Abstract
Multi-walled carbon nanotubes (MWCNTs) are known for their transient inflammatory and progressive fibrotic pulmonary effects; however, the mechanisms underlying these pathologies are unknown. In this study, we used time-series microarray data of global lung mRNA and miRNA expression isolated from C57BL/6J mice exposed by pharyngeal aspiration to vehicle or 10, 20, 40, or 80 µg MWCNT at 1, 7, 28, or 56 days post-exposure to determine miRNA and mRNA regulatory networks that are potentially involved in MWCNT-induced inflammatory and fibrotic lung etiology. Using a non-negative matrix factorization method, we determined mRNAs and miRNAs with expression profiles associated with pathology patterns of MWCNT-induced inflammation (based on bronchoalveolar lavage score) and fibrosis (based on Sirius Red staining measured with quantitative morphometric analysis). Potential binding targets between pathology-related mRNAs and miRNAs were identified using Ingenuity Pathway Analysis and the miRTarBase, miRecords, and TargetScan databases. Using these experimentally validated and predicted binding targets, we were able to build molecular signaling networks that are potentially reflective of and play a role in MWCNT-induced lung inflammatory and fibrotic pathology. As understanding the regulatory networks between mRNAs and miRNAs in different disease states would be beneficial for understanding the complex mechanisms of pathogenesis, these identified genes and pathways may be useful for determining biomarkers of MWCNT-induced lung inflammation and fibrosis for early detection of disease.
Keywords: multi-walled carbon nanotubes, mRNA, microRNA
The transcriptome undergoes rapid restructuring during the cellular lifecycle and in response to external stimuli (Brown, 2006). Protein-coding mRNAs are under constant regulation by non-coding RNAs, such as ribosomal RNAs, transfer RNAs, and miRNAs, during nearly all stages of gene expression (Brown, 2006; Fatica and Bozzoni, 2014). The regulatory relationship between mRNA and miRNA has been heavily studied since its first discovery (Lee et al., 1993). Mature miRNAs are more than 22 nt in length and bind to complementary nucleotide sequences in the 3′-UTR of their target mRNAs, directing the mRNA for degradation or translational repression, dependent upon the level of complimentarity (Bartel, 2004; Beezhold et al., 2010). Transcriptomic analysis of the overall mRNA and miRNA expression profiles of a particular cell type or disease state has become a promising method when choosing appropriate drug treatments or determining potential biomarkers of disease (Ferte et al., 2013; Karn, 2013).
Computational toxicology, the integration of global cellular transcriptomic analysis with advanced computer modeling for high throughput, in silico toxicology testing, aims to evaluate the potential human health effects of the ever growing number of toxicants (EPA, 2014) and is of particular interest in the burgeoning nanotoxicology field. Nanomaterials, materials with at least 1 dimension in the 1–100 nm range, have different physicochemical characteristics and toxicological effects than their fine-sized counterparts. Due to their small size, nanomaterials and nanoparticles pose a significant pulmonary exposure threat during their production and use, with the potential to damage the lung and translocate beyond the respiratory tract (Kendall and Holgate, 2012). In particular, multi-walled carbon nanotubes (MWCNTs) have raised concern for potential pulmonary toxicity due to their high level of production and use in various industrial settings. Consisting of concentric cylinders of carbon, MWCNT possesses immense strength and a long, fibrous phenotype (Iijima, 1991). In vivo pulmonary studies of both aspiration and inhalation exposure to MWCNT resulted in transient inflammation after aspiration and persistent inflammation after inhalation, with progressive fibrosis and translocation of the MWCNT to various other organs, including the diaphragm, brain, liver, and kidney (Mercer et al., 2013a, 2013c; Porter et al., 2010, 2013). Fibrosis in the lung is an irreversible restructuring of tissue that results in loss of basement membrane integrity, inappropriate extracellular matrix deposition without reestablishment of normal alveolar structures, and eventual end-stage lung disease (Strieter, 2008). The mechanisms leading to fibrosis are unclear, and early detection remains difficult; therefore, the need to determine predictive biomarkers of lung fibrosis is of utmost importance (Putman et al., 2014).
In addition to the discovery of the regulatory role between miRNA and mRNA comes the potential for these relationships to be used for understanding the complex mechanisms of disease pathogenesis (Cho et al., 2011; Nana-Sinkam et al., 2009; Sessa and Hata, 2013). Understanding the regulatory interactions between mRNA and miRNA in fibrosis would be a powerful analytical tool; however, although large amounts of mRNA gene expression data are available, the underlying regulatory miRNA networks are largely unknown (Cheng and Li, 2008). Traditional methods for studying this regulatory relationship typically include integrated methods of negative correlation and up- and down-regulation, but these methods do not take into account disease pathology patterns. Dynamic analysis of the miRNA/mRNA relationship in a disease state needs to be considered over the time course of response, allowing for the evaluation of miRNA/mRNA pairs that are consistently in opposition at most time points, but may exhibit divergence from this trend at key times post-exposure (Dymacek and Guo, 2014). In this study, we used an integrated analysis of miRNA and mRNA time series microarray data to identify miRNA/mRNA regulatory relationships reflective of lung inflammatory and fibrotic pathologies after in vivo mouse exposure by pharyngeal aspiration to dispersion media (DM) or 10, 20, 40, or 80 µg MWCNT at 7 or 56 days post-exposure (peak time points of transient inflammation and progressive fibrosis, respectively) (Porter et al., 2008, 2010). We suggest that these regulatory networks may be important for determining the biological processes underlying lung inflammatory and fibrotic pathologies, as well as serving as potential biomarkers for early detection of MWCNT-induced lung disease.
MATERIALS AND METHODS
MWCNT
Bulk MWCNT: The MWCNT used in this study were obtained from Mitsui & Company (MWCNT-7, Lot No. 05072001K28) and have been previously characterized (Porter et al., 2010). Briefly, the bulk MWCNT exhibited a distinctive crystalline structure, with the number of walls ranging from 20 to 50 as determined by high-resolution transmission electron microscopy (TEM). Overall, MWCNT trace metal contamination was 0.78%, including sodium (0.41%) and iron (0.32%), with no other trace metal contamination over 0.02%. Endotoxin contamination was below the level of detection. A survey-scan spectrum obtained by x-ray photoelectron spectroscopy measurement had a dominant C 1s peak (284.6 eV), with a small amount of oxygen and no other elements detected.
MWCNT in DM: TEM micrographs of MWCNT dispersed in DM demonstrated significant dispersion of MWCNT in DM (Porter et al., 2008, 2010). Quantitative analysis of the TEM micrographs determined the median length of the MWCNT sample to be 3.86 µm (geometric standard deviation (GSD) 1.94) and the count mean width to be 49 ± 13.4 (standard deviation [SD]) nm. The zeta potential of the MWCNT in DM was −11 mV. The MWCNTs used in this study were analyzed by electron spin resonance (ESR) and were found to decrease the ESR signal when added to the ESR reaction mixture, suggesting that MWCNT could scavenge •OH produced by the Fenton reaction. Additionally, when the MWCNTs were substituted for the Fe2+ in the Fenton reaction, no •OH was detected, suggesting that the iron present in the MWCNT was not capable of inducing reactive oxygen species (Porter et al., 2010).
Animals
The animal study described here was previously reported (Porter et al., 2010). Briefly, male C57BL/6J mice (7 weeks old) were obtained from Jackson Laboratories (Bar Harbor, Maine). Individual mice were housed 1 per cage in polycarbonate isolator ventilated cages and provided high-efficiency particulate absorption (HEPA)-filtered air with fluorescent lighting from 07:00 to 19:00 h. Autoclaved Alpha-Dri virgin cellulose chips and hardwood Beta-chips were used as bedding. Mice were monitored to be free of adventitous viral pathogens, parasites, mycoplasms, Helicobacter, and CAR Bacillus. Mice were maintained on Harlan Teklad Rodent Diet 7913 (Indianapolis, Indiana), and tap water was provided ad libitum. Animals were allowed to acclimate for at least 5 days before use. All animals in this study were housed in an AAALAC-accredited, specific pathogen-free, and environmentally controlled facility. All animal studies and procedures were approved by the National Institute for Occupational Safety and Health ACUC.
MWCNT pharyngeal aspiration exposure
Suspensions of MWCNT were prepared in DM (Ca2+ and Mg2+-free phosphate-buffered saline, pH 7.4, supplemented with 5.5mM d-glucose, 0.6 mg/ml mouse serum albumin, and 0.01 mg/ml 1,2-dipalmitoyl-sn-glycero-3-phosphocholine) and administered as previously described (Porter et al., 2008, 2010). In brief, each treatment group (DM or 10, 20, 40, or 80 µg MWCNT) consisted of 8 mice, which were anesthetized with isoflurane (Abbott Laboratories, North Chicago, Illinois). When fully anesthetized, the mouse was positioned with its back against a slant board and suspended by the incisor teeth using a rubber band. The mouth was opened and the tongue gently pulled aside from the oral cavity. A 50 µl aliquot of sample was pipetted at the base of the tongue, and the tongue was restrained until at least 2 deep breaths were completed (but not for longer than 15 s). Following release of the tongue, the mouse was gently lifted off the board, placed on its left side, and monitored for recovery from anesthesia. Comparative studies have shown that well-dispersed suspensions of MWCNT given by aspiration exposure can result in similar lung distribution patterns as MWCNT administered by inhalation exposure, and there is similar structure between MWCNT prepared for aspiration and those prepared for inhalation (Mercer et al., 2010, 2013a, 2013b; Porter et al., 2010, 2013).
At 1, 7, 28, and 56 days post-exposure, mice were euthanized by an intraperitoneal injection of sodium pentobarbital (>100 mg/kg body weight). Deep anesthesia was confirmed when the mouse no longer responded to a toe pinch. Transection of the abdominal aorta was completed to provide exsanguination. Lungs were rapidly removed, placed into RNAlater, and frozen at −80°C for future use.
Tissue RNA extraction
Total RNA was extracted from frozen mouse lung tissue samples (−80°C) in RNAlater using an RNeasy Fibrous Tissue Mini Kit according to manufacturer’s protocol (Qiagen) (Pacurari et al., 2011), followed by DNase treatment with a TURBO DNA-free kit (Ambion). Total RNA was eluted in RNase-free water and stored at −80°C until further analysis. The concentration of each RNA sample was determined using a NanoDrop-1000 Spectrophotometer (NanoDrop Tech, Germany).
mRNA microarray expression profiling and preprocessing
For mRNA profiling, extracted RNA was analyzed for expression profiling using Agilent Mouse Whole Genome Arrays (Agilent, Santa Clara, California) by the Marshall University Microarray Facility (Huntington, West Virginia). A universal reference design was employed using Stratagene Universal Mouse Reference RNA—Cat. No. 740100 (Agilent) as the reference RNA. Total RNA quality was determined on an Agilent 2100 Bioanalyzer, with all samples having RNA integrity numbers greater than 8. Total RNA (250 ng) was labeled using the QuickAmp labeling kit (Agilent). RNA extracted from each mouse was labeled with cyanine (Cy)-3-CTP (PerkinElmer, Waltham, Massachusetts), and reference RNA was labeled with (Cy)-5-CTP. Following purification of labeled cRNAs, 825 ng of Cy3- and Cy5-labeled cRNAs were combined and hybridized for 17 h at 65°C in an Agilent hybridization oven. Microarrays were washed and scanned using an Agilent DNA Microarray Scanner.
Data were exported from the Agilent DNA Microarray Scanner using Feature Extraction v10 as tab-delimited text files after background subtraction, log transformation, and lowess normalization and reported as log or relative expression of the sample compared with the universal reference. Data were read from each file into R using a custom script. For each array, values for control spots, spots which were saturated on either channel, and spots which were not well above background on at least 1 channel were considered unreliable and/or uninformative and replaced by “NA.” Values were collated into a single table, and probes for which fewer than 10 present values were available were removed. For probes spotted multiple times on the array, values were averaged across replicate probes. The resulting table is available as a series matrix in the NCBI Gene Expression Omnibus repository with accession number GSE29042 (Guo et al., 2012).
miRNA expression profiling and preprocessing
For miRNA profiling, RNA samples were sent to Ocean Ridge Biosciences (Palm Beach Gardens, Florida) for analysis using custom multi-species microarrays containing 704 mouse miRNA probes covering 714 mature mouse miRNAs present in miRBase version 15. The sensitivity of the microarray is such that it could detect as low as 20 amoles of synthetic miRNA being hybridized along with each sample. The microarrays were produced by Microarrays Inc (Huntsville, Alabama) and consisted of epoxide glass substrates that had been spotted in triplicate with each probe. Quality control of the total RNA samples was assessed using UV spectrophotometry and agarose gel electrophoresis. The samples were DNAse digested, and low-molecular weight (LMW) RNA was isolated by ultrafiltration through YM-100 columns (Millipore) and subsequently purified using an RNeasy MinElute Clean-Up Kit (Qiagen). The LMW RNA samples were 3′-end labeled with Oyster-550 fluorescent dye using a Flash Taq RNA Labeling Kit (Genisphere). Labeled LMW RNA samples were hybridized to the miRNA microarrays according to conditions recommended in the Flash Taq RNA Labeling Kit manual. The microarrays were scanned on an Axon Genepix 4000B scanner, and data were extracted from images using GenePix V4.1 software.
Spot intensities were obtained for the 5916 features on each microarray by subtracting the median local background from the median local foreground for each spot. Detection thresholds (T) for each array were determined by calculating the 10% trim mean intensity of the negative control spots and adding 5× the SD of the background (non-spot area). The spot intensities and the T were transformed by taking the log (base 2) of each value. The normalization factor (N) for each microarray was determined by obtaining the 20% trim mean of the spot intensities for the mouse probes above threshold in all samples. The log2-transformed spot intensities for all 5916 features were normalized by subtracting N from each spot intensity and scaled by adding the grand mean of N across all microarrays. The mean probe intensities for each of the 1936 mouse probes on each of the 160 arrays (1 sample per array) were then determined by averaging the triplicate spot intensities. Spots flagged as poor quality during data extraction were omitted prior to averaging. The 704 mouse non-control, log2-transformed, normalized, and averaged probe processing intensities were filtered to obtain a list of 484 mouse miRNA probes showing probe intensity above T in all samples from at least 1 treatment group.
Identifying mRNA associated with MWCNT-induced pathological responses
The MEGPath (Dymacek and Guo, 2011) system was used to identify mRNAs with expression transcriptionally concordant with inflammatory and fibrotic pathologies in mice exposed to DM or 10, 20, 40, or 80 µg MWCNT by aspiration for 1, 7, 28, or 56 days (Porter et al., 2008, 2010) as previously described (Snyder-Talkington et al., 2013). Inflammatory pathology was defined by the analysis of bronchoalveolar lavage fluid at Day 7, Dose 40 µg as previously reported (Porter et al., 2010). Fibrotic pathology was defined by the analysis of Sirius Red staining of collagen in mouse lungs at Day 56, Dose 80 µg as previously reported (Porter et al., 2010), and was quantified with morphometric analysis (Mercer et al., 2011). Using non-negative matrix factorization (NMF) (Lee and Seung, 1999), biological patterns related to the inflammatory and fibrotic pathologies were identified, with the expression profiles of the significant genes and pathology patterns as constraints. Identified patterns were then used to find genome-wide coefficients that related each gene to the different pathological patterns and identified sets of genes from the curated Molecular Signatures Database (MSigDB) (Subramanian et al., 2005) whose expression matched the inflammatory and fibrotic pathological patterns, allowing for annotations for each gene function and biological processes and pathways that are significantly related to the pathology patterns. Genes could be contained in multiple biological processes and pathways. The gene sets were functionally related and belonged to the same biological process, but were not required to be coexpressed (Snyder-Talkington et al., 2013).
Identifying miRNA associated with MWCNT-induced pathological responses
As there are currently no available resources comprehensively annotating the involvement of miRNA in biological processes and pathways, we used 2 approaches to identify miRNA associated with MWCNT-induced pathological responses. In the first approach, miRNA with transcriptional profiles that matched MWCNT-induced pathological patterns were selected using the NMF algorithm. In the second approach, all significant miRNA in the in vivo animal study were analyzed with Ingenuity Pathway Analysis (IPA, Ingenuity Systems, www.ingenuity.com), and those functionally involved in inflammation and fibrosis were selected.
In the first approach, a constrained NMF algorithm was used to relate the constraint pathology to the miRNA. Patterns and each probe’s corresponding coefficients were computed from time series miRNA microarray data. The first pattern was constrained to match the pathology. A probe’s error was calculated as the absolute difference between the normalized, reconstructed probe expression and the normalized original probe expression. Next, coefficients corresponding to the constrained pathological pattern were considered for significance testing. Error values were subtracted from the coefficients to eliminate probes with high reconstruction error and reduce the opportunity for false positives generated from probes with noisy data. The modified genome-wide coefficients were then plotted to visually check for a normal distribution. A normal distribution was fit to the modified coefficients, and significant probes (P ≤ 0.05) were kept for further analysis. Only the coefficients for the pathological pattern were used as constraints in the NMF modeling.
In the second approach, all significant miRNAs were input into IPA. Inflammatory response—Inflammatory response and organismal injury and abnormalities—fibrosis disease overlays were used to determine miRNAs experimentally shown to be involved in inflammation and fibrosis, respectively.
The time and dose-dependent expression changes of each miRNA in mice following MWCNT exposure can be viewed at http://www.mwcnttranscriptome.org/.
Integrated mRNA/miRNA analysis
After mRNA and miRNA associated with MWCNT-induced pathological patterns were identified in the previous steps, known potential miRNA targets and both mRNA and miRNA expression data were utilized to select mRNA/miRNA pairs that were potentially involved in miRNA-mediated post-transcriptional regulation. Potential mRNA/miRNA target pairs were identified from the miRTarBase (Hsu et al., 2014), miRecords (Xiao et al., 2009), and TargetScan (Lewis et al., 2005) databases. Both validated human and mouse miRNA/mRNA target pairs were kept for further analysis. The miRBase (Griffiths-Jones et al., 2006) website was used to translate a probe’s name into its most recent form. Potential target pairs were filtered according to the gene expression data by using the second derivatives of the mRNA and miRNA fold change (Dymacek and Guo, 2014). Specifically, a miRNA/mRNA pair was considered a target pair if the second derivatives at the same time point were of opposite signs, indicating a divergence in expression during miRNA-mediated post-transcriptional regulation.
Ingenuity pathway analysis
mRNAs involved in inflammation and fibrosis were determined as previously described (Snyder-Talkington et al., 2013) through the use of IPA. A network/My pathway is a graphical representation of the molecular relationships between molecules. Molecules are represented as nodes, and the biological relationship between 2 nodes is represented as an edge (line). All edges are supported by at least 1 reference from the literature, from a textbook, or from canonical information stored in the Ingenuity Knowledge Base. Human, mouse, and rat orthologs of a gene are stored as separate objects in the Ingenuity Knowledge Base but are represented as a single node in the network. Nodes are displayed using various shapes that represent the functional class of the gene product.
To create connections between significant mRNAs and miRNAs, the Build-Connect tool of IPA was used, with only direct relationships shown. The relationships between mRNAs and miRNAs were also assessed using miRecords, mirTarBase, and TargetScan (Hsu et al., 2014; Lewis et al., 2005; Xiao et al., 2009). Direct relationships confirmed by IPA are shown by a solid black line. If a confirmed regulatory relationship was found in miRecords or mirTarBase that was not found in IPA, the mRNA and miRNA were connected with a solid line without arrow. If a predicted relationship was found in TargetScan that was not found in IPA, the mRNA and miRNA were connected using a dotted line.
A Core Analysis was run on each set of inflammation, fibrosis (NFM), and fibrosis (IPA) mRNAs and miRNAs. The top 5 canonical pathways represented by the mRNAs and miRNAs in the given set were determined. The P-value determined by IPA is a measure of significance based on the number of genes/molecules that map to a biological function, pathway, or network.
RESULTS
Identifying Candidate mRNAs and miRNAs for Integrated Analysis
Identifying mRNAs associated with inflammatory and fibrotic pathological patterns
A set of mRNAs significantly related to MWCNT-induced inflammatory and fibrotic pathological patterns in mice exposed by pharyngeal aspiration to DM or 10, 20, 40, or 80 µg MWCNT at 7 and 56 days post-exposure was determined as previously described (Snyder-Talkington et al., 2013). These sets of mRNAs were newly analyzed for this study using the IPA Winter Release 2013. A total of 724 mRNAs significantly matched the inflammatory pathological patterns (inflammation leading set), and 833 mRNAs matched the fibrotic pathological patterns (fibrosis leading set). The inflammation and fibrosis leading sets were then analyzed with IPA to determine those mRNAs with expression transcriptionally related to the MWCNT-induced inflammatory or fibrotic pathologies that were also functionally related to inflammation or fibrosis according to the curated Ingenuity Knowledge Base. A total of 134 and 89 genes was found to be functionally involved in inflammation (Supplementary Table 1) and fibrosis (Supplementary Table 2), respectively, according to IPA.
Identifying miRNAs associated with inflammation and fibrosis pathology patterns
Expression of total miRNA isolated from mice exposed to DM or 10, 20, 40, or 80 µg MWCNT at 1, 7, 28, and 56 days post-exposure was analyzed by microarray and Significance Analysis of Microarrays (SAM) analysis. The expression of 92 miRNAs was significantly (FDR <5%) up- or down-regulated when compared with the DM control (Table 1), and there were no significant miRNAs identified at the 10 µg dose. To identify miRNAs associated with inflammatory and fibrotic pathologies in mice, only miRNAs significantly changed at the time points of peak inflammation (Day 7 post-exposure) and fibrosis (Day 56 post-exposure) were analyzed with the corresponding pathological patterns. The NMF algorithm was run using the 80 µg dose fibrotic time series pathology constraints and the 40 µg dose time series inflammatory pathology constraints to determine the fibrotic and inflammatory patterns, respectively. For both analyses, 3 patterns were found for each dose over the 4 time points. The coefficients relating miRNA to the constraint pathology pattern were modified by subtracting the reconstruction error to penalize noisy gene expression in the reconstruction.
TABLE 1.
All miRNAs Significantly Changed from DM Controls Identified from Mice Exposed to 10, 20, 40, or 80 µg MWCNT at 1, 7, 28, and 56 Days Post-exposure
| Dose 20 | Dose 40 | Dose 80 | |
|---|---|---|---|
| Day 1 post-exposure | miR-15a*, miR-16*, miR-223, miR-720 | miR-125b-3p, miR-1306, miR-223, miR-296-3p, miR-29b*, miR-341, miR-382, miR-711 | |
| Day 7 post-exposure | miR-125b-3p, miR-146b, miR-341 | miR-125b-3p, miR-1306, miR-142-5p, miR-146b, miR-16*, miR-1904, miR-30c-1*, miR-323-5p, miR-341, miR-382, miR-449a, miR-714, miR-720 | miR-125b-3p, miR-146b, miR-341, miR-449a, miR-92a* |
| Day 28 post-exposure | miR-125b-3p, miR-149, miR-1932, miR-1935, miR-1937c, miR-2132, miR-2133, miR-2140, miR-296-3p, miR-30c-1*, miR-322, miR-486, miR-720 | miR-125b-3p, miR-129-5p, miR-1306, miR-146b, miR-188-5p, miR-1935, miR-1937c, miR-1951, miR-196b, miR-1982*, miR-21, miR-296-3p, miR-29b*, miR-30c-1*, miR-323-5p, miR-327, miR-330, miR-341, miR-370, miR-382, miR-673-3p, miR-714, miR-744 | miR-125b-5p, miR-1937c, miR-21, miR-328, miR-486, miR-720 |
| Day 56 post-exposure | miR-125b-3p, miR-146b, miR-147, miR-16*, miR-296-3p, miR-341, miR-3473, miR-449c, miR-450a-3p, miR-669a, miR-679, miR-696 | miR-103, miR-125b-3p, miR-142-5p, miR-146a, miR-146b, miR-147, miR-15a, miR-15b, miR-188-5p, miR-1897-5p, miR-1982*, miR-199a-3p; mmu-miR-199b, miR-199a-5p, miR-200b, miR-221, miR-26b, miR-297a, miR-297c, miR-30c, miR-30e, miR-31, miR-3473, miR-34b-3p, miR-434-3p, miR-449c, miR-450a-3p, miR-466h, miR-467e, miR-467h, miR-471, miR-669a, miR-669e, miR-679, miR-696 | miR-103, miR-1188, miR-125b-3p, miR-126-3p, miR-1306, miR-130b, miR-132, miR-142-5p, miR-146a, miR-146b, miR-15a, miR-15a*, miR-16, miR-16*, miR-1892, miR-18b, miR-1902, miR-1903, miR-1906, miR-195, miR-196a, miR-199a-3p; mmu-miR-199b, miR-199a-5p, miR-200a, miR-200b, miR-21, miR-22, miR-221, miR-222, miR-26b, miR-296-3p, miR-30c, miR-30e, miR-31, miR-341, miR-342-3p, miR-3470b, miR-3473, miR-34a, miR-34b-3p, miR-34c, miR-429, miR-434-3p, miR-449b, miR-466b-5p, miR-669a, miR-669e, miR-679 |
Note: There were no significant miRNAs identified at Dose 10.
A normal distribution (mean: 0.271, SD: 0.0849) was fit to the modified inflammation coefficients. miRNAs with a coefficient greater than 0.4108 (P < 0.05) were considered to be significantly associated with the inflammation pathology and are listed in Table 2. Likewise, a normal distribution was fit to the modified fibrosis coefficients (mean: 0.318, SD: 0.128). miRNAs with a coefficient greater than 0.5285 (P < 0.05) were considered significantly associated with the fibrotic pathology and are listed in Table 3. These identified miRNAs have expression profiles highly concordant with MWCNT-induced pathological patterns, including lung inflammation and fibrosis, indicating a possible involvement in MWCNT-induced pathogenesis. These miRNA were selected for further integrative miRNA-mRNA analysis.
TABLE 2.
miRNAs Associated with the Inflammatory Pathology and Their Experimentally Confirmed and Predicted mRNA Binding Partners
| miRBase ID | Accession | Experimentally Confirmed Targets (miRTarBase/miRecords) | Predicted Targets (TargetScan, Highly Conserved) | IPA Relationships (TargetScan, Poorly Conserved) |
|---|---|---|---|---|
| mmu-miR-1224-5p | MIMAT0005460 | |||
| mmu-miR-147-3p | MIMAT0004857 | vegfa (Ye et al., 2008) | ||
| mmu-miR-188-5p | MIMAT0000217 | |||
| mmu-miR-290a-5p | MIMAT0000366 | |||
| mmu-miR-327 | MIMAT0004867 | |||
| mmu-miR-3474 | MIMAT0015646 | |||
| mmu-miR-380-3p | MIMAT0000745 | |||
| mmu-miR-449a-5p | MIMAT0001542 | gnai2, serpine1 | ||
| mmu-miR-494-3p | MIMAT0003182 | tnfrsf9 | serpine1, wnt5a | |
| mmu-miR-551b-3p | MIMAT0003890 | |||
| mmu-miR-667-3p | MIMAT0003734 | |||
| mmu-miR-696 | MIMAT0003483 | |||
| mmu-miR-703 | MIMAT0003493 | |||
| mmu-miR-877-5p | MIMAT0004861 | |||
| mmu-miR-881-5p | MIMAT0004845 | |||
| mmu-miR-92a-2-5p | MIMAT0004635 | mcl1, wnt5a |
Note: Significantly changed miRNAs are highlighted in bold.
TABLE 3.
miRNAs Associated with the Fibrotic Pathology and Their Experimentally Confirmed and Predicted mRNA Binding Partners
| miRBase ID | Accession | Experimentally Confirmed Targets (miRTarBase/miRecords) | Predicted Targets (TargetScan, Highly Conserved) | IPA Relationships (TargetScan, Poorly Conserved) |
|---|---|---|---|---|
| mmu-let-7c-5p | MIMAT0000523 | ago1 (Helwak et al., 2013), ptk2 (Helwak et al., 2013) | il6, rgs16, tnfaip3, tnfrsf1b | hbegf, fas, rgs16, tnfrsf1b, il6 (Sugimura et al., 2012) |
| mmu-miR-205-5p | MIMAT0000238 | vegfa (Ye et al., 2008) | il1r1, ptx3 | il1r1, smad4 |
| mmu-miR-23b-3p | MIMAT0000125 | smad4 (Rogler et al., 2009) | ednrb, fas, gsk3b, il11 | il11, fas, tnfaip3, gsk3b |
| mmu-miR-31-5p | MIMAT0000538 | sele (Suarez et al., 2010), hif1a (Shen et al., 2008) | hbegf | hbegf, ednrb, il1r1 |
| mmu-miR-326-3p | MIMAT0000559 | ago1 (Helwak et al., 2013) | rassf1 | tnfaip3, rassf1 |
| mmu-miR-328-3p | MIMAT0000565 | ago1 (Helwak et al., 2013) | tnfrsf1b | |
| mmu-miR-330-3p | MIMAT0000569 | vegfa (Ye et al., 2008), ago1 (Helwak et al., 2013) | rassf1 | bmpr2 |
| mmu-miR-34c-3p | MIMAT0004580 | pdgfra, serpine1, smad4 | ||
| mmu-miR-375-3p | MIMAT0000739 | kcnn4 (Tsukamoto et al., 2010) | bmpr2, ednrb, rgs16 | bmpr2, rgs16 |
| mmu-miR-455-3p | MIMAT0003742 | ago1 (Helwak et al., 2013) | ||
| mmu-miR-652-3p | MIMAT0003711 | ago1 (Helwak et al., 2013) | ||
| mmu-miR-92b-3p | MIMAT0004899 | ednrb, bmpr2 (Brock et al., 2009) |
Note: Significantly changed miRNAs are highlighted in bold.
Significantly Changed miRNAs Functionally Involved in Inflammation and Fibrosis
In addition to the NMF analysis, IPA was used to find miRNAs functionally related to lung inflammation or fibrosis from those miRNAs significantly (FDR < 5%; SAM analysis) altered after MWCNT exposure (Table 1), regardless of their association to the inflammatory or fibrotic pathologies. All significant miRNAs were input into IPA, and inflammatory response—inflammatory response and organismal injury and abnormalities—fibrosis disease overlays were used to determine miRNAs involved in fibrosis or inflammation, respectively, according to the Ingenuity Knowledge Base. IPA determined 10 miRNA to be functionally related to fibrosis (Table 4) and no significant miRNA to be related to inflammation.
TABLE 4.
miRNAs Associated with Fibrosis Based on IPA Analysis and Their Experimentally Confirmed and Predicted mRNA Binding Partners
| miRBase ID | Accession | Experimentally Confirmed Targets (miRTarBase/miRecords) | Predicted Targets (TargetScan, Highly Conserved) | IPA Relationships (TargetScan, Poorly Conserved) |
|---|---|---|---|---|
| mmu-miR-125-5p | MIMAT0000136 | il1rn (Hofmann et al., 2009), tnf (Tili et al., 2007), tnfaip3 (Kim et al., 2012), mmp13 (Xu et al., 2012) | bmpr2, smad4, tnfrsf1b, vegfa | bmpr2, tnfrsf1b, smad4 |
| mmu-miR-126a-3p | MIMAT0000138 | vegfa (Zhu et al., 2011) | ||
| mmu-miR-141-3p | MIMAT0000153 | csf3 | hmgcs1, tnfaip3, egfr | |
| mmu-miR-16-5p | MIMAT0000527 | vegfa (Karaa et al., 2009), kncc4 (Selbach et al., 2008), egfr (Selbach et al., 2008), vim (Selbach et al., 2008), smurf2 (Bockhorn et al., 2013) | cxcl1 | ptgs2 (Selbach et al., 2008), ago1, igf1 |
| mmu-miR-18a-3p | MIMAT0004626 | hif1a (Hafner et al., 2010), smad4 (Dews et al., 2010), hmgcs1 (Hafner et al., 2010) | igf1, tnfaip3 | |
| mmu-miR-199a-5p | MIMAT0000229 | hif1a (Rane et al., 2009), smad4 (Zhang et al., 2012b) | gsk3b, serpine1, vegfa | vegfa, ago1, ednrb, gsk3b, serpine1 |
| mmu-miR-21a-5p | MIMAT0000530 | fas (Sayed et al., 2010), bmpr2 (Papagiannakopoulos et al., 2008), egfr (Zhou et al., 2010), plat (Terao et al., 2011), ptx3 (Terao et al., 2011), tnfaip3 (Terao et al., 2011), ccr1 (Terao et al., 2011), vegfa (Liu et al., 2011), mmp9 (Moriyama et al., 2009), ptk2 (Gabriely et al., 2008), arid4a (Gabriely et al., 2008) | smurf2, tnf (Zhang et al., 2012a) | |
| mmu-miR-26a-5p | MIMAT0000533 | egr1 (Chi et al., 2009), smad4 (Dey et al., 2012), gsk3b (Mohamed et al., 2010), il6 (Yang et al., 2013), ago1 (Helwak et al., 2013) | mmp14, ptgs2, ptx3 | igf1 |
| mmu-miR-30c-5p | MIMAT0000514 | socs1 (Zhang et al., 2013), vim (Bockhorn et al., 2013), ago1 (Helwak et al., 2013) | actc1, arid4a, ednrb, igf1 | igf1, actc1, arid4a, serpine1 |
| mmu-miR-322-5p | MIMAT0000548 | cx3cl1, kcnn4, ptgs2, smurf2, vegfa |
Note: Significantly changed miRNAs are highlighted in bold.
Integrated mRNA and miRNA Analysis
mRNAs significantly associated with MWCNT-induced inflammatory and fibrotic pathological patterns and functionally involved in lung inflammation and fibrosis in IPA analysis were used to identify miRNA targets and mRNA/miRNA regulatory networks. Similarly, the set of miRNAs significantly changed after MWCNT exposure and functionally involved in inflammation and fibrosis and the set of miRNAs associated with pathological patterns were used in the integrated miRNA/mRNA analysis. Potential mRNA targets of significant miRNAs were identified using the miRTarBase (Hsu et al., 2014), miRecords (Xiao et al., 2009), and TargetScan (Lewis et al., 2005) databases. To be considered as a potential mRNA/miRNA target pair, the expression profiles of each mRNA/miRNA pair needed to have a differing second derivative in at least 1 time point, indicating a divergence in expression during miRNA regulated post-transcriptional activities. The integrated pathways of identified miRNAs and mRNAs were then analyzed and visualized with IPA. The results are provided in Tables 2–4, listing miRNA/mRNA regulations identified through our analysis that were either experimentally confirmed (Column 3 in Tables 2–4) or predicted as highly conserved target pairs (Column 4 in Tables 2–4). In comparison, the miRNA-mediated regulations retrieved from the IPA database are listed in the last column in Tables 2–4. It is worth noting that the regulations stored in the IPA database may or may not overlap with the results identified with our algorithms.
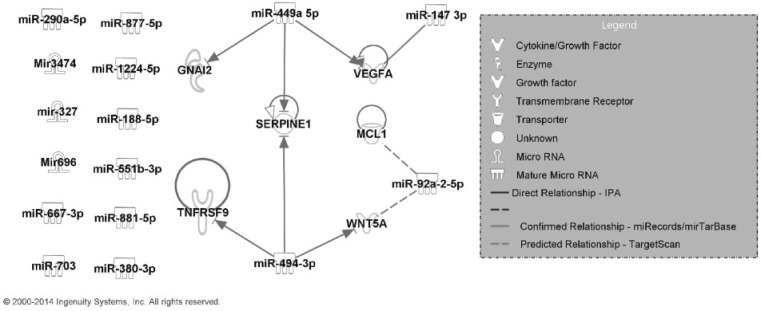
In total, 16 miRNAs were associated with the inflammatory pathology, 5 of which (miR-147-3p, miR-188-5p, miR-327, miR-449a-5p, and miR-696) were significantly up- or down-regulated after MWCNT exposure. Four miRNAs, miR-147-3p, miR-449a-5p, miR-494-3p, and miR-92a-2-5p, had predicted or experimentally confirmed mRNA targets that were also associated with the inflammatory pathology (Table 2). The integrated inflammation pathway based on miRNAs and mRNAs in Table 2 is shown in Figure 1. The identified relations highlighted with a solid/dash line without arrow were not available in the IPA database. Specifically, a regulatory relationship between miR-147-3p and vegfa identified in our analysis was experimentally confirmed (Ye et al., 2008) (designated as a solid line without arrow in Fig. 1). Our analysis also predicted highly conserved target pairs between miR-92a-2-5p and mcl1 and wnt5a (dashed lines in Fig. 1). The gene expression direction (up- or down-regulation relative to control) at doses 10, 20, 40, or 80 µg MWCNT on post-exposure Day 7 for each mRNA and miRNA is summarized in Supplementary Figure 1. This time point was chosen because quantitative bronchoalveolar lavage scores from post-exposure Day 7 (Mercer et al., 2011; Porter et al., 2010) were the basis for the inflammatory pathological pattern.
FIG. 1.
Regulatory network of mRNAs and miRNAs transcriptionally related to the Day 7 post-exposure bronchoalveolar lavage inflammatory pathological pattern.
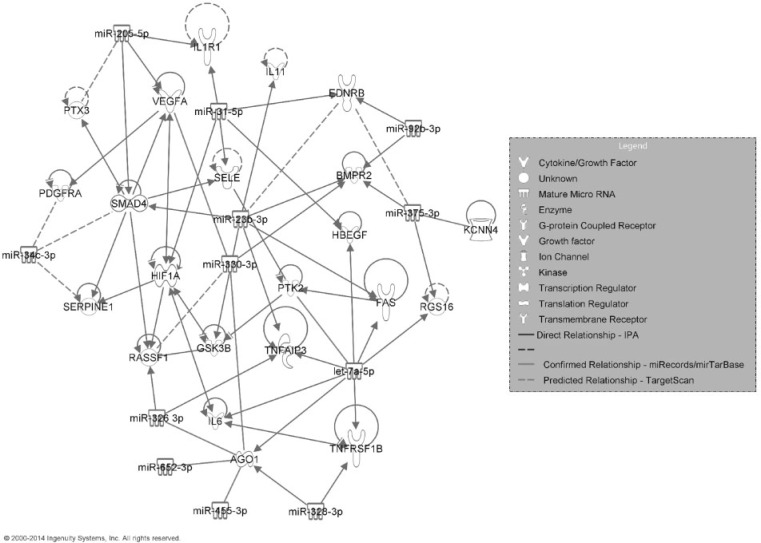
Among the 12 miRNAs associated with the fibrotic patterns, 2 miRNAs (miR-31-5p and miR-328-3p) were significantly (FDR <5%; SAM analysis) up- or down-regulated after MWCNT exposure (Table 3). All 12 miRNAs had at least 1 mRNA target based on the miRTarBase, miRecords, TargetScan, or IPA databases (Table 3). The integrated fibrotic pathway based on miRNAs and mRNAs in Table 3 is shown in Figure 2. In addition to the functional relationships found by IPA, our system identified experimentally confirmed regulations between ago1 and let-7c-5p, miR-455-3p, miR-652-3p, miR-326-3p, miR-328-3p, and miR-330-3p (Helwak et al., 2013) and between vegfa and miR-330-3p (Ye et al., 2008). Our system predicted highly conserved target pairs between ednrb and miR-375-3p and miR-23b-3p; rassf1 and miR-330-3p; and miR-34c-3p and pdgfr, smad4, and serpine1. The gene expression direction (up- or down-regulation relative to control) at doses 10, 20, 40, or 80 µg MWCNT at post-exposure Day 56 for each mRNA and miRNA is summarized in Supplementary Figure 2. This time point was chosen because quantitative morphometric analysis of Sirius Red staining for collagen at Day 56 (Mercer et al., 2011) was used as the basis to determine the fibrotic pattern.
FIG. 2.
Regulatory network of mRNAs and miRNAs transcriptionally related to the Day 56 post-exposure Sirius Red staining fibrotic pathological pattern.
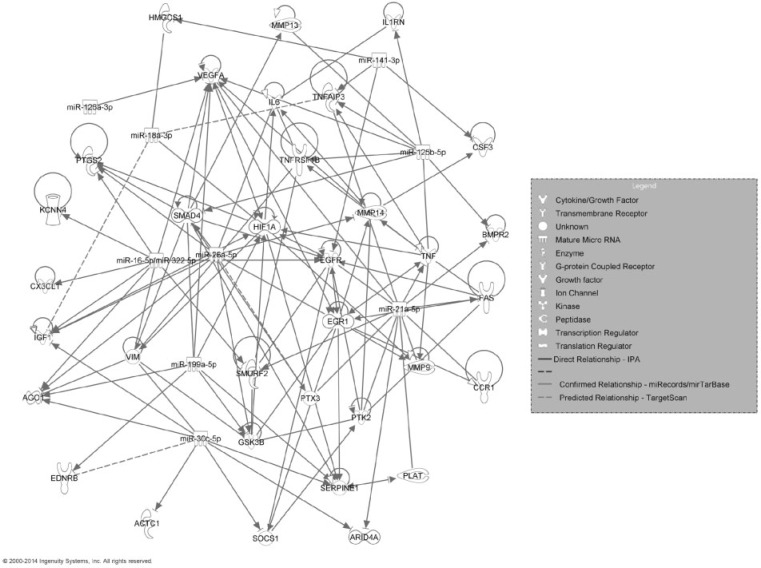
Next, we used the set of mRNAs associated with the MWCNT-induced fibrotic pathological patterns and functionally involved with fibrosis in IPA analysis to identify their miRNA regulators. A total of 10 miRNAs were identified as potentially involved in MWCNT-induced fibrosis (Table 4). Among them, 7 miRNAs (miR-125b-5p, miR-126a-3p, miR-16-5p, miR-199a-5p, miR-21-5p, miR-30c-5p, and miR-322) had a significant (FDR <5%; SAM analysis) expression change after MWNCT exposure. All 10 miRNAs had at least 1 mRNA target based on the miRTarBase, miRecords, TargetScan, or IPA databases. The integrated fibrotic pathway for the miRNAs and mRNAs listed in Table 4 is shown in Figure 3. In addition to the functional relationships retrieved with IPA, our analysis identified numerous post-transcriptional regulations that have been previously experimentally confirmed, including regulatory relationships between miR-125-5p and il1rn (Hofmann et al., 2009), tnf (Tili et al., 2007), tnfaip3 (Kim et al., 2012), and mmp13 (Xu et al., 2012); miR-126a-3p and vegfa (Zhu et al., 2011); miR-18a-3p and hif1a (Hafner et al., 2010), smad4 (Dews et al., 2010), and hmgcs1 (Hafner et al., 2010); miR-26a-5p andegr1 (Chi et al., 2009), smad4 (Dey et al., 2012), gsk3b (Mohamed et al., 2010), il6 (Yang et al., 2013), ago1 (Helwak et al., 2013); miR-21a-5p and fas (Sayed et al., 2010), bmpr2 (Papagiannakopoulos et al., 2008), egfr (Zhou et al., 2010), plat (Terao et al., 2011), ptx3 (Terao et al., 2011), tnfaip3 (Terao et al., 2011), ccr1 (Terao et al., 2011), vegfa (Liu et al., 2011), mmp9 (Moriyama et al., 2009), ptk2 (Gabriely et al., 2008), arid4a (Gabriely et al., 2008), among many others listed in Table 4 (highlighted with solid lines without arrows in Fig. 3). Our analysis predicted highly conserved target pairs between miR-18a-3p and igf1 and tnfaip3; miR-322-5p and ptgs2, vegfa, kcnn4, cxcl1, and smurf2; miR-30c-5p and ednrb; and miR-26a-5p and ptx3 (designated by dashed lines in Fig. 3). The gene expression direction (up- or down-regulation relative to control) at doses 10, 20, 40, or 80 µg MWCNT at post-exposure Day 56 for the mRNAs and miRNAs in Figure 3 is summarized in Supplementary Figure 3.
FIG. 3.
Regulatory network of mRNAs transcriptionally related to the Day 56 post-exposure Sirius Red staining fibrotic pathological pattern and miRNAs experimentally validated to be involved in fibrosis according to the Ingenuity Knowledge Base.
These results indicate that our algorithms could identify miRNA-mediated post-transcriptional regulations in MWCNT-treated mice, either experimentally confirmed interactions or highly conserved target pair predictions, many of which were not available in the Ingenuity Knowledge Base. The identified miRNAs and mRNAs were significantly associated with MWCNT-induced pathological patterns and/or significantly up- or down-regulated in the mouse lung following MWCNT exposure, indicating their potential involvement in pathogenesis and utility as biomarkers for disease. The integrated pathway analysis of miRNA and mRNA and the revealing of their regulatory interactions further elucidated their functional roles in molecular disease mechanisms.
Potential Signaling Pathways in MWCNT-Induced Lung Inflammation and Fibrosis
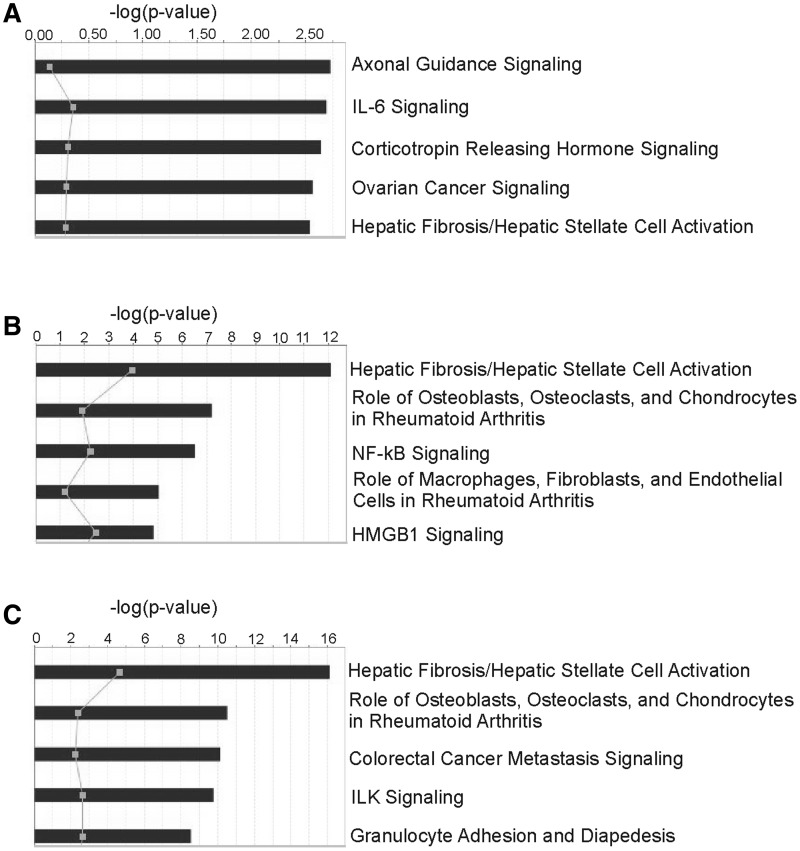
To determine signaling pathways potentially involved in the inflammatory and fibrotic pathological responses to MWCNT exposure, all functionally related mRNAs and miRNAs were analyzed by IPA using a Core Analysis. The top 5 canonical pathways significant to the mRNAs and miRNAs found in the integrated inflammation analysis (Table 2) were Axonal Guidance Signaling, IL-6 Signaling, Corticotropin Releasing Hormone Signaling, Ovarian Cancer Signaling, and Hepatic Fibrosis/Hepatic Stellate Cell Activation (Fig. 4A). The top 5 canonical pathways significant to the mRNAs and miRNAs found in the integrated fibrosis analysis using the NMF algorithm (Table 3) were Hepatic Fibrosis/Hepatic Stellate Cell Activation; Role of Osteoblasts, Osteoclasts, and Chondrocytes in Rheumatoid Arthritis; NF-κB Signaling; Role of Macrophages, Fibroblasts, and Endothelial Cells in Rheumatoid Arthritis; and HMGB1 Signaling (Fig. 4B). The top 5 canonical pathways significant to the mRNAs and miRNAs found in the integrated fibrosis (IPA) analysis (Table 4) were Hepatic Fibrosis/Hepatic Stellate Cell Activation; Role of Osteoblasts, Osteoclasts, and Chondrocytes in Rheumatoid Arthritis; Colorectal Cancer Metastasis Signaling; ILK Signaling; and Granulocyte Adhesion and Diapedesis (Fig. 4C). These results suggest that the miRNA and mRNA regulatory networks identified in our analysis are reflective of the inflammatory response and tissue remodeling biological processes during the onset and progression of fibrosis following MWCNT exposure.
FIG. 4.
Top 5 canonical pathways of mRNAs and miRNAs transcriptionally related to the inflammatory pathological pattern (A), the fibrotic pathological pattern (B), or experimentally validated to be involved in fibrosis by IPA (C).
DISCUSSION
MWCNT are an important class of engineered nanomaterials with broad applications in many industries (Pacurari et al., 2010). Concerns over potential MWCNT-induced toxicity have emerged, particularly due to the structural similarity between asbestos and MWCNT (Donaldson et al., 2006; Kobayashi et al., 2010; Muller et al., 2005). Previous studies have shown that MWCNT induce lung damage, including inflammatory granulomas and substantial interstitial lung fibrosis (Mercer et al., 2011; Porter et al., 2010). Pulmonary fibrosis has a poor clinical outcome (Bjoraker et al., 1998; Douglas et al., 1998; Flaherty et al., 2001; Schwartz et al., 1994) and may be a potential precursor to lung cancer (Auerbach et al., 1979; Peretz et al., 2006; Shiels et al., 2011; Yu et al., 2008). However, there are no clinically applicable biomarkers for early detection and no effective treatment for pulmonary fibrosis due to its late diagnosis and poorly understood molecular mechanisms for initiation (Homer et al., 2011; Hoo and Whyte, 2012; Walter et al., 2006).
Recently, there is emerging interest in exploring miRNAs as potential therapeutic targets and biomarkers for diagnosis and prognosis. Advantages of miRNA biomarkers include their presence in various bodily fluids (Chen et al., 2012; Mitchell et al., 2008) and greater stability in prepared tissue samples, including formalin fixation, relative to mRNA (Jung et al., 2010; Mraz et al., 2009; Xi et al., 2007). The use of miRNA markers in the selection of appropriate treatments has the possibility for improving patient outcomes by determining the best application of existing drugs. Additionally, identifying miRNA biomarkers may aid in the development of novel treatments through the elucidation of new pathways (Avraham and Yarden, 2012; Iorio and Croce, 2012).
This study used an integrated analysis of miRNA and mRNA time series microarray data to determine miRNA/mRNA regulatory relationships and potential signaling pathways either reflective of lung inflammatory and fibrotic pathologies, or functionally related to inflammation and fibrosis according to the Ingenuity Knowledge Base, after in vivo mouse exposure by pharyngeal aspiration to DM or 10, 20, 40, or 80 µg MWCNT at 7 or 56 days post-exposure (Porter et al., 2008, 2010). We have previously described a novel computational model capable of using mRNA expression data from mouse lungs exposed to MWCNT to determine biological processes that were strongly associated with lung inflammatory and fibrotic pathologies (Snyder-Talkington et al., 2013). In the current study, we used a similar approach to determine significant miRNAs associated with MWCNT-induced pathologies from time series microarray data and performed an integrated analysis, using both mRNA and miRNA expression, to determine integrated expression networks suggestive of potential biomarkers and signaling pathways reflective of lung inflammation and fibrosis after MWCNT exposure.
We have previously described a system (MEGPath) for determining a set of genes reflective of lung inflammatory and fibrotic pathologies that have been shown to be functionally involved in inflammation and fibrosis through IPA (Dymacek and Guo, 2011; Snyder-Talkington et al., 2013). The MEGPath system was designed to identify sets of genes from the MSigDB that, as a group, have transcriptional expression significantly resembling a known pathological pattern. These sets consist of genes belonging to the same biological processes or pathways. Genes sets that were transcriptionally associated with the inflammatory and fibrotic pathologies following MWCNT exposure (fold change >1.5, FDR <1% in SAM analysis) (Snyder-Talkington et al., 2013) were identified at each dose and time condition, and analyzed with IPA to determine those mRNAs detected by our system that were also shown in the scientific literature to be functionally involved in inflammation and fibrosis, respectively.
Although sets of mRNAs reflective of the pathological patterns could be curated from the MSigDB, there are currently no functional annotation databases for miRNA similar to the MSigDB, necessitating a different approach for miRNA analysis in this study. Here, 2 methods were used to determine miRNAs relevant to MWCNT-induced pathologies. First, we used a constrained NMF algorithm to determine significant miRNAs whose expressions were associated with the inflammatory and fibrotic pathologies in mouse lungs following MWCNT exposure. In the NMF analysis, miRNA expressions matched the inflammatory and fibrotic pathologies, but did not need to be experimentally validated in inflammation and fibrosis with IPA. Secondly, all significantly up- or down-regulated miRNAs were analyzed with IPA, and only those miRNAs shown to be experimentally validated in inflammation and fibrosis by the scientific literature were kept for further analysis. Both methods were conducted to ensure that all relevant miRNAs, those described in the scientific literature and those identified by our system, related to inflammatory and fibrotic pathologies were included for further analysis.
After identifying significant mRNAs and miRNAs related to in vivo inflammatory and fibrotic pathologies following MWCNT exposure, an integrated mRNA/miRNA analysis was performed to identify miRNA-regulated, post-transcriptional mechanisms involved in MWCNT-induced pathogenesis. Historically, paired miRNA and mRNA expressions are considered regulatory if they are predicted binding partners and have opposite expression directions. Therefore, traditionally, a negative correlation analysis is performed when determining regulatory pairs, as the miRNA and mRNA expressions should be in opposite directions to signify regulation. However, each miRNA may target multiple mRNAs; hence, over the course of time, a miRNA’s expression may be changing to regulate multiple mRNAs and not be “opposite” of a targeted mRNA’s expression at the same time point. Therefore, the discrete time points of time series data may miss critical moments when a miRNA’s expression changes. In this study, we used a dynamic, second derivative analysis of mRNA and miRNA expression over time to determine potential mRNA/miRNA pairs that had expression in opposition at most time points, but may diverge from this trend at a few. This method may be more reflective of the regulatory feedback between mRNA and miRNA expression that changes over time but may be lost when only considering expression at a discrete time point.
After we used the second derivative analysis of expression profiles to identify potential regulatory miRNA/mRNA pairs, mRNA and miRNA functional relationships were assessed through the Ingenuity Knowledge Base, as well as by the miRTarBase (Hsu et al., 2014), miRecords (Xiao et al., 2009), and TargetScan (Lewis et al., 2005) databases. The Ingenuity Knowledge Base is based on the collated findings of patient phenotypes and disease, cellular, molecular, and sequence mechanisms, and all connections between miRNAs and mRNAs are supported by at least 1 reference in the scientific literature. The TargetScan database provided predicted regulatory mRNA/miRNA pairs, whereas both miRTarBase and miRecords provided a mix of published, experimentally validated, and predicted mRNA and miRNA pairings. In our analysis, only those binding relationships considered to be highly conserved by TargetScan were considered to be “predicted targets” in our analysis (Tables 2–4). IPA considers all relationships, both highly and poorly conserved; therefore, additional predicted relationships were found through the TargetScan database using IPA that were not identified through our second derivative analysis. It is worth noting that vegfa was experimentally confirmed in the fibrosis analysis to be an experimentally confirmed target of the tumor suppressor miRNA miR-205-5p (Wu et al., 2009) and the oncogenic miRNA miR-330-3p (Ye et al., 2008). In our previous study, vegfa was predicted to be involved in both MWCNT-induced lung inflammation and fibrosis using both in vivo and in vitro mRNA and protein assays (Snyder-Talkington et al., 2013). The interactions with these potential miRNA regulators could provide new insights into post-transcriptional regulatory mechanisms involved in MWCNT-induced lung angiogenesis, inflammation, and fibrosis. In addition, miR-23b-3p was experimentally confirmed as a regulator of smad4 (Rogler et al., 2009), and miR-34c-3p was identified as a potential regulator of smad4 in MWCNT-induced fibrosis (Table 3), revealing potential involvement of the TGF-β signaling pathway.
The regulatory networks of mRNA and miRNA determined by both IPA analysis and predicted binding due to sequence similarity in this study give a detailed view of potential regulatory networks and signaling pathways involved in MWCNT-induced inflammation and fibrosis. Core analysis of the significant integrated inflammatory mRNA and miRNA detected pathways involved cytoskeletal remodeling, acute-phase and stress responses, cell adhesion and growth, and the accumulation of ECM proteins. Core analysis of the integrated fibrotic mRNA and miRNA (both NMF and IPA) detected pathways involved in the accumulation of ECM protein, chronic inflammation, innate and acquired immunity, cellular transcription and metastasis, integrin/ECM signaling, and leukocyte migration, suggesting that the miRNA and mRNA regulatory networks determined by our analysis are reflective of the inflammatory response and tissue remodeling that takes place during MWCNT exposure and the onset of fibrosis.
Although the gross response to MWCNT exposure has been well characterized, the underlying mechanisms behind the switch from a transient inflammatory response to progressive fibrosis are unknown. By analyzing both miRNA and mRNA in a dynamic integrated analysis, we determined regulatory signaling networks that are significantly related to inflammatory and fibrotic disease pathologies. We suggest that these regulatory networks may be important for determining the biological processes underlying lung inflammatory and fibrotic pathologies and may serve as potential biomarkers for early detection.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
FUNDING
National Institutes of Health (R01ES021764 and R56LM009500 to N.L.G.); and National Science Foundation training grant (to J.D.).
Supplementary Material
REFERENCES
- Auerbach O., Garfinkel L., Parks V. R. (1979). Scar cancer of the lung: increase over a 21 year period. Cancer 43, 636–642. [DOI] [PubMed] [Google Scholar]
- Avraham R., Yarden Y. (2012). Regulation of signalling by microRNAs. Biochem. Soc. Trans. 40, 26–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297. [DOI] [PubMed] [Google Scholar]
- Beezhold K. J., Castranova V., Chen F. (2010). Microprocessor of microRNAs: regulation and potential for therapeutic intervention. Mol. Cancer 9, 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjoraker J. A., Ryu J. H., Edwin M. K., Myers J. L., Tazelaar H. D., Schroeder D. R., Offord K. P. (1998). Prognostic significance of histopathologic subsets in idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 157, 199–203. [DOI] [PubMed] [Google Scholar]
- Bockhorn J., Yee K., Chang Y. F., Prat A., Huo D., Nwachukwu C., Dalton R., Huang S., Swanson K. E., Perou C. M., et al. (2013). MicroRNA-30c targets cytoskeleton genes involved in breast cancer cell invasion. Breast Cancer Res. Treat. 137, 373–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock M., Trenkmann M., Gay R. E., Michel B. A., Gay S., Fischler M., Ulrich S., Speich R., Huber L. C. (2009). Interleukin-6 modulates the expression of the bone morphogenic protein receptor type II through a novel STAT3-microRNA cluster 17/92 pathway. Circ. Res. 104, 1184–1191. [DOI] [PubMed] [Google Scholar]
- Brown T. A. (2006). Genomes , 3rd ed pp. 14–18 Garland Science, New York. [Google Scholar]
- Chen X., Hu Z., Wang W., Ba Y., Ma L., Zhang C., Wang C., Ren Z., Zhao Y., Wu S., et al. (2012). Identification of ten serum microRNAs from a genome-wide serum microRNA expression profile as novel noninvasive biomarkers for nonsmall cell lung cancer diagnosis. Int. J. Cancer 130, 1620–1628. [DOI] [PubMed] [Google Scholar]
- Cheng C., Li L. M. (2008). Inferring microRNA activities by combining gene expression with microRNA target prediction. PLoS One 3, e1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi S. W., Zang J. B., Mele A., Darnell R. B. (2009). Argonaute HITS-CLIP decodes microRNA–mRNA interaction maps. Nature 460, 479–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J. H., Gelinas R., Wang K., Etheridge A., Piper M. G., Batte K., Dakhallah D., Price J., Bornman D., Zhang S., et al. (2011). Systems biology of interstitial lung diseases: integration of mRNA and microRNA expression changes. BMC Med. Genomics 4, 8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Dews M., Fox J. L., Hultine S., Sundaram P., Wang W., Liu Y. Y., Furth E., Enders G. H., El-Deiry W., Schelter J. M., et al. (2010). The myc-miR-17∼92 axis blunts TGFβ signaling and production of multiple TGFβ-dependent antiangiogenic factors. Cancer Res. 70, 8233–8246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey B. K., Gagan J., Yan Z., Dutta A. (2012). miR-26a is required for skeletal muscle differentiation and regeneration in mice. Genes Dev. 26, 2180–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson K., Aitken R., Tran L., Stone V., Duffin R., Forrest G., Alexander A. (2006). Carbon nanotubes: a review of their properties in relation to pulmonary toxicology and workplace safety. Toxicol. Sci. 92, 5–22. [DOI] [PubMed] [Google Scholar]
- Douglas W. W., Ryu J. H., Swensen S. J., Offord K. P., Schroeder D. R., Caron G. M., DeRemee R. A. (1998). Colchicine versus prednisone in the treatment of idiopathic pulmonary fibrosis. A randomized prospective study. Members of the Lung Study Group. Am. J. Respir. Crit. Care Med. 158, 220–225. [DOI] [PubMed] [Google Scholar]
- Dymacek J., Guo N. L. (2011). Systems approach to identifying relevant pathways from phenotype information in dose-dependent time series microarray data. Proceedings of 2011 IEEE International Conference on Bioinformatics and Biomedicine, pp. 290–293. IEEE Computer Society, Washington, DC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dymacek J., Guo N. L. (2014). Integrated miRNA and mRNA analysis of time series microarray data. Proceedings of the 5th ACM Conference on Bioinformatics, Computational Biology, and Health Informatics. pp. 122–127 ACM New York, NY, USA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EPA. (2014). United States environmental protection agency computational toxicology research. http://www.epa.gov/ncct/, Accessed June 6, 2014. [Google Scholar]
- Fatica A., Bozzoni I. (2014). Long non-coding RNAs: new players in cell differentiation and development. Nat. Rev. Genetics 15, 7–21. [DOI] [PubMed] [Google Scholar]
- Ferte C., Trister A. D., Huang E., Bot B. M., Guinney J., Commo F., Sieberts S., Andre F., Besse B., Soria J. C., et al. (2013). Impact of bioinformatic procedures in the development and translation of high-throughput molecular classifiers in oncology. Clin. Cancer Res. 19, 4315–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty K. R., Travis W. D., Colby T. V., Toews G. B., Kazerooni E. A., Gross B. H., Jain A., Strawderman R. L., Flint A., Lynch J. P., et al. (2001). Histopathologic variability in usual and nonspecific interstitial pneumonias. Am. J. Respir. Crit. Care Med. 164, 1722–1727. [DOI] [PubMed] [Google Scholar]
- Gabriely G., Wurdinger T., Kesari S., Esau C. C., Burchard J., Linsley P. S., Krichevsky A. M. (2008). MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol. Cell. Biol. 28, 5369–5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S., Grocock R. J., van Dongen S., Bateman A., Enright A. J. (2006). miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 34, D140–D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo N. L., Wan Y. W., Denvir J., Porter D. W., Pacurari M., Wolfarth M. G., Castranova V., Qian Y. (2012). Multiwalled carbon nanotube-induced gene signatures in the mouse lung: potential predictive value for human lung cancer risk and prognosis. J. Toxicol. Environ. Health Part A 75, 1129–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M., Landthaler M., Burger L., Khorshid M., Hausser J., Berninger P., Rothballer A., Ascano M., Jr, Jungkamp A. C., Munschauer M., et al. (2010). Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141, 129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helwak A., Kudla G., Dudnakova T., Tollervey D. (2013). Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell 153, 654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann M. H., Heinrich J., Radziwill G., Moelling K. (2009). A short hairpin DNA analogous to miR-125b inhibits C-Raf expression, proliferation, and survival of breast cancer cells. Mol. Cancer Res. 7, 1635–1644. [DOI] [PubMed] [Google Scholar]
- Homer R. J., Elias J. A., Lee C. G., Herzog E. (2011). Modern concepts on the role of inflammation in pulmonary fibrosis. Arch. Pathol. Lab. Med. 135, 780–788. [DOI] [PubMed] [Google Scholar]
- Hoo Z. H., Whyte M. K. (2012). Idiopathic pulmonary fibrosis. Thorax 67, 742–746. [DOI] [PubMed] [Google Scholar]
- Hsu S. D., Tseng Y. T., Shrestha S., Lin Y. L., Khaleel A., Chou C. H., Chu C. F., Huang H. Y., Lin C. M., Ho S. Y., et al. (2014). miRTarBase update 2014: an information resource for experimentally validated miRNA-target interactions. Nucleic Acids Res. 42, D78–D85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima S. (1991). Helical microtubules of graphitic carbon. Nature 354, 56–58. [Google Scholar]
- Iorio M. V., Croce C. M. (2012). microRNA involvement in human cancer. Carcinogenesis 33, 1126–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung M., Schaefer A., Steiner I., Kempkensteffen C., Stephan C., Erbersdobler A., Jung K. (2010). Robust microRNA stability in degraded RNA preparations from human tissue and cell samples. Clin. Chem. 56, 998–1006. [DOI] [PubMed] [Google Scholar]
- Karaa Z. S., Iacovoni J. S., Bastide A., Lacazette E., Touriol C., Prats H. (2009). The VEGF IRESes are differentially susceptible to translation inhibition by miR-16. RNA 15, 249–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karn T. (2013). High-throughput gene expression and mutation profiling: current methods and future perspectives. Breast Care 8, 401–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall M., Holgate S. (2012). Health impact and toxicological effects of nanomaterials in the lung. Respirology 17, 743–758. [DOI] [PubMed] [Google Scholar]
- Kim S. W., Ramasamy K., Bouamar H., Lin A. P., Jiang D., Aguiar R. C. (2012). MicroRNAs miR-125a and miR-125b constitutively activate the NF-kappaB pathway by targeting the tumor necrosis factor alpha-induced protein 3 (TNFAIP3, A20). Proc. Natl Acad. Sci. U.S.A. 109, 7865–7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi N., Naya M., Ema M., Endoh S., Maru J., Mizuno K., Nakanishi J. (2010). Biological response and morphological assessment of individually dispersed multi-wall carbon nanotubes in the lung after intratracheal instillation in rats. Toxicology 276, 143–153. [DOI] [PubMed] [Google Scholar]
- Lee D. D., Seung H. S. (1999). Learning the parts of objects by non-negative matrix factorization. Nature 401, 788–791. [DOI] [PubMed] [Google Scholar]
- Lee R. C., Feinbaum R. L., Ambros V. (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854. [DOI] [PubMed] [Google Scholar]
- Lewis B. P., Burge C. B., Bartel D. P. (2005). Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120, 15–20. [DOI] [PubMed] [Google Scholar]
- Liu L. Z., Li C., Chen Q., Jing Y., Carpenter R., Jiang Y., Kung H. F., Lai L., Jiang B. H. (2011). MiR-21 induced angiogenesis through AKT and ERK activation and HIF-1alpha expression. PLoS One 6, e19139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer R. R., Hubbs A. F., Scabilloni J. F., Wang L., Battelli L. A., Friend S., Castranova V., Porter D. W. (2011). Pulmonary fibrotic response to aspiration of multi-walled carbon nanotubes. Part. Fibre Toxicol. 8, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer R. R., Hubbs A. F., Scabilloni J. F., Wang L., Battelli L. A., Schwegler-Berry D., Castranova V., Porter D. W. (2010). Distribution and persistence of pleural penetrations by multi-walled carbon nanotubes. Part. Fibre Toxicol. 7, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer R. R., Scabilloni J. F., Hubbs A. F., Battelli L. A., McKinney W., Friend S., Wolfarth M. G., Andrew M., Castranova V., Porter D. W. (2013a). Distribution and fibrotic response following inhalation exposure to multi-walled carbon nanotubes. Part. Fibre Toxicol. 10, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer R. R., Scabilloni J. F., Hubbs A. F., Wang L., Battelli L. A., McKinney W., Castranova V., Porter D. W. (2013b). Extrapulmonary transport of MWCNT following inhalation exposure. Part. Fibre Toxicol. 10, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P. S., Parkin R. K., Kroh E. M., Fritz B. R., Wyman S. K., Pogosova-Agadjanyan E. L., Peterson A., Noteboom J., O'Briant K. C., Allen A., et al. (2008). Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. U.S.A. 105, 10513–10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed J. S., Lopez M. A., Boriek A. M. (2010). Mechanical stretch up-regulates microRNA-26a and induces human airway smooth muscle hypertrophy by suppressing glycogen synthase kinase-3beta. J. Biol. Chem. 285, 29336–29347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama T., Ohuchida K., Mizumoto K., Yu J., Sato N., Nabae T., Takahata S., Toma H., Nagai E., Tanaka M. (2009). MicroRNA-21 modulates biological functions of pancreatic cancer cells including their proliferation, invasion, and chemoresistance. Mol. Cancer Ther. 8, 1067–1074. [DOI] [PubMed] [Google Scholar]
- Mraz M., Malinova K., Mayer J., Pospisilova S. (2009). MicroRNA isolation and stability in stored RNA samples. Biochem. Biophys. Res. Commun. 390, 1–4. [DOI] [PubMed] [Google Scholar]
- Muller J., Huaux F., Moreau N., Misson P., Heilier J. F., Delos M., Arras M., Fonseca A., Nagy J. B., Lison D. (2005). Respiratory toxicity of multi-wall carbon nanotubes. Toxicol. Appl. Pharmacol. 207, 221–231. [DOI] [PubMed] [Google Scholar]
- Nana-Sinkam S. P., Hunter M. G., Nuovo G. J., Schmittgen T. D., Gelinas R., Galas D., Marsh C. B. (2009). Integrating the MicroRNome into the study of lung disease. Am. J. Respir. Crit. Care Med. 179, 4–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacurari M., Castranova V., Vallyathan V. (2010). Single- and multi-wall carbon nanotubes versus asbestos: are the carbon nanotubes a new health risk to humans? J . Toxicol. Environ. Health Part A 73, 378–395. [DOI] [PubMed] [Google Scholar]
- Pacurari M., Qian Y., Porter D. W., Wolfarth M., Wan Y., Luo D., Ding M., Castranova V., Guo N. L. (2011). Multi-walled carbon nanotube-induced gene expression in the mouse lung: association with lung pathology . Toxicol Appl Pharmacol 255, 18–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papagiannakopoulos T., Shapiro A., Kosik K. S. (2008). MicroRNA-21 targets a network of key tumor-suppressive pathways in glioblastoma cells. Cancer Res. 68, 8164–8172. [DOI] [PubMed] [Google Scholar]
- Peretz A., Checkoway H., Kaufman J. D., Trajber I., Lerman Y. (2006). Silica, silicosis, and lung cancer. Isr. Med. Assoc. J. 8, 114–118. [PubMed] [Google Scholar]
- Porter D., Sriram K., Wolfarth M., Jefferson A., Schwegler-Berry D., Andrew M., Castranova V. (2008). A biocompatible medium for nanoparticle dispersion. Nanotoxicology 2, 144–154. [Google Scholar]
- Porter D. W., Hubbs A. F., Chen B. T., McKinney W., Mercer R. R., Wolfarth M. G., Battelli L., Wu N., Sriram K., Leonard S., et al. (2013). Acute pulmonary dose-responses to inhaled multi-walled carbon nanotubes. Nanotoxicology 7, 1179–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter D. W., Hubbs A. F., Mercer R. R., Wu N., Wolfarth M. G., Sriram K., Leonard S., Battelli L., Schwegler-Berry D., Friend S., et al. (2010). Mouse pulmonary dose- and time course-responses induced by exposure to multi-walled carbon nanotubes. Toxicology 269, 136–147. [DOI] [PubMed] [Google Scholar]
- Putman R. K., Rosas I. O., Hunninghake G. M. (2014). Genetics and early detection in idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 189, 770–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rane S., He M., Sayed D., Vashistha H., Malhotra A., Sadoshima J., Vatner D. E., Vatner S. F., Abdellatif M. (2009). Downregulation of miR-199a derepresses hypoxia-inducible factor-1alpha and Sirtuin 1 and recapitulates hypoxia preconditioning in cardiac myocytes. Circ. Res. 104, 879–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogler C. E., Levoci L., Ader T., Massimi A., Tchaikovskaya T., Norel R., Rogler L. E. (2009). MicroRNA-23b cluster microRNAs regulate transforming growth factor-beta/bone morphogenetic protein signaling and liver stem cell differentiation by targeting Smads. Hepatology 50, 575–584. [DOI] [PubMed] [Google Scholar]
- Sayed D., He M., Hong C., Gao S., Rane S., Yang Z., Abdellatif M. (2010). MicroRNA-21 is a downstream effector of AKT that mediates its antiapoptotic effects via suppression of Fas ligand. J. Biol. Chem. 285, 20281–20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D. A., Helmers R. A., Galvin J. R., Van Fossen D. S., Frees K. L., Dayton C. S., Burmeister L. F., Hunninghake G. W. (1994). Determinants of survival in idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 149, 450–454. [DOI] [PubMed] [Google Scholar]
- Selbach M., Schwanhausser B., Thierfelder N., Fang Z., Khanin R., Rajewsky N. (2008). Widespread changes in protein synthesis induced by microRNAs. Nature 455, 58–63. [DOI] [PubMed] [Google Scholar]
- Sessa R., Hata A. (2013). Role of microRNAs in lung development and pulmonary diseases. Pulm. Circ. 3, 315–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J., Yang X., Xie B., Chen Y., Swaim M., Hackett S. F., Campochiaro P. A. (2008). MicroRNAs regulate ocular neovascularization. Mol. Ther. 16, 1208–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiels M. S., Albanes D., Virtamo J., Engels E. A. (2011). Increased risk of lung cancer in men with tuberculosis in the alpha-tocopherol, beta-carotene cancer prevention study. Cancer Epidemiol. Biomarkers Prev. 20, 672–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder-Talkington B. N., Dymacek J., Porter D. W., Wolfarth M. G., Mercer R. R., Pacurari M., Denvir J., Castranova V., Qian Y., Guo N. L. (2013). System-based identification of toxicity pathways associated with multi-walled carbon nanotube-induced pathological responses. Toxicol. Appl. Pharmacol. 272, 476–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strieter R. M. (2008). What differentiates normal lung repair and fibrosis? Inflammation, resolution of repair, and fibrosis. Proc. Am. Thorac. Soc. 5, 305–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez Y., Wang C., Manes T. D., Pober J. S. (2010). Cutting edge: TNF-induced microRNAs regulate TNF-induced expression of E-selectin and intercellular adhesion molecule-1 on human endothelial cells: feedback control of inflammation. J. Immunol. 184, 21–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimura K., Miyata H., Tanaka K., Hamano R., Takahashi T., Kurokawa Y., Yamasaki M., Nakajima K., Takiguchi S., Mori M., et al. (2012). Let-7 expression is a significant determinant of response to chemotherapy through the regulation of IL-6/STAT3 pathway in esophageal squamous cell carcinoma. Clin. Cancer Res. 18, 5144–5153. [DOI] [PubMed] [Google Scholar]
- Terao M., Fratelli M., Kurosaki M., Zanetti A., Guarnaccia V., Paroni G., Tsykin A., Lupi M., Gianni M., Goodall G. J., et al. (2011). Induction of miR-21 by retinoic acid in estrogen receptor-positive breast carcinoma cells: biological correlates and molecular targets. J. Biol. Chem. 286, 4027–4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tili E., Michaille J. J., Cimino A., Costinean S., Dumitru C. D., Adair B., Fabbri M., Alder H., Liu C. G., Calin G. A., et al. (2007). Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J. Immunol. 179, 5082–5089. [DOI] [PubMed] [Google Scholar]
- Tsukamoto Y., Nakada C., Noguchi T., Tanigawa M., Nguyen L. T., Uchida T., Hijiya N., Matsuura K., Fujioka T., Seto M., et al. (2010). MicroRNA-375 is downregulated in gastric carcinomas and regulates cell survival by targeting PDK1 and 14-3-3zeta. Cancer Res. 70, 2339–2349. [DOI] [PubMed] [Google Scholar]
- Walter N., Collard H. R., King T. E., Jr (2006). Current perspectives on the treatment of idiopathic pulmonary fibrosis. Proc. Am. Thorac. Soc. 3, 330–338. [DOI] [PubMed] [Google Scholar]
- Wu H., Zhu S., Mo Y. Y. (2009). Suppression of cell growth and invasion by miR-205 in breast cancer. Cell Res. 19, 439–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Y., Nakajima G., Gavin E., Morris C. G., Kudo K., Hayashi K., Ju J. (2007). Systematic analysis of microRNA expression of RNA extracted from fresh frozen and formalin-fixed paraffin-embedded samples. RNA 13, 1668–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F., Zuo Z., Cai G., Kang S., Gao X., Li T. (2009). miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res. 37, D105–D110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N., Zhang L., Meisgen F., Harada M., Heilborn J., Homey B., Grander D., Stahle M., Sonkoly E., Pivarcsi A. (2012). MicroRNA-125b down-regulates matrix metallopeptidase 13 and inhibits cutaneous squamous cell carcinoma cell proliferation, migration, and invasion. J. Biol. Chem. 287, 29899–29908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Liang L., Zhang X. F., Jia H. L., Qin Y., Zhu X. C., Gao X. M., Qiao P., Zheng Y., Sheng Y. Y., et al. (2013). MicroRNA-26a suppresses tumor growth and metastasis of human hepatocellular carcinoma by targeting interleukin-6-Stat3 pathway. Hepatology 58, 158–170. [DOI] [PubMed] [Google Scholar]
- Ye W., Lv Q., Wong C. K., Hu S., Fu C., Hua Z., Cai G., Li G., Yang B. B., Zhang Y. (2008). The effect of central loops in miRNA:MRE duplexes on the efficiency of miRNA-mediated gene regulation. PLoS One 3, e1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y. Y., Pinsky P. F., Caporaso N. E., Chatterjee N., Baumgarten M., Langenberg P., Furuno J. P., Lan Q., Engels E. A. (2008). Lung cancer risk following detection of pulmonary scarring by chest radiography in the prostate, lung, colorectal, and ovarian cancer screening trial. Arch. Int. Med. 168, 2326–2332; discussion 2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Daucher M., Armistead D., Russell R., Kottilil S. (2013). MicroRNA expression profiling in HCV-infected human hepatoma cells identifies potential anti-viral targets induced by interferon-alpha. PLoS One 8, e55733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Ng W. L., Wang P., Tian L., Werner E., Wang H., Doetsch P., Wang Y. (2012a). MicroRNA-21 modulates the levels of reactive oxygen species by targeting SOD3 and TNFalpha. Cancer Res. 72, 4707–4713. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Zhang Y., Fan K. J., Sun Q., Chen A. Z., Shen W. L., Zhao Z. H., Zheng X. F., Yang X. (2012b). Functional screening for miRNAs targeting Smad4 identified miR-199a as a negative regulator of TGF-beta signalling pathway. Nucleic Acids Res. 40, 9286–9297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Ren Y., Moore L., Mei M., You Y., Xu P., Wang B., Wang G., Jia Z., Pu P., et al. (2010). Downregulation of miR-21 inhibits EGFR pathway and suppresses the growth of human glioblastoma cells independent of PTEN status. Lab. Invest. 90, 144–155. [DOI] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Xie H., Zhou Z., Chen H., Hu T., Bai Y., Shen Y., Yuan W., Jing Q., Qin Y. (2011). Endothelial-specific intron-derived miR-126 is down-regulated in human breast cancer and targets both VEGFA and PIK3R2. Mol. Cell. Biochem. 351, 157–164. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.