Abstract
The inhibitory role of p53 in DNA double-strand break (DSB) repair seems contradictory to its tumor-suppressing property. The p53 isoform Δ113p53/Δ133p53 is a p53 target gene that antagonizes p53 apoptotic activity. However, information on its functions in DNA damage repair is lacking. Here we report that Δ113p53 expression is strongly induced by γ-irradiation, but not by UV-irradiation or heat shock treatment. Strikingly, Δ113p53 promotes DNA DSB repair pathways, including homologous recombination, non-homologous end joining and single-strand annealing. To study the biological significance of Δ113p53 in promoting DNA DSB repair, we generated a zebrafish Δ113p53M/M mutant via the transcription activator-like effector nuclease technique and found that the mutant is more sensitive to γ-irradiation. The human ortholog, Δ133p53, is also only induced by γ-irradiation and functions to promote DNA DSB repair. Δ133p53-knockdown cells were arrested at the G2 phase at the later stage in response to γ-irradiation due to a high level of unrepaired DNA DSBs, which finally led to cell senescence. Furthermore, Δ113p53/Δ133p53 promotes DNA DSB repair via upregulating the transcription of repair genes rad51, lig4 and rad52 by binding to a novel type of p53-responsive element in their promoters. Our results demonstrate that Δ113p53/Δ133p53 is an evolutionally conserved pro-survival factor for DNA damage stress by preventing apoptosis and promoting DNA DSB repair to inhibit cell senescence. Our data also suggest that the induction of Δ133p53 expression in normal cells or tissues provides an important tolerance marker for cancer patients to radiotherapy.
Keywords: p53 isoform, Δ113p53/Δ133p53, NHEJ, HR, SSA, cell death, senescence
Introduction
The genetic material DNA is frequently attacked by both endogenous (cellular metabolic processes) and exogenous (environmental) factors. DNA double-strand breaks (DSBs) are the most catastrophic form of genotoxic insult that a cell can encounter. If not repaired, DNA DSBs can lead to chromosome loss and/or cell death. If improperly repaired, they can give rise to genetic mutations and chromosomal rearrangements, which can predispose an organism to immunodeficiency, neurological damage and cancer1. Organisms have evolved three efficient DNA DSB repair mechanisms, homologous recombination (HR), non-homologous end joining (NHEJ) and single-strand annealing (SSA), to minimize the effects of toxic insults on their DNA2,3,4. A number of environmental factors, such as ionizing radiation and various chemical agents (e.g., methyl methanesulfonate and bleomycin), can cause DNA DSBs1. To survive in such DNA damage stress conditions, it is very important for an organism to decide which cells are non-repairable and thus can be induced to die and which cells are repairable and thus can survive after DNA damage repair. However, how these decisions are made in response to DNA DSBs remains unexplored.
A central part of the DNA damage response is the activation of the tumor repressor gene, p53. Upon activation, p53 upregulates or represses the expression of a large number of downstream genes. The promoters of genes activated by p53 usually contain a consensus sequence of two pairs (half-sites) of pentamers arranged head-to-head, 5′-RRRC(A/T)(A/T)GYYY-3′ (R: purine, Y: pyrimidine), separated by 0-38 nucleotides. The promoters of genes repressed by p53 usually contain a consensus sequence of two pairs of pentamers arranged end-to-head, 5′-RRRC(A/T)(N)RRRC(A/T)-3′ or 5′-(A/T)GYYY(N)(A/T)GYYY-3′ (N: purine or pyrimidine), separated by 0-13 nucleotides5,6. The expression of p53 downstream genes triggers cell cycle arrest, DNA damage repair, apoptosis and/or senescence to ensure genome stability7,8. Intriguingly, p53 protein appears to promote only some DNA damage repair pathways, such as base excision repair, mismatch repair and nucleotide excision repair9,10,11, but inhibit DNA DSB repair pathways, including the HR, NHEJ and SSA pathways12,13,14. It has been demonstrated that p53 exerts a direct effect on DNA DSB repair, as mutations in p53 that impair or even abolish its transcriptional activity and cell cycle regulatory capacity do not significantly affect its inhibition of HR15,16,17. Further experiments have shown that the p53 protein is able to interact with repair proteins to prevent repair complex formation, such as RAD51 (a recombinase for HR) and replication protein A (RPA; a single-strand DNA-interacting protein required for stabilizing processed DNA ends)16,18,19. In contrast, there is also evidence that p53 transcriptionally inhibits the expression of repair genes, such as RAD5120. Recent studies have shown that the p53 protein relies on dynamic changes in its levels to control cell fate in response to DNA DSB stress, such as γ-irradiation, which is quite different from a single p53 pulse induced by UV irradiation21,22. Therefore, although full-length p53 inhibits DNA DSB repair, it is not clear how the p53 signal pathway regulates DNA DSB repair in response to DNA DSB stress.
The zebrafish protein Δ113p53 and its human counterpart Δ133p53 are N-terminally truncated forms of p53 with deletion of both the MDM2-interacting motif and the transactivation domain, together with partial deletion of the DNA-binding domain23,24,25. Δ113p53/Δ133p53 is a p53 target gene, which is transcribed by an alternative p53 promoter in intron 4. It is strongly induced by DNA damage stress to antagonize p53-mediated apoptosis26,27,28. Our previous studies showed that Δ113p53 does not act on p53 in a dominant-negative manner, but rather interferes with p53 function by differentially modulating p53 target gene expression to protect cells from apoptosis26. Δ133p53 also represses cell replication senescence29 and promotes angiogenesis and tumor progression30. However, knowledge of its function in DNA DSB repair is lacking.
In this study, we demonstrate that Δ113p53/Δ133p53 is strongly accumulated at the later stage in response to DNA DSB signals, such as γ-irradiation, to promote all three DNA DSB repair pathways in both zebrafish and human cells. We also demonstrate that Δ113p53/Δ133p53 regulates DNA DSB repair by transcriptionally upregulating the expression of RAD51, LIG4 and RAD52, independent of full-length p53. Our findings provide an important clue to unravel the perplex of p53 in the DSB repair.
Results
Zebrafish Δ113p53 expression was strongly induced by γ-irradiation, but not UV irradiation and heat shock treatment
We showed previously that Δ113p53 expression is induced by γ-irradiation26. In the current study, we examined the expression of Δ113p53 in zebrafish embryos after UV irradiation and heat shock treatment. We found that although upregulation of full-length p53 expression reached a similar level upon different treatments, the expression of Δ113p53 was only induced by 16 gray of γ-irradiation and was not, or only weakly, induced by other treatments (Figure 1A and Supplementary information, Figure S1A). This induction appears to be a specific outcome of γ-irradiation treatment, because there was no, or only a low-level, induction of Δ113p53 expression even when embryos were exposed to harsher UV or higher temperature conditions that caused most embryos to die at 32 hours post treatment (hpt). In contrast, almost 100% of embryos treated with 16 gray of γ-irradiation survived at 32 hpt (Supplementary information, Figure S1B). Upon exposure to γ-irradiation, p53 levels peaked as early as 4 hours post irradiation (hpi), whereas Δ113p53 levels peaked later, at 24 hpi (Figure 1B). As the main difference in the damage induced by the different treatments was that only γ-irradiation led to genome-wide DNA DSBs, we speculated whether the high level of Δ113p53 induced by γ-irradiation might play a role in DNA DSB repair.
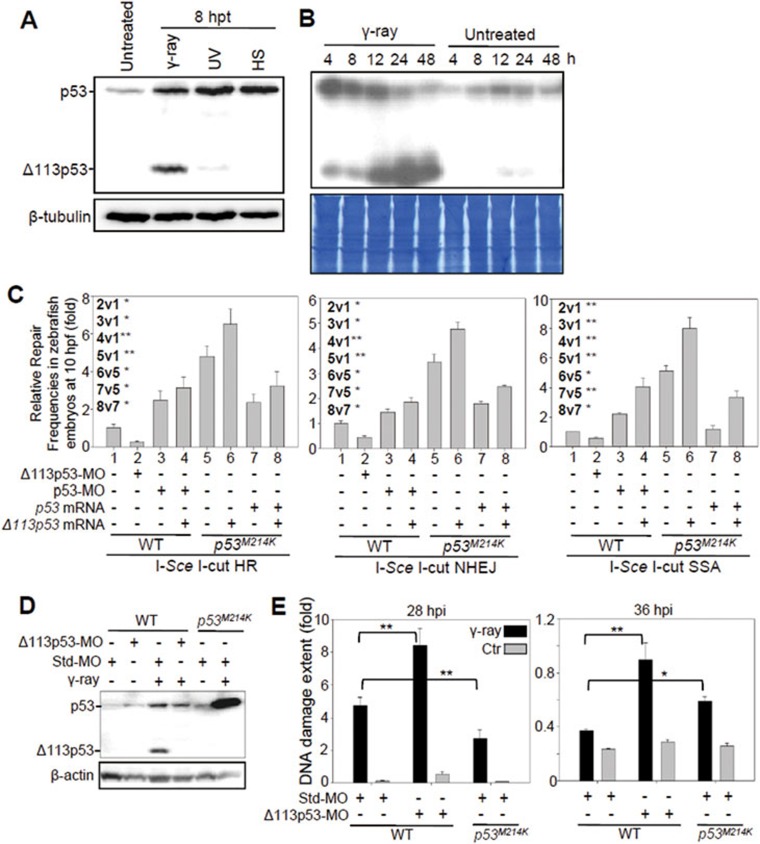
Figure 1.
Zebrafish Δ113p53 promotes DSB repair. (A) Western blot of zebrafish p53 and Δ113p53 from the untreated control (untreated) and embryos treated with γ-ray, UV irradiation (UV) or heat shock (HS) at 8 hpt using the A7-C10 monoclonal antibody against zebrafish p53. β-tubulin was used as the protein loading control. (B) Kinetics of p53 and Δ113p53 protein expression in zebrafish embryos treated with 16 gray of γ-ray irradiation or untreated. Total protein stained with Coomassie blue was used as the loading control. h: hours after treatments. (C) Effects of zebrafish p53 and Δ113p53 on HR, NHEJ and SSA repair frequencies. The average repair frequencies were measured using a qPCR analysis of the repair assay constructs (Supplementary information, Figure S2) from three repeat experiments at 10 hpf. Different lanes are numbered; v: versus, t-test between two lanes. (D) Western blot of p53 and Δ113p53 in different embryos as indicated. Proteins were extracted from non-irradiated and irradiated embryos at 8 hpi. (E) Assessment of DNA DSB with a comet assay in different embryos as indicated. Individual cells were dissociated at 28 and 36 hpi and used in the comet assay. 130-900 cells from each sample were randomly chosen to measure the extent of DNA damage (Supplementary information, Figure S6). All statistically significant differences between treatments were assessed with the independent samples t-test (*P < 0.05, **P < 0.01).
Zebrafish Δ113p53 promotes DNA DSB repair
To test our hypothesis, we used three Egfp-repairing-aided visual-plus-quantitative analysis reporter systems to measure HR, NHEJ and SSA repairs31 (Supplementary information, Figure S2). The corresponding plasmids were linearized with I-SceI, and then co-injected with p53 morpholino (p53-MO, which targets the ATG of full-length p53 mRNA to block its translation), Δ113p53 morpholino (Δ113p53-MO, which specifically targets the 5′-UTR of Δ113p53 mRNA)26 or a p53-MO-plus-Δ113p53 mRNA mix into zebrafish wild-type (WT) embryos. The linearized plasmid DNA was also co-injected into p53M214K mutant embryos (p53M214K carries an M214-to-K214 substitution in the DNA-binding domain32) with p53 mRNA, Δ113p53 mRNA or a p53-plus-Δ113p53 mRNA mix (Supplementary information, Figure S3). Protein analysis showed that injection of linearized plasmid alone activated the p53 pathway, which further induced Δ113p53 expression in WT embryos (Supplementary information, Figure S4). We confirmed DSB repair in each treatment at 8 hours post fertilization (hpf), by either EGFP fluorescence intensity measurement or quantitative real-time PCR (qPCR) analysis of the repaired Egfp DNA fragments. Our results showed that zebrafish p53, like human p53, inhibited all three DNA DSB repair pathways at 8 hpf (Figure 1C, lanes 3 vs 1 and 7 vs 5, and Supplementary information, Figure S5). Knockdown of Δ113p53 significantly enhanced the inhibitory effect of the endogenous p53 on DSB repair (Figure 1C, lanes 2 vs 1, and Supplementary information, Figure S5). In contrast, the overexpression of Δ113p53 promoted all three DSB repair pathways in p53 mutant embryos (Figure 1C, lanes 6 vs 5, and Supplementary information, Figure S5). To investigate whether p53M214K and Δ113p53M214K mutant proteins have a gain-of-function effect on DNA DSB repairs, we co-injected the linearized repair plasmids with either p53-MO or Δ113p53-MO into p53M214K mutant embryos (Supplementary information, Figure S6). The qPCR analysis of the repaired Egfp DNA fragments showed that knockdown of either p53M214K or Δ113p53M214K mutant protein had little effects on HR, NHEJ and SSA repairs, suggesting that both mutant proteins do not have a gain-of-function effect on DNA DSB repairs.
We next investigated the influence of Δ113p53 on DNA DSB repair of genomic DNA using a comet assay (single cell gel electrophoresis) by analyzing the genomic DNA damage induced by γ-irradiation in zebrafish embryos (Supplementary information, Figure S7). WT and p53M214K mutant embryos were injected with either the standard control morpholino (Std-MO, against human β-globin) or Δ113p53-MO. The injected WT and p53M214K mutant embryos were treated with 16 gray of γ-irradiation (Figure 1D). A TUNEL assay showed that apoptosis decreased to the basal level after 24 hpi (Supplementary information, Figure S8). We thus used 28-hpi and 36-hpi irradiated embryos to detect the levels of DNA DSB, minimizing the interference of apoptosis in the assay. Our results showed that the extent of DNA damage in WT embryos with Δ113p53 knockdown was ∼2-fold of that in the irradiated control embryos at either 28 hpi or 36 hpi (Figure 1E). Very interestingly, the extent of DNA damage dropped faster in WT embryos (from 4.76 at 28 hpi to 0.37 at 36 hpi, 12.86-fold) than in the p53M214K mutants (from 2.72 at 28 hpi to 0.59 at 36 hpi, ∼4.6-fold), which correlated well with the presence of Δ113p53 accumulation in WT and its absence in p53M214K embryos induced by γ-irradiation (Figure 1D and 1E). Notably, the extent of DNA damage in the irradiated WT embryos (4.76) was significantly higher than that in the irradiated p53M214K embryos (2.72) at 28 hpi. In contrast, at 36 hpi, the extent of DNA damage was significantly lower in the irradiated WT (0.37) than in the irradiated p53M214K embryos (0.59). One possible explanation for this observation is that full-length p53 is induced to a high level at the early stage (Figure 1B) in WT embryos after irradiation, which could guide the cells with severe DNA damage towards apoptosis while repressing DNA DSB repair in the surviving cells. On the other hand, due to the lack of bioactive p53, the DNA-damaged cells in the p53M214K mutant were still able to undergo the DNA DSB repair. Hence, we observed that the extent of DNA damage was higher in WT than that in p53M214K at 28 hpi. At 36 hpi, the expression of Δ113p53 in WT embryos accumulated to a high level, which in turn blocked apoptosis and promoted DNA DSB repair in the surviving cells. This resulted in a drastic drop in the extent of DNA damage in these WT cells. However, in the irradiated p53M214K embryos, although the DNA-damaged cells were able to undergo DNA DSB repair, the repair efficiency was low due to the absence of Δ113p53 expression (Figure 1D). Furthermore, the irradiated p53M214K embryos contained a large number of non-repairable cells with severe DNA damage, which escaped apoptosis in the absence of the bioactive p53. As a result, cells in p53M214K embryos exhibited significantly higher levels of DNA damage than those in WT embryos at 36 hpi. These results demonstrate the importance of the coordination of p53 and Δ113p53 functions at the organismal level to minimize DNA damage upon DNA DSB stress.
Generation of zebrafish Δ113p53M/M mutant
To study the biological significance of Δ113p53 in DNA DSB repair, we generated a zebrafish Δ113p53Mutation/mutation(M/M) knockout mutant. As the coding sequence of Δ113p53 is completely overlapped with the full-length p53, we chose to knock out Δ113p53 by targeting its promoter. One of our previous studies showed that the Δ113p53 promoter is located in the fourth intron of the full-length p53 gene and contains three putative p53 response elements (REs)26 (Figure 2A). A subsequent study showed that the third p53 RE is required for Δ113p53 expression (unpublished data). Therefore, we generated a Δ113p53 mutant by targeting the third p53 RE in its promoter with the transcription activator-like effector nuclease (TALEN) technique. One mutant was obtained with an 11-bp deletion, which includes an 8-bp sequence within the third p53 RE (Figure 2A). Western blot showed that the induction of Δ113p53 expression was almost completely blocked, whereas the activation of full-length p53 was unaffected in the Δ113p53M/M mutants in response to γ-irradiation (Figure 2B).
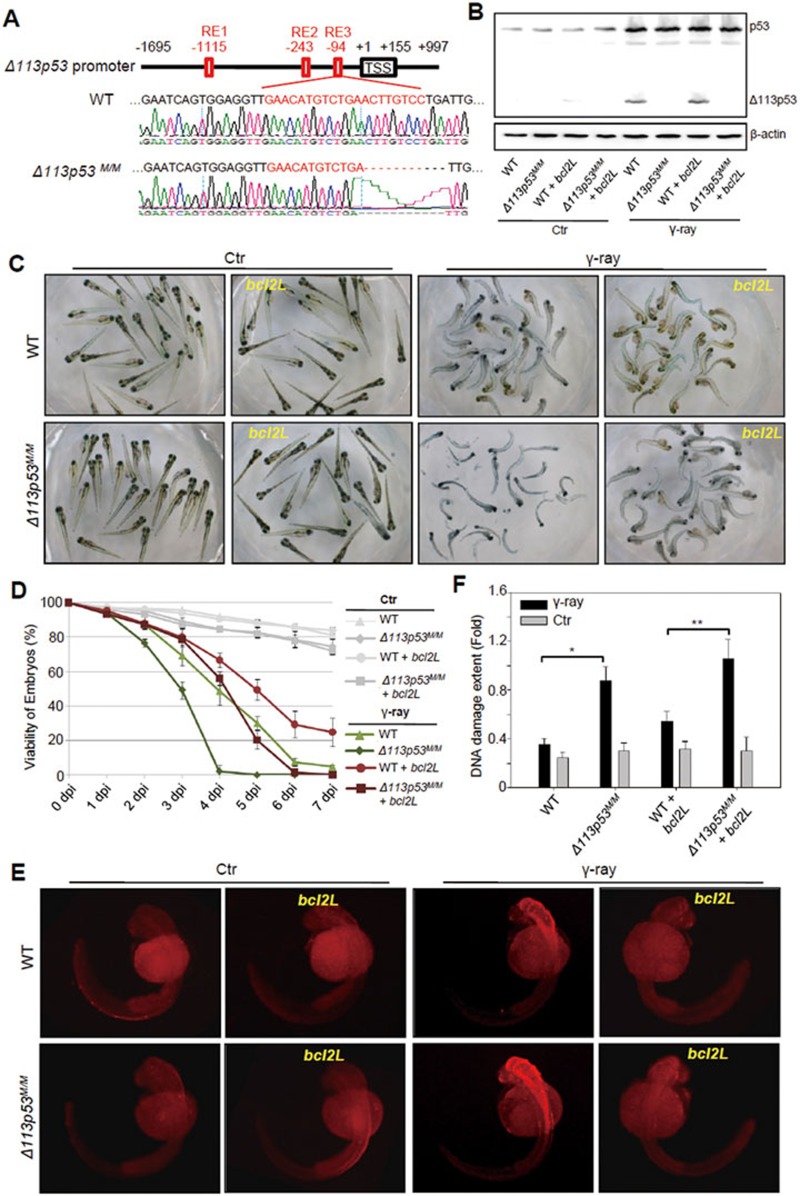
Figure 2.
Zebrafish Δ113p53M/M mutant is more sensitive to γ-irradiation. (A) Diagram showing the Δ113p53 promoter and an 11-bp deletion in the promoter of Δ113p53M/M mutant. TSS: transcription start site of Δ113p53. RE: p53 response element. The numbers indicate the positions in the Δ113p53 promoter. Out of the deleted 11 bp, 8 bp are within the RE3. (B) Western blot analysis of p53 activation and Δ113p53 induction in WT and Δ113p53M/M embryos injected or uninjected with bcl2L mRNA, followed by 16 gray of γ-ray irradiation. (C, D) WT and Δ113p53M/M embryos with or without injection of bcl2L mRNA at 1 dpf were treated with γ-irradiation. The pictures were taken at 5 dpi (C). The average viabilities of embryos with different treatments were taken from three repeats from 1 to 7 dpi as indicated (D). (E) A TUNEL assay was used to examine apoptotic cells in embryos with different treatments at 8 hpi as indicated. Approximately 20 embryos from each treatment were sampled at each time point. (F) Assessment of DNA DSB by a comet assay in embryos with different treatments as indicated at 2 dpi.
Zebrafish Δ113p53M/M mutant is more sensitive to γ-irradiation due to loss of functions in anti-apoptosis and promoting DNA DSB repair
The Δ113p53M/M mutant fish grows to adulthood normally in standard growth conditions. To test whether three DNA DSB repair pathways are affected in the mutant, the I-SceI-linearized HR, NHEJ or SSA plasmid was injected into WT and Δ113p53M/M embryos, and was co-injected with Δ113p53 mRNA into Δ113p53M/M embryos. Results showed that the efficiency of the three DNA DSB repair pathways was significantly decreased in Δ113p53M/M embryos (Supplementary information, Figure S9), which is similar to that observed in the Δ113p53-MO-injected embryos (Figure 1C). The efficiency of all three repair pathways was restored by Δ113p53 mRNA co-injection (Supplementary information, Figure S9), demonstrating that the decrease of DNA DSB repair efficiency in Δ113p53M/M embryos was due to the absence of Δ113p53.
We then treated WT and Δ113p53M/M embryos with γ-irradiation. Assessment of embryo viability revealed that the Δ113p53M/M embryos (all of which died at 5 dpi) were much more susceptible to γ-irradiation than WT embryos (∼30% of which was viable at 5 dpi; Figure 2C and 2D). Two main functions of Δ113p53 have been demonstrated, i.e., to antagonize the pro-apoptotic function of p53 and to promote DNA DSB repair. To determine the contribution of Δ113p53's DSB repair function to the high mortality rate in the mutant embryos in response to γ-irradiation, we blocked cell apoptosis by injecting bcl2L (anti-apoptotic protein)26 mRNA into WT and Δ113p53M/M embryos. Western blot showed that bcl2L mRNA injection did not influence the induction of Δ113p53 (Figure 2B). Similar to the results in embryos injected with Δ113p53-MO26, more apoptotic cells were observed in Δ113p53M/M embryos than in WT embryos upon γ-irradiation (Figure 2E). However, irradiation-induced apoptosis was almost completely inhibited by bcl2L mRNA injection in both WT and Δ113p53M/M embryos (Figure 2E). The viability of irradiated mutant embryos injected with bcl2L mRNA (∼20% at 5 dpi) was significantly lower than that of WT embryos (∼50%) with the same treatment, and even lower than that of irradiated WT embryos (∼30%) without bcl2L mRNA injection (albeit with abundant apoptotic cells; Figure 2C and 2D). Comet assay results showed that bcl2L mRNA injection slightly increased the extent of DNA damage in both irradiated WT and Δ113p53M/M embryos at a similar scale. This increase occurred possibly because Bcl2L overexpression prevented cells with severe DNA damage from apoptosis in both irradiated WT and Δ113p53M/M embryos (Figure 2F). Conversely, Δ113p53 mRNA injection restored the viability of irradiated mutant embryos to the WT level upon γ-irradiation (Supplementary information, Figure S10). Taken together, these results suggest that loss of both functions of Δ113p53 (i.e., anti-apoptosis and promotion of DNA DSB repair) renders Δ113p53M/M embryos more sensitive to γ-irradiation.
The promotion of DNA DSB repair is conserved in human Δ133p53
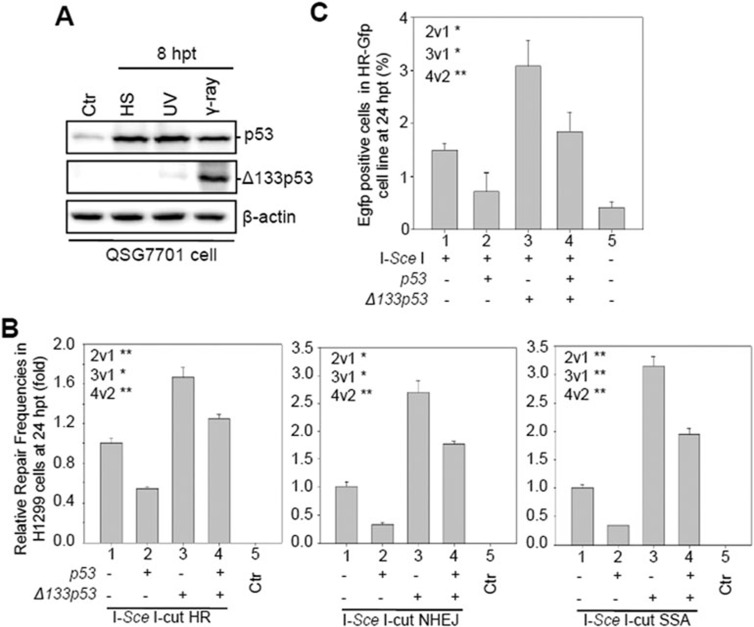
We treated human QSG-7701 cells (a non-cancerous liver epithelial cell line containing WT p53) with γ-irradiation, UV irradiation and heat shock, and analyzed the function of the human ortholog, Δ133p53, in DNA DSB repair. Both Δ133p53 transcript and protein were strongly induced by γ-irradiation only (Figure 3A; Supplementary information, Figure S11A-S11C). We then transfected the H1299 cells (which lack the endogenous p53 gene) with each of the three visual-plus-quantitative assay constructs, along with Δ133p53, p53 or p53-plus-Δ133p53 mRNA (Supplementary information, Figure S12). Both qPCR analysis of the repaired Egfp DNA fragments and fluorescence-activated cell sorting (FACS) analysis of EGFP-positive cells revealed that, apart from neutralizing the DSB repair inhibitory effect of full-length p53, Δ133p53 also almost doubled the efficiency of all the three DNA DSB repair pathways in a p53-independent manner, compared to their corresponding controls (Figure 3B and Supplementary information, Figure S13). To study the function of endogenous Δ133p53 in DNA DSB repair, we co-transfected each of the three repair assay constructs with either a non-specific siRNA control (siNS) or two Δ133p53 siRNAs, siRNA1 (Δ133p53i-1) or siRNA2 (Δ133p53i-2; both targeting 5′-UTR of Δ133p53 located in the intron 4 of full-length p53)29 into QSG-7701 cells (Supplementary information, Figure S14A). The qPCR analysis showed that the knockdown of Δ133p53 significantly decreased the efficiencies of the three DNA DSB repair pathways (Supplementary information, Figure S14B). The positive role of Δ133p53 in DNA DSB repair was also observed in U2OS cells (Figure 3C), which harbor WT p53 and stably express HR-GFP33.
Figure 3.
Human Δ133p53 promotes HR, NHEJ and SSA repair pathways. (A) Western blot of human p53 and Δ133p53 from human QSG7701 cells treated as indicated using a monoclonal antibody DO-1 and a polyclonal antibody CM1, respectively. β-actin was used as the protein loading control. (B) Effect of human Δ133p53 on HR, NHEJ and SSA repair frequencies. Relative DNA DSB repair frequencies for HR, NHEJ and SSA were measured by qPCR at 24 hpt. (C) Effect of Δ133p53 on HR repair frequency in the U2OS (HR-GFP) cell line. CMV-I-SceI plasmid was transfected or co-transfected with CMV-p53, CMV-Δ133p53 or CMV-p53-plus-CMV-Δ133p53 plasmids into HR-GFP cells as indicated. The transfected cells were harvested at 24 hpt for the FACS analysis. The average frequency was calculated from three repeat experiments. Different lanes are numbered; v: versus, t-test between two lanes.
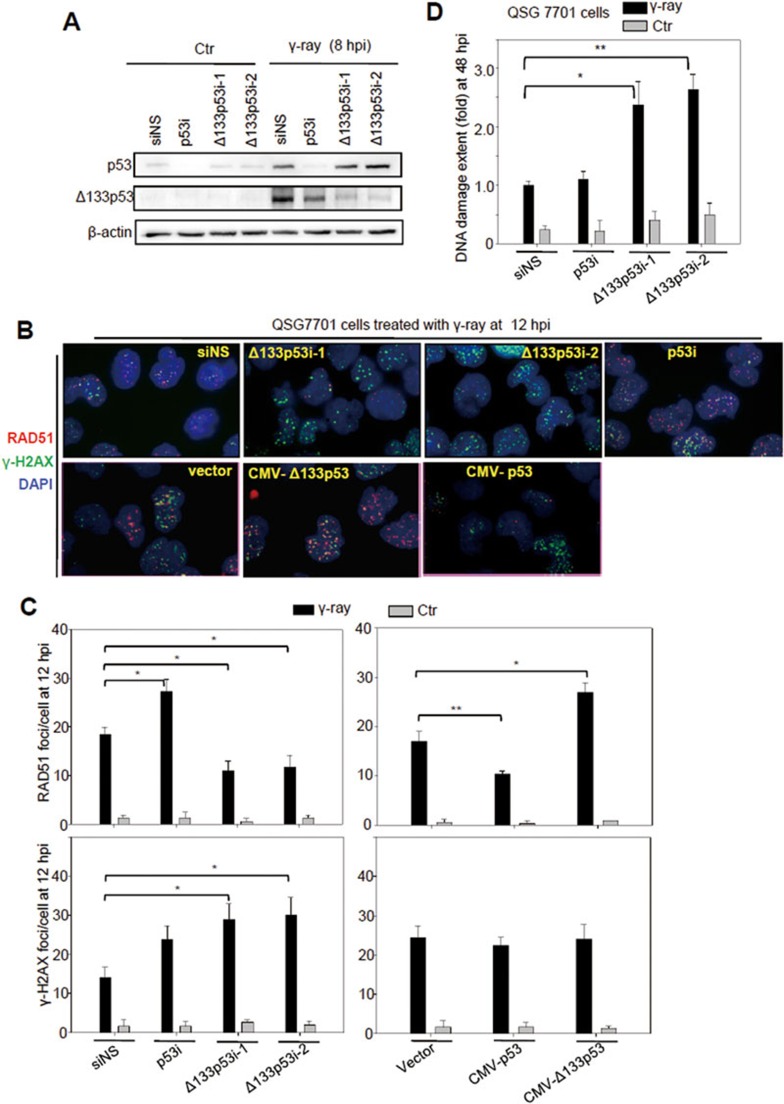
It has been reported that human p53 inhibits RAD51 foci formation in response to DNA damage34,35. We used QSG-7701 cells to study the function of Δ133p53 in the formation of the DNA DSB repair foci of phosphorylated H2AX (γH2AX; which is one of the early DNA DSB repair markers) and RAD51 upon -irradiation. QSG-7701 cells were transfected with either a non-specific siRNA control (siNS), a p53 siRNA (p53i; targeting exon 4 of full-length p53)27, or two Δ133p53 siRNAs, Δ133p53i-1 and Δ133p53i-2, and treated with 10 gray of -irradiation (Figure 4A). Our results confirmed that p53 has a negative influence on RAD51 foci formation (Figure 4B and 4C; Supplementary information, Figure S15). In contrast, overexpression of Δ133p53 significantly increased RAD51 foci formation at 12 hpi upon γ-irradiation, whereas knockdown of endogenous Δ133p53 significantly decreased foci formation under the same conditions (Figure 4B, 4C and Supplementary information, Figure S15). Our results also showed that the formation of γH2AX foci was not significantly affected by Δ133p53 or p53 overexpression, suggesting that Δ133p53 and p53 may not have a significant effect on the early steps of DNA DSB repair (Figure 4B, 4C and Supplementary information, Figure S15).
Figure 4.
Δ133p53 promotes RAD51 foci formation and DNA DSB repair in QSG-7701 cells upon ionizing irradiation. (A) Western blot analysis of p53 activation and Δ133p53 induction in QSG-7701 cells transfected with non-specific siRNA (siNS), p53 interference RNA (p53i) or two Δ133p53 interference RNAs, Δ133p53i-1 and Δ133p53i-2, followed by 10 gray of γ-ray irradiation. (B) Co-immunostaining of RAD51 (in red) and γH2AX (in green) in QSG-7701 cells with different treatments as indicated. The specific monoclonal antibodies were used to determine the RAD51 and H2AX foci formation at 12 hpi as indicated. DAPI was used to stain the nuclear DNA (blue). (C) Statistical analysis of the average number of RAD51 and γH2AX foci per cell in different samples, as shown in B. At least 100 cells from each sample were randomly chosen for counting RAD51 and γH2AX foci. (D) Assessment of DNA DSB with a comet assay at 48 hpi in QSG-7701 cells with different treatments, as indicated. About 100 cells from each sample were randomly chosen to measure the extent of DNA damage. A statistical analysis was performed based on the data from three repeat experiments.
FACS analysis revealed, as expected, that the number of apoptotic cells (sub-G0 summit) was decreased by p53 knockdown from 8 to 24 hpi and was increased by Δ133p53 knockdown from 4 to 24 hpi (Supplementary information, Figure S16)23,26. However, apoptosis decreased to the basal level by 36 hpi in all cases (Supplementary information, Figure S16). Therefore, we performed the comet assay at 48 hpi to test whether the decrease in the number of RAD51 foci upon Δ133p53 knockdown was accompanied by an increase in DNA damage. Comet assay results showed ∼1.5-fold greater damage in cells transfected with the Δ133p53 siRNAs than in the irradiated control cells (Figure 4D). These results demonstrate that Δ133p53 plays a positive role in genomic DNA DSB repair upon γ-irradiation. However, the extent of DNA damage in irradiated control cells (1.0) was only slightly lower than that in irradiated p53-knockdown cells (1.1) at 48 hpi (Figure 4D), which differed from the comet assay results obtained from irradiated zebrafish WT and p53M214K embryos at 36 hpi (Figure 1E). One likely explanation is that in embryos, apoptotic cells are cleared away by other cells in vivo, while in cell culture conditions, there is no such system to remove the apoptotic cells, which may interfere with the comet assay carried out in cultured cells.
Knockdown of Δ133p53 in human cells inhibits cell proliferation through arresting cell cycle at the G2 phase and promoting cell senescence upon γ-irradiation
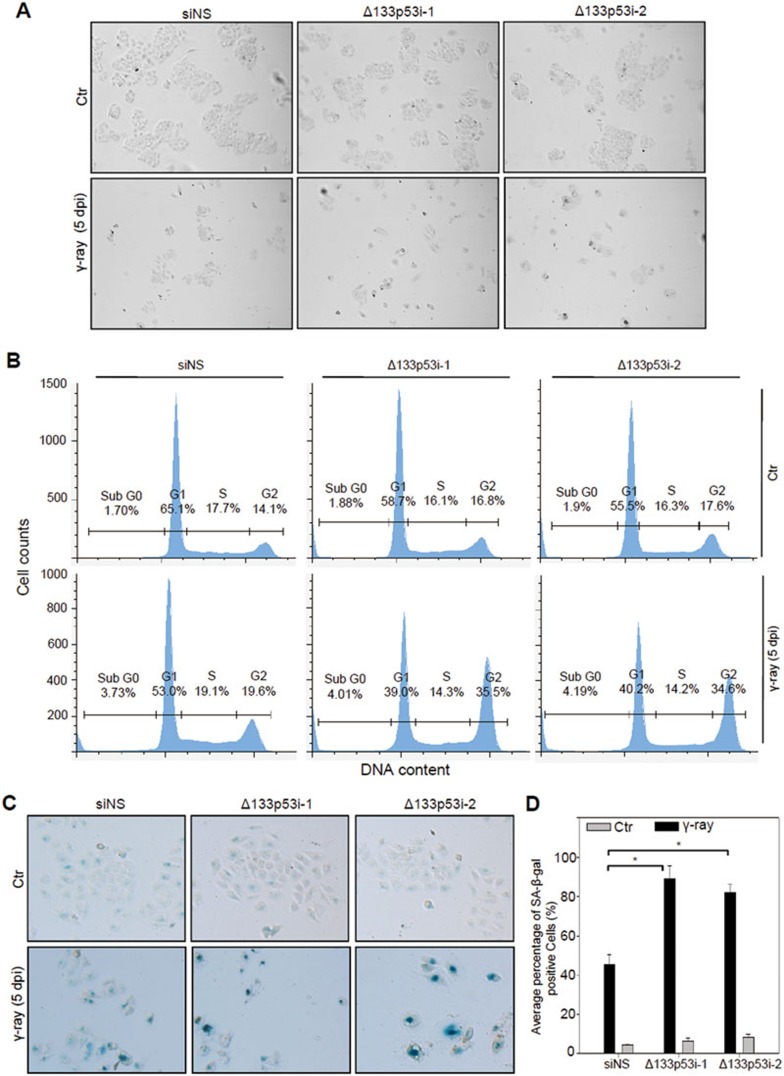
To study the consequence of increased DNA damage at the cellular level, we transfected QSG-7701 cells with siNS, Δ133p53i-1, or 133p53i-2 and treated them with 10 gray of γ-irradiation. As described above, apoptosis decreased to the basal level at 36 hpi (Supplementary information, Figure S14). We washed away apoptotic cells at 2 dpi and replaced with a new culture medium to allow the remaining cells to grow under normal conditions. At 5 dpi, total cell number and colony size (which showed flattened cell morphology) were observably decreased by the treatment of γ-irradiation, compared to those of unirradiated controls (Figure 5A). Interestingly, after γ-irradiation fewer cell numbers and a smaller colony size were observed in cells transfected with Δ133p53 siRNA compared with the siNS-transfected control (Figure 5A), which correlates well with the extent of DNA damage observed (Figure 4D). FACS analysis of cells at 5 dpi showed that the proportion of cells at the G2 phase increased slightly, from 14.1% to 19.6%, in siNS-transfected cells, but almost doubled from 16.8% to 35.5% in Δ133p53i-1- and from 17.6% to 34.6% in Δ133p53i-2-transfected cells (Figure 5B). In contrast, there was little difference in the proportion of cells at the S phase between the irradiated cells and untreated controls (Figure 5B). These results suggest that a high level of DNA damage results in cell cycle arrest at the G2 phase.
Figure 5.
Knockdown of Δ133p53 arrests cell growth at the G2 phase and promotes cell senescence upon γ-irradiation. (A) Cell colony formation of irradiated cells. QSG-7701 cells transfected with non-specific siNS, Δ133p53i-1, or Δ133p53i-2 siRNA were treated with 10 gray of γ-ray irradiation. The pictures were taken at 5 dpi. (B) FACS analysis of the percentage of cells at different cell cycle phases based on Propidium Iodide (PI) staining. QSG-7701 cells transfected with siNS, Δ133p53i-1, or Δ133p53i-2 siRNA at 5 dpi as indicated. (C) SA-β-gal staining to analyze the senescence status in the QSG-7701 cells with different treatments as described in B. (D) Statistical analysis of the senescent cells in different samples shown in C.
Next, cell senescence analysis was performed with senescence-associated β-galactosidase (SA-β-gal) staining. The occurrence of positive cells (about 89% in Δ133p53i-1- and 80% in Δ133p53i-2-transfected cells) at 5 dpi was significantly increased by Δ133p53 knockdown upon γ-irradiation, compared to that in the irradiated siNS control (about 40%; Figure 5C, 5D and Supplementary information, Figure S17). Taken together, loss of function of Δ133p53 increased DNA DSBs upon γ-irradiation, which in turn inhibited cell proliferation by arresting cell cycle at the G2 phase, finally resulting in cell senescence.
Δ133p53 does not form a complex with either RAD51 or RPA
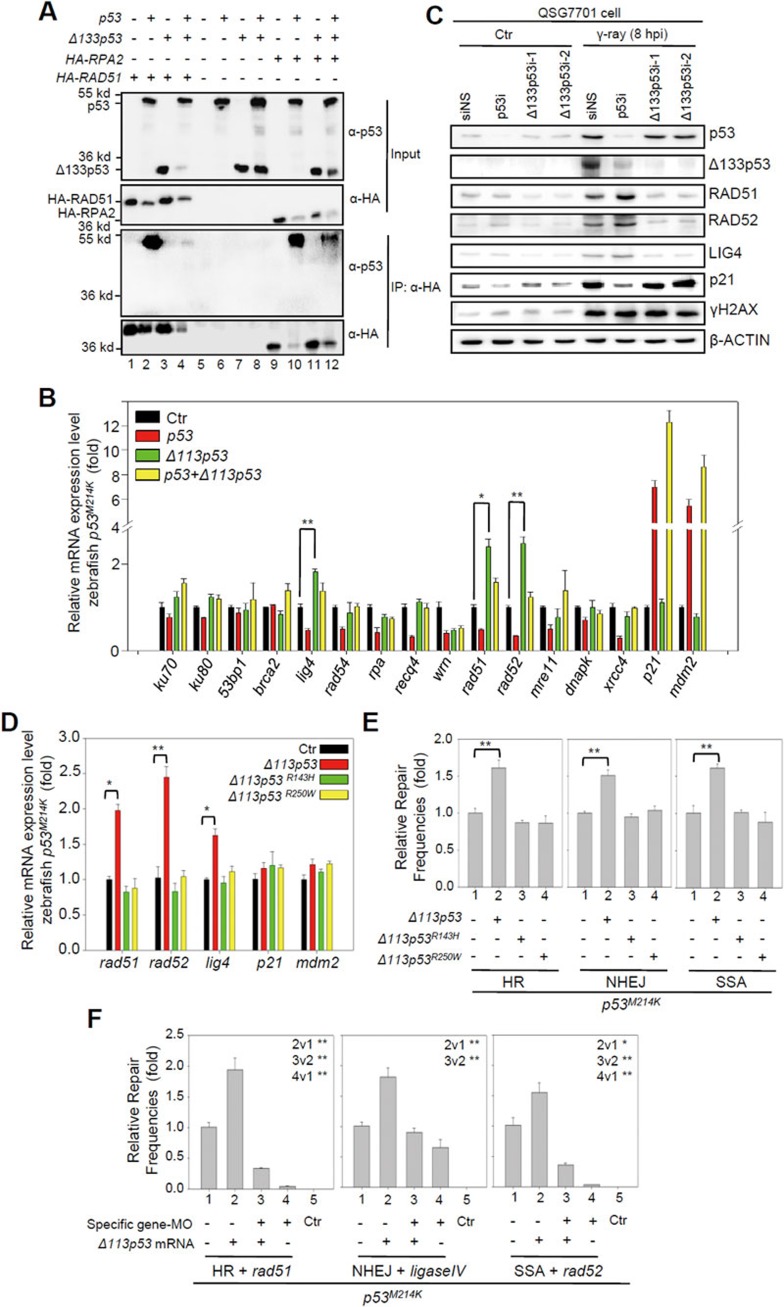
It was proposed that the p53 protein directly interacts with either RAD5118 or RPA19 to inhibit DNA DSB repair complex formation. Previous studies have shown that the DNA-binding core domain (94-312) of p53 is required for p53-RAD51 interactions, and its N-terminal domain (37-57) is required for p53-RPA interactions36,37, which suggests that Δ133p53 may not be able to form a complex with these two proteins. We performed a co-immunoprecipitation (co-IP) experiment to test this hypothesis by co-transfecting HA-RAD51 or HA-RPA2 with p53, Δ133p53 or both, into H1299 cells. The results showed that full-length p53 (Figure 6A, lanes 2 and 10), but not Δ133p53 (Figure 6A, lanes 3 and 11) formed a complex with either HA-RAD51 or HA-RPA2. It was observed that the protein level of RAD51, RPA2 or Δ133p53 was dramatically decreased when it was co-expressed with full-length p53 in the experiments, but the reason is currently not known.
Figure 6.
Δ113p53/Δ133p53 promotes DNA DSB repair by upregulating the expression of Rad51, Rad52 and Lig4. (A) Co-IP analysis of the interaction between p53 or Δ133p53 with HA-RAD51 or HA-RPA2 in H1299 cells. An anti-HA antibody was used in an immunoprecipitation. Proteins from co-IP were detected with a p53 CM1 antibody (third panel) and the HA antibody (fourth panel). The 10% of input from each sample was used as a control: top panel p53 (CM1); second panel: HA. (B) Relative mRNA expression of the listed genes in zebrafish p53M214K mutant embryos overexpressing Δ113p53, p53 or both p53 and Δ113p53 measured by qRT-PCR at 8 hpf. Gene expression was normalized against 18S rRNA and expressed as the fold change compared to the injection control. (C) Western blot analysis of proteins in human QSG7701 cells with different treatments as indicated. (D) Relative mRNA expression of the listed genes in zebrafish p53M214K mutant embryos overexpressing Δ113p53, Δ113p53R143H or Δ113p53R250W measured by qRT-PCR. (E) Effects of Δ113p53, Δ113p53R143H and Δ113p53R250W on HR, NHEJ and SSA repair frequencies. The average repair frequencies were measured by qPCR analysis of different repaired assay constructs from three repeat experiments at 10 hpf. (F) The activity of rad51, lig4 and rad52 was required for zebrafish Δ113p53-meadited HR, NHEJ and SSA repairs. The rad51-MO, lig4-MO or rad52-MO was used to knock down its corresponding gene expression in the HR, NHEJ or SSA analysis. The average repair frequencies were measured with a qPCR analysis of the repaired assay constructs from three repeat experiments at 10 hpf. Different lanes are numbered; v: versus, t-test between two lanes.
Δ113p53/Δ133p53 upregulates the expression of key DNA DSB repair genes
We investigated the molecular mechanisms by which Δ113p53/Δ133p53 promotes DNA DSB repair independent of p53. We co-injected a linearized plasmid (to mimic DNA DSB stress) with either p53, Δ113p53 or p53-plus-Δ113p53 mRNA into p53M214K mutant embryos and analyzed the expression of DSB- and p53-response genes by quantitative reverse transcription PCR (qRT-PCR). Unlike two p53-responsive genes, p21 (a cell cycle inhibitor) and mdm2 (an E3 ligase), the expression of 8 out of 14 DNA DSB repair genes (including lig4, rad54, recq4, wrn, rad51, rad52, mre11 and xrcc4) was significantly downregulated by p53. Δ113p53 suppressed the inhibitory effect of p53 on the expression of all of these genes except for wrn (Figure 6B), which may explain Δ133p53's ability to neutralize the inhibitory effect of full-length p53 on DSB repair.
Strikingly, Δ113p53 alone promoted the expression of rad51 (required for HR repair38), lig4 (required for NHEJ repair39), and rad52 (required for SSA repair40) (Figure 6B). We examined the transcriptional activity of human Δ133p53 by transfecting QSG7701 cells with siNS, p53i, 133p53i-1 or Δ133p53i-2 and then treating them with γ-irradiation. The results from both qRT-PCR and protein analyses showed that the expression levels of RAD51, LIG4 and RAD52 were all upregulated at 12 hpi (Figure 6C; Supplementary information, Figure S18). The upregulation of these genes after γ-irradiation was attenuated by knockdown of Δ133p53 and enhanced by knockdown of p53 (Figure 6C; Supplementary information, Figure S18).
We generated two Δ113p53 mutants to test whether the function of Δ113p53 in facilitating DNA DSB repair is dependent on its transcriptional activity, Δ113p53R143H and Δ113p53R250W (the number denotes the mutation's position in the full-length zebrafish p53). Δ113p53R143H and Δ113p53R250W correspond to the R175H and R282W mutations in full-length human p53, respectively, which are known to lose their DNA binding capacity41. qRT-PCR results showed that, unlike WT Δ113p53, the two Δ113p53 mutants did not upregulate the expression of rad51, lig4 and rad52 (Figure 6D). Further experiments demonstrated that the two mutants also failed to promote HR, NHEJ and SSA repairs (Figure 6E).
Next, we used zebrafish p53M214K mutant embryos to investigate the roles of rad51, lig4 and rad52 in the DNA DSB repair pathways, in the context of Δ113p53. Specific MOs were used to knock down rad51, lig4 or rad52 under different conditions in embryos overexpressing Δ113p53 and an HR, NHEJ or SSA reporter construct. Our results revealed that knockdown of rad51, lig4 and rad52 significantly attenuated the effect of Δ113p53 on promoting DNA DSB repair in the corresponding pathway (Figure 6F). All of these data suggested that Δ113p53's transcriptional activity is important for DNA DSB repair.
Δ113p53 binds to a novel p53 RE in the promoters of rad51, lig4 and rad52
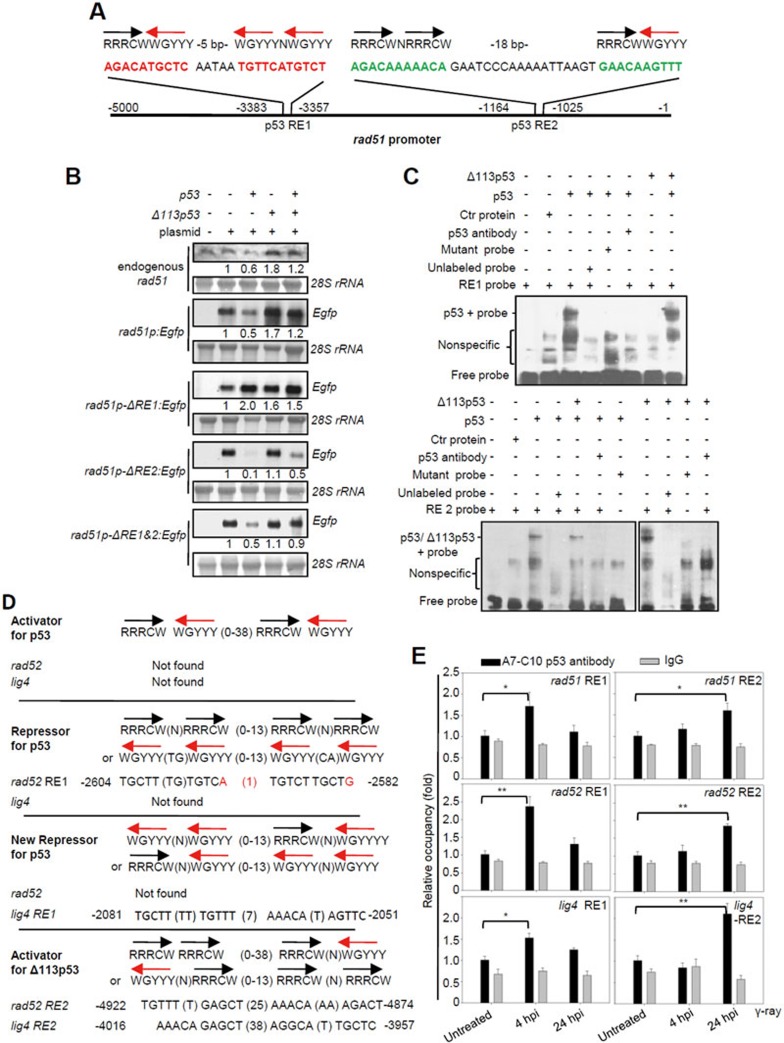
A previous study showed that human p53 repressed RAD51 transcription by directly binding to its promoter20. We tested whether Δ113p53 also has a direct role in rad51 transcription by cloning the zebrafish rad51 promoter of 5 kb upstream of the rad51 transcriptional start site and generating the rad51p:Egfp reporter construct (Figure 7A). This 5-kb fragment recapitulates the pattern of endogenous rad51 expression in response to p53 and Δ113p53 expression (Figure 7B). Two putative p53 REs were found within the promoter region of rad51 at positions −3 384 and −1 165 nucleotide (Figure 7A). Interestingly, the arrangements of four pentamers found in both of the REs are novel compared to those reported previously (Figure 7A). We found that the deletion of RE1 switched the effect of p53 from repressing to promoting Egfp expression. The deletion of RE2 abrogated the effect of Δ113p53 but enhanced the suppressing effect of p53 (Figure 7B). A gel retardation experiment revealed that both p53 and Δ113p53 could bind to RE2, whereas only p53 could bind to RE1 (Figure 7C). These results suggest that p53 first binds to RE1 to suppress rad51 expression. In the absence of RE1, p53 binds to RE2 to promote rad51 expression, and RE2 serves as the sole site for Δ113p53 binding to promote rad51 expression.
Figure 7.
Δ113p53 upregulates the expression of rad51, rad52 and lig4 by directly binding to a new type of p53 RE in their promoter regions. (A) The rad51 promoter. The black and red arrows correspond to the orientations of the quarter sites. R = A or G, W = A or T, Y = C or T. The positions of two p53 REs in the rad51 promoter are indicated. (B) Northern blot analysis of the transcription levels of endogenous rad51 and Egfp in p53M214K mutant embryos injected with rad51p:Egfp, rad51p-ΔRE1:Egfp (with a 26-bp deletion of RE1), rad51p-ΔRE2:Egfp (with a 39-bp deletion of RE2) and rad51p-ΔRE1+2:Egfp (with double deletions in RE1 and RE2) plasmids, or co-injected with these plasmids and p53, Δ113p53 or p53-plus-Δ113p53 mRNAs, as indicated. 28S rRNA was used as the loading control. The numbers between the panels are the relative gene expression levels normalized against 28S rRNA in each experiment. (C) EMSA was performed to detect p53 and Δ113p53 interactions with RE1 and RE2 in the rad51 promoter. The 26-bp DNA fragments of RE1 and an RE1 mutant with 6 bp mutated (AGAAATACAC AATAA TTTTCATTTAT; mutations are underlined), and 39-bp DNA fragments of RE2 and an RE2 mutant with 6 bp mutated (ATATAAAAATA GAATCCCAAAAATTAAGT GAAAAATTAT; mutations are underlined) of the rad51 promoter were labeled with biotin to form probes. Nuclear proteins were extracted from zebrafish p53M214K mutant embryos injected with different mRNAs as indicated. Labeled probes were incubated with different protein extracts, with unlabeled probes and zebrafish A7-C10 antibody, as indicated. (D) p53 and Δ113p53 REs in rad52 and lig4 promoters compared to other p53 REs. Mismatch nucleotides are labeled red. The positions of p53 REs in the respective promoters are indicated. (E) ChIP of RE1 and RE2 in rad51, rad52 and lig4 promoters in the irradiated embryos at 4 and 24 hpi. WT embryos were treated with γ-irradiation and sampled at 4 and 24 hpi, respectively. The A7-C10 p53 antibody was used to co-immunoprecipitate the protein-DNA complex, while IgG was used as a non-specific binding control. Specific primer pairs were designed to amplify the corresponding REs. DNA was normalized with a pair of negative control primers for β-actin exon. The results are presented as the relative occupancies of different REs. Statistics were obtained from three repeat experiments.
Further analysis showed that the p53-repressing RE (RE1) and Δ113p53-activating RE (RE2) were also present in zebrafish rad52 and lig4 promoters (Figure 7D) and in human RAD51, LIG4 and RAD52 promoters (Supplementary information, Figure S19). A chromatin immunoprecipitation (ChIP) assay was performed to study whether p53 and Δ113p53 bind to their respective REs in the promoters of three DNA DSB repair genes in vivo upon γ-irradiation. As shown in Figure 1B, expression of full-length p53 reached its peak level at 4 hpi, while Δ113p53 expression peaked at 24 hpi. Based on this, we used untreated embryos as the controls and sampled irradiated embryos at 4 and 24 hpi. We used the A7-C10 zebrafish p53 monoclonal antibody, recognizing both p53 and Δ113p53, to perform ChIP experiment. First, we validated our ChIP products by analyzing the occupancy of p53 on the two known p53 REs in the promoter of Δ113p53 by qPCR. The enrichment of both p53 RE1 and RE3 of the Δ113p53 promoter in the ChIP products was nicely correlated with the dynamic expression levels of p53 at 4 and 24 hpi (Supplementary information, Figure S18A). Next, we examined the occupancy of p53 and Δ113p53 in the promoters of rad51, rad52 and lig4. The qPCR analysis showed that RE1 sequences (p53-repressing RE) of rad51, rad52 and lig4 were all enriched in the ChIP products from the 4-hpi samples (Figure 7E). As the expression level of p53 peaked at 4 hpi (Figure 1B), this result suggests that occupancy of RE1 in these promoters by p53 at this stage locks the expression of these genes at a repressive status. In contrast, RE2 sequences (Δ113p53-activating RE) of rad51, rad52 and lig4 were all enriched in the ChIP products from the 24-hpi samples (Figure 7E). As the level of Δ113p53 greatly exceeds that of p53 at 24 hpi (Figure 1B), these results demonstrate that the promoters of the three genes are switched from a status of repression by p53 at RE1 to a status of activation by Δ113p53 at RE2 in vivo. This occurs as a consequence of the dynamic change of expression levels of p53 and Δ113p53, from 4 to 24 hpi.
To analyze whether the binding of Δ113p53 to RE2 of these three DNA DSB repair gene promoters is independent of full-length p53, we overexpressed HA-p53 and HA-Δ113p53 in p53−/− mutants. An HA monoclonal antibody was used to perform the ChIP assay. The assay demonstrated that RE1 was enriched in the ChIP products from the sample overexpressing HA-p53, whereas the sample overexpressing HA-Δ113p53 showed enrichment at RE2 in the promoters of zebrafish lig4, rad52 and rad51 (Supplementary information, Figure S20), further confirming the ChIP assay results performed with irradiated zebrafish embryos. These results demonstrate that Δ113p53 upregulates the expression of rad51, lig4 and rad52 by binding to a novel type of p53 REs in their promoters.
Discussion
Up to 13 human p53 isoforms have been identified, and these isoforms are generated through alternative initiation of translation, use of an internal promoter or alternative splicing42. p53 isoforms can modulate p53 functions either synergistically or antagonistically, depending on the isoform's structure and the target genes affected42. However, how these isoforms affect DNA damage repair is rarely studied. Many studies have demonstrated that full-length p53 inhibits DNA DSB repair12,13,14. A recent study using human cells has shown that, in response to γ-irradiation treatment, p53 pulses induce apoptosis at the early stage and postpone DNA DSB repair to the later stage22. Here, we found that the p53 isoform Δ113p53/Δ133p53 is strongly induced by γ-irradiation, but not by UV irradiation and heat shock treatment. Interestingly, we observed that, upon γ-irradiation, the levels of full-length p53 and Δ113p53p53 proteins in the treated zebrafish embryos were differentially expressed. Full-length p53 protein level peaked early, at 4 hpi, whereas Δ113p53p53 protein level peaked later, at 24 hpi. We showed previously that Δ113p53/Δ133p53 is a p53 target gene and inhibits p53-mediated apoptosis by modulating the expression of p53 target genes26. All of our findings imply that Δ113p53/Δ133p53 may coordinate with full-length p53 to regulate cell death and DNA DSB repair in response to DNA DSB stress. Through Egfp-repairing-aided visual-plus-quantitative analysis reporter systems, comet assay and repair foci analysis, we demonstrated that Δ113p53/Δ133p53 promotes all three DNA DSB repair pathways in both zebrafish and human cells in a p53-independent manner. Further experiments with γ-irradiated zebrafish embryos showed that the proportion of apoptotic cells peaked around 8 hpi and dropped to the basal level at 24 hpi, which correlated well with the level of full-length p53 protein. In contrast, the extent of DNA damage decreased rapidly after 28 hpi, corresponding to the level of Δ113p53 protein. We revealed how changes in the levels of p53 and Δ113p53 proteins regulate cell death and DNA DSB repair in response to DNA damage. To minimize DNA DSBs as the first defense at the early stage of DNA damage response, full-length p53 is induced to a high level to guide cells with severe DNA damage to undergo apoptosis. The subsequent expression of Δ113p53, as the second wave of defense, inhibits apoptosis in the remaining cells with repairable DNA damage and, at the same time, promotes DNA DSB repair. Our findings demonstrate that Δ113p53/Δ133p53 is a pro-survival factor and may also imply possible roles of the other p53 isoforms in different DNA damage repair pathways.
The importance of Δ113p53/Δ133p53 for cell survival and its significance to the survival of a whole organism is further demonstrated in the Δ113p53M/M mutant. Although the Δ113p53M/M mutant zebrafish grows normally in standard growth conditions, it is sensitive to γ-irradiation. No mutant embryos were able to survive longer than 5 days after irradiation, while irradiated WT embryos exhibited a survival rate of about 30%. Sensitization to γ-irradiation is due to an increase in both apoptotic activity and the extent of DNA damage in the Δ113p53M/M mutant embryos upon irradiation. The fact that the mortality of irradiated Δ113p53M/M mutant embryos was much higher than that of irradiated WT embryos, even when apoptosis was inhibited by bcl2 mRNA injection, strongly suggests that in addition to its anti-apoptosis activity, the function of promoting DSB damage repair of Δ113p53 is crucial in protecting an organism from DNA damage. Similarly, in human cells the ratios of cells at the G2 phase and SA-β-gal-positive cells were significantly higher in irradiated Δ133p53-knockdown cells, which eventually resulted in smaller colony sizes and fewer colonies. A previous study reported that the basal expression of Δ133p53 inhibits p53-mediated replicative senescence through downregulating the expression of p21WAF1 and miR-34a in normal human fibroblasts29. Δ133p53 knockdown-induced senescence was accompanied by the attenuation of BrdU (bromo-deoxyuridine) incorporation, which suggests that the cell senescence was due to cell cycle arrest at the G1 phase29. In this study, we showed that knockdown of Δ133p53 in cells exposed to DNA DSB stress also resulted in cell senescence. However, this senescence was caused by unrepaired DNA DSBs and accompanied by the increase of cells at the G2 phase. These results suggest that Δ133p53 regulates cell replicative senescence in the normal condition and cell senescence upon a DNA damage stress by different mechanisms.
One important rationale for p53 inhibition of DNA DSB repair is its direct interactions with repair proteins, such as RAD51 and RPA, to prevent repair complex formation. The key residues in human p53's DNA binding core domain (including residues 102, 103, 105, 114, 115, 122 and 126) are required for interactions with RAD51, and those in the N-terminal motif (residues 37-57) are required for interactions with RPA. These key amino acid residues are absent in the Δ133p53 protein36,37. This might be the reason that Δ133p53 was not co-immunoprecipitated with RAD51 and RPA in this study. However, Δ133p53 may interrupt the interaction between p53 and HA-Rad51 or HA-RPA2, which was probably due to Δ133p53's ability to form a hetero-complex with p5328,43, which may allow it to neutralize the DSB repair inhibitory effect of full-length p53.
Δ113p53/Δ133p53 is an N-terminally truncated protein without the transactivation domain. Our previous studies showed that, although co-expression of Δ113p53 and p53 alters the expression patterns of p53 downstream genes such as p21, mdm2 and bcl2L, expressing Δ113p53 alone results in little transcriptional activity on these genes in the p53M214K mutant background26. Surprisingly, here we found that Δ113p53 upregulates the expression of the DNA DSB repair genes rad51, lig4 and rad52, independent of full-length p53. The transcriptional activity of Δ113p53 is required for its positive effect on DNA DSB repair as, apart from impairing its transcriptional activity, mutations in its DNA-binding domain also abolished its ability to promote DNA DSB repair. Through promoter functional analysis, gel shift and ChIP assays, we demonstrated that Δ113p53 binds to a novel type of p53 RE in the promoters of zebrafish rad51, lig4 and rad52 genes. A similar type of RE was also found in the promoter regions of human RAD51, LIG4 and RAD52. It is currently unclear how Δ113p53/Δ133p53 lacking the transactivation domain of full-length p53 exerts a transcriptional activity independent of full-length p53. A recent study showed that p53 isoforms, including Δ133p53, differentially regulate p73 transcriptional activities by protein interactions44, which suggests that Δ113p53/Δ133p53 may interact with p73 or its isoforms to achieve its transcriptional activity.
From an evolutionary point of view, given a DNA damage stress condition, the first, crucial action taken by an organism is to survive under such environment. The second action is to minimize genetic insults to avoid genetic diseases during the course of development and reproduction. Here, we demonstrate that the Δ113p53/Δ133p53 is a pro-survival factor for DNA damage stress, and induction of its expression prevents apoptosis and promotes DNA DSB repair, thus inhibiting cell senescence. However, whether Δ113p53/Δ133p53 also plays a role in preventing diseases in response to DNA damage needs to be further explored. It would be very interesting to know whether the Δ113p53M/M mutant exhibits a shortened life-span and high frequency of tumorigenesis in response to low dosage of γ-irradiation.
About 60% of all cancer patients are treated with radio-therapy alone or in combination with other anticancer treatments, including surgery45,46. Most patients can tolerate radiation treatment well, with 5%-10% suffering severe side effects in normal tissue. This radio-sensitivity is partly genetically determined. A few molecular markers have been successfully applied to predict the radio-sensitivity in individual patients47. Here, we demonstrate that Δ133p53 is strongly induced by ionizing radiation and protects cells from death and senescence through preventing apoptosis and promoting DNA DSB repair, which suggests that the induction of Δ133p53 expression in normal cells and tissues provides a potential marker to assess a patient's tolerance to radiation treatment.
Materials and Methods
Zebrafish husbandry
Zebrafish was raised and maintained in standard zebrafish units at Zhejiang University. The p53−/− mutant allele p53M214K line32 was provided by professor Thomas Look at Harvard Medical School (Boston, USA).
Cell culture
H1299 (TCHu160) and QSG-7701 (GNHu7) cells were purchased from the cell bank of the Chinese Academy of Sciences (Shanghai, China). HR-U2OS33 was a gift from professor Huang Jun at Zhejiang University (Hangzhou, China). Plasmids and siRNAs were transfected into cells with FuGENE HD (Roche) and Lipofectamine 2000 transfection reagents, respectively.
HR, NHEJ and SSA assays
The construction of the HR, SSA and NHEJ visual-plus-quantitative assay systems and analyzing procedures were performed as described previously31 (Supplementary information, Figure S2). The primers used in qPCR are listed in Supplementary information, Table S1.
The H1299 cell line was used for HR, SSA, and NHEJ assays in human cells. 1.5 μg of I-SceI-cut HR, 0.5 μg of I-SceI-cut NHEJ or 0.5 μg of I-SceI-cut SSA plasmid DNA was co-transfected with 0.5 μg of CMV-p53, 1.5 μg of CMV-Δ133p53 or 0.5 μg of CMV-p53 with 1.5 μg of CMV-Δ133p53 into 1 × 106 H1299 cells. An uncut plasmid was transfected as the negative control. Transfected cells were cultivated for 24 h at 37 °C and subsequently subjected to FACS analysis with a FACS Calibur Flow Cytometer (BD Biosciences). A minimum of 10 000 cells per sample were analyzed. DNA was also extracted at 24 hpt for qPCR analysis, as described above.
γ-irradiation, UV-irradiation and heat shock treatments
Zebrafish embryos at 24 hpf were irradiated with a dose of 16 gray of γ-ray from a 137Cs source. For UV-irradiation treatment, embryos at 24 hpf were treated with a total energy of 75 mJ/cm2 UV irradiation by a UV source (UV-CL-1000 Ultraviolet Crosslinker) emitting 254 nm light (UVP, USA). For heat shock treatment, 24-hpf embryos, growing at 28.5 °C, were transferred to a 38 °C growth chamber until protein extraction.
For γ-irradiation in human cell lines, untreated or transfected cells at 24 hpt were irradiated with a dose of 10 gray of γ-ray. For UV-irradiation treatment, cells were treated with a total energy of 30 mJ/cm2 UV. For heat shock treatment, cells cultured at 37 °C were transferred to a 42 °C growth chamber for 8 h and then returned to 37 °C until protein extraction.
Comet assay
For the comet assay in zebrafish, ∼100 irradiated or un-irradiated control embryos were sampled at 28 and 36 hpi, and subjected to cell dissociation in ice-cold PBS containing 20 mM EDTA (without Mg2+ and Ca2+). The comet assay was performed with a OxiSelectTM comet assay kit (3-well slides, Cell Biolabs Inc.) according to the manufacturer's recommendations. Embedded cells were treated with a lysis buffer at pH 7 without alkaline treatment to release the double-stranded DNA. For data processing, each comet picture was measured with the software ImageJ 1.45 (National Institutes of Health)48 and the extent of damage in individual cells was calculated as described in Supplementary information, Figure S6.
For the comet assay in the human cell line, QSG7701 cells were transfected with siRNAs, followed by γ-irradiation, as described in the apoptosis and cell cycle assay. The irradiated cells were fixed in 70% ethanol at 48 hpi and subjected to the comet assay, as described in the zebrafish comet assay.
Construction of overexpression plasmids
Zebrafish p53, Δ113p53 and bcl2L and human CMV-p53 and CMV-Δ113p53 were constructed as described previously49. Human CMV-HA-RAD51 was amplified using the primer pair HA-HuRad51-BamHI-For and HA-HuRad51-XbaI-Rev. Human CMV-HA-RPA2 was amplified using the primer pair HA-HuRPA2-For-BamHI and HA-HuRPA2-Rev-EcoRI. The primer sequences are provided in Supplementary information, Table S1.
Generation of zebrafish Δ113p53M/M mutant with the TALEN technique
The Δ113p53 promoter is located in the fourth intron of the full-length p53 gene24,26. The third p53 RE in the Δ113p53 promoter (5′-cagtggaggttGAACATGTCTGAACTTGTCCtgattgagcagtggggg-3′ the sequence of p53 RE is shown in upper case) was chosen for the TALEN targeting site50. We placed the third p53 RE at the spacer region where indels often occur. The two TALEN plasmids with the target binding sites (shown in red letters in Figure 2A) were ordered from ViewSolid Biotech. The two TALEN mRNAs were prepared and co-injected into WT embryos at one-cell stage according to the manufacturer's recommendations.
The TALEN-injected embryos were raised to adulthood and outcrossed with WT fish. The F1 embryos were used to identify mutant founders. The tail of F1 adult fish was used to identify heterozygous mutants. To identify the genetic mutants, a pair of primers (5′-GGCAGTCTAGCTTATGTGT-3′ and 5′-GCTTGACTGTCCAGCACTA-3′) flanking the target site, were used to amplify a 400-bp DNA fragment from genomic DNA. The PCR product contains a digestion site of the restriction enzyme Hpy188III around the third p53 RE. The PCR fragment from WT can be digested into two 200-bp bands, while the PCR fragment from a mutant remains as a 400-bp band. The fragment deletions were subsequently confirmed by sequencing.
SA-β-gal staining
For SA-β-gal staining, QSG7701 cells were transfected with siRNAs followed by γ-irradiation, as described in the apoptosis and cell cycle assays. At 48 hpi, the irradiated cells were fixed in 4% PFA and subjected to SA-β-gal staining with Cell Senescence SA-β-Gal Staining Kit (Beyotime, C0602). Statistics was obtained from three repeat experiments.
rad51 promoter reporter assay
A 5.0-kb DNA fragment upstream of the transcriptional start site of rad51 (Figure 7A) was amplified from genomic DNA (AB strain WT zebrafish) with the primer pair rad51pro-XhoI-For and rad51pro-BamHI-Rev, and cloned into the pEgfp-1 vector to generate the plasmid rad51p:Egfp. The single motif deletion promoters rad51p-RE1:Egfp or rad51p-RE2:Egfp (Figure 7B) were amplified from the rad51p:Egfp plasmid using their respective primer pairs. The primers sequences used are listed in Supplementary information, Table S1. The promoter rad51p-ΔRE1&2:Egfp, with a double-deletion, was generated from the single deletion plasmid.
RNA analysis
For northern blot hybridization, full-length Egfp and 21-760-bp DNA fragment of rad51 cDNA were labeled with Digoxigenin (DIG) to form probes. qRT-PCR in zebrafish was performed as described previously26. The primer sequences and accession numbers of the analyzed genes are listed in Supplementary information, Table S1.
Electrophoretic mobility shift assay (EMSA)
Twenty-six-bp DNA fragments of RE1 and an RE1 mutant with 6 bp mutated, and 39 bp of RE2 and an RE2 mutant with 6 bp mutated of the rad51 promoter (Figure 7C) were artificially synthesized and labeled with biotin as probes (Shanghai Sangon). Nuclear proteins were extracted from injected embryos at 8 hpf with a nuclear protein and cytoplasm protein extraction kit (Beyotime, P0027). Forty fmol of labeled probe was incubated with 2 μg of extracted nuclear protein for 20 min. To specifically block band shift, 8 pmol of unlabeled probe or 200 ng of A7-C10 zebrafish p53 monoclonal antibody was added to the mixture and incubated for 20 min. Labeled biotin was analyzed with a light shift chemiluminescent EMSA kit (Pierce, 20148), according to the manufacturer's instructions.
ChIP assay
ChIP assays were performed as described previously26. For immunoprecipitation of endogenous p53 and Δ113p53, WT embryos were treated with 16 gray of γ-ray. Untreated embryos, and irradiated embryos at 4 and 24 hpi were sampled. Chromatin was sheared into 200-800-bp fragments with Cole-Parmer sonicator equipped with a 2-mm tip. The A7-C10 zebrafish p53 antibody was used to perform immunoprecipitation with the sonicated DNA-protein complex solutions, while IgG was used as the non-specific binding control with the same amount of the sonicated solution. Primers used in qPCR are listed in Supplementary information, Table S1. Total pulled down DNA was normalized with a pair of non-specific primers for the β-actin exon. The specific primers for p53 RE1 and RE3 of the Δ113p53 promoter were used as a p53-binding positive control26.
To immunoprecipitate ectopically expressed HA-p53 and HA-Δ113p53, ∼40 pg of pGEMT plasmid was injected alone, or co-injected with 50 pg of HA-p53 mRNA and 300 pg of HA-Δ113p53 mRNA, into one-cell-stage embryos. At 7 hpf, injected embryos from each treatment were sampled. HA antibody matrix (Abmart) was used for immunoprecipitation. Total DNA was normalized with exon-specific primers. Meanwhile, p53, RE1, and RE3 of Δ113p53 promoter were used as p53-binding positive control.
Western blot, co-IP and immunofluorescence staining
Western blotting was performed as described previously49. Zebrafish p53 monoclonal antibody (A7-C10) was generated as described49.
For co-IP analysis, transfected cells were cultivated for 24 h at 37 °C, followed by protein extraction. An HA antibody matrix (Abmart) was used for immunoprecipitation. For western blot, the HA monoclonal antibody was used to detect HA-RAD51 and HA-RPA. p53 polyclonal antibody CM1 was used to detect p53 and Δ133p53.
For immunofluorescence staining of the cultured cells, cells were plated onto coverslips placed in six-well plates. To analyze RAD51 and γH2AX foci formation, cells were collected and washed with hES culture medium, plated on a Coverglass for Growth (Fisher Scientific, FIS12-545-82) covered with gelatin. After being cultured for 6 h, cells were subjected to immunofluorescence staining as previously described49. At least 100 cells from each sample were randomly chosen for counting RAD51 and γH2AX foci. All antibodies are listed in Supplementary information, Table S2.
FACS
To assay Egfp expression for the determination of DNA DSB repair frequency, transfected cells were cultured for 24 h at 37 °C and subsequently subjected to FACS analysis. The percentage of Egfp-positive cells was counted to represent the DSB repair frequency. A minimum of 8 000 cells per sample were analyzed.
To assay apoptosis and cell cycle, transfected cells at 24 hpt were treated with 10 gray of -irradiation. The irradiated cells were fixed with 70% ethanol at 4, 8, 12, 24, 36 and 48 hpi. The fixed cells were stained with propidium iodide (PI) and subsequently subjected to FACS. Cells in sub-G0 phase were counted as apoptotic cells. A minimum of 10 000 cells per sample were analyzed.
Morpholinos and siRNA
Morpholinos were purchased from GeneTools (Philomath, USA). p53-MO, Δ113p53-MO, rad51-MO, rad52-MO and lig4-MO were designed as previously described26,31.
siRNAs and a negative control duplex (non-specific control siRNA, siNS) were ordered from Invitrogen (Carlsbad, CA, USA). p53i, Δ133p53i1 and Δ133p53i2 were as described previously27,29.
Acknowledgments
This work was supported by the National Basic Research Program of China (973 Program; 2012CB944500), the International Science and Technology Cooperation Program of China (2013DFG32910), the National Natural Science Foundation of China (31371491 and 30971677), and Zhejiang Provincial Natural Science Foundation of China (LZ13C120001).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
(A) Western blot of zebrafish p53 and Δ113p53 proteins in an untreated control (ctr) and in embryos treated with γ-ray, UV irradiation (UV) and heat shock (HS) at 4 and 24 h post treatment (hpt), using A7-C10 zebrafish p53 monoclonal antibody.
Visual-plus-quantitative assay systems for homologous recombination (HR), non-homologous end joining (NHEJ) and single-strand annealing (SSA) repairs.
Knockdown of p53 and Δ113p53 proteins by p53-MO and Δ113p53-MO, or the overexpression of p53 and Δ113p53 by p53 and Δ113p53 mRNA injection in embryos injected with linearized plasmid DNA.
The activation of p53 and induction of Δ113p53 proteins in zebrafish WT embryos injected with a linearized plasmid.
Fluorescence imaging of HR, SSA and NHEJ repairs from zebrafish embryos injected with different reagents as indicated at 10 hpf.
The induced p53M214K mutant protein and basal expression of Δ113p53p53M214K protein do not have a gain-of-function on DNA DSB repairs.
Comet assay to assess the extent of DNA double-strand breaks (DSB).
A TUNEL assay was used to examine apoptotic cells in Δ113p53-MO or Std-MO injected WT embryos or uninjected p53M214K mutant embryos, which were either treated with γ-ray irradiation or untreated, at 8, 16 and 24 hour post irradiation (hpi) as indicated.
A TUNEL assay was used to examine apoptotic cells in Δ113p53-MO or Std-MO injected WT embryos or uninjected p53M214K mutant embryos, which were either treated with γ-ray irradiation or untreated, at 8, 16 and 24 hour post irradiation (hpi) as indicated.
(A) Δ113p53 mRNA was injected into Δ113p53M/M mutant embryos at the one cell stage.
Similar to zebrafish Δ113p53, human Δ133p53 was also induced only by γ-irradiation, but not by UV and heat shock.
Western blot was performed to show the overexpression of p53 and Δ133p53 in H1299 cells.
DNA DSB repair frequencies for HR, NHEJ and SSA were measured using Egfp positive cells sorted by a FACS machine at 24 hpt.
The knockdown of Δ133p53 significantly decreased the efficiencies of HR, NHEJ and SSA DNA DBS repair pathways.
Fluorescence images of γH2AX (green), RAD51 (red) and DAPI (blue) staining were taken individually and used to construct the merged picture shown in Figure 4B.
FACS analysis of the percentage of cells at different cell cycle phases, based on propidium iodide (PI) staining of QSG-7701 cells transfected with siNS, p53i, Δ133p53i1 or Δ133p53i2 siRNA at different time points after 10 gray of γ-ray irradiation, as indicated.
Large views for senescence-associated β-galactosidase (SA-β-gal) staining in Figure 5C to show that cell colony size was negatively correlated with cell senescence.
Transcriptional expression of the indicated genes in human GSG7701 cells.
A comparison of p53 responsive elements in human RAD51, RAD52 and LIG4 promoters with the known p53-repressive or -activating consensus sequences.
ChIP of the p53 and Δ113p53 REs in rad51, rad52 and lig4 promoters in the absence and presence of HA-p53 and HA-Δ113p53.
PCR Primers
Antibody Information
References
- Dudas A, Chovanec M. DNA double-strand break repair by homologous recombination. Mutat Res. 2004;566:131–167. doi: 10.1016/j.mrrev.2003.07.001. [DOI] [PubMed] [Google Scholar]
- Hakem R. DNA damage repair; the good, the bad, and the ugly. EMBO J. 2008;27:589–605. doi: 10.1038/emboj.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiom K. Coping with DNA double strand breaks. DNA Repair (Amst) 2010;9:1256–1263. doi: 10.1016/j.dnarep.2010.09.018. [DOI] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoh J, Jin S, Parrado T, et al. The p53MH algorithm and its application in detecting p53-responsive genes. Proc Natl Acad Sci USA. 2002;99:8467–8472. doi: 10.1073/pnas.132268899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amson R, Pece S, Lespagnol A, et al. Reciprocal repression between P53 and TCTP. Nat Med. 2012;18:91–99. doi: 10.1038/nm.2546. [DOI] [PubMed] [Google Scholar]
- Levine AJ, Oren M. The first 30 years of p53: growing ever more complex. Nat Rev Cancer. 2009;9:749–758. doi: 10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- Meek DW. Tumour suppression by p53: a role for the DNA damage response. Nat Rev Cancer. 2009;9:714–723. doi: 10.1038/nrc2716. [DOI] [PubMed] [Google Scholar]
- Helton ES, Chen X. p53 modulation of the DNA damage response. J Cell Biochem. 2007;100:883–896. doi: 10.1002/jcb.21091. [DOI] [PubMed] [Google Scholar]
- Gatz SA, Wiesmuller L. p53 in recombination and repair. Cell Death Differ. 2006;13:1003–1016. doi: 10.1038/sj.cdd.4401903. [DOI] [PubMed] [Google Scholar]
- Mekeel KL, Tang W, Kachnic LA, et al. Inactivation of p53 results in high rates of homologous recombination. Oncogene. 1997;14:1847–1857. doi: 10.1038/sj.onc.1201143. [DOI] [PubMed] [Google Scholar]
- Akyuz N, Boehden GS, Susse S, et al. DNA substrate dependence of p53-mediated regulation of double-strand break repair. Mol Cell Biol. 2002;22:6306–6317. doi: 10.1128/MCB.22.17.6306-6317.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keimling M, Wiesmuller L. DNA double-strand break repair activities in mammary epithelial cells ― influence of endogenous p53 variants. Carcinogenesis. 2009;30:1260–1268. doi: 10.1093/carcin/bgp117. [DOI] [PubMed] [Google Scholar]
- Willers H, McCarthy EE, Wu B, et al. Dissociation of p53-mediated suppression of homologous recombination from G1/S cell cycle checkpoint control. Oncogene. 2000;19:632–639. doi: 10.1038/sj.onc.1203142. [DOI] [PubMed] [Google Scholar]
- Linke SP, Sengupta S, Khabie N, et al. p53 interacts with hRAD51 and hRAD54, and directly modulates homologous recombination. Cancer Res. 2003;63:2596–2605. [PubMed] [Google Scholar]
- Boehden GS, Akyuz N, Roemer K, Wiesmuller L. p53 mutated in the transactivation domain retains regulatory functions in homology-directed double-strand break repair. Oncogene. 2003;22:4111–4117. doi: 10.1038/sj.onc.1206632. [DOI] [PubMed] [Google Scholar]
- Buchhop S, Gibson MK, Wang XW, et al. Interaction of p53 with the human Rad51 protein. Nucleic Acids Res. 1997;25:3868–3874. doi: 10.1093/nar/25.19.3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanova LY, Willers H, Blagosklonny MV, Powell SN. The interaction of p53 with replication protein A mediates suppression of homologous recombination. Oncogene. 2004;23:9025–9033. doi: 10.1038/sj.onc.1207982. [DOI] [PubMed] [Google Scholar]
- Arias-Lopez C, Lazaro-Trueba I, Kerr P, et al. p53 modulates homologous recombination by transcriptional regulation of the RAD51 gene. EMBO Rep. 2006;7:219–224. doi: 10.1038/sj.embor.7400587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purvis JE, Karhohs KW, Mock C, et al. p53 dynamics control cell fate. Science. 2012;336:1440–1444. doi: 10.1126/science.1218351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XP, Liu F, Cheng Z, Wang W. Cell fate decision mediated by p53 pulses. Proc Natl Acad Sci USA. 2009;106:12245–12250. doi: 10.1073/pnas.0813088106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourdon JC, Fernandes K, Murray-Zmijewski F, et al. p53 isoforms can regulate p53 transcriptional activity. Genes Dev. 2005;19:2122–2137. doi: 10.1101/gad.1339905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Ruan H, Ng SM, et al. Loss of function of def selectively upregulates {Delta}113p53 expression to arrest expansion growth of digestive organs in zebrafish. Genes Dev. 2005;19:2900–2911. doi: 10.1101/gad.1366405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Peng J. p53 isoform delta113p53 in zebrafish. Zebrafish. 2009;6:389–395. doi: 10.1089/zeb.2009.0598. [DOI] [PubMed] [Google Scholar]
- Chen J, Ng SM, Chang C, et al. p53 isoform delta113p53 is a p53 target gene that antagonizes p53 apoptotic activity via BclxL activation in zebrafish. Genes Dev. 2009;23:278–290. doi: 10.1101/gad.1761609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcel V, Vijayakumar V, Fernandez-Cuesta L, et al. p53 regulates the transcription of its Delta133p53 isoform through specific response elements contained within the TP53 P2 internal promoter. Oncogene. 2010;29:2691–2700. doi: 10.1038/onc.2010.26. [DOI] [PubMed] [Google Scholar]
- Aoubala M, Murray-Zmijewski F, Khoury MP, et al. p53 directly transactivates Delta133p53alpha, regulating cell fate outcome in response to DNA damage. Cell Death Differ. 2011;18:248–258. doi: 10.1038/cdd.2010.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita K, Mondal AM, Horikawa I, et al. p53 isoforms Delta133p53 and p53beta are endogenous regulators of replicative cellular senescence. Nat Cell Biol. 2009;11:1135–1142. doi: 10.1038/ncb1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard H, Garmy-Susini B, Ainaoui N, et al. The p53 isoform, delta133p53alpha, stimulates angiogenesis and tumour progression. Oncogene. 2013;32:2150–2160. doi: 10.1038/onc.2012.242. [DOI] [PubMed] [Google Scholar]
- Liu J, Gong L, Chang C, et al. Development of novel visual-plus quantitative analysis systems for studying DNA double-strand break repairs in zebrafish. J Genet Genomics. 2012;39:489–502. doi: 10.1016/j.jgg.2012.07.009. [DOI] [PubMed] [Google Scholar]
- Berghmans S, Murphey RD, Wienholds E, et al. tp53 mutant zebrafish develop malignant peripheral nerve sheath tumors. Proc Natl Acad Sci USA. 2005;102:407–412. doi: 10.1073/pnas.0406252102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia B, Sheng Q, Nakanishi K, et al. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell. 2006;22:719–729. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- Susse S, Janz C, Janus F, Deppert W, Wiesmuller L. Role of heteroduplex joints in the functional interactions between human Rad51 and wild-type p53. Oncogene. 2000;19:4500–4512. doi: 10.1038/sj.onc.1203809. [DOI] [PubMed] [Google Scholar]
- Yoon D, Wang Y, Stapleford K, Wiesmuller L, Chen J. P53 inhibits strand exchange and replication fork regression promoted by human Rad51. J Mol Biol. 2004;336:639–654. doi: 10.1016/j.jmb.2003.12.050. [DOI] [PubMed] [Google Scholar]
- Bochkareva E, Kaustov L, Ayed A, et al. Single-stranded DNA mimicry in the p53 transactivation domain interaction with replication protein A. Proc Natl Acad Sci USA. 2005;102:15412–15417. doi: 10.1073/pnas.0504614102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedler A, Veprintsev DB, Rutherford T, von Glos KI, Fersht AR. Binding of Rad51 and other peptide sequences to a promiscuous, highly electrostatic binding site in p53. J Biol Chem. 2005;280:8051–8059. doi: 10.1074/jbc.M411176200. [DOI] [PubMed] [Google Scholar]
- Sung P, Krejci L, Van KS, Sehorn MG. Rad51 recombinase and recombination mediators. J Biol Chem. 2003;278:42729–42732. doi: 10.1074/jbc.R300027200. [DOI] [PubMed] [Google Scholar]
- Mills KD, Ferguson DO, Essers J, et al. Rad54 and DNA ligase IV cooperate to maintain mammalian chromatid stability. Genes Dev. 2004;18:1283–1292. doi: 10.1101/gad.1204304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov EL, Sugawara N, Fishman-Lobell J, Haber JE. Genetic requirements for the single-strand annealing pathway of double-strand break repair in Saccharomyces cerevisiae. Genetics. 1996;142:693–704. doi: 10.1093/genetics/142.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bykov VJ, Issaeva N, Shilov A, et al. Restoration of the tumor suppressor function to mutant p53 by a low-molecular-weight compound. Nat Med. 2002;8:282–288. doi: 10.1038/nm0302-282. [DOI] [PubMed] [Google Scholar]
- Marcel V, Dichtel-Danjoy ML, Sagne C, et al. Biological functions of p53 isoforms through evolution: lessons from animal and cellular models. Cell Death Differ. 2011;18:1815–1824. doi: 10.1038/cdd.2011.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou Z, Yin L, Chang C, Peng J, Chen J. Protein interaction between p53 and delta113p53 is required for the anti-apoptotic function of delta113p53. J Genet Genomics. 2014;41:53–62. doi: 10.1016/j.jgg.2014.01.001. [DOI] [PubMed] [Google Scholar]
- Zoric A, Horvat A, Slade N. Differential effects of diverse p53 isoforms on TAp73 transcriptional activity and apoptosis. Carcinogenesis. 2013;34:522–529. doi: 10.1093/carcin/bgs370. [DOI] [PubMed] [Google Scholar]
- Barnett GC, West CM, Dunning AM, et al. Normal tissue reactions to radiotherapy: towards tailoring treatment dose by genotype. Nat Rev Cancer. 2009;9:134–142. doi: 10.1038/nrc2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriquez-Hernandez LA, Bordon E, Pinar B, et al. Prediction of normal tissue toxicity as part of the individualized treatment with radiotherapy in oncology patients. Surg Oncol. 2012;21:201–206. doi: 10.1016/j.suronc.2011.12.002. [DOI] [PubMed] [Google Scholar]
- Mayer C, Popanda O, Greve B, et al. A radiation-induced gene expression signature as a tool to predict acute radiotherapy-induced adverse side effects. Cancer Lett. 2011;302:20–28. doi: 10.1016/j.canlet.2010.12.006. [DOI] [PubMed] [Google Scholar]
- Vrekoussis T, Chaniotis V, Navrozoglou I, et al. Image analysis of breast cancer immunohistochemistry-stained sections using imageJ: an RGB-based model. Anticancer Res. 2009;29:4995–4998. [PubMed] [Google Scholar]
- Tao T, Shi H, Guan YH, et al. Def defines a conserved nucleolar pathway that leads p53 to proteasome-independent degradation. Cell Res. 2013;23:620–634. doi: 10.1038/cr.2013.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, Zhu Z, Lin S, Zhang B. Reverse genetic approaches in zebrafish. J Genet Genomics. 2012;39:421–433. doi: 10.1016/j.jgg.2012.07.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Western blot of zebrafish p53 and Δ113p53 proteins in an untreated control (ctr) and in embryos treated with γ-ray, UV irradiation (UV) and heat shock (HS) at 4 and 24 h post treatment (hpt), using A7-C10 zebrafish p53 monoclonal antibody.
Visual-plus-quantitative assay systems for homologous recombination (HR), non-homologous end joining (NHEJ) and single-strand annealing (SSA) repairs.
Knockdown of p53 and Δ113p53 proteins by p53-MO and Δ113p53-MO, or the overexpression of p53 and Δ113p53 by p53 and Δ113p53 mRNA injection in embryos injected with linearized plasmid DNA.
The activation of p53 and induction of Δ113p53 proteins in zebrafish WT embryos injected with a linearized plasmid.
Fluorescence imaging of HR, SSA and NHEJ repairs from zebrafish embryos injected with different reagents as indicated at 10 hpf.
The induced p53M214K mutant protein and basal expression of Δ113p53p53M214K protein do not have a gain-of-function on DNA DSB repairs.
Comet assay to assess the extent of DNA double-strand breaks (DSB).
A TUNEL assay was used to examine apoptotic cells in Δ113p53-MO or Std-MO injected WT embryos or uninjected p53M214K mutant embryos, which were either treated with γ-ray irradiation or untreated, at 8, 16 and 24 hour post irradiation (hpi) as indicated.
A TUNEL assay was used to examine apoptotic cells in Δ113p53-MO or Std-MO injected WT embryos or uninjected p53M214K mutant embryos, which were either treated with γ-ray irradiation or untreated, at 8, 16 and 24 hour post irradiation (hpi) as indicated.
(A) Δ113p53 mRNA was injected into Δ113p53M/M mutant embryos at the one cell stage.
Similar to zebrafish Δ113p53, human Δ133p53 was also induced only by γ-irradiation, but not by UV and heat shock.
Western blot was performed to show the overexpression of p53 and Δ133p53 in H1299 cells.
DNA DSB repair frequencies for HR, NHEJ and SSA were measured using Egfp positive cells sorted by a FACS machine at 24 hpt.
The knockdown of Δ133p53 significantly decreased the efficiencies of HR, NHEJ and SSA DNA DBS repair pathways.
Fluorescence images of γH2AX (green), RAD51 (red) and DAPI (blue) staining were taken individually and used to construct the merged picture shown in Figure 4B.
FACS analysis of the percentage of cells at different cell cycle phases, based on propidium iodide (PI) staining of QSG-7701 cells transfected with siNS, p53i, Δ133p53i1 or Δ133p53i2 siRNA at different time points after 10 gray of γ-ray irradiation, as indicated.
Large views for senescence-associated β-galactosidase (SA-β-gal) staining in Figure 5C to show that cell colony size was negatively correlated with cell senescence.
Transcriptional expression of the indicated genes in human GSG7701 cells.
A comparison of p53 responsive elements in human RAD51, RAD52 and LIG4 promoters with the known p53-repressive or -activating consensus sequences.
ChIP of the p53 and Δ113p53 REs in rad51, rad52 and lig4 promoters in the absence and presence of HA-p53 and HA-Δ113p53.
PCR Primers
Antibody Information