Abstract
Discovery of individualized therapies to address resistance to tyrosine kinase inhibitors (TKIs) has been hampered by the inability to test drug combinations on patient samples before and after TKI resistance. A recent study published in Science by Crystal et al. describes a methodology for pharmacological screening using a panel of 76 targeted agents and cell lines made directly from patient biopsies.
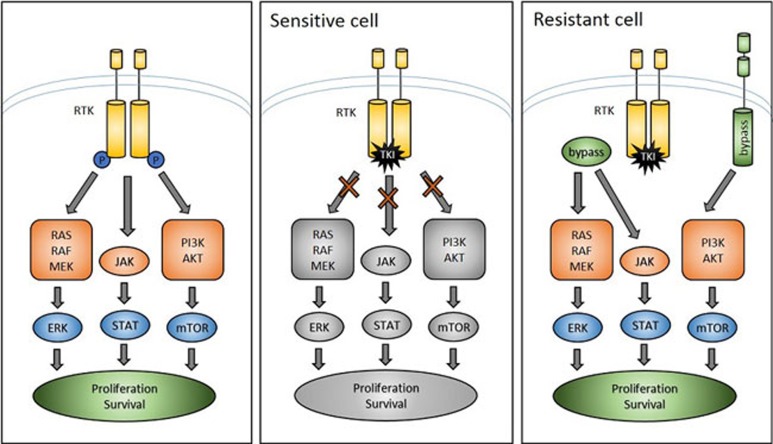
Genetic characterization of tumors and the subsequent use of TKIs as targeted therapies has become a mainstream treatment modality for a subset of non-small cell lung cancer (NSCLC). Patients with NSCLCs harboring EGFR activating mutations or ALK translocations often respond initially to TKIs but, in what has become a common storyline of many oncogene-driven cancers, varied mechanisms of resistance develop and relapse inevitably occurs. Resistance usually occurs in one of two ways: 1) as a result of a secondary mutation in the oncogene that prevents inhibition by the targeted agent (e.g., EGFR T790M1,2 and ALK L1196M3,4) or 2) through accessing a compensatory signaling pathway that bypasses the necessity of the oncogene for proliferation and survival5 (Figure 1). An understanding of these “bypass tracks” is driving current efforts to develop drug combinations that overcome resistance in NSCLC. These efforts have historically taken two forms: recapitulating the development of resistance in vitro using cell lines, and performing genetic analysis on biopsies to identify new drivers of resistance. While the identification of resistance mechanisms that develop in vitro provides insight into the range of such mechanisms, such discoveries are of limited clinical relevance for individual patients. On the other hand, genetic analysis of individual patient biopsies can reveal post-resistance mutations that can be treated with specific second- and third-generation inhibitors, which can produce high response rates6.
Figure 1.
RTK-driven cancers are dependent on signaling through an oncogenic RTK for proliferation and survival. Upon treatment with a targeted TKI, sensitive cells undergo senescence and apoptosis. In resistant cells, there exist secondary “bypass” mechanisms that reactivate signaling downstream of the inhibited RTK, allowing proliferation and survival to continue in the presence of targeted therapies.
In a recent study published in Science, Crystal et al.7 describe a methodology to screen individual patient biopsies for drug combinations that overcome clinically-developed resistance. The authors take advantage of methods recently described in Liu et al.8 that allowed them to create cell lines directly from patient specimens with a 50% success rate, which conferred the power of cell culture models to patient-derived tumor cells. They tested a panel of 76 targeted agents (in various phases of FDA approval) in the presence and absence of the original TKI against these patient-derived resistant lines, (11 EGFR-resistant and 9 ALK-resistant) and in vitro derived resistant lines, and determined the resultant change in cell viability and growth using in vitro proliferation assays. To compare the information yielded by genetic analysis to that yielded by their panel, they analyzed each of the patient-derived models by next-generation sequencing.
The extensive drug panel includes agents that target other receptor tyrosine kinases (RTKs; e.g., ErbB2, Flt3, FGFR), components of the original target's signaling pathway (e.g., PI3K, BRAF) and master regulators of proliferation and survival (e.g., STAT, Survivin, AKT). The panel also includes regulators of apoptosis, transcription, protein-folding machinery and DNA damage sensors. These drug targets represent critical components of pathways that could be activated in lung cancers to provide resistance to the original targeted therapy. As proof of concept, the authors performed the screen on five previously-established cell lines with known mechanisms of resistance, and found that drugs that target those mechanisms were among those producing the greatest responses. Among the 60 models screened, a total of 201 “hits” were identified, and at least one hit was identified in 50 of 60 lines. Hits were identified for 83% (8/11) of the EGFR-TKI resistant models and 89% (8/9) of the ALK-TKI resistant models. The screen demonstrated considerable robustness; drugs known to have overlapping specificity were found to have overlapping activity across cell lines, and consistent with previously published reports, EGFR inhibitors tended to be hits in both ALK- and MET-driven resistant lines. Additionally, three patient-derived and two in vitro derived lines were tested as xenografts and demonstrated substantial tumor regression only when treated with the drug combination revealed through this screen.
Importantly, the authors identified SRC family kinase inhibitors as hits across several patient-derived ALK-positive resistance models, for the first time implicating SRC signaling as a potential mechanism of resistance to ALK inhibition. Overexpression of kinase-dead SRC and shRNA-mediated knockdown of SRC recapitulated the effects of pharmacological SRC inhibitors, validating SRC as the relevant target. In subsequent experiments, they showed that both direct inhibition of ALK and inhibition of its downstream pathways in these cells resulted in robust upregulation of SRC activity. Suppression of redundant tyrosine kinases by an oncogene-driven cancer (and their subsequent reactivation upon oncogenic inhibition) is a well-established mechanism of intrinsic TKI resistance in EGFR-driven cancers9. Their findings here support a model in which ALK activity broadly suppresses SRC activity in ALK-driven cancers.
Clinical protocols at many sites call for targeted genetic typing of resistant biopsies. Importantly, this study demonstrated the inadequacy of genetic profiling in identifying effective drug combinations. Despite the fact that multiple SRC family kinase inhibitors were effective across several patient-derived resistant models, next-generation sequencing failed to identify mutations in these genes or other known regulators of SRC activity. Additionally, a MEK inhibitor, selumetinib was revealed to be a potent hit in one line derived from an ALK-positive cancer that had become resistant to ceritinib (a second-generation ALK inhibitor). Next-generation sequencing revealed no mutations in MEK, but did reveal a MAP2K1 mutation and an activating JAK3 mutation (V722I). MAP2K1 (an activator of MEK10) has not previously been described in the setting of resistance to ALK inhibition and thus would likely not have registered as an actionable mutation from the genetic screen. Of note, the JAK3 inhibitor tofacitinib was not a hit in the screen, and the cells did not express a high level of JAK3. This was also the first report of the use of a MEK inhibitor to resensitize ALK-positive cancers to ALK inhibitors and was observed only in one patient-derived line, illustrating the potential for this approach to identify efficacious, patient-specific drug combinations.
In summary, this study presents a potentially interesting methodology that can contribute to both the identification of effective drug combinations in TKI-resistant NSCLCs and the discovery of novel mechanisms of resistance, though some additional issues need to be addressed. Despite the innovations of the authors' approach, there remain questions of this platform's applicability in the clinic. It took 2-6 months to establish the biopsies as cell lines, a timeline which may not allow for fast enough turnaround to inform clinical decisions. Further studies comparing clinical outcomes based on strategies derived from relapsed tumors to those from the method presented here are needed before it is clear how these approaches can affect patient outcomes.
References
- Regales L, Gong Y, Shen R, et al. J Clin Invest. 2009. pp. 3000–3010. [DOI] [PMC free article] [PubMed]
- Chong CR, Jänne PA. Nat Med. 2013. pp. 1389–1400. [DOI] [PMC free article] [PubMed]
- Katayama R, Shaw AT, Khan TM, et al. Sci Transl Med. 2012. p. 120ra17. [DOI] [PMC free article] [PubMed]
- Roebele RC, Pilling AB, Aisner DL, et al. Clin Cancer Res. 2012. pp. 1472–1482. [DOI] [PMC free article] [PubMed]
- Nidederst MJ, Engelman JA. Sci Signal. 2013. p. re6. [DOI] [PMC free article] [PubMed]
- Sequist LV, Waltman BA, Dias-Santagata D, et al. Sci Transl Med. 2011. p. 75ra26. [DOI] [PMC free article] [PubMed]
- Crystal AS, Shaw AT, Sequist LV, et al. Science. 2014. pp. 1480–1486. [DOI] [PMC free article] [PubMed]
- Liu X, Ory V, Chapman S, et al. Am J Pathol. 2012. pp. 599–607. [DOI] [PMC free article] [PubMed]
- Nguyen KS, Kobayashi S, Costa DB. Clin Lung Cancer. 2009. pp. 281–289. [DOI] [PMC free article] [PubMed]
- Marks JL, Gong Y, Chitale D, et al. Cancer Res. 2008. pp. 5524–5528. [DOI] [PMC free article] [PubMed]