riezman I., et al. , 2009. Protection of C. elegans from anoxia by HYL-2 ceramide synthase. Science 324: 381–384. [DOI] [PubMed] [Google Scholar]
- Miller D. L., Roth M. B., 2009. C. elegans are protected from lethal hypoxia by an embryonic diapause. Curr. Biol. 19: 1233–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu W., Lu Z. J., Zhong M., Sarov M., Murray J. I., et al. , 2011. Diverse transcription factor binding features revealed by genome-wide ChIP-seq in C. elegans. Genome Res. 21: 245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt S. L., Fairfax K. A., Kallies A., 2007. BLIMP1 guides the fate of effector B and T cells. Nat. Rev. Immunol. 7: 923–927. [DOI] [PubMed] [Google Scholar]
- Nystul T. G., Goldmark J. P., Padilla P. A., Roth M. B., 2003. Suspended animation in C. elegans requires the spindle checkpoint. Science 302: 1038–1041. [DOI] [PubMed] [Google Scholar]
- Padilla P. A., Nystul T. G., Zager R. A., Johnson A. C., Roth M. B., 2002. Dephosphorylation of cell cycle-regulated proteins correlates with anoxia-induced suspended animation in Caenorhabditis elegans. Mol. Biol. Cell 13: 1473–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilla, P. A., J. M. Goy, and V. A. Hajeri, 2012 Anoxia-induced suspended animation in Caenorhabditis elegans, pp 25–58 in Anoxia, edited by P. A. Padilla. InTech, Rijeka, Croatia. [Google Scholar]
- Pocock R., Hobert O., 2008. Oxygen levels affect axon guidance and neuronal migration in Caenorhabditis elegans. Nat. Neurosci. 11: 894–900. [DOI] [PubMed] [Google Scholar]
- Powell-Coffman J. A., 2010. Hypoxia signaling and resistance in C. elegans. Trends Endocrinol. Metab. 21: 435–440. [DOI] [PubMed] [Google Scholar]
- Semenza G. L., 2011. Oxygen sensing, homeostasis, and disease. N. Engl. J. Med. 365: 537–547. [DOI] [PubMed] [Google Scholar]
- Semenza G. L., 2014. Hypoxia-inducible factor 1 and cardiovascular disease. Annu. Rev. Physiol. 76: 39–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Z., Zhang Y., Powell-Coffman J. A., 2009. Two distinct roles for EGL-9 in the regulation of HIF-1-mediated gene expression in Caenorhabditis elegans. Genetics 183: 821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C., Nettleton D., Jiang M., Kim S. K., Powell-Coffman J. A., 2005. Roles of the HIF-1 hypoxia-inducible factor during hypoxia response in Caenorhabditis elegans. J. Biol. Chem. 280: 20580–20588. [DOI] [PubMed] [Google Scholar]
- Shen C., Shao Z., Powell-Coffman J. A., 2006. The Caenorhabditis elegans rhy-1 gene inhibits HIF-1 hypoxia-inducible factor activity in a negative feedback loop that does not include vhl-1. Genetics 174: 1205–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solari F., Bateman A., Ahringer J., 1999. The Caenorhabditis elegans genes egl-27 and egr-1 are similar to MTA1, a member of a chromatin regulatory complex, and are redundantly required for embryonic patterning. Development 126: 2483–2494. [DOI] [PubMed] [Google Scholar]
- Sulston, J., and J. Hodgkin, 1988 Methods, pp. 587–606 in The Nematode Caenorhabditis Elegans, edited by W.B. Wood. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Treinin M., Shliar J., Jiang H., Powell-Coffman J. A., Bromberg Z., et al. , 2003. HIF-1 is required for heat acclimation in the nematode Caenorhabditis elegans. Physiol. Genomics 14: 17–24. [DOI] [PubMed] [Google Scholar]
- Vengellur A., Woods B. G., Ryan H. E., Johnson R. S., LaPres J. J., 2003. Gene expression profiling of the hypoxia signaling pathway in hypoxia-inducible factor 1alpha null mouse embryonic fibroblasts. Gene Expr. 11: 181–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vengellur A., Phillips J. M., Hogenesch J. B., LaPres J. J., 2005. Gene expression profiling of hypoxia signaling in human hepatocellular carcinoma cells. Physiol. Genomics 22: 308–318. [DOI] [PubMed] [Google Scholar]
- Zhong M., Niu W., Lu Z. J., Sarov M., Murray J. I., et al. , 2010. Genome-wide identification of binding sites defines distinct functions for Caenorhabditis elegans PHA-4/FOXA in development and environmental response. PLoS Genet. 6: e1000848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman S. M., Kim S. K., 2014. The GATA transcription factor/MTA-1 homolog egr-1 promotes longevity and stress resistance in Caenorhabditis elegans. Aging Cell 13: 329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.